Abstract
Laser-capture microdissection (LCM) allows for the one-step procurement of large homogeneous populations of cells from tissue sections. In mammals, LCM has been used to conduct cDNA microarray and proteomics studies on specific cell types. However, LCM has not been applied to plant cells, most likely because plant cell walls make it difficult to separate target cells from surrounding cells and because ice crystals can form in the air spaces between cells when preparing frozen sections. By fixing tissues, using a cryoprotectant before freezing, and using an adhesive-coated slide system, it was possible to capture large numbers (>10,000) of epidermal cells and vascular tissues (vascular bundles and bundle sheath cells) from ethanol:acetic acid–fixed coleoptiles of maize. RNA extracted from these cells was amplified with T7 RNA polymerase and used to hybridize a microarray containing ∼8800 maize cDNAs. Approximately 250 of these were expressed preferentially in epidermal cells or vascular tissues. These results demonstrate that the combination of LCM and microarrays makes it feasible to conduct high-resolution global gene expression analyses of plants. This approach has the potential to enhance our understanding of diverse plant cell type–specific biological processes.
INTRODUCTION
The growth and development of plants depends to a large degree on the organ- and/or cell type–specific expression of genes. Although in situ RNA hybridization experiments can provide gene expression data at high resolution, this technique is not suitable for the study of large numbers of genes. The recent development of genome-wide gene expression profiling has advanced our understanding of the regulatory networks that control plant growth and development (reviewed by Cushman and Bohnert, 2000; Breyne and Zabeau, 2001). cDNA microarrays (reviewed by Richmond and Somerville, 2000; Aharoni and Vorst, 2001) make it possible to examine the expression of large numbers of genes whose transcript levels change between two “states” and to develop hypotheses regarding how these genes function and how their gene products interact. However, it can be difficult to collect enough of a specific tissue to isolate the 1 to 2 μg of mRNA typically required to conduct microarray hybridization (Duggan et al., 1999; Richmond and Somerville, 2000). Furthermore, it is difficult to specifically collect single cell types from plant organs. Thus, many microarray studies use RNA extracted from a mixture of tissues and/or cell types (Girke et al., 2000). These limitations make it difficult to analyze the expression of large numbers of plant genes at high resolution (i.e., in specific cell types). High-resolution gene expression analysis of specific plant tissues or cells promises to provide more information on diverse biological processes in individual cell types.
At least two approaches to collecting specific cell types from plant tissues have been reported (Karrer et al., 1995; Brandt et al., 1999; Kehr, 2001). The first is to sample single cells from living plant tissues using microcapillaries (Karrer et al., 1995; Brandt et al., 1999). Karrer et al. (1995) reported a method for the isolation of mRNA and the preparation of a cDNA library from tomato leaf epidermal or guard cells by aspiration with microcapillaries. Linear amplification of RNA (e.g., via T7 RNA polymerase–based [T7-based] RNA amplification) (Van Gelder et al., 1990; Eberwine et al., 1992) from a small amount of RNA is needed for DNA microarrays. However, the amplification of RNA extracted by this method often fails (Karrer et al., 1995), which makes it difficult to apply this method to microarray analysis. Furthermore, this method is limited to surface cells, because it is difficult to identify specific cell types in the center of intact organs. To identify nonsurface cells, Brandt et al. (1999) developed transgenic plants that carry a nonsurface cell (e.g., companion cell)–specific promoter fused to a gene for green fluorescent protein. In this method, an RNA sample is collected from companion cells labeled by the expression of green fluorescent protein (Brandt et al., 1999). However, a disadvantage of this method is that it requires the use of cell-specific promoters, and relatively few such promoters are available. A second way to collect specific cell types is to use protoplasts. In general, however, the preparation of protoplasts from intact plant tissues is time consuming. Of greater concern is the fact that gene expression patterns change during protoplast preparation (Grosset et al., 1990). Thus, this method is unlikely to be useful for monitoring gene expression in specialized cell types.
In mammals, laser-capture microdissection (LCM) has been used to obtain populations of specific cell types from tissue sections and to conduct microarray experiments (Emmert-Buck et al., 1996; Simone et al., 1998; Luo et al., 1999). Briefly, a tissue section that contains the cell type of interest is placed on a microscope stage and then covered with a transparent transfer film (a thermoplastic polymer film) carrier. A laser beam then is targeted on the cells of interest. This activates the transfer film and causes it to fuse to the target cells. The fused cells then are separated from the rest of the tissue. To increase the number of cells acquired, this process can be repeated many times using new tissue sections. RNA is extracted from the captured cells and amplified using the T7-based RNA amplification method. It is possible to conduct microarray experiments with the mRNA isolated from as few as 1000 captured cells (Luo et al., 1999).
The application of the LCM technique to plant cells has not been reported, most likely because of their small sizes, the presence of air spaces between them, and their rigid cell walls (Kehr, 2001). If plant tissues are frozen before fixation, large ice crystals often form in the air spaces between tissues and cells, damaging cell morphology (see http://www.arctur.com/support/faq_pixcellIIe.htm). Moreover, plant cells are connected by strong cell walls (Carpita and McCann, 2000), which cause surrounding cells to remain attached to the target cells during LCM. To solve these technical challenges, specimens were fixed and treated with a cryoprotectant under vacuum before freezing in combination with an adhesive-coated slide system. Here, we report the application of LCM to specific cell types of maize. The cells were obtained from the epidermis and vascular tissues (vascular bundles and bundle sheath cells) of coleoptiles from maize seedlings. RNA extracted from the captured cells was amplified using a highly reproducible T7-based RNA amplification method. The amplified RNA then was hybridized to a maize cDNA microarray.
RESULTS
LCM and RNA Amplification
To obtain reliable microarray results, LCM must produce a sufficient amount of RNA of high integrity. LCM conditions were optimized for the epidermis and vascular tissues of the coleoptiles of maize seedlings.
Two types of chemical fixatives are used widely for the preservation of tissues (Ruzin, 1999). The first type are coagulating fixatives (e.g., alcohol and acetone), which denature proteins and transform protoplasm into an artificially interconnected network. The other type are cross-linking (noncoagulating) fixatives (e.g., aldehydes), which chemically cross-link protoplasmic components (proteins and lipids), fixing them in situ. Cross-linking fixatives are better at preserving cell structure. The effects of fixative type on the quantity and quality of RNA extracted from LCM samples were compared. The morphologies of tissue sections fixed with 37–40% formaldehyde/glacial acid/95% ethyl alcohol (10%:5%:50%, v/v) (FAA), a cross-linking fixative, were better than those fixed with 75% ethanol and 25% acetic acid (data not shown). However, fixation with the coagulating fixative (75% ethanol and 25% acetic acid) resulted in much better RNA yields than fixation with FAA (data not shown). It appears that the RNA extracted from FAA-fixed tissues was degraded, because reverse transcriptase–mediated PCR products were obtained from the RNAs extracted from LCM tissues fixed with 75% ethanol and 25% acetic acid but not from LCM tissues fixed with FAA (data not shown).
Tissues were frozen because frozen mouse liver tissues yield more RNA and reverse transcriptase–mediated PCR products than do LCM samples from paraffin-embedded tissues (Goldsworthy et al., 1999). However, if plant tissues are frozen before fixation, the presence of air spaces between cells and tissues may facilitate ice crystal formation, which damages cell morphology. To overcome this problem, maize seedlings were gently infiltrated and fixed under vacuum and treated with Suc solutions (see Methods). This greatly improved cell morphologies in the sections (data not shown).
The existence of the cell wall complicates the application of LCM to plants. Thus, the next hurdle was to minimize contamination with undesired cells surrounding the target cells. An adhesive-coated slide system was used to attach tissue sections tightly to slides (see Methods). The use of the adhesive required LCM to be conducted under relatively strong conditions (see Methods) to make the target cells fuse to the transfer film firmly and separate the LCM cells from other cells efficiently.
Of the conditions tested, freezing damage was minimized and PCR amplification was maximized using 6-μm-thick sections fixed in 75% ethanol and 25% acetic acid (Figure 1). As shown in Table 1, two sets of epidermal cells and two sets of vascular tissues, each containing >10,000 cells, were obtained. Each set yielded ∼40 ng of RNA, corresponding to a few picograms of RNA per cell. After the second round of T7-based RNA amplification, RNA was amplified >60,000-fold.
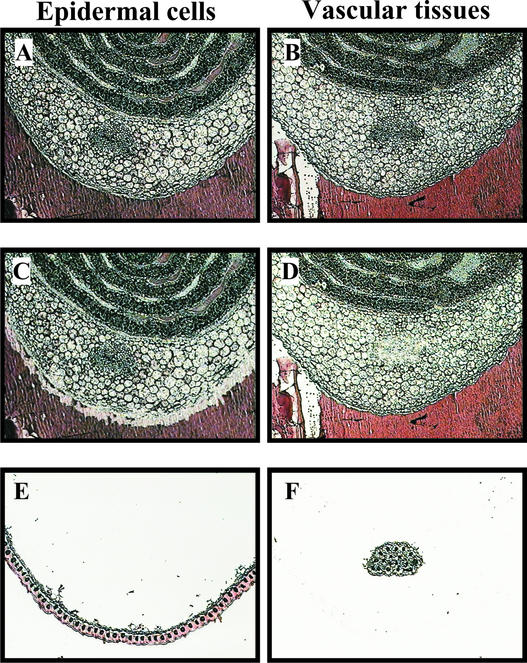
Figure 1.
LCM-Mediated Isolation of Epidermal Cells and Vascular Tissues from 6-μm-Thick Sections of Maize Coleoptiles.
Sections before LCM ([A] and [B]), sections after LCM ([C] and [D]), and captured cells on the cap ([E] and [F]).
Table 1.
Yields of Amplified RNA from Laser-Captured Maize Coleoptile Cells
| Second-Round Amplification
|
||||||
|---|---|---|---|---|---|---|
| Experiment | Cell Source | Estimated No. of Captured Cells | RNA Yield (ng) | RNA per Cell (pg) | Yield (μg) | Amplificationa |
| 1 | Epidermis | 11,100 | 39.5 | 3.56 | 24.5 | ×62,000 |
| Vascular tissues | 11,700 | 43.0 | 3.67 | 46.9 | ×109,000 | |
| 2 | Epidermis | 16,100 | 35.2 | 2.19 | 28.6 | ×81,300 |
| Vascular tissues | 18,700 | 39.0 | 2.08 | 25.4 | ×65,100 | |
Calculated based on the assumption that poly (A+) RNA is 1% of total RNA (Sugiura and Takeda, 2000).
Reproducibility of RNA Amplification
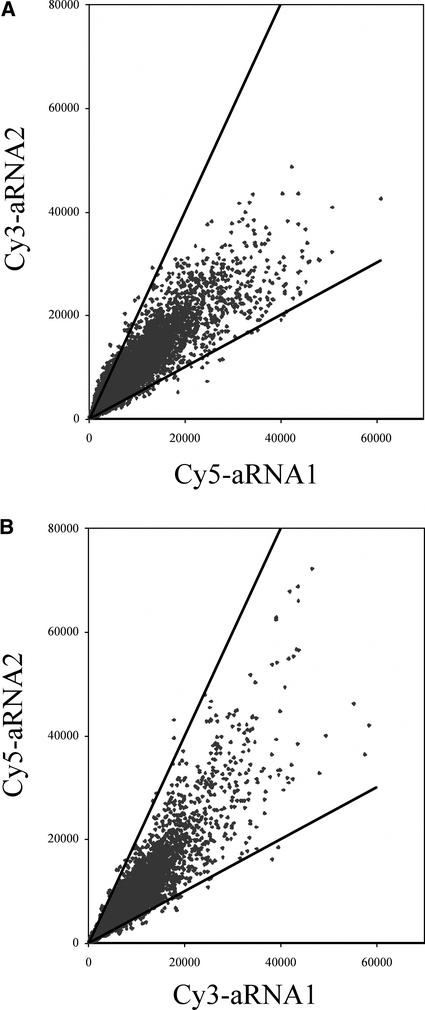
A microarray experiment was performed to assess the fidelity of T7-based RNA amplification. Two independent second-round T7-based RNA amplifications were conducted (aRNA1 and aRNA2) using 40 ng of RNA from coleoptiles. This is the same amount of RNA obtained in an LCM experiment (Table 1). Amplified RNA was labeled individually with Cy3 or Cy5 dye. Two microarray hybridizations were performed, one using Cy3-aRNA1 and Cy5-aRNA2 and another using Cy5-aRNA1 and Cy3-aRNA2. A small percentage (3.08%) of the spots in the Cy5-aRNA1 versus Cy3-aRNA2 comparison and 2.79% of the spots in the Cy3-aRNA1 versus Cy5-aRNA2 comparison exhibited aRNA1/aRNA2 signal ratio differences of >2 or <0.5. By contrast, only 0.24% of the spots exhibited aRNA1/aRNA2 signal ratio differences of >2 or <0.5 in both experiments.
As shown in Figure 2, a linear relationship was observed between the ratios of signal intensities for aRNA1 and aRNA2 in both experiments. The correlation coefficient between the Cy5-aRNA1 and Cy3-aRNA2 data sets was 0.93, and that between the Cy3-aRNA1 and Cy5-aRNA2 data sets was 0.94. Thus, these results demonstrate that independent T7-based RNA amplifications from the same RNA sample are quite reproducible.
Figure 2.
Evaluation of the Fidelity of T7-Based RNA Amplification.
Independent amplifications (aRNA1 and aRNA2) were performed using a single source of RNA isolated from coleoptiles. The amplified RNAs were compared with each other by microarray analysis. ESTs with greater than twofold differences in signal intensity between the two amplified RNAs are plotted outside the angle.
(A) Cy5-aRNA1 versus Cy3-aRNA2.
(B) Cy3-aRNA1 versus Cy5-aRNA2.
Identification of Genes Expressed Differentially in Epidermis or Vascular Tissues
RNA was extracted from epidermal cells and vascular tissues collected via LCM and from whole coleoptiles. RNA from the second round of T7-based RNA amplification was reverse-transcribed and labeled with Cy3 or Cy5. Microarray analyses were used to compare global patterns of gene expression between epidermal cells and vascular tissues, between epidermal cells and whole coleoptiles, and between vascular tissues and whole coleoptiles. Each of the three comparisons was based on four hybridizations involving two independently isolated RNA samples and a dye swap. In addition, each cDNA was duplicate spotted on the microarray. A gene was deemed to be differentially expressed only if at least six of the eight corresponding spots exhibited at least a twofold statistically significant difference in signal intensities between the two RNA samples.
In the comparisons involving epidermal cells versus vascular tissues and epidermal cells versus coleoptiles, 130 of the ESTs that met the signal intensity requirement (see Methods) were expressed at levels at least twofold higher in epidermal cells (Table 2); 21 were expressed at higher levels in epidermal cells in both comparisons. In the two comparisons involving vascular tissues (vascular tissues versus epidermal cells and vascular tissues versus coleoptiles), 137 ESTs that met the signal intensity requirement were expressed at levels at least twofold higher in vascular tissues; 30 were expressed at higher levels in vascular tissues in both comparisons.
Table 2.
Number of ESTs Expressed Preferentially in Epidermal Cells or Vascular Tissues
| Epidermal Cells
|
Vascular Tissues
|
|||||
|---|---|---|---|---|---|---|
| Measurement | E > V | E > C | E > V and E > C |
V > E | V > C | V > E and V >C |
| No. of ESTs | 61 | 48 | 21 | 48 | 59 | 30 |
ESTs with expression ratios of >2 in at least six of eight replicates (2 biological replications × 2 technical replications [dye swaps] × 2 spots per slide) and that are statistically significant based on one-tailed Student's t tests (95% confidence level) are listed. E, epidermal cells; V, vascular tissues; C, coleoptiles.
The 130 ESTs that were expressed preferentially in epidermal cells represent 125 unique genes, including 61 that do not have any statistically significant matches in GenBank. The genes that were expressed preferentially in epidermal cells and for which functions can be predicted are listed in Table 3. They were grouped into the following categories according to their functions: (1) shikimate pathway and secondary metabolism; (2) defense; and (3) others. The supplemental data online include a listing of novel genes that are expressed preferentially in epidermal cells.
Table 3.
Genes Whose Transcripts Accumulated to at Least Twofold Higher Levels in Epidermal Cells Than in Vascular Tissues or Total Coleoptilesabc
| EST Clone No. (Contig No.)a |
Accession No. |
Relative Expression Levels (Ratio of Signal Intensities)b |
Putative Gene Identification [Species] (Accession No.) | e Value |
|---|---|---|---|---|
| Shikimate pathway and secondary metabolism (phenylpropanoid/flavonoid/lignin) | ||||
| 486051D10 | AI586463 | E > V (3.62) | O-Methyltransferase ZRP4 [Zea mays] (P47917) | 4e-41 (X)c |
| 606005D06 | AI666136 | E > C (2.98) | Cinnamic acid 4-hydroxylase (C4H) [Sorghum bicolor] (AY034143) | 0 (N) |
| 614022H05 | AW036957 | E > V (12.33) E > C (3.49) |
Herbicide safener binding protein1 (O-methyltransferase) [Zea mays] (AF033496) | 2e-60 (N) |
| 614032C03 | AW053102 | E > V (3.77) E > C (2.25) |
Cytosolic acetyl-CoA carboxylase (cyt-ACCase) [Triticum aestivum] (U10187) | 2e-19 (N) |
| 614039F12 | AI944262 | E > V (2.77) | Putative O-methyltransferase ZRP4 [Oryza sativa] (AAL31649) | 1e-26 (X) |
| 614063A04 (1) | AW017751 | E > V (30.21) E > C (2.68) |
Herbicide safener binding protein1 (O-methyltransferase) [Zea mays] (AF033496) Orcinol O-methyltransferase [Rosa hybrid cultivar] (AF502434) |
0 (N) 2e-26 (X) |
| 683003F08 | AW066599 | E > V (3.64) E > C (2.36) |
Cinnamyl alcohol dehydrogenase (CAD2) [Lolium perenne] (AF472592) | 1e-06 (N) |
| 707081G08 | AW447887 | E > V (3.63) E > C (2.95) |
Phe ammonia-lyase (PAL) [Zea mays] (L77912) | 7e-19 (N) |
| MEST11-D08 | BG840749 | E > V (2.62) | Chorismate synthase, chloroplast precursor (CS) [Corydalis sempervirens] (P27793) Gmlip15 gene [Zea mays] (D63956) |
3e-19 (X) 2e-29 (N) |
| MEST23-D02 | BG841585 | E > V (2.59) | N-Hydroxycinnamoyl/benzoyltransferase [Ipomoea batatas] (BAA87043) | 1e-17 (X) |
| MEST42-C07 (1) | BG873760 | E > V (23.53) E > C (2.46) |
Herbicide safener binding protein1 (O-methyltransferase) [Zea mays] (AF033496) Orcinol O-methyltransferase [Rosa hybrid cultivar] (AF502434) |
0 (N) 7e-20 (X) |
| Defense | ||||
| 603036E09 | AI948150 | E > V (2.72) | Putative lectin [Hordeum vulgare] (CAB40792) High light–induced mRNA for putative lectin (18 kD) [Hordeum vulgare] (AJ237754) |
5e-27 (X) 2e-31 (N) |
| 707018B02 | AW355997 | E > V (3.44) | Elicitor-responsive gene3 (ERG3) [Oryza sativa] (AF090698) | 2e-34 (N) |
| 707030G07 (2) | AW330962 | E > C (3.12) | Cystatin II [Zea mays] (D38130) | 0 (N) |
| 945030B10 | BE056195 | E > V (3.10) | Permatin precursor [Avena sativa] (U57787) | 8e-56 (N) |
| MEST22-A05 (2) | BG841472 | E > V (6.25) E > C (3.03) |
Cystatin II [Zea mays] (D38130) | 0 (N) |
| Others | ||||
| 486030B08 | AI621976 | E > C (2.79) | Ribosomal protein S7 [Zea mays] (NC_001666) | 7e-11 (X) |
| 486030E08 | AI621992 | E > V (10.65) E > C (2.49) |
Lipid transfer protein [Setaria italica] (AF439446) | 5e-76 (N) |
| 486032E01 | AI622068 | E > V (2.37) | Cellulose synthase1 (CesA-1) [Zea mays] (AF200525) | 0 (N) |
| 486067F01 | AI600714 | E > C (3.42) | Ipomoea nil leaf protein–like protein [Oryza sativa] (AP003434) | 4e-26 (X) |
| 486071G02 | AI600437 | E > C (3.01) | Nt-iaa2.3–deduced protein [Nicotiana tabacum] (AF123504) | 2e-14 (X) |
| 486073B02 | AI619079 | E > C (4.11) | Thioredoxin-like protein [Arabidopsis thaliana] | 1e-41 (X) |
| 486077B02 | AI649641 | E > C (2.33) | Putative (Helianthus annuus) SF16 protein [Arabidopsis thaliana] (NM_128176) | 1e-37 (X) |
| 486077D05 | AI649657 | E > C (3.46) | Chloroplast phosphoglycerate kinase (EC 2.7.2.3) [Triticum aestivum] (X15233) | e-115 (N) |
| 486094E02 | AI615198 | E > C (2.27) | Chloroplast ribosomal protein S1 (rpS1) [Spinacia oleracea] (M82923) | 1e-26 (N) |
| 496018E01 | AI966896 | E > C (2.16) | Putative mitotic control protein dis3 [Arabidopsis thaliana] (NM_127305) | 2e-21 (X) |
| 496028C12 | AI977814 | E > C (3.77) | Germin-like protein1 (GLP1) [Zea mays] (AF261941) | 0 (N) |
| 603009H05 | AI855200 | E > V (3.41) | Putative peptide transporter [Oryza sativa] (AC018727) | 2e-42 (X) |
| 603017D05 | AI861175 | E > V (3.22) | Glutathione S-transferase IV (GSTIV) [Zea mays] (U12679) | 0 (N) |
| 603018E10 | AI861289 | E > C (3.45) E > V (6.18) |
Contains similarity to chalcone-flavonone isomerase (chalcone isomerase) [Arabidopsis thaliana] (AB010692) |
5e-07 (X) |
| 603025G05 | AI947637 | E > V (4.36) | Putative Tyr phosphatase [Oryza sativa] (AF272978) | 8e-18 (N) |
| 603026D08 | AI947671 | E > C (2.98) | Putative Caulobacter crescentusd-isomer–specific 2-hydroxyacid dehydrogenases family protein [Oryza sativa (japonica cultivar group)] (AP003434) | 7e-44 (X) |
| 605014A07 | AI665644 | E > C (2.46) | Chloroplast rpl22 gene for ribosomal protein L22 [Zea mays] (Y00329) | 0 (N) |
| 606030D03 | AI734376 | E > C (2.67) | Chloroplast gene encoding ATP synthase CF-O subunits I and III [Triticum aestivum] (X02595) | e-105 (N) |
| 606031G01 | AI734462 | E > V (2.72) | Lipid transfer protein [Hordeum vulgare] (AF109195) | 4e-24 (N) |
| 606032F05 | AI734526 | E > C (2.52) | OCL5 protein [Zea mays] (AJ250987) | 6e-32 (N) |
| 606054E06 | AI770574 | E > C (2.18) | Similar to putative pyruvate dehydrogenase E1 β-subunit [Zea mays] (AF457974) | 0 (N) |
| 606055H10 | AI770679 | E > V (4.30) | Putative protein phosphatase [Arabidopsis thaliana] (AY065090) | 4e-71 (X) |
| 614009B10 | AI795389 | E > V (3.58) | Β-d-Glucan exohydrolase–like protein [Arabidopsis thaliana] (NM_114567) | 7e-76 (X) |
| 614013G03 | AI861421 | E > V (2.71) | 1-Aminocyclopropane-1-carboxylate oxidase–like protein [Arabidopsis thaliana] (NM_125345) | 2e-38 (X) |
| 614017E07 | AW000557 | E > V (2.60) | Putative wall-associated protein kinase [Oryza sativa] (AC079037) | 9e-29 (X) |
| 614025B02 | AW037041 | E > V (2.73) | Putative GT-1–like transcription factor, predicted mRNA [Arabidopsis thaliana] (NM_129382) | 9e-07 (N) |
| 614025G04 | AW037091 | E > V (2.75) | Small G protein [Pisum sativum] (Z49899) | 4e-48 (N) |
| 614027E04 | AW042353 | E > V (3.15) | 2-Hydroxy-6-oxohepta-2,4-dienoate hydrolase [Nostoc sp PCC 7120] (NC_003272) | 9e-30 (X) |
| 614030G08 | AW053077 | E > V (2.31) | Acyl-ACP thioesterase (FatB1) [Iris tectorum] (AF213479) | 2e-10 (N) |
| 614032B05 | AW053098 | E > V (2.45) E > C (2.31) |
H-β 58 homolog [Sorghum bicolor] (AAD27568) Vacuolar sorting protein–like; embryogenesis protein H-β 58–like protein [Arabidopsis thaliana] (NM_124733) |
2e-94 (X) 4e-27 (N) |
| 614041D02 | AI978191 | E > V (2.55) | Nt-iaa2.3–deduced protein [Nicotiana tabacum] (AF123504) | 3e-07 (X) |
| 614055B10 (3) | AI987532 | E > V (4.50) | PHYT I gene for acidic phytase [Zea mays] (AJ223470) | 4e-17 (N) |
| 614056B10 | AW000597 | E > V (5.17) | Long chain fatty acid–CoA ligase-like protein [Arabidopsis thaliana] (AB012247) | 3e-48 (X) |
| 614058D04 | AW065683 | E > V (5.28) | 23-kD jasmonate-induced protein [Hordeum vulgare] (P32024) | 1e-08 (X) |
| 614064C12 (3) | AW017851 | E > V (3.17) | PHYT I gene for acidic phytase [Zea mays] (AJ223470) | 2e-19 (N) |
| 614067E05 | AW090847 | E > V (3.49) | Putative mannose-6-phosphate isomerase [Oryza sativa (japonica cultivar group)] (AP002860) | 7e-81 (X) |
| 614078B01 | AW146686 | E > V (2.55) | Putative casein kinase I [Oryza sativa (japonica cultivar group)] (AP003273) | 4e-55 (X) |
| 687043E10 | AW216232 | E > C (4.18) | RNA polymerase II second largest subunit (RPB2) [Hordeum vulgare] (AF020839) | e-129 (N) |
| 687043G12 | AW331612 | E > V (2.44) | Putative adenosine kinase [Oryza sativa] | 2e-39 (X) |
| 687063H02 | AW231465 | E > C (2.49) | Putative transmembrane protein [Oryza sativa] (AC016781) | 1e-60 (X) |
| 707018F01 | AW424582 | E > C (6.04) | 18S small subunit ribosomal RNA gene [Zea mays] (AF168884) | 1e-76 (N) |
| 707033E11 | AW331031 | E > C (2.32) | UDP-Glc 6-dehydrogenase [Glycine max] (Q96558) | 3e-30 (X) |
| 707056G08 | AW400251 | E > V (6.12) | Polygalacturonase [Zea mays] (X64408) | e-149 (N) |
| 707062C09 | AW400376 | E > V (14.38) E > C (3.16) |
Pollen allergen–like protein [Arabidopsis thaliana] (NP_173813) | 4e-18 (X) |
| 707074G06 | AW438153 | E > C (2.71) | Putative AP2 domain transcriptional regulator [Oryza sativa (japonica cultivar group) (AC107314) | 4e-08 (X) |
| 707087F03 | AW681259 | E > V (4.42) | Putative copia-like retrotransposon Hopscotch polyprotein [Zea mays] (AF464738) | 1e-29 (X) |
| 707090G04 | AW455731 | E > V (2.70) | Abscisic acid– and ripening-inducible–like protein [Zea mays] (U09276) | 0 (N) |
| 945001G09 | AW787706 | E > V (3.52) | Putative ABC transporter [Arabidopsis thaliana] (NM_128248) | 7e-36 (X) |
| 945029A08 | BE129659 | E > V (9.35) | Xet1 protein [Festuca pratensis] (AJ295943) | 4e-08 (N) |
| 945032B01 | BE129888 | E > C (2.92) | Hypothetical protein A_IG005I10.20 [Arabidopsis thaliana] (T01536) | 3e-43 (X) |
| MEST12-C02 | BG840818 | E > V (2.96) E>C (3.30) |
VsaA-like protein [Oryza sativa] (BAB90075) | 1e-07 (X) |
| MEST14-E02 | BG841003 | E > V (2.94) | Putative Pro-rich protein APG isolog [Cicer arietinum] (AJ276421) | 4e-22 (X) |
| MEST40-A10 | BG842787 | E > V (2.85) | Hypothetical protein similar to transcriptional activator [Oryza sativa (japonica cultivar group)] (AP004330) | 1e-23 (X) |
(Continued)
Significant differences (P < 0.05) were determined by one-tailed Student's t test. E, epidermal cells; V, vascular tissues; C, coleoptiles.
ESTs with the same contig number cluster with CAP3 (Huang and Madan, 1999).
Ratio of mean signal intensity of channel 1 in eight spots to mean signal intensity of channel 2 in eight spots (duplicate spots in four hybridizations).
N, BLASTN (Basic Local Alignment Search Tool N); X, BLASTX.
The 137 ESTs that were overexpressed in vascular tissues represent 134 unique genes, including 66 that are novel. The genes expressed preferentially in vascular tissues and for which functions can be predicted are listed in Table 4. They were grouped into the following functional categories: (1) transporters and aquaporins; (2) metal binding and metal homeostasis; (3) lignin biosynthesis; (4) proteolysis; and (5) others. The supplemental data online include a listing of novel genes that are expressed preferentially in vascular tissues.
Table 4.
Genes Whose Transcripts Accumulated to at Least Twofold Higher Levels in Vascular Tissues than in Epidermal Cells or Total Coleoptilesabc
| EST Clone No. (Contig No.)a |
Accession No. |
Relative Expression Levels (Ratio of Signal Intensities)b |
Putative Gene Identification [Species] (Accession No.) | e Value |
|---|---|---|---|---|
| Transporter and aquaporins | ||||
| 496021E01 | AI967075 | V > E (3.69) | Sulfate transporter protein [Sporobolus stapfianus] (X96761) | 6e-60 (N)c |
| 603023D01 | AI947544 | V > E (2.84) | Plasma membrane integral protein ZmPIP1-4 [Zea mays] (AF326488) | 0 (N) |
| 603028B08 | AI947770 | V > E (3.31) | Plasma membrane integral protein ZmPIP2-6 [Zea mays] (AF326495) | 0 (N) |
| 606028H07 | AI691297 | V > C (6.48) | Putative membrane transporter [Arabidopsis thaliana] (AAM20397) | 1e-58 (X) |
| 606039D11 | AI737357 | V > E (3.85) V > C (5.25) |
NOD26-like membrane integral protein ZmNIP3-1 [Zea mays] (AF326486) | 0 (N) |
| Metal binding and metal homeostasis | ||||
| 496008B02 | AI649699 | V > E (6.15) | Metallothionein-like protein [Zea mays] (S57628) | 6e-70 (N) |
| 496030C04 | AI974911 | V > E (3.47) V > C (8.63) |
Nicotianamine synthase3 (ZmNAS3) [Zea mays] (AB042551) | 1e-12 (N) |
| 606021A06 | AI714806 | V > E (6.89) | Metallothionein-like protein type 2 (OsMT-2) [Oryza sativa] (U43530) Anther-specific protein [Zea mays] (AJ006700) |
4e-24 (N) 7e-35 (N) |
| 618002E08 | AI901370 | V > E (2.46) | Copper homeostasis factor CCH [Arabidopsis thaliana] (NP_191183) | 4e-16 (X) |
| Lignin biosynthesis | ||||
| 614011B10 | AI783384 | V > E (3.55) V > C (3.35) |
Cinnamyl alcohol dehydrogenase [Zea mays] (AJ005702) | 0 (N) |
| Proteolysis | ||||
| 606029E11 (1) | AI734319 | V > E (11.03) V > C (9.34) |
Cys protease XCP1 [Arabidopsis thaliana] (NP_567983) | 4e-49 (X) |
| 606032C10 | AI734503 | V > E (2.37) | Ubiquitin-specific protease12 (UBP12) [Arabidopsis thaliana] (NP_568171) | 2e-18 (X) |
| 606054C10 | AI770559 | V > E (2.06) | Cys protease oryzain β [Oryza sativa] (D90407) Partial mRNA for putative Cys protease (pCYSPROT gene) [Hordeum vulgare] (AJ278817) |
6e-32 (N) 1e-33 (N) |
| 606061G09 | AI770385 | V > E (2.12) | Ubiquitin-conjugating enzyme (UBC) [Zea mays] (AF034946) | 3e-56 (N) |
| 606067D11 | AI834390 | V > E (2.61) | Putative E2, ubiquitin-conjugating enzyme [Arabidopsis thaliana] (AAL66989) | 4e-10 (X) |
| MEST19-E10 (1) | BG841265 | V > E (2.88) V>C (4.57) |
Cys protease XCP1 [Arabidopsis thaliana] (NP_567983) | 4e-53 (X) |
| Others | ||||
| 486019D06 | AI621758 | V > E (5.06) | RNA binding protein–like [Arabidopsis thaliana] (NM_124996) | 2e-22 (X) |
| 486020D11 | AI491665 | V > C (2.70) | Glutathione S-transferase GST 12 [Zea mays] (AF244677) | 0 (N) |
| 486027D11 | AI621860 | V > E (5.50) | Putative chloroplast 50S ribosomal protein L6 [Arabidopsis thaliana] (NM_100397) | 3e-46 (X) |
| 486027G08 | AI621892 | V > E (2.12) | Pyruvate kinase, cytosolic isozyme [Nicotiana tabacum] (Q42954) | 2e-18 (X) |
| 486030A02 | AI621965 | V > E (2.60) | Putative translation initiation factor IF-2 [Arabidopsis thaliana] (NM_117186) | 5e-39 (X) |
| 486040A05 | AI622299 | V > C (2.46) | Receptor protein kinase–like protein [Arabidopsis thaliana] (T50006) | 4e-17 (X) |
| 486068B10 | AI649601 | V > E (6.01) | Putative protein kinase SPK-3 [Oryza sativa] (BAB64101) | 2e-93 (X) |
| 486068D09 | AI600772 | V > E (4.51) | Lipid transfer protein precursor [Oryza sativa] (U29176) | 2e-10 (N) |
| 486068H10 | AI649633 | V > E (5.38) | Putative zinc finger protein [Oryza sativa (japonica cultivar group)] (AP002525) | 2e-07 (X) |
| 486093A08 | AI657290 | V > E (4.30) | Clone R2R3MYB-domain protein [Zea mays] (AF099392) | 2e-65 (N) |
| 486093D03 | AI657322 | V > E (5.93) | Mitochondrial carrier protein–like [Arabidopsis thaliana] (NM_125896) | 2e-06 (X) |
| 496008D11 | AI649714 | V > C (6.32) | γ-Tocopherol methyltransferase [Perilla frutescens] (AF213481) | 5e-46 (X) |
| 496008H11 | AI649739 | V > E (2.94) | ABC transporter–like [Arabidopsis thaliana] (NM_125893) | 2e-37 (X) |
| 603009H06 (2) | AI855201 | V > C (3.27) | Ubiquinol–cytochrome c reductase complex 14-kD protein (CR14) [Solanum tuberosum] (P48502) |
3e-39 (X) |
| 603015D08 | AI855368 | V > E (6.56) V > C (5.76) |
Phosphoenolpyruvate carboxykinase [Zea mays] (AB018744) | 1e-152 (N) |
| 603016C07 | AI855411 | V > E (2.08) | Gs1-5, glutamine synthatase [Zea mays] (X65930) | 0 (N) |
| 603026D08 | AI947671 | V > C (3.29) | Putative Caulobacter crescentusd-isomer–specific 2-hydroxyacid dehydrogenases family protein [Oryza sativa (japonica cultivar group)] (AP003434) |
7e-44 (X) |
| 603029D08 | AI947839 | V > C (3.05) | Water stress–inducible protein (WSI) [Oryza sativa] (AF010584) | 8e-12 (N) |
| 603034C08 | AI948085 | V > C (3.50) | 3-Ketoacyl-CoA thiolase; acetyl-CoA acyltransferase [Cucumis sativus] (X67696) | 2e-10 (X) |
| 496034E08 | AI977867 | V > E (11.53) V >C (5.12) |
50S ribosomal protein L10 (rpl10) [Oryza sativa] (AF095709) | 1e-144 (N) |
| 605018D08 | AI668223 | V > C (3.79) | Proteinase inhibitor precursor [Capsicum annuum] (AF442388) | 9e-09 (X) |
| 606003B05 | AI665879 | V > C (5.02) | Cytochrome c oxidase subunit2 (cox2) gene, mitochondrial gene encoding mitochondrial protein, partial primary transcript [Zea perennis] (U16993) |
0 (N) |
| 606021C04 | AI714822 | V > C (2.25) | HSP18–18-kD heat shock protein (microsporogenesis-specific) [Zea mays, Oh43, clone cMHSP18-1, mRNA, 790 nucleotides] (S59777) |
e-164 (N) |
| 606021G04 | AI714865 | V > E (3.26) | Aluminum-induced protein–like [Arabidopsis thaliana] (NM_123750) | 2e-40 (X) |
| 606028F12 | AI691285 | V > E (8.19) V > C (9.59) |
Putative gag-pol precursor [Zea mays] (AF464738) | 0 (N) |
| 606029B01 | AI734289 | V > E (6.26) V > C (9.82) |
Cystathionine γ-synthase (CGS1) [Zea mays] (AF007785) | 0 (N) |
| 606029D11 | AI734310 | V > E (9.24) V > C (8.30) |
Putative DNA binding protein [Arabidopsis thaliana] (NP_181718) | 2e-09 (X) |
| 606037F05 | AI737228 | V > C (2.07) | Putative 6-phosphogluconolactonase (6PGL) [Brassica carinata] (AY030298) | 9e-10 (N) |
| 606046A10 | AI738135 | V > C (2.66) | Gpa1 gene for glyceraldehyde-3-phosphate dehydrogenase subunit A [Zea mays] (X15408) | 6e-32 (N) |
| 606059D06 | AI770858 | V > C (4.48) | Novel protein, osr40c1 [Oryza sativa] (X95402) | 0 (N) |
| 606066D09 | AI834006 | V > E (5.50) | Putative thiolase [Oryza sativa (japonica cultivar group)] (AC034258) | 7e-62 (X) |
| 614001E04 | AI795691 | V > E (3.52) | ZmRR6, response regulator 6 [Zea mays] (AB042268) | 0 (N) |
| 614006G05 | AI782943 | V > E (2.56) | Cactin gene product [Drosophila melanogaster] (AE003571) | 1e-11 (X) |
| 614010C09 (3) | AI783310 | V > E (2.24) | Putative ubiquinol–cytochrome c reductase complex ubiquinone binding protein (QP-C) [Arabidopsis thaliana] (NM_111923) | 2e-28 (X) |
| 614034F08 | AI944069 | V > E (2.15) | Glyceraldehyde-3-phosphate dehydrogenase [Zea mays] (L13431) | 5e-61 (N) |
| 614038A07 | AW065225 | V > C (2.88) | Plasma membrane H+ ATPase–like gene [Zea mays] (AF480431) | 3e-49 (N) |
| 614043B02 | AI977968 | V > E (3.24) V > C (3.75) |
von Hippel–Lindau binding protein–like protein [Arabidopsis thaliana] (NP_199762) | 1e-55 (X) |
| 614074A04 | AW052810 | V > E (2.52) V > C (5.36) |
Putative abscisic acid–responsive protein [Oryza sativa] (AAL31061) | 3e-77 (X) |
| 614077D10 | AW052986 | V > C (3.98) | Putative nucleoside triphosphatase [Arabidopsis thaliana] (NM_126349) | 3e-10 (X) |
| 614084A02 | AW056312 | V > C (3.18) | SEC5a [Arabidopsis thaliana] (AAL87121) | 6e-61 (X) |
| 687043E11 | AW216233 | V > E (2.94) | Alcohol dehydrogenase1 (adh1) gene, adh1-F allele [Zea mays] (AF123535) | 1e-14 (N) |
| 687064H07 | AW257962 | V > E (2.21) | Nuclear pore protein–like [Arabidopsis thaliana] (NM_123412) | 8e-08 (X) |
| 687064H10 (2) | AW257964 | V > C (2.55) | Ubiquinol–cytochrome c reductase complex 14-kD protein (CR14) [Solanum tuberosum] (P48502) |
2e-39 (X) |
| 687067C04 | AW231567 | V > C (2.39) | Putative RNA-directed RNA polymerase [Arabidopsis thaliana] (NM_117183) | 2e-25 (X) |
| 707018A04 (3) | AW424554 | V > E (2.05) | Putative ubiquinol–cytochrome c reductase complex ubiquinone binding protein (QP-C) [Arabidopsis thaliana] (NM_111923) |
3e-28 (X) |
| 707018F01 | AW424582 | V > C (3.78) | 18S ribosomal RNA gene [Zea mays] (U42796) | 2e-81 (N) |
| 707035F11 | AW330705 | V > C (2.42) | DP-like protein (dpb gene) [Arabidopsis thaliana] (AJ294532) | 9e-19 (N) |
| 707047A12 | AW331087 | V > C (5.40) | Chloride channel ABP [Homo sapiens] (AF034607) | 9e-07 (N) |
| 707058E10 | AW424682 | V > C (2.36) | Polygalacturonase (clone PG2) [Zea mays] (X57628) | 0 (N) |
| 707061D01 | AW400064 | V > C (4.03) | Heat shock protein 101 kD (HSP101) [Nicotiana tabacum] (AF083343) | 5e-11 (N) |
| 707074G06 | AW438153 | V > C (2.57) | Putative AP2 domain transcriptional regulator [Oryza sativa (japonica cultivar group)] (AC107314) |
4e-08 (X) |
| 945033E03 | BE129947 | V > C (2.79) | Translation initiation factor [Pisum sativum] (Y17186) | 8e-16 (N) |
| 945034A12 | BE129972 | V > C (3.74) | Glutamine synthetase [Zea mays] (D14577) | 0 (N) |
| MEST13-H01 | BG840953 | V > E (3.06) V > C (3.09) |
Acyl-CoA binding protein (ACBP) [Oryza sativa] (AY121834) | 7e-57 (N) |
| MEST28-E06 | BG842238 | V > E (3.03) V > C (5.53) |
Carbonic anhydrase (CAH1) [Arabidopsis thaliana] (NP_566971) | 5e-10 (X) |
(Continued)
Significant differences (P < 0.05) were determined by one-tailed Student's t test. E, epidermal cells; V, vascular tissues; C, coleoptiles.
ESTs with the same contig number cluster with CAP3 (Huang and Madan, 1999).
Ratio of mean signal intensity of channel 1 in eight spots to mean signal intensity of channel 2 in eight spots (duplicate spots in four hybridizations).
N, BLASTN (Basic Local Alignment Search Tool N); X, BLASTX.
DISCUSSION
Application of LCM to Plant Tissues
LCM has been used to study cell type–specific expression in animal tissues (Luo et al., 1999; Sgroi et al., 1999; Trogan et al., 2002), but it has not been used to facilitate the microarray analysis of plant tissues. This report demonstrates that LCM-mediated cDNA microarray analyses can be used to conduct global profiling of gene expression on individual types of plant cells and tissues. We report here LCM-based methods to isolate epidermal cells and vascular tissues of maize free of detectable contamination with intact nontarget cells (Figure 1). RNA samples isolated from these cells were amplified using a highly reproducible procedure (Figure 2) and then hybridized to a maize cDNA microarray. Approximately 1.5% (130 of 8791) of the cDNAs were expressed preferentially in epidermal cells compared with vascular tissues and/or total coleoptiles. A similar percentage (137 of 8791) were expressed preferentially in vascular tissues compared with epidermal cells and/or total coleoptiles. Because the coleoptile consists of multiple cell types, those genes that were expressed preferentially in epidermal cells or vascular tissues compared with total coleoptiles are the best candidates for having cell- or tissue-specific expression. The finding (see below) that some genes found in this study to be expressed preferentially in epidermal cells or vascular tissues were reported previously to be expressed predominantly in epidermal cells or vascular tissues of one or more plant species (including maize) validates the procedures reported here as being reliable and meaningful for high-resolution, global gene expression analyses of plant systems.
One of the greatest challenges of the postgenomic era is to define the functions of the thousands of novel genes discovered by genome sequencing projects. One important clue to a gene's physiological function is its expression pattern. This report provides cell type–specific expression data for >120 novel maize genes (see supplemental data online). As such, it suggests one means by which the plant biology community can further its goal of defining the functions of all of the genes in Arabidopsis by 2010.
It needs to be noted, however, that it will be necessary to optimize LCM conditions for each target cell type (e.g., other species or other maize cell types). For example, the conditions used in this study might not be appropriate for the capture of cells whose diameters are <10 μm, because the area fused by the laser pulse was >10 μm (data not shown). Similarly, although minimal contamination with nontarget cells was observed in the 6-μm-thick sections used in this study, thicker sections (i.e., 10-μm-thick sections) were contaminated with nontarget cells (data not shown).
Genes Expressed Preferentially in Epidermis
Shikimate Pathway and Secondary Metabolism
This study identified a gene that encodes a protein involved in the shikimate pathway, chorismate synthase (MEST11-D08), and several genes that encode proteins that function in the phenylpropanoid and phenylpropanoid acetate pathways as being expressed preferentially in epidermal cells of coleoptiles (Table 3). These include Phe ammonia-lyase (707081G08), cinnamic acid 4-hydroxylase (606005D06), and cinnamyl alcohol dehydrogenase (CAD; 683003F08). These findings are consistent with the observation that flavonoids often are present in the epidermal cells of leaves and petals (reviewed by Winkel-Shirley, 2002) and that some phenylpropanoid derivatives function as UV light protectants, phytoalexins, and wound protectants in the outer layers, including the epidermis (Croteau et al., 2000; Hammond-Kosack and Jones, 2000).
Acetyl-CoA carboxylase (ACCase) catalyzes the ATP-dependent carboxylation of acetyl-CoA to form malonyl-CoA in the plastid or cytosol (Konishi et al., 1996; Rawsthorne, 2002). In most nongraminaceous plants, two forms of ACCase have been found: one is a prokaryotic-type enzyme (plastid), and the other is a eukaryotic-type enzyme (cytosol). By contrast, in graminaceous plants, such as maize, both plastidic ACCase and cytosolic ACCase are eukaryotic-type enzymes (Konishi et al., 1996). The plastidic ACCase plays an important role in fatty acid biosynthesis in the plastid (Rawsthorne, 2002), whereas the cytosolic ACCase is thought to provide malonyl-CoA for the biosyntheses of flavonoids and cuticular wax (Fatland et al., 2000). This study identified the cDNA clone that encodes cytosolic ACCase (614032C03) as being expressed preferentially in epidermal cells. This finding is consistent with the report that cytosolic ACCase activity was detected in epidermis lysates of pea leaves (Alban et al., 1994).
The gene for N-hydroxycinnamoyl/benzoyltransferase (HCBT; MEST23-D02) also was found among genes that exhibited epidermis-enhanced expression (Table 3). HCBT catalyzes the first committed reaction in the biosynthesis of phytoalexin (i.e., the formation of N-benzoylanthranilate from benzoyl-CoA and anthranilate) (Yang et al., 1997, 1998). Benzoyl-CoA is supplied by the β-oxidation of cinnamoyl-CoA, whereas anthranilate is formed from chorismate, which is produced by the action of chorismate synthase in the shikimate pathway (Yang et al., 1997, 1998). These results suggest that HCBT is localized preferentially in the epidermis.
Enzymatic O-methylations of many compounds, such as phenylpropanoids and flavonoids, are catalyzed by O-methyltransferases (OMTs), which differ in the specificities of their substrates (Ibrahim et al., 1998). As shown in Table 3, two classes of cDNA clones that encode OMT were identified as having epidermis-enhanced expression. One is the OMT characterized as a maize herbicide safener binding protein1 (SBP1) (Scott-Craig et al., 1998). SBP1 is most abundant in the coleoptiles and least abundant in the leaves (Scott-Craig et al., 1998). The OMTs (614039F12 and 486051D10) that exhibit epidermis-enhanced expression are similar to, but distinct from, the maize ZRP4 protein (Held et al., 1993). The zrp4 mRNA accumulates mainly in the endodermis, with lower levels in the root exodermis (Held et al., 1993). Unlike ZRP4, the OMT identified in this study appears to be expressed preferentially in the epidermis of maize coleoptiles.
Defense
Several genes that are expressed preferentially in epidermal cells and that encode proteins involved in defense systems were identified (lectin [603036E09], elicitor-responsive gene [707018B02], cystain II [707030G07 and MEST22-A05], and permatin [945030B10]) (Table 3). Because the epidermis protects plants from pathogens and insects (Hammond-Kosack and Jones, 2000), it is not surprising that a variety of genes related to plant defense are expressed preferentially in epidermal cells.
Other Proteins
Although the precise biological roles of lipid transfer proteins are not known, they may be involved in somatic embryogenesis (Coutos-Thevenot et al., 1993), wax assembly (Hollenbach et al., 1997), and defense against pathogens (reviewed by Blein et al., 2002). ESTs (486030E08 and 606031G01) that encode two lipid transfer proteins were identified as being expressed preferentially in epidermal cells (Table 3). This finding is consistent with the report that the ltp 7a2b gene that encodes barley lipid transfer protein is expressed preferentially in the leaf epidermis (Hollenbach et al., 1997).
Genes Expressed Preferentially in Vascular Tissues
Transporters and Aquaporins
The uptake of inorganic nutrients, such as potassium, nitrogen, phosphate, and sulfate, and divalent cations, such as calcium, iron, copper, and zinc, is initiated by roots. Their long-distance transport depends on vascular tissues (Fisher, 2000). As shown in Table 4, genes that encode a sulfate transporter, aquaporins (water channels), and a membrane transporter were identified as being expressed preferentially in vascular tissues.
Aquaporins are water channel proteins that belong to the major intrinsic protein superfamily and that transport water and neutral solutes across membranes (reviewed by Johansson et al., 2000; Chaumont et al., 2001). Plant aquaporins are classified as plasma membrane intrinsic proteins (PIPs), tonoplast intrinsic proteins, NOD26-like intrinsic proteins (NIPs), and small and basic intrinsic proteins (Chaumont et al., 2001). Here, genes that encode ZmPIP1-4 (603023D01), ZmPIP2-6 (603028B08), and ZmNIP3-1 (606039D11) were shown to be expressed preferentially in vascular tissues (Table 4). Many aquaporins, especially PIPs and tonoplast intrinsic proteins, are expressed in vascular tissues (e.g., xylem parenchyma) in stems and leaves (Yamada et al., 1995; Barrieu et al., 1998).
Metal Binding and Metal Homeostasis
Micronutrients such as iron, zinc, and copper are essential to plant growth, but in excess they can be toxic. Therefore, intracellular levels of free metals are regulated tightly by the action of metalloproteins (Kochian, 2000; Mira et al., 2001). Metallothioneins are low molecular mass, Cys-rich, metal binding proteins (reviewed by Cobbett and Goldsbrough, 2002). Metallothionein genes are highly expressed in the vascular tissues of some plants (García-Hernández et al., 1998; Cobbett and Goldsbrough, 2002), suggesting that they play a role in metal ion transport (Cobbett and Goldsbrough, 2002). A maize gene that encodes a metallothionein-like protein is expressed abundantly in bundle sheath strands (Furumoto et al., 2000). The identification in this study of metallothionein-like protein genes (496008B02 and 606021A06) as having preferential expression in vascular tissues is consistent with the report of Furumoto et al. (2000).
The copper chaperone CCH is a homolog of the yeast Atx1 protein. Atx1 functions in intracellular copper homeostasis by binding copper ions and delivering them to a copper transporter (Harrison et al., 2000; Mira et al., 2001). The cDNA clone 618002E08 encodes a protein with high similarity to the Arabidopsis CCH protein, which accumulates in sieve elements of stems and leaves and also exists in phloem exudates (Mira et al., 2001).
Graminaceous plants, such as maize, acquire iron ions through roots from soil by strategy II, which involves the secretion from the roots of iron-chelating mugineic acid family phytosiderphores to solubilize iron in the rhizosphere (reviewed by Mori, 1999). Nicotianamine synthase is a key enzyme in the synthetic pathway for mugineic acids and catalyzes the trimerization of S-adenosyl Met (SAM) to form nicotianamine (Higuchi et al., 1999). Nicotianamine is thought to function not only as an intermediate in the synthesis of mugineic acid family phytosiderphores but also as an endogenous chelator of divalent metal cations (e.g., Fe2+, Zn2+, Cu2+, and Mn2+) in xylem and phloem (Stephan et al., 1994; Higuchi et al., 1996). Consistent with this view, a maize nicotianamine synthase (496030C04) is expressed preferentially in the vascular tissues of coleoptiles. Similarly, OsNAS1 is expressed at high levels in the vascular bundles of rice (S. Mori and N.K. Nishizawa, personal communication).
Lignin Biosynthesis
During the differentiation of tracheary elements, after the elongation of the primary wall, the lignified secondary wall is laid down (reviewed by Fukuda, 1997; Ye, 2002). Here, a gene (614011B10) that encodes CAD, which is involved in the biosynthesis of lignin (Humphreys and Chapple, 2002), was shown to be expressed preferentially in vascular tissues. This finding is consistent with a report that CAD is expressed during the tracheary element differentiation of zinnia (Fukuda, 1997; Milioni et al., 2001). The maize CAD gene (614011B10; Table 4) that is expressed preferentially in vascular tissues is distinct (44.1% amino acid identity) from the CAD gene (683003F08; Table 3) that is expressed preferentially in epidermal cells. Hence, maize contains at least two differentially expressed CAD genes.
Proteolysis
Genes that encode two kinds of Cys proteases are expressed preferentially in vascular tissues. One (MEST19-E10 and 606029E11) is homologous with the Arabidopsis xylem-specific papain-like proteases XCP1 and XCP2 (Zhao et al., 2000); the other (606054C10) is homologous with Cys proteases from barley and rice (Watanabe et al., 1991).
Genes that encode ubiquitin-conjugating enzyme (UBC) (606061G09 and 606067D11) and ubiquitin-specific protease12 (UBP12) (606032C10) are expressed preferentially in maize vascular tissues. UBC and UBP12 are involved in the ubiquitin-proteasome system (reviewed by Vierstra, 1996). It has been reported that ubiquitin and the proteasome accumulate in the vascular tissues as well as in the shoot or root apical meristem of sunflower (Ingvardsen et al., 2001). Similarly, UBCs are highly expressed in vascular tissues of Arabidopsis (Thoma et al., 1996).
Other Proteins
A gene that encodes phosphoenolpyruvate carboxykinase (603015D08) was found to exhibit preferential expression in vascular tissue. Phosphoenolpyruvate carboxykinase is a bundle sheath cell–specific enzyme that plays a role in the decarboxylation of aspartate and in C4 photosynthesis in bundle sheath cells (Furumoto et al., 1999, 2000; Wingler et al., 1999).
Another gene (MEST28-E06) that is expressed preferentially in the vascular tissues is homologous with the Arabidopsis carbonic anhydrase CAH1. There are three subfamilies of carbonic anhydrases (α, β, and γ; reviewed by Hewett-Emmett and Tashian, 1996). In maize, a β-type carbonic anhydrase, which catalyzes the conversion of CO2 to HCO3− and plays a key role in CO2 fixation during photosynthesis, is localized in the cytosol of mesophyll cells (reviewed by Ku et al., 1996). The protein encoded by the maize gene MEST28-E06 and the Arabidopsis CAH1 belong to the α subfamily of carbonic anhydrases.
A maize cystathionine γ-synthase (606029B01) gene also is expressed preferentially in vascular tissues of coleoptiles. Cystathionine γ-synthase is a key enzyme in the biosynthesis of Met, and its expression is regulated post-transcriptionally in Arabidopsis (Chiba et al., 1999; Amir et al., 2002). Met is not only necessary for protein synthesis but also is required as a precursor for the biosynthesis of SAM, which is important for the production of plant metabolites such as ethylene, polyamines, vitamin B1, lignin, and the iron chelator mugineic acid (Amir et al., 2002). The gene for SAM synthetase (e.g., SAMS1), which catalyzes the conversion of Met to SAM, is expressed primarily in vascular tissues (Peleman et al., 1989).
Conclusion
The combined use of LCM and cDNA microarray techniques will make it feasible to produce databases that contain gene expression profiles for a large number of specific cell types (Luo et al., 1999) and, subsequently, in combination with genomic sequences, to identify a variety of cell type–specific regulatory sequences. Access to these regulatory sequences will provide plant researchers with greater flexibility to design transgenic plants that are not dangerous to humans or the environment. In addition, by providing cell-specific expression patterns for thousands of genes, these analyses promise to enhance our understanding of plant growth and development.
METHODS
Plant Materials, Growth, and Fixation for Laser-Capture Microdissection
Maize (Zea mays inbred line B73) kernels were placed on germination paper (Anchor Paper, Hudson, WI), rolled up in the paper, placed in a beaker with water, and incubated in the light at 25°C for 7 days, as described by Wen and Schnable (1994).
Five-micrometer-thick cross-sections of these seedlings were fixed immediately on ice in 75% (v/v) ethanol and 25% (v/v) acetic acid. This fixative was infiltrated into the sections under vacuum (400 mm of Hg) for 15 min on ice. The vials containing sections in the fixative were swirled on a rotator at 4°C for 1 h. The vacuum infiltration/swirl steps were repeated twice. To minimize the formation of ice crystals, sections were transferred to 10% (w/v) Suc, which was prepared with diethyl pyrocarbonate–treated PBS buffer (137 mM NaCl, 8.01 mM Na2HPO4, 2.68 mM KCl, and 1.47 mM KH2PO4, pH 7.3). The Suc solution was infiltrated into the sections under vacuum (400 mm of Hg) on ice for 15 min. The vials were swirled on the rotator at 4°C for 1 h. The Suc solution was exchanged for 15% (w/v) Suc (in diethyl pyrocarbonate–treated PBS buffer). The vacuum infiltration/swirl step was conducted as described above. The sections were embedded in TissueTek OCT medium (Sakura Finetek USA, Torrance, CA), frozen in liquid nitrogen, and stored at −80°C.
Cryosectioning and Dehydration of Sections
The tissues were sectioned at 6 μm in a cryostat and mounted on an adhesive-coated slide using the CryoJane Tape-Transfer System (Instrumedics, Hackensack, NJ) at −20°C. Briefly, tissue sections were captured on an adhesive tape window (Instrumedics) as they were being cut. The sections then were laminated to an adhesive-coated slide. The sections were anchored tightly on the slide by a flash of UV light (360 nm) and the tape was removed, leaving the sections on the slide. The frozen sections were treated immediately in 70% (v/v) ethanol at −20°C for 1 min, followed by dehydration steps of 0.5 to 1 min each in 95% (v/v) ethanol and 100% (v/v) ethanol and dehydration for 2 min in xylene twice. Sections were kept in xylene until used for laser-capture microdissection (LCM) (for 0.5 to 1 h).
LCM
The PixCell II LCM system (Arcturus, Mountain View, CA) was used for LCM. Once tissue sections were air-dried, the target cells were laser-capture microdissected according to the manufacturer's instructions. LCM conditions were as follows: laser spot size of <7.5 μm, laser power of 100 mW, and laser pulse duration of 3 ms. Each laser pulse was estimated to capture three cells from the epidermis and eight cells from the vascular tissues. These capture rates were multiplied by the number of laser pulses used in a capture experiment to estimate the number of cells captured.
RNA Extraction and Isolation of Poly(A+) RNA
RNA was extracted from LCM samples using the Absolutely RNA Microprep Kit (Stratagene, La Jolla, CA) according to the manufacturer's protocol. RNA was extracted from whole maize coleoptiles (4 g) using Trizol reagent (Invitrogen, Carlsbad, CA) according to the reagent manual. RNA concentrations were measured using a sensitive fluorescent nucleic acid stain (RiboGreen RNA Quantitation Reagent; Molecular Probes, Eugene, OR) on a model F-2000 fluorescence spectrophotometer (Hitachi, Tokyo, Japan) (excitation wavelength of 480 nm, emission wavelength of 520 nm).
T7 RNA Polymerase–Based RNA Amplification
T7 RNA polymerase–based (T7-based) RNA amplification was performed according to the method of Luo et al. (1999) except that T4 gene 32 protein was added to the reaction of the first-strand synthesis (Baugh et al., 2001). For first-round T7-based RNA amplification, total RNA (35 to 43 ng) extracted from the captured samples or whole coleoptiles was mixed with 0.5 μg of T7-oligo(dT) primer (5′-TCTAGTCGACGGCCAGT-GAATTGTAATACGACTCACTATAGGGCGTTTTTTTTTTTTTTTTTTTTT-3′). The mixture (10.5 μL) was incubated at 65°C for 10 min, chilled on ice for 5 min, and incubated at 42°C for 5 min. Then, 4 μL of 5× first-strand reaction buffer (250 mM Tris-HCl, pH 8.3, 375 mM KCl, 15 mM MgCl2), 2 μL of 0.1 M DTT, 1 μL of 10 mM deoxynucleotide triphosphates (dNTPs), 1 μL of RNase inhibitor (40 units/μL; Roche Diagnostics, Indianapolis, IN), 0.5 μL of T4 gene 32 protein (5 μg/μL; United States Biochemical, Cleveland, OH), and 1 μL of Superscript II (200 units/μL; Invitrogen) were added and incubated at 42°C for 1 h. Subsequently, 15 μL of 10× Escherichia coli DNA polymerase I buffer (New England Biolabs, Beverly, MA), 3 μL of 10 mM dNTPs, 15 μL of 260 μM β-NAD+, 4 μL of E. coli DNA polymerase I (10 units/μL), 1 μL of RNase H (2 units/μL; Invitrogen), 1 μL of E. coli DNA ligase (10 units/μL; New England Biolabs), and 91 μL of water were added and incubated at 16°C. After 2 h, 2 μL of T4 DNA polymerase (3 units/μL; New England Biolabs) was added, and the reaction mixture was incubated at 16°C for 10 min.
After extraction with phenol:chloroform (1:1) and chloroform, cDNA was purified on a Microcon YM-30 column (Millipore, Bedford, MA) and then dried down to 8 μL. In vitro transcription was performed using the MEGAscript T7 kit (Ambion, Austin, TX). Two microliters of 10× transcription buffer, 2 μL each of 75 mM ATP, CTP, GTP, and UTP, and 2 μL of the T7 RNA polymerase enzyme mixture were added to 8 μL of the double-stranded cDNA. The reaction mixture was incubated at 37°C. After 5 h, 1 μL of RNase-free DNase I (2 units/μL; Ambion) was added, and the reaction mixture was incubated at 37°C for 15 min. The amplified RNA was purified on a Microcon YM-30 column and dried down to 10 μL.
For second-round T7-based RNA amplification, 10 μL of amplified RNA was mixed with 1 μL of 1 μg/μL random hexamers (Roche Diagnostics). The mixture was incubated at 70°C for 10 min, followed by treatment on ice for 5 min and at room temperature for 10 min. Four microliters of 5× first-strand reaction buffer, 2 μL of 0.1 M DTT, 1 μL of 10 mM dNTPs, 0.5 μL of RNase inhibitor (40 units/μL), 0.5 μL of T4 gene 32 protein (5 μg/μL), and 1 μL of Superscript II (200 units/μL) were added, and the mixture was incubated at 37°C for 1 h. One microliter of RNase H (5 units/μL) was added and incubated at 37°C for 30 min. The mixture was incubated at 95°C for 2 min and chilled on ice for 5 min. Subsequently, 1 μL of 0.5 μg/μL T7-oligo(dT) primer was added. The mixture was incubated at 70°C for 5 min and then at 42°C for 10 min. The reaction mixture, which consisted of 22 μL of first-strand cDNA/T7-oligo(dT) primer mixture, 15 μL of 10× E. coli DNA polymerase I buffer, 3 μL of 10 mM dNTPs, 15 μL of 260 μM β-NAD+, 4 μL of E. coli DNA polymerase I (10 units/μL), 1 μL of RNase H (2 units/μL), and 90 μL of water, was incubated at 16°C for 2 h. Two microliters of T4 DNA polymerase (3 units/μL) was added, and the reaction mixture was incubated at 16°C for 10 min. After extraction with phenol:chloroform (1:1) and chloroform, cDNA was purified on a Microcon YM-30 column (Millipore) and then dried down to 8 μL. In vitro transcription was conducted as described above. The amplified RNA was purified with an RNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions.
Preparation of the Microarray
ESTs used on the microarray were derived from two sources: 8832 cDNA clones from Unigene Set 1 developed by the National Science Foundation maize genome project at Stanford University (www.zmdb.iastate.edu), and an additional 1152 cDNAs from libraries (ISUM3 and ISUM4) prepared as part of a National Science Foundation Plant Genome Project at Iowa State University (http://www.pslab.agron.iastate.edu/research/genomics/htp_est/libraries.pdf).
The inserts from the cDNA clones were amplified by PCR from E. coli cultures using appropriate vector primers in 100-μL PCR procedures. The final PCR concentrations were as follows: 0.2 mM dNTP, 0.2 μM each primer, 1× PCR buffer (200 mM Tris-HCl, pH 8.4, and 500 mM KCl), 2.5 mM MgCl2, and 0.5 units of Taq polymerase (GIBCO). The PCR program was as follows: 35 cycles of 95°C for 3 min, 95°C for 30 s, 54°C for 30 s, 72°C for 2 min, and 72°C for 2 min. Five microliters of each PCR product was analyzed by 1% agarose gel electrophoresis to examine amplification quality and quantity. Based on these analyses, 8791 cDNAs were amplified successfully. Hybridization signals associated with microarray spots corresponding to reactions that resulted in no product or a smear were not analyzed.
PCR products were purified using Millipore 96-well multiscreen filter plates (LSKC09601) according to the manufacturer's manual, dried in a Speed-Vac (Savant Instruments, Holbrook, NY), and resuspended in 10 μL of double-distilled water, resulting in DNA concentrations of 200 to 800 ng/μL. Purified PCR products (2.5 μL) were transferred to 384-well plates containing 2.5 μL of DMSO and stored at 4°C until they were needed for printing.
PCR-amplified cDNA inserts were printed in duplicate in an 18- × 54-mm grid consisting of 48 subgrids on aminosilane-coated microscope slides (Corning, Corning, NY). Printing was conducted using a PixSys 5500 arraying robot (Cartesian Technologies, Irvine, CA) equipped with 16 ChipMaker3 pins (TeleChem International, San Jose, CA) according to published methods (Hegde et al., 2000). Each PCR product was arrayed in adjacent duplicate spots. After printing, slides were dried, UV cross-linked with a Stratalinker (Stratagene) at 150 mJ, and stored in a slide box in a desiccator at room temperature until they were needed for hybridization.
Synthesis of Fluorescent Probes
Three micrograms of amplified RNA or mRNA was mixed with 3 μg of random hexamers in a total volume of 15.5 μL, denatured at 70°C for 10 min, chilled on ice, added to a 14.5-μL reaction mixture with a final concentration of 1× Superscript II buffer, 10 μM DTT, 500 μM each of dATP, dGTP, and dCTP, 200 μM 5-(3-aminoallyl)-2′-deoxyuridine 5′-triphosphate, 300 μM dTTP, and 400 units of SuperScript II reverse transcriptase, and incubated at 42°C for 1 h. Then, 1.5 μL of 20 mM EDTA was added to stop the reaction. Subsequently, 15 μL of 0.1 N NaOH was added and allowed to incubate for 20 min at 65°C to degrade RNA. After the addition of 15 μL of 0.1 N HCl to neutralize the NaOH, the sample was purified on a Microcon 30 column (Millipore) and concentrated to 10 μL, and 0.5 μL of 1 M sodium bicarbonate, pH 9.0, was added. The sample was mixed with 0.1 mg of monofunctional N-hydroxysuccinimide-ester Cy3 or Cy5 dye (Amersham) and incubated at room temperature in the dark. After 1 h, 4.5 μL of 4 M hydroxylamine (Sigma) was added, and incubation was continued for 15 min at room temperature in the dark to quench the unreacted Cy dye. Cy3 and Cy5 probes were pooled according to the experiment design, purified on Microcon 30 columns, dried under vacuum, and resuspended in 45 μL of hybridization solution (5× SSC [1× SSC is 0.15 M NaCl and 0.015 M sodium citrate], 0.1% SDS, 20 μg of yeast tRNA [Life Technologies, Rockville, MD], 20 μg of poly[A] RNA [Sigma], and 25% formamide).
Hybridization
The slides were prehybridized according to published methods (Hegde et al., 2000). The fluorescent probe mixture was heated at 100°C for 3 min, centrifuged at 13,000 rpm for 5 min, applied to a prehybridized microarray slide, and covered with a cover slip. The array was placed in a hybridization chamber (TeleChem) and hybridized at 42°C overnight. The next morning, the array was washed at room temperature in 1× SSC and 0.2% SDS for 4 min, in 0.1× SSC and 0.2% SDS for 4 min, and in 0.1× SSC for 4 min and then centrifuged briefly.
Microarray Analysis
Arrays were scanned with a ScanArray 5000 (Packard, Meriden, CT). Both Cy3 and Cy5 channels were scanned at a resolution of 10 μm. The photomultiplier tube and laser power settings were adjusted during scanning so that the ratios of signal intensities for the two channels for the majority of the spots were as close to 1 as possible. Fluorescent signal intensities were determined using ImaGene software (Biodiscovery, Marina Del Rey, CA). The intensity of each channel for each spot was calculated by determining the median value from all of the pixels in that spot and then subtracting the intensity of the local background. Spots that had intensity values below the local background, or below 100 in both channels, were excluded from further analysis.
Microarray data are linked at http://www.plantgenomics.iastate.edu/microarray/.
Upon request, all novel materials described in the article will be made available in a timely manner for noncommercial research purposes.
NOTE ADDED IN PROOF
Since the submission of this article, Asano et al. (2002) has also reported the application of LCM to plants (Asano, T., Masumura, T., Kusano, H., Kikuchi, S., Kurita, A., Shimada, H., and Kadowaki, K.-i. (2002). Construction of a specialized cDNA library from plant cells isolated by laser capture microdissection: Toward comprehensive analysis of the genes expressed in the rice phloem. Plant J. 32, 401–408.)
Supplementary Material
Acknowledgments
The authors express their appreciation to H. Miyake, S. Mori, N.K. Nishizawa, Mark Erlander, A.-P. Hsia, and T.-J. Wen for stimulating discussions, and in the case of T.-J. Wen, for conducting some preliminary LCM experiments and helping to establish the maize array. The Iowa State University (ISU) Center for Plant Genomics Microarray Facility, the ISU Veterinary Pathology Laboratory, M. Carter (ISU Image Analysis Facility), Xiaowu Gai (ISU Baker Center for Bioinformatics and Biological Statistics), and Marianne Smith provided technical support and advice. The Microarray Facility and the Veterinary Pathology Laboratory are supported by the Plant Sciences Institute and the Biotechnology Council. M.N. was supported by a Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad. This research was supported by competitive grants to P.S.S. from the U.S. Department of Agriculture National Research Initiative program (Award 0201414) and the Iowa Corn Promotion Board and by Hatch Act and State of Iowa funds.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.008102.
Footnotes
Online version contains Web-only data.
References
- Aharoni, A., and Vorst, O. (2001). DNA microarrays for functional plant genomics. Plant Mol. Biol. 48, 99–118. [DOI] [PubMed] [Google Scholar]
- Alban, C., Baldet, P., and Douce, R. (1994). Localization and characterization of two structurally different forms of acetyl-CoA carboxylase in young pea leaves, of which one is sensitive to aryloxyphenoxypropionate herbicides. Biochem. J. 300, 557–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir, R., Hacham, Y., and Galili, G. (2002). Cystathionine γ-synthase and threonine synthase operate in concert to regulate carbon flow towards methionine in plants. Trends Plant Sci. 7, 153–156. [DOI] [PubMed] [Google Scholar]
- Barrieu, F., Chaumont, F., and Chrispeels, M.J. (1998). High expression of the tonoplast aquaporin ZmTIP1 in epidermal and conducting tissues of maize. Plant Physiol. 117, 1153–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh, L.R., Hill, A.A., Brown, E.L., and Hunter, C.P. (2001). Quantitative analysis of mRNA amplification by in vitro transcription. Nucleic Acids Res. 29, E29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blein, J.-P., Coutos-Thévenot, P., Marion, D., and Ponchet, M. (2002). From elicitins to lipid-transfer proteins: A new insight in cell signaling involved in plant defense mechanisms. Trends Plant Sci. 7, 293–296. [DOI] [PubMed] [Google Scholar]
- Brandt, S., Kehr, J., Walz, C., Imlau, A., Willmitzer, L., and Fisahn, J. (1999). A rapid method for detection of plant gene transcripts from single epidermal, mesophyll and companion cells of intact leaves. Plant J. 20, 245–250. [DOI] [PubMed] [Google Scholar]
- Breyne, P., and Zabeau, M. (2001). Genome-wide expression analysis of plant cell cycle modulated genes. Curr. Opin. Plant Biol. 4, 136–142. [DOI] [PubMed] [Google Scholar]
- Carpita, N., and McCann, M. (2000). The cell wall. In Biochemistry and Molecular Biology of Plants, B.B. Buchanan, W. Gruissem, and R.L. Jones, eds (Rockville, MD: American Society of Plant Physiologists), pp. 52–108.
- Chaumont, F., Barrieu, F., Wojcik, E., Chrispeels, M.J., and Jung, R. (2001). Aquaporins constitute a large and highly divergent protein family in maize. Plant Physiol. 125, 1206–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba, Y., Ishikawa, M., Kijima, F., Tyson, R.H., Kim, J., Yamamoto, A., Nambara, E., Leustek, T., Wallsgrove, R.M., and Naito, S. (1999). Evidence for autoregulation of cystathionine gamma-synthase mRNA stability in Arabidopsis. Science 286, 1371–1374. [DOI] [PubMed] [Google Scholar]
- Cobbett, C., and Goldsbrough, P. (2002). Phytochelatins and metallothioneins: Roles in heavy metal detoxification and homeostasis. Annu. Rev. Plant Biol. 53, 159–182. [DOI] [PubMed] [Google Scholar]
- Coutos-Thevenot, P., Jouenne, T., Maes, O., Guerbette, F., Grosbois, M., Le Caer, J.P., Boulay, M., Deloire, A., Kader, J.C., and Guern, J. (1993). Four 9-kDa proteins excreted by somatic embryos of grapevine are isoforms of lipid-transfer proteins. Eur. J. Biochem. 217, 885–889. [DOI] [PubMed] [Google Scholar]
- Croteau, R., Kutchan, T.M., and Lewis, N.G. (2000). Natural products (secondary metabolites). In Biochemistry and Molecular Biology of Plants, B.B. Buchanan, W. Gruissem, and R.L. Jones, eds (Rockville, MD: American Society of Plant Physiologists), pp. 1250–1318.
- Cushman, J.C., and Bohnert, H.J. (2000). Genomic approaches to plant stress tolerance. Curr. Opin. Plant Biol. 3, 117–124. [DOI] [PubMed] [Google Scholar]
- Duggan, D.J., Bittner, M., Chen, Y., Meltzer, P., and Trent, J.M. (1999). Expression profiling using cDNA microarrays. Nat. Genet. 21, 10–14. [DOI] [PubMed] [Google Scholar]
- Eberwine, J., Yeh, H., Miyashiro, K., Cao, Y., Nair, S., Finnell, R., Zettel, M., and Coleman, P. (1992). Analysis of gene expression in single live neurons. Proc. Natl. Acad. Sci. USA 89, 3010–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmert-Buck, M.R., Bonner, R.F., Smith, P.D., Chuaqui, R.F., Zhuang, Z., Goldstein, S.R., Weiss, R.A., and Liotta, L.A. (1996). Laser capture microdissection. Science 274, 998–1001. [DOI] [PubMed] [Google Scholar]
- Fatland, B., Anderson, M., Nikolau, B.J., and Wurtele, E.S. (2000). Molecular biology of cytosolic acetyl-CoA generation. Biochem. Soc. Trans. 28, 593–595. [PubMed] [Google Scholar]
- Fisher, D.B. (2000). Long distance transport. In Biochemistry and Molecular Biology of Plants, B.B. Buchanan, W. Gruissem, and R.L. Jones, eds (Rockville, MD: American Society of Plant Physiologists), pp. 730–784.
- Fukuda, H. (1997). Tracheary element differentiation. Plant Cell 9, 1147–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furumoto, T., Hata, S., and Izui, K. (1999). cDNA cloning and characterization of maize phosphoenolpyruvate carboxykinase, a bundle sheath cell-specific enzyme. Plant Mol. Biol. 41, 301–311. [DOI] [PubMed] [Google Scholar]
- Furumoto, T., Hata, S., and Izui, K. (2000). Isolation and characterization of cDNAs for differentially accumulated transcripts between mesophyll cells and bundle sheath strands of maize leaves. Plant Cell Physiol. 41, 1200–1209. [DOI] [PubMed] [Google Scholar]
- García-Hernández, M., Murphy, A., and Taiz, L. (1998). Metallothioneins 1 and 2 have distinct but overlapping expression patterns in Arabidopsis. Plant Physiol. 118, 387–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girke, T., Todd, J., Ruuska, S., White, J., Benning, C., and Ohlrogge, J. (2000). Microarray analysis of developing Arabidopsis seeds. Plant Physiol. 124, 1570–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsworthy, S.M., Stockton, P.S., Trempus, C.S., Foley, J.F., and Maronpot, R.R. (1999). Effects of fixation on RNA extraction and amplification from laser capture microdissected tissue. Mol. Carcinog. 25, 86–91. [PubMed] [Google Scholar]
- Grosset, J., Marty, I., Chartier, Y., and Meyer, Y. (1990). mRNAs newly synthesized by tobacco mesophyll protoplasts are wound-inducible. Plant Mol. Biol. 15, 485–496. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack, K., and Jones, J.D.G. (2000). Responses to plant pathogens. In Biochemistry and Molecular Biology of Plants, B.B. Buchanan, W. Gruissem, and R.L. Jones, eds (Rockville, MD: American Society of Plant Physiologists), pp. 1102–1156.
- Harrison, M.D., Jones, C.E., Solioz, M., and Dameron, C.T. (2000). Intracellular copper routing: The role of copper chaperones. Trends Biochem. Sci. 25, 29–32. [DOI] [PubMed] [Google Scholar]
- Hegde, P., Qi, R., Abernathy, K., Gay, C., Dharap, S., Gaspard, R., Hughes, J.E., Snesrud, E., Lee, N., and Quackenbush, J. (2000). A concise guide to cDNA microarray analysis. Biotechniques 29, 548–556. [DOI] [PubMed] [Google Scholar]
- Held, B.M., Wang, H., John, I., Wurtele, E.S., and Colbert, J.T. (1993). An mRNA putatively coding for an O-methyltransferase accumulates preferentially in maize roots and is located predominantly in the region of the endodermis. Plant Physiol. 102, 1001–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewett-Emmett, D., and Tashian, R.E. (1996). Functional diversity, conservation, and convergence in the evolution of the α-, β-, and γ-car-bonic anhydrase gene families. Mol. Phylogenet. Evol. 5, 50–77. [DOI] [PubMed] [Google Scholar]
- Higuchi, K., Nishizawa, N., Römheld, V., Marschner, H., and Mori, S. (1996). Absence of nicotianamine synthase activity in the tomato mutant ‘chloronerva.’ J. Plant Nutr. 19, 1235–1239. [Google Scholar]
- Higuchi, K., Suzuki, K., Nakanishi, H., Yamaguchi, H., Nishizawa, N.K., and Mori, S. (1999). Cloning of nicotianamine synthase genes, novel genes involved in the biosynthesis of phytosiderophores. Plant Physiol. 119, 471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbach, B., Schreiber, L., Hartung, W., and Dietz, K.-J. (1997). Cadmium leads to stimulated expression of the lipid transfer protein genes in barley: Implications for the involvement of lipid transfer proteins in wax assembly. Planta 203, 9–19. [DOI] [PubMed] [Google Scholar]
- Huang, X., and Madan, A. (1999). CAP3: A DNA sequence assembly program. Genome Res. 9, 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys, J.M., and Chapple, C. (2002). Rewriting the lignin roadmap. Curr. Opin. Plant Biol. 5, 224–229. [DOI] [PubMed] [Google Scholar]
- Ibrahim, R.K., Bruneau, A., and Bantignies, B. (1998). Plant O-methyltransferases: Molecular analysis, common signature and classification. Plant Mol. Biol. 36, 1–10. [DOI] [PubMed] [Google Scholar]
- Ingvardsen, C., Veierskov, B., and Joshi, P.A. (2001). Immunohistochemical localization of ubiquitin and the proteasome in sunflower (Helianthus annuus cv. Giganteus). Planta 213, 333–341. [DOI] [PubMed] [Google Scholar]
- Johansson, I., Karlsson, M., Johanson, U., Larsson, C., and Kjellbom, P. (2000). The role of aquaporins in cellular and whole plant water balance. Biochim. Biophys. Acta 1465, 324–342. [DOI] [PubMed] [Google Scholar]
- Karrer, E.E., Lincoln, J.E., Hogenhout, S., Bennett, A.B., Bostock, R.M., Martineau, B., Lucas, W.J., Gilchrist, D.G., and Alexander, D. (1995). In situ isolation of mRNA from individual plant cells: Creation of cell-specific cDNA libraries. Proc. Natl. Acad. Sci. USA 92, 3814–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehr, J. (2001). High resolution spatial analysis of plant systems. Curr. Opin. Plant Biol. 4, 197–201. [DOI] [PubMed] [Google Scholar]
- Kochian, L.V. (2000). Molecular physiology of mineral nutrient acquisition, transport, and utilization. In Biochemistry and Molecular Biology of Plants, B.B. Buchanan, W. Gruissem, and R.L. Jones, eds (Rockville, MD: American Society of Plant Physiologists), pp. 1204–1249.
- Konishi, T., Shinohara, K., Yamada, K., and Sasaki, Y. (1996). Acetyl-CoA carboxylase in higher plants: Most plants other than Gramineae have both the prokaryotic and the eukaryotic forms of this enzyme. Plant Cell Physiol. 37, 117–122. [DOI] [PubMed] [Google Scholar]
- Ku, M.S.B., Kano-Murakami, Y., and Matsuoka, M. (1996). Evolution and expression of C4 photosynthesis genes. Plant Physiol. 111, 949–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, L., Salunga, R.C., Guo, H., Bittner, A., Joy, K.C., Galindo, J.E., Xiao, H., Rogers, K.E., Wan, J.S., Jackson, M.R., and Erlander, M.G. (1999). Gene expression profiles of laser-captured adjacent neuronal subtypes. Nat. Med. 5, 117–122. [DOI] [PubMed] [Google Scholar]
- Milioni, D., Sado, P.-E., Stacey, N.J., Domingo, C., Roberts, K., and McCann, M.C. (2001). Differential expression of cell-wall-related genes during the formation of tracheary elements in the Zinnia mesophyll cell system. Plant Mol. Biol. 47, 221–238. [PubMed] [Google Scholar]
- Mira, H., Martínez-García, F., and Peñarrubia, L. (2001). Evidence for the plant-specific intercellular transport of the Arabidopsis copper chaperone CCH. Plant J. 25, 521–528. [DOI] [PubMed] [Google Scholar]
- Mori, S. (1999). Iron acquisition by plants. Curr. Opin. Plant Biol. 2, 250–253. [DOI] [PubMed] [Google Scholar]
- Peleman, J., Boerjan, W., Engler, G., Seurinck, J., Botterman, J., Alliotte, T., Van Montagu, M., and Inzé, D. (1989). Strong cellular preference in the expression of a housekeeping gene of Arabidopsis thaliana encoding S-adenosylmethionine synthetase. Plant Cell 1, 81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawsthorne, S. (2002). Carbon flux and fatty acid synthesis in plants. Prog. Lipid Res. 41, 182–196. [DOI] [PubMed] [Google Scholar]
- Richmond, T., and Somerville, S. (2000). Chasing the dream: Plant EST microarrays. Curr. Opin. Plant Biol. 3, 108–116. [DOI] [PubMed] [Google Scholar]
- Ruzin, S.E. (1999). Plant Microtechnique and Microscopy. (New York: Oxford University Press).
- Scott-Craig, J.S., Casida, J.E., Poduje, L., and Walton, J.D. (1998). Herbicide safener-binding protein of maize: Purification, cloning, and expression of an encoding cDNA. Plant Physiol. 116, 1083–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgroi, D.C., Teng, S., Robinson, G., LeVangie, R., Hudson, J.R., Jr., and Elkahloun, A.G. (1999). In vivo gene expression profile analysis of human breast cancer progression. Cancer Res. 59, 5656–5661. [PubMed] [Google Scholar]
- Simone, N.L., Bonner, R.F., Gillespie, J.W., Emmert-Buck, M.R., and Liotta, L.A. (1998). Laser-capture microdissection: Opening the microscopic frontier to molecular analysis. Trends Genet. 14, 272–276. [DOI] [PubMed] [Google Scholar]
- Stephan, U.W., Schmidke, I., and Pich, A. (1994). Phloem translocation of Fe, Cu, Mn and Zn in Ricinus seedlings in relation to the concentrations of nicotianamine, an endogenous chelator for divalent metal ions, in different seedling parts. Plant Soil 165, 181–188. [Google Scholar]
- Sugiura, M., and Takeda, Y. (2000). Nucleic acids. In Biochemistry and Molecular Biology of Plants, B.B. Buchanan, W. Gruissem, and R.L. Jones, eds (Rockville, MD: American Society of Plant Physiologists), pp. 260–310.
- Thoma, S., Sullivan, M.L., and Vierstra, R.D. (1996). Members of two gene families encoding ubiquitin-conjugating enzymes, AtUBC1-3 and AtUBC4-6, from Arabidopsis thaliana are differentially expressed. Plant Mol. Biol. 31, 493–505. [DOI] [PubMed] [Google Scholar]
- Trogan, E., Choudhury, R.P., Dansky, H.M., Rong, J.X., Breslow, J.L., and Fisher, E.A. (2002). Laser capture microdissection analysis of gene expression in macrophages from atherosclerotic lesions of apolipoprotein E-deficient mice. Proc. Natl. Acad. Sci. USA 19, 2234–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gelder, R.N., von Zastrow, M.E., Yool, A., Dement, W.C., Barchas, J.D., and Eberwine, J.H. (1990). Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc. Natl. Acad. Sci. USA 87, 1663–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra, R.D. (1996). Proteolysis in plants: Mechanisms and functions. Plant Mol. Biol. 32, 275–302. [DOI] [PubMed] [Google Scholar]
- Watanabe, H., Abe, K., Emori, Y., Hosoyama, H., and Arai, S. (1991). Molecular cloning and gibberellin-induced expression of multiple cysteine proteinases of rice seeds (oryzains). J. Biol. Chem. 266, 16897–16902. [PubMed] [Google Scholar]
- Wen, T.-J., and Schnable, P.S. (1994). Analyses of mutants of three genes that influence root hair development in Zea mays (Gramineae) suggest that root hairs are dispensable. Am. J. Bot. 81, 833–842. [Google Scholar]
- Wingler, A., Walker, R.P., Chen, Z.-H., and Leegood, R.C. (1999). Phosphoenolpyruvate carboxykinase is involved in the decarboxylation of aspartate in the bundle sheath of maize. Plant Physiol. 120, 539–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley, B. (2002). Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 5, 218–223. [DOI] [PubMed] [Google Scholar]
- Yamada, S., Katsuhara, M., Kelly, W.B., Michalowski, C.B., and Bohnert, H.J. (1995). A family of transcripts encoding water channel proteins: Tissue-specific expression in the common ice plant. Plant Cell 7, 1129–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Q., Grimmig, B., and Matern, U. (1998). Anthranilate N-hydroxycinnamoyl/benzoyltransferase gene from carnation: Rapid elicitation of transcription and promoter analysis. Plant Mol. Biol. 38, 1201–1214. [DOI] [PubMed] [Google Scholar]
- Yang, Q., Reinhard, K., Schiltz, E., and Matern, U. (1997). Characterization and heterologous expression of hydroxycinnamoyl/benzoyl-CoA:anthranilate N-hydroxycinnamoyl/benzoyltransferase from elicited cell cultures of carnation, Dianthus caryophyllus L. Plant Mol. Biol. 35, 777–789. [DOI] [PubMed] [Google Scholar]
- Ye, Z.-H. (2002). Vascular tissue differentiation and pattern formation in plants. Annu. Rev. Plant Biol. 53, 183–202. [DOI] [PubMed] [Google Scholar]
- Zhao, C., Johnson, B.J., Kositsup, B., and Beers, E.P. (2000). Exploiting secondary growth in Arabidopsis: Construction of xylem and bark cDNA libraries and cloning of three xylem endopeptidases. Plant Physiol. 123, 1185–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.