Abstract
At the onset of mitosis, plant cells form a microtubular preprophase band that defines the plane of cell division, but the mechanism of its formation remains a mystery. Here, we describe the use of mammalian yellow fluorescent protein–tagged CLIP170 to visualize the dynamic plus ends of plant microtubules in transfected cowpea protoplasts and in stably transformed and dividing tobacco Bright Yellow 2 cells. Using plus-end labeling, we observed dynamic instability in different microtubular conformations in live plant cells. The interphase plant microtubules grow at 5 μm/min, shrink at 20 μm/min, and display catastrophe and rescue frequencies of 0.02 and 0.08 events/s, respectively, exhibiting faster turnover than their mammalian counterparts. Strikingly, during preprophase band formation, the growth rate and catastrophe frequency of plant microtubules double, whereas the shrinkage rate and rescue frequency remain unchanged, making microtubules shorter and more dynamic. Using these novel insights and four-dimensional time-lapse imaging data, we propose a model that can explain the mechanism by which changes in microtubule dynamic instability drive the dramatic rearrangements of microtubules during preprophase band and spindle formation in plant cells.
INTRODUCTION
Microtubules play a prominent role in plant morphogenesis (Kost et al., 1999; Kost and Chua, 2002). Unlike other eukaryote cells, vacuolated and cell wall–confined plant cells follow a specialized mode of cytokinesis, exhibiting four major microtubular arrays at different cell cycle stages with a variety of functions (Goddard et al., 1994). During interphase, a cortical array consisting of parallel microtubules oriented perpendicular to the cell expansion axis assists cellulose deposition (Cyr, 1994) and responds to stimuli (Wymer et al., 1996). At the onset of mitosis, the cortical array is replaced by a densely packed ring of microtubules encircling the nucleus called the preprophase band (PPB), which defines the location of the cell plate formed during cytokinesis (Mineyuki, 1999). Subsequently, the PPB is replaced by a mitotic spindle apparatus consisting of bundles of kinetochore microtubules separating the duplicated chromosomes in daughter cells (Yu et al., 2000). Finally, a phragmoplast containing two oppositely directed rings of microtubules running outward from the cell center provides tracks for vesicles that carry new cell wall–depositing material at the division plane (Otegui and Staehelin, 2000).
As an important component of cellular morphogenesis, plant microtubules have been shown to be essential for embryo development (Steinborn et al., 2002), organ formation (Whittington et al., 2001), organ twisting (Thitamadee et al., 2002), trichome development (Mathur and Chua, 2000), maintaining the growth direction of elongating root hairs (Bibikova et al., 1999), pollen tube cytoplasmic organization (Taylor and Hepler, 1997), and stomatal movement (Marcus et al., 2001). To understand the role of plant microtubules in all of these vital processes, we need to know how microtubules change from one conformation to another in terms of space and time. The dynamic behavior of microtubules growing by polymerization and shrinking by depolymerization, which is known as “dynamic instability,” is considered the basic determinant of changes in microtubular conformation (Desai and Mitchison, 1997). Dynamic instability is characterized by four parameters: growth rate, shrinkage rate, frequency of transition from growth to shrinkage (catastrophe frequency), and frequency of transition from shrinkage to growth (rescue frequency) (Mitchison and Kirschner, 1984). By altering dynamic instability parameters, cells can rearrange the microtubular network and quickly respond to stimuli, regulate cellular morphogenesis, and control cell division (Kirschner and Mitchison, 1986; Belmont et al., 1990; Desai and Mitchison, 1997). To date, the dynamic instability of plant microtubules in vivo has not been quantified.
The PPB represents the transition of microtubular conformation from interphase to mitosis. In plant cells, the formation of the PPB plays a pivotal role in cell division by marking the division plane and organizing the spindle axis, and in the absence of the PPB, plant development is affected severely (Traas et al., 1995; McClinton and Sung, 1997). In the last three decades, ample information has become available regarding the timing of appearance and importance of PPB (Mineyuki, 1999; Granger and Cyr, 2001), but the mechanism that drives the formation of the PPB remains unknown. This motivated us to investigate the dynamic instability parameters of plant microtubules in vivo, giving special attention to the PPB formation process.
Two studies addressed plant microtubule dynamics previously: one used fluorescence recovery after photobleaching (FRAP) of microinjected fluorescently labeled pig brain tubulin in live Tradescantia virginiana stamen hair cells (Hush et al., 1994); the other used plant microtubules assembled in vitro on sea urchin axonemes (Moore et al., 1997). Still, quantitative in vivo studies of plant microtubule dynamic instability are lacking. The use of green fluorescent protein (GFP)–tagged tubulin or GFP-tagged microtubule-associated proteins provides a noninvasive way to monitor individual microtubule dynamics in vivo. GFP-tagged Arabidopsis α-Tubulin6 (TUA6) (Ueda et al., 1999) and GFP-tagged Microtubule-Associated Protein4 (MAP4) from mouse (Olson et al., 1995) used in plants (Marc et al., 1998; Mathur and Chua, 2000; Camilleri et al., 2002) have been reported to decorate microtubules in live plant cells, but studies using these proteins to visualize the dynamic instability of plant microtubules have not appeared. In addition to using GFP-TUA6 and GFP-MAP4, we applied an additional strategy to visualize microtubule dynamics that used Cytoplasmic Linker Protein170 (CLIP170) isolated from HeLa cells (Pierre et al., 1992), which when attached to GFP, has been shown to bind to the plus ends of microtubules in mammalian cells (Perez et al., 1999).
Here, we present data showing that mammalian CLIP170 binds to plant microtubular plus ends in vivo with conserved domain functionality compared with mammalian cells. We used CLIP170 microtubular plus-end labeling to quantify the dynamic instability of individual microtubules in live and dividing plant cells compared with TUA6 and MAP4 and report the parameters of in vivo plant microtubule dynamic instability. Strikingly, we observed a marked change in these parameters during the transition from interphase to PPB formation at the onset of mitosis, providing novel insights into the mechanism that drives the formation of the PPB.
RESULTS
Transiently Expressed CLIP170 Binds Specifically to Plant Microtubules in Vivo
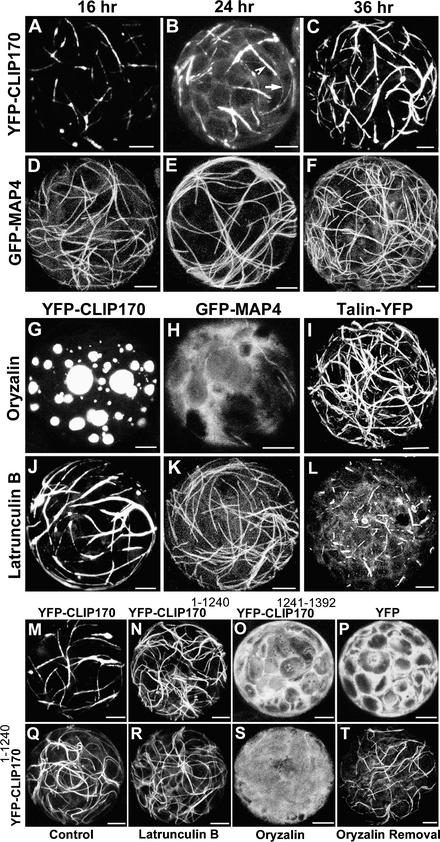
To investigate whether CLIP170 is able to specifically decorate plant microtubule growing ends, we constructed a chimeric gene that encodes a yellow fluorescent protein (YFP)–CLIP170 fusion protein and analyzed its localization in live plant cells by fluorescence microscopy. In a transient expression using transfected cowpea mesophyll protoplasts, small stretches of YFP-CLIP170 appeared at 16 h after transfection (Figure 1A), resembling specific labeling of microtubular ends. With increased expression levels (24 h after transfection; Figure 1B), YFP-CLIP170 showed a more patchy appearance, with extended fine thin labeling along the remaining parts of microtubules. At later time points (36 h after transfection; Figure 1C), YFP-CLIP170 extensively labeled plant microtubules almost along their entire length. These observations in plant protoplasts closely resemble the previously reported CLIP170 expression pattern in HeLa cells (Pierre et al., 1994). By contrast, a well-established plant microtubule binding protein, GFP-MAP4 (Marc et al., 1998; Mathur and Chua, 2000; Camilleri et al., 2002), labeled plant microtubules along their entire length irrespective of time and expression level (Figures 1D to 1F). When Figures 1A to 1C and 1D to 1F are compared, it is evident that after prolonged high expression of YFP-CLIP170, the length of the labeled microtubule stretch increased (an effect also observed in mammalian cells; Perez et al., 1999). In the transfections, no adverse effect of the expression of either YFP-CLIP170 or GFP-MAP4 was noted, based on the normal shape and viability of the protoplasts compared with the control transfection (our unpublished results).
Figure 1.
YFP-CLIP170 and YFP-CLIP1701-1240 but Not YFP-CLIP1701241-1392 Bind to Microtubules in Living Cowpea Protoplasts and Are Responsive to Microtubule Drugs.
(A) to (C) Confocal laser scanning microscopy (CLSM) micrographs of the localization of YFP-CLIP170 in cowpea protoplasts at the indicated times after transfection. In (B), the arrowhead shows labeling of a stretch of a microtubule end, and the arrow shows faint labeling of the remaining microtubule.
(D) to (F) CLSM micrographs of GFP-MAP4 in cowpea protoplasts at the indicated times after transfection.
(G) to (I) Effect of the microtubule-depolymerizing agent oryzalin (10 μM) on YFP-CLIP170 (G), GFP-MAP4 (H), and Talin-YFP (I) localization in cowpea protoplasts.
(J) to (L) Effect of the actin-depolymerizing agent latrunculin B (20 μM) on YFP-CLIP170 (J), GFP-MAP4 (K), and Talin-YFP (L) localization in cowpea protoplasts.
(M) to (T) Domain analysis of CLIP170 in cowpea protoplasts. Protoplasts expressing YFP-CLIP170 (M), YFP-CLIP1701-1240 ([N] and [Q] to [T]), YFP-CLIP1701241-1392 (O), and nontargeted YFP (P) are shown. (Q) shows a control sample, (R) shows a sample after treatment with latrunculin B (20 μM) for 30 min, (S) shows a sample after treatment with oryzalin (10 μM) for 30 min, and (T) shows a sample 20 min after oryzalin was washed away.
All images shown are maximum projections of 30 confocal slices covering a depth of 15 μm and denoting approximately a half-hemisphere of each protoplast. Bars = 5 μm in the xy plane.
To test the specificity of YFP-CLIP170 binding to plant microtubules using actin filaments as a negative control, we treated protoplasts transfected with YFP-CLIP170, GFP-MAP4, and Talin-YFP (the latter labels the actin network) (Kost et al., 1998) with the plant microtubule-depolymerizing drug oryzalin and the actin filament–depolymerizing drug latrunculin B. As shown in Figures 1G to 1I, within 30 min, oryzalin induced a massive redistribution of YFP-CLIP170, forming large spherical cytosolic aggregates; GFP-MAP4 induced a diffuse cytosolic GFP fluorescence, confirming microtubule depolymerization; and there was no effect on Talin-YFP–labeled actin filaments. Interestingly, YFP-CLIP170 aggregates look exactly like those formed in CLIP170-transfected and nocodazole-treated HeLa cells (Pierre et al., 1994). As shown in Figures 1J to 1L, latrunculin B treatment had no effect on YFP-CLIP170– and GFP-MAP4–labeled microtubules, whereas the actin filaments stained with Talin-YFP were strongly affected. These observations indicate that YFP-CLIP170 (like GFP-MAP4) binds specifically to the plant microtubules.
CLIP1701-1240 Labels Plant Microtubules Like MAP4, Whereas CLIP1701241-1392 Remains Cytosolic
The 1392–amino acid CLIP170 protein has three domains (Pierre et al., 1992). The basic N-terminal domain (350 amino acids) contains a tandem repeat of the cytoskeleton-associated protein Gly-conserved domain, which is essential for the microtubular binding of CLIP170. The middle long coiled-coil domain (890 amino acids) contains heptad repeats that likely are involved in dimerizing CLIP170. The C-terminal domain (152 amino acids) is predicted to allow the interaction of CLIP170 with other proteins or membranes (Rickard and Kreis, 1996). It has been shown in mammalian cells that a truncated version of CLIP170 lacking the C terminus (hereafter referred to as CLIP1701-1240) no longer binds specifically at the plus end but rather binds uniformly to microtubules, like MAP4 (Pierre et al., 1994). To investigate the domain functionality of CLIP170 in plant cells, we introduced the truncated fusion proteins YFP-CLIP1701-1240 and YFP-CLIP1701241-1392 into cowpea protoplasts. As shown in Figures 1N and 1Q, YFP-CLIP1701-1240 bound to the entire length of microtubules (like GFP-MAP4), completely abolishing the patchy appearance of YFP-CLIP170 (cf. Figures 1M and 1N). This finding shows the sufficiency of CLIP1701-1240 for microtubule labeling and the importance of the C-terminal domain for specific targeting of CLIP170 to microtubule ends. No variation in CLIP1701-1240 microtubular labeling with time or expression level was noted (our unpublished results). As shown in Figure 1O, YFP-CLIP1701241-1392 remained completely cytosolic, like nontargeted YFP (Figure 1P). Surprisingly, after oryzalin-induced microtubule depolymerization, no cytosolic aggregates of YFP-CLIP1701-1240 were observed (Figure 1S), suggesting that the C-terminal domain is necessary for cytosolic aggregation. Additionally, within 20 min after oryzalin removal, YFP-CLIP1701-1240 reappeared along the entire length of newly formed microtubules (Figure 1T), reconfirming the requirement of the C-terminal domain for CLIP170 plant microtubular end–specific targeting. As expected, latrunculin B did not have any effect on YFP-CLIP1701-1240 microtubular labeling (Figure 1R).
In Vivo Colocalization of CLIP170 and CLIP1701-1240 with MAP4 on Plant Microtubules
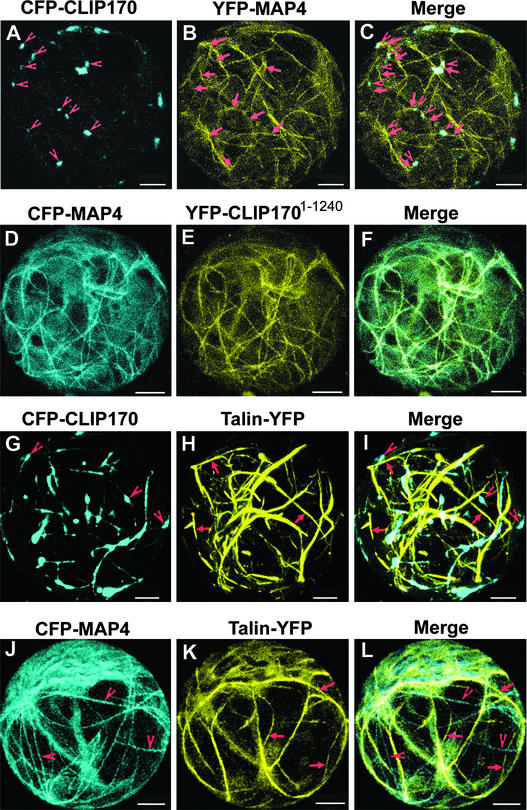
With the availability of different spectral variants of GFP, it has become possible to visualize intracellular locations and interactions of two (or more) different GFP-labeled proteins in a single living cell (Ellenberg et al., 1999). Because CLIP170 forms dimers (Scheel et al., 1999), we did not attempt to visualize colocalization between CLIP170 and CLIP1701-1240. Instead, we used cyan fluorescent protein (CFP)–CLIP170 and YFP-CLIP1701-1240 in combinations with YFP-MAP4 and CFP-MAP4 to visualize their spatial localizations on plant microtubules. In addition, we also used CFP-MAP4 and CFP-CLIP170 in combination with Talin-YFP to investigate possible colocalization between the microtubular and actin networks. As shown in Figures 2A to 2C, in cotransfected protoplasts at early time points of expression (<12 h), CFP-CLIP170 was associated specifically with the ends of plant microtubules (decorated by YFP-MAP4). By contrast, YFP-CLIP1701-1240 bound to the entire length of plant microtubules and colocalized perfectly with CFP-MAP4 (Figures 2D to 2F). Protoplasts coexpressing Talin-YFP and CFP-CLIP170 (Figures 2G to 2I) or CFP-MAP4 (Figures 2J to 2L) did not show any colocalization of YFP and CFP labels, indicating no interaction of CLIP170- or MAP4-labeled microtubules with Talin-decorated actin filaments.
Figure 2.
CLIP170 and CLIP1701-1240 Colocalize with MAP4-Decorated Microtubules but Not with Talin-Decorated Actin Filaments in Living Cowpea Protoplasts.
(A) to (C) CLSM micrographs of a cowpea protoplast cotransfected with CFP-CLIP170 and YFP-MAP4.
(D) to (F) CLSM micrographs of a cowpea protoplast expressing both CFP-MAP4 and YFP-CLIP1701-1240.
(G) to (I) CLSM micrographs of a cowpea protoplast expressing both CFP-CLIP170 and Talin-YFP.
(J) to (L) CLSM micrographs of a cowpea protoplast expressing both CFP-MAP4 and Talin-YFP.
(A), (D), (G), and (J) show CFP fluorescence; (B), (E), (H), and (K) show YFP fluorescence; and (C), (F), (I), and (L) show superimposed CFP and YFP fluorescence. Arrowheads in (A) and arrows in (B) show that CFP-CLIP170 labels the ends of the corresponding microtubules that are labeled with YFP-MAP4. Arrowheads in (G) depict stretches of microtubules labeled with CFP-CLIP170, and arrows in (H) indicate actin filaments labeled with Talin-YFP. (I) clearly shows the lack of colocalization of the indicated structures. Similarly, microtubules decorated with CFP-MAP4 (arrowheads in [J]) and actin filaments decorated with Talin-YFP (arrows in [K]) are not colocalized in (L). All images shown are maximum projections of 30 confocal slices covering a depth of 15 μm and denoting approximately a half-hemisphere of each protoplast. Bars = 5 μm in the xy plane.
YFP-CLIP170 Binds Differentially to Plant Microtubules According to the Cell Maturity Status in Stably Transformed Tobacco Bright Yellow 2 Cells
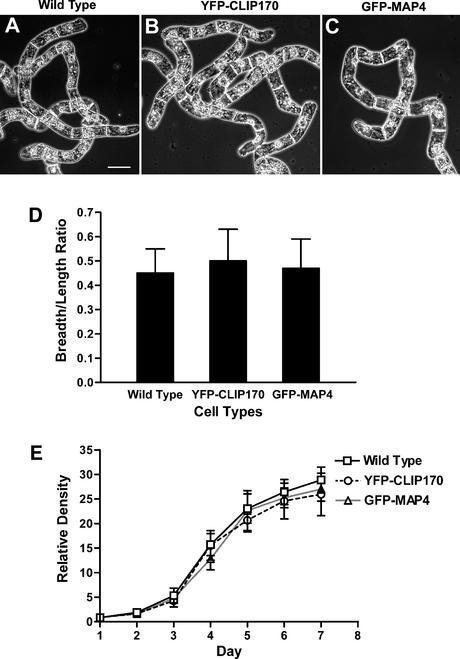
To determine whether YFP-CLIP170 and YFP-CLIP1701-1240 label plant microtubules in continuously dividing tobacco Bright Yellow 2 (BY-2) cells, we introduced them into BY-2 cells by Agrobacterium tumefaciens–mediated stable cell transformations (see Methods). Although some of the YFP-CLIP170–transformed cell lines appeared round in shape with a tendency to form clumps, we selected cell lines that appeared healthy. To determine whether the selected YFP-CLIP170–transformed cell lines grow and divide normally, cell shape and cell culture growth characteristics were analyzed and compared with those of wild-type and GFP-MAP4–transformed cell lines (Figures 3A to 3C). At half-time of subculturing (4-day-old cells), cell shape (analyzed as breadth-to-length ratio) was found to be similar in wild-type, YFP-CLIP170–, and GFP-MAP4–transformed cell lines (Figure 3D). Both transgenic and wild-type cells exhibited similar sigmoidal growth curves, as inferred from the relative cell density profile of cells subcultured weekly (Figure 3E). This finding suggests no interference of YFP-CLIP170 expression on either microtubular or cellular functioning.
Figure 3.
Cytology of Wild-Type, YFP-CLIP170–Transformed, and GFP-MAP4–Transformed BY-2 Cells.
(A) Wild type BY-2 cells observed with phase-contrast microscopy.
(B) YFP-CLIP170–expressing BY-2 cells observed with phase-contrast microscopy.
(C) GFP-MAP4–expressing BY-2 cells observed with phase-contrast microscopy.
(D) Comparison of cell shapes expressed as breadth-to-length ratio of wild-type, YFP-CLIP170–transformed, and GFP-MAP4–transformed BY-2 cells (n = 4 replicates, 77 to 89 cells per replicate).
(E) Relative cell numbers of wild type, YFP-CLIP170–transformed, and GFP-MAP4–transformed BY-2 cells (n = 3 replicates).
For (D) and (E), means and standard deviations are indicated. Bar in (A) = 50 μm for (A) to (C).
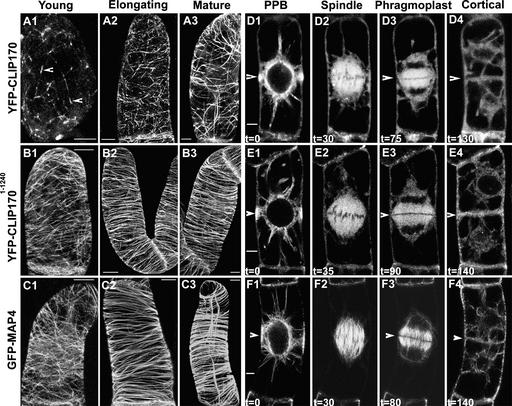
The analysis of microtubular labeling in transformed cells was performed with fluorescence microscopy. Strikingly, in young BY-2 cells (Figure 4A1), YFP-CLIP170 marked ends of microtubules as distinct small dashes (<5 μm). As the cells progressed toward elongation, YFP-CLIP170 decorated more extended stretches (∼5 μm) at microtubule ends (Figure 4A2). These stretches extended up to 10 to 20 μm in full-grown mature cells (Figure 4A3). By contrast, there was no variation in YFP-CLIP1701-1240 (Figures 4B1 to 4B3) and GFP-MAP4 (Figures 4C1 to 4C3) microtubule-labeling patterns according to cell maturity status. YFP-CLIP170–elongated stretches (Figure 4A3) appeared much shorter than the total microtubule length visualized with either YFP-CLIP1701-1240 or GFP-MAP4 (Figures 4B2, 4B3, 4C2, and 4C3). This finding indicates that in young cells (just after cell division), YFP-CLIP170 binds to the ends of microtubules, probably as a result of the lower levels of protein in the daughter cells. Because the daughter cells increase protein synthesis during growth and elongation, YFP-CLIP170 decorates more extended stretches at the microtubule ends. Upon complete maturation (e.g., in interphase arrested cells), YFP-CLIP170 labels much longer stretches of the microtubules, which might be attributable to the accumulated protein levels. Interestingly, a similar dependence of microtubule-labeling patterns on CLIP170 protein expression levels has been observed previously in mammalian cells (Perez et al., 1999).
Figure 4.
YFP-CLIP170 Binds Differentially to Plant Microtubules in Stably Transformed BY-2 Cells and Labels All Four Microtubule Arrays.
(A) and (D) YFP-CLIP170 expression.
(B) and (E) YFP-CLIP1701-1240 expression.
(C) and (F) GFP-MAP4 expression.
BY-2 cells are shown at different time points after cytokinesis (young, ∼1 h; elongating, ∼6 h; mature, >24 h) and at different cell cycle stages. (A) to (C) show maximum projections of 40 confocal slices covering a depth of 20 μm and denoting approximately a half-hemi-cylinder of each cell, and (D) to (F) show confocal median sections of ∼2 μm thickness. The arrowheads in (A1) show YFP-CLIP170–labeled microtubule ends. The arrowheads in (D1), (E1), and (F1) indicate the center of the PPB. At the corresponding positions (arrows in [D3], [E3], [F3], [D4], [E4], and [F4]), the phragmoplast contacts the border of the cell and a new cell wall is formed. The time is indicated in minutes. Bars = 5 μm in the xy plane.
YFP-CLIP170 and YFP-CLIP1701-1240 Decorate All Four Plant Microtubular Arrays
As shown in Figure 4, YFP-CLIP170 (Figures 4D1 to 4D4) and YFP-CLIP1701-1240 (Figures 4E1 to 4E4) labeled all four plant microtubular arrays—PPB, spindle, phragmoplast, and interphase cortical microtubules—comparable to GFP-MAP4–decorated (Figures 4F1 to 4F4) and GFP-TUA6–decorated arrays (our unpublished results). To capture more cross-sectional areas to visualize different microtubular arrays in single cell cycle follow-up studies, the pinhole of the confocal microscope was kept wider (>2 airy disk units). This setup did not permit us to visualize individual microtubules or their ends, making it difficult to visualize YFP-CLIP170 fine microtubular labeling within these arrays. Interestingly, the time points of formation of the four successive microtubular arrays are similar in BY-2 cells expressing YFP-CLIP170, YFP-CLIP1701-1240, and GFP-MAP4. This finding indicates that YFP-CLIP170 and YFP-CLIP1701-1240 expression and labeling of plant microtubules had no adverse effect on the cell cycle, validating the use of YFP-CLIP170 as an authentic reporter to visualize plant microtubule dynamics at different cell cycle stages.
CLIP170 Highlights Plus Ends of Microtubules in Stably Transformed Tobacco BY-2 Cells
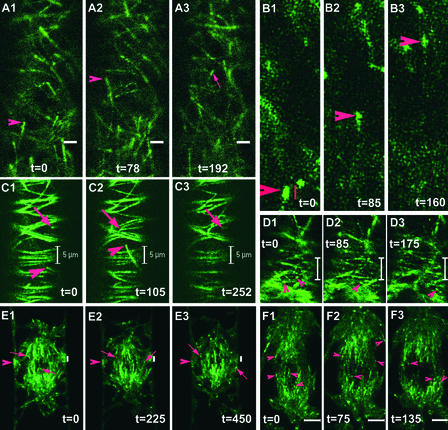
To visualize the dynamic behavior of CLIP170 in plant cells, we studied young cells exhibiting a small dash-like appearance of YFP-CLIP170 on microtubule ends. As in mammalian cells, the dynamic properties of CLIP170 in plant cells are striking. In young cells, the dynamic labeling patterns of YFP-CLIP170 resemble “moving comets” that highlight microtubule plus ends (Figures 5A1 to 5A3; see also supplemental data online). The YFP-CLIP170 fluorescence dash varies in length, with an average size of ∼1 μm, as inferred from Figures 5B1 to 5B3. The labeling appeared sharp at the front end and diffuse at the rear end of the fluorescent dash. As mentioned above, in relatively more mature cells with higher CLIP-170 expression levels, the labeling was extended to a longer stretch at the plus ends (Figures 5C1 to 5C3; see also supplemental data online). The velocities of the movement of the microtubular ends exhibiting comet-like labeling or an extended labeling pattern were similar, suggesting no influence of the extent of YFP-CLIP170 labeling on plant microtubular dynamics. The YFP-CLIP170 labeling pattern with a strong dash-like appearance at microtubule plus ends with additional fine thin labeling on the remaining parts of microtubules or labeling of more extended stretches provides an opportunity to visualize not only the growth but also the shrinkage, catastrophes, and rescues: the four parameters of microtubule dynamic instability. Additionally, we visualized CLIP170-labeled microtubule plus-end dynamics in the PPB (Figures 5D1 to 5D3; see also supplemental data online), on the nuclear envelope (NE) (Figures 5E1 to 5E3; see also supplemental data online), and in the spindle (Figures 5F1 to 5F3; see also supplemental data online), creating the possibility to analyze microtubule dynamics at these different stages. For the spindle structure, the specific plus-end labeling clearly showed the polarity of this microtubule array, with plus ends moving away form the two microtubule organizing centers (see supplemental data online).
Figure 5.
Plant Microtubule Dynamics Visualized with YFP-CLIP170 Microtubule Plus-End Labeling in Different Microtubular Arrays.
(A1) to (A3) YFP-CLIP170–decorated microtubule plus-end dynamics in BY-2 cells. The arrowheads in (A1) and (A2) denote the YFP-CLIP170–labeled growing microtubule plus end of one microtubule, whereas the arrow in (A3) represents the YFP-CLIP170–decorated plus end of another microtubule. Bars = 2 μm. The movie of this sequence (see supplemental data online) has been accelerated ∼20 times (total duration in real time of 192 s).
(B1) to (B3) YFP-CLIP170 specifically binds to the plus ends of plant microtubules, exhibiting a comet-like appearance. The arrowheads represent a single microtubule plus end as highlighted by YFP-CLIP170 labeling. Bar = 1 μm. The movie of this sequence (see supplemental data online) has been accelerated ∼20 times (total duration in real time of 160 s).
(C1) to (C3) YFP-CLIP170–labeled interphase cortical microtubules in BY-2 cells. The arrowheads represent a microtubule showing growth, and arrows denote a microtubule exhibiting growth and shrinkage. Bars = 5 μm. The movie of this sequence (see supplemental data online) has been accelerated ∼25 times (total duration in real time of 252 s).
(D1) to (D3) YFP-CLIP170–decorated PPB microtubules. The arrowheads represent single microtubule plus ends. Bars = 5 μm. The movie of this sequence (see supplemental data online) has been accelerated ∼20 times (total duration in real time of 175 s).
(E1) to (E3) YFP-CLIP170–labeled NE-originating microtubules. The arrowheads denote the gradual disappearance of PPB, and the arrows represent microtubule plus ends labeled by YFP-CLIP170. Bars = 2 μm. The movie of this sequence (see supplemental data online) has been accelerated ∼20 times (total duration in real time of 450 s).
(F1) to (F3) YFP-CLIP170–labeled spindle microtubules. Arrowheads represent YFP-CLIP170–labeled microtubule plus ends. Bars = 5 μm. The movie of this sequence (see supplemental data online) has been accelerated ∼20 times (total duration in real time of 135 s).
The time measurements in all panels represent seconds.
CLIP170 Exhibits in Vivo Dynamic Instability of Plant Microtubules
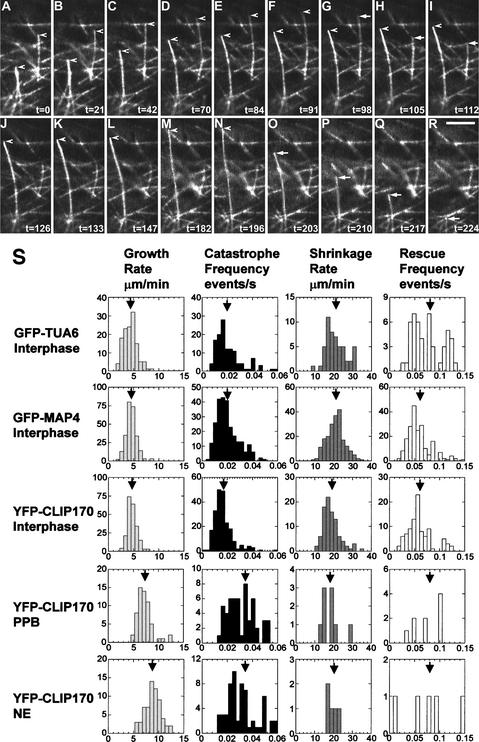
To visualize the dynamics of individual microtubules, single microtubule ends were located and then tracked for several minutes. As shown in Figures 6A to 6R and 7, YFP-CLIP170–labeled microtubules can be seen in growing or shrinking states displaying dynamic instability. Life history plots of microtubule end–tracking data displayed the dynamic instability—expressed as growth, catastrophe, shrinkage, and rescue—with noticeable rare pauses (indicating sudden transitions between growth and shrinkage). To study whether the expression of YFP-CLIP170 changes dynamic instability behavior, we compared the dynamic instability parameters of microtubules in BY-2 cells expressing GFP-TUA6, GFP-MAP4, and YFP-CLIP170 at interphase. Four parameters of microtubule dynamic instability (growth rate, shrinkage rate, catastrophe frequency, and rescue frequency) were determined (Table 1, Figure 6S). From Table 1, it can be inferred that plant microtubules grow at a speed of ∼5 μm/min and shrink four times faster, at a speed of ∼20 μm/min. The parameters for GFP-TUA6, GFP-MAP4, and YFP-CLIP170 were remarkably similar for interphase cortical microtubules. It is important to note that each parameter was determined for three different cell lines (obtained by independent transformation events) for each fusion protein. This large similarity demonstrates the robustness of the chosen approach of using different GFP fusion probes to visualize plant microtubule dynamics, but above all, it stresses that YFP-CLIP170 accurately reports microtubule dynamic instability in plant cells.
Figure 6.
Plant Microtubule Dynamic Instability in BY-2 Cells.
(A) to (R) CLSM micrographs of the dynamics of YFP-CLIP170–labeled cortical microtubules in BY2 cells. Arrowheads show growing microtubules, and arrows show shrinking microtubules, which have grown previously. Time measurements in all panels represent seconds. All images are confocal sections (0.5 μm thick) taken near the cell surface. Bar in (R) = 5 μm for (A) to (R).
(S) From left to right: histograms of growth rate, estimated catastrophe frequency, shrinkage rate, and estimated rescue frequency obtained from different BY-2 cells. From top to bottom: BY-2 cells expressing GFP-TUA6 (interphase microtubules), GFP-MAP4 (interphase microtubules), and YFP-CLIP170 (interphase, PPB, and NE microtubules). Arrows indicate mean values (see also Table 1).
Figure 7.
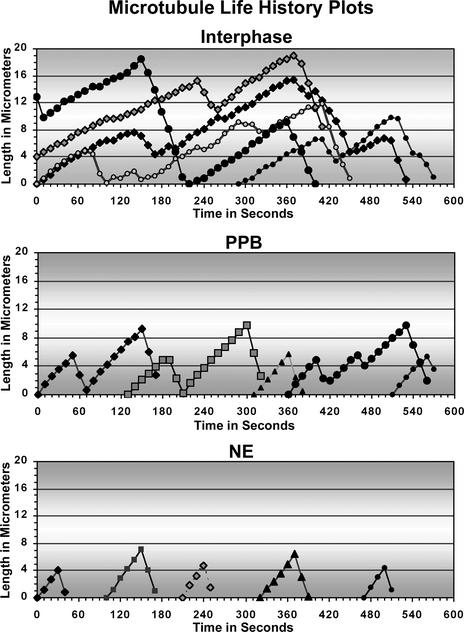
Life History Plots of Individual Plant Microtubules from Different Arrays.
Five individual microtubules were tracked for several minutes at the interphase cortical array, PPB, and NE as described in Methods. Each microtubule is represented by a different symbol.
Table 1.
Plant Microtubule Dynamic Instability Parameters
| Proteins Used and Cell Stage | Growtha Rate (μm/min) |
Catastrophea Frequency (events/s) |
nb | Shrinkagea Rate (μm/min) |
Rescuea Frequency (events/s) |
nc |
|---|---|---|---|---|---|---|
| GFP-TUA6, interphase | 4.3 ± 1.3 4.15 |
0.02 ± 0.01 0.018 |
127 | 21 ± 6 19.50 |
0.08 ± 0.04 0.067 |
53 |
| GFP-MAP4, interphase | 4.5 ± 1.1 4.59 |
0.020 ± 0.009 0.018 |
258 | 21 ± 5 20.83 |
0.06 ± 0.03 0.053 |
206 |
| YFP-CLIP170, interphase | 4.7 ± 0.9 4.59 |
0.017 ± 0.007 0.015 |
217 | 19 ± 4 18.36 |
0.06 ± 0.02 0.051 |
94 |
| YFP-CLIP170, PPB | 7.2 ± 1.5 6.88 |
0.034 ± 0.014 0.029 |
59 | 18 ± 5 17.89 |
0.08 ± 0.03 0.065 |
9 |
| YFP-CLIP170, NE | 8.8 ± 1.5 8.56 |
0.034 ± 0.015 0.029 |
63 | 20 ± 2 19.19 |
0.08 ± 0.05 0.077 |
5 |
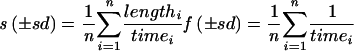 |
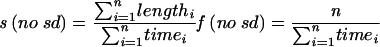 |
Number of microtubules evaluated to calculate the growth rate and catastrophe frequency.
Number of microtubules evaluated to calculate the shrinkage rate and rescue frequency.
Dynamic Instability Parameters Are Altered during the Formation of PPB
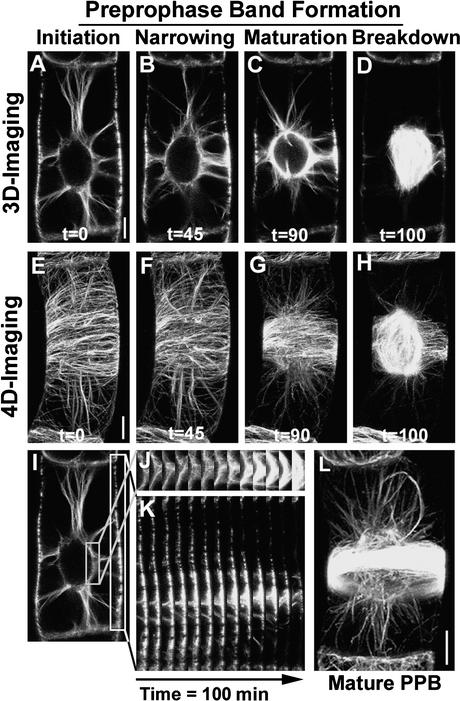
Three-dimensional (Figures 8A to 8D) and four-dimensional (Figures 8E to 8H) imaging of GFP-MAP4–expressing BY-2 cells allowed us to resolve PPB formation into four stages: PPB initiation, PPB narrowing, PPB maturation, and PPB breakdown. In BY-2 cells, PPB initiation to maturation took ∼90 min and PPB breakdown required 5 to 10 min, consistent with previous observations (Mineyuki, 1999). During PPB formation, first, microtubules formed a broad ring covering almost two-thirds of the cell length, and this structure gradually narrowed to a ring structure with a width of 2 to 5 μm (Figure 8). In the region where the PPB matures and in the region of the NE, GFP-MAP4 fluorescence intensity increased (Figures 8I to 8K). This finding suggests a flow of tubulin (or microtubules) from the cell cortex toward the region of PPB maturation and the NE. In addition, the length of NE-originated microtubules appeared to be reduced sequentially (cf. Figures 8A to 8C and Figures 8E to 8G). In the case of GFP-MAP4 (Figure 8L) and GFP-TUA6 (our unpublished results), because of their intense labeling along the entire microtubule length, it became difficult to resolve individual microtubules during the formation of compact PPB. This greatly complicates the use of these two proteins to analyze plant microtubule dynamic instability during PPB formation. Because YFP-CLIP170 tends to accumulate at the plus ends (with fine thin labeling on other microtubular parts), we still were successful in resolving individual microtubules during the transition from interphase to PPB and also on the NE (at the condensed state of the PPB) before its breakdown to form spindle.
Figure 8.
CLSM Micrographs of GFP-MAP4–Transformed BY-2 Cells Visualized with Three-Dimensional (xy and Time) and Four-Dimensional (xyz and Time) Imaging Displaying Four Phases of PPB Formation.
The phases are PPB initiation ([A] and [E]), PPB narrowing ([B] and [F]), PPB maturation ([C] and [G]), and PPB breakdown ([D] and [H]). The sections at right of (I) of the NE (J) and the cell cortex (K) are from the three-dimensional imaged cell from (A) to (D) and represent a pseudo x-time plot. (L) shows a completely mature and condensed PPB visualized with GFP-MAP4. Bars = 5 μm in the xy plane.
As shown in Figure 7, in YFP-CLIP170–transformed cell lines, microtubule end tracking still was possible at the microtubule-dense regions of the PPB and NE. In Table 1 and Figure 6S, the four parameters of individual plant microtubule dynamic instability are described for the interphase array, developing PPB, and prespindle NE-originated microtubules (at the condensed state of the PPB). As inferred from Figures 6S and 7 and Table 1, the microtubules in the PPB and on the NE display a marked and step-wise increase in growth rate, with constant shrinkage rate, and an increased catastrophe frequency, with almost unchanged rescue frequency. Additionally, the life history plots of YFP-CLIP170–labeled microtubules indicate that the average length of the microtubules from interphase to NE decreased (Figure 7). This finding highlights the importance of altered microtubule dynamic instability in driving the formation of the PPB that is plant cell specific and morphogenetically important. It also suggests that the PPB microtubules are more dynamic and shorter (as a result of higher growth rate and increased catastrophe frequency) than the interphase microtubules.
DISCUSSION
CLIP170 Binding to Plant Microtubule Plus Ends
To date, CLIP170 is the best-characterized microtubule plus-end binding protein from HeLa cells (Pierre et al., 1992; Perez et al., 1999). Our data provide clear evidence that CLIP170 of mammalian origin also binds to plant microtubules based on three criteria: the filamentous pattern displayed is typical of the spatial arrangement of plant microtubules in the cell cortex; the fluorescent pattern is sensitive to the anti-microtubular herbicide oryzalin (and not to the actin-depolymerizing drug latrunculin B); and CLIP170 labels all four plant microtubular arrays. That CLIP170 accumulates at plant microtubule plus ends is evident by two additional criteria: first, at low expression levels, CLIP170 appears in small dashes in protoplasts and the dash-like appearance displays dynamic behavior in stably transformed cells, highlighting the plus ends of microtubules; second, cotransfection experiments show that CLIP170 binds to the ends of MAP4-labeled plant microtubules. Apparently, as in mammalian cells, plant microtubules possess a GTP cap (Hyman et al., 1992) or specific structural features at the plus ends recognized by CLIP170, such as individual protofilaments of αβ-tubulin heterodimers that are not fully merged into the microtubule structure (Schuyler and Pellman, 2001).
As in mammalian cells (Pierre et al., 1994), CLIP170 lacking the 152–amino acid C-terminal part (CLIP1701-1240) decorates plant microtubules along their entire length (like MAP4). By contrast, the C-terminal domain remains cytosolic without any microtubule labeling. This finding suggests that CLIP1701-1240 is sufficient for plant microtubular labeling of CLIP170, whereas the C-terminal domain is essential but not sufficient for the targeting of CLIP170 to the plus ends of plant microtubules. This conserved similarity of CLIP170 functional domains in plant and animal cells suggests that the three-dimensional interaction site of microtubules to which CLIP170 binds is highly similar in the two different cell systems.
Biological Significance of the Altered Microtubule Dynamic Instability during PPB Formation
Dynamic instability of individual microtubules plays a pivotal role in cell morphogenesis (Kirschner and Mitchison, 1986) because it allows the microtubules to rapidly explore the three-dimensional cytoplasmic space and to rearrange and make specific configurations (Desai and Mitchison, 1997). Two previous reports (Hush et al., 1994; Moore et al., 1997) have shown some differences in plant and mammalian microtubule dynamics. Still, to our knowledge, the parameters of in vivo microtubule dynamic instability have not yet been reported for plant cells. Using GFP-tagged tubulin or GFP-tagged microtubule-associated proteins in stably transformed plant cells, we were able to study these parameters in living plant cells. Because there are no reference conditions, we analyzed the dynamic instability parameters obtained by plant microtubule labeling of three different fluorescent fusion proteins. In an interphase cortical array, the shrinkage rates (∼20 μm/min) are four times faster than the growth rates (5 μm/min). For mammalian cell microtubules, growth rates (11.5 μm/min) are similar to the shrinkage rates (13.1 μm/min) (Rusan et al., 2001), suggesting that plant microtubules grow slower but shrink faster than mammalian microtubules. By contrast, the catastrophe frequency (∼0.02 events/s) of plant microtubules is similar to that of mammalian microtubules (0.026 events/s), and the rescue frequency of plant microtubules (∼0.08 events/s) is half that of mammalian microtubules (0.175 events/s). These findings indicate that the plant microtubules have a faster polymer turnover rate than the mammalian microtubules, which makes them more dynamic, in agreement with data from a FRAP analysis (Hush et al., 1994).
Only the YFP-CLIP170 construct enabled us to monitor microtubule dynamic instability at the microtubule-dense regions during PPB formation and on the NE (PPB-condensed state). Strikingly, during the transition from interphase to PPB, the growth rate increased (4.7 to 7.2 μm/min) with an unchanged shrinkage rate (∼20 μm/min), whereas the catastrophe frequency doubled (0.017 to 0.034 events/s) with a similar rescue frequency (∼0.08 events/s). Such a change in microtubule dynamics was not observed in the previous FRAP analysis (Hush et al., 1994). On the one hand, this finding could suggest that there are cell type–related differences in the parameters. On the other hand, it is important to note that the fit for the PPB microtubule data from the FRAP study was not completely satisfactory according to the authors (Hush et al., 1994). Interestingly, at the condensed state of the PPB, the NE-originated microtubules became more dynamic considering their growth rate (8.8 μm/min), shrinkage rate (20 μm/min), catastrophe frequency (0.034 events/s), and rescue frequency (0.08 events/s). This finding provides strong evidence that the individual plant microtubule dynamic instability parameters (which should be controlled by the upstream regulators) are important determinants for the transition of the cortical interphase to the compact PPB microtubules surrounding the nucleus.
The Mechanism Responsible for PPB Formation
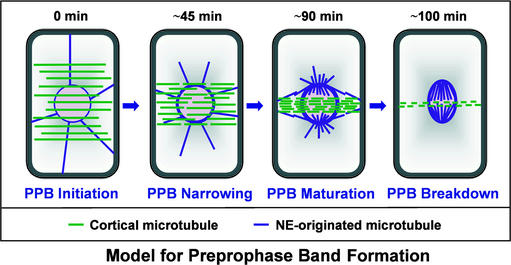
Together, these results prompt us to propose a model to elucidate the mechanism governing the microtubular events that occur during the formation of the PPB (Figure 9). At the onset of the transition from interphase to mitosis during PPB initiation, a broad PPB forms, overlapping almost two-thirds of the cell length (Figures 8A and 8E). At this stage, the catastrophe frequency increases gradually (possibly as a result of the inactivation of microtubule-stabilizing proteins or the activation of microtubule-destabilizing proteins), destroying the microtubules and creating an enhanced free tubulin pool that is used for the formation of new microtubules. The PPB contains newly formed microtubules, which is inferred from their ability to incorporate labeled tubulin (Cleary et al., 1992), the fact that PPB microtubules repolymerize after cold treatment–induced depolymerization (Murata and Wada, 1991), and the finding that the microtubule-stabilizing drug taxol abolishes PPB formation (Panteris et al., 1995). We hypothesize that the observed increased growth rate in the PPB is caused by an increased free tubulin concentration (Mitchison and Kirschner, 1984). During PPB narrowing, the cycle of gradual increase in catastrophe frequency and subsequently increased growth rate makes microtubules increasingly dynamic.
Figure 9.
Model Depicting the Formation of the PPB by the Alteration of Microtubule Dynamic Instability.
The previously described colocalization of γ-tubulin with the narrowing PPB and the enhanced staining of the NE by the γ-tubulin antibody (Liu et al., 1993) suggests a simultaneous increase in the microtubule organizing centers at the microtubule areas that display enhanced dynamics. However, it is noteworthy that the role of γ-tubulin in plant microtubule organizing centers has not been proven. Also, electron microscopy studies showed this increased number of microtubules in the PPB (Nogami et al., 1996), which is consistent with the observed increase in GFP-MAP4 labeling (Figures 8I to 8K) during PPB initiation to maturation. Moreover, the PPB still can form in cells whose de novo protein (or RNA) synthesis is inhibited by drugs, indicating that PPB formation occurs without new tubulin synthesis (Mineyuki, 1999) but rather using the existing tubulin pool. Because the same tubulin is used for the formation of more microtubules in the PPB and on the NE and the growth rate and catastrophe frequency are higher for those microtubules, PPB-originating microtubules must be shorter according to this model. Interestingly, in an early electron microscopy study, it was shown by serial sectioning that PPB microtubules are shorter in length than interphase microtubules (Hardham and Gunning, 1978), supporting our model. Additionally, the life history plots of YFP-CLIP170–decorated microtubules displayed a gradual reduction in average length, indicating shorter microtubules in the PPB and at the NE compared with interphase microtubules.
It has been shown that the actin-depolymerizing drug cytochalasin D affects the narrowing of the PPB (Eleftheriou and Palevitz, 1992; Granger and Cyr, 2001) and that the PPB region is depleted of actin (Cleary et al., 1992), suggesting a role of actin in the maturation of the PPB. We did not succeed in following the dynamic instability parameters of the PPB microtubules at its most condensed state, but we were able to follow the microtubule dynamics on the NE. Our idea is that during the final breakdown of the PPB, the catastrophe frequency reaches an even higher level (probably the result of the complete inactivation of microtubule-stabilizing proteins or the complete use of free tubulin by microtubules at the PPB and on the NE) that cannot be overcome by the increased growth rate of the PPB microtubules, resulting in its collapse. Apparently, the growth rate of NE-originating microtubules remains higher (or some NE-specific microtubule-stabilizing proteins still remain active) and they withstand the maximum catastrophe rate, survive, and use the new tubulin pool available from the destruction of the PPB to form the spindle microtubules.
Candidates That Regulate Microtubule Dynamic Instability during PPB Formation
The PPB formation process must be highly timed and regulated to yield the proper changes in the microtubular network. Possible candidates for the regulation of dynamic instability are microtubule-stabilizing proteins (such as XMAP215) (Tournebize et al., 2000) and microtubule-severing proteins (such as katanin) (Quarmby, 2000). In a physiological reconstitution of microtubule dynamics using purified components (Kinoshita et al., 2001), XMAP215 has been shown to be the key component of the regulation of microtubule dynamics (Kinoshita et al., 2002). The recently identified plant homolog of XMAP215 (MOR1) (Hussey and Hawkins, 2001; Whittington et al., 2001) is required in Arabidopsis to maintain long cortical microtubules during interphase. Consequently, mor1 mutant plants display highly stunted growth as a result of the short interphase microtubules. Interestingly, although the interphase microtubules are affected by the mor1 mutation, the PPB and spindle microtubules remain normal, being less affected by this mutation, supporting our model with shorter microtubules in these structures. The isolated plant homolog of mammalian katanin from Arabidopsis (AtKTN1) (Burk et al., 2001) does not show any defect in the PPB in its mutant, and the katanin-like p60 subunit–containing protein from Arabidopsis does not label the PPB by immunofluorescence (McClinton et al., 2001), suggesting no influence of microtubule severing in the formation of PPB, again strengthening the significance of altered microtubule dynamic instability.
Other candidates for the regulation of microtubule dynamics are cyclin-dependent kinases (cdc), which have been proposed to phosphorylate microtubule-stabilizing or catastrophe-inhibiting proteins, making them inactive, and proteins involved in the dephosphorylation of microtubule-severing proteins, making them active (Belmont et al., 1990; Verde et al., 1992). In addition, kinase inhibitors have been shown to inhibit the development and disappearance of the PPB (Katsuta and Shibaoka, 1992). It has been shown that microinjection of cdc2 in plant cells induces rapid degradation of the PPB (Hush et al., 1996), and GFP-labeled cdc2 forms a narrow and bright equatorial band resembling the late PPB (Weingartner et al., 2001). In Xenopus egg extracts, type 2A phosphatases were shown to be essential for maintaining the short steady state length of microtubules by regulating the level of microtubule catastrophe (Tournebize et al., 1997). Recently, it was reported that the Arabidopsis TONNEAU2 gene encodes a putative novel protein phosphatase 2A regulatory subunit. In a mutant of this gene, the plant cells become unable to form the PPB, finally resulting in completely hindered growth and development (Camilleri et al., 2002). Additionally, it has been shown that the inhibition of Ser/Thr-specific protein phosphatases causes premature activation of cdc2 kinase at the G2/M transition and early mitotic microtubule organization in plants (Ayaydin et al., 2000). Moreover, the nuclear cycle and the microtubule transition from interphase to prophase are coupled in BY-2 cells, as indicated by the inhibition of PPB formation as well as the S-phase progression by aphidicolin (a DNA polymerase–inhibiting drug) and the restart of PPB formation after aphidicolin removal (Katsuta et al., 1990). This could link (as in mammalian cells) the cdc regulation of plant microtubule dynamic instability responsible for the transition from interphase to mitotic microtubular conformation. Future research should address the different plant factors (including MOR1, TONNEAU2, cdc2, and AtKTN1) that control the dynamic instability responsible for microtubular rearrangements in various cellular processes, and the use of YFP-CLIP170 to decorate the plus ends of microtubules in plant cells will be of great help in such studies.
METHODS
Construction of Reporter Genes
CLIP170 Constructs
Wild-type green fluorescent protein (GFP) from the wild-type GFP-CLIP170 fusion protein (Perez et al., 1999) in mammalian cell expression vector pCB6 was replaced with human codon–optimized enhanced yellow fluorescent protein (YFP) (S65G, S72A, and T203Y) and enhanced cyan fluorescent protein (CFP) (F64L, S65T, Y66W, N146I, M153T, V163A, and N212K) (Miyawaki et al., 1997) by a multiple-step cloning procedure performed in pBluescript KS+ vector (Stratagene). Plant-specific Cauliflower mosaic virus double 35S promoter and nopaline synthase (NOS) terminator were cloned into this vector to give the complete plant expression cassettes pBluescript-double 35S-Myc-EYFP-CLIP170-tNOS and pBluescript-double 35S-Myc-ECFP-CLIP170-tNOS, which were used for protoplast transfections. Vector pBluescript-double 35S-Myc-EYFP-CLIP1701-1240-tNOS was created by replacing CLIP170 with the PCR-amplified fragment of CLIP1701-1240. Similarly, pBluescript-double 35S-Myc-EYFP-CLIP1701241-1392-tNOS was made by replacing CLIP170 with the CLIP1701241-1392 fragment. For stable tobacco (Nicotiana tabacum) Bright Yellow 2 (BY-2) cell transformations, we subcloned double 35S-Myc-EYFP-CLIP170-tNOS and double 35S-Myc-EYFP-CLIP1701-1240-tNOS from pBluescript into the plant transformation vector pBINPLUS.
MAP4 Constructs
We constructed reporter genes by fusing human codon–optimized EGFP (S65T and F64L), EYFP, and ECFP to full-length MAP4 cDNA (Olson et al., 1995) by a PCR-based cloning method, resulting in vector pMON-double 35S-EGFP-MAP4-tNOS, pMON-double 35S-EYFP-MAP4-tNOS, and pMON-double 35S-ECFP-MAP4-tNOS, which were used for transient expression assays. For stable transformation of BY-2 cells, double 35S-EGFP-MAP4-tNOS was transferred to plant transformation vector pBINPLUS.
α-Tubulin Construct
Vector pBI121 containing SmRSGFP-TUA6 (Ueda et al., 1999) was used for stable transformations of BY-2 cells.
Talin Construct
Vector pMON-double 35S-Talin-EYFP-tNOS containing the actin binding domain of Dictyostelium Talin was used for protoplast transfections.
In preparation for all of these constructs, PCR amplification was performed with either the Expand High-Fidelity PCR system or the Expand Long-Template PCR system (Boehringer Mannheim). PCR-amplified fragments were purified using the High Pure PCR product purification kit (Boehringer Mannheim). The sequences of all of the PCR-amplified fragments and the linkers were confirmed by DNA sequencing. Plasmid DNA isolations were performed with either the High Pure Plasmid isolation kit (Boehringer Mannheim) or the Plasmid Midi kit (Qiagen, Valencia, California). Additional details regarding cloning are available in the supplemental data online.
Protoplast Preparation and Transfection
Cowpea (Vigna unguiculata) mesophyll leaf-originating protoplast isolation and transfection were performed as described by van Bokhoven et al. (1993).
Transformation of Tobacco BY-2 Cells
Stable transformations of BY-2 cells were performed using Agrobacterium tumefaciens strain LBA4404, which was transformed with plant transformation vector pBINPLUS containing double 35S-Myc-EYFP-CLIP170-tNOS, double 35S-Myc-EYFP-CLIP1701-1240-tNOS, and double 35S-EGFP-MAP4-tNOS constructs and vector pBI121 containing 35S-SmRSGFP-TUA6-tNOS. A 1-mL aliquot of 4-day-old BY-2 cells was inoculated with 50 μL of LBA4404 log-phase culture. After 3 days of incubation at 25°C on a stable surface in the dark, the cells were washed four times in 10 mL of BY-2 medium (3% Suc, 4.3 g/L Murashige and Skoog [1962] salts, 100 mg/L myo-inositol, 1 mg/L thiamine HCl, 0.2 mg/L 2,4-D, and 255 mg/L KH2PO4, pH 5.8) containing cefotaxime sodium (250 mg/L) to eliminate agrobacteria and plated on solid BY-2 medium (with 0.8% Japanese agar) containing cefotaxime sodium (250 mg/L) and kanamycin (100 mg/L). The plates were incubated at 25°C in the dark for 3 weeks. Within 3 weeks, the transformed cells were grown to form calluses on plates, which were tested with a Leica DMR stereomicroscope (Wetzlar, Germany) equipped with an excitation mercury lamp and emission filters for GFP and YFP to detect their fluorescence levels. Calli exhibiting sufficient levels of fluorescence were transferred to new BY-2 plates containing kanamycin (100 mg/L) three to four times and then were cultured independently into BY-2 liquid medium containing kanamycin (100 mg/L). Stably transformed and nontransformed BY-2 cell lines were maintained independently by culturing them every week (1:50) in BY-2 medium in 250-mL flasks and shaking at 25°C and 130 rpm in an Innova 4330 temperature-controlled rotary shaker (New Brunswick Scientific, Edison, NJ).
Measurement of Cell Shapes and Cell Numbers
Phase-contrast images were obtained with a Zeiss Axiovert inverted microscope (Jena, Germany) coupled to a Photometrics Coolsnap-fx charge-coupled device camera (Roper Scientific, Trenton, NJ). Images were obtained with a ×10 Plan Neofluar phase-contrast objective (numerical aperture of 0.3). All of the images were analyzed with Adobe Photoshop version 5.0 (Mountain View, CA). The cell shapes were determined using the breadth-to-length measurement function of the Object-Image 2.08 program (http://simon.bio.uva.nl/object-image.html) developed especially by N. Vischer from our laboratory. To measure transgenic and wild-type BY-2 cell numbers, both cell types were incubated briefly with digesting enzymes (1% cellulase, 0.1% pectinase, and 0.35 M mannitol) to disrupt cell clumps (Granger and Cyr, 2000). Appropriate dilutions were made, and cells were counted using a hemocytometer. Relative cell numbers were calculated based on initial numbers at day 1. Means and standard deviations were calculated, and the graphs were obtained using Excel (Microsoft, Redmond, WA).
Fluorescence Microscopy
Samples were prepared in NUNC chambers (Nunc, Inc., Naperville, IL) made especially for microscopy. To acquire images of transfected cowpea protoplasts and transformed tobacco BY-2 cells with different fluorescent constructs, we used confocal laser scanning microscopy based on a Zeiss LSM510 microscopy system composed of an Axiovert inverted microscope equipped with an argon ion laser as an excitation source. Protoplasts and BY-2 cells expressing GFP were excited with a 488-nm laser line, and GFP emission was detected using a 505- to 530-nm band-pass filter. Protoplasts and BY-2 cells expressing EYFP were excited with a 514-nm laser line, and EYFP emission was detected with a 535- to 590-nm band-pass filter. Protoplasts expressing ECFP were excited with a 458-nm laser line, and ECFP emission was captured with a 470- to 500-nm band-pass filter. In cotransfection experiments, both ECFP/EYFP were scanned simultaneously using a 458/514 main dichroic splitter and a 515 secondary dichroic splitter operating in the multi-tracking imaging mode. This procedure effectively eliminates bleeding of CFP fluorescence into the YFP channel and vice versa. A ×40 oil-immersion objective (numerical aperture of 1.3) was used to scan the samples. The images were captured using LSM510 image-acquisition software (Zeiss). For each sample, at least 30 optical sections spaced 0.5 μm apart were taken to cover a half-hemisphere of a cell. For analysis of individual plant microtubule dynamics, cortical and nuclear surface cell regions were scanned with an image-acquisition time of 1 s and 2- to 5-s intervals between two successive scans for several minutes, yielding clear movies of the movements of microtubule plus ends.
Image Processing and Quantification of Microtubule Dynamic Instability Parameters
Acquired images were processed using LSM510 image browser version 3.0 (Zeiss). Maximum projections were obtained from serial optical sections spaced 0.5 μm apart and were exported as TIFF files. For time series scans, all of the images were exported as time series TIFF files. All of the exported images were processed with Adobe Photoshop version 5. For individual plant microtubule dynamic instability measurements, all of the time scans were analyzed in the animation mode of LSM510 image browser 3.0 (Zeiss) by marking the single ends of individual microtubules in each image using a zoom function and tracking the microtubule ends for several minutes. The shortest resolvable displacement of the plus ends that was measurable by this analysis was 0.1 μm. The time values were obtained from individual frame times in time-lapse experiments. Thereafter, the data were transferred manually into Excel files and processed. Life history plots of individual microtubules were obtained from these tracking data. Growth and shrinkage velocities were calculated by dividing the distance grown or shortened by the time spent in that event. The frequency of catastrophe was calculated from the inverse of the mean time spent in elongation, and the frequency of rescue was obtained from the inverse of the mean time spent in shrinkage, as described previously (Cassimeris et al., 1988). The movies were assembled by exporting confocal laser scanning microscopy data files to the Videomach 2.3.4 program (Gromada, Berkeley, CA).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Supplementary Material
Acknowledgments
We thank F. Perez (Curie Institute, Centre National de la Recherche Scientifique, Paris, France) for the generous gift of vector pCB6 containing wild-type GFP-CLIP170. We are grateful to J. Olmsted (University of Rochester, NY) for kindly providing MAP4 cDNA, T. Hashimoto (Nara Institute of Science and Technology, Nara, Japan) for the gift of SmRSGFP-TUA6, and G. van der Krogt (Wageningen University, The Netherlands) for kindly supplying Talin-YFP, EGFP, EYFP, and ECFP. We are thankful to T. Bisseling and T. Ruttink (Wageningen University) for their support in the initial phase of this research, to M. Hink, J.-W. Borst (Wageningen University), and W. Takkenberg (University of Amsterdam, The Netherlands) for the CLSM facilities, and to E. van Munster and N. Vischer for assistance with phase-contrast microscopy. We are grateful to M. Dogterom, J. Carette, C. Woldringh, N. Nanninga, and J. Goedhart for stimulating discussions and critical reading of the manuscript. The authors were supported by the Netherlands Organization for Scientific Research (NWO 805.47.012).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.008961.
Footnotes
Online version contains Web-only data.
References
- Ayaydin, F., Vissi, E., Meszaros, T., Miskolczi, P., Kovacs, I., Feher, A., Dombradi, V., Erdodi, F., Gergely, P., and Dudits, D. (2000). Inhibition of serine/threonine-specific protein phosphatases causes premature activation of cdc2MsF kinase at G2/M transition and early mitotic microtubule organisation in alfalfa. Plant J. 23, 85–96. [DOI] [PubMed] [Google Scholar]
- Belmont, L.D., Hyman, A.A., Sawin, K.E., and Mitchison, T.J. (1990). Real-time visualization of cell cycle-dependent changes in microtubule dynamics in cytoplasmic extracts. Cell 62, 579–589. [DOI] [PubMed] [Google Scholar]
- Bibikova, T.N., Blancaflor, E.B., and Gilroy, S. (1999). Microtubules regulate tip growth and orientation in root hairs of Arabidopsis thaliana. Plant J. 17, 657–665. [DOI] [PubMed] [Google Scholar]
- Burk, D.H., Liu, B., Zhong, R., Morrison, W.H., and Ye, Z.H. (2001). A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13, 807–827. [PMC free article] [PubMed] [Google Scholar]
- Camilleri, C., Azimzadeh, J., Pastuglia, M., Bellini, C., Grandjean, O., and Bouchez, D. (2002). The Arabidopsis TONNEAU2 gene encodes a putative novel protein phosphatase 2A regulatory subunit essential for the control of the cortical cytoskeleton. Plant Cell 14, 833–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassimeris, L., Pryer, N.K., and Salmon, E.D. (1988). Real-time observations of microtubule dynamic instability in living cells. J. Cell Biol. 107, 2223–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary, A.L., Gunning, B.E., Wasteneys, G.O., and Hepler, P.K. (1992). Microtubule and F-actin dynamics at the division site in living Tradescantia stamen hair cells. J. Cell Sci. 103, 977–988. [Google Scholar]
- Cyr, R.J. (1994). Microtubules in plant morphogenesis: Role of the cortical array. Annu. Rev. Cell Biol. 10, 153–180. [DOI] [PubMed] [Google Scholar]
- Desai, A., and Mitchison, T.J. (1997). Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13, 83–117. [DOI] [PubMed] [Google Scholar]
- Eleftheriou, E.P., and Palevitz, B.A. (1992). The effect of cytochalasin D on preprophase band organization in root tip cells of Allium. J. Cell Sci. 103, 989–998. [Google Scholar]
- Ellenberg, J., Lippincott-Schwartz, J., and Presley, J.F. (1999). Dual-colour imaging with GFP variants. Trends Cell Biol. 9, 52–56. [DOI] [PubMed] [Google Scholar]
- Goddard, R.H., Wick, S.M., Silflow, C.D., and Snustad, D.P. (1994). Microtubule components of the plant cell cytoskeleton. Plant Physiol. 104, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger, C., and Cyr, R. (2000). Microtubule reorganization in tobacco BY-2 cells stably expressing GFP-MBD. Planta 210, 502–509. [DOI] [PubMed] [Google Scholar]
- Granger, C., and Cyr, R. (2001). Use of abnormal preprophase bands to decipher division plane determination. J. Cell Sci. 114, 599–607. [DOI] [PubMed] [Google Scholar]
- Hardham, A.R., and Gunning, B.E. (1978). Structure of cortical microtubule arrays in plant cells. J. Cell Biol. 77, 14–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hush, J., Wu, L., John, P.C., Hepler, L.H., and Hepler, P.K. (1996). Plant mitosis promoting factor disassembles the microtubule preprophase band and accelerates prophase progression in Tradescantia. Cell Biol. Int. 20, 275–287. [DOI] [PubMed] [Google Scholar]
- Hush, J.M., Wadsworth, P., Callaham, D.A., and Hepler, P.K. (1994). Quantification of microtubule dynamics in living plant cells using fluorescence redistribution after photobleaching. J. Cell Sci. 107, 775–784. [DOI] [PubMed] [Google Scholar]
- Hussey, P.J., and Hawkins, T.J. (2001). Plant microtubule-associated proteins: The HEAT is off in temperature-sensitive mor1. Trends Plant Sci. 6, 389–392. [DOI] [PubMed] [Google Scholar]
- Hyman, A.A., Salser, S., Drechsel, D.N., Unwin, N., and Mitchison, T.J. (1992). Role of GTP hydrolysis in microtubule dynamics: Information from a slowly hydrolyzable analogue, GMPCPP. Mol. Biol. Cell 3, 1155–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuta, J., Hashiguchi, Y., and Shibaoka, H. (1990). The role of the cytoskeleton in positioning of the nucleus in premitotic tobacco BY-2 cells. J. Cell Sci. 95, 413–422. [Google Scholar]
- Katsuta, J., and Shibaoka, H. (1992). Inhibition by kinase inhibitors of the development and disappearance of the preprophase band of microtubules in tobacco BY-2 cells. J. Cell Sci. 103, 397–405. [Google Scholar]
- Kinoshita, K., Arnal, I., Desai, A., Drechsel, D.N., and Hyman, A.A. (2001). Reconstitution of physiological microtubule dynamics using purified components. Science 294, 1340–1343. [DOI] [PubMed] [Google Scholar]
- Kinoshita, K., Habermann, B., and Hyman, A.A. (2002). XMAP215: A key component of the dynamic microtubule cytoskeleton. Trends Cell Biol. 12, 267–273. [DOI] [PubMed] [Google Scholar]
- Kirschner, M., and Mitchison, T. (1986). Beyond self-assembly: From microtubules to morphogenesis. Cell 45, 329–342. [DOI] [PubMed] [Google Scholar]
- Kost, B., and Chua, N.H. (2002). The plant cytoskeleton: Vacuoles and cell walls make the difference. Cell 108, 9–12. [DOI] [PubMed] [Google Scholar]
- Kost, B., Mathur, J., and Chua, N.H. (1999). Cytoskeleton in plant development. Curr. Opin. Plant Biol. 2, 462–470. [DOI] [PubMed] [Google Scholar]
- Kost, B., Spielhofer, P., and Chua, N.H. (1998). A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16, 393–401. [DOI] [PubMed] [Google Scholar]
- Liu, B., Marc, J., Joshi, H.C., and Palevitz, B.A. (1993). A gamma-tubulin-related protein associated with the microtubule arrays of higher plants in a cell cycle-dependent manner. J. Cell Sci. 104, 1217–1228. [DOI] [PubMed] [Google Scholar]
- Marc, J., Granger, C.L., Brincat, J., Fisher, D.D., Kao, T., McCubbin, A.G., and Cyr, R.J. (1998). A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 10, 1927–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus, A.I., Moore, R.C., and Cyr, R.J. (2001). The role of microtubules in guard cell function. Plant Physiol. 125, 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur, J., and Chua, N.H. (2000). Microtubule stabilization leads to growth reorientation in Arabidopsis trichomes. Plant Cell 12, 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClinton, R.S., Chandler, J.S., and Callis, J. (2001). cDNA isolation, characterization, and protein intracellular localization of a katanin-like p60 subunit from Arabidopsis thaliana. Protoplasma 216, 181–190. [DOI] [PubMed] [Google Scholar]
- McClinton, R.S., and Sung, Z.R. (1997). Organization of cortical microtubules at the plasma membrane in Arabidopsis. Planta 201, 252–260. [DOI] [PubMed] [Google Scholar]
- Mineyuki, Y. (1999). The preprophase band of microtubules: Its function as a cytokinetic apparatus in higher plants. Int. Rev. Cytol. 187, 1–49. [Google Scholar]
- Mitchison, T., and Kirschner, M. (1984). Dynamic instability of microtubule growth. Nature 312, 237–242. [DOI] [PubMed] [Google Scholar]
- Miyawaki, A., Llopis, J., Heim, R., McCaffery, J.M., Adams, J.A., Ikura, M., and Tsien, R.Y. (1997). Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388, 882–887. [DOI] [PubMed] [Google Scholar]
- Moore, R.C., Zhang, M., Cassimeris, L., and Cyr, R.J. (1997). In vitro assembled plant microtubules exhibit a high state of dynamic instability. Cell Motil. Cytoskeleton 38, 278–286. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Murata, T., and Wada, M. (1991). Re-formation of the preprophase band after cold-induced depolymerization of microtubules in Adiantum protonemata. Plant Cell Physiol. 32, 1145–1151. [Google Scholar]
- Nogami, A., Suzaki, T., Shigenaka, Y., Nagahama, Y., and Mineyuki, Y. (1996). Effects of cycloheximide on preprophase bands and prophase spindles in onion (Allium cepa L.) root tip cells. Protoplasma 192, 109–121. [Google Scholar]
- Olson, K.R., McIntosh, J.R., and Olmsted, J.B. (1995). Analysis of MAP 4 function in living cells using green fluorescent protein (GFP) chimeras. J. Cell Biol. 130, 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui, M., and Staehelin, L.A. (2000). Cytokinesis in flowering plants: More than one way to divide a cell. Curr. Opin. Plant Biol. 3, 493–502. [DOI] [PubMed] [Google Scholar]
- Panteris, E., Apostolakos, P., and Galatis, B. (1995). The effect of taxol on Triticum preprophase root cells: Preprophase microtubule band organization seems to depend on new microtubule assembly. Protoplasma 186, 72–78. [Google Scholar]
- Perez, F., Diamantopoulos, G.S., Stalder, R., and Kreis, T.E. (1999). CLIP-170 highlights growing microtubule ends in vivo. Cell 96, 517–527. [DOI] [PubMed] [Google Scholar]
- Pierre, P., Pepperkok, R., and Kreis, T.E. (1994). Molecular characterization of two functional domains of CLIP-170 in vivo. J. Cell Sci. 107, 1909–1920. [DOI] [PubMed] [Google Scholar]
- Pierre, P., Scheel, J., Rickard, J.E., and Kreis, T.E. (1992). CLIP-170 links endocytic vesicles to microtubules. Cell 70, 887–900. [DOI] [PubMed] [Google Scholar]
- Quarmby, L. (2000). Cellular samurai: Katanin and the severing of microtubules. J. Cell Sci. 113, 2821–2827. [DOI] [PubMed] [Google Scholar]
- Rickard, J.E., and Kreis, T.E. (1996). CLIPs for organelle-microtubule interactions. Trends Cell Biol. 6, 178–183. [DOI] [PubMed] [Google Scholar]
- Rusan, N.M., Fagerstrom, C.J., Yvon, A.M., and Wadsworth, P. (2001). Cell cycle-dependent changes in microtubule dynamics in living cells expressing green fluorescent protein-alpha tubulin. Mol. Biol. Cell 12, 971–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel, J., Pierre, P., Rickard, J.E., Diamantopoulos, G.S., Valetti, C., van der Goot, F.G., Haner, M., Aebi, U., and Kreis, T.E. (1999). Purification and analysis of authentic CLIP-170 and recombinant fragments. J. Biol. Chem. 274, 25883–25891. [DOI] [PubMed] [Google Scholar]
- Schuyler, S.C., and Pellman, D. (2001). Microtubule “plus-end-tracking proteins”: The end is just the beginning. Cell 105, 421–424. [DOI] [PubMed] [Google Scholar]
- Steinborn, K., Maulbetsch, C., Priester, B., Trautmann, S., Pacher, T., Geiges, B., Kuttner, F., Lepiniec, L., Stierhof, Y.D., Schwarz, H., Jurgens, G., and Mayer, U. (2002). The Arabidopsis PILZ group genes encode tubulin-folding cofactor orthologs required for cell division but not cell growth. Genes Dev. 16, 959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, L.P., and Hepler, P.K. (1997). Pollen germination and tube growth. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 461–491. [DOI] [PubMed] [Google Scholar]
- Thitamadee, S., Tuchihara, K., and Hashimoto, T. (2002). Microtubule basis for left-handed helical growth in Arabidopsis. Nature 417, 193–196. [DOI] [PubMed] [Google Scholar]
- Tournebize, R., Andersen, S.S., Verde, F., Doree, M., Karsenti, E., and Hyman, A.A. (1997). Distinct roles of PP1 and PP2A-like phosphatases in control of microtubule dynamics during mitosis. EMBO J. 16, 5537–5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournebize, R., Popov, A., Kinoshita, K., Ashford, A.J., Rybina, S., Pozniakovsky, A., Mayer, T.U., Walczak, C.E., Karsenti, E., and Hyman, A.A. (2000). Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat. Cell Biol. 2, 13–19. [DOI] [PubMed] [Google Scholar]
- Traas, J., Bellini, C., Nacry, P., Kronenberger, J., Bouchez, D., and Caboche, M. (1995). Normal differentiation patterns in plants lacking microtubular preprophase bands. Nature 375, 676–677. [Google Scholar]
- Ueda, K., Matsuyama, T., and Hashimoto, T. (1999). Visualization of microtubules in living cells of transgenic Arabidopsis thaliana. Protoplasma 206, 201–206. [Google Scholar]
- van Bokhoven, H., Verver, J., Wellink, J., and van Kammen, A. (1993). Protoplasts transiently expressing the 200K coding sequence of cowpea mosaic virus B-RNA support replication of M-RNA. J. Gen. Virol. 74, 2233–2241. [DOI] [PubMed] [Google Scholar]
- Verde, F., Dogterom, M., Stelzer, E., Karsenti, E., and Leibler, S. (1992). Control of microtubule dynamics and length by cyclin A- and cyclin B-dependent kinases in Xenopus egg extracts. J. Cell Biol. 118, 1097–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingartner, M., Binarova, P., Drykova, D., Schweighofer, A., David, J.P., Heberle-Bors, E., Doonan, J., and Bogre, L. (2001). Dynamic recruitment of Cdc2 to specific microtubule structures during mitosis. Plant Cell 13, 1929–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittington, A.T., Vugrek, O., Wei, K.J., Hasenbein, N.G., Sugimoto, K., Rashbrooke, M.C., and Wasteneys, G.O. (2001). MOR1 is essential for organizing cortical microtubules in plants. Nature 411, 610–613. [DOI] [PubMed] [Google Scholar]
- Wymer, C.L., Wymer, S.A., Cosgrove, D.J., and Cyr, R.J. (1996). Plant cell growth responds to external forces and the response requires intact microtubules. Plant Physiol. 110, 425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H.G., Hiatt, E.N., and Dawe, R.K. (2000). The plant kinetochore. Trends Plant Sci. 5, 543–547. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.