Abstract
The DEFORMED ROOTS AND LEAVES1 (DRL1) gene is single copy in the Arabidopsis genome, and based on overall amino acid similarity and conservation of functional domains, the DRL1 protein is homologous with yeast TOT4/KTI12. TOT4/KTI12 associates with Elongator, a multisubunit complex that binds the RNA polymerase II transcription elongation complex. Recessive mutations at the DRL1 locus caused defective organ formation indicative of disorganized shoot, inflorescence, flower, and root meristems. DRL1 is a putative ATP/GTP binding protein; in addition, calmodulin binding activity was demonstrated in vitro for the C terminus of the DRL1 protein. Phenotypic and genetic data position DRL1 relative to regulatory loci for leaf development, in which it acts early. We identified Arabidopsis homologs for the six Elongator components and hypothesize that DRL1 regulates transcription elongation through a putative plant Elongator. Upregulation of the ANGUSTIFOLIA transcript in the strong drl1-2 allele supports this model.
INTRODUCTION
In plants, developmental processes occur primarily after germination, because the embryonic body is rather rudimentary. The apical-basal axis of the embryo is delineated by apical meristems that determine the future growth direction of the organism, whereas the radial axis specifies the identity and arrangement of tissues in concentric layers. During development, pattern formation, growth, and differentiation are overlapping rather than consecutive events. These processes are reiterated throughout the life cycle upon the formation of every new organ.
In Arabidopsis, the shoot apical meristem (SAM) is essential for the formation of the vegetative plant body. It is organized in zones with different rates of cell division and different functions (Medford et al., 1992). The central zone consists of “stem” cells that maintain indeterminate growth and produce daughter cells for the neighboring peripheral and rib zones (Laux et al., 1996; Long et al., 1996; Mayer et al., 1998; Moussian et al., 1998). From the peripheral and rib zones, lateral organs and the pith tissues are produced, respectively. Progenitor tissue layers are radially organized in the Arabidopsis SAM. In the leaf primordia, the L1 layer is the progenitor of the epidermis, the L2 layer contributes to the outer leaf mesophyll tissue, and the L3 layer forms the inner part of the leaf (Sussex, 1989). Upon disruption of the cell balance between the different layers and zones, the size of the SAM is altered, which affects lateral organ number and size (Clark et al., 1993, 1997; Laufs et al., 1998; Fletcher et al., 1999; Trotochaud et al., 1999; Brand et al., 2000; Schoof et al., 2000). Common mechanisms for meristem maintenance, structure, organization, and partitioning exist for the SAM, the inflorescence meristem, and the floral meristem (Clark et al., 1993). As a consequence, functional defects in meristems often are visible as defects in both leaf and flower formation. The root apical meristem produces the primary root, which is organized radially with initials that produce cell files in concentric layers. These cells enter the elongation zone, in which they continue to divide and start expanding and differentiating.
In Arabidopsis, leaves initiate postembryonically at specific positions at the periphery of the SAM according to a radial pattern (Reinhardt et al., 2000). The first two rosette leaves are in opposite positions; the third leaf is perpendicular to the axis between the first two leaves; and the fourth and subsequent leaves are at angles of ∼137° in a so-called spiral phyllotaxis. Regulated cell division activity and changes in the orientation of cell plates in the L2 layer precede the initiation of leaf primordia (Medford et al., 1992). The repression of the homeobox gene SHOOT MERISTEMLESS and the activation of the myb gene ASYMMETRIC LEAVES1 (AS1) are crucial for leaf initiation (Long et al., 1996; Byrne et al., 2000). AS1 imposes a dorsoventral asymmetry on the radial symmetry of the leaf primordium (Byrne et al., 2000). In the leaf blade, dorsal identity is pro-moted by PHABULOSA (PHAB) and PHAVOLUTA (McConnell et al., 2001) and ventral identity is promoted by FILAMENTOUS FLOWER (FIL)/YABBY (YAB) and KANADI (Sawa et al., 1999; Siegfried et al., 1999; Eshed et al., 2001; Kerstetter et al., 2001). Leaves grow by cell division and cell expansion mainly along their length and width according to gradients (Pyke et al., 1991; Van Lijsebettens and Clarke, 1998; Donnelly et al., 1999). Growth is restricted along the thickness (dorsoventral axis) direction because of the pattern formation in tissue layers. Four tissues are specified along the dorsoventral axis: the upper epidermis and palisade parenchyma, with dorsal identity; the spongy parenchyma; and the lower epidermis, with ventral identity. Later growth occurs mainly by polar and nonpolar cell expansion processes. ANGUSTIFOLIA (AN) is a transcriptional corepressor that regulates lamina width by polar expansion of palisade cells (Tsuge et al., 1996; Kim et al., 1998, 2002a). Pattern formation in lateral growth results in the distinction between the lamina and the petiole (van der Graaff et al., 2000).
Regulation of gene expression at the transcriptional level is an important and universal mechanism for the control of developmental programs. Classes of specific transcription factors recognize upstream promoter boxes in specific sets of genes. Through direct or indirect interaction with the general transcription factors, the RNA polymerase II (RNAPII) transcription initiation complex is either activated or repressed. Specific transcription factors are activated by environmental or developmental stimuli that are transduced from the cell plasma membrane into the nucleus. Evidence in yeast and humans is accumulating that the expression of gene sets also is controlled by transcription elongation. The RNAPII transcription elongation complex forms the unfolded structure of transcribing nucleosomes (Walia et al., 1998). The elongation reaction is stimulated by a large variety of factors, of which some prevent pausing or stalling of the RNAPII complex and others model the chromatin for transcription. The degree of chromatin condensation is modulated by histone acetyltransferases and deacetylases (Walia et al., 1998; Wittschieben et al., 1999). The elongating RNAPII holoenzyme is copurified with a multisubunit complex, Elongator, whose stable interaction depends on the hyperphosphorylated state of the RNAPII C-terminal domain (Otero et al., 1999). In yeast, the Elongator complex consists of two subcomplexes: one consists of ELP1 (Otero et al., 1999), ELP2 (a WD40 repeat protein) (Fellows et al., 2000), and ELP3 (a histone acetyltransferase) (Wittschieben et al., 1999); the other consists of ELP4, ELP5, and ELP6 (Krogan and Greenblatt, 2001; Li et al., 2001; Winkler et al., 2001). Phenotypes of elpΔ mutants in yeast include slow growth adaptation, delayed gene activation, and temperature sensitivity, demonstrating that the ELP genes activate inducible genes to adapt to new growth conditions. In addition, the elpΔ mutants also are delayed in the G1 phase of the cell cycle and are hypersensitive to calcofluor white, 6-azauracil, and caffeine (Otero et al., 1999; Wittschieben et al., 1999; Fellows et al., 2000; Krogan and Greenblatt, 2001; Winkler et al., 2001).
We identified the DEFORMED ROOT AND LEAVES1 (DRL1) gene, a homolog of the yeast TOT4/KTI12 gene (Butler et al., 1994; Frohloff et al., 2001; Fichtner et al., 2002), upon screening a Dissociation (Ds)-mutagenized Arabidopsis population for leaf mutants. TOT genes were identified in a search for yeast mutants resistant to the zymocin toxin of Kluyveromyces lactis. TOT1, TOT2, and TOT3 are isoallelic to ELP1, ELP2, and ELP3; hence, TOT corresponds to Elongator. To understand the mode of action of this toxin, another screen was performed to isolate killer toxin–insensitive (kti) mutants (Butler et al., 1994). The described KTI genes (KTI11, KTI12, and KTI13) encoded proteins that were not Elongator components but that associate with Elongator and possibly regulate its function (Fichtner et al., 2002; Fichtner and Schaffrath, 2002). Thus, TOT4/KTI12 encodes a protein that associates with the Elongator complex (Frohloff et al., 2001; Fichtner et al., 2002). The tot4 mutant displays phenotypes similar to those of deficient-elongator mutants.
We investigated the function of DRL1 in higher plants by mutational analyses. drl1 alleles showed defects in meristem activity and organ growth. The positions of the mutations identified additional functional domains in the protein. Calmodulin binding of the DRL1 protein was demonstrated, indicating that the plant protein activity is Ca2+ dependent, in contrast to that of the yeast homolog, which does not contain a calmodulin binding site. We also provide evidence for the existence of Elongator in plants.
RESULTS
drl1-2 Leaf Phenotype
The drl1-2 leaf mutant was identified when screening 250 F2 populations derived from a cross between the DsB1 line containing the Ds element cloned in the leader of the p35S-streptomycin phosphotransferase gene and marked by a p35S-hygromycin phosphotransferase II gene (Bancroft et al., 1992) and the AcTn25 line containing an Activator (Ac) element with a p35S-Ac transposase (Swinburne et al., 1992). drl1-2 mutants were obtained in the F2 population of the cross as full green individuals on streptomycin/hygromycin-containing selective medium. A mutant individual was crossed to Arabidopsis wild-type Landsberg erecta (Ler), and F2 plants were analyzed: 702 wild-type and 217 drl1-2 plants were obtained, showing that drl1-2 is a nuclear recessive mutation [χ2 (3:1) = 0.87; P > 0.05]. Three additional drl1 alleles were identified. drl1-1 (Bancroft et al., 1993) and drl1-3 (R. Simon, unpublished results) were isolated after independent Ds transactivation experiments starting from the DsB1 parental line (Table 1). The plant genomic sequences flanking the transposed Ds (tDs) in these alleles were derived from the same gene as that flanking the tDs in drl1-2. An ethyl methanesulfonate–induced leaf mutant, elongata4 (elo4) (Berná et al., 1999; Robles and Micol, 2001), was shown to be allelic to drl1 and is designated drl1-4 hereafter (Table 1). The four alleles are recessive.
Table 1.
drl1 Alleles
| Locus | Alleles | Ecotype | Mutagen | Position of Mutation (Amino Acids) | Phenotypic Strength | Reference |
|---|---|---|---|---|---|---|
| DRL1 | drl1-1 | Ler | tDs | 38 | Strong | Bancroft et al. (1993) |
| drl1-2 | Ler | tDs | 256 | Strong | This report | |
| drl1-3 | Ler | tDs | 262 | Strong | R. Simon | |
| drl1-4 (= elo4) | Ler | EMSa | 194 | Weak | This report |
EMS, ethyl methanesulfonate.
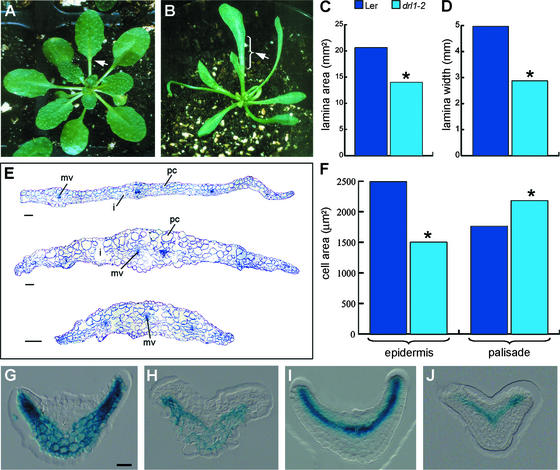
drl1-2 was isolated as a mutant with narrow leaves compared with the wild type (Figures 1A and 1B). The lamina length varied enormously among different drl1-2 individuals. The drl1-4 mutants had a less severe phenotype, with significantly narrower leaf lamina, normal leaf length, and number (seven to eight) of rosette leaves comparable to that in the wild type; this number varied from four to nine in drl1-2. The lamina width and area of the first and second expanded rosette leaves of a subpopulation of drl1-2 individuals with normal leaf length were measured by image analysis and found to be reduced significantly compared with the wild type (Figures 1C and 1D). The pattern formation of lateral growth along the length axis of the leaf results in a certain ratio between lamina length and petiole length. This ratio was affected in the subpopulation of drl1-2 individuals with normal leaf length (i.e., drl1-2 mutants had increased lamina length and reduced petiole length). In some mutant individuals, no clear transition between lamina and petiole was seen (Figures 1A and 1B).
Figure 1.
drl1-2 Leaf Phenotype.
(A) and (B) Fully grown rosettes of wild-type (A) and drl1-2 (B) plants. Arrowheads indicate the transition between the lamina and the petiole.
(C) and (D) Mean values of lamina area (C) and lamina width (D) of first and second expanded leaves of normal length. Asterisks indicate statistically significant differences between mutant and wild-type samples (t test, P ≤ 0.05).
(E) Transverse sections at the widest locations of expanded lamina of first leaves of wild-type (top) and drl1-2 (middle and bottom) plants. i, intercellular space; mv, midvein; pc, palisade cells.
(F) Mean values of cell area of upper epidermis and palisade cells in cleared expanded first and second leaves. Asterisks indicate statistically significant differences between mutant and wild-type samples (t test, P ≤ 0.05).
(G) and (H) GUS activity of the pFIL-GUS ventral marker in wild-type (G) and drl1-2 (H) plants.
(I) and (J) GUS activity of the pREV-GUS dorsal marker in wild-type (I) and drl1-2 (J) plants.
Bars = 50 μm for (G) to (J) and 100 μm for (E).
In serial sections through expanded first and second leaves of drl1-2 (35-day-old seedlings), palisade cells were larger and more irregularly shaped than in the wild type, and intercellular spaces were present next to the adaxial epidermis (Figure 1E). In addition, lateral growth was reduced severely, the lamina was thicker, and the midvein was less pronounced (Figure 1E). These features may indicate ventralization of the leaf. The number of palisade cells in serial sections of an expanded leaf blade was taken as a measure of lateral growth (Tsuge et al., 1996). There were 53.4 ± 3.4 cells in the drl1-2 mutant at the largest width (n = 3), 104.2 ± 14.1 in the drl1-4 mutant (n = 4), and 112.0 ± 5.4 in Ler (n = 3); thus, the number of palisade cells was reduced by 50% in drl1-2 and was reduced slightly in drl1-4. Epidermal and palisade cells of cleared expanded first and second leaves were visualized with differential interference contrast microscopy and the images were analyzed. Statistically significantly smaller cells were present in the dorsal epidermis of drl1-2 (Figure 1F) and drl1-4 (data not shown). The palisade layer contained significantly larger cells in drl1-2 (Figure 1F) and drl1-4 (data not shown). Analysis showed that drl1-2 and drl1-4 are strong and weak alleles, respectively (Table 1).
To investigate the polarity in leaves, the dorsal markers pPHAB-GUS and pREV(DLUTA)-GUS and the ventral markers pFIL-GUS and pYAB3-GUS (kindly provided by J. Bowman, University of California, Davis) were introgressed into drl1-2. These marker lines displayed promoter activity in the dorsal part of the leaf primordium, including the vascular bundles, and in the abaxial part of the leaf primordia, excluding vascular bundles, respectively (Figures 1G and 1I; data not shown for pPHAB-GUS and pYAB3-GUS). Serial transverse sections of leaf primordia from F2 drl1-2 mutants containing the pREV-GUS, pPHAB-GUS, pFIL-GUS, and pYAB3-GUS markers showed β-glucuronidase (GUS) activity in either the dorsal or the ventral side, similar to the parental marker lines (Figures 1H and 1J; data not shown for pPHAB-GUS and pYAB3-GUS). These results demonstrate that the pattern of polarity for these genes was not altered in the drl1-2 mutant leaves, indicating that dorsal and ventral identity was maintained. This feature was confirmed by the normal polarity in vascular bundles with adaxial xylem and abaxial phloem and the normal functional differentiation of the palisade cells, which was visible in the number of chloroplasts.
drl1-2 Meristematic Defects
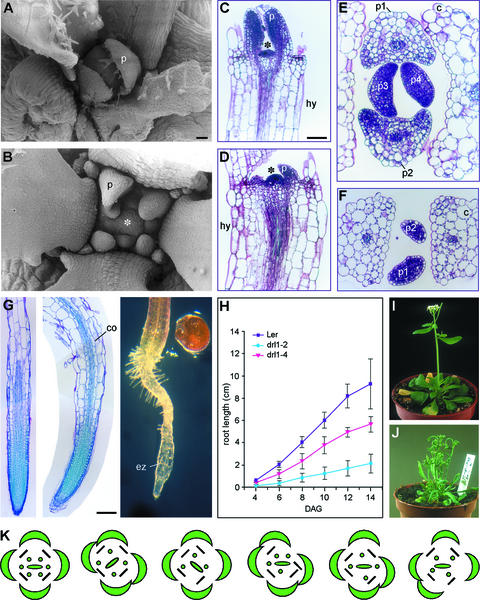
Germination of drl1-2 seeds was affected severely. Of 168 seeds sown onto germination medium, 74 did not germinate (44%), 39 were seedling lethal (23%), and only 55 grew further to maturation (33%). Scanning electron microscopy and sections showed that upon emergence from the SAM, the leaf primordia were much smaller than in the wild type (Figures 2A, 2B, 2E, and 2F). This effect is apparent also from transverse sections through the SAM. In addition, the mutant leaf primordia emerged more slowly than did the wild type primordia (Figures 2C to 2F). Longitudinal sections through the SAM confirmed that it was more dome shaped in the mutant than in the wild type (Figures 2C and 2D). Transverse sections of the SAM showed that the phyllotaxis of leaves 1 and 2 of drl1-2 was not opposite, but oblique, indicating that the pattern of leaf initiation was defective in the mutant (Figures 2E and 2F). The more dome-shaped SAM, the smaller leaf primordia, and the aberrant phyllotaxis indicate that the organization of the SAM was defective in the drl1-2 mutant.
Figure 2.
drl1-2 Meristematic Defects.
(A) and (B) Scanning electron micrographs of wild-type (A) and drl1-2 (B) SAM.
(C) Longitudinal section through a 6-day-old SAM of a wild-type plant.
(D) Longitudinal section through a 9-day-old drl1-2 SAM.
(E) and (F) Transverse sections through 12-day-old shoot apices of wild-type (E) and drl1-2 (F) plants.
(G) Longitudinal section through 12-day-old primary roots of wild-type (left) and drl1-2 (middle) plants and whole-mount section of a 12-day-old primary root of a drl1-2 plant (right).
(H) Primary root growth kinetics.
(I) and (J) Inflorescences of wild-type (I) and drl1-2 (J) plants.
(K) Floral diagrams of wild-type (left) and drl1-2 (five individuals) plants.
Asterisks indicate the SAM. c, cotyledon; co, cortex; DAG, days after germination; ez, elongation zone; hy, hypocotyl; p, leaf primordium; p1 to p4, first to fourth leaf primordia. Bar in (A) = 25 μm for (A) and (B); bars in (C) and (G) = 50 μm for (C) to (G).
Kinetics of primary root growth demonstrated that it was affected severely in drl1-2 and was less defective in drl1-4 (Figure 2H). The reduced root growth probably was related to root apical meristem defects, as illustrated in longitudinal sections of 12-day-old primary roots of several drl1-2 mutant individuals (Figure 2G). These sections also demonstrated that the cortex cells in the elongation zone were more expanded in the mutant than in the wild type (Figure 2G). Hypocotyl elongation was reduced significantly in the mutant, not because of a reduction in cell size (hypocotyl cells were even larger in the mutant) (Figures 2C and 2D), but probably because of a smaller number of cell divisions.
Flowering in drl1-2 was delayed by 1 week. Mutant inflorescences were fasciated and their size was one-third that of the wild type (Figures 2I and 2J), indicating that the inflorescence meristem activity was defective. Flowers consisted of normal floral organs, but their arrangement was abnormal and the number of stamens was reduced: 4.36 ± 0.73 in drl1-2 (n = 22 flowers) compared with 6 in the wild type (Figure 2K). These defects relate to floral meristem organization.
DRL1 Gene Isolation and Complementation
After inverse PCR of genomic DNA from the drl1-2 mutant, sequences were determined for 682 bp of plant DNA flanking the 3′ end of the tDs and 585 bp of plant DNA flanking the 5′ end of the tDs: 100% identity was found, with a 250-bp genomic fragment, designated DRL1, flanking a tDs in the drl1-1 mutant obtained after independent transactivation of the Ds from the DsB1 line (Bancroft et al., 1993). The full genomic DRL1 sequence revealed one continuous open reading frame of 302 amino acids. In addition, 100% identity was found between the DRL1 genomic sequence and a full-length cDNA; hence, the DRL1 gene is intronless. Upon Ds insertion into drl1-2, no target site duplicated in the plant DNA. The 3′ end of the tDs element was deleted by 22 bp, including the terminally inverted repeat, four bases of plant DNA were deleted, and one extra C was added to the 5′ side of the tDs. As a consequence of these changes, no reversion events were determined for this allele.
The tDs insertion corresponded with amino acids 38, 256, and 262 in the protein sequences of drl1-1 (Bancroft et al., 1993), drl1-2, and drl1-3, respectively (Table 1). In the drl1-2 mutant, the open reading frame extended 40 amino acids within the tDs. In drl1-4, a single base change introduced a premature stop codon at amino acid 194 (a C-to-T change at the nucleotide level at position 579). DNA gel blot analysis of wild-type and mutant DNA showed that the DRL1 gene is single copy in the Arabidopsis genome, which was confirmed by Basic Local Alignment Search Tool X (BLASTX) analysis of the Arabidopsis genome (http://www.arabidopsis.org/home.html) with the DRL1 genomic sequence.
Partial or complete reversion events of the mutant phenotype to the wild type were determined for the drl1-1 allele. DNA sequence analysis of the tDs footprints showed excision events of the Ds element from the DRL1 gene (Bancroft et al., 1993). We introduced the wild-type DRL1 gene, with its 1240-bp promoter fragment, into the homozygous drl1-2 mutant using a T-DNA construct containing the bialaphos acetyltransferase selectable marker gene that confers resistance to phosphinothricin. Seventeen independent T1 transgenic lines were obtained that segregated fully restored wild-type and mutant seedlings in 3:1 or 15:1 ratios, indicating one or two T-DNA loci, respectively. Of the 320 T2 wild-type seedlings that were tested for phosphinothricin resistance, all were resistant; 33 drl1-2 seedlings were phosphinothricin sensitive, indicating that the T-DNA containing the wild-type DRL1 gene was present in the wild-type seedlings and that complementation had occurred. Our data demonstrate that the drl1-2 phenotype is attributable to a Ds insertion into the DRL1 gene. Complementation analysis showed that the 1240-bp promoter fragment contained all of the regulatory information needed to direct correct gene activity throughout the plant's life cycle.
DRL1 Is a Universal ATP/GTP Binding Protein
The DRL1 protein (AtDRL1) shares a high level of homology with the TOT4/KTI12 protein of baker's yeast (Saccharomyces cerevisiae) (P34253) (Butler et al., 1994; Frohloff et al., 2001). The TOT4 protein copurifies with the Elongator complex, which is important for the regulation of transcription elongation of RNAPII (Frohloff et al., 2001). Full-length genomic sequences homologous with DRL1 were obtained in Schizosaccharomyces pombe, Caenorhabditis elegans, Drosophila melanogaster, Mus musculus, Anopheles gambiae, human, Oryza sativa, and Methanopyrus kandleri. An alignment presented by Fichtner et al. (2002) showed the homology between the S. cerevisiae, S. pombe, C. elegans, D. melanogaster, M. musculus, Arabidopsis, and human sequences. Thus, DRL1 is not only conserved among eukaryotes: homologs also are found in archaea, suggesting that DRL1 is a universal and ancient protein. Putative DRL1 orthologs also were identified in EST collections of many plant species (dicots, monocots, mosses, and conifers) and other organisms. An overview of the actual DRL1 homologs is given in Table 2.
Table 2.
Homology of DRL1 with proteins of other phyla
| Class | Species | Accession Number | Genomic/EST | Identity (%) | Similarity (%) |
|---|---|---|---|---|---|
| Eukaryota | |||||
| Plants, dicots | Arabidopsis thaliana | AJ428870 | Genomic | 100 | 100 |
| Medicago truncatula | AW560006 | EST | 76 | 87 | |
| Glycine max | BH021350 | EST | 68 | 82 | |
| Gossypium arboreum | BF276634 | EST | 64 | 76 | |
| Euphorbia esula | BE231335 | EST | 70 | 82 | |
| Lotus japonicus | AV420673 | EST | 74 | 88 | |
| Mesembryanthemum crystallinum | BF480675 | EST | 72 | 83 | |
| Lycopersicon esculentum | BI930978 | EST | 72 | 84 | |
| Plants, monocots | Zea mays | AI920610 | EST | 68 | 82 |
| Triticum aestivum | BF428902 | EST | 62 | 76 | |
| Oryza sativa | cld000341.4 | Genomic | 66 | 76 | |
| Hordeum vulgare | BF616626 | EST | 62 | 77 | |
| Plants, conifers | Pinus taeda | BE451838 | EST | 66 | 80 |
| Plants, mosses | Physcomitrella patens | AW145049 | EST | 65 | 73 |
| Birds | Gallus gallus | BG713512 | EST | 44 | 68 |
| Fish | Ictalurus punctatus | IpHdk02331 | EST | 47 | 67 |
| Molluscs | Crassostrea virginica | BG624862 | EST | 41 | 55 |
| Amphibians | Xenopus laevis | AW643264 | EST | 39 | 62 |
| Nematodes | Caenorhabditis elegans | Z99281 | Genomic | 33 | 54 |
| Fungi | Schizosaccharomyces pombe | CAB66461 | Genomic | 31 | 50 |
| Saccharomyces cerevisiae | Z28110 | Genomic | 29 | 46 | |
| Flies | Drosophila melanogaster | O46079 | Genomic | 27 | 46 |
| Anopheles gambiae | agCP15124 | Genomic | 28 | 48 | |
| Mammals | Mus musculus | BAB22635 | Genomic | 26 | 43 |
| Homo sapiens | AAH12173 | Genomic | 27 | 42 | |
| Archaea | |||||
| Methanopyrus | Methanopyrus kandleri | NP_614962 | Genomic | 25 | 41 |
The proteins were identified either as genomic sequences or as partial cDNAs (EST). For the partial cDNAs, identity and similarity are given for the cDNA, which encodes that part of the protein with the highest homology with DRL1.
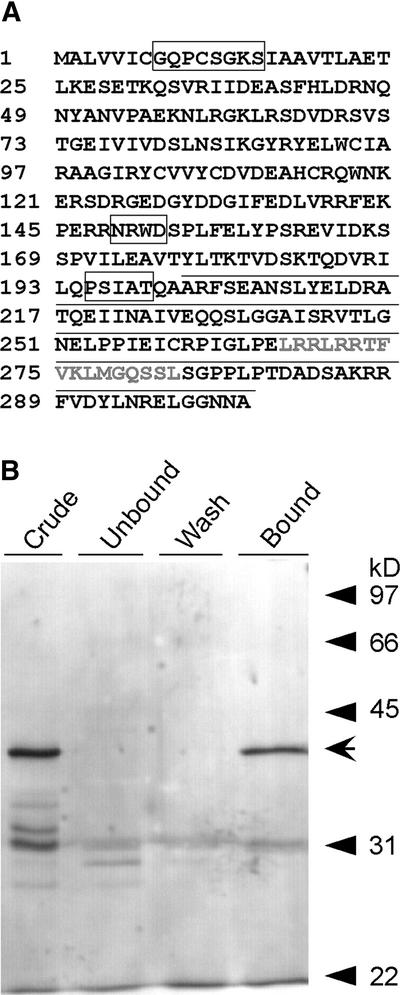
The DRL1 protein contains a conserved ATP/GTP binding domain (P-loop: PDOC00017 in PROSITE) ([AG]-x(4)-G-K-[ST]) spanning amino acids 8 to 15 (Figure 3A). This domain is conserved among the homologs of TOT4, as described by Fichtner et al. (2002). This P-loop is one of the four highly conserved sequence motifs that are required for guanine nucleotide binding and GTP hydrolysis in GTP binding proteins (Kaziro et al., 1991). DRL1 also contains an N[KR]XD box (amino acids 148 to 152), another conserved box of the GTP binding proteins that is important for direct interaction with the guanine ring. The other two highly conserved boxes of GTP binding proteins are not present in the DRL1 protein. A highly conserved region among DRL1 and its homologs (amino acids 194 to 199, PXX[AS]T) is found in many ATPs and in enzymes using GTP (http://www.expasy.ch/tools/scanprosite/) (Figure 3A).
Figure 3.
Deduced Protein Sequence of DRL1.
(A) The DRL1 protein (302 amino acids) is presented with its functional domains. ATP/GTP binding motifs are boxed, and amino acids that contain calmodulin binding sites are underlined. The predicted calmodulin binding site is shown in gray.
(B) In vitro calmodulin binding assay. Elution profiles of proteins from a calmodulin-Sepharose affinity column fractionated on a 15% acrylamide gel containing SDS. The “crude” starting material consisted of glutathione S-transferase–DRL1 (indicated by arrow) produced in E. coli TOP 10F′ and purified on glutathione agarose. Equal proportions of the unbound, wash, and elution fractions were separated by SDS-PAGE, and proteins were detected by silver staining.
DRL1 Binds Calmodulin in a Calcium-Dependent Manner
An in vitro assay demonstrated that the C-terminal 100 amino acids of the DRL1 protein bound calmodulin in a calcium-dependent manner (Figure 3B). O'Neil and DeGrado (1990) showed that the binding of calmodulin to its targets represents a sequence-independent recognition of amphiphilic α-helices. We found a stretch of 17 amino acids within the C-terminal 100 amino acids of DRL1 (amino acids 257 to 273) that very probably is the calmodulin binding domain. The prediction program of the calmodulin target database (http://calcium.oci.utoronto.ca) identified the same stretch as a putative calmodulin binding site. Because this stretch was the only predicted calmodulin binding site in the C-terminal 100 amino acids of the DRL1 protein, these amino acids very likely constitute the calmodulin binding site.
Reported calmodulin binding domains were compared to identify the critical elements required in the binding process. Based on the conserved hydrophobic residues within these motifs, two related motifs for calcium-dependent binding, termed 1-8-14 and 1-5-10, were described (Rhoads and Friedberg, 1997). In our proposed stretch, the motif LXXXFX-XLXXXXXL and the net charge of +5 were found, according to the characteristics of a 1-8-14 calmodulin binding motif of type A.
Because the calmodulin binding site is sequence independent, the prediction program of the calmodulin target database (http://calcium.oci.utoronto.ca) was used to search for putative calmodulin binding sites in the DRL1 homologs. No calmodulin binding sites were predicted in the human, mouse, or yeast homologs. In the fruit fly homolog, a putative calmodulin binding site also is predicted at the C-terminal end of the protein. In the rice homolog, the predicted calmodulin binding site also shares sequences homology with the putative calmodulin binding site in DRL1. These data indicate that the regulation of the DRL1 protein is conserved among plants through the binding of calmodulin.
DRL1 Promoter Activity Is Regulated during Development
DRL1 gene expression was analyzed by reverse transcription (RT) PCR followed by DNA gel blot hybridization with total RNA isolated from wild-type roots, hypocotyls, cotyledons, shoot apices, stems, inflorescence apices, and leaves and flowers at different developmental stages (data not shown). Because the mRNA of DRL1 was detected in every plant organ investigated in the wild type, DRL1 expression was not organ specific. DRL1 also was expressed at different growth stages of Arabidopsis cell suspension cultures. DRL1 transcript was detected in drl1-2, drl1-3, and drl1-4 mutants (data not shown). Semiquantitative RT-PCR on total RNA from shoot apices showed that the level of DRL1 transcript was reduced slightly (twofold) in the weak drl1-4 allele compared with that in the wild type. In drl1-4, the single-nucleotide change (C replaced by T at position 579 of the DRL1-coding sequence) causes a truncated protein that may have reduced autoregulatory activity. In the strong drl1-2 allele, the DRL1 transcript level was reduced significantly (ninefold), probably because of the removal of the 3′ end of the DRL1 transcript in the mutant, with instability of the mRNA as a result and a truncated DRL1 protein if any is synthesized at all (see next section). mRNA in situ hybridizations were performed repeatedly with a radioactive DRL1 probe on shoot apices and developing leaves, but no above-background signal was obtained at different exposure times. Therefore, we switched to an artifact-free GUS reporter gene assay (De Block and Debrouwer, 1992) that is much more sensitive to score DRL promoter activity at the cellular level.
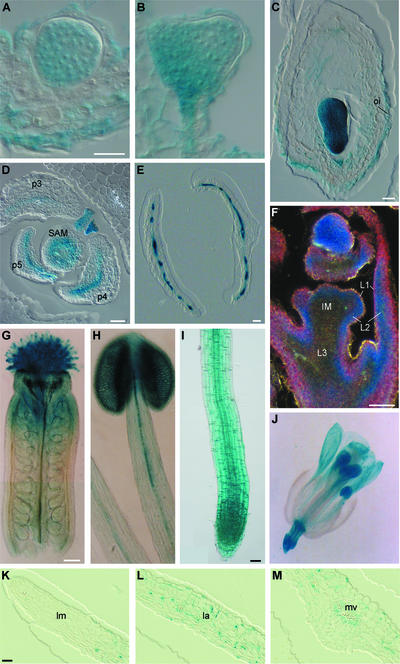
A SphI-NcoI DRL1 promoter fragment of 1240 bp was generated and fused at the start codon of the GUS-coding sequence. This promoter fragment was used in a complementation test of the drl1-2 mutant, and it contained all of the sequences necessary to direct complete gene activity. DRL1 promoter activity was analyzed at the cellular level in several transgenic lines transformed with the pDRL1-GUS chimeric construct using histochemical analysis of serial sections through plastic-embedded tissues (De Block and Debrouwer, 1992; De Block and Van Lijsebettens, 1998). Homogeneous GUS activity was detected in globular-, heart-, and torpedo-stage embryos (Figures 4A to 4C). High GUS activity was present in the outer integuments of ovules (Figure 4C) and in the funiculi (data not shown). Other reproductive tissues were negative for GUS activity. In transverse sections of 8- to 12-day-old SAMs (n = 6), staining was ring shaped, indicating GUS activity in the peripheral zone of the SAM (Figure 4D). The section more apical to the SAM showed circular 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-Gluc) staining that coincided with the central zone. The sections more distal from the SAM had X-Gluc staining only at the periphery of the vascular bundle. In longitudinal sections through shoot and inflorescence apices, GUS activity was most prominent in the L2 layer (blue), less prominent in the L1 layer (pink), and absent in the L3 layer (black) (n = 3) (Figure 4F).
Figure 4.
GUS Activity in pDRL1-GUS Transgenic Plant Lines.
Activity was determined according to De Block and Van Lijsebettens (1998) ([A] to [F] and [K] to [M]) and Jefferson et al. (1987) ([G] to [J]).
(A) Late globular stage.
(B) Heart stage.
(C) Torpedo stage.
(D) Transverse section through a 12-day-old shoot apex.
(E) Transverse section through young leaves.
(F) Longitudinal section through an inflorescence meristem.
(G) Whole-mount section of gynoecium.
(H) Whole-mount section of stamen.
(I) Whole-mount section of primary root.
(J) Whole-mount section of flower.
(K) to (M) Half a lamina of an expanding leaf sectioned transversally.
IM, inflorescence meristem; L1, L2, and L3, first, second, and third progenitor tissue layers, respectively; la, lamina; lm, leaf margin; mv, midvein; oi, outer integument; p3 to p5, third to fifth leaf primordia, respectively. Bars = 10 μm for (A) to (C), 50 μm for (D) to (F), (I), and (K) to (M), 100 μm for (G) and (H), and 200 μm for (J).
In young leaf primordia, GUS activity occurred as a linear area representing the basal part of the dorsal side. In older leaf primordia, X-Gluc staining was apparent as a continuous blue linear area, including vascular bundles and the mesophyll between the vascular bundles (Figure 4E). This pattern was identical to the pattern of the dorsal marker lines pPHAB-GUS and pREV-GUS in leaf primordia (Figure 1I). In transverse serial sections of expanding leaves, GUS activity was patchy: staining was seen in individual palisade and spongy mesophyll parenchyma cells. GUS activity was not observed in the mesophyll cells at the leaf tip, at the margin of the distal part of the leaf lamina, and at the ventral mesophyll of the midrib (Figures 4K to 4M). These sections correspond exactly to the first parts of the leaf in which cell division arrests. In the leaf epidermis, GUS activity was restricted to the stomatal guard cells, which are generated by cell division from epidermal meristemoids after cell division has ceased in the epidermal pavement cells. GUS activity also was observed typically around the vascular bundles. The pDRL1-GUS pattern coincided with the patchy pattern of expression of pcyc1at-GUS during leaf development (Donnelly et al., 1999) (cyc1at is equivalent to Arath;CycB1;1).
Whole-mount X-Gluc staining (Jefferson et al., 1987) was performed on flowers and primary roots. Young flower organs stained completely blue, whereas fully developed sepals and petals did not, in analogy with fully developed leaves. A gradient of GUS activity was observed in the stamens and carpels, with the highest activity at the tip of the organs (Figures 4G, 4H, and 4J). The DRL1 promoter was active in the meristematic and elongation zones of the primary root and in the vascular bundle of the differentiation zone (Figure 4I).
Genetic Interaction between DRL1 and AN
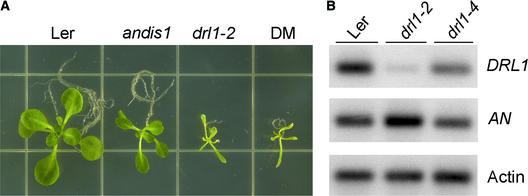
Double mutant analysis was performed between drl1-2 and angustifolia distorted trichomes1 (andis1), a mutant with a leaf phenotype similar to that of drl1-2 in addition to a trichome mutation, dis1, in the an background. Compared with the drl1-2 mutants, the an mutants have normal roots, flowers, and inflorescences but wrinkled siliques. These four phenotypic differences between the two mutants were used to analyze the characteristics of the double mutant. Homozygous an F2 plants that were heterozygous for drl1-2 (selected for hygromycin resistance) were self-fertilized and analyzed in the F3 population: one double mutant with a drl1-2 phenotype segregated to three an homozygous plants. Figure 5A shows the vegetative phenotypes of Ler, drl1-2, andis1, and the drl1-2 andis1 double mutant, clearly demonstrating that drl1-2 is epistatic to an.
Figure 5.
Genetic Interaction between DRL1 and AN.
(A) Double mutant (DM) analysis.
(B) Semiquantitative RT-PCR. Transcript levels of DRL1 and AN were compared in Ler, drl1-2, and drl1-4. Actin transcript was used as an internal control. Three independent repeats were performed.
The level of AN transcript in a drl1-2 background was assessed by means of semiquantitative RT-PCR. AN expression levels were compared with the amount of actin transcript (Figure 5B). In the strong drl1-2 allele with severely reduced DRL1 transcript, a significant upregulation of the AN transcript (up to twofold) was seen. In the weak drl1-4 allele with slightly reduced DRL1 transcript, the level of AN transcript was comparable to that in the wild type. The upregulation of AN transcript in the drl1-2 allele suggests that DRL1 acts as a repressor for AN expression.
DISCUSSION
DRL1 shares overall amino acid sequence homology and conserved functional domains with yeast TOT4, a protein that associates with Elongator (Fichtner et al., 2002). Moreover, DRL1 is a single-copy gene, and mutants at this locus are affected in their growth processes. These findings indicate that DRL1 is the Arabidopsis homolog of TOT4.
In yeast, Elongator was described as a histone acetyltransferase complex associated with the elongating form of RNAPII to facilitate transcription elongation (Otero et al., 1999). The RNAPII transcription elongation complex is believed to control gene expression by remodeling the chromatin through histone acetylation (Winkler et al., 2002) and by regulating movement along the DNA (Kim et al., 2002b). Although Elongator was not detected on promoters or open reading frames in vivo (Pokholok et al., 2002), numerous data suggest a role for Elongator in transcriptional regulation. Originally, Elongator was found stoichiometrically associated with the elongating form of RNAPII and to bind preferentially to the hyperphosphorylated form of RNAPII in vitro (Otero et al., 1999). Mutations in transcription elongation machinery confer increased sensitivity to the drug 6-azauracil, as seen in the tot mutants (Shaw and Reines, 2000). Moreover, microarray analysis using deletion mutants of Elongator components (ELP1, ELP2, ELP4, and ELP6) revealed that subsets of genes were downregulated or upregulated, indicating that Elongator is important in regulating the expression of specific sets of genes (Krogan and Greenblatt, 2001). We propose that in Arabidopsis, Elongator regulates the transcription of several genes involved in specific processes during development. In the drl1-2 mutant, the AN transcript is upregulated, indicating an inhibitory function of DRL1 during AN transcription. Several of the drl1 phenotypes also suggest an activating or inhibitory role of DRL1 in specific processes, as discussed below.
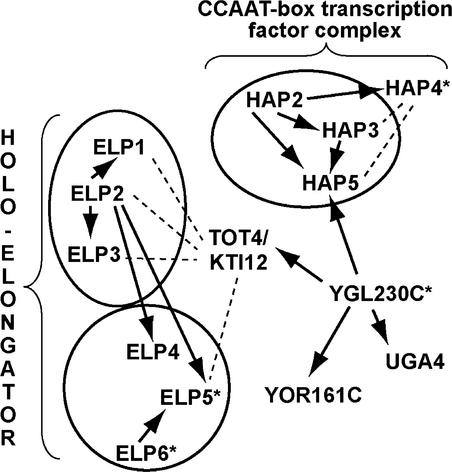
In addition to DRL1, putative homologs of the Elongator components ELP1, ELP2, ELP3, ELP4, ELP5, and ELP6 also are found in the Arabidopsis genome (Figure 6), suggesting that a functional equivalent of the Elongator complex is present in plants. Recently, the presence of Elongator (ELP1, ELP2, ELP3, and ELP4) was shown in humans (Winkler et al., 2001; Hawkes et al., 2002). A mutation in the human IKAP protein, the homolog of yeast ELP1, causes familial dysautonomia, which is a severe human recessive neuronal disorder (Hawkes et al., 2002). Together, these findings suggest that an equivalent of Elongator functions in multicellular organisms. Yeast two-hybrid interactions have shown an indirect link between KTI12 and the transcription initiation process through YGL230c and the CCAAT binding transcription factor complex (Figure 6). At present, no other experimental data are available to support this link between KTI12 and transcription initiation through the CCAAT binding complex.
Figure 6.
Protein Interactions of TOT4/KTI12 in Yeast.
Two-hybrid interactions are represented by arrows. Protein interactions detected by other methods are indicated by dashed lines. Homologs in Arabidopsis were identified using BLASTP; those detected by PSI-BLAST are indicated with asterisks.
The similar phenotypes of the different elp deletion mutants in yeast suggest that all subunits of Elongator are involved equally in its function (Krogan and Greenblatt, 2001). The viability of the tot mutants showed that none of the components of the Elongator complex, except ELP5, is essential. Similarly, in the drl1-2 mutant, levels of DRL1 transcript are dramatically low, yet 30.5% of the plants still are viable. However, the severe pleiotropic phenotype of the drl1 mutants implies that, although the DRL1 protein might not be essential, it plays a crucial role in meristem organization and organ growth, which ultimately may influence reproductive success and viability.
The superfamily of GTP binding proteins consists of several members, including translation factors, tubulin, heterotrimeric GTP binding proteins, Ras proteins, and other low molecular mass GTP binding proteins. All GTP binding proteins share four highly conserved sequence motifs, which are required for guanine nucleotide binding and GTP hydrolysis (Kaziro et al., 1991). The DRL1 protein contains the first and fourth conserved boxes of GTP binding proteins, but because the other two boxes, which are important for the hydrolysis of GTP, are not present in the amino acid sequence, DRL1 presumably is not a GTP binding protein. This notion contradicts the previously suggested role of KTI13 as a guanine nucleotide exchange factor for TOT4. Guanine nucleotide exchange factors are proteins that catalyze the reactivation of GTP binding proteins by exchanging GDP for GTP (Fichtner and Schaffrath, 2002). The P-loop consensus motif is found not only in GTP–binding proteins but also in many nucleotide triphosphate–using enzymes. Although this region probably is involved directly in the hydrolytic process (Kaziro et al., 1991), no domains for hydrolytic activity can be detected in the DRL1 protein based on its sequence. The importance of the P-loop for TOT4 function was shown by Fichtner et al. (2002). An N-terminally truncated TOT4 protein that lacked the P-loop motif (amino acids 4 to 19) was functionally inactive because it could not complement the zymocin resistance phenotype. Because another conserved box, PXX[AS]T, is present in many proteins that use ATP or GTP, DRL1 seems to be involved at least in the binding of a nucleotide triphosphate, but not necessarily in its hydrolysis. Thus, DRL1 and its homologs may either exert their function upon ATP/GTP binding or transfer ATP/GTP to another protein, allowing it to become functional.
DRL1 binds calmodulin in a calcium-dependent manner. Calmodulin binding domains often overlap an autoinhibitory region. These two regions are thought to regulate enzyme activity through a fine balance between internal binding and binding of calmodulin, causing inhibition and activation, respectively (James et al., 1995). This finding implies that the activity of the protein itself is regulated at the calmodulin binding site, although DRL1 is expressed only in specific cells and tissues. Several calmodulin binding proteins in which the calmodulin binding site was deleted were shown to be fully functional, meaning that without a calmodulin binding site these enzymes are constitutively active (Wang et al., 1989). Knowledge of the functional domains provides an insight into the strength of the different alleles of DRL1. The phenotypes of the four alleles are similar, but in the drl1-4 mutants the phenotype is weaker. At the transcriptional level, the DRL1 transcript decreased dramatically in drl1-2 mutants, whereas the level of DRL1 expression in drl1-4 mutants was more comparable to that of the wild type. This finding correlates with the phenotypes in both alleles. Presumably, very few proteins (or none) are formed in drl1-2, with its dramatic phenotype; by contrast, the drl1-4 mutants, with a weak phenotype, form proteins lacking the Pxx[AS]T box and the calmodulin binding site because of a premature stop codon. This observation suggests that the drl1-4 phenotype is attributable either to a constitutively active DRL1 protein or to the loss of the Pxx[AS]T box. Because the phenotypes of drl1-4 and drl1-2 are similar, a constitutively active protein probably would not phenocopy low-abundance proteins. Moreover, a constitutively active protein would not correlate with the recessive mutation of drl1-4, suggesting that the PXX[AS]T box is essential for the function of the DRL1 protein.
A number of phenotypes in drl1-2 relate to meristematic defects: the number of lateral organs is reduced (leaves and stamens), their positioning (phyllotaxis) is defective, and primary root growth is reduced severely. The drl1-2 shoot phenotypes resemble those of mutants affected in SAM organization, such as clavata1, clavata2, and clavata3 with an enlarged central zone and mgoun1 and mgoun2 with an enlarged peripheral zone (Laufs et al., 1998). In these mutants, the balance in cell division between the different zones and/or layers of the meristem is disturbed. Some of these loci could be target genes for DRL1 regulation, but this possibility remains to be tested.
In drl1-2 leaves, lateral growth is reduced severely and leaf length is reduced as well, the palisade cells resemble the spongy parenchyma cells in size and shape, and the upper epidermal cell area is reduced. These phenotypes were observed in the so-called polarity mutants, such as kan, fil, and the fil yab3 double mutant (Sawa et al., 1999; Siegfried et al., 1999; Kerstetter et al., 2001). A model has been proposed in which dorsoventral patterning in leaf primordia is a prerequisite to lateral outgrowth of the lamina (Waites and Hudson, 1995; Eshed et al., 2001). However, the introgression of the PHAB, REV, FIL, and YAB3 molecular markers in the drl1-2 mutant showed that the dorsal and ventral identities of the leaf blade were maintained. Thus, the DRL1 gene does not act upstream of the polarity genes. Other phenotypes in the mutant leaf, such as the preservation of polarity in the vascular tissue and the presence of many chloroplasts in the enlarged palisade cells, confirm that polarity is maintained in the mutant. There is a striking similarity in the promoter activities of the DRL1 gene and the dorsal identity genes PHAB and REV, as shown by GUS reporter gene analyses. The activity in leaf primordia is situated mainly in the vascular bundles and in the mesophyll between them. All three genes may be part of a common regulatory mechanism, and DRL1 may not act downstream of the dorsal identity genes. The drl1-2 mutation may identify an early pathway for leaf lateral growth that acts in parallel with that for dorsoventrality identified by the polarity mutations. Double mutant analysis showed that DRL1 acts upstream of AN, a transcriptional corepressor that regulates polar expansion of palisade cells, probably by controlling the arrangement of cortical microtubuli (Kim et al., 2002a). The upregulation in drl1-2 shoot apices of AN transcript could explain the increase in palisade cell size in the drl1-2 mutant without affecting the polarity. Indeed, recessive mutation at the AN locus results in a reduction of palisade cell size (Tsuge et al., 1996).
Increased palisade cell size in the drl1-2 mutant could be the result of a compensatory mechanism for the severe reduction in cell number in the mutant to restore the intrinsic size of the organ. Mizukami and Fischer (2000) suggested such a mechanism to explain the regulation of lateral organ growth by the AP2-type transcription factor AINTEGUMENTA (ANT). The ANT gene promotes cell proliferation, and in the ant1 mutant, the reduced cell number in organs also is compensated for by an increase in cell size. This phenomenon has been observed in transgenic plants that contain an overexpression construct of a cell cycle inhibitor (De Veylder et al., 2001). The relationship between DRL1 gene function and cell cycle regulation could explain the reduction in leaf cell number upon recessive mutation. In fact, one of the yeast phenotypes of the tot4 deletion mutant is a retardation of the G1-to-S transition of the cell cycle (Fichtner et al., 2002). The expression of a number of cell cycle genes has been investigated by semiquantitative RT-PCR in young seedlings, but to date, no gene has been identified with altered expression levels in the drl1-2 and drl1-4 mutant lines (data not shown).
In addition to alterations in cell expansion in the epidermis (L1 derived) and the palisade (L2 derived), altered cell numbers also have been measured in drl1-2 leaves in these two cell layers. The phenotypes relate to DRL1 promoter activity, which predominates in the L2 layer is less active in the L1 layer, and is not active in the L3 layer of the SAM. The resulting reduction in the size and shape of the leaf lamina in drl1-2 confirms previous observations in which the importance of the L2 and L1 layers in the determination of leaf size and shape has been reported (Dolan and Poethig, 1998).
Our study on the function of DRL1 adds to the accumulating evidence that leaf growth is controlled genetically by components that act on the transcriptional regulation of sets of genes. Indeed, two transcriptional corepressors have been identified that regulate lateral growth in leaves: AN, which promotes polar cell expansion (Tsuge et al., 1996; Kim et al., 2002a); and ROTUNDA2 (=LEUNIG) (G. Cnops, personal communication). The drl1-2 leaf defects are similar to those described for struwwelpeter (Autran et al., 2002). STRUWWELPETER is a component of Mediator, a coactivator complex of the RNAPII transcription initiation complex (Boube et al., 2002), which was described first in yeast. The identification of the target genes of these transcriptional regulators will further prove this concept and will be essential to understand the biology of organ growth regulation in plants.
METHODS
Plant Material and Growth Conditions
The drl1-1 and drl1-3 alleles were kindly provided by I. Bancroft (John Innes Centre) and R. Simon (University of Cologne, Germany), respectively. The elo4 mutant was provided by J.L. Micol (Universidad Miguel Hernández, Alicante, Spain), and the Arabidopsis thaliana Landsberg erecta (Ler) marker lines pFIL-GUS, pYAB3-GUS, pPHAB-GUS, and pREV-GUS were provided by J. Bowman (University of California, Davis). The N2 line homozygous for the andis1 mutations in a Ler background was obtained from the Nottingham Arabidopsis Stock Centre (Nottingham, UK). drl1-2 has been registered at the Nottingham Seed Stock Centre.
Plants were grown in a soil:vermiculite (3:1) mixture in a 16-h-light/ 8-h-dark regime at 22°C, with light intensity of 100 μmol·m−2·s−1 and 70% RH. In vitro, plants were grown on germination medium (Valvekens et al., 1988) under the same growth conditions.
Genetic Linkage Analysis
Genomic DNA of nine independent drl1-2 mutants was digested with HindIII and hybridized with an Ac probe. None of these lines contained the parental Ds band of ∼14 kb (Bancroft et al., 1993). Instead, they all contained a new band of 5.8 kb, showing that germinal transposition of the Ds had occurred. The transposed Ds (tDs) had inserted into a HindIII fragment. No reversion of the leaf phenotype to the wild type was observed in subsequent progeny; therefore, genetic linkage analysis was performed between the drl1-2 mutation and the tDs in this F2 population. No recombination was found in 919 F2 and 38 F3 individuals of the drl1-2 hygromycin-resistant class, of which the maximum genetic distance was calculated as 6.6 ± 3.3 centimorgan (F2 data) or 1.3 ± 1.2 centimorgan (F3 data) (Koornneef and Stam, 1987).
Isolation of DRL1 Genomic DNA
Total genomic DNA of the drl1-2 mutant was prepared according to Pruitt and Meyerowitz (1986) and digested with HindIII. Among the pool of fragments, a 5.8-kb HindIII fragment was generated containing the intact Ds and flanking plant DNA. This pool was ligated under conditions that promoted the formation of monomeric circles. PCR was performed with Ds-specific primers that were pointed outward (5′-CGGGATTTT-CCCATCCTACTTTCATCCCTG-3′ and 5′-TTCGTTTCCGTCCCGCAA-GTTAAATA-3′) and degenerate primers. A fragment of 1.4 kb was amplified and cloned in the pGEM-T vector (Promega, Madison, WI). A HindIII digest confirmed the presence of a corresponding HindIII site. The DNA sequence was determined from 682 bp of plant DNA flanking the 3′ end of Ds and from 585 bp of plant DNA flanking the 5′ end of Ds. Identity (100%) was found with a 250-bp genomic fragment flanking a tDs in the drl1-1 mutant, which was obtained after transactivation of the Ds from the DsB1 line (Bancroft et al., 1993).
A DRL1 genomic fragment containing the intronless coding region was amplified on wild-type DNA (Ler ecotype) with primer 5′-TTTTGTAGG-CAGTGTGTTTA-3′ at the 5′ end of the 250-bp published DRL1 sequence (Bancroft et al., 1993) and primer 5′-TCGTCGTTTTATGATTTTAT-3′ at the 3′ end of the gene and cloned in pGEM-T to create the plasmid pGEMT-DRL1. The rice genomic sequence homologous with DRL1 was kindly provided by Syngenta Research and Technology (San Diego, CA).
Mapping of the DRL1 Gene
A restriction fragment length polymorphism was found between the genomes of the Ler and Columbia ecotypes using a BclI restriction enzyme digest and the DRL1 genomic clone as a probe. The DRL1 gene was mapped using a set of 100 recombinant inbred lines (http://nasc.nott.ac.uk/new_ri_map.html). The map position of DRL1 was determined on the recombinant inbred map at the top half of chromosome 1 between markers g12080 and 0818 (http://nasc.nott.ac.uk/new_ri_map.html). The 0818 marker corresponded to the plant DNA flanking the Ds-containing T-DNA in the DsB1 parental line (C. Dean, personal communication). The DRL1 gene, identified after the transactivation of Ds from the DsB1 line, maps at 0.06 centimorgan from the 0818 marker. Thus, the Ds transposed over a short distance of only 12 kb, a clear example of targeted tagging.
Calmodulin Binding in Vitro
A glutathione S-transferase–DRL1 C terminus was fused to Escherichia coli TOP 10F′, and the crude extract was purified on glutathione agarose. After the protein solution was adjusted to 1 mM CaCl2, it was mixed batch-wise for 15 min at room temperature with calmodulin-Sepharose equilibrated with binding buffer (40 mM Tris-HCl, pH 7.5, 50 mM NaCl, 3 mM MgCl2, 0.2 mM CaCl2, and 0.1 mM DTT) according to Liao and Zielinski (1995). The slurry was packed into a column, the buffer was drained, and the column was washed with 5 bed volumes of binding buffer. Bound proteins were eluted in buffer containing 4 mM Tris-HCl, pH 7.5, 200 mM NaCl, 1 mM MgCl2, 2 mM EGTA, and 0.1 mM DTT. Equal proportions of the unbound, wash, and elution fractions were separated by SDS-PAGE, and proteins were detected by silver staining.
DRL1 Gene Expression Analysis by Reverse Transcription PCR
The QuickPrep Micro mRNA Purification kit (Amersham Biosciences, Little Chalfont, UK) was used to isolate mRNA. The mRNA samples were treated with DNaseI for 1 h to ensure the absence of genomic DNA, because DRL1 is an intronless gene. From each sample, 60 to 90 ng of mRNA was reverse transcribed with the SuperScript Preamplification System for First-Strand cDNA Synthesis (Invitrogen, Gaithersburg, MD). cDNA synthesis was started from primer 5′-AGCCCCAAAATATGTTTGCATTA-3′ (p5), which, together with primer 5′-CTAGACCGCAACCAAAACTATGC-3′ (p7), was used to amplify the cDNA. The absence of genomic DNA was checked by PCR on the DNaseI-treated mRNA samples under the same conditions used for the cDNA. All samples were blotted onto a nitrocellulose membrane and hybridized with a DRL1 probe, a PCR product amplified with primers p5 and p7. To perform semiquantitative reverse transcription PCR, total RNA was extracted from the shoot apices using TRIzol (Invitrogen). Total RNA (2 μg) was used as a template to synthesize the cDNA using the SuperScript First-Strand Synthesis system for reverse transcription PCR (Invitrogen). To assess the levels of RNA in each sample, actin cDNA was amplified with primers 5′-GTGCCAATCTACGCGGGTTTC-3′ and 5′-CAATGGGACTAAAACGCAAAA-3′ and hybridized. The DRL1 gene was amplified using primers 5′-TCGCGTTGATGATTTCTTGTGTC-3′ and p7. The AN gene was amplified with primers 5′-TGAGACGGTGCCGTGGTATGG-3′ and 5′-GTTGCCTACTGGTGGATTCC-3′. The amplification of the cDNAs was terminated in the exponential phase of the PCR (18 cycles). The intensity of the hybridized fragments were measured with ImageQuant version 4.1b (Amersham Biosciences).
The pDRL1-GUS Chimeric Construct and Histochemical Analysis
A promoter fragment of 1240 bp of the DRL1 gene, defined as pDRL1, was amplified from genomic Ler DNA with the modified primer (5′-ACTAGCGCCATGGGTTTTTAAAC-3′) containing a SphI restriction site and a modified primer (5′-TAGTTACTTGGCATGCAGGTTATCTG-3′) containing a NcoI restriction site. The amplified sequence was cloned into the pGUS1 plasmid (kindly provided by J. Botterman, Bayer Bioscience) as a SphI-NcoI fragment to create a translational GUS gene fusion. The pDRL1-GUS cassette was cloned as a PvuII fragment in the SmaI site of the pGSV4 plant transformation vector containing a kanamycin resistance marker (also provided by J. Botterman) and transformed into E. coli JM109. The pGSV4-pDRL1-GUS plasmid was transferred to the Agrobacterium tumefaciens strain C58C1RifR(pGV2260) (Deblaere et al., 1985) by triparental mating with the helper strain HB101(pRK2013) according to Van Haute et al. (1983). Transgenic plants containing the pDRL1-GUS construct were obtained after root explant transformation of Ler plants using kanamycin selection (Valvekens et al., 1988). Histo-chemical staining using 5-bromo-4-chloro-3-indolyl-β-d-glucuronide was used to assay DRL1 promoter activity in intact seedlings (Jefferson et al., 1987) and on semithin sections (15 μm) of plastic-embedded tissue (De Block and Van Lijsebettens, 1998).
Complementation Analysis Using drl1-2
The promoter-coding sequence of DRL1 was amplified from Ler DNA with Pfu polymerase using primers 5′-AAGGAGAACCAAAGCCAT-TAGT-3′ and 5′-GCATTAGCGATTAATGAAGCTG-3′. The fragment (2576 bp) was cloned in the EcoRV site of a pGEM-5Zf(+) vector (Promega) and transformed into E. coli JM109. The DRL1 genomic sequence was cloned as a NotI-NcoI fragment in the plasmid pAUX3133 (Goderis et al., 2002) and transferred subsequently to the pMODUL3337 plasmid containing a bialaphos acetyltransferase selectable marker gene (Goderis et al., 2002) by endonuclease PI-PspI cloning. The pMODUL3337-DRL1 plasmid was transferred to Agrobacterium strain C58C1RifR(pGV2260) (Deblaere et al., 1985) by triparental mating (Van Haute et al., 1983). The DRL1 gene was transformed into drl1-2 root explants (Valvekens et al., 1988), and transgenic shoots were selected on phosphinothricin at 15 mg/L. The progeny of these transgenic shoots were germinated on germination medium, and the seedlings were scored for the restoration of the wild-type phenotype. Root explants of wild-type T1 seedlings were tested for phosphinothricin resistance in a tissue culture assay. They were incubated for 4 days on callus-inducing medium containing 15 mg/L phosphinothricin and then transferred for 2 weeks on shoot-inducing medium containing 15 mg/L phosphinothricin (Valvekens et al., 1988). Resistant root explants were covered with shoots, whereas the sensitive controls did not develop callus or shoots.
Morphological and Cellular Characterization
For morphological and cellular analyses, the expanded first two leaves of drl1-2 (35 days) and Ler (28 days) were harvested. The whole-mounted leaves were fixed in 100% methanol and cleared in 90% lactic acid. Measurements of palisade and epidermal cell areas were obtained from digitized camera lucida drawings made from the adaxial leaf surfaces using differential interference contrast optics on a Diaplan microscope (Leitz, Wetzlar, Germany). Image analyses were performed with the public domain Image program (version β-3b; Scion Corp., Frederick, MD).
The statistical significance of the mean differences (P ≤ 0.05) was analyzed by t test using the Statistical Package for the Social Sciences (release 10.0.5) (SPSS, Inc., Chicago, IL) on normally distributed data sets. In cases of skewed distribution, the data were transformed to logarithmic values to normalize it. Shoot apices, first expanded leaves, and roots were fixed in formaldehyde/acetic acid/ethanol, embedded in historesin, cut serially into 5-μm sections with a Ralph glass knife (Reichert Jung, Nussloch, Germany), and stained with toluidine blue (0.05%). Seeds were germinated in vitro on germination medium (Valvekens et al., 1988) and solidified with phytagel (0.35%) in the vertical position; every 2 days, the position of the root tip was marked on the plate (Ler and drl1-4) or the plates were scanned (drl1-2). Root growth was measured over a period of 14 days. Shoot apices of 14-day-old seedlings were fixed (mixture of 5 mL of 40% formaldehyde, 5 mL of acetic acid, and 90 mL of 50% ethanol) and dehydrated (step wise with 95% ethanol). The explants were transferred to OsO4 (2% in 50 mM cacodylate buffer, pH 7), sputter-coated with gold, and stored under vacuum. Photographs were taken with a scanning electron microscope (JSM-840; JEOL, Tokyo, Japan) at a magnification of ×3000.
Interacting Proteins of Yeast TOT4/KTI12 and Their Homologs in Arabidopsis
In yeast, protein–protein interactions by two-hybrid analysis were performed on a large scale to define molecular networks. Two World Wide Web sites containing this information (http://mips.gsf.de/proj/yeast/CYGD/db/index.html/ and http://yeast.cellzome.com/) were used to determine the network of proteins that interact with TOT4/KTI12. An analysis of protein–protein interactions also was described by Uetz et al. (2000). TOT4/KTI12 interacted with YGL230c as bait; in addition, the latter interacted with three more proteins, UGA4, YOR161, and HAP5. UGA4 (amino acid permeability) and YOR161 (unknown function) are not discussed further. The Basic Local Alignment Search Tool P (BLASTP) program of the TAIR World Wide Web site (http://www.arabidopsis.org/) was used to find the Arabidopsis homologs of the yeast Elongator components and the TOT4/KTI12-interacting proteins. Besides DRL1 (At1g13870), which is a homolog of TOT4/KTI12, Arabidopsis homologs were identified: At5g13680 (ELP1), At1g49540 (ELP2), At5g50320 (ELP3), At3g11220 (ELP4, with a high E value of 0.005), HAP2-3-5 homologs (Edwards et al., 1998), At1g25500 (YOR161c), and At2g01170 (UGA4) (Figure 6). Using the PSI-BLAST program (Altschul et al., 1997), Arabidopsis homologs were found for ELP5 (At2g18410), ELP6 (At4g10090) (Ponting, 2002), YGL230c (At4g23860), and HAP4 (At5g25820) (Figure 6, asterisks). PSI-BLAST for HAP4 was performed with Kluyveromyces lactis homolog AF072675.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
The authors thank John Bowman, John Emery, and Rita Khodosh (University of California, Davis) for the pREV-GUS, pFIL-GUS, pPHAB-GUS, and pYAB3-GUS lines; José Luis Micol (Universidad Miguel Hernández, Alicante, Spain) for the elo4 mutant line; John Bowman, Gerda Cnops, Lieven De Veylder, and Marc Zabeau for critical reading of the manuscript and suggestions; Stefaan Royaert, Dimitri Tack, Els Cochuyt, Ingrid Lambein, Tom Van Acker, Marianne Van de Wiele, Janice de Almeida-Engler, and Wilson Ardiles-Diaz for technical help; and Rebecca Verbanck and Martine De Cock for help in preparing the manuscript. This research was supported by grants from the Fund for Scientific Research (Flanders) (G.0075.97), the Interuniversity Poles of Attraction Programme (Belgian State, Prime Minister's Office, Federal Office for Scientific, Technical, and Cultural Affairs; P4/15), and the European Research Training Network HPRN-CT-2002-00267. H.N. is indebted to the Instituut voor de Aanmoediging van Innovatie door Wetenschap en Technologie in Vlaanderen for a predoctoral fellowship.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.007062.
References
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autran, D., Jonak, C., Belcram, K., Beemster, G.T.S., Kornenberger, J., Grandjean, O., Inzé, D., and Traas, J. (2002). Cell numbers and leaf development in Arabidopsis: A functional analysis of the STRUWWELPETER gene. EMBO J. 21, 6036–6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft, I., Bhatt, A.M., Sjodin, C., Scofield, S., Jones, J.D.G., and Dean, C. (1992). Development of an efficient two-element transposon tagging system in Arabidopsis thaliana. Mol. Gen. Genet. 233, 449–461. [DOI] [PubMed] [Google Scholar]
- Bancroft, I., Jones, J.D.G., and Dean, C. (1993). Heterologous transposon tagging of the DRL1 locus in Arabidopsis. Plant Cell 5, 631–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berná, G., Robles, P., and Micol, J.L. (1999). A mutational analysis of leaf morphogenesis in Arabidopsis thaliana. Genetics 152, 729–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boube, M., Joulia, L., Cribbs, D.L., and Bourbon, H.-M. (2002). Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell 110, 143–151. [DOI] [PubMed] [Google Scholar]
- Brand, U., Fletcher, J.C., Hobe, M., Meyerowitz, E.M., and Simon, R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619. [DOI] [PubMed] [Google Scholar]
- Butler, A.R., White, J.H., Folawiyo, Y., Edlin, A., Gardiner, D., and Stark, M.J.R. (1994). Two Saccharomyces cerevisiae genes which control sensitivity to G1 arrest induced by Kluyveromyces lactis toxin. Mol. Cell. Biol. 14, 6306–6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne, M.E., Barley, R., Curtis, M., Arroyo, J.M., Dunham, M., Hudson, A., and Martienssen, R.A. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408, 967–971. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Running, M.P., and Meyerowitz, E.M. (1993). CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119, 397–418. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Williams, R.W., and Meyerowitz, E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585. [DOI] [PubMed] [Google Scholar]
- Deblaere, R., Bytebier, B., De Greve, H., Deboeck, F., Schell, J., Van Montagu, M., and Leemans, J. (1985). Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene transfer to plants. Nucleic Acids Res. 13, 4777–4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Block, M., and Debrouwer, D. (1992). In-situ enzyme histochemistry on plastic-embedded plant material: The development of an artefact-free β-glucuronidase assay. Plant J. 2, 261–266. [Google Scholar]
- De Block, M., and Van Lijsebettens, M. (1998). β-Glucuronidase enzyme histochemistry on semithin sections of plastic-embedded Arabidopsis explants. Methods Mol. Biol. 82, 397–407. [DOI] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., Beemster, G.T.S., Krols, L., Terras, F., Landrieu, I., Van Der Schueren, E., Maes, S., Naudts, M., and Inzé, D. (2001). Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13, 1653–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan, L., and Poethig, R.S. (1998). The okra leaf shape mutation in cotton is active in all cell layers of the leaf. Am. J. Bot. 85, 322–327. [PubMed] [Google Scholar]
- Donnelly, P.M., Bonetta, D., Tsukaya, H., Dengler, R.E., and Dengler, N.G. (1999). Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215, 407–419. [DOI] [PubMed] [Google Scholar]
- Edwards, D., Murray, J.A.H., and Smith, A.G. (1998). Multiple genes encoding the conserved CCAAT-box transcription factor complex are expressed in Arabidopsis. Plant Physiol. 117, 1015–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S.F., Perea, J.V., and Bowman, J.L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol. 11, 1251–1260. [DOI] [PubMed] [Google Scholar]
- Fellows, J., Erdjument-Bromage, H., Tempst, P., and Svejstrup, J.Q. (2000). The Elp2 subunit of Elongator and elongating RNA polymerase II holoenzyme is a WD40 repeat protein. J. Biol. Chem. 275, 12896–12899. [DOI] [PubMed] [Google Scholar]
- Fichtner, L., Frohloff, F., Bürkner, K., Larsen, M., Breunig, K.D., and Schaffrath, R. (2002). Molecular analysis of KTI12/TOT4, a Saccharomyces cerevisiae gene required for Kluyveromyces lactis zymocin action. Mol. Microbiol. 43, 783–791. [DOI] [PubMed] [Google Scholar]
- Fichtner, L., and Schaffrath, R. (2002). KTI11 and KTI13, Saccharomyces cerevisiae genes controlling sensitivity to G1 arrest induced by Kluyveromyces lactis zymocin. Mol. Microbiol. 44, 865–875. [DOI] [PubMed] [Google Scholar]
- Fletcher, J.C., Brand, U., Running, M.P., Simon, R., and Meyerowitz, E.M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot systems. Science 283, 1911–1914. [DOI] [PubMed] [Google Scholar]
- Frohloff, F., Fichtner, L., Jablonowski, D., Breunig, K.D., and Schaffrath, R. (2001). Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. EMBO J. 20, 1993–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goderis, I.J.W.M., De Bolle, M.F.C., François, I.E.J.A., Wouters, P.F.J., Broekaert, W.F., and Cammue, B.P.A. (2002). A set of modular plant transformation vectors allowing flexible insertion of up to six expression units. Plant Mol. Biol. 50, 17–27. [DOI] [PubMed] [Google Scholar]
- Hawkes, N.A., Otero, G., Winkler, G.S., Marshall, N., Dahmus, M.E., Krappmann, D., Scheidereit, C., Thomas, C.L., Schiavo, G., Erdjument-Bromage, H., Tempst, P., and Svejstrup, J.Q. (2002). Purification and characterization of the human Elongator complex. J. Biol. Chem. 277, 3047–3052. [DOI] [PubMed] [Google Scholar]
- James, P., Vorherr, T., and Carafoli, E. (1995). Calmodulin-binding domains: Just two faced or multi-faceted? Trends Biochem. Sci. 20, 38–42. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaziro, Y., Itoh, H., Kozasa, T., Nakafuku, M., and Satoh, T. (1991). Structure and function of signal-transducing GTP-binding proteins. Annu. Rev. Biochem. 60, 349–400. [DOI] [PubMed] [Google Scholar]
- Kerstetter, R.A., Bollman, K., Taylor, R.A., Bomblies, K., and Poethig, R.S. (2001). KANADI regulates organ polarity in Arabidopsis. Nature 411, 706–709. [DOI] [PubMed] [Google Scholar]
- Kim, G.-T., Shoda, K., Tsuge, T., Cho, K.-H., Uchimiya, H., Yokoyama, R., Nishitani, K., and Tsukaya, H. (2002. a). The ANGUSTIFOLIA gene of Arabidopsis, a plant CtBP gene, regulates leaf-cell expansion, the arrangement of cortical microtubules in leaf cells and expression of a gene involved in cell-wall formation. EMBO J. 21, 1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, G.-T., Tsukaya, H., and Uchimiya, H. (1998). The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev. 12, 2381–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.-H., Lane, W.S., and Reinberg, D. (2002. b). Human Elongator facilitates RNA polymerase II transcription through chromatin. Proc. Natl. Acad. Sci. USA 99, 1241–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., and Stam, P. (1987). Procedure for mapping by using F2 and F3 populations. Arabidopsis Inf. Serv. 25, 35–40. [Google Scholar]
- Krogan, N.J., and Greenblatt, J.F. (2001). Characterization of a six-subunit holo-Elongator complex required for the regulated expression of a group of genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 21, 8203–8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs, P., Dockx, J., Kronenberger, J., and Traas, J. (1998). MGOUN1 and MGOUN2: Two genes required for primordium initiation at the shoot apical and floral meristems in Arabidopsis thaliana. Development 125, 1253–1260. [DOI] [PubMed] [Google Scholar]
- Laux, T., Mayer, K.F.X., Berger, J., and Jürgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122, 87–96. [DOI] [PubMed] [Google Scholar]
- Li, Y., Takagi, Y., Jiang, Y., Tokunaga, M., Erdjument-Bromage, H., Tempst, P., and Kornberg, R.D. (2001). A multiprotein complex that interacts with RNA polymerase II Elongator. J. Biol. Chem. 276, 29628–29631. [DOI] [PubMed] [Google Scholar]
- Liao, B., and Zielinski, R.E. (1995). Production of recombinant plant calmodulin and its use to detect calmodulin-binding proteins. Methods Cell Biol. 49, 487–494. [DOI] [PubMed] [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379, 66–69. [DOI] [PubMed] [Google Scholar]
- Mayer, K.F., Schoof, H., Haecker, A., Lenhard, M., Jürgens, G., and Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713. [DOI] [PubMed] [Google Scholar]
- Medford, J.I., Behringer, F.J., Callos, J.D., and Feldmann, K.A. (1992). Normal and abnormal development in the Arabidopsis vegetative shoot apex. Plant Cell 4, 631–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami, Y., and Fischer, R.L. (2000). Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. USA 97, 942–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussian, B., Schoof, H., Haecker, A., Jürgens, G., and Laux, T. (1998). Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J. 17, 1799–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neil, K.T., and DeGrado, W.F. (1990). How calmodulin binds its targets: Sequence independent recognition of amphiphilic α-helices. Trends Biochem. Sci. 15, 59–64. [DOI] [PubMed] [Google Scholar]
- Otero, G., Fellows, J., Li, Y., de Bizemont, T., Dirac, A.M.G., Gustafsson, C.M., Erdjument-Bromage, H., Tempst, P., and Svejstrup, J.Q. (1999). Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell 3, 109–118. [DOI] [PubMed] [Google Scholar]
- Pokholok, D.K., Hannett, N.M., and Young, R.A. (2002). Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9, 799–809. [DOI] [PubMed] [Google Scholar]
- Ponting, C.P. (2002). Novel domains and orthologues of eukaryotic transcription elongation factors. Nucleic Acids Res. 30, 3643–3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt, R.E., and Meyerowitz, E.M. (1986). Characterization of the genome of Arabidopsis thaliana. J. Mol. Biol. 187, 169–183. [DOI] [PubMed] [Google Scholar]
- Pyke, K.A., Marrison, J.L., and Leech, R.M. (1991). Temporal and spatial development of the cells of the expanding first leaf of Arabidopsis thaliana (L.) Heynh. J. Exp. Bot. 42, 1407–1416. [Google Scholar]
- Reinhardt, D., Mandel, T., and Kuhlemeier, C. (2000). Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12, 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads, A.R., and Friedberg, F. (1997). Sequence motifs for calmodulin recognition. FASEB J. 11, 331–340. [DOI] [PubMed] [Google Scholar]
- Robles, P., and Micol, J.L. (2001). Genome-wide linkage analysis of Arabidopsis genes required for leaf development. Mol. Genet. Genomics 266, 12–19. [DOI] [PubMed] [Google Scholar]
- Sawa, S., Watanabe, K., Goto, K., Kanaya, E., Morita, R.H., and Okada, K. (1999). FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 13, 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof, H., Lenhard, M., Haecker, A., Mayer, K.F.X., Jürgens, G., and Laux, T. (2000). The stem cell population of Arabidopsis shoot meristem is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644. [DOI] [PubMed] [Google Scholar]
- Shaw, R.J., and Reines, D. (2000). Saccharomyces cerevisiae transcription elongation mutants are defective in PUR5 induction in response to nucleotide depletion. Mol. Cell. Biol. 20, 7427–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried, K.R., Eshed, Y., Baum, S.F., Otsuga, D., Drews, G.N., and Bowman, J.L. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126, 4117–4128. [DOI] [PubMed] [Google Scholar]
- Sussex, I.M. (1989). Developmental programming of the shoot meristem. Cell 56, 225–229. [DOI] [PubMed] [Google Scholar]
- Swinburne, J., Balcells, L., Scofield, S.R., Jones, J.D.G., and Coupland, G. (1992). Elevated levels of Activator transposase mRNA are associated with high frequencies of Dissociation excision in Arabidopsis. Plant Cell 4, 583–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotochaud, A.E., Hao, T., Wu, G., Yang, Z., and Clark, S.E. (1999). The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signaling complex that includes KAPP and a Rho-related protein. Plant Cell 11, 393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuge, T., Tsukaya, H., and Uchimiya, H. (1996). Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development 122, 1589–1600. [DOI] [PubMed] [Google Scholar]
- Uetz, P., et al. (2000). A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627. [DOI] [PubMed] [Google Scholar]
- Valvekens, D., Van Montagu, M., and Van Lijsebettens, M. (1988). Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. USA 85, 5536–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaff, E., Den Dulk-Ras, A., Hooykaas, P.J.J., and Keller, B. (2000). Activation tagging of the LEAFY PETIOLE gene affects leaf petiole development in Arabidopsis thaliana. Development 127, 4971–4980. [DOI] [PubMed] [Google Scholar]
- Van Haute, E., Joos, H., Maes, M., Warren, G., Van Montagu, M., and Schell, J. (1983). Intergeneric transfer and exchange recombination of restriction fragments cloned in pBR322: A novel strategy for the reversed genetics of Ti plasmids of Agrobacterium tumefaciens. EMBO J. 2, 411–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lijsebettens, M., and Clarke, J. (1998). Leaf development in Arabidopsis. Plant Physiol. Biochem. 36, 47–60. [Google Scholar]
- Waites, R., and Hudson, A. (1995). phantastica: a gene required for dorsoventrality of leaves in Antirrhinum majus. Development 121, 2143–2154. [Google Scholar]
- Walia, H., Chen, H.Y., Sun, J.-M., Holth, L.T., and Davie, J.R. (1998). Histone acetylation is required to maintain the unfolded nucleosome structure associated with transcribing DNA. J. Biol. Chem. 273, 14516–14522. [DOI] [PubMed] [Google Scholar]
- Wang, K.K.W., Villalobo, A., and Roufogalis, B.D. (1989). Calmodulin-binding proteins as calpain substrates. Biochem. J. 262, 693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, G.S., Kristjuhan, A., Erdjument-Bromage, H., Tempst, P., and Svejstrup, J.Q. (2002). Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc. Natl. Acad. Sci. USA 99, 3517–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, G.S., Petrakis, T.G., Ethelberg, S., Tokunaga, M., Erdjument- Bromage, H., Tempst, P., and Svejstrup, J.Q. (2001). RNA polymerase II Elongator holoenzyme is composed of two discrete subcomplexes. J. Biol. Chem. 276, 32743–32749. [DOI] [PubMed] [Google Scholar]
- Wittschieben, B.Ø., Otero, G., de Bizemont, T., Fellows, J., Erdjument- Bromage, H., Ohba, R., Li, Y., Allis, C.D., Tempst, P., and Svejstrup, J.Q. (1999). A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell 4, 123–128. [DOI] [PubMed] [Google Scholar]