Abstract
Plants contain two types of α(1→6) glucan hydrolase (starch-debranching enzyme [DBE]). Mutations that affect the pullulanase-type DBE have not been described, although defects in isoamylase-type DBE, known in many plant species, indicate a function in starch biosynthesis. We describe a null mutation of a pullulanase-type DBE gene, a Mutator insertion in maize Zpu1. Plants homozygous for the zpu1-204 mutation are impaired in transient and storage starch degradation. Thus, hydrolytic activity of pullulanase-type DBE contributes to starch catabolism. Developing zpu1-204 endosperm accumulates branched maltooligosaccharides not found in the wild type and is deficient in linear maltooligosaccharides, indicating that the pullulanase-type DBE functions in glucan hydrolysis during kernel starch formation. Furthermore, in a background deficient in isoamylase-type DBE, zpu1-204 conditions a significant accumulation of phytoglycogen in the kernel that is not seen in the wild type. Therefore, pullulanase-type DBE partially compensates for the defect in isoamylase-type DBE, suggesting a function during starch synthesis as well as degradation.
INTRODUCTION
The central role served by starch in the metabolic strategy of plants requires that its production and degradation be strictly coordinated events. Starch accumulates in photosynthetic tissues when light is available, but in the dark, the metabolic flux changes so that starch is degraded and serves as a source of reduced carbon. Distinct controls operate in reproductive and perenniating tissues (e.g., seeds and tubers), which synthesize starch continuously throughout the diurnal cycle. Degradation commences in such storage tissues during germination and sprouting to supply metabolites and energy to the developing plant before it attains photoautotrophy. Starch is involved in many other physiological processes. In the gravitropic responses of roots and shoots, for example, the sedimentation of amyloplasts is believed to be the sensing mechanism for the orientation of the plant with respect to gravity (Chen et al., 1999). Therefore, the metabolism of starch is essential to the development and fitness of plants. Most of the molecular details regarding the biochemical mechanisms that accomplish and control starch anabolism and catabolism remain to be discerned.
All plants characterized to date contain two remarkably conserved types of α(1→6) glucan hydrolase (i.e., starch-debranching enzyme [DBE]) with as yet undefined function(s) in starch metabolism. The two classes, pullulanase-type DBE (also known as R-enzyme or limit-dextrinase) and isoamylase-type DBE, are defined by sequence similarity and in vitro substrate specificity. These enzymes appear to have been conserved separately during the evolution of prokaryotes and higher plants, because each is more closely related to a bacterial protein than to the other type in the same plant species. Therefore, each DBE type likely has a unique function in the processes of carbohydrate storage and utilization. In maize, the pullulanase-type DBE, termed ZPU1, is encoded by the Zpu1 gene (Beatty et al., 1999). At least three isoamylase-type DBE genes are present in the maize genome. Sugary1 (Su1) codes for the major activity present in maize endosperm (James et al., 1995; Rahman et al., 1998), and two additional isoamylase-type DBE coding sequences are defined by the Iso2 and Iso3 cDNAs (J. Dinges, unpublished results).
The DBEs are part of a large network of proteins that par-ticipate in starch metabolism, which includes ADP-Glc pyrophosphorylase, starch synthase, starch branching enzyme (SBE), disproportionating enzyme, α-amylase, β-amylase, starch phosphorylase, and α-glucosidase. The complexity of starch metabolism is evident from the fact that multiple isoforms exist in plants for all of these classes of enzymes. The Arabidopsis genome, for example, encodes >30 such proteins. Understanding of the functional contribution of each enzyme isoform to starch assembly and disassembly will help explain why so many proteins are needed for this process in plants compared with the few required for glycogen metabolism in other kingdoms.
A straightforward hypothesis based on the hydrolytic activity of DBEs is that they function in the degradation of starch. If so, then mutations that affect such enzymes might be identified through starch excess phenotypes (e.g., the accumulation of abnormally high levels in plants that fail to fully degrade leaf starch in the dark). Examples from Arabidopsis are sex4, dpe1, and sex1, which affect, respectively, an amylase (Zeeman et al., 1998a), a D enzyme (Critchley et al., 2001), and the R1 homolog, a dikinase that is speculated to control the rate of starch degradation (Yu et al., 2001; Ritte et al., 2002). Mutations in other starch degradative enzymes (e.g., Arabidopsis ram1, which codes for the major form of β-amylase) have no noticeable effect on starch synthesis or degradation (Laby et al., 2001). To date, a mutation affecting pullulanase-type DBE has not been identified in starch excess screens (or any other screens), so clues to its function are not available from genetic analysis. The lack of a known pullulanase-type DBE mutation cannot be explained by gene duplication, because in maize the gene Zpu1 is present in only one copy (Beatty et al., 1999), and the complete rice and Arabidopsis genome sequences also contain only one such gene (Tabata et al., 2000; Yu et al., 2002).
In contrast to pullulanase-type DBE, mutations that affect an isoamylase-type DBE are observed frequently in many plant species, including maize (James et al., 1995), rice (Nakamura et al., 1996b), barley (Burton et al., 2002), Chlamydomonas (Mouille et al., 1996), and Arabidopsis (Zeeman et al., 1998b). The function of this isoamylase-type DBE is not evident from analysis of the mutant phenotypes. Contrary to the expectation of excess starch resulting from the loss of catabolic function, there is a deficiency in starch production and a concomitant accumulation of phytoglycogen, which do not occur in wild-type plants. Phytoglycogen resembles glycogen in structure and is similar in many respects to amylopectin, but it lacks the structural organization that allows crystallization into insoluble granules. One hypothesis to explain this phenotype is that this DBE is involved directly in the determination of the architectural structure of amylopectin (Ball et al., 1996; Myers et al., 2000). Another hypothesis proposes that isoamylase-type DBE degrades soluble glucans that would otherwise interfere with amylopectin synthesis by drawing off enzymes and substrate from the productive pathway (Zeeman et al., 1998b; Smith, 2001). A third model proposes that DBE activity is required for proper starch granule initiation (Burton et al., 2002). Thus, the function of isoamylase-type DBE in starch biosynthesis remains unclear. Furthermore, the fact that mutation of an isoamylase-type DBE affects starch formation does not exclude the possibility that the enzyme also functions in starch degradation.
Even less is known about the physiological function of the pullulanase-type DBE. Owing to the lack of a defined mutation, it was not known whether this enzyme also participates in starch assembly, is involved only in catabolism, or has a dual function. To address this question, we applied reverse genetics to identify maize plants that contain a null mutation of the Zpu1 gene. The mutant plants completely lack pullulanase-type DBE activity, a condition that has not been reported previously. ZPU1 is required for normal rates of starch degradation in leaves and seeds and thus clearly plays a catabolic role. A function for ZPU1 during starch anabolism also is indicated based on the observation that the mutation conditions the accumulation of phytoglycogen in endosperm tissue in a specific genetic background. Thus, the pullulanase-type DBE appears to function both in the biosynthesis of starch and in the degradation of granules.
RESULTS
Isolation and Characterization of zpu1 Mutants
The Trait Utility System for Corn (TUSC; Pioneer Hi-Bred International) (Bensen et al., 1995) was used to identify Mutator (Mu) transposable element insertions in the Zpu1 gene (Beatty et al., 1999). Pool screening was conducted using the Zpu1-specific primers pulA and pulB (Figure 1A) and the degenerate Mu terminal inverted repeat primer 9242. Specific PCR amplification products requiring one Zpu1 primer and the Mu primer were identified in nine TUSC F1 leaf DNA pools. PCR screening in a successive generation (F2) revealed that three of these Mu insertions in Zpu1 are transmissible through the germ line. These mutations were designated zpu1-204, zpu1-225, and zpu1-849, with the allele number indicating the TUSC pool from which each mutation was identified originally.
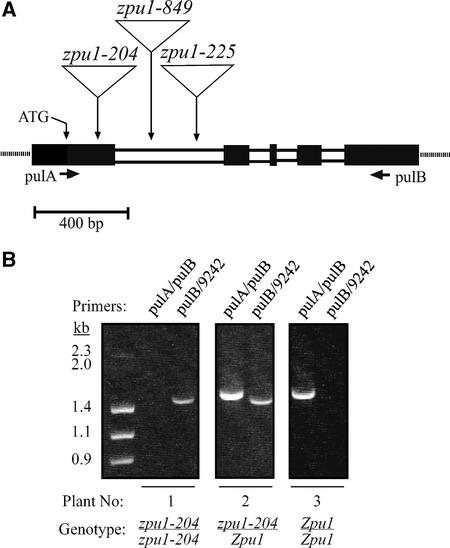
Figure 1.
Molecular Structure of zpu1 Mutant Alleles.
(A) Relative positions of Mu transposable elements within the Zpu1 gene. The allele zpu1-204 contains a Mu element in the first exon of the gene, downstream of the ATG start codon. The alleles zpu1-225 and zpu1-849 contain insertions in the first intron of the gene. The positions of the PCR primers pulA and pulB are indicated by arrows. Exons are indicated by black boxes.
(B) Genotypes of plants were determined using PCR. The pulA/pulB primer pair amplifies the wild-type Zpu1 allele from genomic DNA (∼1.6 kb), whereas the pulB/9242 primer pair amplifies the mutant zpu1-204 allele (∼1.4 kb). Data from three representative plants are shown. Plant 1 is homozygous zpu1-204, plant 2 is heterozygous zpu1-204/Zpu1, and plant 3 is homozygous Zpu1.
The location of the Mu transposable element within the zpu1 locus was determined by sequencing of the pulB/9242 and pulA/9242 PCR products. First, the pulA/pulB PCR amplification product from wild-type (W64A) genomic DNA (Figure 1B) was sequenced. Comparison of this genomic sequence with the Zpu1 cDNA sequence revealed four introns within the 5′ region of the Zpu1 gene (Figure 1A). The sequences of the pulB/9242 and pulA/9242 fragments immediately flanking the Mu termini then were compared with the wild-type genomic sequence to identify the transposon insertion sites. The alleles zpu1-225 and zpu1-849 arise from Mu element insertions in the first intron. However, the zpu1-204 mutation contains a Mu element inserted in the first exon at nucleotide 189 in the Zpu1 cDNA. This mutation was analyzed further because of the likelihood that disruption of the coding sequence would inactivate Zpu1 function.
The zpu1-204 mutation exhibited simple Mendelian inheritance. Amplification of a 1.6-kb fragment with the pulA/pulB primer pair revealed the presence of the wild-type allele Zpu1, and production of a 1.45-kb fragment using the pulB/9242 primer pair indicated that the mutant allele zpu1-204 was present. Figure 1B shows the genotypes of three representative plants grown from randomly selected kernels produced by self-pollination of a heterozygous zpu1-204/Zpu1 plant. Hundreds of progeny plants from such crosses have been examined, and the expected ratio of 25% Zpu1 homozygotes, 25% zpu1-204 homozygotes, and 50% Zpu1/zpu1-204 heterozygotes was closely approximated in the actual PCR genotype data.
zpu1-204 Is a Null Allele
Gene expression from the zpu1 locus was characterized at the level of mRNA accumulation using reverse transcriptase PCR. Total RNA was isolated from developing wild-type or zpu1-204 homozygous kernels harvested 20 days after pollination (DAP). Reverse transcriptase PCR performed using the pulA/pulB primer pair yielded a 0.7-kb fragment from the kernel RNA of both wild-type plants analyzed (Figure 2A), as expected from the sequence of the Zpu1 cDNA. The same procedure failed to amplify any Zpu1-specific fragment from kernels of two different zpu1-204 mutant plants. The integrity of the total RNA samples from the wild-type and mutant plants was tested by ethidium bromide staining after agarose gel electrophoresis, and no differences were detected (data not shown). Thus, Zpu1 mRNA cannot be detected in zpu1-204 mutants by this sensitive method.
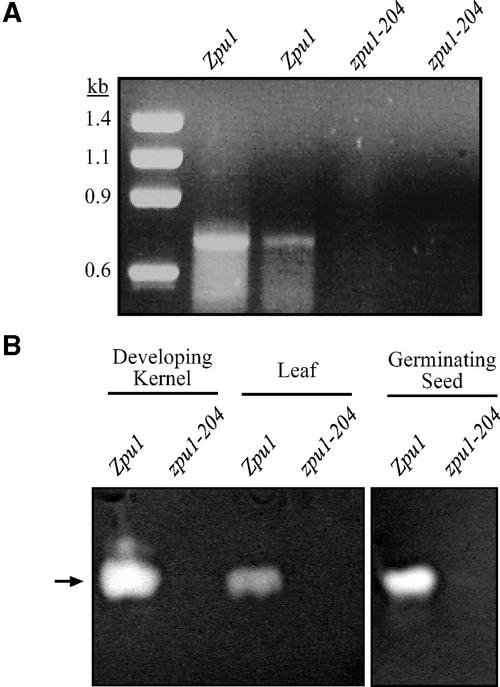
Figure 2.
zpu1-204 Is a Null Allele.
(A) Reverse transcriptase PCR was performed on total RNA isolated from kernels at 20 DAP using the pulA/pulB primer pair. Two independent samples from the wild type (lanes 1 and 2) and the zpu1-204 mutant (lanes 3 and 4) are shown.
(B) Zymogram analysis of wild-type Zpu1 (W64A) and zpu1-204 developing kernels, leaves, and germinating seeds. Pullulanase activity (indicated by the arrow) was visualized by clearing of the pullulan azure substrate. Approximately 50 μg of sample was loaded in all cases, except for Zpu1 (W64A) germinating seeds, for which 25 μg of protein was loaded to reduce the intensity of the band.
Pullulanase-type DBE activity was undetectable in leaves, 20-DAP developing kernels, or germinating seeds of zpu1-204 plants (Figure 2B). Proteins in total soluble extracts were separated by native PAGE and then transferred to another polyacrylamide gel containing pullulan azure. This dyed polysaccharide polymer is a highly specific substrate of pullulanase-type DBE, so that enzyme activity is indicated by clear bands that result from the cleavage of α(1→6) linkages and diffusion of the maltotriose products out of the gel. Extracts from wild-type endosperm, leaves, and germinating seeds showed specific bands of pullulanase-type DBE activity in which the dyed substrate had been cleared. However, the same tissues from the zpu1-204 mutant all failed to show any such activity. Together, these data demonstrate that zpu1-204 is a null mutation that conditions the complete elimination of pullulanase-type DBE.
Effects of ZPU1 Deficiency on Leaf Starch Metabolism
The zpu1-204 mutation, when homozygous, does not condition any readily noticeable plant phenotype. Under field conditions, mutant plants grow at a normal rate after germination and show no obvious differences from the wild-type standard line in morphology or flowering time.
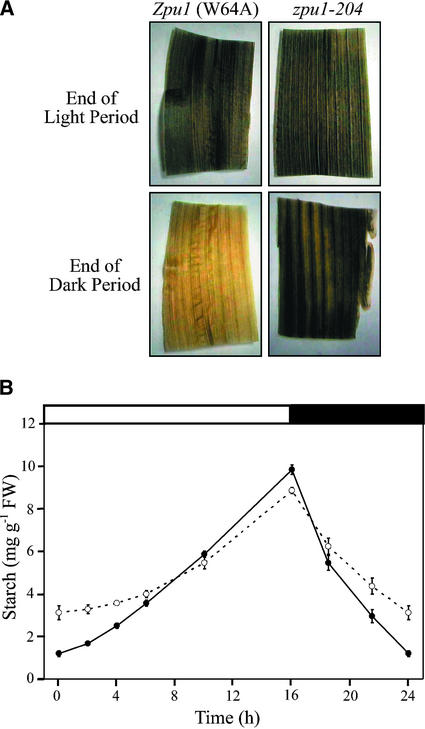
To specifically examine the effects of zpu1-204 on transient starch synthesis and degradation, leaf starch content was measured over a diurnal cycle of 16 h of light and 8 h of dark. Tissue samples were collected from adult maize leaves (∼3 weeks after planting, before anthesis) at various times in the cycle. The leaves were decolorized in boiling ethanol (80%, v/v) and then stained with I2/KI solution to reveal qualitatively the presence or absence of starch. The intensity of staining was similar in mutant and wild-type leaves at the end of the light period (Figure 3A), which indicates that starch accumulates to apparently normal levels in the absence of pullulanase-type DBE activity when plants are grown under these conditions. As expected, at the end of the dark period, the iodine stain failed to produce a deep brown color in wild-type leaves. However, the leaves of zpu1-204 mutants still stained brown at the end of the dark period (Figure 3A), suggesting a defect in starch catabolism. Thus, zpu1-204 conditions a starch excess phenotype.
Figure 3.
Leaf Starch Content during the Diurnal Cycle.
(A) Leaves from Zpu1 (W64A) and the zpu1-204 mutant were harvested at the end of a 16-h light period and the end of an 8-h dark period, decolorized in boiling 80% (v/v) ethanol, and stained with I2/KI solution. A qualitative measure of starch amount is indicated by dark brown staining.
(B) Starch was extracted from leaves of individual plants and quantified by measuring the Glc released after complete digestion with amyloglucosidase. Closed circles represent the wild type, and open circles represent zpu1-204. Each point represents the mean ± se of 15 plants. Where absent, error bars are smaller than the symbols. FW, fresh weight.
Quantification of leaf starch content during the diurnal cycle revealed that the rate of starch degradation was slower in zpu1-204 plants (Figure 3B). The starch content exhibited a statistically significant reduction in zpu1-204 leaves compared with wild-type leaves (Student's two-sample t test, P < 0.05) at 21.5 and 24 h in the cycle. Removing plants from the diurnal cycle and extending the dark period to 16 h resulted in a reduction of the starch level in zpu1-204 leaves to the same level seen in the wild type (data not shown). Thus, pullulanase-type DBE affects the rate of starch degradation but is not absolutely required for this metabolic process.
At the end of the light period, the level of accumulated starch approached that of wild-type leaves under normal diurnal conditions (Figure 3B, 16 h). Because the quantity of starch present at the beginning of the light period was greater in the mutant, the net amount of starch synthesis was reduced compared with that in the wild type. One possible explanation for these data is that the zpu1-204 mutation causes a defect in starch accumulation. Alternatively, the reduced starch accumulation could be an indirect effect. For example, the Arabidopsis dpe1 mutant (Critchley et al., 2001), which lacks disproportionating enzyme, is impaired in starch degradation and accumulates relatively large quantities of maltooligosaccharides during the night. Mutant plants subsequently synthesize less starch than wild-type plants during the day. Critchley et al. (2001) speculate that the surplus starch and/or maltooligosaccharides present at the end of the dark period negatively influence synthesis in the sub-sequent light period.
To test for such carryover effects from the preceding dark period, wild-type and zpu1-204 plants were placed in the dark for 32 h to allow the complete degradation of glucan. The destarched plants then were allowed to grow in the light for 16 h, after which time leaf starch was extracted, quantified, and analyzed with respect to starch composition and chain length distribution (see below). Activity gel analysis showed that wild-type destarched leaves possessed normal levels of pullulanase activity compared with untreated controls (data not shown). In these conditions, wild-type plants accumulated almost twofold more starch than zpu1-204 plants (Table 1). Plants bearing the sbe2a::Mu mutation (Blauth et al., 2001), which lacks the SBE isoform SBEIIa, also showed a similarly reduced starch content. This analysis was necessary because of the fact, shown below, that zpu1-204 conditions a reduction of SBEIIa activity as a secondary pleiotropic effect. Leaves of zpu1-204 and sbe2a::Mu also showed a relative increase in the amount of Suc (Table 1), which is consistent with there being an altered partitioning between Suc and starch as a result of the defect in starch synthesis.
Table 1.
Carbohydrate Content of Destarched Leaves
| Metabolite (mg/g fresh wt)
|
|||||
|---|---|---|---|---|---|
| Genotype | Collection Time | Samples | Starch | WSPa | Suc |
| Zpu1 (W64A) | End of light period | 4 | 10.31 ± 0.16 | 0.84 ± 0.06 | 2.17 ± 0.28 |
| zpu1-204 | End of light period | 4 | 5.23 ± 0.11 | 0.77 ± 0.15 | 4.59 ± 0.32 |
| sbe2a::Mu | End of light period | 2 | 6.35 ± 0.04 | 1.03 ± 0.18 | 5.52 ± 0.23 |
| Zpu1 (W64A) | End of dark period | 4 | 1.11 ± 0.03 | 0.19 ± 0.05 | 0.56 ± 0.10 |
| zpu1-204 | End of dark period | 4 | 1.83 ± 0.03 | 0.26 ± 0.04 | 0.96 ± 0.07 |
| sbe2a::Mu | End of dark period | 2 | 1.19 ± 0.06 | 0.22 ± 0.04 | 1.36 ± 0.11 |
Plants were destarched by extending the dark period to 32 h and transferred to the light for 16 h, followed by an 8-h dark period. Results are means ± se for the number of samples indicated.
Water-soluble polysaccharide (WSP) is defined as the quantity of Glc equivalents liberated from polymers after treatment of total soluble extract with amyloglucosidase.
Effects of ZPU1 Deficiency on Leaf Starch Structure
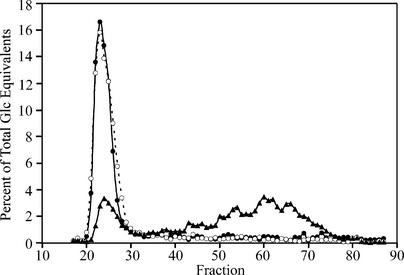
The composition of the leaf starch was analyzed by Sepharose CL2B chromatography. The amylopectin and apparent amylose contents were essentially identical in granules from the wild type and the zpu1-204 mutant (Figure 4). By contrast, starch from the sbe2a::Mu mutant was decreased markedly in the relative amount of normal amylopectin and accordingly increased in lower molecular mass that could be amylose or a combination of amylose and altered amylopectin (Figure 4).
Figure 4.
Sepharose CL2B Separation of Leaf Starch.
Starch was extracted from leaves of wild-type W64A (closed circles, solid line), zpu1-204 (open circles, dashed line), and sbe2a::Mu (closed triangles, solid line) at the end of the photoperiod and applied to a 100-mL column. Fractions were eluted with 10 mM NaOH. Results shown are averages of two samples from each genotype. The data were normalized by dividing the amount of Glc equivalents in each fraction by the sum total of all fractions.
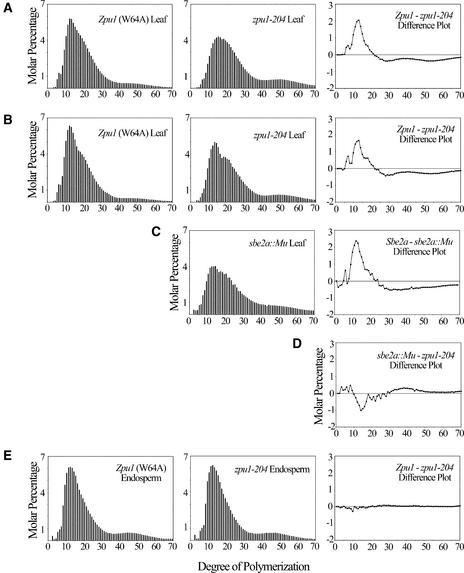
The structure of amylopectin in leaf starch was characterized with regard to the distribution of linear chain lengths. Fractionated amylopectin from Sepharose CL2B chromatography was completely debranched by treatment with a bacterial isoamylase. Linear glucan chains in the resultant mixture were labeled with a fluorophore at the reducing end, separated by capillary electrophoresis, and quantified by fluorescence spectroscopy. Comparison of the distributions indicated that amylopectin from zpu1-204 leaves contained significantly fewer smaller chains (8 to 15 degrees of polymerization [DP]) than that from nonmutant leaves and a slightly greater proportion of longer chains (Figures 5A and 5B). A similar amylopectin chain length profile was observed from sbe2a::Mu leaves (Figure 5C). However, comparison between the profiles for zpu1-204 and sbe2a::Mu showed that zpu1-204 had slightly more short chains (10 to 20 DP) than sbe2a::Mu (Figure 5D).
Figure 5.
Chain Length Distribution of Amylopectin from Leaves and Endosperm.
Amylopectin from pooled Sepharose CL2B chromatography fractions of endosperm and leaf starch was debranched with Pseudomonas amyloderamosa isoamylase. The reducing ends of the linear chains were labeled with the fluorophore 8-amino-1,3,6-pyrenetrisulfonic acid and separated by capillary electrophoresis. Individual chains were normalized to total peak area, and differences in chain lengths are shown in subtraction plots. Results shown are averages of four independent analyses.
(A) and (B) Amylopectin from wild-type and zpu1-204 leaves harvested at the end of the photoperiod from growth chamber–grown (A) and field-grown (B) plants.
(C) Chain length distribution of sbe2a::Mu leaf amylopectin (compared with the same wild-type profile shown in [B]).
(D) Difference plot comparing the chain length distribution of zpu1-204 and sbe2a::Mu.
(E) Amylopectin chain length distribution of wild-type and zpu1-204 endosperm at 20 DAP.
Effects of ZPU1 Deficiency on Endosperm Starch Metabolism
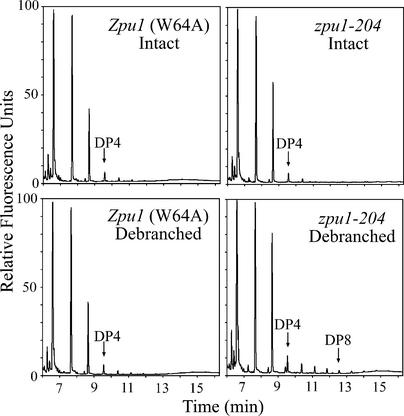
The contents of Glc, Suc, Fru, WSP, and starch in endosperm tissue harvested at 20 DAP were compared in isogenic zpu1-204 plants and the nonmutant control line W64A (Table 2). The levels of Suc and the monosaccharides, as well as the amount of starch, were statistically indistinguishable between mutant and wild-type endosperm. However, a statistically significant difference was observed in the level of WSP (i.e., the glucan polymer present in the soluble phase after centrifugation at 10,000g), which was reduced in the mutant compared with the wild-type line. WSP from wild-type endosperm was characterized by capillary electrophoresis and shown to consist almost entirely of maltose, maltotriose, and maltotetraose (Figure 6, Intact). Treatment with a mixture of bacterial α(1→6) glucan hydrolases failed to alter the electrophoretic profile, indicating that small, branched polymers are not present in the wild type. By contrast, the WSP from zpu1-204 endosperm contained significant quantities of low molecular mass, α(1→6)-linked polyglucans, as indicated by the increase after debranching in the abundance of chains composed of five to eight Glc units (Figure 6). This is a distinctly different profile from that observed after the debranching of phytoglycogen (Dinges et al., 2001), in which the most abundant chain lengths were 8 to 12 DP. These results suggest that ZPU1 functions during periods of endosperm starch accumulation to hydrolyze α(1→6) linkages in small, branched polysaccharides.
Table 2.
Endosperm Carbohydrate Content
| Genotype
|
|||||
|---|---|---|---|---|---|
|
Zpu1 (W64A)
|
zpu1-204
|
||||
| Metabolite | mg/g dry wt | % | mg/g dry wt | % | P Valuea (Zpu1 versus zpu1-204) |
| Glc | 4.48 ± 0.59 | 0.5 | 3.51 ± 0.40 | 0.4 | 0.07 |
| Fru | 2.82 ± 0.29 | 0.3 | 2.01 ± 0.36 | 0.2 | 0.11 |
| Suc | 51.1 ± 2.80 | 6.1 | 48.7 ± 1.97 | 5.9 | 0.50 |
| WSP | 14.6 ± 0.83 | 1.7 | 9.97 ± 0.45 | 1.2 | 0.001 |
| Starch | 766 ± 31 | 91.3 | 768 ± 32 | 92.3 | 0.99 |
Results are means ± se of 12 total assays. Four ears of each genotype were harvested at 20 DAP, and three independent assays were performed on separate pools of kernels taken from each ear.
Determined using Student's two sample t test.
Figure 6.
Analysis of WSP from zpu1-204 Endosperm.
Water-soluble glucan was labeled with 8-amino-1,3,6-pyrenetrisulfonic acid and subjected to capillary electrophoresis before and after treatment with a mixture of bacterial pullulanase and isoamylase. The profile of Zpu1 WSP did not change significantly after debranching. However, the profile of zpu1-204 WSP showed a relative increase in chains of >3 Glc units, indicating the presence of small, branched polysaccharides. The DP of each peak was assigned by comparing the retention time with that of known standards.
Endosperm starch composition and structure were compared in the wild-type and zpu1-204 lines. The relative proportions of the amylose and amylopectin components of starch granules were similar, as determined by gel-permeation chromatography on Sepharose CL2B (data not shown). The chain length distributions of purified amylopectin were determined for zpu1-204 and wild-type endosperm, and essentially identical results were observed (Figure 5E). Therefore, in endosperm tissue, seemingly normal starch accumulated to the wild-type level in the absence of any pullulanase-type DBE activity.
Effects of ZPU1 Deficiency on Starch Degradation during Germination
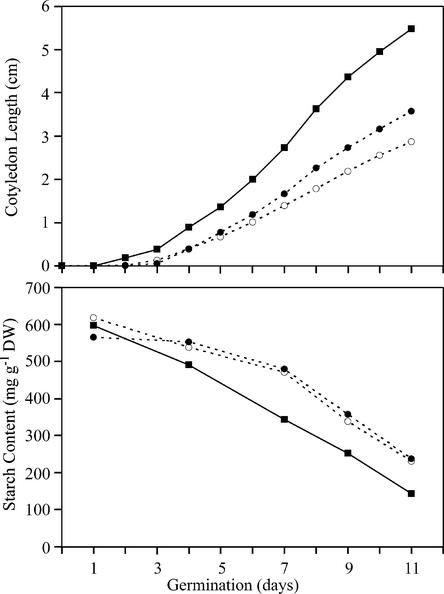
Pullulanase-type DBE is expressed de novo within the aleurone layer during the germination of cereal grains, which suggests a role in the degradation of storage starch. In barley, for example, the same pullulanase-type DBE that is present in endosperm during development also is expressed during germination, and maximal mRNA accumulation is achieved at day 5 after planting (Longstaff and Bryce, 1993; Schroeder and MacGregor, 1998; Burton et al., 1999). The availability of the ZPU1-deficient maize mutant allowed a direct test of whether pullulanase-type DBE is required for kernel germination. Mature, dried wild-type and zpu1-204 mutant kernels were hydrated in the dark at 30°C. The time of cotyledon emergence was noted, and cotyledon length was recorded over successive days. The percentage of mutant and wild-type kernels that germinated was nearly the same (data not shown). Cotyledons were first visible in the wild-type seeds 1 to 2 days earlier than in the mutants, and the average length of the mutant cotyledons was significantly less than that of the wild-type plants at all time points recorded over 2 weeks (Figure 7). This effect was obvious in kernels of self-pollinated ears from two different zpu1-204 homozygous plants. Therefore, in the absence of ZPU1, seeds take longer to germinate and the rate of plant growth before attaining photoautotrophy is decreased compared with that in the wild type.
Figure 7.
Germination Analysis of zpu1-204 Kernels.
Kernels from Zpu1 (squares) and two different zpu1-204 homozygotes (circles) were germinated on moist filter paper at 30°C. The lengths of the cotyledons were measured on successive days during the incubation period (each point represents the mean of 15 cotyledons). Starch content in the germinating endosperm was determined by removing the roots and cotyledons from three kernels at each time point and measuring Glc equivalents of insoluble α-glucan polymer.
The amount of starch remaining in the endosperm of germinating kernels was measured periodically after cotyledon emergence to test the hypothesis that retarded growth results from a reduced rate of starch degradation. Kernels from two different zpu1-204 plants showed a slower rate of starch degradation until approximately day 7 of germination compared with the wild type (Figure 7). After that time, the rate of starch degradation matched that of wild-type kernels. These results indicate that ZPU1 contributes to starch hydrolysis during the early stages of seed germination.
Effects of ZPU1 Deficiency on Other Starch-Metabolizing Enzymes
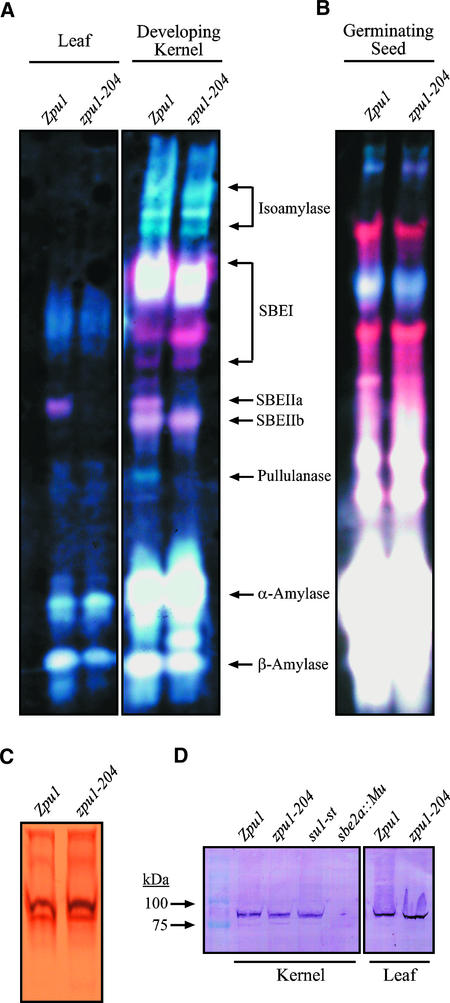
To draw any conclusions about the direct effects of ZPU1 deficiency on starch metabolism, it is necessary to determine whether other biosynthetic or degradative enzymes are affected pleiotropically by zpu1-204. This is particularly relevant to DBEs in rice and maize, because pleiotropic effects on both SBE and pullulanase-type DBE are known to result from mutations that affect the isoamylase-type DBE SU1 (Beatty et al., 1999; Kubo et al., 1999; Dinges et al., 2001). Zymogram assays were applied to broadly characterize the starch-metabolizing activities present in total soluble extracts from wild-type and zpu1-204 mutant kernels harvested at 20 DAP as well as from wild-type and mutant leaves collected at the end of the light and dark phases of the standard diurnal cycle. Proteins were separated by native PAGE and then transferred electrophoretically to a gel containing 0.3% (w/v) potato starch. Staining of the gels after incubation resulted in a series of colored bands indicative of specific enzyme activities (Figure 8A), many of which have been identified using mutant analysis, partial purification, and immunological characterization (our unpublished results).
Figure 8.
Activity Gel Analysis of Starch-Modifying Enzymes.
(A) and (B) Approximately 100 μg of total protein extracts from wild-type W64A and zpu1-204 leaves (A), 20-DAP kernels (A), and germinating seeds (B) was separated on a 15-cm native polyacrylamide gel and electroblotted to a starch-containing gel. Starch-modifying enzyme activities were visualized by staining with I2/KI.
(C) To detect starch synthase activities, total leaf extracts were separated on a native gel containing 0.2% glycogen, incubated for 16 h in the presence of ADP-Glc, and stained with iodine.
(D) Immunoblot from SDS-PAGE of total protein extracts developed with antiserum against SBEIIa.
The zpu1-204 mutation clearly had a secondary effect on the activity of the branching enzyme isoform SBEIIa, which was undetectable in kernel extracts and reduced significantly in leaf and germinating seed extracts of mutant plants (Figure 8A). A pleiotropic effect also was observed on an amylolytic activity presumed to be a β-amylase (Figure 8A). A novel β-amylase activity band was observed in ZPU1-deficient kernels, and the intensity of the normal band appeared to be reduced slightly compared with the wild type in endosperm and light-harvested leaves (Figure 8A). No defects were observed in other starch-metabolizing enzymes, particularly isoamylase-type DBE, SBEIIb, and SBEI (Figure 8A). Isoamylase-type DBEs were characterized in greater detail to reveal the major activity band that was obscured by SBEI. After fractionation of crude extracts by anion-exchange chromatography, zymogram analysis showed the presence of three characteristic blue bands in extracts from both wild-type and zpu1-204 kernels (data not shown). These data confirm that the loss of pullulanase-type DBE does not pleiotropically affect the isoamylase-type DBE. A separate zymogram analysis was performed to observe starch synthase activities. Four starch synthase activity bands were apparent in both zpu1-204 and wild-type W64A leaf extracts from tissue harvested at the end of the light phase of the standard diurnal cycle, and no differences were discernible between the two lines (Figure 8C).
Immunoblot analysis was used to determine whether the lack of SBEIIa activity in zpu1-204 leaves and kernels resulted from the failure to accumulate the protein. Polypeptides in total soluble extracts were separated by SDS-PAGE and then probed with a polyclonal antibody that recognizes SBEIIa (Figure 8D). The antibody detected a protein of the expected molecular mass, ∼85 kD, that was not present in a control sbe2a::Mu mutant line (Blauth et al., 2001). Furthermore, the protein was present in approximately equal abundance in both wild-type and zpu1-204 plants in developing kernel and leaf tissue. The su1-st mutation, in the gene that encodes the major isoamylase-type DBE, has the same pleiotropic result of eliminating SBEIIa activity (Dinges et al., 2001). In this mutant, the SBEIIa protein also accumulates to the same level as in the wild type. Therefore, secondary effects on SBEIIa activity are exerted at the post-translational level by mutation of either the pullulanase-type DBE ZPU1 or the isoamylase-type DBE SU1.
WSP Accumulation Conditioned by zpu1-204
A null mutation in the Su1 gene, which encodes an isoamylase-type DBE, conditions a reduction of 80% in the amount of starch produced in endosperm tissue and also the appearance of significant quantities of phytoglycogen. The reason that some starch granules form even in the complete absence of SU1 is not clear. Activity provided by the Iso2 and Iso3 genes may account for the residual starch, but there is no direct evidence that these enzymes contribute to starch metabolism in maize endosperm. Another plausible explanation is that the pullulanase-type enzyme provides some of the DBE function required for normal starch accumulation. Double mutants containing both zpu1-204 and a su1 mutation were constructed to test this hypothesis, with the expectation that the overlapping functions of the two DBEs would result in a more severe starch biosynthesis defect than that conditioned by either mutation alone. The allele su1-st (Dahlstrom and Lonnquist, 1964; Dinges et al., 2001) was used because it causes only a modest reduction in the amount of starch, accumulation of relatively small amounts of phytoglycogen, and a mild kernel phenotype compared with the severely shrunken and translucent kernels that result from the su1 null mutation. In this genetic background, another defect in the process that accounts for phytoglycogen accumulation and granular starch deficiency would be readily detectable.
Plants homozygous for zpu1-204 were crossed to a homozygous su1-st line, and the resulting double heterozygotes were self-pollinated. Three distinct kernel phenotypes were distinguishable on the F2 ears from this cross (Figure 9). In addition to kernels with wild-type appearance, the moderate but clearly recognizable kernel phenotype conditioned by su1-st was present in approximately three-sixteenths of the progeny. A third distinct phenotype, obviously more severe than that of the su1-st single mutant, appeared at the approximate frequency of one-sixteenth, and these kernels were expected to be the zpu1-204;su1-st double mutants. Like zpu1-204, su1-st can be detected definitively by PCR analysis (Dinges et al., 2001). This technique was applied to DNA of 45 seedlings grown from severely mutant kernels. The PCR data in all instances confirmed that these individuals were su1-st; zpu1-204 double homozygotes. Thus, the ears derived from self pollination of the double heterozygote allow comparison between sibling kernels: wild-type (Su1/Su1 or Su1/su1-st; Zpu1/Zpu1, Zpu1/zpu1-204, or zpu1-204/zpu1-204), su1-st single mutant (su1-st/su1-st; Zpu1/Zpu1 or Zpu1/zpu1-204), or double mutant (su1-st/su1-st; zpu1-204/zpu1-204).
Figure 9.
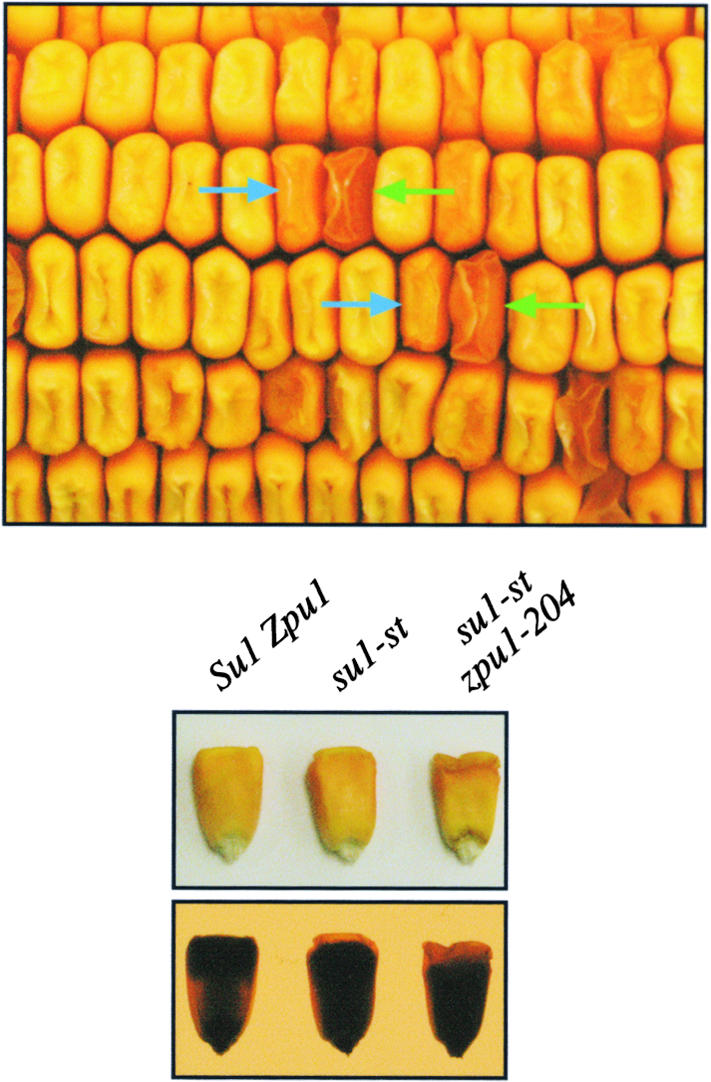
Synthetic Phenotypes of DBE Double Mutants.
Crosses were made between zpu1-204 and the isoamylase-type DBE mutant su1-st. A portion of an ear resulting from the self-pollination of the double heterozygote is shown. Approximately three-sixteenths of the kernels possessed the su1-st phenotype (blue arrows), having a slightly wrinkled, translucent crown but an opaque, starchy central and basal region (observed in the side view of the kernels at bottom, as seen with overhead light and on a lightbox). A markedly more severe phenotype appeared in approximately one-sixteenth of the kernels (green arrows). These kernels were characterized by a more severe wrinkling of the crown, extending farther into the central region of the kernel, compared with su1-st homozygotes. PCR genotyping of these kernels indicated that they were homozygous for the zpu1-204 allele, but singly homozygous ears for this allele showed no phenotype.
Single mutants homozygous for su1-st displayed slight wrinkling on the crown of the kernel, and the zpu1-204 homozygous mutant kernels, as mentioned previously, appeared to be identical to wild-type kernels. The double mutants were easily distinguished from either single mutant by severe wrinkling at the crown and a fully translucent appearance on the ear (Figure 9). Such an appearance typically is indicative of significant amounts of phytoglycogen in the affected kernels. Side views of kernels removed from the ear showed that the extent of the apparent phytoglycogen-containing region was much greater in the double-mutant kernels than in the su1-st single mutants (Figure 9). Carbohydrates were extracted from kernels of the different phenotypes, and starch and WSP were quantified (Table 3). The double-mutant kernels contained five times the WSP quantity of the su1-st single mutants and were reduced by 15% in the content of granular starch. Like phytoglycogen, the WSP from su1-st; zpu1-204 double mutants was precipitable in 5 volumes of 100% (v/v) ethanol. From these results, we conclude that ZPU1 is capable of participating in the process of starch biosynthesis by providing some functional overlap with the isoamylase-type DBE SU1 and/or by contributing to the total amount of α(1→6) hydrolytic activity in the developing endosperm. This is despite the fact that ZPU1's biosynthetic function is not essential for the production of normal quantities of endosperm starch or amylopectin with the wild-type distribution of linear glucan chain lengths.
Table 3.
Endosperm Carbohydrate Content in DBE Double Mutants
| Phenotype
|
||||||
|---|---|---|---|---|---|---|
| Wild-Type Segregants
|
su1-st
|
su1-st zpu1-204
|
||||
| Metabolite | mg/g dry wt | % | mg/g dry wt | % | mg/g dry wt | % |
| Glc | 9.4 ± 0.4 | 1.3 | 20.6 ± 3.0 | 2.8 | 43.9 ± 9.0 | 4.7 |
| Fru | 3.7 ± 0.5 | 0.5 | 5.5 ± 1.3 | 0.8 | 8.6 ± 1.4 | 0.9 |
| Suc | 4.0 ± 0.6 | 0.5 | 5.7 ± 0.6 | 0.8 | 16.5 ± 3.7 | 1.8 |
| WSP | 12.3 ± 3.1 | 1.7 | 56.1 ± 10.9 | 7.7 | 310 ± 44 | 33 |
| Starch | 704 ± 49 | 96 | 646 ± 43 | 88 | 549 ± 21.5 | 59 |
Kernels resulting from the self-pollination of Su1/su1-st Zpu1/zpu1-204 plants were selected based on their phenotype. Segregants with a wild-type phenotype may have the genotype Zpu1 or zpu1-204. Five mature, imbibed kernels were pooled for each assay. Measurements are means ± se from four independent assays.
Starch Granule Size and Number in DBE Mutants
Mutational analysis has shown that a defect in an isoamylase-type DBE of maize and barley causes an increase in the number of starch granules in endosperm tissue compared with that in the wild type (Creech, 1968; Burton et al., 2002). The mutant granules are compound in nature and are smaller than those produced in wild-type endosperm (Creech, 1968; Burton et al., 2002). These observations led to the hypothesis that the isoamylase-type DBE is important for proper starch granule initiation (Burton et al., 2002), functioning as a negative factor. To determine whether pullulanase-type DBE affects granule initiation, starch granules extracted from endosperm at mid development (20 DAP) were counted using a hemacytometer (Shannon et al., 1996), and their size distribution was measured using a Coulter Counter. Wild-type and zpu1-204 endosperm contained a similar number of granules per kernel (Table 4), and their size distribution was similar. From these data, there is no suggestion that ZPU1 functions as a negative regulator of granule initiation.
Table 4.
Starch Granule Characteristics of DBE Mutants
| Granules per Endosperm (Millions) |
Granule Size Distribution (μm)a |
|||
|---|---|---|---|---|
| Sample | 10% | 50% | 90% | |
| Zpu1 (W64A) | 9.90 ± 0.27 | 1.88 | 5.29 | 12.3 |
| zpu1-204 | 10.02 ± 0.25 | 2.06 | 5.08 | 11.5 |
| su1-st | 17.14 ± 1.18 | 1.64 | 2.86 | 6.25 |
| su1-st zpu1-204 | 40.51 ± 2.71 | 1.57 | 2.42 | 5.62 |
The endosperm starch of three kernels harvested at 20 DAP was extracted together. Results are means ± se of three replicate aliquots of the starch suspension. Each aliquot was sampled six times and counted using a hemacytometer.
Determined using a Coulter Counter.
However, an effect of ZPU1 deficiency on granule number was observed when isoamylase-type DBE function also was compromised. As expected, su1-st conditioned a significant increase in the number of granules per kernel compared with the wild type, and these granules were smaller than normal (Table 4). The su1-st; zpu1-204 double mutant showed a further increase in granule number per kernel, approximately twofold more than the su1-st single mutant (Table 4). Therefore, ZPU1 is capable of functioning as a factor that determines granule number, and again, the two different types of DBE appear to overlap functionally in the process of starch biosynthesis.
DISCUSSION
To investigate the function(s) of the pullulanase-type DBE in starch metabolism, a reverse genetics approach was used to identify transposable element insertions in the maize Zpu1 gene. The mutation zpu1-204 was shown to be a null allele by analysis of the gene structure and by the complete lack of detectable mRNA transcript or enzymatic activity. These results demonstrate that the protein encoded by Zpu1 provides the only pullulanase-type DBE activity in maize, and they are consistent with the fact that the gene that encodes this enzyme is unique (Beatty et al., 1999). Despite the complete loss of pullulanase-type DBE activity, plants homozygous for zpu1-204 did not exhibit any obvious morphological differences in the leaf or kernel. This fact accounts for the lack of mutations in pullulanase-type DBE detected in any forward genetic screen. However, the application of reverse genetics revealed that the loss of ZPU1 activity alters various aspects of starch metabolism.
Pullulanase-type DBE is required for normal starch catabolism. Germinating mutant seeds showed a decreased rate of starch degradation and a slower rate of cotyledon growth compared with wild-type seeds. This effect occurred in the presence of very high levels of amylolytic activity and apparently normal levels of isoamylase-type DBE (Figure 8B). Leaves of zpu1-204 also exhibited a slower rate of starch catabolism, as indicated by an increased amount of granular starch at the end of the dark period compared with that in wild-type leaves. This effect was observed in leaves operating under a diurnal cycle (Figure 3B) and in destarched leaves (Table 1). One possible explanation for this finding is that the total level of α(1→6) glucosidase activity (i.e., both pullulanase-type and isoamylase-type DBE) may be limiting in germinating zpu1-204 seeds and leaves. Alternatively, pullulanase-type DBE might serve a specific catalytic function in starch catabolism that cannot be filled by any other glucan hydrolases. Accordingly, isoamylase-type DBE would not fully compensate for the loss of pullulanase-type DBE. In vitro studies of recombinant maize pullulanase-type DBE show clear differences in substrate preference compared with isoamylase-type DBE, with pullulanases generally preferring shorter chains (Rahman et al., 1998; Wu et al., 2002). This fact, along with the high degree of evolutionary conservation of both classes of DBEs, is consistent with the hypothesis that enzyme specificity is the reason that starch degradation is reduced in zpu1-204 mutants.
The observed reduction in starch degradation raises the possibility that altered carbon partitioning could cause a deficiency in general plant growth characteristics. Obvious qualitative defects were not observed in kernel size, ear development, plant height, or other features, although further characterization is required to determine whether any quantitative defects on traits such as yield might result from mutations that affect the zpu1 locus.
In addition to the demonstrated and anticipated role of ZPU1 in starch degradation, this study revealed that the pullulanase-type DBE also is capable of functioning in starch anabolism. Expression of pullulanase-type DBE in the developing seeds of many plant species, such as maize (Beatty et al., 1999), rice (Nakamura et al., 1996a), and pea (Zhu et al., 1998), is consistent with a role during starch synthesis. Evidence that pullulanase-type DBE might be involved in starch synthesis is shown here by the fact that in some genetic backgrounds, zpu1-204 can condition a “sugary” kernel phenotype. This condition includes an increase in starch granule number, a reduced amount of granular starch, and significant amounts of phytoglycogen in developing endosperm. Pleiotropic effects on SBEIIa activity can be excluded as the causative agent of this phenotype in the su1-st background because su1-st itself conditions a loss of SBEIIa activity. Thus, we conclude that the enhanced starch biosynthesis defect observed in the su1-st; zpu1-204 double mutant is a specific effect of the loss of ZPU1.
This effect of zpu1-204 was observed only in the presence of a mutation that affects isoamylase-type DBE, not in the wild-type genetic background. Therefore, the two classes of DBE appear to overlap to some extent in their biosynthetic function, despite the fact that each enzyme class has been conserved separately throughout the evolution of bacteria and plants. The simplest explanation for these findings is that ZPU1 functions during the normal starch biosynthesis process; however, from the available data, the possibility cannot be excluded that this function arises only in the abnormal conditions engendered by the alteration of isoamylase-type DBE. The specific function of pullulanase-type DBE in the isoamylase-compromised background is not yet known. Both direct and indirect functions are consistent with the data presented here, including preamylopectin processing, a role in the metabolism of maltooligosaccharides, and involvement in starch granule initiation.
A previous analysis of DBE activities in isoamylase-type DBE mutants of rice indicated a correlation between regions of the endosperm that accumulated significant amounts of pullulanase-type DBE activity and the presence of starch granules in those particular cells (Nakamura et al., 1997). From these data, the hypothesis was suggested that pullulanase-type DBE, not isoamylase-type DBE, may be the primary determinant of phytoglycogen accumulation. This possibility can now be excluded based on the fact that zpu1-204 endosperm tissue exhibits normal starch structure and quantity in the absence of any pullulanase-type DBE activity. Rather, the principal factor appears to be the combined activity of the two DBEs. A critical level of isoamylase-type DBE activity clearly is required to prevent the accumulation of phytoglycogen. When this enzyme is defective, pullulanase-type DBE may become limiting in determining the amount of phytoglycogen versus granular starch produced. The incomplete ability of ZPU1 to fully compensate for the loss of isoamylase-type DBE may be explained by the limitation of ZPU1's ability to hydrolyze select substrates. This interpretation is entirely consistent with the results of a study in rice (Nakamura et al., 1997), because both isoamylase- and pullulanase-type DBE are affected by the su1 mutations in the lines that were analyzed.
In genetic backgrounds with intact isoamylase-type DBE, pullulanase-type DBE still performs a metabolic function. Small amounts of free glucan polymer in the form of WSP are present in developing wild-type endosperm during the period of starch accumulation (Figure 6), and the concentration of this material is reduced to a statistically significant extent as a result of zpu1-204 (Table 2). Therefore, pullulanase-type DBE activity appears to produce maltooligosaccharides through the hydrolysis of branch linkages in a precursor polymer. The source of the precursor polymer that gives rise to the maltooligosaccharides is not known. One possibility is that trimming of preamylopectin by DBEs produces these polymers (Ball et al., 1996; Myers et al., 2000). A population of glucan chains at the surface of the granule, which is not present in mature starch, has been shown to exist transiently in Arabidopsis leaves (Nielsen et al., 2002). If similar structures exist in developing maize endosperm, ZPU1 could be involved in cleaving some of these chains to generate maltooligosaccharides.
Within the soluble material in the mutant, there is a detectable quantity of α(1→6)-branched polymer with chains ranging from 3 to 10 DP that is not present in the wild type (Figure 6). This observation is consistent with the suggestion that branched oligosaccharides exist transiently in the soluble phase and are cleared by degradative activities, including α(1→6) glucan hydrolase (Zeeman et al., 1998b). Some of the necessary hydrolytic activities are likely to be specific to pullulanase-type DBE, because isoamylase-type DBE is present in zpu1-204 endosperm at apparently normal levels, yet the branched soluble polymer still accumulates.
Starch biosynthesis also was clearly affected in the leaves of the zpu1-204 mutant, as indicated by the altered amylopectin structure and the reduced accumulation of starch in the light phase (Figures 5A and 5B, Table 1). Both of these effects might be explained by a direct role of the ZPU1 protein in this process or by one or more pleiotropic effects. In particular, the pleiotropic effect of zpu1-204 on SBEIIa activity must be considered. Loss of SBEIIa, which is a major form of branching enzyme in the maize leaf, is known to affect the composition and structure of starch (Blauth et al., 2001) (Figures 4 and 5C). The effects of zpu1-204 on the chain length distribution of leaf amylopectin were similar to, but not as severe as, those of the sbe2a::Mu mutation (Figure 5D). These effects are not universal, because amylopectin structure in endosperm starch is not affected by zpu1-204. This fact could be explained by differences in the isoform components of the biosynthetic systems (i.e., starch synthases and SBEs) in these two tissues. Thus, although the zpu1-204 mutation clearly affects leaf starch synthesis, it cannot be concluded that this effect is attributable directly to the loss of ZPU1.
However, there are distinct differences in the effects of zpu1-204 and sbe2a::Mu on chain length distribution that were not expected given the fact that SBEIIa activity is reduced in both instances. Furthermore, the sbe2a::Mu mutation had severe effects on starch composition (Figure 4), whereas zpu1-204 did not. Possibly, the lack of pullulanase-type DBE causes an altered starch structure by itself, which is distinct from that caused by the SBEIIa deficiency. Alternatively, the effect that the zpu1-204 mutation has on SBEIIa, in which an apparently inactive polypeptide is present, might result in a different amylopectin structure than that observed when there is a complete lack of SBEIIa protein.
Analysis of zpu1-204 mutant plants provides additional examples of the numerous pleiotropic interactions that occur among starch biosynthetic enzymes, which seem to involve particularly the DBEs. In maize and rice, molecular defects in isoamylase-type DBE have long been known to secondarily reduce the activity of the pullulanase-type DBE (Pan and Nelson, 1984; Nakamura et al., 1996b). The converse effect, in which the loss of pullulanase-type DBE negatively affects the activity of SU1, does not appear to occur. However, the loss of ZPU1 does cause a defect in the activity of SBEIIa in leaves and endosperm. The same deficiency in SBEIIa also is a secondary effect of a specific mutation in the isoamylase-type DBE, which provides further indirect evidence to support the functional interaction between the two types of DBE. Transcriptional or translational regulation does not account for the effects on SBEIIa, because the polypeptide accumulates to seemingly normal levels in both the zpu1-204 and su1-st mutants. Multiple-subunit complexes must be considered as a possible explanation for allele-specific defects of this type, although little direct information is available at present.
The metabolic processes of starch catabolism and anabolism require that the plant evaluate environmental, temporal, and developmental cues and then regulate an entire suite of enzymes based on the information contained in those signals. This investigation of the zpu1-204 mutation has shown that some enzymes of starch metabolism may have a dual function, being required for periods of both net synthesis and degradation. In addition to the starch phenotypes described here, the specific pleiotropic effects of zpu1-204 on both a starch biosynthetic enzyme (SBEIIa) and a degradative enzyme (β-amylase) indicate that ZPU1 interacts with components of both the assembly and disassembly machineries. Only a small part of the regulation of starch synthesis and degradation involves the control of gene expression at the transcriptional or translational level. Rather, the transitions between the processes of synthesis and degradation likely involve more subtle and responsive changes to signaling mechanisms, perhaps mediated by altered direct interactions between enzymes that recognize Glc homopolymers as substrates.
METHODS
Genetic Analysis and Generation of Plant Material
Plants used for biochemical and molecular analyses were of the BC3S2 generation, which was produced as follows. TUSC (Trait Utility System for Maize) F1 maize (Zea mays) seeds were generated by Pioneer Hi-Bred International (Johnston, IA) by crossing a Mu-active line (i.e., possessing a high Mutator copy number and the autonomous element, MuDR) to a non-Mutator line. PCR screening of DNA pools (see below) was performed on DNA isolated from the leaves of the F1 plants, and they were self-pollinated to give F2 seeds. The F2 seeds were provided by Pioneer, and PCR analysis of leaf DNA samples from progeny plants identified Zpu1/zpu1 heterozygotes. These were crossed to inbred line W64A, and the identification of heterozygotes by PCR and crossing to W64A was repeated three additional times to generate BC3 ears. A heterozygote grown from a BC3 seed was self-pollinated, and seeds from the resulting BC3S1 ear were planted. Homozygous zpu1-204 plants (representing one-fourth of the progeny) were identified by PCR analysis. Self-pollination of those plants generated the BC3S2 kernels used for the analysis of endosperm tissue, and plants grown from those kernels were used for the analysis of leaf tissue. The wild-type controls for both leaves and developing endosperm were from inbred W64A plants. Immature kernels or leaf samples were quick-frozen in liquid nitrogen and stored at −80°C until further analysis.
Plants used for the analysis of leaf starch were grown in a growth chamber in a diurnal cycle that consisted of alternating periods of 8 h of darkness at 25°C and 16 h of light at 28°C with ∼280 μmol·m−2·s−1 mixed fluorescent and incandescent light. The sixth, seventh, eighth, and ninth vegetative leaves (V6 to V9) were harvested at ∼3 weeks after planting for the analysis of starch structure and content. Other plants were grown under field conditions in Ames, Iowa, or Molokai, Hawaii.
The su1-st mutation was characterized in detail in a previous report (Dinges et al., 2001). All su1-st lines used in this study were derived from crosses to the W64A inbred line for a minimum of five generations. The sbe2a::Mu mutation was described in a previous report (Blauth et al., 2001). Adult leaves (after anthesis) from greenhouse-grown plants homozygous for sbe2a::Mu were provided by Mark Guiltinan (Pennsylvania State University, University Park). The sbe2a:: Mu mutation was reported originally to cosegregate with a leaf-senescence phenotype (Blauth et al., 2001). After repeated backcrossing, this phenotype was separated from sbe2a::Mu, and the mutant leaves lacking SBEIIa appeared normal (Blauth et al., 2002).
Genotype Determination and Gene Expression Analysis by PCR
DNA was isolated from fresh or lyophilized maize leaves using cetyl-trimethyl-ammonium bromide extraction (Saghai-Maroof et al., 1984). Total RNA was isolated from endosperm harvested at 20 days after pollination using the TRIzol reagent (catalog No. 15596-026; Invitrogen, Carlsbad, CA) according to the supplier's instructions. TUSC pool screening was performed by Pioneer Hi-Bred International (Bensen et al., 1995). One of the oligonucleotide primers was taken from the Zpu1 cDNA sequence, either pulA (5′-AATCCAAACGCGGACGCAAATGTT-GCTC-3′) (nucleotides 22 to 50 on the forward strand) or pulB (5′-ACTTACATCCTGTGCTGTAGGAGCCCA-3′) (nucleotides 727 to 701 on the reverse strand). The second PCR primer in each reaction was the Mu terminal inverted repeat sequence 9242 (5′-AGAGAAGCCAACGCCA[AT]-CGCCTC[CT]ATTTCGTC-3′).
The primers pulA, pulB, and 9242 also were used for the genotyping of individual plants. The parameters of these PCR procedures were 95°C for 3 min followed by 35 cycles of 95°C for 1 min and 68°C for 2 min. The reactions were performed in the presence of 5% (v/v) DMSO using Platinum Taq polymerase (catalog No. 11304-029; Invitrogen) in the buffer conditions suggested by the supplier with a final MgCl2 concentration of 3.5 mM. Reverse transcriptase PCR amplification of endosperm RNA was performed using the Superscript One-Step reverse transcriptase PCR system (catalog No. 10928-034; Invitrogen) and the pulA/pulB primer pair according to the manufacturer's instructions.
Carbohydrate Extraction and Quantification
Carbohydrate extraction from developing kernels harvested at 20 days after pollination and quantification of Glc, Fru, Suc, granular starch, and water-soluble polysaccharide (WSP) were performed as described previously (Dinges et al., 2001). Mature, dried kernels were analyzed by the same procedure, except that they were first soaked overnight at 50°C in 0.3% (w/v) sodium bisulfite and pericarp and embryo were removed by manual dissection. Leaf starch content over a diurnal cycle was determined according to a previously described procedure (Zeeman et al., 1998a), with slight modifications. Approximately 500 mg of leaf tissue was harvested, weighed, and immediately boiled in 50 mL of 80% (v/v) ethanol. The decolorized leaves then were homogenized in 80% (v/v) ethanol using a mortar and pestle. The homogenate was centrifuged at 1500g for 10 min at room temperature, and the pellet was extracted again with 80% (v/v) ethanol. The pelleted material was suspended in 4 mL of water and boiled for 30 min. Total α-glucan polysaccharide, presumed to be derived nearly entirely from granular starch, was quantified using a commercial assay kit that measures Glc released after digestion with amyloglucosidase (catalog No. E0207748; R-Biopharm, Darmstadt, Germany).
To isolate leaf starch for structural analysis, 5 g of tissue was ground with a mortar and pestle in 500 mM 3-(N-morpholino)-propanesulfonic acid, pH 7.5, 5 mM EDTA, and 10% ethylene glycol. The homogenate was filtered through Miracloth (Calbiochem) and centrifuged at 5000g for 10 min. The supernatant was boiled and stored at −20°C. The pellet was suspended in 1 mL of extraction buffer and 9 mL of Percoll (catalog No. 17-0891-02; Amersham Biosciences, Piscataway, NJ). The Percoll gradient established by centrifugation at 10,000g for 30 min yielded a starch pellet largely free of other cell components. Starch granules were solubilized by boiling in 90% (v/v) DMSO for 30 min and then stored at −20°C until further analysis.
The size distribution of isolated starch grains was estimated using a Coulter Counter (Multisizer II; Beckman Coulter, Fullerton, CA) equipped with a 70-μm aperture. The starch pellet was thoroughly suspended in water. An aliquot was added to the diluent and immediately placed in the instrument.
Amylopectin Structural Analysis
Amylopectin and amylose in the granular starch fraction were separated by gel-permeation chromatography as described previously (Dinges et al., 2001). Chain length distribution was determined by fluorophore-assisted carbohydrate electrophoresis according to a previously published procedure (O'Shea et al., 1998) with slight modifications. The lyophilized amylopectin fraction was suspended at 4 mg Glc equivalents/mL in 30% DMSO and boiled for 10 min. A 10-μL aliquot, containing 200 μg of Glc equivalents, was diluted to a final volume of 50 μL with 50 mM sodium acetate, pH 4.5. Pseudomonas amyloderamosa isoamylase (1 μL, 0.3 units) (catalog No. E-ISAMY; Megazyme International, Bray, Ireland) was added, and the reaction was incubated overnight at 42°C. The mixture was heated in boiling water for 5 min and then centrifuged for 2 min at full speed in a microfuge. A 10-μL sample of the reaction, containing 4 μg of Glc equivalents, was evaporated to dryness in a Speed-Vac (Savant Instruments, Holbrook, NY).
The reducing ends of the liberated oligosaccharide chains were derivatized with the fluorescent compound 8-amino-1,3,6-pyrenetrisulfonic acid (APTS) (catalog No. 09341; Sigma-Aldrich, St. Louis, MO) as follows. The dried sample was suspended in 2 μL of 1 M sodium cyanoborohydride in tetrahydrofuran (catalog No. 29681-3; Sigma-Aldrich) and 2 μL of APTS (0.1 mg/μL in 15% acetic acid). The reaction was incubated overnight at 42°C, diluted with 46 μL of water, vortexed, and centrifuged briefly in a microfuge. A 5-μL aliquot, containing 0.4 μg of Glc equivalents, was added to 195 μL of purified water, and this sample was applied to a Beckman P/ACE capillary electrophoresis instrument. The sample injection parameters were 5 s at 0.5 p.s.i. Separation was accomplished at 23.5 kV in an uncoated capillary using Carbohydrate Separation Gel Buffer N (catalog Nos. 338451 and 477623; Beckman Coulter). Results shown are average chain length distributions of amylopectin isolated from four separate plants. Differences between samples were statistically insignificant.
The structure of WSP from kernels was determined similarly. The only differences were that some samples of WSP were not treated with any debranching enzyme before APTS labeling, whereas others were digested with a combination of the isoamylase described above and 0.05 units of Klebsiella pneumoniae pullulanase (catalog No. P-5420; Sigma-Aldrich).
Germination Analysis
Seeds were surface-sterilized by immersion in 1% sodium hypochlorite for 5 min and then washed three times with sterile water. Fifteen seeds from each of three ears were placed in Petri dishes containing three layers of moist Whatman paper and incubated at 30°C in the dark. The lengths of the 15 cotyledons of each genotype were measured on successive days throughout the incubation period. To measure the amount of starch, roots, cotyledons, and pericarps were removed from three kernels, and the endosperms were dissected and ground with a mortar and pestle. The ground tissue was washed in water, centrifuged at 1500g for 10 min, and suspended in 4 mL of water. For normalization of samples, 1 mL of this suspension was dried and weighed. The remaining 3 mL of the suspension was boiled for 30 min, and the total glucan polysaccharide in the soluble phase was quantified as described above for leaf tissue.
Protein Analysis
To isolate proteins from kernels, leaves, and germinating seeds, 5 g of tissue was ground to a fine powder in liquid nitrogen with a mortar and pestle, and the tissue was suspended in 5 to 7 mL of buffer containing 50 mM Tris acetate, pH 7.5, 10 mM EDTA, and 20 mM DTT. The homogenate was centrifuged at 100,000g for 1 h at 4°C. The supernatant was stored at −80°C. Starch zymogram analysis was performed as described (Dinges et al., 2001). For the pullulan azure activity gel, the resolving gel contained 7% (w/v) acrylamide (29:1 acrylamide:bisacrylamide) and 375 mM Tris-HCl, pH 8.8. The stacking gel contained 4% (w/v) acrylamide and 63 mM Tris-HCl, pH 6.8. Electrophoresis was conducted using ∼50 μg of sample (except where noted) at 4°C and 25 V/cm for 2 h in a Mini-Protean II cell (Bio-Rad) in an electrode buffer of 25 mM Tris, 192 mM Gly, pH 8.8, and 2 mM DTT. At the end of the run, the gel was electroblotted to a polyacrylamide gel of the same size containing 7% (w/v) acrylamide, 1% (w/v) pullulan azure (catalog No. P-0588; Sigma-Aldrich), and 375 mM Tris-HCl, pH 8.8. The transfer was performed overnight at 20 V in the electrode buffer at room temperature.
For zymogram analysis of starch synthase activities, protein samples were separated on a native 6% (w/v) polyacrylamide gel containing 0.2% (w/v) rabbit glycogen (catalog No. G-8876; Sigma-Aldrich) for 3 h at 4°C, incubated overnight, and stained as described (Cao et al., 1999). Gels were photographed immediately after staining or transfer. SDS-PAGE/ immunoblot analysis was performed as described (Beatty et al., 1999). Crude anti-SBEIIa was diluted 1:5000. Anion-exchange fractionation of total soluble kernel extracts was performed using a MonoQ HR 5/5 column (catalog No. 17-0546-01; Amersham Biosciences). The proteins were separated using a linear gradient of 0 to 0.4 M NaCl.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
The GenBank accession numbers for the genes mentioned in this article are AF030882 (Su1), AY172633 (Iso2), AY172634, (Iso3), and AF080567 (Zpu1).
Acknowledgments
We thank Bob Meeley and Pioneer Hi-Bred International for TUSC pool screening and for providing the original zpu1 mutant seeds, Marna Yandeau for mapping the insertion position of several zpu1 alleles, Sam Zeeman and Alison Smith for helpful discussions regarding leaf starch analyses, and Rebekah Marsh for preparation of the anti-SBEIIa antiserum. This work was supported by U.S. Department of Agriculture National Research Initiative Award 99-35304-8642 to M.G.J. and A.M.M, by a U.S. Department of Agriculture National Needs Fellowship in Plant Biotechnology (Grant 98-38420-5838) to J.R.D., and by the Pioneer Maize Genome Fellowship Program (Pioneer Hi-Bred International).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.007575.
References
- Ball, S., Guan, H.P., James, M., Myers, A., Keeling, P., Mouille, G., Buleon, A., Colonna, P., and Preiss, J. (1996). From glycogen to amylopectin: A model for the biogenesis of the plant starch granule. Cell 86, 349–352. [DOI] [PubMed] [Google Scholar]
- Beatty, M.K., Rahman, A., Cao, H., Woodman, W., Lee, M., Myers, A.M., and James, M.G. (1999). Purification and molecular genetic characterization of ZPU1, a pullulanase-type starch-debranching enzyme from maize. Plant Physiol. 119, 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensen, R.J., Johal, G.S., Crane, V.C., Tossberg, J.T., Schnable, P.S., Meeley, R.B., and Briggs, S.P. (1995). Cloning and characterization of the maize An1 gene. Plant Cell 7, 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blauth, S.L., Kim, K.-N., Klucinec, J.D., Shannon, J.C., Thompson, D.B., and Guiltinan, M.J. (2002). Identification of Mutator insertional mutants of starch-branching enzyme 1 (sbe1) in Zea mays L. Plant Mol. Biol. 48, 287–297. [DOI] [PubMed] [Google Scholar]
- Blauth, S.L., Yao, Y., Klucinec, J.D., Shannon, J.C., Thompson, D.B., and Guiltinan, M.J. (2001). Identification of Mutator insertional mutants of starch-branching enzyme 2a (BEIIa) in corn. Plant Physiol. 125, 1396–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton, R.A., Jenner, H., Carrangis, L., Fahy, B., Fincher, G.B., Hylton, C.M., Laurie, D.A., Parker, M., Waite, D., van Wegen, S., Verhoeven, T., and Denyer, K. (2002). Starch granule initiation and growth are altered in barley mutants that lack isoamylase activity. Plant J. 31, 97–112. [DOI] [PubMed] [Google Scholar]
- Burton, R.A., Zhang, X.Q., Hrmova, M., and Fincher, G.B. (1999). A single limit dextrinase gene is expressed both in the developing endosperm and in germinated grains of barley. Plant Physiol. 119, 859–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Imparl-Radosevich, J., Guan, H., Keeling, P.L., James, M.G., and Myers, A.M. (1999). Identification of the soluble starch synthase activities of maize endosperm. Plant Physiol. 120, 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, R., Rosen, E., and Masson, P.H. (1999). Gravitropism in higher plants. Plant Physiol. 120, 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creech, R.G. (1968). Carbohydrate synthesis in maize. Adv. Agron. 20, 275–322. [Google Scholar]
- Critchley, J.H., Zeeman, S.C., Takaha, T., Smith, A.M., and Smith, S.M. (2001). A critical role for disproportionating enzyme in starch breakdown is revealed by a knock-out mutation in Arabidopsis. Plant J. 26, 89–100. [DOI] [PubMed] [Google Scholar]
- Dahlstrom, D.E., and Lonnquist, J.H. (1964). A new allele at the sugary-1 locus in maize. J. Hered. 55, 242–246. [Google Scholar]
- Dinges, J.R., Colleoni, C., Myers, A.M., and James, M.G. (2001). Molecular structure of three mutations at the maize sugary1 locus and their allele-specific phenotypic effects. Plant Physiol. 125, 1406–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, M.G., Robertson, D.S., and Myers, A.M. (1995). Characterization of the maize gene sugary1, a determinant of starch composition in kernels. Plant Cell 7, 417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo, A., Fujita, N., Harada, K., Matsuda, T., Satoh, H., and Nakamura, Y. (1999). The starch-debranching enzymes isoamylase and pullulanase are both involved in amylopectin biosynthesis in rice endosperm. Plant Physiol. 121, 399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laby, R.J., Kim, D., and Gibson, S.I. (2001). The ram1 mutant of Arabidopsis exhibits severely decreased β-amylase activity. Plant Physiol. 127, 1798–1807. [PMC free article] [PubMed] [Google Scholar]
- Longstaff, M.A., and Bryce, J.H. (1993). Development of limit dextrinase in germinated barley (Hordeum vulgare L.). Plant Physiol. 101, 881–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouille, G., Maddelein, M.-L., Libessart, N., Talaga, P., Decq, A., Belrue, B., and Ball, S. (1996). Preamylopectin processing: A mandatory step for starch biosynthesis in plants. Plant Cell 8, 1353–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, A.M., Morell, M.K., James, M.G., and Ball, S.G. (2000). Recent progress toward understanding the biosynthesis of the amylopectin crystal. Plant Physiol. 122, 989–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, Y., Kubo, A., Shimamune, T., Matsuda, T., Harada, K., and Satoh, H. (1997). Correlation between activities of starch de-branching enzyme and alpha-polyglucan structure in endosperms of sugary-1 mutants of rice. Plant J. 12, 143–153. [Google Scholar]
- Nakamura, Y., Umemoto, T., Ogata, N., Kuboki, Y., Yano, M., and Sasaki, T. (1996. a). Starch debranching enzyme (R-enzyme or pullulanase) from developing rice endosperm: Purification, cDNA and chromosomal localization of the gene. Planta 199, 209–218. [DOI] [PubMed] [Google Scholar]
- Nakamura, Y., Umemoto, T., Takahata, T., Komae, K., Amano, E., and Satoh, H. (1996. b). Changes in structure of starch and enzyme activities affected by sugary mutations in developing rice endosperm: Possible role of starch debranching enzyme (R-enzyme) in amylopectin biosynthesis. Physiol. Plant. 97, 491–498. [Google Scholar]
- Nielsen, T.H., Baunsgaard, L., and Blennow, A. (2002). Intermediary glucan structures formed during starch granule biosynthesis are enriched in short side chains: A dynamic pulse labeling approach. J. Biol. Chem. 277, 20249–20255. [DOI] [PubMed] [Google Scholar]
- O'Shea, M.G., Samuel, M.S., Konik, C.M., and Morell, M.K. (1998). Fluorophore-assisted carbohydrate electrophoresis (FACE) of oligosaccharides: Efficiency of labelling and high-resolution separation. Carbohydr. Res. 307, 1–12. [Google Scholar]
- Pan, D., and Nelson, O.E., Jr. (1984). A debranching enzyme deficiency in endosperms of the sugary1 mutants of maize. Plant Physiol. 74, 324–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman, A., Wong, K., Jane, J., Myers, A.M., and James, M.G. (1998). Characterization of SU1 isoamylase, a determinant of storage starch structure in maize. Plant Physiol. 117, 425–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritte, G., Lloyd, J.R., Eckermann, N., Rottmann, A., Kossmann, J., and Steup, M. (2002). The starch-related R1 protein is an alpha-glucan, water dikinase. Proc. Natl. Acad. Sci. USA 99, 7166–7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghai-Maroof, M.A., Soliman, K.M., Jorgensen, R.A., and Allard, R.W. (1984). Ribosomal DNA spacer-length polymorphisms in barley. Proc. Natl. Acad. Sci. USA 81, 8014–8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, S.W., and MacGregor, A.W. (1998). Synthesis of limit dextrinase in germinated barley kernels and aleurone tissues. J. Am. Soc. Brew. Chem. 56, 32–37. [Google Scholar]
- Shannon, J.C., Pien, F.-M., and Liu, K.-C. (1996). Nucleotides and nucleotide sugars in developing maize endosperms. Plant Physiol. 110, 835–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, A.M. (2001). The biosynthesis of starch granules. Biomacromolecules 2, 335–341. [DOI] [PubMed] [Google Scholar]
- Tabata, S., et al. (2000). Sequence and analysis of chromosome 5 of the plant Arabidopsis thaliana. Nature 408, 823–826. [DOI] [PubMed] [Google Scholar]
- Wu, C., Colleoni, C., Myers, A.M., and James, M.G. (2002). Enzymatic properties of ZPU1, the maize pullulanase-type starch debranching enzyme. Arch. Biochem. Biophys. 406, 21–32. [DOI] [PubMed] [Google Scholar]
- Yu, J., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296, 79–92. [DOI] [PubMed] [Google Scholar]
- Yu, T.S., et al. (2001). The Arabidopsis sex1 mutant is defective in the R1 protein, a general regulator of starch degradation in plants, and not in the chloroplast hexose transporter. Plant Cell 13, 1907–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman, S.C., Northrop, F., Smith, A.M., and Rees, T. (1998. a). A starch-accumulating mutant of Arabidopsis thaliana deficient in a chloroplastic starch-hydrolysing enzyme. Plant J. 15, 357–365. [DOI] [PubMed] [Google Scholar]
- Zeeman, S.C., Umemoto, T., Lue, W.L., Au-Yeung, P., Martin, C., Smith, A.M., and Chen, J. (1998. b). A mutant of Arabidopsis lacking a chloroplastic isoamylase accumulates both starch and phytoglycogen. Plant Cell 10, 1699–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Z.P., Hylton, C.M., Rossner, U., and Smith, A.M. (1998). Characterization of starch-debranching enzymes in pea embryos. Plant Physiol. 118, 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]