Abstract
The COP9 signalosome (CSN) is an evolutionarily conserved protein complex that resembles the lid subcomplex of proteasomes. Through its ability to regulate specific proteasome-mediated protein degradation events, CSN controls multiple aspects of development. Here, we report the cloning and characterization of AtCSN2, the last uncharacterized CSN subunit from Arabidopsis. We show that the AtCSN2 gene corresponds to the previously identified FUS12 locus and that AtCSN2 copurifies with CSN, confirming that AtCSN2 is an integral component of CSN. AtCSN2 is not only able to interact with the SCFTIR1 subunit AtCUL1, which is partially responsible for the regulatory interaction between CSN and SCFTIR1, but also interacts with AtCUL3, suggesting that CSN is able to regulate the activity of other cullin-based E3 ligases through conserved interactions. Phylogenetic analysis indicated that the duplication and subsequent divergence events that led to the genes that encode CSN and lid subunits occurred before the divergence of unicellular and multicellular eukaryotic organisms and that the CSN subunits were more conserved than the lid subunits during evolution. Comparative analyses of the subunit interaction of CSN revealed a set of conserved subunit contacts and resulted in a model of CSN subunit topology, some aspects of which were substantiated by in vivo cross-link tests.
INTRODUCTION
The COP9 signalosome (CSN) is an evolutionarily conserved protein complex that was identified initially in Arabidopsis as a repressor of light-regulated development (Wei and Deng, 1992; Wei et al., 1994; Chamovitz et al., 1996). Genetic screening aimed at identifying Arabidopsis mutants that display an altered developmental response to light led to the identification of the class of mutants known as pleiotropic cop/det/fus mutants (Hardtke and Deng, 2000). These mutants are characterized by a constitutive photomorphogenic phenotype, regardless of the light condition used, and in the light accumulate anthocyanin, which is responsible for their purple color (fusca) (Wei and Deng, 1996). Furthermore, the severe cop/det/fus mutations lead to growth arrest and lethality after the seedling stage, indicating that their corresponding genes control, in addition to photomorphogenesis, other essential developmental programs. At present, 10 distinct pleiotropic COP/DET/FUS loci have been identified and characterized, 5 of which have been shown to encode CSN subunits (Wei et al., 1994; Staub et al., 1996; Karniol et al., 1999; Serino et al., 1999; Peng et al., 2001b). CSN contains a total of eight subunits, is expressed ubiquitously in plant tissues, and is remarkably conserved throughout evolution, indicating that it plays a fundamental role in higher eukaryotes (Wei and Deng, 1999). This conclusion is supported by reports that CSN loss-of-function mutations lead to cell cycle arrest in fission yeast and to postembryonic growth arrest in Drosophila (Freilich et al., 1999; Mundt et al., 1999).
An increasing amount of evidence suggests that CSN is involved in the regulation of the ubiquitin-proteasome system, one of the major cellular pathways responsible for protein degradation (Schwechheimer and Deng, 2001). A key step on this pathway is the attachment of a polyubiquitin chain to the substrate that will be degraded. This occurs through an enzymatic cascade that includes three separate enzymatic activities designated E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme), and E3 (ubiquitin ligase) (Hershko and Ciechanover, 1998). The E3 class of enzymes is very diverse and is responsible for both selecting the appropriate substrate and promoting the formation of the ubiquitin chain. Once the substrate is ubiquitylated, it is recognized and degraded by the proteasome. The proteasome is composed of a 20S catalytic particle and of a 19S regulatory particle. The 19S regulatory particle consists of two subcomplexes, the lid and the base (Glickman et al., 1998). The lid is necessary for the recognition of the ubiquitin chain and the subsequent deubiquitylation of the substrate (Verma et al., 2001), which is unfolded by the base and injected into the 20S catalytic particle for proteolysis (Glickman, 2000).
Remarkably, CSN exhibits a striking homology, in both complex composition and subunit sequence, with the lid of the proteasome (Wei et al., 1998). In addition, CSN is necessary for the regulation of E3 activity. For example, CSN interacts with the plant ubiquitin ligase SCFTIR1 and is necessary for the cleavage of the ubiquitin-like protein RUB1/NEDD8 from the CUL1 subunit of SCF ubiquitin ligases in human and plants (Lyapina et al., 2001; Schwechheimer et al., 2001, 2002; Cope et al., 2002). The RUB1/NEDD8 modification enhances SCF activity, probably through the recruitment of its cognate E2 (Kawakami et al., 2001). The CUL1 subunit of SCF belongs to the cullin family, which contains six members in human (Kipreos et al., 1996). Only hCUL1, hCUL2, and hCUL5 have been shown to be part of functional ubiquitin ligase complexes (Lyapina et al., 1998; Stebbins et al., 1999; Querido et al., 2001). However, all six cullins have been reported to interact with the same ring finger protein, RBX1, and to have the potential of being modified by RUB1/NEDD8, suggesting that all of them could form ubiquitin ligase complexes (Hori et al., 1999; Ohta et al., 1999). Interestingly, in human, all six cullins can copurify with CSN (Lyapina et al., 2001). By removing RUB1/NEDD8 from the cullin, CSN could trigger the termination of the polyubiquitylation process and the release of the substrate from a wide array of ubiquitin ligases. This finding is in agreement with the recent report that CSN mutants accumulate higher amounts of polyubiquitylated proteins (Peng et al., 2001a).
Together, these results suggest that CSN acts as a central regulator of the protein degradation process, ensuring that a specific substrate will be modified by ubiquitin and properly degraded by the proteasome.
To reveal the molecular mechanism of CSN action, a thorough analysis of its composition and structure is needed. Here, we report the cloning and characterization of subunit 2 (AtCSN2) of the COP9 signalosome. We show that AtCSN2 is encoded by FUS12, one of the Arabidopsis COP/DET/FUS loci. Furthermore, we report that AtCSN2 can make direct contact with distinct members of the cullin family from Arabidopsis. We propose that this is a conserved interaction module through which the COP9 signalosome can regulate multiple cullin-based E3 ligases. This study concluded the characterization of the Arabidopsis CSN subunits. It allowed us to investigate the phylogenetic relationships between CSN and the structurally related lid of the proteasome and to perform a comprehensive interaction analysis among all subunits.
RESULTS
Cloning and Molecular Characterization of the AtCSN2 Gene
To clone the gene that encodes subunit 2 of the Arabidopsis CSN, we used the CSN2 peptide sequences obtained from the purified cauliflower CSN (Serino et al., 1999) to search the Arabidopsis database (http://www.Arabidopsis.org). A partial EST clone (H4H12T7) was retrieved. Using specific primers, we performed a 5′ rapid amplification of cDNA ends analysis to obtain the 5′ portion of the gene (see Methods). Meanwhile, we were able to retrieve a full-length genomic clone encompassing the complete open reading frame for AtCSN2 from the Arabidopsis database. According to the sequence of the genomic clone and our 5′ rapid amplification of cDNA ends analysis, we amplified by reverse transcription PCR the full-length cDNA for AtCSN2.
The full coding region of the cDNA for AtCSN2 is 1317 bp long and encodes a protein of 439 amino acids with a predicted molecular mass of 51 kD. This molecular mass is slightly smaller than our predicted size for the cauliflower CSN2, which runs in SDS-PAGE with an apparent molecular mass of 54 kD (Serino et al., 1999). Two possible explanations for this discrepancy can be hypothesized: as a result of its conformation, the cauliflower CSN2 protein could run abnormally in SDS-PAGE. Alternatively, the cauliflower CSN2 protein could be modified post-translationally and thus have a molecular mass higher than predicted. Indeed, potential phosphorylation and glycosylation sites are found within the AtCSN2 amino acid sequence.
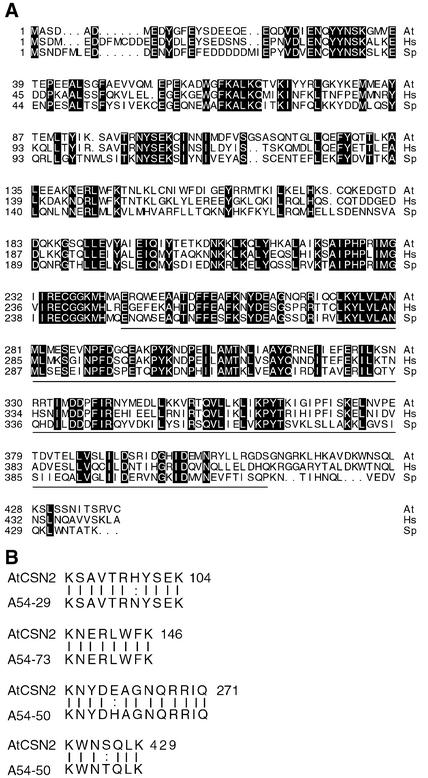
The AtCSN2 protein is closely related to its homologs from human (hCSN2; 60% identity) and Schizosaccharomyces pombe (SpCSN2; 46% identity). In fact, CSN2 and CSN5 are the two most conserved subunits of the complex (Figure 1A). Interestingly, like all other CSNs with the exception of CSN5, we found no AtCSN2 homolog in the genome of Saccharomyces cerevisiae. As shown in Figure 1A, the identity among CSN2 subunits is particularly high in the C-terminal half of the protein, except for the last few amino acids. This conserved region includes the PCI domain (Proteasome-COP9 complex-eIF3), which encompasses amino acids 244 to 409. The PCI domain is a protein motif found in the other five CSN subunits and in components of the lid and the eIF3 complex (Hofmann and Bucher, 1998). Arabidopsis and human CSN2 also contain a sequence that has homology with a TPR region (amino acids 42 to 96), which could be a site of protein–protein interaction (Blatch and Lassle, 1999). Four peptides sequenced from the cauliflower CSN2 display almost exact matches with AtCSN2 (Figure 1B).
Figure 1.
Comparison of the AtCSN2 Sequence with Its Homologs from Other Organisms.
(A) Protein sequences of AtCSN2 (At) and its homologs from human (Hs) and S. pombe (Sp). Boxed residues indicate amino acid identity. The underlined sequence corresponds to the PCI domain of AtCSN2. Numbers at left indicate the positions of the first amino acid residues in each row.
(B) Comparison of peptide sequences from cauliflower CSN2 with the corresponding AtCSN2 protein sequences. Vertical bars indicate identity. Numbers at left of the peptide sequences indicate HPLC fraction numbers. The numbers for the Arabidopsis peptides indicate the last amino acid positions in each row.
The AtCSN2 Gene Maps to the FUS12 Locus, Whose Mutants Contain Molecular Lesions in the Gene
Mutations in seven pleiotropic COP/DET/FUS loci result in the apparent absence of CSN in Arabidopsis (Kwok et al., 1998; Wei and Deng, 1999). Five of those loci (COP9, FUS6/COP11, FUS5, COP8/FUS4, and FUS11) have been demonstrated to encode subunits of Arabidopsis CSN (Wei et al., 1994; Staub et al., 1996; Karniol et al., 1999; Serino et al., 1999; Peng et al., 2001b). Only two COP/DET/FUS genes (COP16 and FUS12) remain to be cloned, and these do not accumulate detectable CSN (Kwok et al., 1998). Therefore, it is possible that one of the two could encode AtCSN2. Indeed, the genomic clone that contains AtCSN2 is very close to the FUS12 locus, which has been mapped to position 61.2 centimorgan on chromosome II of the Arabidopsis genome (Kwok et al., 1996).
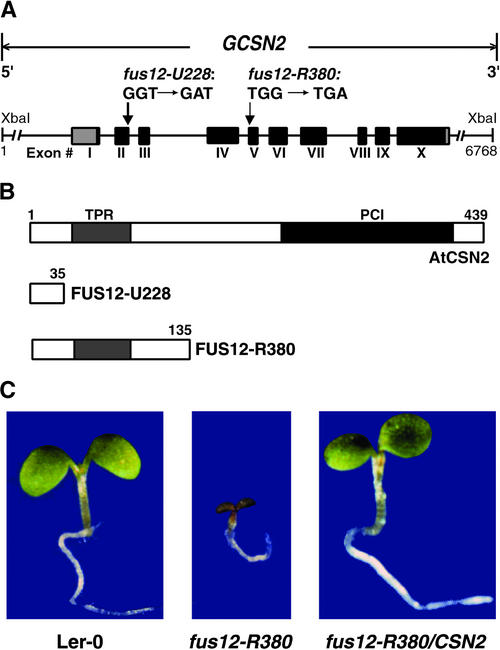
To determine whether the FUS12 gene encodes AtCSN2, we sought to identify mutations in the AtCSN2 sequence in two available alleles of fus12: fus12-R380 and fus12-U228 (Miséra et al., 1994; Kwok et al., 1998). The cDNA and genomic sequences of the AtCSN2 gene were amplified by PCR from the homozygous mutants and their corresponding wild-type ecotype (see Methods). Sequence analysis of the PCR products revealed a point mutation (transition from G to A) in the AtCSN2 gene of the fus12-U228 mutant (Figure 2A). This mutation removes the splice site between exon II and intron III, leaving the unspliced intron III in the mature mRNA and leading to premature termination of the open reading frame. The resulting mutant FUS12-U228 protein has only the first N-terminal 35 amino acids followed by a premature stop codon (Figure 2B). Similarly, sequencing of the fus12-R380 allele identified a single base pair substitution at the last nucleotide position of intron IV, introducing a premature stop. The predicted mutant version of the protein is only 135 amino acids long (FUS12-R380; Figure 2B).
Figure 2.
The FUS12 Locus from Arabidopsis Corresponds to the Gene That Encodes AtCSN2.
(A) Genomic structure of the AtCSN2 locus and molecular nature of the mutations found in the fus12-U228 and fus12-R380 alleles. Introns are shown as lines; boxes and Roman numerals indicate exons. The 5′ and 3′ untranslated regions are shown by gray boxes, and the protein-encoding region is denoted by black boxes. A genomic fragment containing the complete genomic sequence of AtCSN2 together with its own promoter is indicated at top (GCSN2) and was used for the genetic complementation experiment. The two inserts are the mutated sequences found in fus12-R380 and fus12-U228. Numbers indicate base pairs
(B) Wild-type and mutant versions of AtCSN2 proteins. The light gray boxes indicate the TPR homology region; the dark gray box indicates the location of the PCI domain. Numbers indicate amino acid positions.
(C) Seedling phenotype of Landsberg wild type (Ler-0), fus12R380, and fus12-R380/CSN2, a representative line obtained by transforming the fus12-R380 mutant with GCSN2, indicating complementation of the mutant phenotype. All seedlings were grown in the light for 5 days.
Both fus12 alleles used for this analysis are lethal after the seedling stage and do not accumulate CSN (Kwok et al., 1998), indicating that the PCI domain is critical for the stability of AtCSN2 and of the whole complex.
Expression of AtCSN2 Rescues the Phenotype of the fus12 Mutant
To further confirm that AtCSN2 is encoded by the FUS12 gene, a genomic fragment containing the complete AtCSN2 open reading frame together with its own promoter was introduced in the fus12-R380 background to test functional complementation (GCSN2; Figure 2A). Because the homozygous fus12-R380 mutant is lethal, the transgene was transformed into heterozygous plants.
The stably transformed plants heterozygous for both fus12-R380 and the transgene were selected, and their progeny were examined. In the absence of phenotype rescue, a 3:1 ratio of wild-type to mutant seedlings would be expected from the progeny of a selfed plant heterozygous for the fus12-R380 mutation and the transgene. If the AtCSN2 transgene rescues the mutant phenotype, the ratio of wild-type to mutant plants in the progeny should be 15:1 rather than 3:1, provided that the transgene is at a single locus unlinked to FUS12. Table 1 summarizes the segregation analyses of the T2 generation for six selected lines with a single T-DNA insertion per genome (3:1 ratio of resistant to sensitive). The wild-type to mutant ratios obtained are very similar to the 15:1 expected in the case of genetic complementation. Figure 2C shows that the phenotype of the fus12 mutant was rescued completely by the introduction of the AtCSN2 wild-type genomic clone. This result confirms that FUS12 encodes AtCSN2.
Table 1.
Phenotype Complementation of the fus12 Mutation by Genomic AtCSN2
| Line No. | Resistant | Sensitive | Resistant:Sensitive Ratio | Wild Type | fusca | Wild-Type:fusca Ratio |
|---|---|---|---|---|---|---|
| T2.1 | 90 | 30 | 3.0:1 | 90 | 7 | 14.8:1.1 |
| T2.3 | 80 | 28 | 2.9:1 | 110 | 9 | 14.7:1.2 |
| T2.4 | 94 | 32 | 2.9:1 | 150 | 11 | 14.9:1.0 |
| T2.6 | 85 | 27 | 3:0.8 | 90 | 8 | 14.6:1.3 |
| T2.10 | 105 | 36 | 2.9:1 | 115 | 9 | 14.8:1.1 |
| T2.11 | 94 | 31 | 3.0:1 | 120 | 10 | 14.7:1.2 |
The segregation ratios were calculated on T2 seedlings. Resistant and sensitive ratios were calculated according to the ability of the seedlings to survive on gentamycin-containing Murashige and Skoog (1962) medium (see Methods). The mutant (fusca) and wild-type phenotypes were scored at 10 days after germination on Murashige and Skoog (1962) medium without antibiotics (see Methods).
AtCSN2 Is an Integral Component of the COP9 Signalosome
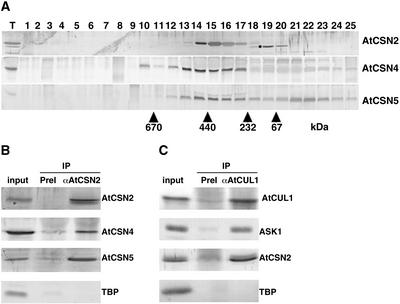
Although CSN can accumulate stably only if all eight subunits are present, some of its single subunits can be stable as monomers, as has been demonstrated for CSN5 and CSN7 (Karniol et al., 1998; Kwok et al., 1998). CSN1, CSN3, CSN4, CSN6, and CSN8 appear to be stable only within the complex (Wei et al., 1994; Staub et al., 1996; Serino et al., 1999; Peng et al., 2001a, 2001b). To ascertain whether AtCSN2 cofractionates with the CSN and whether it exists in any other form, we raised polyclonal antibodies against the full-length AtCSN2 protein and performed gel-filtration analysis of the total protein extracts from light-grown seedlings (see Methods). The elution profiles of AtCSN2 and other representative CSN subunits are shown in Figure 3A. Like most CSN subunits, AtCSN4 is present only in the complex form, whereas AtCSN5 exists in both monomeric and complex forms. AtCSN2 cofractionates with AtCSN4 and with the complexed form of AtCSN5, indicating that AtCSN2 is a component of CSN and that it is stable only when associated with the complex.
Figure 3.
AtCSN2 Is a Subunit of the COP9 Signalosome.
(A) Gel filtration analysis of AtCSN2 and comparison with the elution profiles of two other CSN subunits, AtCSN4 and AtCSN5. Total soluble protein extracts from 5-day-old light-grown Columbia seedlings were separated by gel filtration and subjected to immunoblot analysis with antibodies to AtCSN2 (top gel; the dot indicates a cross-reacting protein), AtCSN4 (middle gel), and AtCSN5 (bottom gel). Lane T indicates the total extract; lanes 1 to 25 indicate the gel filtration elution fractions. Numbers below the arrowheads indicate the positions of molecular mass markers (kD).
(B) AtCSN2 coimmunoprecipitates with other subunits of the COP9 signalosome. Total soluble protein extracts were mixed with AtCSN2 antibodies coupled to protein A beads (see Methods), and the immunoprecipitates (IP) were separated by SDS-PAGE. Antibodies used for immunoprecipitation are indicated at top, and antibodies used in the immunoblots are listed at right. PreI represents the preimmune serum from the same rabbit before immunization.
(C) AtCSN2 coimmunoprecipitates with subunits of SCFTIR1. SCF complexes and their associated proteins were immunoprecipitated from total Arabidopsis protein extracts with AtCUL1 antibodies and subjected to immunoblot analysis with the antibodies listed at right. TBP antibodies were used to verify the specificity of the interactions.
To further substantiate that AtCSN2 is part of CSN, we immunoprecipitated AtCSN2 from crude cell extracts and examined the immunoprecipitated protein by immunoblot analysis for the presence of other CSN subunits. As shown in Figure 3B, the AtCSN2 antibody, but not the preimmune serum, was able to immunoprecipitate AtCSN4 and AtCSN5 and, in a reciprocal experiment, was immunoprecipitated by AtCSN4 antibodies (data not shown). In addition, an AtCUL1 antibody immunoprecipitated the SCFTIR1 subunit ASK1 and AtCSN2 (Figure 3C). These results indicate that AtCSN2 is associated physically with CSN in vivo and that it is present only in the complex form.
AtCSN2 represents the last subunit of Arabidopsis CSN to be characterized, so this study concludes the genetic and molecular characterization of all eight AtCSN subunits. Table 2 contains a summary of all eight AtCSN subunits and their identities to their respective human counterparts.
Table 2.
Summary of the Eight AtCSN Subunits and Their Corresponding Genes
| Subunit Number | Designated Name | Gene/Locus Name | Identity to Human (%) | Reference |
|---|---|---|---|---|
| 1 | AtCSN1 | FUS6/COP11 | 44.7 | Staub et al. (1996) |
| 2 | AtCSN2 | FUS12/COP12 | 61.1 | This work |
| 3 | AtCSN3 | FUS11/COP13 | 42.0 | Peng et al. (2001b) |
| 4 | AtCSN4 | COP8/FUS4 | 49.5 | Serino et al. (1999) |
| 5 | AtCSN5 | AJH1 and AJH2 | 62.0 | Kwok et al. (1998) |
| 6 | AtCSN6 | AtCSN6a and AtCSN6b | 39.8 | Peng et al. (2001a) |
| 7 | AtCSN7 | FUS5/COP15 | 34.4 | Karniol et al. (1999) |
| 8 | AtCSN8 | COP9/FUS7/FUS8 | 32.3 | Wei et al. (1994) |
AtCSN2 Interacts Directly with Arabidopsis Cullin3
As mentioned above, it has been shown that CSN can copurify with all six human cullins and that hCSN2 interacts directly with hCUL1 (Lyapina et al., 2001). Because AtCSN2 has been demonstrated to interact with AtCUL1 in the two-hybrid system (Schwechheimer et al., 2001), we hypothesized that, like human CSN, Arabidopsis CSN might be able to bind distinct cullin-containing complexes through a direct cullin–AtCSN2 interaction.
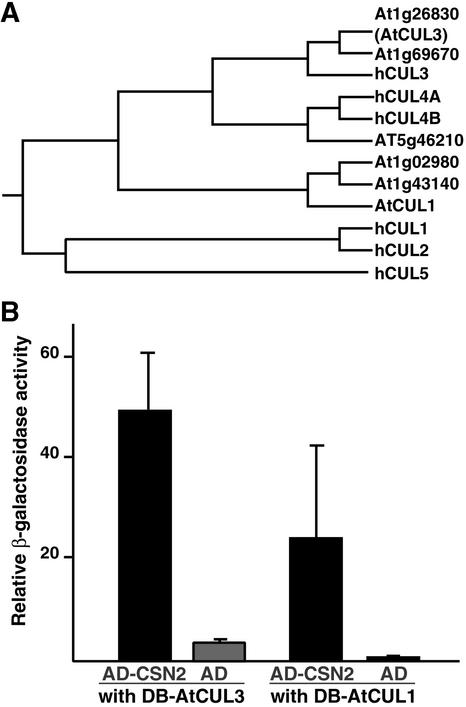
To test this hypothesis, we searched the Arabidopsis genome database (http://www.Arabidopsis.org) for Arabidopsis homologs of the human cullins. A recent sequence similarity search revealed that the Arabidopsis genome contains 11 cullin-related genes (Shen et al., 2002). In our analysis, we selected the six clones that displayed the closest homology with the six human cullins, according to a Basic Local Alignment Search Tool (BLAST) search using the human cullin sequences as queries. Figure 4A illustrates the results of our phylogenetic comparison between human and Arabidopsis cullins. The phylogenetic tree divides into three major branches. The first branch contains the human cullins hCUL1, hCUL2, and hCUL5, which do not seem to have any close homolog in Arabidopsis. The second branch contains only three Arabidopsis cullins: the previously characterized AtCUL1 (Gray et al., 1999) and two close homologs. The third branch is divided into two subgroups: the first contains the human cullins hCUL4A and hCUL4B and their Arabidopsis homolog, and the second contains hCUL3 and two highly homologous Arabidopsis proteins. We decided to focus our studies on the Arabidopsis CUL3 homolog At1g26830 (renamed AtCUL3) and went on to test its possible interaction with AtCSN2. We chose AtCUL3 because of its sequence divergence from AtCUL1 and its close homology with hCUL3, which has been shown to coimmunoprecipitate with the human CSN (Lyapina et al., 2001). We took advantage of a LexA-based yeast two-hybrid assay to assess possible interactions between AtCUL3 and AtCSN2 (see Methods). As a positive control, we used AtCUL1, which has been reported to interact with AtCSN2. These results are shown in Figure 4B. Remarkably, AtCUL3 interacted with AtCSN2 more strongly than AtCUL1. Therefore, CSN binds, and possibly regulates, different cullin-based E3 ligases in a conserved manner through direct contacts between AtCSN2 and cullins.
Figure 4.
AtCSN2 Interacts with Different Members of the Cullin Family.
(A) Phylogenetic relationship of the six human cullins and their putative Arabidopsis homologs. The tree was generated using the CLUSTAL algorithm in the Megalign program (DNASTAR).
(B) AtCSN2 interacts with AtCUL1 and AtCUL3. The interactions between AD-AtCSN2 and AtCUL1 or AtCUL3 fused to the DB are shown by black bars. The background controls (AtCUL1 or AtCUL3 interacting with AD alone) are indicated with gray bars. Numbers on the y axis are average values of at least six independent transformants. Error bars represent sd.
Phylogenetic Analysis of CSN and Lid Subunits
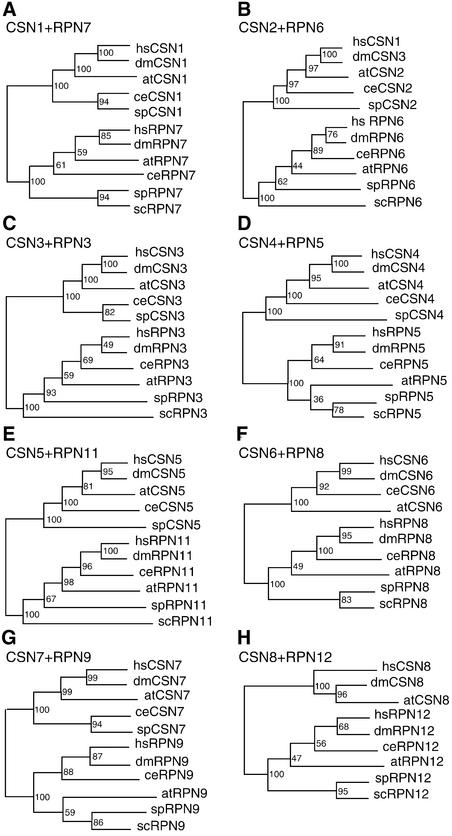
CSN exhibits a striking similarity, in terms of subunit composition and overall structure, to the lid subcomplex of the proteasome (Glickman et al., 1998). Furthermore, each CSN subunit is similar to a specific lid subunit. Therefore, it has been suggested that the two complexes might have evolved from a common ancestor. The availability of complete sets of mammalian and plant CSN subunits prompted us to investigate the evolutionary relationships of the lid and the CSN. Therefore, we conducted a phylogenetic analysis of all eight CSN subunits and their lid paralogs by comparing their amino acid sequences from representative organisms. The sequences used for the analysis are indicated in the supplemental data online, and the results obtained from this analysis are shown in Figure 5. The phylogenetic trees for each CSN subunit and its corresponding lid subunit (RPN) always consist of two clades. It is important to note that, for every tree, the subunits from one of the two protein complexes, although from different organisms, including yeast, are grouped in the same clade. Therefore, the duplication and subsequent divergence events that led to the genes that encode CSN and lid subunits occurred before the divergence of unicellular and multicellular eukaryotic organisms. In our analysis, every tree reveals the same branching pattern and thus reflects the evolutionary history of the organisms selected. For instance, the yeast sequences (budding yeast and/or fission yeast), which are the most diverged, always lie at the base of the clade, whereas the sequences from Drosophila and mammals are grouped together at the top of the clade. The sequences from Caenorhabditis elegans, however, are not positioned stably in all of the trees, probably because their amino acid substitution rates for each subunit differ from those of other organisms.
Figure 5.
Evolutionary Relationships of CSN and Lid Subunits (RPN) from Selected Organisms.
Numbers above the branches indicate support values. The GenBank accession numbers for the proteins listed are given in the supplemental data online. at, Arabidopsis thaliana; ce, Caenorhabditis elegans; dm, Drosophila melanogaster; hs, Homo sapiens; sc, Saccharomyces cerevisiae; sp, Schizosaccharomyces pombe.
We also noticed that the branching patterns among the eight RPN subunits of the lid differ slightly. For example, the budding and fission yeast are clustered together as a basal group in the RPN7, RPN8, and RPN12 trees, whereas only budding yeast sequences are found at the base of the branch in the RPN3, RPN6, and RPN11 trees. In RPN5 and RPN9, Arabidopsis is clustered together with budding and fission yeast. These differences might be attributable to a slightly uneven evolutionary rate among the sequences of individual subunits, possibly reflecting their different functions. One important point from our sequence comparison is that the CSN subunits are more conserved than the lid subunits, which implies that CSN subunits either originated later or were exposed to greater selective pressure than the lid subunits. More sequence information from other organisms, especially from the basal unicellular organisms, is needed to reveal the complete evolutionary history of CSN and lid subunits.
Pair-Wise Yeast Two-Hybrid Interaction Analysis of the Eight COP9 Signalosome Subunits from Arabidopsis and Mammals
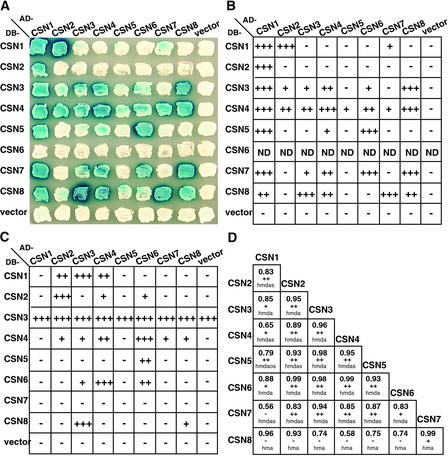
A key issue lacking from our current understanding of CSN is its full complement of subunit–subunit contacts and the three-dimensional topology of the complex. In an effort to map CSN topology, several attempts have been made that used electron microscopy, the yeast two-hybrid assay, and in vitro binding assays (Kapelari et al., 2000; Fu et al., 2001). Previous yeast two-hybrid studies have been used successfully to reveal interactions among several known CSN subunits. For instance, we have shown that AtCSN4, AtCSN5, and AtCSN6 are capable of interactions with other specific CSN subunits, whereas an extensive interaction study among selected CSN subunits has revealed the existence of a central core within the complex composed of CSN4, CSN5, CSN6, and CSN7 (Kwok et al., 1998; Serino et al., 1999; Fu et al., 2001; Peng et al., 2001a). With the cloning of the last subunit, we performed a comprehensive pair-wise interaction screen of all eight CSN subunits from Arabidopsis (Figure 6A). In our assay, all eight subunits were fused to both the LexA DNA binding domain (DB) and the activation domain (AD). The constructs then were transformed into yeast strains suitable for the interaction assay (see Methods).
Figure 6.
Pair-wise Interaction Analysis among CSN Subunits.
(A) Arabidopsis CSN subunit interactions detected by yeast two-hybrid assay. The eight CSN subunits from Arabidopsis were tested for potential interactions in both configurations (DB and AD). Dark blue color indicates lacZ production and thus protein interaction.
(B) Colonies that tested positive for interactions in (A) were subjected to quantitative assays to verify the strength of the binding. − and + indicate the fold increase of β-galactosidase activity over the empty vector: −, 1-fold; +, 2- to 9-fold; ++, 10- to 49-fold; +++, 50- to 99-fold. ND, not determined.
(C) An experiment similar to the one shown in (B) was performed with mammalian CSN subunits.
(D) Correlation analysis of CSN subunits. Numbers indicate the Pearson's correlation coefficient. The signs below the numbers represent the statistical significance of the correlation: −, no significant correlation (P > 0.05); +, significant correlation (P < 0.05); ++, highly significant correlation (P < 0.01). Letters indicate the organisms used for the analysis: a, Arabidopsis thaliana; d, Drosophila melanogaster; h, Homo sapiens; o, Oryza sativa; s, Schizosaccharomyces pombe.
Subunit interactions were quantified using a lacZ reporter gene. Positive interactions in the plate assay (Figure 6A) were tested again with a liquid β-galactosidase activity assay (Figure 6B) (see Methods). All of the protein constructs examined, with the exception of DB-AtCSN6, expressed the predicted fusion proteins in yeast at similar levels, as determined by immunoblot analysis (data not shown). DB-AtCSN6 was found to be lethal to yeast; the only colonies recovered from the DB-AtCSN6 transformation plate were white in all assays and did not express any LexA fusion protein. Therefore, with the exception of DB-AtCSN6, the observed differences in reporter expression should indicate the relative strength of the interaction. In summary, 23 interacting pairs were identified from this comprehensive assay: CSN1–CSN1, CSN1–CSN2, CSN1–CSN3, CSN1–CSN4, CSN1–CSN5, CSN1–CSN7, CSN1–CSN8, CSN2–CSN3, CSN2–CSN4, CSN3–CSN3, CSN3–CSN4, CSN3–CSN6, CSN3–CSN7, CSN3–CSN8, CSN4–CSN4, CSN4–CSN5, CSN4–CSN6, CSN4–CSN7, CSN4–CSN8, CSN5–CSN6, CSN6–CSN7, CSN7–CSN8, and CSN8–CSN8. Based on these data, we confirmed all previously reported results (Serino et al., 1999; Fu et al., 2001; Peng et al., 2001a) and detected six new interactions. For instance, we were able to test the binding ability of AtCSN2 for the first time and found that it interacts very strongly with AtCSN1 and, less intensely but very reproducibly, with AtCSN4. Although the AtCSN1–AtCSN2 interaction seems to be very well conserved, because it has been found in fission yeast and human as well (Mundt et al., 1999; Tsuge et al., 2001), the interaction between AtCSN4 and AtCSN2 is novel and has not been found for the corresponding subunits of any other organism tested. Remarkably, some of the interactions were demonstrated in only one of the two possible configurations. This finding could be attributable to misfolding of the proteins when fused to either the DB or the AD moiety, resulting in fusion proteins that cannot function in the yeast interaction assay.
To examine the evolutionary conservation of CSN interactions, we performed a similar experiment with the mammalian CSN subunits and tested all of their possible pair-wise interactions (see Methods). The average results from the liquid β-galactosidase activity assays for all combinations are shown in Figure 6C. Because the DB-CSN3 fusion protein showed transactivation activity by itself, the interaction of CSN3 with other subunits was tested only in the AD-CSN3 configuration. For the other seven subunits, the pair-wise interactions were examined in both vector configurations. All of the proteins were expressed at comparable levels in yeast, as judged by immunoblot analysis of the total yeast protein extracts (data not shown). The results from this comprehensive yeast two-hybrid assay confirmed 11 pair-wise interactions and 2 self-interactions already found for the Arabidopsis CSN subunits and revealed 2 new self-interactions. In summary, the subunit–subunit interactions among mammalian CSN subunits are as follows: CSN1–CSN2, CSN1–CSN3, CSN1–CSN4, CSN2–CSN4, CSN2–CSN6, CSN3–CSN4, CSN3–CSN6, CSN3–CSN8, CSN4–CSN6, CSN4–CSN7, CSN4–CSN8, and CSN5–CSN6. In addition, self-interactions were found for four subunits: CSN2, CSN4, CSN6, and CSN8, suggesting that they could form dimers or oligomers within the complex or mediate potential CSN–CSN interactions. It is worth noting that, although the CSN2 self-interaction was not detected for AtCSN2, it was clearly detected in mammals. In conclusion, even though we found fewer positive interacting pairs with the mammalian subunits, the results from the two yeast two-hybrid matrices complement and largely overlap each other.
Interacting Pairs among CSN Subunits Can Be Identified by Correlation Analysis of Their Evolutionary Rates
The data obtained for the interactions of mammalian and Arabidopsis subunits demonstrate that the topological architecture of CSN likely remained conserved throughout evolution. This finding suggests that it would be feasible to compare the interacting pairs among different organisms to ascertain their conservation and coevolution. Proteins that interact are known to coevolve, because molecular interactions require precise structures that impose constraints on the evolutionary rate (Fraser et al., 2002). If two proteins are correlated in their functions, the changes of amino acids in one protein will cause changes in the other. Therefore, to substantiate the CSN subunit–subunit interactions, we measured the correlation of the evolutionary rates of the genes that encode CSN subunits. A similar approach has been used to identify interacting proteins in other systems (He and Wu, 1993). To this end, we compared the ratio of nonsynonymous and synonymous substitution rates (dN/dS) of the CSN subunits from selected organisms (see Methods). dN and dS represent the number of nonsynonymous and synonymous substitutions per site, respectively, between two DNA sequences. The results of this analysis are shown in Figure 6D, where the numbers correspond to the Pearson's correlation coefficient between two given subunits based on dN/dS analysis (see legend for details). Interaction among two proteins can be hypothesized if the correlation coefficient is >0.8 and the statistical significance is high. According to these criteria, a strong correlation was found for the following 19 CSN pairs: CSN1–CSN2, CSN1–CSN3, CSN1–CSN6, CSN2–CSN3, CSN2–CSN4, CSN2–CSN5, CSN2–CSN6, CSN2–CSN7, CSN3–CSN4, CSN3–CSN5, CSN3–CSN6, CSN3–CSN7, CSN4–CSN5, CSN4–CSN6, CSN4–CSN7, CSN5–CSN6, CSN5–CSN7, CSN6–CSN7, and CSN7–CSN8.
Not all of these interacting pairs were found by the yeast two-hybrid interaction analyses described above. For example, the interaction between CSN3 and CSN5, although predicted by coevolution analysis, was not detected by the two-hybrid assay. This discrepancy could be caused by several factors. First, this experimental procedure calculates protein–protein interaction on the basis of evolutionary rate. Thus, it is possible that, because the two proteins are subunits of the same complex, they may tend to coevolve even if they are not physically in contact. Second, our correlation analysis could reveal interactions that require more than two proteins to take place, which would not be detected by a pair-wise two-hybrid interaction system. However, even omitting the interacting pairs supported only by coevolution analysis and the weak two-hybrid interactions, more than one set of evidence supported the following 17 pair-wise interactions: CSN1–CSN2, CSN1–CSN3, CSN1–CSN4, CSN1–CSN7, CSN2–CSN4, CSN3–CSN4, CSN3–CSN6, CSN3–CSN8, CSN4–CSN4, CSN4–CSN5, CSN4–CSN6, CSN4–CSN7, CSN4–CSN8, CSN5–CSN6, CSN6–CSN7, CSN7–CSN8, and CSN8–CSN8. Our results also confirm the strong interaction cluster among subunits 4, 5, 6, and 7 that has been reported previously (Fu et al., 2001).
AtCSN5 and AtCSN6 Contact Each Other Directly within CSN in Vivo
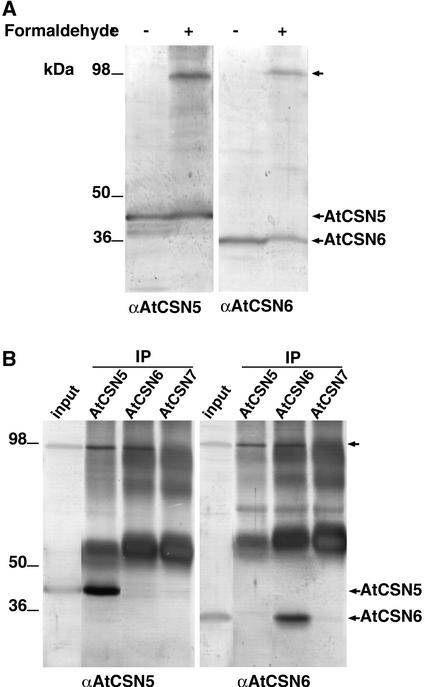
To verify some of the interactions found with the indirect techniques described above, we examined the in vivo subunit interaction by biochemical cross-linking. We used the chemical cross-linker formaldehyde to covalently preserve in vivo interactions among CSN components. To this aim, cauliflower inflorescences were treated for 30 min with 1% formaldehyde before protein extraction and analysis. As shown in Figure 7, after treatment with formaldehyde, a novel high molecular mass band that was recognized by antibodies to either CSN5 or CSN6 (Figures 7A and 7B, top arrows) was observed after SDS-PAGE separation of the cauliflower protein extract. This novel protein band could be coimmunoprecipitated by antibodies to either CSN5 or CSN6 after denaturing treatment of the protein extracts with 8 M urea (Figure 7B). Furthermore, this band vanished after the cross-linking treatment was reversed (data not shown). The efficient covalent cross-linking of CSN5 and CSN6 after brief treatment with the chemical cross-linker formaldehyde suggests a close proximity between the two, confirming their physical interaction and contact within CSN.
Figure 7.
CSN5 and CSN6 Contact Each Other Physically within CSN.
(A) The formaldehyde treatment of cauliflower inflorescences generates a high molecular mass protein complex (top arrow at right) on SDS-PAGE that is recognized by the antibodies to AtCSN5 and AtCSN6 (indicated at bottom).
(B) The high molecular mass complex (top arrow at right) can be immunoprecipitated by antibodies to both AtCSN5 and AtCSN6 but not by antibodies to AtCSN7. Antibodies used for immunoprecipitation (IP) are indicated at top. The immunoblot analysis was performed with the antibodies indicated at bottom.
DISCUSSION
Here, we report the molecular identity of subunit 2 of the COP9 signalosome (AtCSN2) as FUS12, thus bringing to six the number of CSN subunits identified by their corresponding COP/DET/FUS loci. Definitive evidence confirmed that AtCSN2 is a subunit of the COP9 signalosome. Thus, the cloning of AtCSN2 concludes the molecular characterization of the eight AtCSN subunits and made possible an overall topology and interaction analysis of the entire complex.
An Updated Structural Topology Map for CSN
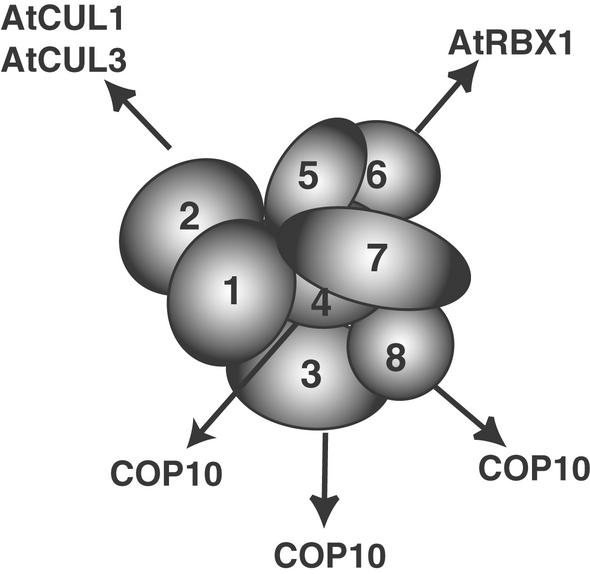
Our comprehensive subunit interaction study also provides further insight into the molecular architecture of the COP9 signalosome. In addition to the two-hybrid system, we used two novel approaches to further define the CSN topology map. First, we made use of a coevolution strategy. This strategy is valuable because it can predict interactions that can be confirmed using other approaches. In this case, it allowed us to confirm and broaden the data from our two-hybrid analysis. It also helped us to conclude that the core CSN subunit interactions are conserved among distantly related organisms, suggesting that the CSN architecture has remained similar during evolution, probably because it is closely related to CSN cellular function. However, the coevolution strategy always needs to be confirmed in vivo. Two-hybrid techniques only partially solve this problem, because they allow the testing of interactions only among a limited number of subunits. Some components of the complex might make contact with more than one subunit within the complex. Thus, we established a procedure to determine the in vivo binding of CSN subunits through cross-linking experiments. In this approach, we used formaldehyde as a cross-linking agent. Formaldehyde is a powerful reversible chemical cross-linker that has been used extensively in cell biology to preserve the cell structure and in protein-DNA cross-linking studies (Orlando, 2000). Our results show that it can be used efficiently to fix the in vivo protein–protein interactions that are barely detected in a regular buffer system and open the way to more complex studies of CSN structure. Together, our data lead to an optimal topology model for CSN subunit interactions, which is shown in Figure 8. Obviously, this model does not imply a three-dimensional structure of the CSN, but it can serve as a starting point for further interaction and structural analyses.
Figure 8.
CSN Regulates Proteins of the Ubiquitin-Proteasome Pathway by Specific Contacts through Distinct Subunits.
CSN subunit contacts were defined by the interaction demonstrated in this work. Potential dimers of the same subunit are not shown, and the subunits are not drawn to scale.
The CSN and the Lid: Evolutionary Implications
Because of the similarity in the composition and sequences of their components, it has been proposed that the lid of the proteasome and CSN might have similar structures and functions. However, data obtained from electron microscopy analysis showed a different surface topology for the two particles (Kapelari et al., 2000), supporting a difference in their biological activities. Our phylogenetic analysis shows that the duplication and divergence events of CSN and the lid from a hypothetical ancestor complex occurred very early, before the divergence of unicellular and multicellular eukaryotic organisms. This finding implies that the two complexes have evolved independently for a long time and may have different functions. Although it has been proposed several times that CSN could replace the lid and form a specialized “COP9 proteasome,” evidence that this is the case is lacking. It is possible that the two complexes, although maintaining a similar internal scaffold, have evolved distinct biological activities. This possibility is suggested as well by recent studies showing clearly different activities, de-neddylation and de-ubiquitylation, respectively, for CSN5 and its lid paralog RPN11 (Verma et al., 2001; Cope et al., 2002). Additional work will be required to fully address this issue.
The CSN as a General Regulator of Diverse Cullin-Based E3 Ligases
We demonstrated previously that in Arabidopsis, CSN interacts with and regulates the ubiquitin ligase SCFTIR1. This interaction is direct, because specific SCF subunits interact with specific CSN subunits. Among those subunits, AtCSN2 has been shown to interact with AtCUL1. We report here that AtCSN2 also can interact with AtCUL3, which is another protein belonging to the cullin family that is distantly related to AtCUL1 (Figure 6). It is possible that part of the reason why CSN2 is one of the two most conserved subunits during evolution is their need to cooperate, through direct physical contacts, with the cullins.
Our new findings are in agreement with the current model, in which the COP9 signalosome regulates several classes of cullin-containing ubiquitin ligases and possibly other classes of E3s as well, probably through contacts between specific CSN subunits and selected members of the protein degradation machinery (Schwechheimer et al., 2001, 2002). This notion is supported by the recent cloning of COP10, a ubiquitin-conjugating enzyme variant that binds to COP1, to a putative E3 ligase, and to specific CSN subunits that are different from those that contact SCF complexes (Figure 8) (Suzuki et al., 2002). Remarkably, mutations in COP10, COP1, and CSN yield a similar seedling phenotype (Kwok et al., 1996). However, although cop1 and cop10 weak alleles are viable and can survive to the adult stage, all of the CSN mutant alleles found to date are lethal. This would be expected if CSN functions as a general regulator of many E3 ligases, which control a variety of developmental processes (Figure 8).
METHODS
Plant Material and Growth Conditions
Plant germination and growth conditions for Arabidopsis thaliana were as described previously (Wei and Deng, 1992). All seedlings were vernalized for 1 week at 4°C before transfer to growth chambers. Unless stated otherwise, the growth chamber was used under a cycling long-day photoperiod (16 h of light at 48 μmol·m−2·s−1 and 8 h of dark) at 22°C. Wild-type plants were in the Columbia or Landsberg background. fus12-R380 and fus12-U228 were in the Landsberg background (Miséra et al., 1994).
Isolation of the AtCSN2 cDNA and Sequence Analysis of AtCSN2 in the fus12-R380 and fus12-U228 Alleles
Four peptide sequences from three different HPLC fractions (A54-29, A54-73, and A54-50; Figure 1B) (Serino et al., 1999) were used to search the Arabidopsis database (http://www.Arabidopsis.org). A partial EST clone (H4H12T7; obtained from the ABRC [Columbus, OH]) was retrieved. The EST sequence and the four peptides also were found to correspond to a completely sequenced BAC clone (T20P8) on chromosome II as part of the Arabidopsis genome-sequencing project.
RNA extraction was performed using Trizol (Invitrogen, Carlsbad, CA). The 5′ end of the gene was determined using the 5′ rapid amplification of cDNA ends kit (Invitrogen) according to the manufacturer's instructions. The complete full-length cDNA clone was obtained by reverse transcription PCR (Advantage RT-for-PCR kit; Clontech, Palo Alto, CA) of RNA extracted from 1-week-old Columbia seedlings using 21-bp primers specific for the 5′ and 3′ ends of the gene, starting from the ATG and the stop codon, respectively. The PCR conditions were as follows: 35 cycles of 1 min at 94°C, 1 min at 54°C, and 2 min at 72°C. Pfu polymerase (Invitrogen) was used. The PCR product was cloned with the TOPO-TA cloning kit in the vector pCR-II (Invitrogen), and the sequence was confirmed by direct sequencing. The first Met residue was determined by comparison with the CSN2 amino acid sequence from its animal counterpart (Figure 1A) and by the observation that a stop codon and a clear TATA box were found 42 and 60 bp, respectively, upstream of the ATG codon.
To determine the mutation of the AtCSN2 gene in the fus12 alleles, 1 μg of total RNA from 1-week-old seedlings homozygous for each allele and their respective wild-type ecotypes was used for reverse transcription PCR (see above). Mutations were confirmed further by PCR and sequencing of the corresponding genomic region. The PCR conditions were the same as those described above. The PCR products were isolated using the QIAEXII gel extraction kit (Qiagen, Chatsworth, CA) and then sequenced directly. For the genomic DNA, at least four independent PCR products for each of the mutant alleles and their corresponding ecotypes were cloned in vector pCRII with the TOPO-TA cloning kit (Stratagene) and sequenced to confirm the mutation. The sequence data were analyzed using LaserGene software (DNASTAR, Madison, WI).
Complementation of the fus12 Mutation by the AtCSN2 cDNA
The BAC clone T2OP8, obtained from the ABRC, was digested with the enzyme XbaI to generate a fragment of 6762 bp that contains the complete AtCSN2 open reading frame together with its own promoter. This fragment was cloned in the binary vector pPZPY122 (Yamamoto et al., 1998) and transformed into fus12-R380 heterozygous plants as described above. Approximately 40 independent lines were generated. The T1 gentamycin-resistant plants were selfed, and the T2 generation of six representative lines was used for the analysis in this study.
Antibody Production, Protein Extraction, Gel Filtration, and Immunoblot Analyses
The full-length cDNA clone for AtCSN2 was cloned in the vector pGEX4T1 (Amersham Pharmacia Biotech). The resulting glutathione S-transferase–AtCSN2 fusion protein was overexpressed in Escherichia coli, purified from inclusion bodies, and used to produce rabbit polyclonal antibodies, as reported previously (Kwok et al., 1998). Other antibodies used were anti-CSN4 (Serino et al., 1999), anti-AtCSN5 (Kwok et al., 1998), anti-TBP (Schwechheimer et al., 2001), anti-AtCUL1, and anti-ASK1 (Gray et al., 1999).
Total soluble protein extracts from fus12-R380, fus12-U228, and wild-type Columbia and Landsberg seedlings were obtained and used for all immunoblot analyses, as described previously (Staub et al., 1996). For gel filtration, the total soluble protein extract was fractionated through a Superose 6 HR 10/30 gel-filtration column (Amersham Pharmacia Biotech) and examined by immunoblot analysis, as described previously (Kwok et al., 1998).
Yeast Two-Hybrid Assay
The full-length cDNA clone for AtCSN2 was cloned as a translational fusion to the LexA DNA binding domain (in pEG202) or to the transcription activation domain (in pJG4-5) (Golemis et al., 1994). The two new plasmids were designated pEG-AtCSN2 and pJG-AtCSN2. For the Arabidopsis CSN two-hybrid pair-wise interaction assay, the other plasmids used in the assay were pEG-AtCSN1 and pJG-AtCSN1 (formerly FUS6/COP11; Kwok et al., 1998); pEG-AtCSN3 and pJG-AtCSN3 (Peng et al., 2001b); pEG-CSN4 and pJG-CSN4 (formerly COP8; Serino et al., 1999); pEG-AtCSN5 and pJG-AtCSN5 (formerly AJH1; Kwok et al., 1998); pEG-AtCSN6 and pJG-AtCSN6 (Peng et al., 2001a); pEG-AtCSN7 and pJG-AtCSN7 (formerly FUS5; Karniol et al., 1999); and pEG-AtCSN8 and pJG-AtCSN8 (formerly COP9; Kwok et al., 1998). For the CSN two-hybrid assay with mammalian subunits, all constructs have been reported previously (Lyapina et al., 2001; Tsuge et al., 2001). For the cullin interaction assay, the full-length cDNA for AtCUL3 was cloned in the pEG vector (pEG-AtCUL3). The other clones used were pEG-AtCUL1 (Schwechheimer et al., 2001) and pJG-AtCSN2 (this study). All of the LexA fusion constructs were transformed in the yeast strain EGY48. The activation domain fusion constructs were transformed into the yeast strain L40 (Invitrogen), which contains a β-galactosidase reporter gene. The transformants were selected and mated (Bendixen et al., 1994), and the resulting diploids were placed in medium selective for the diploids and containing X-Gal as a substrate for detection of the interaction. Selected pair-wise interactions were analyzed further by liquid assay, as described previously (Serino et al., 1999).
Phylogenetic and Correlation Analyses
The CSN sequences were obtained by a Basic Local Alignment Search Tool (BLAST) search of all databases at the National Center for Biotechnology Information World Wide Web site (http://www.ncbi.nlm.nih.gov/). A complete list of the sequences used, together with their GenBank accession numbers, is provided in the supplemental data online. All phylogenetic analyses were performed using maximum likelihood with PAUP (Phylogenetic Analysis Using Parsimony and other methods) version 4 (Rogers and Swofford, 1998).
For ratio of nonsynonymous versus synonymous substitution rate (dN/dS) correlation studies, the confirmed amino acid sequences from different organisms of the same CSN subunit were aligned using CLUSTAL X (Thompson et al., 1997). The DNA sequences of the genes that code for the subunits then were aligned manually according to their deduced amino acid sequences. The YN00 program in PAML version 3.0 (http://abacus.gene.ucl.ac.uk/software/paml.html) was used to calculate the dN/dS between pairs of gene sequences of the same subunit from different organisms. The correlation between pairs of dN/dS was determined using Excel (Microsoft, Redmond, WA) to evaluate the subunit-to-subunit relationship. The statistical significance of the correlation analysis was measured with the Pearson coefficient.
In Vivo Cross-Linking
The top layer tissues of the cauliflower (Brassica oleracea botrytis) inflorescences (∼1 mm) were incubated with 10 volumes of 1% formaldehyde in PBSM buffer (1.76 mM KH2PO4, 10 mM Na2HPO4, 136 mM NaCl, 2.6 mM KCl, 5 mM MgCl2, and 10% glycerol) under vacuum for 30 min. The reaction was stopped by adding Gly to a final concentration of 0.125 M for 5 min. The tissue was washed three times with protein extraction buffer before protein extraction (Peng et al., 2001a). Protein extracts were subjected to SDS-PAGE and immunoblot analysis with antibodies to selected CSN subunits. For immunoprecipitation, the formaldehyde-treated protein extracts were subjected to denaturing treatment with 8 M urea. The urea was removed by gradient buffer exchange, and the soluble proteins were used for coimmunoprecipitation studies.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
The GenBank accession numbers for the sequences mentioned in this article are W43761 (EST clone H4H12T7), AC005623 (BAC clone T20P8), XP_056853 (hCSN2), AF314168 (SpCSN2), and NP180267 (AtCSN2). Accession numbers for the human proteins and Arabidopsis cullins shown in Figure 4 are as follows: NP_003583 (hCUL1), AAD23581 (hCUL2), NP_003581 (hCUL3), AF077188 (hCUL4A), NP_003579 (hCUL4B), NP_003469 (hCUL5), NP174005 (At1g26830), NP176266 (At1g69670), NP199433 (At5g46210), NP171797 (At1g02980), NP175007 (At1g43140), and AAK76704 (AtCUL1).
Supplementary Material
Acknowledgments
We thank Elizabeth Strickland for reading and commenting on the manuscript. This research was supported by National Science Foundation Grant MCD-0077217 and Binational Agricultural Research and Development Fund Grant IS-3123-99 to X.W.D. and by a grant from the National Science Foundation of China (39725002). X.W.D. was a National Science Foundation Presidential Faculty Fellow. Z.P. was the recipient of a National Institutes of Health postdoctoral fellowship.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.009092.
Footnotes
Online version contains Web-only data.
References
- Bendixen, C., Gangloff, S., and Rothstein, R. (1994). A yeast mating-selection scheme for detection of protein–protein interactions. Nucleic Acids Res. 22, 1778–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatch, G.L., and Lassle, M. (1999). The tetratricopeptide repeat: A structural motif mediating protein-protein interactions. Bioessays 21, 932–939. [DOI] [PubMed] [Google Scholar]
- Chamovitz, D.A., Wei, N., Osterlund, M.T., von Arnim, A.G., Staub, J.M., Matsui, M., and Deng, X.W. (1996). The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 86, 115–121. [DOI] [PubMed] [Google Scholar]
- Cope, G.A., Suh, G.S., Aravind, L., Schwarz, S.E., Zipursky, S.L., Koonin, E.V., and Deshaies, R.J. (2002). Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of NEDD8 from CUL1. Science 298, 608–611. [DOI] [PubMed] [Google Scholar]
- Fraser, H.B., Hirsh, A.E., Steinmetz, L.M., Scharfe, C., and Feldman, M.W. (2002). Evolutionary rate in the protein interaction network. Science 296, 750–752. [DOI] [PubMed] [Google Scholar]
- Freilich, S., Oron, E., Kapp, Y., Nevo-Caspi, Y., Orgad, S., Segal, D., and Chamovitz, D.A. (1999). The COP9 signalosome is essential for development of Drosophila melanogaster. Curr. Biol. 9, 1187–1190. [DOI] [PubMed] [Google Scholar]
- Fu, H., Reis, N., Lee, Y., Glickman, M.H., and Vierstra, R.D. (2001). Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO J. 20, 7096–7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman, M.H. (2000). Getting in and out of the proteasome. Semin. Cell Dev. Biol. 11, 149–158. [DOI] [PubMed] [Google Scholar]
- Glickman, M.H., Rubin, D., Coux, O., Wefes, I., Pfeifer, G., Cjeka, Z., Baumeister, W., Fried, V.A., and Finley, D. (1998). A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94, 615–623. [DOI] [PubMed] [Google Scholar]
- Golemis, E.A., Gyuris, J., and Brent, R. (1994). Interaction trap/two-hybrid system to identify interacting proteins. In Current Protocols in Molecular Biology, F.M. Ausubel, R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl, eds (New York: John Wiley & Sons).
- Gray, W.M., del Pozo, J.C., Walker, L., Hobbie, L., Risseeuw, E., Banks, T., Crosby, W.L., Yang, M., Ma, H., and Estelle, M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13, 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke, C.S., and Deng, X.W. (2000). The cell biology of the COP/DET/FUS proteins: Regulating proteolysis in photomorphogenesis and beyond? Plant Physiol. 124, 1548–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, F.C., and Wu, Z.Z. (1993). The principle of concerted evolution between interacting proteins: Darwinian selection at molecular level. Chin. Sci. Bull. 38, 1396–1401. [Google Scholar]
- Hershko, A., and Ciechanover, A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Hofmann, K., and Bucher, P. (1998). The PCI domain: A common theme in three multiprotein complexes. Trends Biochem. 23, 204–205. [DOI] [PubMed] [Google Scholar]
- Hori, T., Osaka, F., Chiba, T., Miyamoto, C., Okabayashi, K., Shimbara, N., Kato, S., and Tanaka, K. (1999). Covalent modification of all members of human cullin family proteins by NEDD8. Oncogene 18, 6829–6834. [DOI] [PubMed] [Google Scholar]
- Kapelari, B., Bech-Otschir, D., Hegerl, R., Schade, R., Dumdey, R., and Dubiel, W. (2000). Electron microscopy and subunit-subunit interaction studies reveal a first architecture of COP9 signalosome. J. Mol. Biol. 300, 1169–1178. [DOI] [PubMed] [Google Scholar]
- Karniol, B., Malec, P., and Chamovitz, D.A. (1999). Arabidopsis FUSCA5 encodes a novel phosphoprotein that is a component of the COP9 complex. Plant Cell 11, 839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karniol, B., Yahalom, A., Kwok, S., Tsuge, T., Matsui, M., and Deng, X.W. (1998). The Arabidopsis homologue of an eIF3 complex subunit associates with the COP9 complex. FEBS Lett. 439, 173–179. [DOI] [PubMed] [Google Scholar]
- Kawakami, T., Chiba, T., Suzuki, T., Iwai, K., Yamanaka, K., Minato, N., Suzuki, H., Shimbara, N., Hidaka, Y., Osaka, F., Omata, M., and Tanaka, K. (2001). NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 20, 4003–4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipreos, E.T., Lander, L.E., Wing, J.P., He, W.W., and Hedgecock, E.M. (1996). Cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell 85, 1–20. [DOI] [PubMed] [Google Scholar]
- Kwok, S.F., Piekos, B., Miséra, S., and Deng, X.W. (1996). A complement of ten essential and pleiotropic Arabidopsis COP/DET/FUS genes is necessary for repression of photomorphogenesis in darkness. Plant Physiol. 110, 731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok, S.F., Solano, R., Tsuge, T., Chamovitz, D.A., Ecker, J.R., Matsui, M., and Deng, X.W. (1998). Arabidopsis homologs of a c-Jun coactivator are present both in monomeric form and in the COP9 complex, and their abundance is differentially affected by the pleiotropic cop/det/fus mutations. Plant Cell 19, 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina, S., Cope, G., Shevchenko, A., Serino, G., Tsuge, T., Zhou, C., Wolf, D., Wei, N., Shevchenko, A., and Deshaies, R.J. (2001). Promotion of NEDD8-CUL1 conjugate cleavage by COP9 signalosome. Science 292, 1382–1385. [DOI] [PubMed] [Google Scholar]
- Lyapina, S., Correll, C.C., Kipreos, E.T., and Deshaies, R.J. (1998). Human CUL1 forms an evolutionarily conserved ubiquitin ligase complex (SCF) with SKP1 and an F-box protein. Proc. Natl. Acad. Sci. USA 95, 7451–7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miséra, S., Müller, A.J., Weiland-Heidecker, U., and Jürgens, G. (1994). The FUSCA genes of Arabidopsis: Negative regulators of light responses. Mol. Gen. Genet. 244, 242–252. [DOI] [PubMed] [Google Scholar]
- Mundt, K.E., Porte, J., Murray, J.M., Brikos, C., Christensen, P.U., Caspari, T., Hagan, I.M., Millar, J.B., Simanis, V., Hofmann, K., and Carr, A.M. (1999). The COP9/signalosome complex is conserved in fission yeast and has a role in S phase. Curr. Biol. 9, 1427–1430. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Ohta, T., Michel, J.J., Schottelius, A.J., and Xiong, Y. (1999). ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell 3, 535–541. [DOI] [PubMed] [Google Scholar]
- Orlando, V. (2000). Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. 25, 99–104. [DOI] [PubMed] [Google Scholar]
- Peng, Z., Serino, G., and Deng, X.W. (2001. a). Molecular characterization of subunit 6 of the COP9 signalosome and its role in multifaceted development processes in Arabidopsis. Plant Cell 13, 2393–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Z., Serino, G., and Deng, X.W. (2001. b). A role of Arabidopsis COP9 signalosome in multifaceted developmental processes revealed by the characterization of its subunit 3. Development 128, 4277–4288. [DOI] [PubMed] [Google Scholar]
- Querido, E., Blanchette, P., Kamura, T., Morrison, M., Boivin, D., Kaelin, W.G., Conaway, R.C., Conaway, J.W., and Branton, P.E. (2001). Degradation of p53 by adenovirus E4orf6 and EIB55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15, 3104–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, J.S., and Swofford, D.L. (1998). A fast method for approximating maximum likelihoods of phylogenetic trees from nucleotide sequences. Syst. Biol. 147, 77–89. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., and Deng, X.W. (2001). COP9 signalosome revisited: A novel mediator of protein degradation. Trends Cell Biol. 11, 420–426. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., Serino, G., Callis, J., Crosby, W.L., Lyapina, S., Deshaies, R.J., Gray, W.M., Estelle, M., and Deng, X.W. (2001). Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science 292, 1379–1382. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., Serino, G., and Deng, X.W. (2002). Multiple ubiquitin ligase-mediated processes require COP9 signalosome and AXR1 function. Plant Cell 14, 2553–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serino, G., Tsuge, T., Kwok, S., Matsui, M., Wei, N., and Deng, X.W. (1999). Arabidopsis cop8 and fus4 mutations define the same gene that encodes subunit 4 of the COP9 signalosome. Plant Cell 11, 1967–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, W.H., Parmentier, Y., Hellmann, H., Lechner, E., Dong, A., Masson, J., Granier, F., Lepiniec, L., Estelle, M., and Genschik, P. (2002). Null mutation of AtCUL1 causes arrest in early embryogenesis in Arabidopsis. Mol. Biol. Cell 13, 1916–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub, J.M., Wei, N., and Deng, X.W. (1996). Evidence for FUS6 as a component of the nuclear-localized COP9 complex in Arabidopsis. Plant Cell 8, 2047–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins, C., Kaelin, W., and Pavletich, N. (1999). Structure of the VHL-elongin C-elongin B complex: Implications for VHL tumor suppressor function. Science 284, 455–461. [DOI] [PubMed] [Google Scholar]
- Suzuki, G., Yanagawa, Y., Kwok, S.F., Matsui, M., and Deng, X.W. (2002). Arabidopsis COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. Genes Dev. 16, 554–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTALX Windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuge, T., Matsui, M., and Wei, N. (2001). The subunit 1 of the COP9 signalosome suppresses gene expression through its N-terminal domain and incorporates into the complex through the PCI domain. J. Mol. Biol. 305, 1–9. [DOI] [PubMed] [Google Scholar]
- Verma, R., Aravind, L., Oania, R., McDonald, W.H., Yates, J.R., III, Koonin, E.V., and Deshaies, R.J. (2001). Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298, 611–615. [DOI] [PubMed] [Google Scholar]
- Wei, N., Chamovitz, D.A., and Deng, X.W. (1994). Arabidopsis COP9 is a component of a novel signaling complex mediating light control of development. Cell 78, 117–124. [DOI] [PubMed] [Google Scholar]
- Wei, N., and Deng, X.W. (1992). COP9: A new genetic locus involved in light-regulated development and gene expression in Arabidopsis. Plant Cell 4, 1507–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, N., and Deng, X.W. (1996). The role of the COP/DET/FUS genes in light control of Arabidopsis seedling development. Plant Physiol. 112, 871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, N., and Deng, X.W. (1999). Making sense of the COP9 signalosome: A regulatory protein complex conserved from Arabidopsis to human. Trends Genet. 15, 98–103. [DOI] [PubMed] [Google Scholar]
- Wei, N., Tsuge, T., Serino, G., Dohmae, N., Takio, K., Matsui, M., and Deng, X.W. (1998). The COP9 complex is conserved between plants and mammals and is related to the 26S proteasome regulatory complex. Curr. Biol. 8, 919–922. [DOI] [PubMed] [Google Scholar]
- Yamamoto, Y.Y., Matsui, M., Ang, L.H., and Deng, X.W. (1998). Role of a COP1 interactive protein in mediating light-regulated gene expression in Arabidopsis. Plant Cell 10, 1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.