Abstract
A large number of resistance specificities to the powdery mildew fungus Blumeria graminis f. sp. hordei map to the barley Mla locus. This complex locus harbors multiple members of three distantly related gene families that encode proteins that contain an N-terminal coiled-coil (CC) structure, a central nucleotide binding (NB) site, a Leu-rich repeat (LRR) region, and a C-terminal non-LRR (CT) region. We identified Mla12, which encodes a CC-NB-LRR-CT protein that shares 89 and 92% identical residues with the known proteins MLA1 and MLA6. Slow Mla12-triggered resistance was altered dramatically to a rapid response by overexpression of Mla12. A series of reciprocal domains swaps between MLA1 and MLA6 identified in each protein recognition domain for cognate powdery mildew fungus avirulence genes (AvrMla1 and AvrMla6). These domains were within different but overlapping LRR regions and the CT part. Unexpectedly, MLA chimeras that confer AvrMla6 recognition exhibited markedly different dependence on Rar1, a gene required for the function of some but not all Mla resistance specificities. Furthermore, uncoupling of MLA6-specific function from RAR1 also uncoupled the response from SGT1, a protein known to associate physically with RAR1. Our findings suggest that differences in the degree of RAR1 dependence of different MLA immunity responses are determined by intrinsic properties of MLA variants and place RAR1/SGT1 activity downstream of and/or coincident with the action of resistance protein–containing recognition complexes.
INTRODUCTION
Intraspecific genetic variation in the capacity of plants to combat microbial attack is confined mainly to disease resistance (R) loci. These can encode a single gene but frequently they are complex, harboring multiple similar and/or dissimilar R genes (reviewed by Ellis et al., 2000). A single R gene has the capacity to recognize one or very few normally unrelated strain-specific pathogen effector molecules (encoded by avirulence [Avr] genes) that are released during pathogenesis. Most R genes encode one of two groups of Leu-rich repeat (LRR)–containing proteins. An intracellular class shares a central nucleotide binding (NB) site and C-terminal LRRs with variable repeat numbers. This is the largest group of known R proteins and can be divided further into subfamilies containing either N-terminal sequences predicted to form a coiled-coil (CC) structure (CC-NB-LRR subfamily) or sequences that are related to the cytoplasmic domain of the Drosophila Toll and human Interleukin1 receptor (TIR-NB-LRR). A second R protein class is membrane-anchored by a single transmembrane helix, consists of variable repeat numbers of extracellular LRRs, and contains at least in one case an intracellular Ser/Thr kinase module (reviewed by Ellis et al., 2000).
Little is known about the molecular mechanics of the R-AVR recognition process. Recent studies suggest that members of the intracellular and membrane-anchored classes assemble in preformed heteromultimeric recognition complexes in the absence of pathogens (Leister and Katagiri, 2000; Holt et al., 2002; Mackey et al., 2002; Rivas et al., 2002a, 2002b). With one exception, there are no documented examples of direct interactions between LRR-containing R and AVR proteins (rice Pi-ta and AVR-Pita from Magnaporthe grisea; Jia et al., 2000). Thus, it seems possible that R proteins function indirectly in the recognition process, which involves the surveillance of a host protein or a complex that is targeted by AVR products (Dangl and Jones, 2001; Mackey et al., 2002).
Approximately 30 genetically characterized barley Mla variants have been described, each triggering immunity responses upon recognition of cognate isolate-specific powdery mildew fungus (Blumeria graminis f. sp. hordei [Bgh]) effector molecules (encoded by AvrMla genes) (Jørgensen, 1994). Some of these variants confer a rapid resistance response resulting in Bgh growth termination at an early stage during pathogenesis, whereas others trigger a delayed response that permits substantial growth of fungal hyphae on the leaf surface (Wise and Ellingboe, 1983; Boyd et al., 1995). Although none of the Bgh AvrMla genes has been isolated to date, their genetic mapping in the powdery mildew genome revealed mainly dispersed and a few linked positions on multiple Bgh chromosomes (Brown and Jessop, 1995; Caffier et al., 1996; Pedersen et al., 2002). The complex Mla locus was located genetically and physically within an interval of ∼250 kb (Wei et al., 1999). A contiguous DNA sequence of the interval in barley cv Morex revealed 32 predicted genes, of which 8 encode CC-NB-LRR resistance gene homologs (RGHs) (Wei et al., 2002). The RGHs belong to three dissimilar families sharing <43% amino acid sequence similarity between families (Wei et al., 1999, 2002). Because Morex lacks a known Mla resistance specificity, the first two identified Mla powdery mildew R genes, Mla1 and Mla6, were isolated from other barley accessions (Halterman et al., 2001; Zhou et al., 2001). The deduced proteins share 91% identical residues and show highest overall similarity to the deduced Morex RGH1bcd family member (83 and 79% identity to MLA1 and MLA6, respectively) (Halterman et al., 2001; Wei et al., 2002).
Mutants of barley Rar1 were isolated originally as suppressors of Mla12 function, and wild-type Rar1 was shown subsequently to be required for the function of a subset of Mla powdery mildew resistance specificities (e.g., Mla6 and Mla12 but not Mla1) (Torp and Jørgensen, 1986; Jørgensen, 1996). Homologs of Rar1 in Arabidopsis and Nicotiana benthamiana play a conserved role in the function of a subset of NB-LRR R proteins that confer resistance to different pathogens (Liu et al., 2002a; Muskett et al., 2002; Tornero et al., 2002). The highly conserved zinc binding RAR1 proteins interact physically with another conserved protein, SGT1, which was demonstrated originally to function in ubiquitin-dependent cell cycle control in yeast (Kitagawa et al., 1999; Shirasu et al., 1999a; Azevedo et al., 2002; Liu et al., 2002b). Genetic evidence for a role of plant SGT1 in R gene–triggered resistance was obtained from Arabidopsis sgt1b mutants and SGT1 gene-silencing experiments in barley and N. benthamiana (Austin et al., 2002; Azevedo et al., 2002; Liu et al., 2002b; Peart et al., 2002; Tör et al., 2002). Barley and N. benthamiana SGT1 associate physically with one or several SCF ubiquitin E3 ligase complexes and the COP9 signalosome (Azevedo et al., 2002; Liu et al., 2002b). Because gene silencing of the core SCF component, SKP1, or the COP9 signalosome compromised R gene–triggered resistance in N. benthamiana, it seems likely that ubiquitin-protein conjugation path-ways play an important role in plant innate immunity responses (Liu et al., 2002b). However, it remains unclear whether ubiquitin-dependent processes occur upstream of, coincident with, or downstream of R protein–containing recognition complexes.
Here, we exploited a high sequence relatedness between identified (Mla1 and Mla6) and other genetically characterized Mla specificities to clone Mla12. Although Mla12 might be an allele of Mla1 and Mla6, it differs from them by belonging to a subgroup of Mla variants that trigger delayed resistance responses (Freialdenhoven et al., 1994; Boyd et al., 1995). Using a single-cell transient gene expression assay (Shirasu et al., 1999b; Zhou et al., 2001), we demonstrate that Mla12 overexpression shifts the slow Mla12-triggered response to a rapid Mla1/Mla6-like resistance. We gained insights into structure-function relationships of MLA proteins by analyzing a series of reciprocal domain swaps between MLA1 and MLA6. This analysis revealed a function for the MLA LRR-CT unit in specificity determination, whereas CC-NB and LRR sequences modulated RAR1 dependence. Moreover, we show that recognition specificity can be uncoupled from both RAR1 and SGT1 dependence. We discuss possible roles of RAR1/SGT1 in folding presumed MLA recognition complexes and in signaling downstream of activated recognition complexes.
RESULTS
Isolation of Mla12 and Characterization of Susceptible Mutant Alleles
To isolate Mla12, we constructed a genomic cosmid library comprising five barley genome equivalents using DNA from cv Sultan 5 containing Mla12 (see Methods). Sixteen cosmid clones were isolated from this library with a DNA probe corresponding to the LRR region of MLA1. Low-pass DNA sequencing of the cosmid clones revealed that all of them contain NB-LRR–type RGHs. Two clones, designated Sp14-1 and Sp14-4, contain identical RGHs showing ∼90% sequence identity to Mla1 and Mla6 in deduced exon and intron sequences. A closer comparison of the NB-LRR gene in Sp14-4 with Mla1 and Mla6 revealed an identical 5′ untranslated small open reading frame of nine amino acids and the same intron-exon structure (Halterman et al., 2001; Zhou et al., 2001). These genes share a simple sequence repeat (AT)n in intron 3, although the exact numbers of the repeats differ (see Figure 6 below). Therefore, we considered the RGH in Sp14-4 a candidate Mla12 gene that encodes a predicted CC-NB-LRR-CT protein of 108 kD sharing 89% identical residues with MLA1 and 92% identical residues with MLA6 (Figure 1).
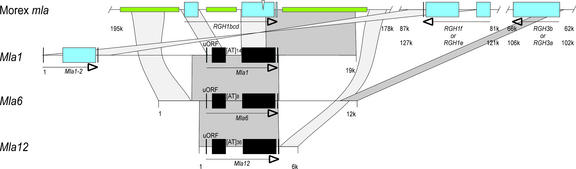
Figure 6.
Schemes of the Morex Mla Locus and Genomic Regions Containing Identified Mla Resistance Genes.
DNA sequences encompassing the Morex Mla locus (261 kb, in reverse orientation) (Wei et al., 2002) are represented schematically and drawn to scale in the top line (relevant sequences only). Available genomic sequences of Mla1, Mla6, and Mla12 and flanking regions are shown below. Coding sequences of functional Mla R genes and RGHs are boxed and highlighted in black and blue, respectively. A conserved upstream open reading frame (uORF) and a simple [AT]n microsatellite are shared among functional Mla R genes. Green boxes denote retrotransposon sequences: a BARE1 solo LTR in intron 3 of RGH1bcd, HORPIA2 immediately 3′ of RGH1bcd, and ALEXANDRA 5′ of RGH1bcd. Dark gray areas denote sequences showing >90% identity, and light gray areas denote sequences showing >75% identity. A possible inversion event could explain the altered relative orientations of homologous genes Mla1-2 and RGH1f as indicated. Note that RGH1e/f and RGH3a/b are extremely similar and located within a 40-kb duplicated region (Wei et al., 2002). For this reason, the indicated homologies exist between RGH1e and RGH1f and between RGH3a and RGH3b. Arrows indicate the relative orientations of genes (5′ to 3′). Borders of Morex sequences are indicated in kb according to accession AF427791.
Figure 1.
Amino Acid Sequence Alignment of Deduced Products of the Mla1, Mla6, and Mla12 Genes.
Residues identical to those in MLA1 are shown as dots, and deletions are shown as hyphens. A predicted CC structure is underlined. The starts of the NB, LRR, and CT regions are indicated with arrows and are operational according to Zhou et al. (2001). Boldface letters in the NB domain indicate amino acid motifs conserved among known NB-LRR proteins. Boxes indicate amino acid exchanges identified in three susceptible Mla12 mutants, and affected residues are shaded in black.
To obtain evidence for the function of the candidate R gene, we took advantage of chemically induced susceptible mutants that were isolated previously from a mutagenized barley M2 population of Sultan 5 harboring Mla12 (Torp and Jørgensen, 1986; Jørgensen, 1988). Genetic analysis indicated that susceptibility in some of the mutants (e.g., mutants M66 and M86) is likely attributable to mutations in Mla12, whereas susceptibility in three other lines (M22, M82, and M100) resulted from extragenic suppressor mutations of Mla12 function. Mutant lines M82 and M100 were demonstrated to contain recessive mutations in Rar1 (rar1-1 and rar1-2, respectively) (Shirasu et al., 1999a), and genetic analysis of mutant M22 suggested another gene required for Mla12 function, designated Rar2 (Jørgensen, 1988, 1996; Freialdenhoven et al., 1994). DNA sequence analysis of the candidate Mla12 in the susceptible mutants M66 and M86 revealed in each a single nucleotide substitution compared with the wild-type gene. The substitutions replace amino acid Leu-631 with Arg in the second LRR of the deduced candidate MLA12 protein in M66 and amino acid Glu-866 with Lys in the CT region in M86, respectively (Figure 1). Thus, we concluded that the Sp14-4–derived candidate gene probably is Mla12.
DNA marker–based mapping of susceptibility conferred by the M22 mutant revealed its location on chromosome 1H at the Mla locus between restriction fragment length polymorphism markers MWG036 and MWG068 (Schüller et al., 1992; our unpublished data). This finding suggested that susceptibility might be caused by a mutation in Mla12 or in a tightly linked gene. DNA sequence analysis of the candidate Mla12 in M22 plants revealed a single nucleotide substitution that replaces amino acid Lys-916 with Met in the CT region (Figure 1). This finding suggests that M22, like M66 and M86, likely is a mutant allele of Mla12 (see below).
Overexpression of Mla12 Alters the Resistance Kinetics but Retains Rar1 Dependence
To test directly the function of the candidate Mla12 gene, Sp14-4 DNA was delivered into epidermal cells of detached barley leaves by particle bombardment (Shirasu et al., 1999b). Transformed cells were tested for their ability to activate race-specific powdery mildew resistance upon inoculation with Bgh conidiospores of isolates expressing or lacking AvrMla12 (isolate A6 harboring AvrMla6 and AvrMla12 and isolate K1 har-boring AvrMla1) (Zhou et al., 2001). Infection phenotypes of transgene-expressing epidermal cells were microscopically inspected at 48 h after inoculation by scoring the presence or absence of intracellular Bgh haustoria at single interaction sites. Unlike control bombardments with cosmid DNA harboring Mla1 or Mla6, which are known to mediate race-specific resistance in the transient gene expression assay (Halterman et al., 2001; Zhou et al., 2001), delivery of Sp14-4 DNA failed to trigger detectable resistance upon inoculation with Bgh strains A6 and K1 (data not shown). This effect may be caused by insufficient 5′ flanking regulatory sequences (∼400 bp upstream of the transcription start) in cosmid Sp14-4, driving expression of the candidate Mla12, or delayed activation of Mla12 compared with Mla1 and Mla6 resistance (see Discussion) (Freialdenhoven et al., 1994; Boyd et al., 1995).
To examine this possibility further, we subcloned the coding region of the Mla12 candidate under the control of the strong maize ubiquitin promoter and the nopaline synthase (Nos) terminator. DNA of this overexpression construct and two similar control overexpression plasmids harboring Mla1 or Mla6 were delivered into leaf epidermal cells of barley cv Ingrid lacking Mla12 and Mlo (Figure 2A). Delivery of each plasmid DNA together with an Mlo-expressing construct resulted in a haustorium index of 2 to 5% upon challenge with the Bgh isolate containing the cognate Avr genes, whereas the control compatible interactions showed an index of ∼80%. Note that the very high level of haustorium incidence found in the compatible interactions likely is the result of cobombardment of the race-nonspecific defense modulator Mlo, which renders transformed epidermal cells supersusceptible to the fungus (Kim et al., 2002). These data provided evidence that the candidate Mla12 gene subcloned from cosmid Sp14-4 triggered AvrMla12-dependent Bgh growth termination. Interestingly, bombardments with empty vector DNA into epidermal cells of Sultan 5, which contains Mla12, resulted in a high haustorium index of 45% when inoculated with the incompatible isolate Bgh A6 (Figure 2B). This finding suggests that Mla12 resistance is not effective before haustorium development, consistent with a previous quantitative inspection of single interaction sites in resistant Mla12 wild-type and susceptible mutant leaves (Freialdenhoven et al., 1994). However, when the putative Mla12 was overexpressed in Sultan 5 using the single cell expression assay, the haustorium index was reduced to ∼2%, similar to the level conferred by Mla6 (Figure 2B). Apparently, overexpression of the candidate Mla12 shifted the resistance response from posthaustorium growth arrest to an abortion of fungal development before haustorium formation.
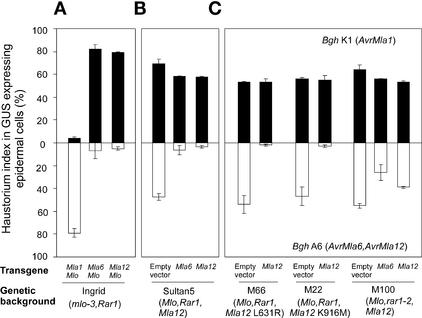
Figure 2.
Complementation of Susceptible Mla12 Mutants by Overexpression of Mla12 Resistance.
Relative single cell resistance/susceptibility upon delivery of various Mla transgenes at 48 h after spore inoculation is indicated by haustorium indices of attacked β-glucuronidase (GUS)–expressing cells (%). Data shown were obtained by bombardment of plasmid DNAs into epidermal cells of detached barley leaves (described by Shirasu et al., 1999b; Zhou et al., 2001). A β-glucuronidase reporter gene was used to identify transformed cells.
(A) The indicated transgenes were tested in detached leaves of barley cv Ingrid harboring mlo-3 Rar1. In this line, broad-spectrum mlo-3 resistance was complemented by cobombardment with a plasmid expressing wild-type Mlo; this renders cells supersusceptible to all tested Bgh isolates (Zhou et al., 2001; Kim et al., 2002). Results obtained with the Bgh isolate K1 (AvrMla1) are shown by closed columns, and results obtained with isolate A6 (AvrMla6 and AvrMla12) are shown by open columns in downward orientation. The data shown are means of at least three independent experiments (sd indicated). Each experiment involved light microscopic examination of at least 100 interaction sites between a single Bgh sporeling and an attacked epidermal cell.
(B) The indicated transgenes and an empty vector control were delivered into epidermal cells of Sultan 5 containing Mla12 Mlo Rar1. Experimental conditions and symbols are identical to those in (A).
(C) Transgene Mla12 or an empty vector control was delivered into epidermal cells of two susceptible Mla12 mutant lines (M66 and M22). Transgene Mla6 or Mla12 or an empty vector control also was delivered into the rar1-2 mutant line M100. Experimental conditions and symbols are identical to those in (A).
To corroborate the function of Mla12, we bombarded the overexpression construct in epidermal cells of mutant lines M66, M22, and M100 (the latter contains the severely defective rar1-2 allele) (Shirasu et al., 1999a) (Figure 2C). In these experiments, full AvrMla12-dependent resistance was restored in both M66 and M22 plants, demonstrating that the mutant phenotypes were complemented by the candidate Mla12. By contrast, neither overexpression of Mla6 nor overexpression of the candidate Mla12 restored full resistance in the rar1-2 mutant line M100. The Mla12 overexpression phenotype was affected more strongly than the Mla6 response in the rar1 mutant background. Together, these data strongly support our claim that the RGH in cosmid Sp14-4 is Mla12.
Context-Dependent Function of Conserved MLA Residues Leu-631 and Lys-916
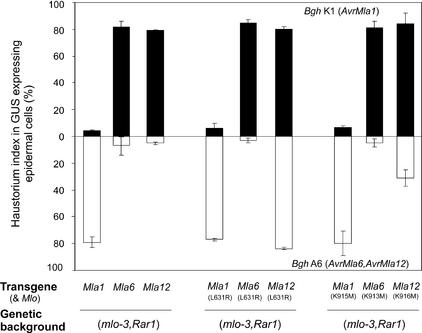
We noted that amino acid substitutions in the susceptible Mla12 mutants M66 (L631R) and M22 (K916M) affect residues that are conserved in MLA1 and MLA6, whereas the substitution in mutant M86 (E866K) changes a nonconserved residue (Figure 1). To investigate the importance of Leu-631 and Lys-916 in Mla1- and Mla6-triggered resistance, the same amino acid substitutions were introduced into Mla1 and Mla6 under the control of the ubiquitin promoter and were reintroduced into Mla12 for comparison and confirmation. Wild-type and mutant variant plants were tested in the transient gene expression system. This analysis showed that Mla12 mutant variant L631R impaired AvrMla12-dependent resistance fully (84%) and K916M impaired it partially (31%), indicating that the MLA12 (K916M) variant protein retains residual activity (Figure 3). This observation is consistent with the fully compromised and partially impaired Mla12 resistance reported for M66 and M22 mutant plants (infection types 4 and 2/3, respectively) (Torp and Jørgensen, 1986) and validates the usefulness of the single-cell assay to evaluate Mla12 activity using the strong ubiquitin promoter. The weakly susceptible infection phenotype of M22 mutant plants likely complicated the scoring of infection phenotypes in progeny derived from M22 test crosses and may explain the apparent misinterpretation of the mutant line as an extragenic suppressor of Mla12 resistance (Jørgensen, 1988, 1996). Surprisingly, despite an overall sequence relatedness of 90% between the tested MLA proteins, none of the amino acid replacements in MLA6 or MLA1 resulted in a detectable change of resistance activity compared with that in the respective wild-type genes (Figure 3). Thus, it is possible that other regions are critical for R protein function in MLA1 and MLA6 (see below). Alternatively, other residues that are absent or polymorphic in MLA12 might compensate for the functional contributions of Leu-631 and Lys-916 in the MLA1/MLA6 substitution mutants.
Figure 3.
Context-Dependent Functions of Conserved MLA Residues Leu-631 and Lys-916.
Mean values of single cell resistance/susceptibility (%) are shown at left after delivery of Mla1, Mla6, or Mla12 into the genetic background of cv Ingrid (mlo-3 Rar1). Results obtained with L631R variants of Mla1, Mla6, and Mla12 are shown in the middle. Results obtained with Mla1, Mla6, and Mla12 variants each containing a K to M substitution at the indicated positions are shown at right. Experimental conditions and designations are identical to those in Figure 2. GUS, β-glucuronidase.
Recognition Specificity Is Determined by the LRR-CT Unit
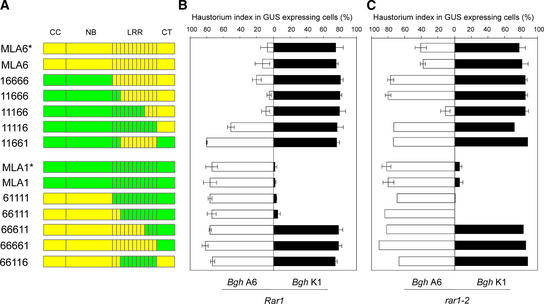
For further analysis of regions that are critical for MLA function, we constructed a series of reciprocal domain swaps between Mla1 and Mla6 (Figure 4A). These two R genes recognize different AvrMla genes and have different requirements for Rar1 and Sgt1 (Halterman et al., 2001; Zhou et al., 2001; Azevedo et al., 2002). The maize ubiquitin promoter drove the expression of each chimeric gene, and their function was tested after bombardment into leaf epidermal cells by spore inoculation with Bgh isolates K1 (AvrMla1) and A6 (AvrMla6) at 15 h after delivery. Recognition specificity and activity of the chimeras were compared with those of the respective Mla1 and Mla6 wild-type genes whose expression was driven by either native regulatory 5′ sequences or the strong ubiquitin promoter (Figure 4B). No significantly different activity was seen using constructs driven by the native or the strong ubiquitin promoter. Full AvrMla6-dependent recognition specificity was retained in chimeras containing the complete MLA1-derived CC-NB domains and in chimeras containing progressively more MLA1-derived N-terminal LRR repeats (constructs 16666, 11666, and 11166; Figure 4B). Activities mediated by chimeras containing only MLA6-derived LRRs 3 to 11 (11661) or only the MLA6-derived C terminus (11116) were inactive or severely impaired, respectively. These data suggest that MLA6 LRRs 9 to 11 act together with the cognate C-terminal domain to confer AvrMla6 recognition specificity.
Figure 4.
Domain Swaps between MLA1 and MLA6 Reveal Determinants for Recognition Specificity and RAR1 Dependence.
(A) Schemes of MLA6 (yellow), MLA1 (green), and 10 chimeras are shown. The relative positions of the CC, NB, LRR, and CT parts are indicated at top, and acronyms for each chimera are shown at left. The stars indicate gene expression driven by native 5′ flanking sequences; the strong ubiquitin promoter drove the expression of all other genes.
(B) All genes shown in (A) were expressed in the Rar1 wild-type background, and mean values of single cell resistance/susceptibility were scored microscopically upon challenge inoculation with Bgh isolates A6 or K1. Experimental conditions and designations are identical to those in Figure 2. GUS, β-glucuronidase.
(C) All genes shown in (A) were expressed in the rar1-2 mutant background, and mean values of single cell resistance/susceptibility were scored microscopically upon challenge inoculation with Bgh isolates A6 or K1. Experimental conditions and designations are identical to those in Figure 2.
Reciprocal domain swaps showed that AvrMla1-dependent activity was retained upon replacement of the entire MLA1 CC-NB domain only and upon additional replacement of LRRs 1 and 2 (constructs 61111 and 66111). Interestingly, longer substitutions up to LRR 8 rendered the 66611 chimera fully inactive, although the reciprocal construct 11166 fully retained AvrMla6-dependent activity. Substitutions containing LRRs 3 to 11 (construct 11661) also compromised AvrMla1 recognition specificity. Because chimeras containing only MLA1-derived LRRs 3 to 11 (66116) or only the MLA1-derived C terminus (66661) were inactive, we conclude that MLA1-derived LRRs 3 to 11 together with the cognate C-terminal domain are required for MLA1 recognition specificity.
Uncoupling MLA6 Recognition Specificity from RAR1 Dependence
Barley Rar1 is required for the function of Mla6 but not Mla1 (Jørgensen, 1996; Halterman et al., 2001; Zhou et al., 2001). This fact prompted us to examine the activities of wild-type MLA1 and MLA6 and the MLA chimeras in the rar1-2 genetic background (Figure 4C). The rar1-2 mutation leads to a transcript-splicing defect, and a RAR1 antiserum fails to detect RAR1 signals on protein gel blots (Azevedo et al., 2002). Delivery of wild-type MLA1 or MLA6 plasmid DNA in rar1-2 leaf epidermal cells led to fully retained or partially compromised resistance (4 and 39% haustorium index, respectively) (Figure 4C). No significant differences were found between wild-type constructs driven by the native and strong ubiquitin promoters. Thus, Mla6 function is compromised partially by the rar1-2 mutation compared with bombardments of the same constructs in the Rar1 background (Figure 4B). Remarkably, delivery of the three chimeras conferring AvrMla6-dependent resistance in Rar1 plants (16666, 11666, and 11166) displayed either full RAR1 dependence (constructs 16666 and 11666, each showing 80% haustorium index) or uncoupled RAR1 dependence from recognition specificity (construct 11166, showing 10% haustorium index) in the rar1-2 background. Neither of the two chimeras that retained AvrMla1-dependent resistance activity (61111 and 66111) was impaired functionally upon delivery in rar1-2 mutant plants. Unless MLA6 accumulation is self-limited, we conclude that RAR1 dependence cannot be overcome by Mla6 overexpression and appears to be modulated by both the CC-NB and LRR regions. Because it was reported that an Arabidopsis rar1 mutant line failed to accumulate a CC-NB-LRR protein, RPM1 (Tornero et al. 2002), we also tested whether MLA6 becomes unstable in the rar1-2 mutant background. At 96 h after delivery, MLA6 remained as active as at 15 h after delivery (39% haustorium index), suggesting that the stability of MLA6 remained unchanged in rar1-2 plants (see below for examples of unstable MLA variants 16666 and 11666).
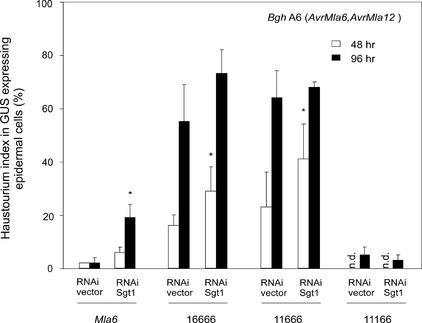
Requirement of Sgt1 for MLA-Mediated Resistance
Barley Sgt1 (HvSgt1) was shown to be required for Mla6- but not Mla1-mediated resistance using double-stranded RNA interference (dsRNAi) gene silencing of HvSgt1 in a single-cell expression system (Azevedo et al., 2002). This technique was used to examine in the Rar1 wild-type background the SGT1 requirement of MLA chimeras that retain MLA6 recognition specificity (constructs 16666, 11666, and 11166 in Figure 5). In these experiments, Bgh spore inoculations were performed at 48 or 96 h after delivery, and the leaf tissue was fixed for microscopic analysis 48 h after spore inoculation. Cobombardment of SGT1 dsRNAi DNA with a plasmid driving wild-type Mla6 from the ubiquitin promoter resulted in a small but significantly increased haustorium index (19% at 96 h after delivery) compared with delivery of an empty dsRNAi vector control (2%). This finding is consistent with previous data (Azevedo et al., 2002). Unexpectedly, the functioning of chimeras 16666 and 11666 was partially impaired at 48 h after delivery in cobombardment experiments with the empty vector dsRNAi control. This phenomenon was time dependent in that the chimeras were almost completely inactive at 96 h after delivery. This finding may indicate that the two chimeric MLA proteins are less stable or that fewer or less active recognition complexes are formed compared with complex formation in the MLA6 wild-type protein. Nevertheless, at 48 h after delivery, cobombardment of plasmids 16666 and 11666 with SGT1 dsRNAi DNA significantly enhanced the haustorium index compared with that in empty vector controls (P < 0.05), indicating at least a partial requirement of the chimeras for Sgt1. By contrast, the 11166 chimeric protein retained full activity upon cobombardment with the empty dsRNAi plasmid control, and its function remained unaffected by Sgt1 silencing even at 96 h after delivery (Figure 5). Unlike wild-type Mla6, AvrMla6-dependent resistance conferred by the 11166 variant appears to be uncoupled from both Rar1 and Sgt1 dependence (Figures 4C and 5).
Figure 5.
Single Cell Silencing of Sgt1 by dsRNAi.
Wild-type Mla6 or chimeras retaining AvrMla6-dependent recognition specificity were coexpressed with a HvSgt1 dsRNAi-silencing plasmid (Azevedo et al., 2002) in the Rar1 wild-type background using a modified single cell transient gene expression assay (Azevedo et al., 2002). After delivery of plasmid DNAs into epidermal cells, detached barley leaves were incubated for 48 h (open bars) or 96 h (closed bars). Subsequently, leaves were inoculated with spores of Bgh isolate A6 (AvrMla6) and incubated for another 48 h. Microscopic scoring of single interaction sites was identical to that described for Figure 2. Asterisks indicate haustorium indices that are significantly different (P < 0.05) from bombardments using empty dsRNAi vector controls. GUS, β-glucuronidase; n.d., not determined.
DISCUSSION
Allelic Variants Encode MLA Powdery Mildew R Proteins
Eight NB-LRR genes are present in a 260-kb interval comprising the Mla locus in barley cv Morex and were classified into three dissimilar families (RGH1, RGH2, and RGH3) with <43% amino acid sequence similarity between families (Wei et al., 2002). Computational analysis of the Morex 260-kb sequence contig suggested that a progenitor Mla locus harbored at >8 million years before the present one member of each RGH family (RGH1bcd, RGH2a, and RGH3a) (Wei et al., 2002). Each of the Mla powdery mildew R genes identified to date shows highest overall sequence similarity to Morex RGH1bcd in coding regions and shares the same exon/intron structure (Figure 6) (Wei et al., 2002). Unlike RGH1bcd, however, Mla1/6/12 each contains a 5′ untranslated open reading frame and, within intron 3, an (AT)n simple sequence repeat consisting of different repeat numbers (Figure 6). Also, Morex RGH1bcd contains a BARE1 solo long terminal repeat in intron 3 that is absent in Mla1/6/12, and the presence of a 29-bp deletion in the LRR region, resulting in a premature stop codon, suggests that it is nonfunctional (Figure 6). Because Morex lacks known Mla powdery mildew resistance specificity, it has been inferred that RGH1bcd is a naturally inactive allele of Mla1 and Mla6 that may have served as a progenitor for the other Morex RGH1 family members (RGH1a, RGH1e, and RGH1f ) (Wei et al., 2002). Closer examination of all possible pair-wise sequence comparisons of the four Morex RGH1 variants and the identified Mla resistance specificities revealed for exon 4 sequences a common cluster that includes genes Mla1/6/12 and RGH1bcd. However, sequences of RGH1bcd exon 3 and intron 3 cluster together with the other RGH1 gene sequences, whereas the identified Mla resistance specificities form a second group (even after the exclusion of the BARE1 long terminal repeat in intron 3 of RGH1bcd; data not shown). Therefore, it is possible that RGH1bcd is the product of a recombination between an ancestral Morex allele of Mla1/6/12 and another more divergent RGH.
DNA gel blot analysis and preliminary sequence information obtained from nearly isogenic barley lines containing other Mla powdery mildew resistance specificities indicate for each line the presence of one candidate gene with high sequence relatedness to MLA1/6/12 (data not shown). Thus, it is possible that many genetically characterized powdery mildew R genes at Mla are variants of the same ancestral RGH1 family member. The presence of the (AT)n microsatellite in all Mla R genes in Bgh identified to date and its absence in currently available Mla RGHs are consistent with our hypothesis, because recent findings indicate that most microsatellites reside in regions predating recent genome expansion in plants (Morgante et al., 2002).
The very high level of DNA sequence conservation in exon and intron sequences of identified Mla R genes (average overall identity of 94 and 93%, respectively) may be indicative of selective constraints acting on both coding and noncoding regions. By contrast, inspection of flanking regions revealed evidence for extensive intralocus recombination events that reshuffled both genes and intergenic regions (Figure 6). For example, a HORPIA2 element was found in the same direction immediately 3′ of RGH1bcd and 3′ of Mla1, whereas 3.7 kb of 3′ flanking sequence of Mla6 showed no significant relatedness to any stretch in the 260-kb Mla Morex contig. Sequences located immediately 3′ of Mla12 were found 5.5 kb downstream of RGH1bcd, indicating an extensive intralocus insertion/deletion event. Morex RGH1f/e exhibited highest sequence relatedness to the Mla1 paralog Mla1-2; their altered relative orientation to RGH1bcd and Mla1, respectively, suggests the occurrence of an intralocus inversion event (Figure 6).
Altering Resistance Response Kinetics by Mla Dosage
Different Mla resistance genes to Bgh show characteristic infection phenotypes that are macroscopically visible by different infection types (Boyd et al., 1995). A quantitative analysis of single interaction sites in nearly isogenic lines containing different Mla genes revealed for Mla1 and Mla6 early termination of Bgh growth coincident with haustorium differentiation (Boyd et al., 1995). By contrast, Mla3 and Mla7 mediated cessation of fungal growth at a later stage of the infection process, permitting the growth of elongating secondary hyphae on the leaf surface in addition to haustorium differentiation. These Mla gene-specific differences correlated with the timing of a cell death response that was either rapid, involving attacked epidermal cells, or slower, including epidermal and subtending mesophyll cells (Boyd et al., 1995). Similarly, delayed cell death–associated resistance is characteristic for lines carrying Mla12, permitting indistinguishable fungal growth for up to 36 h after Bgh spore inoculation and a high haustorium index of ∼60% on both Mla12-resistant and Mla12-susceptible mutant plants (Freialdenhoven et al., 1994). It is possible that differences in the speed of Mla resistance responses are the indirect consequence of different infection stage–specific delivery systems for particular Bgh AVRMLA effector proteins (e.g., delivery of AVRMLA12 only after or coincident with haustorium differen-tiation).
Precedence for this idea is found in the expression of Cladosporium fulvum AVR9, which is induced strongly upon a switch from surface to intercellular growth of the fungus in leaves, which may be cued by fungal nitrogen starvation (Van Kan et al., 1991; Perez-Garcia et al., 2001). Here, we have shown that slow Mla12-triggered resistance was altered dramatically to a rapid response by Mla12 overexpression, leading to almost complete abortion of Bgh attack before haustorium differentiation (Figure 2). Because the rapid response retained AVRMLA12 dependence, the Bgh effector protein must be, like AVRMLA1 and AVRMLA6 (Halterman et al., 2001; Zhou et al., 2001) (Figure 2), delivered before or during the switch from surface to invasive fungal growth. The rapid Mla12 overexpression response suggests that cellular amounts of MLA12 or protein complexes containing MLA12 are rate limiting for the onset or speed of the resistance. This finding is consistent with previous results demonstrating markedly reduced resistance in plants that are heterozygous for Mla12 (Torp and Jørgensen, 1986). In addition, the retained Rar1 dependence of the Mla12 overexpression phenotype corroborates this as an authentic response. Assuming that expression levels of different Mla genes are similar and sustain comparable protein abundance, it remains possible that the gene-specific infection types reflect differences in the activities of presumed MLA-containing recognition complexes or different intrinsic activities of AVRMLA proteins.
Determinants of MLA Recognition Specificity
Functional analysis of reciprocal domain-swap constructs between Mla1 and Mla6 revealed an essential role of the LRR-CT unit in specificity determination (Figure 4B). We found that distinct regions in the LRRs of MLA1 and MLA6 (LRRs 3 to 11 and 9 to 11, respectively) were necessary for cognate AVRMLA perception. This finding is in agreement with LRRs representing the most variable part of MLA and other characterized NB-LRR– type R proteins (Botella et al., 1998; McDowell et al., 1998; Meyers et al., 1998; Ellis et al., 1999; Halterman et al., 2001). It also is consistent with the finding that potentially solvent-exposed residues in MLA LRRs and those of other NB-LRR R proteins are subject to diversifying selection (Botella et al., 1998; McDowell et al., 1998; Meyers et al., 1998; Halterman et al., 2001). One interpretation of these data is that the diversified regions are involved in ligand-specific recognition.
LRRs have been demonstrated to function as specificity determinants of membrane-anchored R proteins (Van der Hoorn et al., 2001; Wulff et al., 2001). Successful domain-swap experiments have been reported only for intracellular TIR-NB-LRR–encoding resistance alleles at the L locus in flax to the flax rust fungus (Ellis et al., 1999; Luck et al., 2000). Both MLA and L proteins exhibit comparable average polymorphisms in corresponding domains (based on four MLA variants, including MLA13 [Halterman et al., 2003], and 11 L variants from flax). Unlike our study involving CC-NB-LRR proteins, the analysis of L chimera functions suggested that both TIR-NB and LRR regions can determine specificity differences (Ellis et al., 1999; Luck et al., 2000). Although it is possible that the CC-NB domain is irrelevant for specificity determination, more diverged CC-NB domains from other MLA proteins must be tested before we can generalize from the observations based on MLA1 and MLA6 chimeras.
Reciprocal swaps of the CT domains between MLA1 and MLA6 resulted in nonfunctional chimeras (11116 and 66661; Figure 4B). Our interpretation that cognate LRR-CT units are required for MLA specificity determination was supported by the finding that two of three single–amino acid replacements in mutant MLA12 variants affect CT amino acids and the third affects an LRR residue (Figure 1). Additional evidence for a role of the MLA CT in specificity determination comes from the identification of a hypervariable region in the middle of this domain (residues 893 to 945 in MLA1). This hypervariable region shows an increased ratio of nonsynonymous (ka = 15.4) to synonymous (ks = 9.6) nucleotide substitutions (based on Mla1, Mla6, Mla12, and Mla13 sequences [Halterman et al., 2003]; significant at P < 0.1%), which is indicative of the operation of diversifying selection. This is like the C-terminal non-LRR domain of P locus genes that encode flax TIR-NB-LRR proteins, which also was found to contain a region that is subject to diversifying selection and might contribute to specificity determination (Dodds et al., 2001).
RAR1/SGT1 May Act Downstream of Presumptive MLA Recognition Complexes
There is strong evidence suggesting a conserved role for RAR1 in R gene–mediated resistance to different pathogen classes and in different plant clades (Shirasu et al., 1999a; Liu et al., 2002b; Muskett et al., 2002; Tornero et al., 2002). RAR1 has been implicated in ubiquitin-protein conjugation pathway(s) together with SGT1 (Azevedo et al., 2002; Liu et al., 2002b). Ubiquitination targets have not been identified to date, and it remains unclear whether RAR1/SGT1 acts upstream of, coincident with, or downstream of R protein recognition complexes. The variation in Rar1 requirement for the function of different Mla resistance specificities (Jørgensen, 1996) is unique with regard to their potential allelism and unusual sequence relatedness. Despite a dramatic shift to a rapid resistance response resulting from the overexpression of Mla12, its Rar1 dependence remained unaltered (Figure 2). Likewise, the partial Rar1 requirement for Mla6 function and the Rar1-independent Mla1 activity remained unchanged upon the expression of both R genes from either the strong ubiquitin promoter or native 5′ flanking regulatory sequences (Figure 4). Thus, RAR1 dependence appears to be conditioned by subtle intrinsic properties of MLA proteins but not by dosage. Consistent with this finding, replacement of MLA6 domains with the corresponding MLA1 parts generated variants conferring AvrMla6-specific immunity that was either fully dependent on or independent of Rar1 (Figure 4C). We were unable to examine this using the reciprocal chimeras because these were either nonfunctional (66611) or mediated Rar1-independent resistance activity (61111 and 66111).
A role for RAR1 in the assembly of preformed R protein–containing recognition complexes may be inferred from the finding that a nonchallenged Arabidopsis rar1 mutant line failed to accumulate the RPM1 CC-NB-LRR protein to Pseudomonas syringae (Tornero et al., 2002). Our study demonstrates that the reliance on RAR1 and SGT1 is not absolute for a given Mla recognition specificity. Successful uncoupling of AvrMla6 recognition from Rar1/Sgt1 dependence implies that RAR1 cannot be required for processes that occur “upstream” from recognition (e.g., in planta processing or transport of AVRMLA6) (see chimera 11166 in Figures 4C and 5). Also, the uncoupling excludes the possibility that MLA6 “guards” RAR1 or SGT1 in presumed MLA-containing recognition complexes. It is possible that the MLA6 CC-NB domain and the LRRs exert antagonistic roles, the former inhibiting and the latter enhancing RAR1-dependent R protein function (cf. constructs 16666, 11166, and wild-type MLA6 in Figure 4C). The observed partial impairment of Mla6 wild-type function in rar1 plants probably is not the result of MLA6 destabilization, because the activity was time independent (unchanged at 15 and 96 h after DNA delivery). This result is consistent with the finding that Mla6 overexpression in the rar1 mutant background did not increase resistance (i.e., the amount of functional recognition complexes) (Figure 4C). Thus, it seems possible that RAR1/SGT1 exerts a function downstream from activated MLA recognition complexes in resistance signaling. Therefore, the observed variation in Rar1/Sgt1 reliance on the function of different MLA wild type or MLA chimeras may be attributable to variation in signal flux set by intrinsic activities of MLA variants in AVRMLA-activated recognition complexes (e.g., by different half-lives of active complexes).
Do MLA Chimeras Affect Folding of MLA Recognition Complexes?
The SGT1 binding function of plant RAR1 proteins has been conserved in monocots and dicots (Azevedo et al., 2002; Liu et al., 2002b). Our data obtained from Sgt1-silencing experiments in cells expressing MLA chimeras that retain AVRMLA6 recognition suggest that RAR1/SGT1 functions in MLA6 resistance are closely linked (Figure 5). For example, the RAR1-independent function of chimera 11166 retained full activity upon Sgt1 silencing; inversely, chimeras showing full RAR1 dependence also retained SGT1 dependence. In addition, the function of Mla12, which requires Rar1, was compromised significantly in Sgt1-silencing experiments (data not shown). Recent experiments using Saccharomyces cerevisiae and Schizosaccharomyces pombe sgt1 mutant strains indicate a potential role of the wild-type protein as a co-chaperone or an assembly factor of diverse regulatory multiprotein complexes, including SCF-type E3 ubiquitin ligases, the structurally related CBF3 kinetochore complex, and the Cyr1p adenylyl cyclase complex (Kitagawa et al., 1999, Dubacq et al., 2002; Garcia-Ranea et al., 2002; Schadick et al., 2002). In this context, it is notable that either of two Arabidopsis SGT1 genes was shown to complement S. cerevisiae sgt1 mutant strains (Azevedo et al., 2002). A central conserved part in SGT1 proteins likely adopts a fold similar to that of the p23 co-chaperone, which is known to interact with the heat-shock protein hsp90 chaperone and participates in the folding of different regulatory proteins (Dubacq et al., 2002; Garcia-Ranea et al., 2002). Therefore, it is possible that the observed variation in RAR1/SGT1 dependence for the function of different Mla resistance specificities or MLA chimeras reflects differences in the degree of folding/activation assistance needed for presumed MLA-containing recognition complexes. In this scenario, the signal flux in downstream signaling pathways might be similar for both RAR1/SGT1-dependent and -independent resistance.
METHODS
Plant and Fungal Material
Sultan 5 is a chromosome-doubled haploid barley (Hordeum vulgare) cultivar containing Mla12. Mla12 mutants (M66 and M86), a rar1-2 mutant allele (M100), and the rar2 mutant (M22) were generated by chemical mutagenesis of Sultan 5 seeds (Torp and Jørgensen, 1986). Ingrid (mlo-3 Rar1) was generated by seven backcrosses with cv Ingrid, and the mlo-29 rar1-2 double mutant was isolated originally from a remutagenized rar1-2 M2 population. The latter line was used to test the Rar1 dependence of MLA chimeras (Figure 4). All barley seedlings were grown at 20°C with 16 h of light and 8 h of darkness. The barley powdery mildew (Blumeria graminis f. sp. hordei [Bgh]) isolate A6 (AvrMla6, AvrMla12, virMla1) was maintained on P01, a nearly isogenic line from cv Pallas containing Mla1. Isolate K1 (AvrMla1 virMla6 virMla12) was maintained on I10, a nearly isogenic line from cv Ingrid containing Mla12. Plants or detached leaves were kept at 18°C and 60% RH with 16 h of light and 8 h of darkness after inoculation with Bgh spores.
Genomic Library Construction and Screening for MLA12-Containing Cosmids
High molecular mass genomic DNA was isolated from Sultan 5 containing Mla12 and partially digested with Sau3AI to produce DNA fragments of 30 to 60 kb. After dephosphorylation, the fragments were ligated to the XbaI-BamHI–linearized SuperCos cosmid vector according to the manufacturer's instructions (Stratagene). A total of 240 pools averaging 4000 clones each were made and kept frozen as glycerol stocks. The library had an average insert size of 25 kb (ranging from 15 to 40 kb) and represents approximately five genome equivalents. DNA preparations were made using the R.E.A.L Prep 96 Plasmid Kit (Qiagen, Valencia, CA) from all pools. For library screening, the plasmid DNA of each pool was digested with HindIII or EcoRI, resolved by 0.8% agarose gel electrophoresis, and blotted onto Hybond-N+ membranes (Amersham Pharmacia Biotech). To identify positive pools containing the Mla12 candidate gene, the DNA gel blots were hybridized with a 32P-labeled probe, which was derived from the Leu-rich repeat region of Mla1 (covering exon 4 of Mla1) (Zhou et al., 2001). Approximately 15,000 colonies of each positive pool were screened by hybridization with the same probe to obtain purified clones. Positive clones were fingerprinted using various restriction enzymes.
Sequencing and Gene Characterization
Plasmid DNA of Mla12-containing clones was isolated using the Midi-Plasmid-DNA Prep Kit (Qiagen), subcloned, and sequenced as described (Zhou et al., 2001). Construction of sequence contigs was performed using the GCG9 and STADEN software packages (University of Wisconsin Genetics Computer Group, Madison). Sequence alignment was performed using a World Wide Web–based program (http://prodes.toulouse.inra.fr/multalin/multalin.html).
Sequencing of Mla12 Mutant Alleles
Genomic DNA was isolated from the Mla12 mutants M86 and M66 and the rar2 mutant M22. The DNA was used as a template for PCR amplification of the respective Mla12 mutant alleles. Mla12-specific primers were designed based on the sequence alignment of Mla12, Mla1, Mla6, Mla1-2, and RGH1a (primer sequences are listed in Table 1). PCR products were purified using the Qiagen PCR Product Purification Kit and then sequenced directly. Mutations were identified by aligning the sequences of PCR products to Mla12 and confirmed by three additional independent PCR procedures and sequencing of plus and minus strands of the mutated region.
Table 1.
Mla12-Specific Primer Sequences
| Primer | Sequence | Position in Mla12 |
|---|---|---|
| Mla12S1a | 5′-CACCTCACCTTCTGTCTCTCTC-3′ | −488 |
| Mla12S1b | 5′-GCATCTTTCTTGCTATTCTGCTC-3′ | −328 |
| Mla12S1c | 5′-TGCCATTTCCAACCTGATTCCC-3′ | 12 |
| Mla12AS1a | 5′-CCTTGTTCCTGTCACGCCTATC-3′ | 34 |
| Mla12AS1b | 5′-CCTTTAATCTTCTCGTATACCGCTC-3′ | 658 |
| Mla12AS1c | 5′-TGTTTAGTGTGAACTGCTTATGCC-3′ | 945 |
| Mla12As1d | 5′-TCTCCCTCTTTCCTTCCTCTCC-3′ | 1228 |
| Mla12S2a | 5′-GATGCTTAATGAGAGTAAGATTATCGAG-3′ | 1705 |
| Mla12S2b | 5′-GGCATCAACTTTGCTTTCTCCAATAG-3′ | 1913 |
| Mla12AS2b | 5′-CGACGACAATTACTCTGTGAAGAC-3′ | 2652 |
| Mla12AS2a | 5′-GAAGGGACAAACGACGACAATTACT-3′ | 2663 |
| Mla12S3a | 5′-TAACAGTTTAGAGGAGATGCGG-3′ | 2366 |
| Mla12S3b | 5′-CTCCCGACTGAGATAGGAAAAC-3′ | 2915 |
| Mla12S3c | 5′-TTGTTGTCCCTTCGTCGTCTCTGG-3′ | 3586 |
| Mla12AS3b | 5′-CACAATAGAGAAGAACAAAGACATC-3′ | 3775 |
| Mla12AS3c | 5′-TGTGCGCCAAAAATCAGTTCTCAC-3′ | 4057 |
| Mla12AS3a | 5′-ATGGAGAAAGGAAGGTAGGTGG-3′ | 4139 |
Construction of Mla-Containing Plasmid Expression Vectors
pUbi-GFP-Nos [maize ubiquitin1 promoter-GFP-Nos poly(A) signal] (Shirasu et al., 1999b) was used as a backbone to subclone Mla1, Mla6, and Mla12. The green fluorescent protein open reading frame was deleted using restriction enzymes PstI and SacI and replaced by an adaptor with a suitable multiple cloning site for Mla genes. The 5′ untranslated region of Mla1 was amplified by PCR using primer pairs MlapstIs1 and MlaAgeIas1, and the product was cloned into pGEM-T vector (Promega) and confirmed by sequencing. The 5′ untranslated regions were subcloned into the pUbi-Adaptor-Nos vector using enzymes HindIII and AgeI. The 3′ untranslated region of Mla1 was amplified with primers MlaEcoRIas1 and MlaBsrDIs1 and cloned into pGEM-T vector. After sequence confirmation, the 3′ untranslated region was subcloned into the pUbi-Adaptor-Nos vector using BsrDI and NotI. The plasmid vector then was linearized with AgeI and BsrDI, and coding regions of Mla1, Mla6, and Mla12, including introns 3 and 4, were inserted. The resulting overexpression plasmids were designated pUbiMla1Nos, pUbiMla6Nos, and pUbiMla12Nos. They served as backbones to generate domain-swap constructs between Mla1 and Mla6 and Mla mutant variant constructs (see below). Plasmids driven by native 5′ flanking Mla promoter sequences were generated by subcloning an 8-kb SacII-XhoI fragment from Mla1 containing cosmid p6-49-15, or an AvrII-PciI fragment of Mla6 containing cosmid 9589-5a, into pBluescript II KS− (Stratagene).
Plasmids 16666 and 61111 were generated by exchanging BbsI-NotI fragments, which were derived from pUbiHEMla1Nos and pUbiHEMla6Nos, respectively. Likewise, plasmids 11666 and 66111 were generated by exchanging Bsu36I-NotI fragments. Plasmids 11166 and 66611 were generated by splicing by overlap extension (SOE) using the forward primer MlaBbSIs, the reverse primer NotIas, and the overlapping primers P10s and P10as. The Bsu36I-NotI enzyme pair was used to digest the SOE products that were inserted into pUbiMla1Nos or pUbiMla6Nos digested with the same enzyme pair, respectively. Plasmids 11661 and 66116 also were generated by SOE with primers P5s/P5as and P12s/P12as covering the swap sites and the flanking primers MlaBbSIs and Mla1EcoRIas1. The BbsI-NotI–digested fragments of the SOE products were inserted into pUbiMla1Nos and pUbiMla6Nos, respectively. Plasmids 11116 and 66661 were generated by subcloning Bsu36I-NotI fragments of plasmids 66116 and 11661 into pUbiMla1Nos and pUbiMla6Nos, respectively.
For the construction of plasmids pUbiMla1(K915M), pUbiMla6 (K913M), and pUbiMla12(K916M), a single amino acid exchange was introduced by SOE using a template of pUbiMla1Nos, pUbiMla6Nos, and pUbiMla12Nos, respectively. Likewise, variants pUbiMla1(L631R), pUbiMla6(L631R), and pUbiMla12(L631R) were generated by SOE reactions using the same template DNAs. Primers used for these reactions are listed in Table 2. For site-directed mutagenesis of the codon leading to the replacement of Lys with Met, two common primers, MLABbSIs and MLABsrDIas1 (for Mla1 and Mla6) and MLA12BsrDIas1 (for Mla12), were used in combination with overlapping primers MLA12DNas2 and Mla12DNs1. The BbSI-BsrDI enzyme pair was used to digest the SOE products, and the resulting fragments were inserted into pUbiMla1Nos, pUbiMla6Nos, and pUbiMla12Nos. For site-directed mutagenesis of the codon leading to the replacement of Leu with Arg, four common primers, P2s, M66-as, M66-s, and Exon-5as, were used for SOE reactions. The SOE products were digested with Bsu36I-SbfI (for Mla1 and Mla12) or Bsu36I-BspEI (for Mla6), and fragments were inserted into pUbiMla1 Nos, pUbiMla6Nos, and pUbiMla12Nos digested with the same enzyme pair, respectively.
Table 2.
Primers Used in SOE Reactions
| Primer | Sequence |
|---|---|
| Exon-5as | 5′-AATCGTCATCATGAGCACCTT-3′ |
| M66-as | 5′-CCAACACCTCCAAAAACTGCCGTTTTCCT-3′ |
| M66-s | 5′-CTGAGATAGGAAAACGGCAGTTT-3′ |
| Mla12BsrDIas1 | 5′-CTGATGCAATGTGAATCCTTGTG-3′ |
| Mla12BsrDIs1 | 5′-ACATTGCATCAGATGTGCTCTG-3′ |
| Mla12DNas2 | 5′-GCTTCCATTGCCTCCCCAACCCT-3′ |
| Mla12DNs1 | 5′-GAGCGAGGGTTGGGGAGGCAATG-3′ |
| Mla1EcoRIas1 | 5′-AAGCGGCCGCGAATTCTAATACTACTAGGA- CTCG-3′ |
| MlaAgeIas1 | 5′-TGGCACCGGTGACAATATCCAT-3′ |
| MlaBbSIs | 5′-TGGGAATAGCATGTCTTCACAG-3′ |
| MlaBsrDIas1 | 5′-TGATGCAATGTGAGTCGCTCTGG-3′ |
| MlaBsrDIs1 | 5′-CTGATCCAGAGCGACTCACATTGC-3′ |
| MlaPstIs1 | 5′-CTTCTGCAGACTGAGTCATCGGCACCTTGC-3′ |
| NotIas | 5′-GCAAGACCGGCAACAGGATTCAA-3′ |
| P10as | 5′-TCGCAGTGCAGAGAGTTGGCT-3′ |
| P10s | 5′-AGCCAACTCTCTGCACTGCGA-3′ |
| P12as | 5′-TCAAACAATATCTGCGTGGCA-3′ |
| P12s | 5′-TGCCACGCAGATATTGTTTGA-3′ |
| P2s | 5′-GCTCGGATTAAATTACTTCAACC-3′ |
| P5as | 5′-CAAGATCCAACACCTCCAAAAACT-3′ |
| P5s | 5′-AGTTTTTGGAGGTGTTGGATCTT-3′ |
Single-Cell Transient Expression Assay
The single-cell transient expression assay was performed essentially according to Shirasu et al. (1999b). Reporter plasmids containing Mlo and β-glucuronidase (GUS) genes (GUS alone in the case of the Mlo genetic background) and the respective effector plasmids were mixed before coating of the particles (molar ratio of 2:1; 5 μg of total DNA). The bombarded leaves were transferred to 1% agar plates supplemented with 85 μM benzimidazole and incubated at 18°C for 15 h before high-density inoculation with Bgh spores. Leaves were stained for GUS, and single leaf epidermal cells attacked by Bgh germlings were evaluated microscopically at 48 h after spore inoculation. In the double-stranded RNA interference single-cell silencing experiments, particles were co-coated with a construct encoding an intron-spliced double-stranded RNA interference construct targeting HvRAR1 or HvSGT1 according to Azevedo et al. (2002) (molar ratio of 1:1:1; 5 μg of total DNA). Note that in the gene-silencing experiments, the bombarded leaves were inoculated at 18°C for 48 or 96 h before high-density inoculation to allow the turnover of preformed RAR1 or SGT1.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Number
The GenBank accession number for the Mla12 genomic sequence is AY196347.
Acknowledgments
We thank Roger Wise and Dennis Halterman for the gift of C.I. 16151 cosmid clone 9589-5a containing Mla6 and for sharing associated sequence data before publication. We thank Jane Parker for providing valuable comments on the manuscript. This work was supported by the Max Planck Society and the Gatsby Charitable Organization.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.009258.
References
- Austin, M.J., Muskett, P., Kahn, K., Feys, B.J., Jones, J.D.G., and Parker, J.E. (2002). Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295, 2077–2080. [DOI] [PubMed] [Google Scholar]
- Azevedo, C., Sadanandom, A., Kitagawa, K., Freialdenhoven, A., Shirasu, K., and Schulze-Lefert, P. (2002). The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295, 2073–2076. [DOI] [PubMed] [Google Scholar]
- Botella, M.A., Parker, J.E., Frost, L.N., Bittner-Eddy, P.D., Beynon, J.L., Daniels, M.J., Holub, E.B., and Jones, J.D.G. (1998). Three genes of the Arabidopsis Rpp1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. Plant Cell 10, 1847–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd, L.A., Smith, P.H., Foster, E.M., and Brown, J.K.M. (1995). The effects of allelic variation at the Mla resistance locus in barley on the early development of Erysiphe graminis f. sp. hordei and host responses. Plant J. 7, 959–968. [Google Scholar]
- Brown, J.K.M., and Jessop, A.C. (1995). Genetics of avirulences in Erysiphe graminis f. sp. hordei. Plant Pathol. 44, 1039–1049. [Google Scholar]
- Caffier, V., Vallavieille-Pope, C., and Brown, J.K.M. (1996). Segregation of avirulences and genetic basis of infection types in Erysiphe graminis f. sp. hordei. Phytopathology 86, 1112–1121. [Google Scholar]
- Dangl, J.L., and Jones, J.D.G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N., Lawrence, G.J., and Ellis, J.G. (2001). Six amino acid changes confined to the leucine-rich repeat β-strand/β-turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell 13, 163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubacq, C., Guerois, R., Courbeyrette, R., Kitagawa, K., and Mann, C. (2002). Sgt1p contributes to cyclic AMP pathway activity and physically interacts with the adenylyl cyclase Cyr1p/Cdc35p in budding yeast. Eukaryot. Cell 1, 568–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, J., Dodds, P., and Pryor, T. (2000). Structure, function and evolution of plant disease resistance genes. Curr. Opin. Plant Biol. 3, 278–284. [DOI] [PubMed] [Google Scholar]
- Ellis, J., Lawrence, G.J., Luck, J.E., and Dodds, P.N. (1999). Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11, 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freialdenhoven, A., Scherag, B., Hollricher, K., Collinge, D.B., Thordal-Christensen, H., and Schulze-Lefert, P. (1994). Nar-1 and Nar-2, two loci required for Mla12-specified race-specific resistance to powdery mildew in barley. Plant Cell 6, 983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ranea, J., Mirey, G., Camonis, J., and Valencia, A. (2002). p23 and HSP20/α-crystallin proteins define a conserved sequence domain present in other eukaryotic protein families. FEBS Lett. 529, 162–167. [DOI] [PubMed] [Google Scholar]
- Halterman, D., Wei, F., and Wise, R.P. (2003). Powdery mildew induced Mla mRNAs are alternatively spliced and contain multiple upstream open reading frames. Plant Physiol. 131, in press. [DOI] [PMC free article] [PubMed]
- Halterman, D., Zhou, F.S., Wei, F., Wise, R.P., and Schulze-Lefert, P. (2001). The MLA6 coiled-coil, NBS-LRR protein confers AvrMla6-dependent resistance specificity to Blumeria graminis f. sp. hordei in barley and wheat. Plant J. 25, 335–348. [DOI] [PubMed] [Google Scholar]
- Holt, B.F., Boyes, D.C., Ellerstrom, M., Siefers, N., Wiig, A., Kauffman, S., Grant, M.R., and Dangl, J.L. (2002). An evolutionarily conserved mediator of plant disease resistance gene function is required for normal Arabidopsis development. Dev. Cell 2, 807–817. [DOI] [PubMed] [Google Scholar]
- Jia, Y., McAdams, S.A., Bryan, G.T., Hershey, H.P., and Valent, B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen, J.H. (1988). Genetic analysis of barley mutants with modifications of powdery mildew resistance gene Ml-a12. Genome 30, 129–132. [Google Scholar]
- Jørgensen, J.H. (1994). Genetics of powdery mildew resistance in barley. Crit. Rev. Plant Sci. 13, 97–119. [Google Scholar]
- Jørgensen, J.H. (1996). Effect of three suppressors on the expression of powdery mildew resistance genes in barley. Genome 39, 492–498. [DOI] [PubMed] [Google Scholar]
- Kim, M.C., Panstruga, R., Elliott, C., Muller, J., Devoto, A., Yoon, H.W., Park, H.C., Cho, M.J., and Schulze-Lefert, P. (2002). Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416, 447–450. [DOI] [PubMed] [Google Scholar]
- Kitagawa, K., Skowyra, D., Elledge, S.J., Harper, J.W., and Hieter, P. (1999). SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol. Cell 4, 21–33. [DOI] [PubMed] [Google Scholar]
- Leister, R.T., and Katagiri, F. (2000). A resistance gene product of the nucleotide binding site-leucine rich repeats class can form a complex with bacterial avirulence proteins in vivo. Plant J. 22, 345–354. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Schiff, M., Marathe, R., and Dinesh-Kumar, S.P. (2002. a). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30, 415–429. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Schiff, M., Serino, G., Deng, X.-W., and Dinesh-Kumar, S.P. (2002. b). Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene–mediated resistance response to Tobacco mosaic virus. Plant Cell 14, 1483–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck, J.E., Lawrence, G.J., Dodds, P.N., Shepherd, K.W., and Ellis, J.G. (2000). Regions outside of the leucine-rich repeats of flax rust resistance proteins play a role in specificity determination. Plant Cell 12, 1367–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey, D., Holt, B.F., Wiig, A., and Dangl, J.L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108, 743–754. [DOI] [PubMed] [Google Scholar]
- McDowell, J.M., Dhandaydham, M., Long, T.A., Aarts, M.G.M., Goff, S., Holub, E.B., and Dangl, J.L. (1998). Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the Rpp8 locus of Arabidopsis. Plant Cell 10, 1861–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B.C., Shen, K.A., Rohani, P., Gaut, B.S., and Michelmore, R.W. (1998). Receptor-like genes in the major resistance locus of lettuce are subject to divergent selection. Plant Cell 10, 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgante, M., Hanafey, M., and Powell, W. (2002). Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat. Genet. 30, 194–200. [DOI] [PubMed] [Google Scholar]
- Muskett, P.R., Kahn, K., Austin, M.J., Moisan, L.J., Sadanandom, A., Shirasu, K., Jones, J.D.G., and Parker, J.E. (2002). Arabidopsis RAR1 exerts rate-limiting control of R gene–mediated defenses against multiple pathogens. Plant Cell 14, 979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart, J.R., et al. (2002). Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl. Acad. Sci. USA 99, 10865–10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen, C., Rasmussen, S., and Giese, H. (2002). A genetic map of Blumeria graminis based on functional genes, avirulence genes, and molecular markers. Fungal Genet. Biol. 35, 235–246. [DOI] [PubMed] [Google Scholar]
- Perez-Garcia, A., Snoeijers, S.S., Joosten, M., Goosen, T., and De Wit, P. (2001). Expression of the avirulence gene Avr9 of the fungal tomato pathogen Cladosporium fulvum is regulated by the global nitrogen response factor NRF1. Mol. Plant-Microbe Interact. 14, 316–325. [DOI] [PubMed] [Google Scholar]
- Rivas, S., Mucyn, T., van den Burg, H.A., Vervoort, J., and Jones, J.D.G. (2002. a). An ∼400 kDa membrane-associated complex that contains one molecule of the resistance protein Cf-4. Plant J. 29, 783–796. [DOI] [PubMed] [Google Scholar]
- Rivas, S., Romeis, T., and Jones, J.D.G. (2002. b). The Cf-9 disease resistance protein is present in an ∼420-kilodalton heteromultimeric membrane-associated complex at one molecule per complex. Plant Cell 14, 689–702. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Schadick, K., Fourcade, H., Boumenot, P., Seitz, J., Morrell, J., Chang, L., Gould, K., Partridge, J., Allshire, R., Kitagawa, K., Hieter, P., and Hoffman, C. (2002). Schizosaccharomyces pombe Git7p, a member of the Saccharomyces cerevisiae Sgt1p family, is required for glucose and cyclic AMP signaling, cell wall integrity, and septation. Eukaryot. Cell 1, 558–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüller, C., Backes, G., Fischbeck, G., and Jahoor, A. (1992). RFLP markers to identify the alleles on the Mla locus conferring powdery mildew resistance in barley. Theor. Appl. Genet. 84, 330–338. [DOI] [PubMed] [Google Scholar]
- Shirasu, K., Lahaye, T., Tan, M.W., Zhou, F.S., Azevedo, C., and Schulze-Lefert, P. (1999. a). A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell 99, 355–366. [DOI] [PubMed] [Google Scholar]
- Shirasu, K., Nielsen, K., Piffanelli, P., Oliver, R., and Schulze-Lefert, P. (1999. b). Cell-autonomous complementation of mlo resistance using a biolistic transient expression system. Plant J. 17, 293–299. [Google Scholar]
- Tör, M., Gordon, P., Cuzick, A., Eulgem, T., Sinapidou, E., Mert-Turk, F., Can, C., Dangl, J.L., and Holub, E.B. (2002). Arabidopsis SGT1b is required for defense signaling conferred by several downy mildew resistance genes. Plant Cell 14, 993–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero, P., Merritt, P., Sadanandom, A., Shirasu, K., Innes, R.W., and Dangl, J.L. (2002). RAR1 and NDR1 contribute quantitatively to disease resistance in Arabidopsis, and their relative contributions are dependent on the R gene assayed. Plant Cell 14, 1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torp, J., and Jørgensen, J.H. (1986). Modification of barley powdery mildew resistance gene Ml-a12 by induced mutation. Can. J. Genet. Cytol. 28, 725–731. [Google Scholar]
- Van der Hoorn, R.A.L., Roth, R., and De Wit, P.J.G. (2001). Identification of distinct specificity determinants in resistance protein Cf-4 allows construction of a Cf-9 mutant that confers recognition of avirulence protein AVR4. Plant Cell 13, 273–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kan, J.A.L., Van den Ackerveken, G.F.J.M., and De Wit, P.J.G.M. (1991). Cloning and characterization of cDNA of avirulence gene avr9 of the fungal pathogen Cladosporium fulvum, causal agent of tomato leaf mold. Mol. Plant-Microbe Interact. 4, 52–59. [DOI] [PubMed] [Google Scholar]
- Wei, F., Gobelman-Werner, K., Morroll, S.M., Kurth, J., Mao, L., Wing, R., Leister, D., Schulze-Lefert, P., and Wise, R.P. (1999). The Mla (powdery mildew) resistance cluster is associated with three NBS-LRR gene families and suppressed recombination within a 240-kb DNA interval on chromosome 5S (1HS) of barley. Genetics 153, 1929–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, F., Wing, R.A., and Wise, R.P. (2002). Genome dynamics and evolution of the Mla (powdery mildew) resistance locus in barley. Plant Cell 14, 1903–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise, R.P., and Ellingboe, A.H. (1983). Infection kinetics of Erysiphe graminis f. sp. hordei on barley with different alleles at the Ml-a locus. Phytopathology 73, 1220–1222. [Google Scholar]
- Wulff, B.B.H., Thomas, C.M., Smoker, M., Grant, M., and Jones, J.D.G. (2001). Domain swapping and gene shuffling identify sequences required for induction of an Avr-dependent hypersensitive response by the tomato Cf-4 and Cf-9 proteins. Plant Cell 13, 255–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, F.S., Kurth, J.C., Wei, F., Elliott, C., Vale, G., Yahiaoui, N., Keller, B., Somerville, S., Wise, R., and Schulze-Lefert, P. (2001). Cell-autonomous expression of barley Mla1 confers race-specific resistance to the powdery mildew fungus via a Rar1-independent signaling pathway. Plant Cell 13, 337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]