Abstract
Mitogen-activated protein kinase (MAPK) cascades play an important role in mediating stress responses in eukaryotic organisms. However, little is known about the role of MAPKs in modulating the interaction of defense pathways activated by biotic and abiotic factors. In this study, we have isolated and functionally characterized a stress-responsive MAPK gene (OsMAPK5) from rice. OsMAPK5 is a single-copy gene but can generate at least two differentially spliced transcripts. The OsMAPK5 gene, its protein, and kinase activity were inducible by abscisic acid as well as various biotic (pathogen infection) and abiotic (wounding, drought, salt, and cold) stresses. To determine its biological function, we generated and analyzed transgenic rice plants with overexpression (using the 35S promoter of Cauliflower mosaic virus) or suppression (using double-stranded RNA interference [dsRNAi]) of OsMAPK5. Interestingly, suppression of OsMAPK5 expression and its kinase activity resulted in the constitutive expression of pathogenesis-related (PR) genes such as PR1 and PR10 in the dsRNAi transgenic plants and significantly enhanced resistance to fungal (Magnaporthe grisea) and bacterial (Burkholderia glumae) pathogens. However, these same dsRNAi lines had significant reductions in drought, salt, and cold tolerance. By contrast, overexpression lines exhibited increased OsMAPK5 kinase activity and increased tolerance to drought, salt, and cold stresses. These results strongly suggest that OsMAPK5 can positively regulate drought, salt, and cold tolerance and negatively modulate PR gene expression and broad-spectrum disease resistance.
INTRODUCTION
Plants are constantly exposed to a variety of biotic (i.e., pathogen infection and insect herbivory) and abiotic (i.e., high or low temperature, drought, and salinity) stresses. To survive these challenges, plants have developed elaborate mechanisms to perceive external signals and to manifest adaptive responses with proper physiological and morphological changes (Bohnert et al., 1995). At the molecular level, the perception of extracellular stimuli and the subsequent activation of defense responses require a complex interplay of signaling cascades, in which reversible protein phosphorylation plays a central role (Yang et al., 1997).
The mitogen-activated protein (MAP) kinase cascade is one of the well-characterized intracellular signaling modules, and it is highly conserved among eukaryotes (Hirt, 1997; Kultz, 1998). This phosphorylation cascade typically consists of three functionally interlinked protein kinases: a MAP kinase kinase kinase (MAPKKK), a MAP kinase kinase (MAPKK), and a MAP kinase (MAPK). In this phosphorylation module, a MAPKKK phosphorylates and activates a particular MAPKK, which in turn phosphorylates and activates a MAPK. Activated MAPK often is imported into the nucleus, where it phosphorylates and activates specific downstream signaling components such as transcription factors (Khokhlatchev et al., 1998). The mammalian MAPKs have been classified into three subgroups based on phylogeny and function (Kultz, 1998). The first subgroup is referred to as extracellular signal-regulated kinases, which are involved in differentiation and cell cycle regulation. The MAPKs in this subgroup are characterized by the specific dual phosphorylation motif TEY (Seger and Krebs, 1995). The other two subgroups are the stress-activated protein kinase/Jun N-terminal kinase subfamily, in which TPY is the phosphorylation motif, and the p38/HOG1 subfamily, which uses TGY as the phosphorylation site (reviewed by Canman and Kastan, 1996; Kyriakis and Avruch, 1996).
In recent years, numerous protein kinases with close sequence similarity to mammalian MAPKs have been identified in plants (reviewed by Stone and Walker, 1995; Hirt, 1997; Mizoguchi et al., 1997; Tena et al., 2001; Zhang and Klessig, 2001; Ichimura et al., 2002). Most plant MAPKs are associated with the subgroup of extracellular signal-regulated kinases based on phylogeny (Kultz, 1998). Increasing evidence has shown that MAPKs play an important role in plant signal transduction related to biotic and abiotic stresses. Activation of MAPKs has been observed in plants exposed to pathogens (Suzuki and Shinshi, 1995; Adam et al., 1997; Ligterink et al., 1997; Zhang and Klessig, 1997, 1998b; He et al., 1999), cold (Jonak et al., 1996), salinity (Munnik et al., 1999; Mikolajczyk et al., 2000), drought (Jonak et al., 1996), and wounding (Seo et al., 1995, 1999; Usami et al., 1995; Bögre et al., 1997; Zhang and Klessig, 1998a; He et al., 1999). Plant MAPKs also can be activated by fungal elicitors (Suzuki and Shinshi, 1995), salicylic acid (Zhang and Klessig, 1997), jasmonic acid (Seo et al., 1999), and abscisic acid (Knetsch et al., 1996; Burnett et al., 2000; Heimovaara-Dijkstra et al., 2000). In addition, considerable progress has been made in cloning and characterizing plant MAPKKs (Morris et al., 1997; Hackett et al., 1998; Hardin and Wolniak, 1998; Ichimura et al., 1998a; Kiegerl et al., 2000; Yang et al., 2001) and MAPKKKs (Ichimura et al., 1998b; Kovtun et al., 2000; Frye et al., 2001).
Although detailed steps of MAPK cascades have yet to be elucidated in a given plant species, specific upstream MAPKKs for a few well-characterized MAPKs have been determined. Among these are NtMEK2 for salicylic acid–induced protein kinase (SIPK)/wounding-induced protein kinase (WIPK) in tobacco (Yang et al., 2001), AtMEK1 for AtMPK4 in Arabidopsis (Huang et al., 2000), and salt stress–induced MAPK kinase for salt stress–induced MAPK in alfalfa (Kiegerl et al., 2000). Most recently, a complete MAPK cascade (MEKK1, MKK4/MKK5, and MPK3/MPK6), together with its upstream receptor kinase FLS2 and downstream WRKY22/WRKY29 transcription factors, was characterized in Arabidopsis (Asai et al., 2002). These findings suggest that MAPKs are important signaling components in plant defense responses and that the cascade of a “three-kinase module” is a general mechanism of defense signal transduction among eukaryotic organisms (reviewed by Ligterink and Hirt, 2000).
To date, most of the reported plant MAPKs have been isolated and characterized from dicot model species such as Arabidopsis and tobacco. However, in economically important monocot species such as rice, very few MAPKs have been identified and characterized. He et al. (1999) reported that a 60-kD MAPK (OsBWMK1) was activated in rice leaves by blast fungus infection and wounding. Most recently, a stress-responsive MAPK gene (variously named OsMAPK5, OsMSRMK2, OsMAPK2, OsMAP1, or OsBIMK1) was identified by at least five laboratories and shown to be induced at the mRNA level by multiple biotic and abiotic stresses (Xiong et al., 2001; Agrawal et al., 2002; Huang et al., 2002; Song and Goodman, 2002; Wen et al., 2002). However, its protein expression, kinase activity, and biological functions were not examined in these studies. Here, we report the molecular characterization and functional analysis of this stress-responsive rice MAPK gene (designated OsMAPK5 based on the order of submitted rice MAPKs in the GenBank database). The OsMAPK5 gene, its protein, and kinase activity were activated by abscisic acid treatment as well as by various biotic (pathogen infection) and abiotic (cold, drought, and salinity) stresses. Most importantly, careful analyses of transgenic plants with overexpression or suppression of OsMAPK5 revealed that this abscisic acid–induc-ible MAPK may inversely modulate broad-spectrum disease resistance and abiotic stress tolerance.
RESULTS
Isolation and Sequence Analysis of OsMAPK5 cDNAs
We previously identified a rice cDNA fragment (clone JBI13) that is inducible by the blast fungus and that shows high homology with plant MAPK genes (Xiong et al., 2001). In this study, the corresponding full-length cDNA clones were isolated from a rice cDNA library using the JBI13 cDNA fragment as a probe. Two full-length cDNAs of OsMAPK5 (designated OsMAPK5a and OsMAPK5b because they are spliced alternatively from a single gene, as shown below) were completely sequenced and analyzed.
The OsMAPK5a cDNA is 1396 bp long and encodes a predicted protein of 369 amino acids (Figure 1A) with an estimated molecular mass of 42.9 kD. The OsMAPK5a protein contains all 11 subdomains that are conserved among all MAPK families (Hirt, 1997) and possesses a dual phosphorylation activation motif (TEY) located between subdomains VII and VIII (Figure 1A). It shares the identical amino acid sequence encoded by OsMSRM2 (Agrawal et al., 2002), OsMAPK2 (Huang et al., 2002), OsMAP1 (Wen et al., 2002), and OsBIMK1 (Song and Goodman, 2002), whose mRNA expression analyses were published during the preparation of this article. The OsMAPK5a protein also shares very high homology with the elicitor-inducible TaWCK1 (91% identity; Takezawa, 1999) from wheat and the wound-inducible NtWIPK (73% identity; Seo et al., 1995) from tobacco. Phylogenetic analysis based on the sequence alignment of the catalytic domain suggests that OsMAPK5a belongs to the A1 subgroup of the plant MAPK family (Figure 1B) (Ichimura et al., 2002). Previous studies indicate that members of the A1 and A2 subgroups frequently are activated by various biotic and abiotic stresses (Zhang and Klessig, 2001).
Figure 1.
Comparison of OsMAPK5a, OsMAPK5b, and Representative MAPKs from Other Higher Plants.
(A) Alignment of deduced amino acid sequences of OsMAPK5a and OsMAPK5b with those of the two most closely related MAPKs, TaWCK1 and NtWIPK. Conserved amino acid residues are shown in the consensus sequence. The 11 subdomains of the protein kinases are indicated above the sequences with Roman numerals. Thr (T) and Tyr (Y), two residues normally phosphorylated for the activation of MAPKs, are marked with asterisks.
(B) Phylogenetic relationship of OsMAPK5a and OsMAPK5b with other plant MAPKs. The dendrogram was constructed using Vector NTI Suite software. For simplicity, representatives from the eight subgroups of plant MAPKs (proposed by Ichimura et al., 2002), including a few putative rice MAPKs, are included in the dendrogram.
The OsMAPK5b cDNA has a nucleotide sequence identical to that of the OsMAPK5a cDNA except that a 312-bp region from positions 285 to 596 is deleted. Thus, OsMAPK5b encodes an incomplete MAPK with the deletion of subdomains III to VI (Figure 1A).
OsMAPK5 Is a Single-Copy Gene with Two Differentially Spliced Transcripts
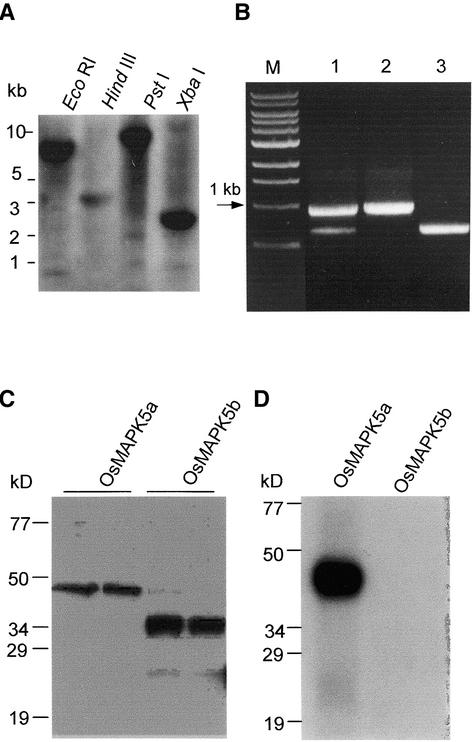
To determine if OsMAPK5a and OsMAPK5b derived from alternative splicing of a single gene, DNA gel blot hybridization was performed with a probe covering an identical region (sequence from position 999 to the 3′ end of OsMAPK5a) of both OsMAPK5a and OsMAPK5b. One strongly hybridizing band was detected in each of the four digestions (EcoRI, HindIII, PstI, and XbaI) of rice genomic DNA (Figure 2A). Genomic PCR with two primers covering the differentiated region also gave rise to a single fragment (data not shown). However, reverse transcriptase–mediated (RT) PCR with the same pair of primers amplified two cDNA fragments from the blast fungus–induced RNA sample. Their sizes (0.9 and 0.6 kb, respectively) match well with the predicted sizes of the cDNA fragments based on the locations of these two primers (Figure 2B). Therefore, OsMAPK5a and OsMAPK5b most likely resulted from the alternative splicing of a single OsMAPK5 gene in rice. There is low-level expression of OsMAPK5 in normal, uninfected leaves, which was detected by RT-PCR (data not shown). In both uninfected and infected leaf tissues, OsMAPK5a was the predominant isoform of OsMAPK5 transcripts.
Figure 2.
Genomic Organization, Alternative Splicing, Recombinant Proteins, and Autophosphorylation Activity of OsMAPK5.
(A) DNA gel blot analysis of the OsMAPK5 gene. Total DNA from cv Drew (4 μg for each lane) was digested individually with EcoRI, HindIII, PstI, and XbaI and hybridized with a gene-specific probe covering the region from nucleotide 999 to the 3′ end of the OsMAPK5a cDNA.
(B) RT-PCR analysis using a primer pair covering the differentiated regions of the OsMAPK5a and OsMAPK5b cDNAs. The blast fungus–induced (2 days after infection) mRNAs from cv Drew were used for RT-PCR analysis (lane 1). The cDNAs of OsMAPK5a and OsMAPK5b also were used for PCR with the same primer pair (lanes 2 and 3). M, DNA size markers.
(C) In vitro expression of OsMAPK5a and OsMAPK5b, and specificity of the OsMAPK5 antibody. One hundred nanograms of total protein from E. coli (left lanes) or 10 ng of affinity-purified fusion protein of His-OsMAPK5a and His-OsMAPK5b (right lanes) was separated by 10% SDS-PAGE and detected with the anti-OsMAPK5 antibody.
(D) In vitro autophosphorylation assay of the affinity-purified fusion proteins His-OsMAPK5a and His-OsMAPK5b.
OsMAPK5a, but Not OsMAPK5b, Possesses the Kinase Activity
To determine if OsMAPK5a and OsMAPK5b encode active MAPKs, the recombinant proteins of both OsMAPK5a and OsMAPK5b were produced and purified from Escherichia coli cells harboring OsMAPK5a and OsMAPK5b coding sequences, respectively, in expression vector pET-28c(+). As expected, OsMAPK5b was 12 kD smaller than OsMAPK5a as a result of a 312-bp (104–amino acid) deletion (Figure 2C). Kinase assays revealed that only OsMAPK5a exhibited autophosphorylation activity (Figure 2D), suggesting that the subdomains that are missing in OsMAPK5b are essential for the kinase activity.
Induction of OsMAPK5, Its Gene Product, and Kinase Activity by Blast Fungus Infection
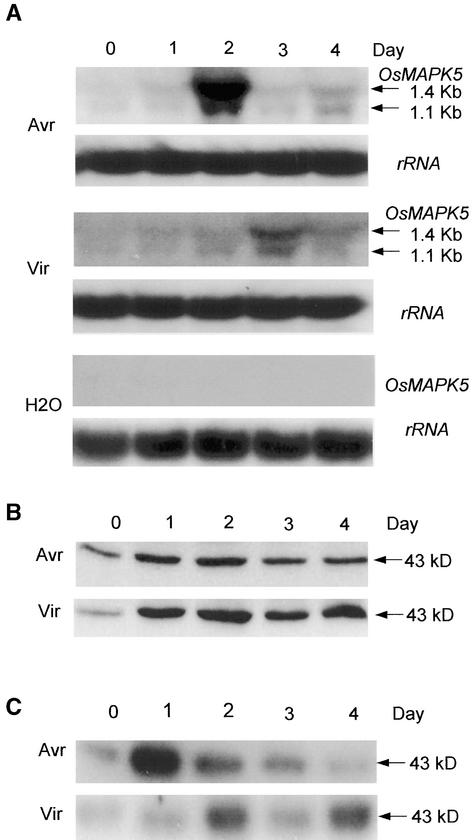
In our previous study, OsMAPK5 was shown to be inducible by the blast fungus (Xiong et al., 2001). To further assess the expression pattern of OsMAPK5 during fungal infection, an avirulent blast isolate (carrying AvrPita) and its virulent mutant (lacking AvrPita) were used to elicit resistant and susceptible reactions, respectively, on rice cv Drew (carrying the Pita resistance gene). RNA gel blots prepared from mock-treated and blast-infected leaves were hybridized with a gene-specific probe of OsMAPK5. Two hybridizing transcripts were found to be induced by the blast fungus (Figure 3A). The sizes of the transcripts were similar to those of the OsMAPK5a (1.4 kb) and OsMAPK5b (1.1 kb) cDNAs. However, the induced level of OsMAPK5b transcripts was significantly lower than that of OsMAPK5a. In the resistant interaction, the mRNA level of OsMAPK5 was induced as early as 1 day after inoculation, peaked on the second day, and then declined (Figure 3A). In the susceptible interaction, the transcripts accumulated slowly but lasted longer than in the resistant interaction. However, the peak level of induced OsMAPK5 was significantly higher in the resistant interaction than in the susceptible interaction. No induction of OsMAPK5 was detected in mock-treated leaves (Figure 3A), indicating that the induction of OsMAPK5 was not attributable to the effect of spray inoculation.
Figure 3.
Activation of OsMAPK5, Its Protein, and Kinase Activity by Inoculation with the Blast Fungus.
Assays were repeated three times using samples from independent experiments. Representative blots are presented.
(A) RNA gel blot analysis of OsMAPK5 expression using the same gene-specific probe used in DNA gel blot analysis. Two-week-old seedlings of cv Drew were spray-inoculated with water (containing 0.02% Tween 20) and with the avirulent (Avr; IC17-18/1) or virulent (Vir; IC17-18/1-2) isolate of blast fungus. Total RNAs were isolated from leaf tissues at the times specified. The two transcripts that resulted from alternative splicing of the OsMAPK5 gene are indicated with arrows, and their sizes are shown at right. Equal loading of total RNAs (20 μg per lane) was verified using rice 25S rRNA as a loading control.
(B) Immunoblot analysis of OsMAPK5. The rat antibody raised against the C terminus of OsMAPK5a recognized both OsMAPK5a and OsMAPK5b. The OsMAPK5b protein was undetectable under the same conditions used to detect OsMAPK5a (5 to 10 min of exposure using the ECL-Plus detection kit). A rather weak band corresponding to OsMAPK5b was detected with an extended exposure time (>1 h). Because neither MBP kinase activity (C) nor autophosphorylation activity (Figure 2D) was detected for OsMAPK5b, only the band corresponding to OsMAPK5a is shown.
(C) MBP in gel kinase assay. The OsMAPK5 protein was immunoprecipitated with the anti-OsMAPK5 antibody from 400 μg of total protein and subjected to in gel kinase assay using MBP as a substrate. Because no activity was detected for OsMAPK5b, only the band corresponding to the activity of OsMAPK5a is shown.
Using anti-OsMAPK5 antibody, a 43-kD protein corresponding to OsMAPK5a (predicted size of 42.9 kD) was detected in rice leaves infected with blast fungus (Figure 3B). Immunoblot analysis indicated that the level of OsMAPK5a protein increased slightly on day 2 after infection with avirulent isolate and then decreased to the base level. In the susceptible reaction, however, much more protein was induced and the induction lasted longer (Figure 3B). The OsMAPK5b protein (predicted size of 31.2 kD) was undetectable using the same experimental conditions. However, a constitutively expressed unknown protein (∼49 kD) cross-reacted with the anti-OsMAPK5 antibody (data not shown).
To determine further if the OsMAPK5 (referring to OsMAPK5a hereafter) kinase activity was induced by blast infection, endogenous OsMAPK5 was immunoprecipitated and subjected to in gel kinase assay using myelin basic protein (MBP) as a substrate. Results showed that OsMAPK5 kinase activity was induced significantly by blast fungus infection. In the resistant interaction, the kinase activity increased 1 day after fungal inoculation and then declined progressively to the base level. In the susceptible interaction, the kinase activity increased after 2 days but remained moderately high until the final stage of infection (Figure 3C). These data suggest that the early transient activation of OsMAPK5 activity probably is related to the resistance response to avirulent blast isolates. On the other hand, the constant activation of OsMAPK5 in the later stage of infection may be related to stress resulting from the development of the disease.
Induction of OsMAPK5, Its Gene Product, and Kinase Activity by Abscisic Acid and Wounding
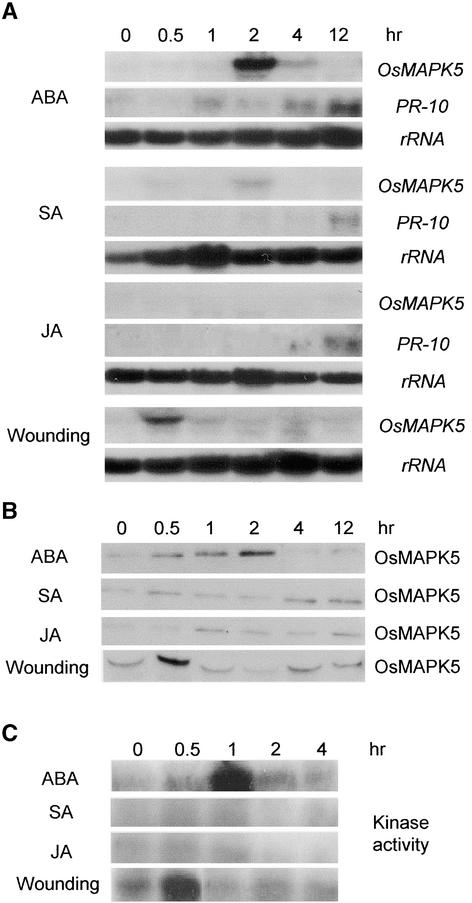
To determine the effects of different signaling molecules on OsMAPK5 activation, 2-week-old rice seedlings were treated with abscisic acid, salicylic acid (SA), and jasmonic acid (JA). RNA gel blot analysis showed that OsMAPK5a was induced significantly in rice leaves treated with 0.1 mM abscisic acid (Figure 4A). Transcripts of OsMAPK5a accumulated quickly to the highest level at 2 h after treatment and then declined. However, OsMAPK5a was induced only slightly, if at all, in leaves treated with 1 mM SA or 0.1 mM JA. Treatments with higher concentrations of SA or JA also did not significantly induce OsMAPK5a (data not shown). By contrast, a defense-related gene, PR10, was induced by SA and JA, as expected (Figure 4A). Expression of OsMAPK5a increased significantly in wounded leaves, peaking at 30 min after wounding and then decreasing rapidly to the base level (Figure 4A). The transcript of OsMAPK5b was not induced by these chemical treatments or by wounding.
Figure 4.
Induction of OsMAPK5, Its Protein, and Kinase Activity by Abscisic Acid and Wounding.
Experiments were repeated twice using samples from independent treatments.
(A) RNA gel blot analysis of OsMAPK5 expression in 2-week-old seedlings treated with 0.1 mM abscisic acid (ABA), 1 mM SA, 0.1 mM JA, or wounding. Total RNAs were extracted at the times specified. The same blots were probed with the cDNA of PBZ1, a rice defense gene that encodes an intracellular PR10 protein (Midoh and Iwate, 1996), to assess the effectiveness of the chemical treatments because PBZ1 is induced by abscisic acid, SA, and JA (Lee et al., 2001).
(B) Immunoblot analysis of OsMAPK5 in 2-week-old seedlings treated with 0.1 mM abscisic acid, 1 mM SA, 0.1 mM JA, or wounding.
(C) MBP in gel kinase activity of immunoprecipitated OsMAPK5 from 2-week-old seedlings treated with 0.1 mM abscisic acid, 1 mM SA, 0.1 mM JA, or wounding.
Immunoblot analysis showed that the OsMAPK5 protein was induced by abscisic acid and wounding but not by SA or JA (Figure 4B). The immunocomplex in gel kinase assay also showed that OsMAPK5 activity was induced by abscisic acid and wounding but not by SA or JA (Figure 4C). After abscisic acid treatment, the peak of OsMAPK5 activity appeared earlier than that of the mRNA and protein. Similar phenomena also were observed after fungal infection (Figure 3) or abiotic treatments (Figure 5). Previously, Seo et al. (1995) reported that the peak of tobacco WIPK activity appeared much earlier than that of its mRNA after mechanical wounding. It is very likely that basal-level OsMAPK5 can be activated very quickly before the accumulation of its mRNA and protein.
Figure 5.
Induction of OsMAPK5, Its Protein, and Kinase Activity by Drought, Salt, and Low Temperature.
Rice tissues from the same time course were used for RNA gel blot, immunoblot, and kinase activity analyses. Experiments were repeated three times using samples from independent treatments.
(A) RNA gel blot analyses of OsMAPK5 expression in 2-week-old seedlings subjected to drought (water withheld for up to 5 days), salt (200 mM NaCl), or cold (4°C) stress. For drought and salt stresses, RNA from both root and leaf tissues was extracted at the times specified. Only leaf tissues were collected for the cold treatment.
(B) Immunoblot analyses of OsMAPK5 under drought (root tissues), salt (root tissues), and cold (leaf tissues) stresses.
(C) MBP in gel kinase activity assay of immunoprecipitated OsMAPK5 under drought (root tissues), salt (root tissues), and cold (leaf tissues) stresses.
Induction of OsMAPK5 by Drought, Salinity, and Low Temperature
Significant induction of OsMAPK5a by abscisic acid (which often mediates abiotic responses) prompted us to investigate the expression pattern of OsMAPK5a in response to abiotic stresses such as drought, salinity, and low temperature. RNA gel blot analysis clearly showed that OsMAPK5a was induced by drought, salinity, and low temperature (Figure 5A). In the drought and salt treatments, OsMAPK5a was induced earlier in roots (within 1 day and 1 h for drought and salinity, respectively) than in leaves (within 4 days and 3 h for drought and salinity, respectively). The transcript of OsMAPK5a remained high throughout the course of drought stress. However, under salt stress, the transcripts declined at 6 h after treatment. The transcript of OsMAPK5a also was inducible within 6 h by low temperature (4°C) treatment (Figure 5A).
Immunoblot analyses showed that the protein level of OsMAPK5 was increased significantly in rice seedlings under drought and salt stresses but was induced slightly by low temperature (Figure 5B). Immunocomplex kinase assay showed that OsMAPK5 activity also was induced by drought, salt, and low temperature (Figure 5C). These results suggest that OsMAPK5 is likely involved in abiotic stress responses in rice plants.
Overexpression and Suppression of OsMAPK5 in Transgenic Rice
To clarify the role of OsMAPK5 (referring to OsMAPK5a hereafter) in biotic and abiotic stress responses, we constitutively increased or suppressed the expression of OsMAPK5 in transgenic rice. The transgenic lines were generated by introducing the overexpression construct (OsMAPK5-OX) or the double-stranded RNA interference construct (OsMAPK5-RI) into cv Nipponbare GA3.
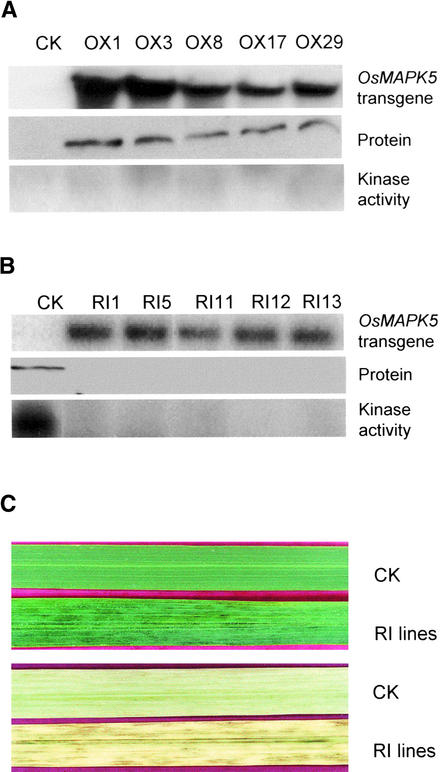
Thirty independent overexpression lines were generated using the OsMAPK5-OX construct. DNA gel blot analysis indicated that 19 OsMAPK5-OX lines contained a single-copy insertion (data not shown). RNA gel blot analysis showed that the OsMAPK5 gene was expressed constitutively in transgenic lines but not in control plants under normal growth conditions (five lines are shown in Figure 6A as examples). As expected, the protein of OsMAPK5 was produced constitutively in the transgenic lines but not in control plants under normal growth conditions (Figure 6A). However, the MBP kinase activity of OsMAPK5 in these lines was not increased significantly (Figure 6A). None of the OsMAPK5-OX lines showed obvious phenotypic changes compared with control plants throughout the life cycle.
Figure 6.
Overexpression and Suppression of OsMAPK5 in Transgenic Rice.
(A) The overexpression construct (OsMAPK5-OX) under the control of the 35S promoter of Cauliflower mosaic virus was introduced into cv Nipponbare by Agrobacterium-mediated transformation. Thirty independent T0 transgenic lines were obtained and examined (results from five representative lines and a control plant [CK] are shown) for OsMAPK5 expression and kinase activity under normal growth conditions. The base level of endogenous OsMAPK5 in control plants (as shown in Figures 3 and 4) was not detected under the optimal exposure time for detecting the overexpressed OsMAPK5.
(B) The dsRNAi construct (OsMAPK5-RI) under the control of the 35S promoter of Cauliflower mosaic virus was introduced into cv Nipponbare by Agrobacterium-mediated transformation. Thirty-eight independent T0 transgenic lines were obtained and analyzed (results from five representative lines and a control plant [CK] are shown) for the expression of transgenic OsMAPK5 (dsRNAi construct of OsMAPK5) and the suppression of endogenous OsMAPK5 in plants. Transgene expression in normally growing T0 plants was detected by an OsMAPK5 cDNA fragment used to make the dsRNAi construct. Endogenous OsMAPK5 protein levels and kinase activities in the transgenic lines were examined using rice leaves infected with the fungal isolate IC17-18/1 at 3 days after spot inoculation
(C) Development of brownish stripes on mature flag leaves of OsMAPK5-RI transgenic lines. Control (CK) and transgenic (RI) rice leaves before (top) and after (bottom) the removal of chlorophyll (overnight soaking in 100% ethanol) are shown.
Thirty-eight independent suppression lines were generated using the OsMAPK5-RI construct. Twenty-four OsMAPK5-RI lines were confirmed by DNA gel blot hybridization to carry a single-copy insertion (data not shown). RNA gel blot analysis showed that the OsMAPK5-RI construct was transcribed constitutively in suppression lines (five lines are shown in Figure 6B as examples). Because the endogenous level of OsMAPK5 in control plants was rather low under normal growth conditions (Figure 3C), the effectiveness of double-stranded RNA interference (dsRNAi) in T0 transgenic lines was examined after the induction of OsMAPK5 by spot inoculation of rice leaves with the blast fungus. Strikingly, the production of endogenous OsMAPK5 protein was blocked almost completely even under the induced condition. In fact, no MBP kinase activity was detected for OsMAPK5 in these transgenic lines (Figure 6B). The suppression of endogenous OsMAPK5 by dsRNAi also was transmitted to T1 transgenic plants, as shown below. None of the OsMAPK5-RI lines showed obvious phenotypic changes from germination to the early vegetative growth stage. However, starting from the late vegetative stage (∼2 months after germination), irregular brownish stripes developed on the mature leaves of OsMAPK5-RI lines (Figure 6C). Nevertheless, each OsMAPK5-RI line proceeded to the reproductive stage and had normal seed setting.
OsMAPK5 Negatively Modulates Broad-Spectrum Host Resistance and PR Gene Expression
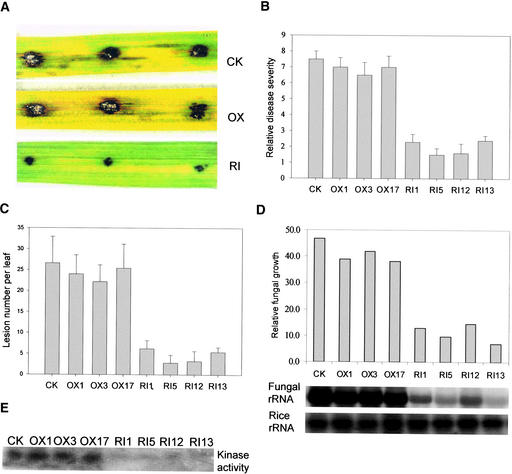
Because OsMAPK5 is inducible by pathogen infection, we examined the effects of the overexpression or suppression of OsMAPK5 on host resistance to fungal and bacterial pathogens. Disease resistance was evaluated initially on first-generation (T0) transgenic lines by spot inoculation of transgenic leaves with a virulent isolate of blast fungus, because single T0 plants were not suitable for spray inoculation. Both control and OsMAPK5-OX T0 lines exhibited the same level of disease susceptibility to blast infection, with average lesion sizes of 7.0 ± 1.2 mm and 6.8 ± 1.6 mm, respectively. But all OsMAPK5-RI T0 lines (20 lines tested) exhibited significantly enhanced resistance, with an average lesion size of 2.8 ± 1.1 mm. Fungal growth (quantified based on relative rRNA contents of blast fungus in inoculated spots) also was reduced by approximately threefold to sixfold in OsMAPK5-RI lines compared with control or OsMAPK5-OX lines.
To confirm the results from the T0 generation, we further evaluated the disease resistance in the second generation (T1) of transgenic rice using three OsMAPK5-OX lines, four OsMAPK5-RI lines, and the control line. As a result of the transgene segregation in the T1 generation, seedlings carrying the OsMAPK5-OX or OsMAPK5-RI constructs first were identified based on hygromycin resistance and positive PCR amplification of the transgene. Approximately 40 2-week-old T1 seedlings from each line (a total of ∼320 seedlings) then were spray-inoculated with the fungal isolate (IC17-18/1). All four OsMAPK5-RI lines showed increased resistance to blast infection, as indicated by significantly reduced disease severity (Figures 7A and 7B), lesion numbers (Figure 7C), and fungal growth (Figure 7D). By contrast, control and OsMAPK5-OX plants were very susceptible to the same fungal isolate. As expected, the normal induction of OsMAPK5 kinase activity by fungal infection was suppressed almost completely in these OsMAPK5-RNAi lines (Figure 7E), suggesting that suppression of OsMAPK5 activity likely led to the enhanced resistance.
Figure 7.
OsMAPK5-RI Lines Exhibited Enhanced Resistance to the Blast Fungus.
Experiments were repeated two times with similar results.
(A) Blast resistance evaluation of T0 transgenic plants by the spot inoculation method. Shown are typical disease symptoms on leaves of control plants (CK) as well as overexpression (OX) and dsRNAi (RI) transgenic plants at 6 days after inoculation with fungal isolate IC17-18/1.
(B) Blast resistance evaluation of T1 transgenic plants based on disease ratings. Two-week-old T1 transgenic seedlings (20 to 40 hygromycin-resistant transgenic seedlings per line were used in each experiment) from three overexpression lines (OX1, OX3, and OX17), four dsRNAi lines (RI1, RI5, RI12, and RI13), and a control line (CK) were spray-inoculated with fungal isolate IC17-18/1. Disease ratings were determined according to Marchetti et al. (1976) at 5 days after inoculation.
(C) Blast resistance evaluation of T1 transgenic plants based on lesion numbers per infected leaf at 5 days after inoculation.
(D) Blast resistance evaluation of T1 transgenic plants based on relative fungal growth. Total RNA from infected leaves at 5 days after inoculation was blotted and hybridized with blast fungus 28S rRNA and rice 25S rRNA. The fungal 28S rRNA hybridization signals were quantified by phosphorimaging and calibrated with the rice 25S rRNA signals for equal loading (Qi and Yang, 2002).
(E) MBP in gel kinase assay of immunoprecipitated OsMAPK5 from leaf tissues of control and transgenic lines at 5 days after inoculation.
To determine whether OsMAPK5-RI lines have broad-spectrum resistance to other pathogens, 4-week-old T1 plants were infected with Burkholderia glumae, a bacterial pathogen that causes rice diseases known as panicle blight, glume blight, and sheath rot complex (Cottyn et al., 1996). Compared with the control or OsMAPK5-OX lines, OsMAPK5-RI lines exhibited significantly increased resistance against the bacterial pathogen, as indicated by reduced lesion size (Figure 8A; see also supplemental data online) and bacterial growth (Figure 8B). OsMAPK5 kinase activity was activated by B. glumae in both control and OsMAPK5-OX plants but again was suppressed in OsMAPK5-RI lines (Figure 8C). These results demonstrate that the suppression of OsMAPK5 activity in rice may result in broad-spectrum resistance to fungal and bacterial pathogens.
Figure 8.
OsMAPK5-RI Lines Exhibited Enhanced Resistance to the Bacterial Pathogen B. glumae.
Leaf sheaths of 1-month-old control and T1 transgenic seedlings were inoculated with B. glumae (1 × 106 colony-forming units). At least 10 hygromycin-positive transgenic seedlings per line were used in each experiment. Experiments were repeated twice with similar results.
(A) Disease resistance evaluation based on lesion size at 7 days after inoculation. See supplemental data online for photographs of disease symptoms.
(B) Disease resistance evaluation based on bacterial growth in planta at 7 days after inoculation.
(C) MBP in gel kinase assay of immunoprecipitated OsMAPK5 from leaf tissues at 7 days after inoculation.
In all of the tests, the control and OsMAPK5-OX plants showed no significant differences in host susceptibility to either blast fungus or B. glumae (Figures 7B and 8B). Although the OsMAPK5 protein was expressed constitutively in the OsMAPK5- OX lines (Figure 6B), the kinase activity was not increased significantly upon infection with either blast fungus or B. glumae (Figures 7E and 8C). Therefore, the levels of disease resistance appear to correlate with the changes of OsMAPK5 kinase activity in rice plants.
Because OsMAPK5-RI lines exhibited increased resistance to fungal and bacterial pathogens, we analyzed the expression of some pathogenesis-related (PR) genes in these lines under normal growth conditions. Interestingly, RNA gel blots showed that two rice PR genes, PR1b and PR10, were expressed constitutively in OsMAPK5-RI T1 transgenic seedlings in the absence of pathogen infection but not in nontransgenic or OsMAPK5-OX seedlings grown under the same conditions (Figure 9). Similar results were obtained in T0 transgenic plants and leaf tissues from different developmental stages (data not shown). These data suggest that OsMAPK5 could negatively modulate (probably through an indirect effect) PR gene expression (at least PR1 and PR10) as well as broad-spectrum disease resistance.
Figure 9.
Constitutive Expression of PR1 and PR10 in OsMAPK5-RI Transgenic Lines.
Total RNA was isolated from 2-week-old control and T1 transgenic seedlings grown under normal conditions. The RNA gel blot (10 μg of RNA per lane) was probed sequentially with the PR1b, PR10, and rice 25S rRNA.
OsMAPK5 Positively Regulates Cold, Drought, and Salt Tolerance
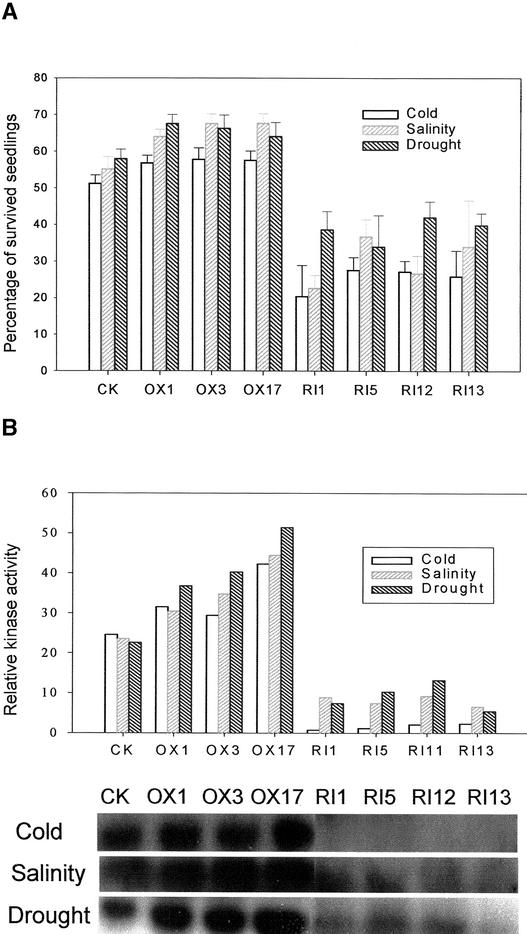
Because OsMAPK5 is inducible by abscisic acid and various biotic stresses (Figure 5), we examined the effects of the overexpression or suppression of OsMAPK5 on the tolerance of transgenic plants to cold, drought, and salt stresses. Stress tolerance was evaluated based on the percentage of seedlings that survived after cold, drought, or salt treatment. Surprisingly, the four OsMAPK5-RI lines with enhanced disease resistance exhibited significantly (P < 0.001) reduced tolerance to cold, drought, and salt stresses (Figure 10A; see also supplemental data online). By contrast, the three OsMAPK5-OX lines showed significantly increased tolerance to salinity (P < 0.005), drought (P < 0.01), and cold (P < 0.05). The kinase activity of OsMAPK5 in transgenic lines also was assayed after the stress treatments. As expected, the normal activation of OsMAPK5 by cold, salinity, and drought was suppressed in OsMAPK5-RI lines, whereas the kinase activity in OsMAPK5-OX lines was higher than that in control plants (Figure 10B). These results suggest that the activation of OsMAPK5 positively regulated plant tolerance to abiotic stresses such as drought, salinity, and low temperature.
Figure 10.
Altered Tolerance of OsMAPK5-OX and OsMAPK5-RI Transgenic Plants to Cold, Salt, and Drought.
(A) Percentage of surviving seedlings after treatment with cold (4°C for 3 days followed by normal growth conditions for recovery), salt (200 mM NaCl for up to 4 days), or drought (withholding water for up to 6 days). At least 40 hygromycin-positive T1 transgenic seedlings were used in each experiment. All experiments were repeated twice. Statistical analysis (t test) was performed to evaluate the levels of cold, salt, and drought tolerance based on the percentage of surviving seedlings in the overexpression or suppression lines versus the control line after the abiotic treatments. See supplemental data online for photographs of rice seedlings after cold, salt, and drought treatments.
(B) MBP in gel kinase assay of immunoprecipitated OsMAPK5 from mixed leaf tissue sampling at different times under cold (6, 12, and 24 h), salinity (6, 12, and 24 h), or drought (2, 3, and 4 day) stresses. The relative MBP kinase activities of control and transgenic lines and the control line were calculated based on phosphorimager quantification of band intensity.
DISCUSSION
As sessile organisms, plants have evolved a complex signaling network that mediates the perception of and responses to different environmental cues. Recent studies have shown that MAPK cascades are evolutionarily conserved signaling modules that play a pivotal role in plant responses to multiple biotic and abiotic stresses. A number of plant MAPK cascade genes have been characterized functionally, and the importance of the three-kinase module in signal transduction has been supported by numerous experimental studies. However, our understanding of the role of MAPK cascades in stress responses remains rather limited, considering the redundancy of cascade components, the antagonism among distinct cascades, and the potential positive and negative regulation of different stress pathways by the same MAPK cascade (Tena et al., 2001).
Here, we have isolated alternatively spliced cDNAs (OsMAPK5a and OsMAPK5b) of a MAPK gene from rice, an important crop and model monocot species. Alternative splicing of heterogeneous nuclear RNA is an important mechanism of gene expression and regulation (Lopez, 1998). In plants, some disease resistance genes (L6, M, RPP5, and N) have been shown to encode two or more transcripts that may have different functions in regulating disease resistance (Anderson et al., 1997; Parker et al., 1997; Ayliffe et al., 1999; Dinesh-Kumar and Baker, 2000). Alternative splicing also was reported in the Arabidopsis MAPKKK gene ANP1 (Nishihama et al., 1997). More recently, alternatively spliced transcripts of a tomato diacylglycerol kinase gene were reported to encode a calmodulin binding isoform and a nonbinding isoform, both of which are catalytically active in vitro (Snedden and Blumwald, 2000). In this study, the intact OsMAPK5a isoform was shown to have kinase activity, but neither autophosphorylation nor MBP kinase activity was detected for the truncated OsMAPK5b isoform. Because subdomain VI contains the catalytic loop of MAPK (Zhang et al., 1994), which is missing in OsMAPK5b, it is not surprising that OsMAPK5b has neither autophosphorylation nor MBP kinase activities. However, the exact role of OsMAPK5b or its product, if any, remains unknown.
When cv Drew carrying the Pita resistance gene was inoculated with an avirulent isolate carrying AvrPita, the OsMAPK5 gene was activated rapidly (1 day after infection) before the appearance of hypersensitive cell death, which normally occurs at 2 to 3 days after infection under our experimental conditions. Interestingly, the kinase activity also declined very quickly to the base level. By contrast, the induction of OsMAPK5 was much slower in the susceptible interactions, and moderate expression accompanied the development of disease lesions. A recent study has suggested that MAPK cascades may relay the signals perceived by upstream R gene products and in turn activate downstream regulatory proteins such as transcription factors (Asai et al., 2002). After the signal is transmitted successfully, MAPK is inactivated quickly by specific phosphatases involved in the incompatible interaction. In plants, both a Ser/Thr protein phosphatase and a Tyr phosphatase have been reported to inactivate MAPKs in in vitro studies (Gupta et al., 1998; Meskiene et al., 1998). We hypothesize that the quick activation and inactivation of OsMAPK5 in the resistant interaction is related to an AvrPita/Pita-mediated host resistant response rather than to wounding or general stress caused by appressorium penetration. However, further experiments are required to determine the role of OsMAPK5 in the AvrPita/Pita-mediated resistance response.
Several lines of evidence reported here indicate that OsMAPK5 negatively regulates the non-race-specific disease resistance and PR gene expression in rice. First, the kinase activity of OsMAPK5 remained quite high during the development of disease lesions in the compatible interaction, whereas the activity decreased to base levels after transient activation in the incompatible interaction. Second, the suppression of OsMAPK5 and its kinase activity significantly increased the levels of disease resistance in the dsRNAi transgenic lines. When infected with the blast fungus, all 20 independent OsMAPK5-RI transgenic lines showed significantly enhanced resistance, suggesting that the increased resistance was not caused by mutations brought on by T-DNA insertion or other random mutations. Four independent lines were tested further in the second (T1) generation and showed significantly enhanced resistance to both fungal (blast fungus) and bacterial (B. glumae) pathogens. In addition, none of OsMAPK5-RI transgenic plants exhibited spontaneous necrotic lesions, which is a common phenotype for many mutants with constitutive expression of PR genes and disease resistance (reviewed by Mittler and Rizhsky, 2000). Although we observed brownish stripes on mature leaves at the late vegetative growth stage, the fungal and bacterial inoculations were performed on 2-week-old seedlings or young plants at least 1 month before the appearance of the brownish stripes. Therefore, it is unlikely that the increased resistance resulted from a pleiotropic effect associated with necrotic lesions caused by the potential disruption of cellular homeostasis (Molina et al., 1999). Third, rice PR genes such as PR1b and PR10, which are involved in disease resistance, were activated constitutively in OsMAPK5-RI transgenic lines (both young seedlings and mature plants) under normal growth conditions. It is likely that OsMAPK5 negatively modulates a specific set of regulatory genes, presumably those encoding transcription factors that activate PR gene expression and broad-spectrum host resistance.
Recent studies demonstrate that MAPKs may positively or negatively regulate defense responses in plants. A number of Arabidopsis MAPK cascade components (e.g., CTR1, EDR1, and MPK4) were shown to negatively regulate PR gene expression and disease resistance. For example, the mpk4 mutant exhibits constitutive expression of PR genes and systemic acquired resistance (Petersen et al., 2000), whereas the edr1 mutant (a mutation in a putative MAPKKK gene) shows enhanced pathogen resistance by a non–systemic acquired resistance mechanism (Frye et al., 2001). By contrast, tobacco NPK1 (a MAPKKK), NtMEK2 (a MAPKK), SIPK, and WIPK appear to positively regulate plant defense responses (Yang et al., 2001; Zhang and Liu, 2001; Jin et al., 2002; Samuel and Ellis, 2002). Based on sequence similarity, OsMAPK5 is closely related to tobacco WIPK, but it may not be the true ortholog of WIPK in rice. Even if OsMAPK5 is the ortholog of WIPK, its function may have evolved after the divergence of dicots and monocots. The abscisic acid–inducible OsMAPK5 appears primarily to mediate abiotic stress, even though it also is activated by biotic stress, such as wounding-associated blast infection. Our transgenic analyses show that OsMAPK5 positively regulates abiotic stress tolerance but negatively modulates PR gene expression and disease resistance. Such an opposite effect could be explained by potential antagonism between distinct MAPK cascades (Tena et al., 2001). Recently, Samuel and Ellis (2002) observed strong and stable activation of WIPK in SIPK-suppressed tobacco lines, but not in overexpression lines, during continuous ozone exposure. When conducting the in gel kinase assay using leaf protein extracts from the blast fungus–infected seedlings, we found that the kinase activity of a 37-kD protein was increased significantly in OsMAPK5-RI transgenic plants but not in the control plants (our unpublished data), suggesting the potential antagonistic effect of OsMAPK5 on an unknown MBP kinase that may positively regulate defense responses in rice.
Signaling pathways involved in plant responses to drought, salt, and cold stresses are largely overlapping and frequently mediated by abscisic acid (Bray, 1997; Leung and Giraudat, 1998). Interestingly, rice OsMAPK5 gene and kinase activity were activated specifically by abscisic acid rather than by SA or JA. To date, a large number of plant MAPK genes have been reported to be induced by abiotic stresses, including drought, salinity, and low temperatures (Jonak et al., 1996; Berberich et al., 1999; Munnik et al., 1999; Mikolajczyk et al., 2000; Agrawal et al., 2002; Huang et al., 2002). However, none of these genes has been characterized functionally and shown to positively regulate the abiotic stresses. It is intriguing to find that the OsMAPK5-suppressed transgenic lines had a significant reduction in drought, salt, and cold tolerance. By contrast, the OsMAPK5 overexpression lines exhibited enhanced tolerance to these abiotic stresses. Furthermore, the reduced and enhanced stress tolerance are in agreement with the suppression and increase of OsMAPK5 kinase activity, respectively. Therefore, based on both genetic and biochemical data, we conclude that OsMAPK5 likely encodes a positive regulator of drought, salt, and cold tolerance.
Interaction between different signaling pathways (e.g., inverse modulation of SA and JA pathways) appears to be very common and important in regulating defense responses against pathogen infection and insect herbivory (Reymond and Farmer, 1998). However, it is unknown if MAPK cascades are involved in the antagonistic regulation between biotic and abiotic stress responses. We have demonstrated that an abscisic acid–inducible rice MAPK is capable of inversely modulating disease resistance and abiotic stress tolerance. On the one hand, overexpression of OsMAPK5 resulted in enhanced plant tolerance to drought, salt, and cold stresses. On the other hand, suppression of OsMAPK5 reduced abiotic stress tolerance but led to constitutive PR gene expression and increased disease resistance. Considering the limited numbers of MAPK genes (e.g., 20 MAPKs, 10 MAPKKs, and 60 MAPKKKs in the Arabidopsis genome), it is not surprising that plants have evolved such integrated signaling and transduction systems to delicately coordinate various physiological activities. Because of these interactions, researchers need to examine both positive and negative effects on different agronomic traits when modifying MAPK components for the genetic improvement of crop plants. By further understanding the MAPK cascades and carefully modifying the components, it should be possible to generate new crop varieties that combine desirable traits such as enhanced disease resistance and abiotic stress tolerance.
METHODS
Isolation and Sequence Analysis of OsMAPK5
A cDNA fragment (JBI13) of OsMAPK5 isolated previously by suppression subtractive hybridization (Xiong et al., 2001) was labeled with α-32P-dCTP and used as a probe to isolate the corresponding full-length cDNA. Approximately 106 plaques from a blast fungus (Magnaporthe grisea)–induced cDNA library (Lee et al., 2001) were screened. The resulting positive clones carrying OsMAPK5 cDNAs were excised in vivo from the λZAP Express vector with the aid of ExAssist helper phage (Stratagene, La Jolla, CA). Subsequently, the full-length OsMAPK5 cDNA clones were sequenced from both directions using a primer-walking approach. Automated sequencing service was provided by the University of Arkansas for Medical Science. Sequence analysis was performed using Vector NTI Suite (Informax, North Bethesda, MD) and Basic Local Alignment Search Tool (BLAST) (Altschul et al., 1990).
Gene Constructs and Rice Transformation
The overexpression construct, OsMAPK5-OX, was constructed by directionally inserting the full cDNA sequence (digested with BamHI and XbaI) into the vector pCAMBIA1300S, which was modified by us based on pCAMBIA1300 and contains a double 35S promoter of Cauliflower mosaic virus and a terminator. To make a double-stranded RNA interference (dsRNAi) construct, antisense and sense fragments were generated by restriction enzyme digestions and PCR from the OsMAPK5 cDNA. The antisense fragment spanning nucleotides 1198 to 1 (also including six bases from the vector pBK-CMV) of OsMAPK5 was obtained by digestion with NcoI and BamHI and inserted into pCAMBIA1300S (digested with NcoI and BamHI) to form the antisense construct pC1300S-A. The sense fragment spanning nucleotides 734 to 1198 of OsMAPK5 was generated by PCR with primers B734 (5′-CGGGATCCGTCGGCTGCA-TCTTCATG-3′; BamHI site was introduced and underlined) and X1198 (5′-GCTCTAGATTCAATCTAGTACCGGA-3′; XbaI site was introduced and underlined). The PCR product was digested with BamHI and XbaI and then inserted into pC1300S-A (digested with BamHI and XbaI) to form the dsRNAi construct, OsMAPK5-RI. Overexpression and dsRNAi constructs of OsMAPK5 were introduced into Agrobacterium tumefaciens (strain EHA105) separately using a freeze-thaw method (Höfgen and Willmitzer, 1988). pCAMBIA1300S also was used for transformation, and the resulting empty vector–transformed plants were used as controls.
Agrobacterium carrying the overexpression or dsRNAi construct was grown overnight in AB induction medium (Winans et al., 1988) containing 50 μg/mL hygromycin and 100 μM acetosyringone. Bacterial cells were collected by centrifugation and resuspended in induction medium to OD600 of 0.1 for transformation. The Agrobacterium-mediated transformation was performed using vigorously growing calli derived from mature embryos of rice (Oryza sativa) cv Nipponbare GA3, a cultivar that is used in the international rice genome sequencing project and that is relatively easy to transform, according to the method of Hiei et al. (1994).
Plant Materials and Pathogen Inoculations
Transgenic rice plantlets (5 to 6 cm in height) from the rooting medium were transplanted into Redi-earth soil mix (Scotts, Marysville, OH) and grown in a 28°C greenhouse with a 14-h-light/10-h-dark cycle. Plants were fertilized with 0.5% ammonium sulfate every 2 weeks until flowering. Self-pollinated seeds from independent transgenic lines were harvested. T1 plants carrying the transgene were selected by germinating seeds on filter paper soaked with 50 μg/mL hygromycin. Nontransgenic seeds of cv Nipponbare did not germinate (0%, 500 seeds tested) in the presence of 50 μg/mL hygromycin. Positive T1 plants were confirmed by PCR or DNA gel blot analysis using primers or a probe corresponding to the 35S promoter and/or the 5′ region of OsMAPK5. In addition to wild-type and transgenic plants of cv Nipponbare, a U.S. rice cultivar (cv Drew) was used for blast fungus infection.
The fungal isolates used in this study belong to the IC-17 pathotype of blast fungus. On cv Drew (carrying the Pita resistance gene), the IC17-18/1 isolate (carrying AvrPita) is avirulent, whereas its race-change mutant (IC17-18/1-2, lacking AvrPita) is virulent (Harp and Correll, 1998). Both isolates are virulent on cv Nipponbare. The fungal infection of T0 transgenic plants was performed using the spot-inoculation method (Jia and Valent, 2001). Briefly, leaf segments (5 to 6 cm long) from the top full-expanded leaf were placed in a Petri dish on a circular filter paper soaked with water. Droplets, each containing ∼50 spores in 0.02% Tween 20, were applied carefully to the leaf surface. The Petri dishes were covered, and the leaf segments were maintained at 24°C under white light (3000 lux). Visual evaluation of disease symptoms and quantification of fungal growth were conducted at 5 or 6 days after inoculation. The fungal infection of 2-week-old T1 and T2 transgenic plants was performed using the typical spray-inoculation method at a concentration of 250,000 spores/mL (Lee et al., 2001). Blast resistance was evaluated based on fungal growth in planta (Qi and Yang, 2002) as well as lesion number and size.
In addition to the blast fungus, control and transgenic plants also were inoculated with a virulent strain of Burkholderia glumae, the causal agent of bacterial sheath rot and panicle blight diseases, by injection of 20 μL of bacterial suspension (∼106 colony-forming units/mL) into sheaths of 1-month-old rice plants. Host resistance to bacterial infection was evaluated based on the severity of disease symptoms and the levels of bacterial growth in planta.
Chemical and Abiotic Treatments
Chemical treatments were conducted on 2-week-old seedlings by spraying with abscisic acid (0.1 mM), jasmonic acid (0.1 mM), or salicylic acid (1 mM) solutions. Mechanical wounding was achieved by crushing rice leaves with a hemostat. Abiotic treatments and evaluations were conducted mainly according to Saijo et al. (2000). Seedlings were grown in large flat trays rather than individual pots to minimize potential variations among different pots. For cold stress, seedlings were transferred to 4°C for 3 days and then returned to normal growth conditions for recovery. Drought stress was induced by withholding water for up to 6 days. Under the greenhouse conditions (28°C and a 14-h-light/8-h-dark cycle) and the age of seedlings (2 weeks old) used in this experiment, leaves began to wilt 3 days after the free water was removed. For salt stress, roots of 2-week-old seedlings were immersed in 200 mM NaCl solution for up to 4 days. The time for returning stressed plants to the normal growth conditions was when approximately half of the control plants became wilted. The levels of cold, drought, and salt tolerance were evaluated based on the percentage of surviving seedlings after a period of recovery.
DNA Gel Blot, RNA Gel Blot, and PCR Analyses
Four micrograms of genomic DNA isolated by the cetyl-trimethyl-ammonium bromide method (Zhang et al., 1992) from cv Drew was digested individually with EcoRI, HindIII, PstI, and XbaI, fractionated on a 0.7% agarose gel, and blotted onto a nylon membrane according to the standard protocol (Sambrook et al., 1989). Total RNA was isolated from rice leaves using TRIzol reagent (Life Technologies, Rockville, MD). Fifteen micrograms of total RNA from each sample was separated on a 1.2% agarose gel containing formaldehyde and then transferred onto a nylon membrane. DNA and RNA ladders (Promega) were added to the gels to estimate the sizes of hybridized bands. DNA and RNA gel blots were hybridized with an α-32P-dCTP–labeled gene-specific probe (sequence from nucleotide 999 to the 3′ end of OsMAPK5a) in PerfectHyb buffer (Sigma). Hybridization and washing conditions were based on the manufacturer's instructions.
Two gene-specific primers, 5′-GAGTTCAGGCCGACGATGAC-3′ (RT-F99) and 5′-ATCGGCGATGTCGTGCAATC-3′ (RT-R1067), were designed to amplify DNA fragments covering the differentiated region of the OsMAPK5a and OsMAPK5b transcripts. Rice genomic DNA and reverse-transcribed cDNAs from the blast fungus–induced total RNA (2 days after infection) were used as templates for the PCR analysis.
Recombinant Protein, Antibody Production, and Autophosphorylation Assay
A BamHI site was introduced into OsMAPK5 at the start codon using Quickchange site-directed mutagenesis (Stratagene). The entire coding region of OsMAPK5 (as determined by digestion of mutagenized plasmid with BamHI and XhoI) then was ligated in frame into the His tag of pET-28c(+) vector (Novagen, Madison, WI). To generate a specific OsMAPK5 antigen, a DNA fragment spanning from nucleotide position 763 to the 3′ end of OsMAPK5 (as determined by digestion of OsMAPK5 with SacI and XhoI) was ligated in frame into the His tag of pET-28a(+). After confirmation with DNA sequencing, recombinant constructs were introduced into Escherichia coli strain BL21 (DE3). The recombinant proteins were induced and purified from E. coli cells according to the manufacturer's instructions (Pierce). Subsequently, polyclonal antisera against a 140–amino acid C-terminal region of OsMAPK5 were raised in rats (service provided by Cocalico Biologicals, Reamstown, PA).
The autophosphorylation assay was conducted according to Huang et al. (2000). Purified recombinant OsMAPK5 protein (300 ng) in reaction buffer (40 mM Hepes, pH 7.5, 20 mM MgSO4, 10 mM MnCl2, 1 mm CaCl2, 200 mM ATP, and 10 μCi of γ-32P-ATP) was incubated for 1 h at room temperature. The reaction mixture was stopped by the addition of SDS sample buffer and heating at 80°C for 10 min. After separation on a 10% SDS-PAGE gel, the phosphorylated product was detected by autoradiography.
Protein Extraction and Immunoblot Analysis
Rice leaf tissues were ground in liquid nitrogen and homogenized in extraction buffer containing 50 mM Tris, pH 8.0, 1 mM EDTA, 6 mM β-mercaptoethanol, 0.5 mM phenylmethylsulfonyl fluoride, and 0.3 μM aprotinin. After centrifugation at 16,000g, aliquots of supernatant were frozen immediately in liquid nitrogen and stored at −80°C. The protein concentration was determined using the protein assay kit (Bio-Rad) with BSA as a standard.
Equal amounts of protein extracts were separated on 12% SDS-polyacrylamide gels and electrotransferred onto nitrocellulose membranes in a transfer buffer (25 mM Tris, 192 mM Gly, and 20% methanol, pH 8.3). Nonspecific binding sites were blocked by incubating the membrane in 1 × TBS-T (25 mM Tris, 140 mM NaCl, and 0.1% Tween 20, pH 7.5) containing 5% nonfat dry milk for 1 h at room temperature. The anti-OsMAPK5 antibody (1:8000 dilution) was added, and the membrane was incubated overnight at 4°C. After rinsing three times (15 min each) with 1 × TBS-T, the membrane was incubated with the horseradish peroxidase–conjugated anti-rat IgG antibody (1:10,000 dilution; Sigma) in TBS-T for 1 h at room temperature. After five washes (15 min each) with TBS-T, the OsMAPK5 protein was detected with the ECL Plus detection system (Amersham) according to the manufacturer's instructions. In addition, biotinylated protein standards were separated in the same gel and detected by avidin–horseradish peroxidase conjugate (Bio-Rad) as a size marker.
Immunoprecipitation and in Gel Kinase Activity Assay
Protein extracts (∼0.4 mg) were incubated with 50 μL of anti-OsMAPK5 antibody at 4°C overnight. Fifty microliters of protein G–agarose beads was added and incubated for 2 h at 4°C. The protein-antibody complex on the beads was collected and washed three times in ice-cold PBS and finally resuspended in protein sample buffer.
The in-gel kinase activity assay was performed essentially as described by Zhang and Klessig (1997) with some modifications. Briefly, 40 μg of total protein, or immunoprecipitate from 400 μg of total protein, was fractionated on a 10% polyacrylamide gel containing 0.1% SDS and 0.25 mg/mL bovine brain myelin basic protein (Sigma). After electrophoresis, the SDS was removed by washing the gel three times (30 min each) at room temperature with buffer containing 25 mM Tris, pH 7.5, 0.5 mM DTT, 0.1 mM Na3VO4, 5 mM NaF, 0.5 mg/mL BSA, and 0.1% Triton X-100. The kinases then were allowed to renature overnight at 4°C with three changes of renaturing buffer (25 mM Tris, pH 7.5, 1 mM DTT, 0.1 mM Na3VO4, and 5 mM NaF). The phosphorylation of myelin basic protein was performed in 30 mL of reaction buffer (25 mM Tris, pH 7.5, 2 mM EGTA, 12 mM MgCl2, 1 mM DTT, and 0.1 mM Na3VO4) with the addition of 0.2 μM ATP and 50 μCi of γ-32P-ATP (3000 Ci/mmol) at room temperature for 60 min. The gel then was transferred into washing buffer (5% trichloroacetic acid and 1% sodium pyrophosphate) at room temperature for at least 5 h with five changes of the buffer. Finally, the gel was dried on filter paper and autoradiographed.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
The accession numbers for the OsMAPK5a and OsMAPK5b cDNAs are AF479883 and AF479884, respectively. The accession numbers for the MAPKs shown in Figure 1 are as follows: AtMPK3, D21839; NtWIPK, D61377; MsMMK4, T09622; TaWCK1, AF079318; OsMAPK5a, AF479883; ZmMPK4, AB016801; AtMPK6, D21842; NtSIPK, U94192; AtMPK4, D21840; AtMPK5, D21841; AtMPK13, AAF75067; AtMPK1, D14713; AtMPK7, D21843; OsMAPK3, AF216317; OsMAPK4, AJ251330; AtMPK8, AB038693; OsBWMK1, AF177392; and OsRMAPK2, AF194416.
Supplementary Material
Acknowledgments
We thank James Correll and Rick Cartright for providing the blast isolates and the bacterial panicle blight strain, respectively. We also thank Patrick Fenn and Benildo de los Reyes for critically reviewing the manuscript. This work was supported in part by the Arkansas Rice Research and Promotion Board.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.008714.
Footnotes
Online version contains Web-only data.
References
- Adam, A.L., Pike, S., Hoyos, E., Stone, J.M., Walker, J.C., and Novacky, A. (1997). Rapid and transient activation of a myelin basic protein kinase in tobacco leaves treated with harpin from Erwinia amylovora. Plant Physiol. 115, 853–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal, G.K., Rakwal, R., and Iwahashi, H. (2002). Isolation of novel rice (Oryza sativa L.) multiple stress responsive MAP kinase gene, OsMSRMK2, whose mRNA accumulates rapidly in response to environmental cues. Biochem. Biophys. Res. Commun. 294, 1009–1016. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Anderson, P.A., Lawrence, G.J., Morrish, B.C., Ayliffe, M.A., Finnegan, E.J., and Ellis, J.G. (1997). Inactivation of the flax rust resistance gene M associated with loss of a repeated unit within the leucine-rich repeat coding region. Plant Cell 9, 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai, T., Tena, G., Plotnikova, J., Willmann, M.R., Chiu, W.L., Gomez-Gomez, L., Boller, T., Ausubel, F.M., and Sheen, J. (2002). MAP kinase signaling cascade in Arabidopsis innate immunity. Nature 415, 977–983. [DOI] [PubMed] [Google Scholar]
- Ayliffe, M.A., Frost, D.V., Finnegan, E.J., Lawrence, G.J., Anderson, P.A., and Ellis, J.G. (1999). Analysis of alternative transcripts of the flax L6 rust resistance gene. Plant J. 17, 287–292. [DOI] [PubMed] [Google Scholar]
- Berberich, T., Sano, H., and Kusano, T. (1999). Involvement of a MAP kinase, ZmMAPK5, in senescence and recovery from low-temperature stress in maize. Mol. Gen. Genet. 262, 534–542. [DOI] [PubMed] [Google Scholar]
- Bögre, L., Ligterink, W., Meskiene, I., Barker, P.J., Heberle-Bors, E., Huskinsson, N.S., and Hirt, H. (1997). Wounding induces the rapid and transient activation of a specific MAP kinase pathway. Plant Cell 9, 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert, H.J., Nelson, D.E., and Jensen, R.G. (1995). Adaptations to environmental stresses. Plant Cell 7, 1099–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray, E.A. (1997). Plant responses to water deficit. Trends Plant Sci. 2, 48–49. [Google Scholar]
- Burnett, E.C., Desikan, R., Moser, R.C., and Neill, S.J. (2000). ABA activation of an MBP kinase in Pisum sativum epidermal peels correlates with stomatal responses to ABA. J. Exp. Bot. 51, 197–205. [DOI] [PubMed] [Google Scholar]
- Canman, C.E., and Kastan, M.B. (1996). Signal transduction: Three paths to stress relief. Nature 384, 213–214. [DOI] [PubMed] [Google Scholar]
- Cottyn, B., van Outryve, M.F., de Cleene, M., Swings, J., and Mew, T.W. (1996). Bacterial diseases of rice. II. Characterization of pathogenic bacteria associated with sheath rot complex and grain discoloration of rice in the Philippines. Plant Dis. 80, 438–445. [Google Scholar]
- Dinesh-Kumar, S.P., and Baker, B.J. (2000). Alternatively spliced N resistance gene transcripts: Their possible role in tobacco mosaic virus resistance. Proc. Natl. Acad. Sci. USA 97, 1908–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye, C.A., Tang, D., and Innes, R.W. (2001). Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc. Natl. Acad. Sci. USA 98, 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, R., Huang, Y., Kieber, J., and Luan, S. (1998). Identification of a dual-specificity protein phosphatase that inactivates a MAP kinase from Arabidopsis. Plant J. 16, 581–589. [DOI] [PubMed] [Google Scholar]
- Hackett, R.M., Oh, S.A., Morris, P.C., and Grierson, D. (1998). A tomato MAP kinase kinase gene differentially regulated during fruit development, leaf senescence, and wounding. Plant Physiol. 117, 1526–1531. [Google Scholar]
- Hardin, S.C., and Wolniak, S.M. (1998). Molecular cloning and characterization of maize ZmMEK1, a protein kinase with a catalytic domain homologous to mitogen- and stress-activated protein kinase kinases. Planta 206, 577–584. [DOI] [PubMed] [Google Scholar]
- Harp, T.L., and Correll, J.C. (1998). Recovery and characterization of spontaneous, selenate-resistant mutants of Magnaporthe grisea, the rice blast pathogen. Mycologia 90, 954–963. [Google Scholar]
- He, C., Fong, S.H., Yang, D., and Wang, G.L. (1999). BWMK1, a novel MAP kinase induced by fungal infection and mechanical wounding in rice. Mol. Plant-Microbe Interact. 12, 1064–1073. [DOI] [PubMed] [Google Scholar]
- Heimovaara-Dijkstra, S., Testerink, C., and Wang, M. (2000). Mitogen-activated protein kinase and abscisic acid signal transduction. Results Probl. Cell Differ. 27, 131–144. [DOI] [PubMed] [Google Scholar]
- Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282. [DOI] [PubMed] [Google Scholar]
- Hirt, H. (1997). Multiple roles of MAP kinases in plant signal transduction. Trends Plant Sci. 2, 11–15. [Google Scholar]
- Höfgen, R., and Willmitzer, L. (1988). Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 16, 9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H.J., Fu, S.F., Tai, Y.H., Chou, W.C., and Huang, D.D. (2002). Expression of Oryza sativa MAP kinase gene is developmentally regulated and stress-responsive. Physiol. Plant. 114, 572–580. [DOI] [PubMed] [Google Scholar]
- Huang, Y., Li, H., Gupta, R., Morris, P.C., Luan, S., and Kieber, J.J. (2000). ATMPK4, an Arabidopsis homolog of mitogen-activated protein kinase, is activated in vitro by AtMEK1 through threonine phosphorylation. Plant Physiol. 122, 1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura, K., Mizoguchi, T., Hayashida, N., Seki, M., and Shinozaki, K. (1998. a). Molecular cloning and characterization of three cDNAs encoding putative mitogen-activated protein kinase kinases (MAPKKs) in Arabidopsis thaliana. DNA Res. 5, 341–348. [DOI] [PubMed] [Google Scholar]
- Ichimura, K., Mizoguchi, T., Irie, K., Morris, P., Giraudat, J., Matsumoto, K., and Shinozaki, K. (1998. b). Isolation of ATMEKK1 (a MAP kinase kinase kinase)-interacting proteins and analysis of a MAP kinase cascade in Arabidopsis. Biochem. Biophys. Res. Commun. 253, 532–543. [DOI] [PubMed] [Google Scholar]
- Ichimura, K., et al. (2002). Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 7, 301–308. [DOI] [PubMed] [Google Scholar]
- Jia, Y., and Valent, B. (2001). Rapid determination of Magnaporthe grisea pathogenicity towards rice. Phytopathology 91, S44 (abstr.). [Google Scholar]
- Jin, H., Axtell, M.J., Dahlbeck, D., Ekwenna, O., Zhang, S., Staskawicz, B., and Baker, B. (2002). NPK1, an MEKK1-like mitogen-activated protein kinase kinase kinase, regulates innate immunity and development in plants. Dev. Cell 3, 291–297. [DOI] [PubMed] [Google Scholar]
- Jonak, C., Kiegerl, S., Ligterink, W., Baker, P.J., Huskisson, N.S., and Hirt, H. (1996). Stress signaling in plants: A mitogen-activated protein kinase pathway is activated by cold and drought. Proc. Natl. Acad. Sci. USA 93, 11274–11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokhlatchev, A.V., Canagarajah, B., Wilsbacher, J., Robinson, M., Atkinson, M., Goldsmith, E., and Cobb, M.H. (1998). Phosphorylation of the MAP kinase ERK2 promotes its homodimerization and nuclear translocation. Cell 93, 605–615. [DOI] [PubMed] [Google Scholar]
- Kiegerl, S., Cardinale, F., Siligan, C., Gross, A., Baudouin, E., Liwosz, A., Eklöf, S., Till, S., Bogre, L., Hirt, H., and Meskiene, I. (2000). SIMKK, a mitogen-activated protein kinase (MAPK) kinase, is a specific activator of the salt stress–induced MAPK, SIMK. Plant Cell 12, 2247–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knetsch, M.L., Wang, M., Snaar-Jagalska, B.E., and Heimovaara-Dijkstra, S. (1996). Abscisic acid induces mitogen-activated protein kinase activation in barley aleurone protoplasts. Plant Cell 8, 1061–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun, Y., Chiu, W.L., Tena, G., and Sheen, J. (2000). Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. USA 97, 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kultz, D. (1998). Phylogenetic and functional classification of mitogen- and stress-activated protein kinases. J. Mol. Evol. 46, 571–588. [DOI] [PubMed] [Google Scholar]
- Kyriakis, J.M., and Avruch, J. (1996). Protein kinase cascades activated by stress and inflammatory cytokines. Bioessays 18, 567–577. [DOI] [PubMed] [Google Scholar]
- Lee, M.-W., Qi, M., and Yang, Y. (2001). A novel jasmonic acid-inducible rice myb gene associates with fungal infection and host cell death. Mol. Plant-Microbe Interact. 14, 527–535. [DOI] [PubMed] [Google Scholar]
- Leung, J., and Giraudat, J. (1998). Abscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 199–222. [DOI] [PubMed] [Google Scholar]
- Ligterink, W., and Hirt, H. (2000). MAP kinase pathways in plants: Versatile signaling tools. Int. Rev. Cytol. 201, 209–258. [DOI] [PubMed] [Google Scholar]
- Ligterink, W., Kroj, T., zur Nieden, U., Hirt, H., and Scheel, D. (1997). Receptor-mediated activation of a MAP kinase in pathogen defense of plants. Science 276, 2054–2057. [DOI] [PubMed] [Google Scholar]
- Lopez, A.J. (1998). Alternative splicing of pre-mRNA: Developmental consequences and mechanisms of regulation. Annu. Rev. Genet. 32, 279–305. [DOI] [PubMed] [Google Scholar]
- Marchetti, M.A., Rush, M.C., and Hunter, W.E. (1976). Current status of rice blast [Pyricularia oryzae] in the southern United States. Plant Dis. Rep. 60, 721–725. [Google Scholar]
- Meskiene, I., Bogre, L., Glaser, W., Balog, J., Brandstotter, M., Zwerger, K., Ammerer, G., and Hirt, H. (1998). MP2C, a plant protein phosphatase 2C, functions as a negative regulator of mitogen-activated protein kinase pathways in yeast and plants. Proc. Natl. Acad. Sci. USA 95, 1938–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midoh, N., and Iwate, M. (1996). Cloning and characterization of probenazole-inducible gene for an intracellular pathogenesis-related protein in rice. Plant Cell Physiol. 37, 9–18. [DOI] [PubMed] [Google Scholar]
- Mikolajczyk, M., Awotunde, O.S., Muszynska, G., Klessig, D.F., and Dobrowolska, G. (2000). Osmotic stress induces rapid activation of a salicylic acid–induced protein kinase and a homolog of protein kinase ASK1 in tobacco cells. Plant Cell 12, 165–178. [PMC free article] [PubMed] [Google Scholar]
- Mittler, R., and Rizhsky, L. (2000). Transgene-induced lesion mimic. Plant Mol. Biol. 44, 335–344. [DOI] [PubMed] [Google Scholar]
- Mizoguchi, T., Ichimura, K., and Shinozaki, K. (1997). Environmental stress response in plants: The role of mitogen-activated protein kinases. Trends Biotechnol. 15, 15–19. [DOI] [PubMed] [Google Scholar]
- Molina, A., Volrath, S., Guyer, D., Maleck, K., Ryals, J., and Ward, E. (1999). Inhibition of protoporphyrinogen oxidase expression in Arabidopsis causes a lesion-mimic phenotype that induces systemic acquired resistance. Plant J. 17, 667–678. [DOI] [PubMed] [Google Scholar]
- Morris, P.C., Guerrier, D., Leung, J., and Giraudat, J. (1997). Cloning and characterisation of MEK1, an Arabidopsis gene encoding a homologue of MAP kinase kinase. Plant Mol. Biol. 35, 1057–1064. [DOI] [PubMed] [Google Scholar]
- Munnik, T., Ligterink, W., Meskiene, I., Calderini, O., Beyerly, J., Musgrave, A., and Hirt, H. (1999). Distinct osmo-sensing protein kinase pathways are involved in signalling moderate and severe hyper-osmotic stress. Plant J. 20, 381–388. [DOI] [PubMed] [Google Scholar]
- Nishihama, R., Banno, H., Kawahara, E., Irie, K., and Machida, Y. (1997). Possible involvement of differential splicing in regulation of the activity of Arabidopsis ANP1 that is related to mitogen-activated protein kinase kinase kinases (MAPKKKs). Plant J. 12, 39–48. [DOI] [PubMed] [Google Scholar]
- Parker, J.E., Coleman, M.J., Szabo, V., Frost, L.N., Schmidt, R., van der Biezen, E.A., Moores, T., Dean, C., Daniels, M.J., and Jones, J.D. (1997). The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the toll and interleukin-1 receptors with N and L6. Plant Cell 9, 879–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, M., et al. (2000). Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 103, 1111–1120. [DOI] [PubMed] [Google Scholar]
- Qi, M., and Yang, Y. (2002). Quantification of Magnaporthe grisea during infection of rice plants using real-time PCR and northern blot/phosphoimaging analysis. Phytopathology 92, 870–876. [DOI] [PubMed] [Google Scholar]
- Reymond, P., and Farmer, E.E. (1998). Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1, 404–411. [DOI] [PubMed] [Google Scholar]
- Saijo, Y., Hata, S., Kyozuka, J., Shimamoto, K., and Izui, K. (2000). Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 23, 319–327. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Samuel, M.A., and Ellis, B.E. (2002). Double jeopardy: Both overexpression and suppression of a redox-activated plant mitogen-activated protein kinase render tobacco plants ozone sensitive. Plant Cell 14, 2059–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seger, R., and Krebs, E.G. (1995). The MAPK signaling cascade. FASEB J. 9, 726–735. [PubMed] [Google Scholar]
- Seo, S., Okamoto, M., Seto, H., Ishizuka, K., Sano, H., and Ohashi, Y. (1995). Tobacco MAP kinase: A possible mediator in wound signal transduction pathways. Science 270, 1988–1992. [DOI] [PubMed] [Google Scholar]
- Seo, S., Sano, H., and Ohashi, Y. (1999). Jasmonate-based wound signal transduction requires activation of WIPK, a tobacco mitogen-activated protein kinase. Plant Cell 11, 289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snedden, W.A., and Blumwald, E. (2000). Alternative splicing of a novel diacylglycerol kinase in tomato leads to a calmodulin-binding isoform. Plant J. 24, 317–326. [DOI] [PubMed] [Google Scholar]
- Song, F., and Goodman, R.M. (2002). OsBIMK1, a rice MAP kinase gene involved in disease resistance response. Planta 215, 997–1005. [DOI] [PubMed] [Google Scholar]
- Stone, J.M., and Walker, J.C. (1995). Plant protein kinase families and signal transduction. Plant Physiol. 108, 451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K., and Shinshi, H. (1995). Transient activation and tyrosine phosphorylation of a protein kinase in tobacco cells treated with a fungal elicitor. Plant Cell 7, 639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takezawa, D. (1999). Elicitor- and A23187-induced expression of WCK-1, a gene encoding mitogen-activated protein kinase in wheat. Plant Mol. Biol. 40, 921–933. [DOI] [PubMed] [Google Scholar]
- Tena, G., Asai, T., Chiu, W.L., and Sheen, J. (2001). Plant mitogen-activated protein kinase signaling cascades. Curr. Opin. Plant Biol. 4, 392–400. [DOI] [PubMed] [Google Scholar]
- Usami, S., Banno, H., Ito, Y., Nishihama, R., and Machida, Y. (1995). Cutting activates a 46-kilodalton protein kinase in plants. Proc. Natl. Acad. Sci. USA 92, 8660–8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, J.-Q., Oono, K., and Imai, R. (2002). Two novel mitogen-activated protein signaling components, OsMEK1 and OsMAP1, are involved in a moderate low-temperature signaling pathway in rice. Plant Physiol. 129, 1880–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winans, S.C., Kerstetter, R.A., and Nester, E.W. (1988). Transcriptional regulation of the virA and virG genes of Agrobacterium tumefaciens. J. Bacteriol. 170, 4047–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L., Lee, M.W., Qi, M., and Yang, Y. (2001). Identification of defense-related rice genes by suppression subtractive hybridization and differential screening. Mol. Plant-Microbe Interact. 14, 685–692. [DOI] [PubMed] [Google Scholar]
- Yang, K.Y., Liu, Y., and Zhang, S. (2001). Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc. Natl. Acad. Sci. USA 98, 741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., Shah, J., and Klessig, D.F. (1997). Signal perception and transduction in plant defense responses. Genes Dev. 11, 1621–1639. [DOI] [PubMed] [Google Scholar]
- Zhang, F., Strand, A., Robbins, D., Cobb, M.H., and Goldsmith, E.J. (1994). Atomic structure of the MAP kinase ERK2 at 2.3 Å resolution. Nature 367, 704–711. [DOI] [PubMed] [Google Scholar]
- Zhang, Q., Saghai Maroof, M.A., Lu, T.Y., and Shen, B.Z. (1992). Genetic diversity and differentiation of indica and japonica rice detected by RFLP analysis. Theor. Appl. Genet. 83, 495–499. [DOI] [PubMed] [Google Scholar]
- Zhang, S., and Klessig, D.F. (1997). Salicylic acid activates a 48-kD MAP kinase in tobacco. Plant Cell 9, 809–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., and Klessig, D.F. (1998. a). The tobacco wounding-activated mitogen-activated protein kinase is encoded by SIPK. Proc. Natl. Acad. Sci. USA 95, 7225–7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., and Klessig, D.F. (1998. b). Resistance gene N–mediated de novo synthesis and activation of a tobacco mitogen-activated protein kinase by tobacco mosaic virus infection. Proc. Natl. Acad. Sci. USA 95, 7433–7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., and Klessig, D.F. (2001). MAPK cascades in plant defense signaling. Trends Plant Sci. 6, 520–527. [DOI] [PubMed] [Google Scholar]
- Zhang, S., and Liu, Y. (2001). Activation of salicylic acid–induced protein kinase, a mitogen-activated protein kinase, induces multiple defense responses in tobacco. Plant Cell 13, 1877–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.