Abstract
Plant defenses against pathogens and insects are regulated differentially by cross-communicating signal transduction pathways in which salicylic acid (SA) and jasmonic acid (JA) play key roles. In this study, we investigated the molecular mechanism of the antagonistic effect of SA on JA signaling. Arabidopsis plants unable to accumulate SA produced 25-fold higher levels of JA and showed enhanced expression of the JA-responsive genes LOX2, PDF1.2, and VSP in response to infection by Pseudomonas syringae pv tomato DC3000, indicating that in wild-type plants, pathogen-induced SA accumulation is associated with the suppression of JA signaling. Analysis of the Arabidopsis mutant npr1, which is impaired in SA signal transduction, revealed that the antagonistic effect of SA on JA signaling requires the regulatory protein NPR1. Nuclear localization of NPR1, which is essential for SA-mediated defense gene expression, is not required for the suppression of JA signaling, indicating that cross-talk between SA and JA is modulated through a novel function of NPR1 in the cytosol.
INTRODUCTION
To effectively combat invasion by microbial pathogens and herbivorous insects, plants are able to activate distinct defense responses that are effective specifically against the invader encountered (Van Loon, 2000). These induced defenses often are expressed not just locally but also in parts distant from the site of primary infection, thereby protecting the plant systemically against subsequent attack. Induced resistance is regulated by a network of interconnecting signal transduction pathways in which salicylic acid (SA) and jasmonic acid (JA) function as key signaling molecules (Reymond and Farmer, 1998; Pieterse and Van Loon, 1999; Glazebrook, 2001; Thomma et al., 2001). SA and JA accumulate in response to pathogen infection or herbivore damage, resulting in the activation of distinct sets of defense-related genes. Mutant and transgenic plants that are affected in SA accumulation often are more susceptible to pathogen infection than wild-type plants (Delaney et al., 1994; Nawrath and Métraux, 1999; Wildermuth et al., 2001). Blocking the response to JA generally renders plants more susceptible to herbivore damage (Howe et al., 1996; McConn et al., 1997), although enhanced susceptibility toward necrotrophic pathogens has been reported as well (Thomma et al., 2001). SA- and JA-dependent defense pathways have been shown to cross-communicate (Felton and Korth, 2000; Feys and Parker, 2000; Pieterse et al., 2001), providing the plant with a regulatory potential to fine-tune the defense reaction depending on the type of attacker encountered.
One of the most studied induced defense responses in plants is systemic acquired resistance (SAR). SAR is triggered after local infection with pathogens, causing hypersensitive necrosis, and is effective against a broad spectrum of plant pathogens (Ryals et al., 1996). The onset of SAR is accompanied by a local and systemic increase in the endogenous levels of SA (Malamy et al., 1990; Métraux et al., 1990) and the concomitant upregulation of a large set of genes (Ward et al., 1991), including genes that encode pathogenesis-related (PR) proteins (Van Loon and Van Strien, 1999). Several PR proteins possess antimicrobial activity and are thought to contribute to the state of resistance attained. Transduction of the SA signal requires the function of NPR1 (also known as NIM1), a regulatory protein that was identified in Arabidopsis through genetic screens for SAR-compromised mutants (Cao et al., 1994; Delaney et al., 1995; Shah et al., 1997). Mutant npr1 plants accumulate normal levels of SA after pathogen infection but are impaired in their ability to express PR genes and to mount a SAR response. The NPR1 gene encodes a protein with a BTB/BOZ domain and an ankyrin-repeat domain (Cao et al., 1997; Ryals et al., 1997; Aravind and Koonin, 1999). Both domains are known to mediate protein–protein interactions and are present in proteins with diverse functions (Bork, 1993; Aravind and Koonin, 1999), including the transcriptional regulator IκB, which mediates animal innate immune responses (Baldwin, 1996). Upon induction of SAR, NPR1 is translocated to the nucleus (Kinkema et al., 2000), where it interacts with members of the TGA/OBF subclass of basic domain/Leu zipper (bZIP) transcription factors (Zhang et al., 1999; Després et al., 2000; Zhou et al., 2000; Subramaniam et al., 2001; Fan and Dong, 2002) that are involved in the SA-dependent activation of PR genes (Lebel et al., 1998; Niggeweg et al., 2000). Physical interaction between NPR1 and TGA transcription factors has been shown to be required for the binding activity of these factors to promoter elements that play a crucial role in the SA-mediated activation of PR genes (Després et al., 2000; Fan and Dong, 2002).
The activation of SAR has been shown to suppress JA signaling in plants, thereby prioritizing SA-dependent resistance to microbial pathogens over JA-dependent defense against insect herbivory (Felton and Korth, 2000; Pieterse et al., 2001). Moreover, pharmacological and genetic experiments have shown that SA is a potent suppressor of JA-inducible gene expression (Doherty et al., 1988; Peña-Cortés et al., 1993; Doares et al., 1995; Harms et al., 1998; Gupta et al., 2000). The antagonistic effect of SA on JA signaling shows a striking resemblance to the effect of the nonsteroidal anti-inflammatory drug acetylsalicylic acid (aspirin), a derivative of SA, on the formation of prostaglandins in animal cells. Prostaglandins are related structurally to JA and play a role in diverse biological processes, such as inflammation at sites of infection or tissue injury (Straus and Glass, 2001). JA and prostaglandins originate biosynthetically from linolenic acid and arachidonic acid, respectively, which are released from cell membranes upon phospholipid hydrolysis. Linolenic acid and arachidonic acid are metabolized rapidly via the octadecanoid pathway, in which the enzymatic reactions leading to JA and prostaglandin formation are similar (Pan et al., 1998). In animal cells, aspirin inhibits the octadecanoid pathway by acetylating the key enzyme cyclooxygenase, ultimately leading to a decrease in prostaglandin formation (Van der Ouderaa et al., 1980).
In a similar process in plants, aspirin has been shown to inhibit the activity of the counterpart of cyclooxygenase, allene oxide synthase, which catalyzes the same step in the octa-decanoid pathway in plants, thereby affecting the formation of JA and the subsequent activation of stress-related gene expression (Pan et al., 1998). Whereas aspirin is able to inhibit prostaglandin and JA biosynthetic enzymes by acetylating them, SA, which lacks the acetyl group, is ineffective in this respect. Indeed, in Arabidopsis and flax plants, no inhibitory effect of SA on allene oxide synthase activity was observed (Harms et al., 1998; Laudert and Weiler, 1998). Thus, given the fact that the acetylated form of SA does not occur naturally in plants (Pierpoint, 1997), it is unlikely that inhibition of the allene oxide synthase activity plays a major role in the cross-communication between SA and JA signaling in plants. Nevertheless, SA is a strong negative regulator of JA-dependent cellular defense responses in plants (Doherty et al., 1988; Doares et al., 1995; Harms et al., 1998; Gupta et al., 2000).
So how does SA negatively regulate JA-dependent cellular defense responses in plants? In animal cells, both aspirin and SA are able to reduce proinflammatory prostaglandin formation by inhibiting the activity of the transcription factor NF-κB (Kopp and Ghosh, 1994; Yin et al., 1998). NF-κB plays a key role in the transcriptional activation of many genes during the innate immune response (Baldwin, 1996; Hatada et al., 2000), including the gene that encodes CYCLOOXYGENASE2, which catalyzes a rate-limiting step in prostaglandin production (Newton et al., 1997). In resting cells, NF-κB is sequestered in the cytoplasm by association with its inhibitory protein IκB. In response to various cellular stress conditions, such as infection by microbial or viral pathogens, IκB kinase is activated and phosphorylates IκB. Subsequently, IκB is ubiquitinated and degraded by the proteasome, releasing NF-κB to migrate into the nucleus and activate gene expression (Baldwin, 1996; Hatada et al., 2000). Both aspirin and SA block the activation of NF-κB by inhibiting IκB kinase, preventing the degradation of IκB and retaining NF-κB in the cytosol (Kopp and Ghosh, 1994; Yin et al., 1998). Interestingly, IκB shares structural similarity with NPR1 in plants (Cao et al., 1997; Ryals et al., 1997). In addition to the ankyrin-repeat domain, the phosphorylated Ser residues important for IκB function also are conserved in the NPR1 protein (Ryals et al., 1997).
Because of the intriguing analogies between the actions of SA/aspirin, prostaglandin, and IκB in animals and SA, JA, and NPR1 in plants, we investigated whether NPR1 plays a role in the SA-mediated negative regulation of JA signaling in Arabidopsis. In contrast to IκB in animal cells, which functions in the cytosol, NPR1 was reported previously to function in the nucleus when acting as a positive regulator of SA-dependent, defense-related gene expression (Kinkema et al., 2000; Subramaniam et al., 2001). Here, we report a novel function of NPR1 in the cytosol and provide evidence that cytosolic NPR1 plays a crucial role in cross-communication between SA- and JA-dependent plant defense responses.
RESULTS
Antagonistic Effect of Pathogen-Induced SA on JA Signaling
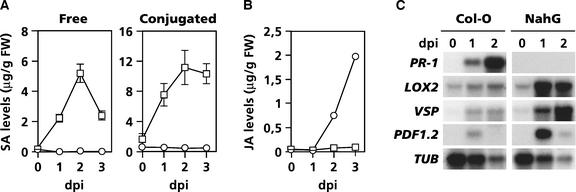
Previously, pharmacological experiments have shown that SA and its derivative aspirin exert an antagonistic effect on JA biosynthesis and JA-responsive gene expression in plants (Doherty et al., 1988; Peña-Cortés et al., 1993). To investigate whether endogenously synthesized SA also functions as a negative regulator of JA signaling during pathogen infection, we analyzed the production of JA and the expression of JA-responsive genes in Arabidopsis wild-type Columbia (Col-0) and transgenic NahG plants after infection with the bacterial speck-inducing pathogen Pseudomonas syringae pv tomato DC3000. Infection of wild-type Col-0 plants with Pseudomonas DC3000 resulted in a strong increase in both free and conjugated SA levels, whereas SA levels in SA hydroxylase–expressing NahG plants remained unchanged (Figure 1A). The expression pattern of the SA-inducible PR-1 gene correlated with the SA levels in infected wild-type and NahG plants (Figure 1C). In Col-0 plants, JA levels increased slightly in response to pathogen infection. However, in NahG plants, JA accumulated to 25-fold higher levels (Figure 1B), suggesting that in wild-type plants JA formation was suppressed by endogenously accumulating SA.
Figure 1.
Enhanced JA Accumulation and JA-Responsive Gene Expression in Pseudomonas DC3000–Infected Arabidopsis NahG Plants.
(A) Endogenous levels of free and conjugated SA in wild-type Col-0 (open squares) and SA-degrading NahG (open circles) plants after inoculation with the bacterial pathogen Pseudomonas DC3000. Error bars represent se (n = 5).
(B) JA levels in Pseudomonas DC3000–infected Col-0 (open squares) and NahG (open circles) plants. The experiment was performed three times with similar results.
(C) RNA gel blot analysis of SA-responsive (PR-1) and JA-responsive (LOX2, VSP, and PDF1.2) genes during pathogen infection.
Plants were inoculated with virulent Pseudomonas DC3000 by dipping the leaves in a bacterial suspension containing 2.5 × 107 colony-forming units/mL. At different days after inoculation (dpi), leaves were harvested for SA, JA, and RNA extraction. To check for equal loading, RNA gel blots were stripped and hybridized with a gene-specific probe for β-tubulin (TUB). Transcript levels of the constitutively expressed TUB gene decreased during the course of the infection process as a result of progressing cell death. FW, fresh weight.
To investigate the effect of pathogen-induced SA on JA-responsive gene expression, we analyzed the expression of three well-characterized Arabidopsis genes involved in various steps of the JA signaling pathway: LOX2 (LIPOXYGENASE2), which encodes LOX2, a key enzyme in the octadecanoid pathway leading to JA biosynthesis (Bell et al., 1995); VSP, which encodes a vegetative storage protein (Berger et al., 1995); and PDF1.2, which encodes a plant defensin with antimicrobial properties (Penninckx et al., 1996). In wild-type plants, LOX2, VSP, and PDF1.2 showed moderate increases in expression upon pathogen infection (Figure 1C). However, in NahG plants, mRNAs of the three JA-responsive genes accumulated to much higher levels. These results indicate that in wild-type plants, pathogen-induced SA accumulation is associated with the suppression of JA-responsive gene expression.
Inhibition of LOX2 Is Sufficient to Suppress Pathogen-Induced JA Production
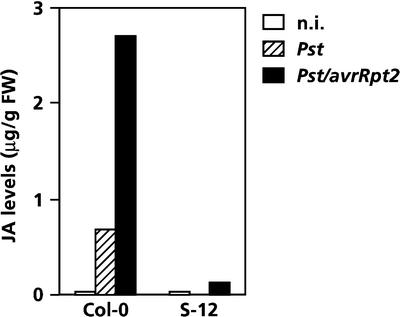
LOX2 is a key enzyme in the octadecanoid pathway leading to the formation of JA. In transgenic Arabidopsis S-12 plants, which have severely reduced levels of LOX2 as a result of cosuppression of the LOX2 gene, the ability to accumulate JA in response to wounding is blocked completely (Bell et al., 1995). To determine whether the SA-mediated suppression of LOX2, as observed during pathogen infection (Figure 1C), could explain the inhibitory effect of SA on JA formation (Figure 1B), JA production was analyzed in infected wild-type Col-0 and transgenic S-12 plants. As shown in Figure 1C, pressure infiltration of wild-type leaves with Pseudomonas DC3000 resulted in increased accumulation of LOX2 transcripts, whereas in S-12 plants, the activation of this gene was blocked completely (data not shown). Furthermore, JA levels increased significantly in wild-type plants after inoculation with virulent Pseudomonas DC3000, but in LOX2-deprived S-12 plants, pathogen-induced accumulation of JA was almost abolished (Figure 2). Note that leaves were inoculated by pressure infiltration instead of dip inoculation, leading to a synchronous infection of virtually all cells and greater JA accumulation in wild-type leaves than that observed in Figure 1B.
Figure 2.
Effect of the Cosuppression of LOX2 on Pathogen-Induced JA Production in Arabidopsis S-12 Plants.
JA levels in wild-type Col-0 and LOX2-deprived S-12 plants at 2 days after inoculation with virulent Pseudomonas DC3000 or avirulent Pseudomonas DC3000/avrRpt2. Plants were inoculated by pressure-infiltrating the leaves with a bacterial suspension containing 107 colony-forming units/mL. FW, fresh weight; n.i., not inoculated.
Compared with inoculation with virulent Pseudomonas DC3000, pressure infiltration of wild-type leaves with avirulent Pseudomonas DC3000/avrRpt2, carrying the avirulence gene avrRpt2 (Kunkel et al., 1993), led to a hypersensitive reaction and fourfold higher JA levels. However, similarly treated S-12 plants showed no significant increase in JA levels. These results demonstrate that LOX2 is required for the pathogen-induced production of JA and that there is a direct correlation between the level of LOX2 gene expression and JA production. During pathogen infection of wild-type plants, endogenously accumulating SA has an inhibitory effect on LOX2 gene expression and JA formation. Therefore, we postulate that the inhibitory effect of SA on JA biosynthesis during infection is regulated at least partly at the transcriptional level, although post-translational effects of SA on JA formation cannot be excluded completely.
NPR1 Controls the Suppression of JA Signaling
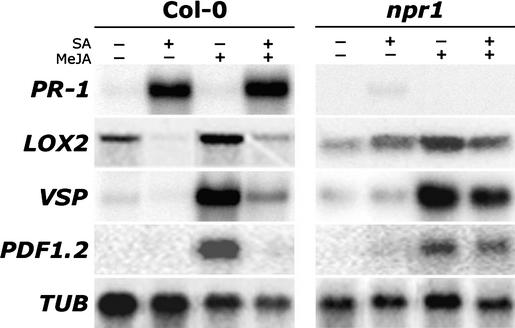
NPR1 is an important transducer of the SA signal in disease resistance. To investigate a possible role of NPR1 in the suppression of JA-responsive gene expression by SA, mutant Arabidopsis npr1-1 plants were tested. Like NahG plants, npr1-1 plants showed enhanced JA-responsive gene expression and increased levels of JA upon infection with Pseudomonas DC3000 (data not shown), suggesting that in wild-type plants NPR1 is involved in the SA-mediated suppression of JA signaling. To investigate the role of NPR1 in cross-talk in more detail, we followed a pharmacological approach. In wild-type Col-0 plants, exogenous application of SA activated PR-1, whereas treatment with methyl jasmonate (MeJA) resulted in the accumulation of LOX2, VSP, and PDF1.2 mRNA (Figure 3). Upon combined treatment with SA and MeJA, MeJA had no effect on SA-induced PR-1 transcript levels. By contrast, both background and MeJA-induced transcript levels of the JA-responsive genes were strongly suppressed by SA, confirming the negative effect of SA on JA-responsive gene expression. Mutant npr1-1 plants, which are compromised in their ability to express PR-1 in response to SA, showed levels of MeJA-induced LOX2, VSP, and PDF1.2 expression that were similar to those observed in wild-type plants. However, in the combined treatment, SA had almost no inhibitory effect on background and MeJA-induced expression of the three JA-responsive genes (Figure 3). Two additional npr1 mutants, npr1-2 and npr1-3, each with a mutation in different domains in the NPR1 protein (Cao et al., 1997), showed similar expression patterns (data not shown), confirming that NPR1 is required for the SA-mediated suppression of JA-responsive gene expression.
Figure 3.
Effect of the npr1 Mutation on the SA-Mediated Suppression of JA-Responsive Gene Expression.
RNA gel blot analysis of SA-responsive (PR-1) and JA-responsive (LOX2, VSP, and PDF1.2) genes after exogenous application of MeJA, SA, or a combination of both in Arabidopsis wild-type Col-0 and mutant npr1 plants. Five-week-old plants were induced by dipping the leaves in a 0.015% (v/v) Silwet L-77 solution containing 1 mM SA, 0.1 mM MeJA, or a combination of both. Two days later, leaves were harvested for RNA extraction. Equal loading of RNA samples was checked using a probe for the constitutively expressed β-tubulin (TUB) gene.
The NPR1 Interactor TGA2 Binds to the TGACG Motif in the PDF1.2 Promoter
NPR1 interacts with members of the TGA/OBF family of bZIP transcription factors, which have been shown to play both positive and negative regulatory roles in plant defense (Niggeweg et al., 2000; Pontier et al., 2001; Fan and Dong, 2002). TGA transcription factors specifically bind to the TGACG motif in target promoters, thereby regulating gene transcription (Zhang et al., 1999; Després et al., 2000; Zhou et al., 2000). The TGACG motif was shown by linker-scanning mutagenesis to be essential for the SA-induced expression of the Arabidopsis PR-1 gene (Lebel et al., 1998), but it has been implicated in JA-responsive gene expression as well (Xiang et al., 1996; Rouster et al., 1997). Interestingly, the JA-responsive genes LOX2, VSP, and PDF1.2 all contain one or more TGACG motifs in their promoters. Therefore, we investigated the possible role of this motif in cross-talk between SA and JA signaling.
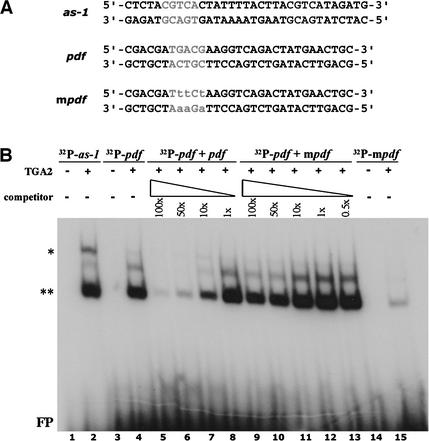
The promoter of the PDF1.2 gene contains a single TGACG sequence at positions −445 to −441 relative to the predicted translational start of the PDF1.2 gene product (Manners et al., 1998). To determine whether TGA transcription factors can bind to this motif, we performed a mobility shift assay with partially purified TGA2 transcription factor protein (Zhang et al., 1999) and TGACG-containing oligonucleotide probes derived from the PR-1 and PDF1.2 promoters (Figure 4A). The as-1 element from the PR-1 promoter contains two inverted TGACG sequences (Lebel et al., 1998). As demonstrated previously (Zhang et al., 1999), TGA2 caused a mobility shift for the as-1 oligonucleotide probe (Figure 4B, lane 2). A similar mobility shift was observed for the PDF1.2 probe (Figure 4B, lane 4). To examine the specificity of the binding of TGA2 to the PDF1.2 probe, a competition experiment was performed using an excess of unlabeled PDF1.2 oligonucleotide probe. Increasing the amount of unlabeled oligonucleotides in the reaction reduced the binding of TGA2 to the labeled probe (Figure 4B, lanes 5 to 8). By contrast, the addition of an unlabeled oligonucleotide PDF1.2 probe with point mutations in the TGACG binding motif (mpdf; Figure 4A) barely affected the binding of TGA2 to the labeled PDF1.2 probe (Figure 4B, lanes 9 to 13), indicating that the binding of TGA2 to the TGACG motif in the PDF1.2 promoter is specific.
Figure 4.
In Vitro Binding of TGA2 to the TGACG Motif in the Promoter of the Arabidopsis PDF1.2 Gene.
(A) Oligonucleotides used in the gel mobility shift assay. The as-1 probe contains two inverted TGACG motifs and represents the SA-responsive as-1 element in the promoter of the Arabidopsis PR-1 gene. The pdf probe resembles the wild-type sequence surrounding the TGACG motif in the JA-responsive PDF1.2 gene. The mpdf probe is similar to the pdf probe but contains three point mutations in the TGACG motif.
(B) Gel mobility shift assay to test the binding of TGA2 to the TGACG motif in the PDF1.2 promoter. Binding reactions contained 7 × 104 cpm of 32P-labeled probe incubated with 1 μg of either a control protein preparation (lanes 1, 3, and 14) or partly purified TGA2 (lanes 2, 4 to 13, and 15). The specificity of the binding of TGA2 to the TGACG motif in the PDF1.2 promoter was tested by the addition of 100×, 50×, 10×, 1×, or 0.5× molar excess amounts of unlabeled pdf (lanes 5 to 8) or mpdf (lanes 9 to 13) competitor probes. The double asterisks indicate specific binding of TGA2 to the oligonucleotide probe, and the single asterisk indicates specific TGA2 binding and dimerization as a result of the presence of two TGACG motifs in the oligonucleotide probe. FP, free probe.
The TGACG Motif Is Not Required for NPR1-Dependent Cross-Talk
To investigate the role of the TGACG motif in the PDF1.2 promoter in the negative regulation of JA-responsive gene expression by SA in planta, we analyzed transgenic Arabidopsis plants containing a series of PDF1.2 promoter deletions fused to the uidA reporter gene in the wild-type background (R. Brown, K. Kazan, D. MacLean, and J.M. Manners. “Book of Abstracts of the 11th International Conference on Arabidopsis Research”, International Conference on Arabidopsis Research, Madison, WI, June 24–28, 2000). Seedlings of six transgenic lines with varying 5′ deletions were selected (Figure 5A) and treated with SA, MeJA, or a combination of SA and MeJA. Consistent with previous findings, lines P1 to P5 showed induced β-glucuronidase (GUS) activity after treatment with MeJA, whereas line P6 showed no GUS activity at all (Figure 5B). MeJA-induced GUS expression was inhibited strongly when SA was included in the medium as well as in lines P4 and P5, which lack the TGACG motif. SA and MeJA had no effect on GUS activity in the constitutive GUS expressor PG15, indicating that neither of these chemical agents affected the activity of the GUS enzyme. Together, these results demonstrate that the TGACG motif in the promoter of PDF1.2 is not essential for the SA-mediated inhibition of JA-responsive gene expression.
Figure 5.
The TGACG Motif Is Not Required for SA-Mediated Suppression of the MeJA-Induced Activation of the Arabidopsis PDF1.2 Promoter.
(A) Scheme of the 5′ deletions of the PDF1.2 promoter fused to the uidA gene. Red bars represent the TGACG motif located at positions −445 to −441 upstream of the predicted translational start of the uidA reporter gene.
(B) Histochemical staining of GUS activity in seedlings of transgenic Arabidopsis lines PG15, which constitutively expresses the uidA gene, and P1 to P6, which contain 5′ deletions of the Arabidopsis PDF1.2 promoter fused to the uidA reporter gene. Twelve-day-old seedlings grown on Murashige and Skoog (1962) medium were transferred to fresh medium containing 0.5 mM SA, 0.02 mM MeJA, or a combination of both and stained for GUS activity 2 days later.
Cross-Talk Is Modulated by Cytosolic NPR1
NPR1 is translocated to the nucleus in response to SA, a process that was shown to be essential for SA-induced PR-1 gene expression (Kinkema et al., 2000). To determine whether the nuclear localization of NPR1 is similarly required for the SA-mediated suppression of JA-responsive gene expression, we used mutant npr1 plants engineered to constitutively express a fusion protein of NPR1 and the hormone binding domain (HBD) of the rat glucocorticoid receptor. The nucleocytoplasmic localization of this fusion protein can be controlled by the steroid hormone dexamethasone (DEX) (Aoyama and Chua, 1997; Kinkema et al., 2000). In the absence of DEX, the NPR1-HBD fusion protein was retained in the cytosol through an association with the heat-shock protein HSP90. In cells treated with DEX, HSP90 dissociated, allowing the NPR1-HBD fusion protein to translocate into the nucleus. As a control, we used mutant npr1 plants constitutively expressing the wild-type NPR1 gene under the control of the 35S promoter of Cauliflower mosaic virus (35S:NPR1).
Figure 6 shows that overexpression of NPR1 in mutant npr1 plants (35S::NPR1) restored the SA-induced activation of PR-1 and the SA-mediated suppression of MeJA-induced PDF1.2 gene expression, both of which were blocked in mutant npr1 plants. In the absence of SA, neither PR-1 nor MeJA-induced PDF1.2 gene expression was affected in NPR1-overexpressing plants, confirming the notion that NPR1 needs to be activated by SA (Cao et al., 1998). Treatment of 35S:NPR1 plants with DEX did not induce PR-1 or PDF1.2 gene expression, nor did it affect the expression of these genes in response to SA or MeJA, indicating that DEX did not affect the NPR1 protein or SA- and JA-responsive gene expression. In 35S:NPR1-HBD plants, the SA-induced expression of PR-1 was restored only when the plants were treated with DEX, confirming previous findings that SA-induced PR-1 expression requires the nuclear localization of NPR1 (Kinkema et al., 2000). Interestingly, treatment of 35S:NPR1-HBD plants with both SA and MeJA suppressed the MeJA-induced expression of PDF1.2, not only in the presence of DEX but also in its absence, when NPR1 was retained in the cytosol. This finding indicates that the nuclear localization of NPR1 is not required to suppress the MeJA-induced expression of PDF1.2 by SA.
Figure 6.
SA-Mediated Suppression of MeJA-Induced PDF1.2 Expression through a Function of NPR1 in the Cytosol.
RNA gel blot analysis of the SA-responsive PR-1 gene and the MeJA-inducible PDF1.2 gene in wild-type Col-0, mutant npr1, and the NPR1-overexpressing transformants 35S:NPR1 (in npr1) and 35S:NPR1-HBD (in npr1). Seedlings were grown for 12 days on Murashige and Skoog (1962) medium with or without DEX (5 μM). Subsequently, the seedlings were transferred to fresh medium with (+) or without (−) 0.5 mM SA and 0.02 mM MeJA. Two days after induction, the seedlings were harvested for RNA gel blot analysis. Equal loading of RNA samples was checked by staining rRNA bands with ethidium bromide.
DISCUSSION
The defense response of plants under attack by microbial pathogens or herbivorous insects is regulated by a network of signal transduction pathways. Cross-communication between defense signaling pathways provides the plant with an elaborate regulatory potential that leads to the activation of the most suitable defense against the invader encountered. In some cases, different defense signal transduction pathways cooperate and enhance resistance against pathogen attack (Van Wees et al., 2000). In other cases, antagonism between pathways allows the defense response to be controlled in a focused manner. For instance, plants that are infected by SAR-inducing pathogens have been shown to suppress JA-dependent defenses against certain herbivorous insects or necrotrophic pathogens (Felton and Korth, 2000; Pieterse et al., 2001), thereby prioritizing SA-dependent defense responses over JA-dependent responses. We report evidence indicating that SA produced during pathogen infection plays an important role in the suppression of both JA biosynthesis and JA-responsive gene expression, which are involved in the reaction of plants to wounding and insect herbivory. Compared with wild-type plants, transgenic NahG plants showed enhanced expression of LOX2 and accordingly synthesized 25-fold higher levels of JA during pathogen infection (Figures 1B and 1C). Moreover, the expression of the JA-responsive genes VSP and PDF1.2 was enhanced strongly in NahG plants, suggesting that in wild-type plants JA signaling is inhibited by SA that accumulates during pathogen infection.
The LOX2 gene, which encodes LOX2 (which is involved in the octadecanoid pathway), is autoregulated by JA and thus controls a feed-forward loop in JA biosynthesis (Bell et al., 1995). Cosuppression of LOX2 gene expression in transgenic S-12 plants appeared to be effective in blocking JA biosynthesis during pathogen infection (Figure 2). Therefore, the inhibition of this JA biosynthetic gene by SA produced during pathogen infection may result in a strong inhibition of JA formation. Other genes involved in JA biosynthesis, such as AOS, which encodes ALLENE OXIDE SYNTHASE, have been shown to be regulated by JA as well (Laudert and Weiler, 1998). Thus, the SA-mediated inhibition of JA formation during pathogen attack might be the result of a coordinated suppression of JA-responsive genes that encode enzymes of the octadecanoid pathway.
NPR1 has been demonstrated to be an important transducer of the SA signal in the SA-mediated activation of PR gene expression and broad-spectrum resistance (Cao et al., 1994; Delaney et al., 1995; Shah et al., 1997). Our study revealed NPR1 as a key regulatory factor in the cross-communication between SA and JA signaling. SA-mediated suppression of the JA-responsive genes LOX2, VSP, and PDF1.2, which was observed in wild type Col-0 plants, was abolished in mutant npr1 plants (Figures 3 and 6), indicating that NPR1 is essential for the inhibition of JA-responsive gene expression by SA. How does SA-activated NPR1 function as a negative regulator of JA-responsive genes? Previously, NPR1 was found to interact with members of the TGA subclass of the bZIP transcription factor family (Zhang et al., 1999; Després et al., 2000; Zhou et al., 2000). TGA factors specifically bind to TGACG motifs and have been shown to play both positive and negative regulatory roles in plant defense (Xiang et al., 1996; Rouster et al., 1997; Lebel et al., 1998; Niggeweg et al., 2000; Pontier et al., 2001; Fan and Dong, 2002). All JA-responsive genes tested in this study contain one or more TGACG motifs in their promoters. Therefore, we hypothesized that NPR1–TGA interactions might play a role in the SA-mediated suppression of JA-responsive gene expression. Experiments indicated that the strong NPR1 interactor TGA2 (Zhang et al., 1999) binds specifically to the TGACG motif in the JA-responsive PDF1.2 promoter in vitro (Figure 4). However, promoter-deletion analysis demonstrated that SA suppressed MeJA-induced expression in all JA-respon-sive transgenic plants, including those that lack the TGACG motif (P4 and P5; Figure 5). These results suggest that the TGACG motif is not essential for the SA-mediated inhibition of JA-responsive gene expression.
In animal cells, it has been demonstrated that SA exerts an inhibitory effect on IκB kinase, thereby preventing IκB from being phosphorylated and subsequently degraded by the proteasome (Kopp and Ghosh, 1994; Yin et al., 1998). As a result, IκB remains associated with the transcription factor NF-κB, retaining NF-κB in the cytosol and preventing it from activating target genes such as the NF-κB–dependent, proinflammatory prostaglandin biosynthesis gene CYCLOOXYGENASE2 (Newton et al., 1997; Gallois et al., 1998). Thus, in animal cells, SA is able to suppress the formation of prostaglandin, the JA counterpart, by affecting the activity of IκB in the cytosol. Because of the structural similarity between IκB and NPR1 (Cao et al., 1997; Ryals et al., 1997), we investigated whether the NPR1-dependent inhibitory effect of SA on JA-responsive gene expression and, consequently, JA production, also functions through a role of NPR1 in the cytosol. Using a DEX-inducible system to control the nucleocytoplasmic localization of NPR1, we demonstrated that nuclear localization of NPR1 is not required for cross-talk between SA and JA signaling (Figure 6). By inference, SA-activated NPR1 must exert its negative effect on JA-responsive gene expression through an unknown function in the cytosol.
How does cytosolic NPR1 control the SA-mediated suppression of JA-responsive gene expression? In the absence of SA, cytosolic NPR1 might be involved in the control of JA-responsive gene expression, either by inhibiting negative regulators of JA-responsive gene expression or by facilitating the delivery of positive regulators of JA-responsive genes to the nucleus. However, in this scenario, npr1 null mutants and NPR1 overexpressors should show an altered JA-responsive phenotype in the absence of SA. This was clearly not the case (data not shown), which makes this possibility unlikely. A more plausible explanation for the cytosolic role of NPR1 is presented in the model shown in Figure 7. In this model, NPR1 is translocated to the nucleus upon activation by SA, where it facilitates the activation of SA-responsive PR genes. In the cytosol, the remaining SA-activated NPR1 pool is involved in the suppression of JA-responsive gene expression, either by facilitating the delivery of negative regulators of JA-responsive genes to the nucleus or by inhibiting positive regulators of JA-responsive gene expression. However, alternative scenarios, such as an effect of SA-activated cytosolic NPR1 on the activity of JA-metabolizing enzymes, cannot be excluded. It is tempting to speculate that, analogous to the effect of SA and aspirin on IκB kinase, (de)phosphorylation of NPR1 plays a role in the process that leads to the SA-mediated activation of NPR1, because the phosphorylated Ser residues important in IκB function are conserved in NPR1 (Ryals et al., 1997).
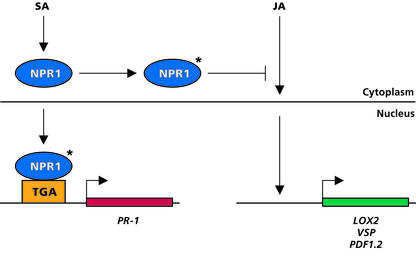
Figure 7.
Proposed Model for Cytosolic NPR1 as a Modulator of Cross-Talk between SA- and JA-Dependent Plant Defense Responses.
In wild-type Col-0 plants, SA accumulates after pathogen infection, resulting in the activation of NPR1 (asterisk). Activated NPR1 then is localized to the nucleus, where it interacts with TGA transcription factors, ultimately leading to the activation of SA-responsive PR genes. In the cytosol, activated NPR1 negatively regulates JA-responsive gene expression, possibly by inhibiting positive regulators of JA-responsive genes or by facilitating the delivery of negative regulators of JA-responsive genes to the nucleus. The suppression of JA-responsive genes that encode enzymes from the octadecanoid pathways, such as LOX2, ultimately results in the inhibition of JA formation.
In conclusion, our results clearly demonstrate the importance of NPR1 in cross-talk between the SA- and JA-dependent signaling pathways in plant defense and reveal a novel function of NPR1 in the cytosol. The striking parallels with processes involved in the animal innate immune response suggest that defense signaling pathways in plants and animals are at least partly conserved.
METHODS
Plant Growth and Pathogen Infection
To grow plants for pathogen infection, seeds of wild-type Arabidopsis thaliana (accession Columbia [Col-0]), transgenic NahG plants harboring the bacterial nahG gene (Gaffney et al., 1993), mutant npr1-1 plants (Cao et al., 1994), and LOX2-cosuppressed S-12 plants harboring the LOX2 cDNA under the control of the 35S promoter of Cauliflower mosaic virus (Bell et al., 1995) were sown in quartz sand. Two-week-old seedlings were transferred to 60-mL pots containing a mixture of sand and potting soil that had been autoclaved twice for 20 min with a 24-h interval. Plants were cultivated in a growth chamber with an 8-h-day (200 μE·m−2·s−1 at 24°C)/16-h-night (20°C) cycle at 70% RH.
The virulent bacterial leaf pathogen Pseudomonas syringae pv tomato DC3000, which causes bacterial speck disease, and the avirulent strain Pseudomonas DC3000/avrRpt2, with the plasmid pV288 carrying the avirulence gene avrRpt2 (Kunkel et al., 1993), were grown overnight at 28°C in liquid King's medium B as described previously (Pieterse et al., 1998). Bacterial cells were collected by centrifugation and resuspended in 10 mM MgSO4 to a final density of 107 or 2.5 × 107 colony-forming units/mL. For leaf dip inoculation of plants, the surfactant Silwet L-77 (Van Meeuwen Chemicals BV, Weesp, The Netherlands) was added to a final concentration of 0.015% (v/v).
Infection of Col-0, NahG, and npr1-1 plants with Pseudomonas DC3000 was performed as described previously (Pieterse et al., 1998). One day before pathogen infection, the plants were placed at 100% RH. Leaves of 5-week-old plants were dipped in a suspension of Pseudomonas DC3000 containing 2.5 × 107 colony-forming units/mL. At different times after inoculation, all of the rosette leaves of 25 plants for each genotype and time point were harvested for RNA extraction and to determine salicylic acid (SA) and jasmonic acid (JA) levels. For the analysis of pathogen-induced JA levels in LOX2-cosuppressed S-12 plants (Bell et al., 1995), a suspension of virulent Pseudomonas DC3000 or avirulent Pseudomonas DC3000/avrRpt2 at a density of 107 colony-forming units/mL was pressure-infiltrated into the leaves as described previously (Pieterse et al., 1998). As a control, Col-0 plants were inoculated similarly.
SA and JA Determination
Leaves were frozen in liquid nitrogen and pulverized with mortar and pestle. For each SA extraction, 0.5 g of ground leaf tissue was transferred to a 1.5-mL microfuge tube, and 100 μL of the internal standard ortho-anisic acid (1 μg/mL) and 0.5 mL of 70% ethanol were added. Subsequently, extraction and quantification of free and conjugated SA were performed as described previously (Meuwly and Métraux, 1993). For each JA extraction, a sample of 1 g was taken from the frozen leaf material, which consisted of at least 20 plants that received the same treatment. Subsequently, the sample was transferred to a 50-mL centrifuge tube. To the frozen samples were added 100 ng of the internal standard 9,10-dihydrojasmonic acid, 10 mL of saturated NaCl solution, 0.5 mL of 1 M citric acid, and 25 mL of diethylether containing 0.005% (w/v) butylated hydroxytoluene as antioxidant. Subsequently, extraction and gas chromatography–mass spectrometry quantification of JA were performed as described (Mueller and Brodschelm, 1994).
RNA Extraction and RNA Gel Blot Analysis
Extraction and electrophoretic separation of RNA, preparation of RNA gel blots, and hybridization of the blots with gene-specific probes for PR-1, LOX2, VSP, PDF1.2, and TUBULIN (TUB) were performed as described previously (Cao et al., 1994; Pieterse et al., 1998). The AGI numbers for the genes studied are At2g14610 (PR-1), At3g45140 (LOX2), At5g24770 (VSP), At4g37750 (PDF1.2), and At5g44340 (TUB).
Chemical Induction
The effect of exogenously applied SA on methyl jasmonate (MeJA)–induced gene expression was studied in wild-type Col-0 plants, mutant npr1-1, npr1-2, and npr1-3 plants (Cao et al., 1994, 1997), 35S:NPR1-H plants overexpressing NPR1 in the wild-type Col-0 background (Cao et al., 1998), and 35S:NPR1 and 35S:NPR1-HBD plants overexpressing NPR1 and NPR1-HBD, respectively, in the mutant npr1 background (Kinkema et al., 2000). Plants were grown in soil as described above or on plates containing Murashige and Skoog (1962) (MS) medium, pH 5.7, supplemented with 20 g/L Suc and 0.8% (w/v) plant agar. Chemical induction of soil-grown plants was performed by dipping the leaves of 5-week-old plants in a solution containing 0.015% (v/v) Silwet L-77 and either 1 mM SA (Mallinckrodt Baker, Deventer, The Netherlands), 0.1 mM MeJA (Serva, Brunschwig Chemie, Amsterdam, The Netherlands), or a combination of these chemicals. Control plants were treated with 0.015% Silwet L-77 only. One day before application of the chemicals, the plants were placed at 100% RH. Chemical induction of plants grown on MS medium was performed by transferring 12-day-old seedlings to fresh MS medium supplemented with 0.5 mM SA, 0.02 mM MeJA, or both chemicals. MeJA was added to the solutions from a 1000-fold concentrated stock in 96% ethanol. To the solutions without MeJA, a similar volume of 96% ethanol was added. To control the nucleocytoplasmic localization of NPR1 in 35S:NPR1-HBD plants, 5 μM dexamethasone (Sigma) was included in the growth medium (before and after induction with SA and/or MeJA). After induction treatment, plants were cultured for 2 days under climate chamber conditions as described above, after which leaf tissue was harvested for RNA extraction.
Analysis of the TGACG Motif in the PDF1.2 Promoter
The gel mobility shift assay was performed with partially purified TGA2 protein as described previously (Zhang et al., 1999). The wild-type and mutant oligonucleotide PDF1.2 probes used were designed according to the sequence surrounding the TGACG motif in the JA-responsive promoter of the Arabidopsis PDF1.2 gene (Manners et al., 1998).
The construction of a series of transgenic Arabidopsis lines (P1 to P6) containing translational fusions of 5′ deletions of the PDF1.2 promoter to the uidA reporter gene was described previously (R. Brown, K. Kazan, D. MacLean, and J.M. Manners. “Book of Abstracts of the 11th International Conference on Arabidopsis Research”, International Conference on Arabidopsis Research, Madison, WI, June 24–28, 2000). Surface-sterilized seeds of homozygous progeny from five independent transformants per line as well as the constitutive uidA-expressing line PG15 were allowed to germinate on MS medium supplemented with 10 g/L Suc and 0.6% (w/v) plant agar, pH 5.7. After 12 days, seedlings were transferred to fresh MS medium containing 0.5 mM SA, 0.02 mM MeJA, or a combination of both, as described above. After 2 days on induction medium, β-glucuronidase activity was assessed by transferring the seedlings to β-glucuronidase staining solution (1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronide, 100 mM NaPi buffer, pH 7.0, 10 mM EDTA, 0.1% [v/v] Triton X-100, 1 mM potassium ferrocyanide, and 1 mM potassium ferricyanide). After overnight incubation at 37°C, the seedlings were destained by repeated washes in 70% ethanol and evaluated for staining intensity.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We thank Stefanie Parchmann and Ruth Imbusch for JA measurements, Weihua Fan and Meenu Kesarwani for technical assistance and valuable discussions, and Peter Bakker and Bas Rutjens for critically reading the manuscript. This research was supported in part by grants from the Schuurman Schimmel–Van Outeren Foundation, the Dr. Hendrik Muller's Vaderlandsch Fonds Foundation, and the Karel Frederik Foundation to S.H.S., by Swiss National Science Foundation Grant 55662.98 to J.-P.M., and by a U.S. Department of Agriculture grant to X.D.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.009159.
References
- Aoyama, T., and Chua, N.H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11, 605–612. [DOI] [PubMed] [Google Scholar]
- Aravind, L., and Koonin, E.V. (1999). Fold prediction and evolutionary analysis of the POZ domain: Structural and evolutionary relationship with the potassium channel tetramerization domain. J. Mol. Biol. 285, 1353–1361. [DOI] [PubMed] [Google Scholar]
- Baldwin, A.S. (1996). The NF-κB and IκB proteins: New discoveries and insights. Annu. Rev. Immunol. 14, 649–683. [DOI] [PubMed] [Google Scholar]
- Bell, E., Creelman, R.A., and Mullet, J.E. (1995). A chloroplast lipoxygenase is required for wound-induced accumulation of jasmonic acid in Arabidopsis. Proc. Natl. Acad. Sci. USA 92, 8675–8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, S., Bell, E., Sadka, A., and Mullet, J.E. (1995). Arabidopsis thaliana Atvsp is homologous to soybean VspA and VspB, genes encoding vegetative storage protein acid phosphatases, and is regulated similarly by methyl jasmonate, wounding, sugars, light and phosphate. Plant Mol. Biol. 27, 933–942. [DOI] [PubMed] [Google Scholar]
- Bork, P. (1993). Hundreds of ankyrin-like repeats in functionally diverse proteins: Mobile modules that cross phyla horizontally? Proteins 17, 363–374. [DOI] [PubMed] [Google Scholar]
- Cao, H., Bowling, S.A., Gordon, A.S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Glazebrook, J., Clarke, J.D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. [DOI] [PubMed] [Google Scholar]
- Cao, H., Li, X., and Dong, X. (1998). Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc. Natl. Acad. Sci. USA 95, 6531–6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney, T.P., Friedrich, L., and Ryals, J.A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA 92, 6602–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney, T.P., Uknes, S., Vernooij, B., Friedrich, L., Weymann, K., Negrotto, D., Gaffney, T., Gur-Rella, M., Kessmann, H., Ward, E., and Ryals, J. (1994). A central role of salicylic acid in plant disease resistance. Science 266, 1247–1250. [DOI] [PubMed] [Google Scholar]
- Després, C., DeLong, C., Glaze, S., Liu, E., and Fobert, P.R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12, 279–290. [PMC free article] [PubMed] [Google Scholar]
- Doares, S.H., Narváez-Vásquez, J., Conconi, A., and Ryan, C.A. (1995). Salicylic acid inhibits synthesis of proteinase inhibitors in tomato leaves induced by systemin and jasmonic acid. Plant Physiol. 108, 1741–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty, H.M., Selvendran, R.R., and Bowles, D.J. (1988). The wound response of tomato plants can be inhibited by aspirin and related hydroxy-benzoic acids. Physiol. Mol. Plant Pathol. 33, 377–384. [Google Scholar]
- Fan, W., and Dong, X. (2002). In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid–mediated gene activation in Arabidopsis. Plant Cell 14, 1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felton, G.W., and Korth, K.L. (2000). Trade-offs between pathogen and herbivore resistance. Curr. Opin. Plant Biol. 3, 309–314. [DOI] [PubMed] [Google Scholar]
- Feys, B.J., and Parker, J.E. (2000). Interplay of signaling pathways in plant disease resistance. Trends Genet. 16, 449–455. [DOI] [PubMed] [Google Scholar]
- Gaffney, T., Friedrich, L., Vernooij, B., Negrotto, D., Nye, G., Uknes, S., Ward, E., Kessmann, H., and Ryals, J. (1993). Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261, 754–756. [DOI] [PubMed] [Google Scholar]
- Gallois, C., Habib, A., Tao, J.C., Moulin, S., Maclouf, J., Mallat, A., and Lotersztajn, S. (1998). Role of NF-κB in the antiproliferative effect of endothelin-1 and tumor necrosis factor-α in human hepatic stellate cells: Involvement of cyclooxygenase-2. J. Biol. Chem. 273, 23183–23190. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (2001). Genes controlling expression of defense re-sponses in Arabidopsis: 2001 status. Curr. Opin. Plant Biol. 4, 301–308. [DOI] [PubMed] [Google Scholar]
- Gupta, V., Willits, M.G., and Glazebrook, J. (2000). Arabidopsis thaliana EDS4 contributes to salicylic acid (SA)-dependent expression of defense responses: Evidence for inhibition of jasmonic acid signaling by SA. Mol. Plant-Microbe Interact. 13, 503–511. [DOI] [PubMed] [Google Scholar]
- Harms, K., Ramirez, I., and Peña-Cortés, H. (1998). Inhibition of wound-induced accumulation of allene oxide synthase transcripts in flax leaves by aspirin and salicylic acid. Plant Physiol. 118, 1057–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatada, E.N., Krappmann, D., and Scheidereit, C. (2000). NF-κB and the innate immune response. Curr. Opin. Immunol. 12, 52–58. [DOI] [PubMed] [Google Scholar]
- Howe, G.A., Lightner, J., Browse, J., and Ryan, C.A. (1996). An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell 8, 2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkema, M., Fan, W., and Dong, X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12, 2339–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp, E., and Ghosh, S. (1994). Inhibition of NF-κB by sodium salicylate and aspirin. Science 265, 956–959. [DOI] [PubMed] [Google Scholar]
- Kunkel, B.N., Bent, A.F., Dahlbeck, D., Innes, R.W., and Staskawicz, B.J. (1993). RPS2, an Arabidopsis disease resistance locus specifying recognition of Pseudomonas syringae strains expressing the avirulence gene AvrRpt2. Plant Cell 5, 865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudert, D., and Weiler, E.W. (1998). Allene oxide synthase: A major control point in Arabidopsis thaliana octadecanoid signalling. Plant J. 15, 675–684. [DOI] [PubMed] [Google Scholar]
- Lebel, E., Heifetz, P., Thorne, L., Uknes, S., Ryals, J., and Ward, E. (1998). Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J. 16, 223–233. [DOI] [PubMed] [Google Scholar]
- Malamy, J., Carr, J.P., Klessig, D.F., and Raskin, I. (1990). Salicylic acid: A likely endogenous signal in the resistance response of tobacco to viral infection. Science 250, 1002–1004. [DOI] [PubMed] [Google Scholar]
- Manners, J.M., Penninckx, I.A.M.A., Vermaere, K., Kazan, K., Brown, R.L., Morgan, A., Maclean, D.J., Curtis, M.D., Cammue, B.P.A., and Broekaert, W.F. (1998). The promoter of the plant defensin gene PDF1.2 from Arabidopsis is systemically activated by fungal pathogens and responds to methyl jasmonate but not to salicylic acid. Plant Mol. Biol. 38, 1071–1080. [DOI] [PubMed] [Google Scholar]
- McConn, J., Creelman, R.A., Bell, E., Mullet, J.E., and Browse, J. (1997). Jasmonate is essential for insect defense in Arabidopsis. Proc. Natl. Acad. Sci. USA 94, 5473–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Métraux, J.-P., Signer, H., Ryals, J., Ward, E., Wyss-Benz, M., Gaudin, J., Raschdorf, K., Schmid, E., Blum, W., and Inverardi, B. (1990). Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science 250, 1004–1006. [DOI] [PubMed] [Google Scholar]
- Meuwly, P., and Métraux, J.-P. (1993). Ortho-anisic acid as internal standard for the simultaneous quantitation of salicylic acid and its putative biosynthetic precursors in cucumber leaves. Anal. Biochem. 214, 500–505. [DOI] [PubMed] [Google Scholar]
- Mueller, M.J., and Brodschelm, W. (1994). Quantification of jasmonic acid by capillary gas chromatography-negative chemical-ionization mass spectrometry. Anal. Biochem. 218, 425–435. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Nawrath, C., and Métraux, J.-P. (1999). Salicylic acid induction–deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11, 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton, R., Kuitert, L.M.E., Bergmann, M., Adcock, I.M., and Barnes, P.J. (1997). Evidence for involvement of NF-κB in the transcriptional control of COX-2 gene expression by IL-1β. Biochem. Biophys. Res. Commun. 237, 28–32. [DOI] [PubMed] [Google Scholar]
- Niggeweg, R., Thurow, C., Kegler, C., and Gatz, C. (2000). Tobacco transcription factor TGA2.2 is the main component of as-1-binding factor ASF-1 and is involved in salicylic acid- and auxin-inducible expression of as-1-containing target promoters. J. Biol. Chem. 275, 19897–19905. [DOI] [PubMed] [Google Scholar]
- Pan, Z.Q., Camara, B., Gardner, H.W., and Backhaus, R.A. (1998). Aspirin inhibition and acetylation of the plant cytochrome P450, allene oxide synthase, resembles that of animal prostaglandin endoperoxide H synthase. J. Biol. Chem. 273, 18139–18145. [DOI] [PubMed] [Google Scholar]
- Peña-Cortés, H., Albrecht, T., Prat, S., Weiler, E.W., and Willmitzer, L. (1993). Aspirin prevents wound-induced gene expression in tomato leaves by blocking jasmonic acid biosynthesis. Planta 191, 123–128. [Google Scholar]
- Penninckx, I.A.M.A., Eggermont, K., Terras, F.R.G., Thomma, B.P.H.J., De Samblanx, G.W., Buchala, A., Métraux, J.-P., Manners, J.M., and Broekaert, W.F. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid–independent pathway. Plant Cell 8, 2309–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierpoint, W.S. (1997). The natural history of salicylic acid: Plant product and mammalian medicine. Interdiscip. Sci. Rev. 22, 45–52. [Google Scholar]
- Pieterse, C.M.J., Ton, J., and Van Loon, L.C. (2001). Cross-talk between plant defence signalling pathways: Boost or burden? AgBiotechNet 3, ABN 068.
- Pieterse, C.M.J., and Van Loon, L.C. (1999). Salicylic acid-independent plant defence pathways. Trends Plant Sci. 4, 52–58. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M.J., Van Wees, S.C.M., Van Pelt, J.A., Knoester, M., Laan, R., Gerrits, H., Weisbeek, P.J., and Van Loon, L.C. (1998). A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10, 1571–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontier, D., Miao, Z.H., and Lam, E. (2001). Trans-dominant suppression of plant TGA factors reveals their negative and positive roles in plant defense responses. Plant J. 27, 529–538. [DOI] [PubMed] [Google Scholar]
- Reymond, P., and Farmer, E.E. (1998). Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1, 404–411. [DOI] [PubMed] [Google Scholar]
- Rouster, J., Leah, R., Mundy, J., and Cameron-Mills, V. (1997). Identification of a methyl jasmonate-responsive region in the promoter of a lipoxygenase 1 gene expressed in barley grain. Plant J. 11, 513–523. [DOI] [PubMed] [Google Scholar]
- Ryals, J., Weymann, K., Lawton, K., Friedrich, L., Ellis, D., Steiner, H.Y., Johnson, J., Delaney, T.P., Jesse, T., Vos, P., and Uknes, S. (1997). The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor IκB. Plant Cell 9, 425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals, J.A., Neuenschwander, U.H., Willits, M.G., Molina, A., Steiner, H.-Y., and Hunt, M.D. (1996). Systemic acquired resistance. Plant Cell 8, 1808–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, J., Tsui, F., and Klessig, D.F. (1997). Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant-Microbe Interact. 10, 69–78. [DOI] [PubMed] [Google Scholar]
- Straus, D.S., and Glass, C.K. (2001). Cyclopentenone prostaglandins: New insights on biological activities and cellular targets. Med. Res. Rev. 21, 185–210. [DOI] [PubMed] [Google Scholar]
- Subramaniam, R., Desveaux, D., Spickler, C., Michnick, S.W., and Brisson, N. (2001). Direct visualization of protein interactions in plant cells. Nat. Biotechnol. 19, 769–772. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P.H.J., Penninckx, I.A.M.A., Cammue, B.P.A., and Broekaert, W.F. (2001). The complexity of disease signaling in Arabidopsis. Curr. Opin. Immunol. 13, 63–68. [DOI] [PubMed] [Google Scholar]
- Van der Ouderaa, F.J., Buytenhek, M., Nugteren, D.H., and Van Dorp, D.A. (1980). Acetylation of prostaglandin endoperoxide synthase with acetylsalicylic acid. Eur. J. Biochem. 109, 1–8. [DOI] [PubMed] [Google Scholar]
- Van Loon, L.C. (2000). Systemic induced resistance. In Mechanisms of Resistance to Plant Diseases, A.J. Slusarenko, R.S.S. Fraser, and L.C. Van Loon, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 521–574.
- Van Loon, L.C., and Van Strien, E.A. (1999). The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 55, 85–97. [Google Scholar]
- Van Wees, S.C.M., De Swart, E.A.M., Van Pelt, J.A., Van Loon, L.C., and Pieterse, C.M.J. (2000). Enhancement of induced disease resistance by simultaneous activation of salicylate- and jasmonate-dependent defense pathways in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97, 8711–8716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, E.R., Uknes, S.J., Williams, S.C., Dincher, S.S., Wiederhold, D.L., Alexander, D.C., Ahl-Goy, P., Métraux, J.-P., and Ryals, J.A. (1991). Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3, 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth, M.C., Dewdney, J., Wu, G., and Ausubel, F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565. [DOI] [PubMed] [Google Scholar]
- Xiang, C.B., Miao, Z.H., and Lam, E. (1996). Coordinated activation of as-1-type elements and a tobacco glutathione S-transferase gene by auxins, salicylic acid, methyl jasmonate and hydrogen peroxide. Plant Mol. Biol. 32, 415–426. [DOI] [PubMed] [Google Scholar]
- Yin, M.J., Yamamoto, Y., and Gaynor, R.B. (1998). The anti-inflammatory agents aspirin and salicylate inhibit the activity of IκB kinase-β. Nature 396, 77–80. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Fan, W., Kinkema, M., Li, X., and Dong, X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. USA 96, 6523–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J.M., Trifa, Y., Silva, H., Pontier, D., Lam, E., Shah, J., and Klessig, D.F. (2000). NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol. Plant-Microbe Interact. 13, 191–202. [DOI] [PubMed] [Google Scholar]