Abstract
Gametophytic self-incompatibility in Rosaceae, Solanaceae, and Scrophulariaceae is controlled by the S locus, which consists of an S-RNase gene and an unidentified “pollen S” gene. An ∼70-kb segment of the S locus of the rosaceous species almond, the S haplotype–specific region containing the S-RNase gene, was sequenced completely. This region was found to contain two pollen-expressed F-box genes that are likely candidates for pollen S genes. One of them, named SFB (S haplotype–specific F-box protein), was expressed specifically in pollen and showed a high level of S haplotype–specific sequence polymorphism, comparable to that of the S-RNases. The other is unlikely to determine the S specificity of pollen because it showed little allelic sequence polymorphism and was expressed also in pistil. Three other S haplotypes were cloned, and the pollen-expressed genes were physically mapped. In all four cases, SFBs were linked physically to the S-RNase genes and were located at the S haplotype–specific region, where recombination is believed to be suppressed, suggesting that the two genes are inherited as a unit. These features are consistent with the hypothesis that SFB is the pollen S gene. This hypothesis predicts the involvement of the ubiquitin/26S proteasome proteolytic pathway in the RNase-based gametophytic self-incompatibility system.
INTRODUCTION
Self-incompatibility (SI) in flowering plants is a genetic system that prevents self-fertilization by enabling the pistil to reject pollen from genetically related individuals, thus promoting outcrossing. Genetic studies have shown that most SI systems are controlled by a single multiallele locus called the S locus. When an S allele of pollen matches that of the pistil, the pollen is recognized as “self” and rejected by the pistil (de Nettancourt, 2001). However, recent studies on pistil- or pollen-specific self-compatible mutants (Thompson et al., 1991; Sassa et al., 1997; Golz et al., 1999) and transformation experiments with solanaceous species (Lee et al., 1994; Murfett et al., 1994; Dodds et al., 1999) demonstrate that SI phenotypes of pistil and pollen are determined by different genes, called pistil S and pollen S genes, respectively. Based on the findings that the S locus is a multigene complex, the term “haplotype” has been adopted to denote variants of the locus, and the term “allele” is used to denote variants of a given polymorphic gene at the S locus (McCubbin and Kao, 2000).
The families Rosaceae, Solanaceae, and Scrophulariaceae display gametophytic self-incompatibility (GSI) and share the same pistil S gene product, called the S-RNase (for review, see McCubbin and Kao, 2000). S-RNase is a basic glycoprotein with RNase activity (McClure et al., 1989). RNase activity of S-RNase is required for the pistil to reject self-pollen (Huang et al., 1994; Royo et al., 1994; McCubbin et al., 1997). The rRNA of incompatible pollen of Nicotiana alata is degraded in the style, consistent with a model postulating that the S-RNases act as intracellular cytotoxins (McClure et al., 1990). On the other hand, the pollen S gene of the RNase-based GSI remains to be identified. Circumstantial evidence suggests two conceivable models for the action of pollen S product: the gatekeeper model and the inhibitor model (Thompson and Kirch, 1992; McCubbin and Kao, 2000). In the gatekeeper model, the pollen S product is assumed to be a gatekeeper located at the plasma membrane of the pollen tube. The gatekeeper interacts specifically with cognate S-RNase, assisting in its uptake into the pollen cytoplasm, leading to the degradation of pollen RNA and the arrest of self-pollen tube elongation. In the inhibitor model, all S-RNases enter the pollen tube regardless of their S haplotypes; however, the pollen S product inhibits the activity of all S-RNases except for the cognate S-RNase. As a consequence, the cognate S-RNase remains active in the pollen tube, leading to the degradation of self-pollen RNA. Golz et al. (1999)(2001) produced pollen-part self-compatible mutants of N. alata by incompatible pollination with irradiated pollen and found that a deletion mutant for the pollen S gene could not be recovered by this screen. This finding suggests that the pollen S product is essential for pollen tube elongation, which is consistent with the inhibitor model but in conflict with the gatekeeper model. Immunocytological experiments showing the entry of the S-RNase into the compatible pollen tube support the inhibitor model (Luu et al., 2000).
The identification and characterization of the pollen S gene is essential to understanding the molecular mechanisms of the RNase-based GSI. The pollen S gene is expected to have the following features: first, tight linkage to the S-RNase gene; second, a high level of S haplotype–specific sequence polymorphism; third, pollen-specific expression. The tight genetic linkage between the pollen S gene and the S-RNase gene suggested that chromosome walking would be one of the feasible approaches for the identification of the pollen S gene. However, conventional chromosome walking in Solanaceae is difficult because the solanaceous S locus is located close to the centromere, a region abundant with repetitive sequences (Coleman and Kao, 1992; Matton et al., 1995; Bernacchi and Tanksley, 1997; Entani et al., 1999). McCubbin et al. (2000) screened a BAC library of the Petunia inflata genome with 13 molecular markers linked to the S-RNase gene and walked an S locus of Petunia from multiple starting points. Although they obtained 51 BAC clones spanning a >2-Mb region around the S locus region, they did not construct a contig encompassing the region that encodes S specificity, which is delimited by recombination breakpoints. Therefore, it is not clear whether the genomic clones contain the pollen S gene (McCubbin et al., 2000).
We previously conducted chromosome walking on the Sc haplotype of almond, which belongs to the Rosaceae, and constructed the contigs covering an ∼200-kb region around the Sc-RNase gene (Ushijima et al., 2001). Genomic DNA gel blot analyses revealed that the nucleotide sequence of the ∼70-kb region, which is defined by the two boundary markers NP79R and NP182F and contains the Sc-RNase gene, was highly diverged and specific to each S haplotype. The regions outside of the boundary markers were relatively conserved among different S haplotypes. The structural heteromorphism is the distinctive feature of loci consisting of coadapted gene complexes in different genetic systems, including sporophytic SI of Brassicaceae (Ferris and Goodenough, 1994; Brown and Casselton, 2001; Kusaba et al., 2001). The heteromorphism of the region containing the S specificity genes is believed to maintain the tight association of the pistil S gene and the pollen S gene, suggesting that the pollen S gene of almond is located in the S haplotype–specific region (Ushijima et al., 2001).
Here, we sequenced the ∼70-kb Sc haplotype–specific region using a shotgun strategy. Based on the prediction of open reading frames (ORFs), two pollen-expressed genes were isolated. One of the two genes, named SFB (S haplotype–specific F-box protein), showed a high level of allelic polymorphism and pollen-specific expression. Comparative analysis of the four S haplotypes showed that all SFB alleles are located within ∼30 kb of the S-RNase genes. Distances between genes and markers are highly variable among the S haplotypes, showing the heteromorphism of the region, which might ensure the tight association of SFB and the S-RNase gene. These features of SFB are consistent with those of the expected pollen S gene. SFB encodes an F-box protein that is a component of a class of ubiquitin ligase (SCF complex) (Deshaies, 1999), raising the possibility that the ubiquitin/26S proteasome proteolytic system plays a central role in the discrimination of self/nonself pollen in the RNase-based GSI.
RESULTS
Sequence Analysis of the ∼70-kb Sc Haplotype–Specific Region
We previously predicted the boundaries of the almond S locus and suggested that the pollen Sc gene might be present in an ∼70-kb area of the Sc haplotype–specific region that also contains the Sc-RNase gene (Ushijima et al., 2001). To identify the pollen S gene, the Sc haplotype–specific region was sequenced completely using the shotgun strategy. Three genomic clones covering the region, p79SCU (∼7 kb), pPdC55 (∼35 kb), and pPdC182 (∼36 kb), were used to construct the shotgun library (Figure 1A). A total of ∼1000 sequences, which included PCR products filling gaps between the shotgun contigs, were assem-bled and gave a nucleotide sequence of 71,953 bp. The ∼70-kb sequence was annotated by RiceGAAS (Rice Genome Automated Annotation System [Sakata et al., 2002]; http://ricegaas.dna.affrc.go.jp/index.html), which automatically analyzes large sequences using several programs for the prediction and analysis of protein-coding sequences: BLAST (Basic Local Alignment Search Tool [Altschul et al., 1990]), AutoPredLTR (Sakata et al., 2002), and GENSCAN (Burge and Karlin, 1997).
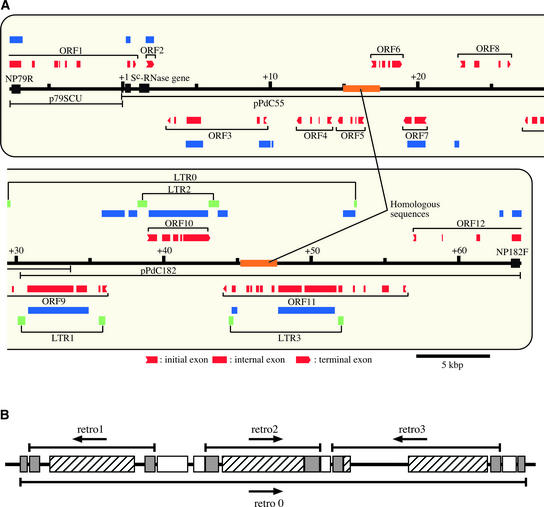
Figure 1.
Sequence Analyses of the Sc Haplotype–Specific Region.
(A) Scheme of the Sc haplotype–specific region. The nucleotide sequence of the ∼70-kb Sc haplotype–specific region was characterized by GENSCAN, BLASTX, and AutoPredLTR search of RiceGAAS (Sakata et al., 2002). The A nucleotide of the putative initiation codon (ATG) of the Sc-RNase gene (Ushijima et al., 1998) is positioned as +1. Exons of the Sc-RNase gene and molecular markers NP79R and NP182F, which define the boundaries of the almond S locus (Ushijima et al., 2001), are represented by black boxes. Red, blue, and green boxes represent the results of GENSCAN, BLAST, and AutoPredLTR searches, respectively. Each pair of LTRs is linked by lines.
(B) Scheme of the retrotransposon-like sequence-rich region (+29 to +53 kb). Gray boxes represent LTR sequences. Hatched boxes represent the region encoding the polyprotein of retro1, retro2, and retro3. White boxes represent the region for the retro0 polyprotein, into which retro1, retro2, and retro3 were inserted.
GENSCAN identified 12 ORFs (ORF1 to ORF12) in the sequenced region (Figure 1A). By this prediction, the three exons of the Sc-RNase gene were misidentified as different genes, ORF1 and ORF2 (Table 1). AutoPredLTR search, which predicted a long terminal repeat (LTR) sequence, showed that the ∼70-kb region contained four pairs of LTRs (Figure 1A). Sequences between the LTRs were found to be similar to retrotransposon-like sequences and were designated retro0 to retro3 (Figure 1B, Table 1). All four retrotransposon-like sequences included several mutations (insertion or deletion), causing their putative polyproteins to be truncated. Retro0 was found to be disrupted by the insertion of retro1 to retro3 (Figure 1B), implying that retro0 was first inserted into the Sc haplotype. Retro3 also was disrupted by the insertion of a 2.5-kb DNA sequence. The inserted sequence showed high similarity to the region at +15 to +18 kb (Figure 1A) but displayed no significant homology with sequences in the public databases.
Table 1.
Summary of Homology Search (BLASTX) Results for the ORFs Predicted by GENSCAN
| Expression
|
||||
|---|---|---|---|---|
| ORF | Homolog (Accession No.) | Pollen | Style | Description |
| ORF1 (1st exon) | Antirrhinum SLF-S2 (AJ297974) | Yes | Yes | PdSLF (Prunus dulcis S locus F-box protein) |
| ORF1 (7th exon) | Almond Sc-RNase (AB011470) | No | Yes | N-terminal region of the Sc-RNase |
| ORF2 | Almond Sc-RNase (AB011470) | No | Yes | C-terminal region of the Sc-RNase |
| ORF3 (1st and 2nd exons) | Arabidopsis putative protein (AL138652) | No | ||
| ORF3 (3rd exon) | Arabidopsis putative F-box protein (AC018907) | Yes | No | SFB (S haplotype–specific F-box protein) |
| ORF4 | Unknown | No | ||
| ORF5 | Unknown | No | ||
| ORF6 | Unknown | No | ||
| ORF7 | Oryza sativa putative protein (AC084763) | No | ||
| ORF8 | Unknown | No | ||
| ORF9 | Zea mays retrotransposon (AF082133) | Designated retro1 in Figure 1 | ||
| ORF10 | Nicotiana retrotransposon (CAA32025) | Designated retro2 in Figure 1 | ||
| ORF11 | Arabidopsis retrotransposon (AC006248) | Designated retro3 in Figure 1 | ||
| ORF12 | Arabidopsis Ser/Thr protein phosphatase (U80922) | No | ||
cDNA Cloning of the Pollen-Expressed Genes Located at the Sc Haplotype
To clone the pollen-expressed genes at the sequenced S locus region, we designed 17 forward and 11 reverse primers based on the prediction of ORFs. The primers were used for rapid amplification of cDNA ends (Frohman et al., 1988) with pollen cDNA. Pollen from almond cv Nonpareil (ScSd) was used, because an Sc homozygous cultivar is not available. For most ORFs, sequences of isolated cDNA clones were not identical to, but were similar to, those of the corresponding ORFs, suggesting that they constitute gene families and that those ORFs are not expressed in pollen. However, sequences of two of the cDNAs completely matched the corresponding ORF sequences (ORF1 and ORF3). The first exon of the predicted ORF1 was isolated as a pollen-expressed gene and is called ORF1 hereafter.
ORF1 was located 6.7 kb upstream of the Sc-RNase gene (Figure 1A). The transcriptional orientations of ORF1 and the Sc-RNase gene were the same. The deduced amino acid sequence of ORF1 showed 29.3% homology with that of SLF (S locus F-box) of Antirrhinum (Figure 2A, Table 1). SLF is located at the Antirrhinum S locus region and encodes a protein with an F-box motif (Lai et al., 2002). We named ORF1 PdSLF for S locus F-box of Prunus dulcis. PdSLF of the Sc haplotype, designated PdSLFc, contained the sequence of the cosmid end probe NP79R, which defined the boundary of the S haplotype–specific region (Ushijima et al., 2001). NP79R sequence has been shown to be conserved in different S haplotypes of almond (Ushijima et al., 2001). PdSLF of the Sd haplotype, designated PdSLFd, was amplified by PCR from the cosmid clone of the Sd haplotype. The deduced amino acid sequences of PdSLFc and PdSLFd were highly similar to each other (95.1% identity; Figure 2B), as were AhSLF-S2 and AhSLF-S2L of Antirrhinum (97.9% identity; Lai et al., 2002), suggesting that they are unlikely to control the S specificity of pollen.
Figure 2.
Amino Acid Sequence Comparisons between SLFs.
Deduced amino acid sequences were compared between PdSLFc and AhSLF-S2 (A) and between PdSLFc and PdSLFd (B). Sites that are conserved or that have only conservative replacements (amino acid groups defined by Dayhoff et al. [1979]: C, STAPG, MILV, HRK, NDEQ, and FYW) are marked with asterisks or dots, respectively.
The ORF3 cDNA also encoded a protein with an F-box motif (Figure 3). ORF3 and the Sc-RNase gene were located in inverse orientation and separated by ∼1.6 kb (Figure 1). ORF3 contained one intron at the 5′ untranslated region (Figure 3). Except for the N-terminal region containing the F-box motif sequence, ORF3 showed no significant homology with known proteins in the public databases. ORF3 has no known localization signal, suggesting that it is a cytoplasmic protein. Genomic DNA gel blot analysis with the ORF3 probe gave an Sc haplotype–specific signal in all cultivars that contained the Sc-RNase gene (Figure 4A). Based on this result, ORF3 was designated SFB (S haplotype–specific F-box protein). Phylogenetic analysis showed that SFB is a member of the A3 subfamily in the plant F-box superfamily defined by Gagne et al. (2002) (data not shown).
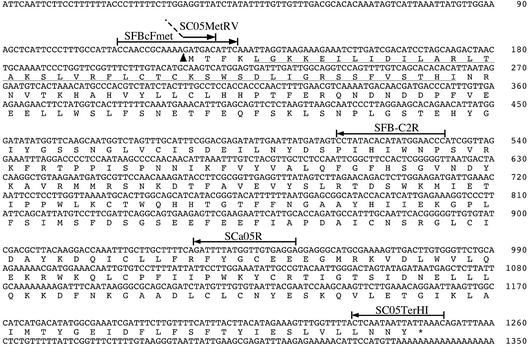
Figure 3.
Nucleotide and Deduced Amino Acid Sequences of the cDNA for SFBc (ORF3).
The deduced amino acid sequence is shown under the nucleotide sequence of the cDNA. The arrowhead represents the position into which an 85-bp intron sequence was inserted. The amino acid sequence of the F-box motif is underlined. Arrows represent the positions and directions of primers that were used to amplify the alleles (see Figure 5) or to characterize organ-specific expression patterns (see Figure 6). The 5′ region of primer SC05MetRV (5′-TGATATCATTTTCTACAGGATGAC-3′) corresponds to the 3′ end of the intron and is represented by a dotted line. Restriction sites for EcoRV and BamHI were added to the 5′ regions of primers SC05MetRV and SC05TerHI (5′-TGGATCCGTTTAATAATTATTGAG-3′), respectively.
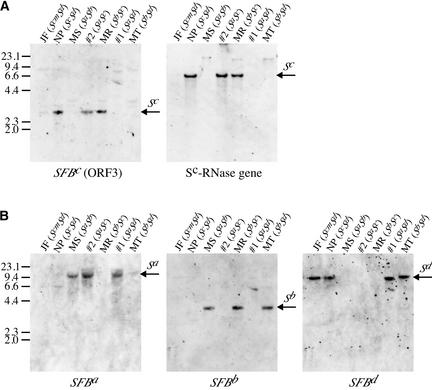
Figure 4.
Genomic DNA Gel Blot Analyses for SFB.
Genomic DNAs from seven almond cultivars were digested with HindIII, separated on 0.8% agarose gels, and transferred to nylon membranes. The blot was probed with the Sc haplotype genes (SFBc and the Sc-RNase gene in [A]) and the homologs of SFBc (SFBa, SFBb, and SFBd in [B]). The arrows Sa, Sb, Sc, and Sd denote the respective S haplotype–specific signals. Abbreviations for almond cultivars are as follows: JF, Jeffries (ScmSd); NP, Nonpareil (ScSd); MS, Mission (SaSb); #2, Sauret No. 2 (SaSc); MR, Merced (SbSc); #1, Sauret No. 1 (SaSd); MT, Monterey (SbSd).
S Haplotype–Specific Sequence Polymorphism of SFB
To clone the alleles of SFB, PCR was conducted with the primer pairs SC05MetRV/SCa05R or SC05MetRV/SFBTerHI (Figure 3). Three clones were isolated from genomic DNA of cv Mission (SaSb), Sauret No. 1 (SaSd), and Monterey (SbSd). Genomic DNA gel blot analyses of the three clones showed the Sa, Sb, and Sd haplotype–specific signals (Figure 4B), suggesting that these SFB clones were derived from the respective S haplotypes.
Because the three clones lacked both ends of the coding region, rapid amplification of cDNA ends was conducted using pollen RNAs from different S haplotypes to obtain full-length cDNA sequences of SFBa, SFBb, and SFBd. Deduced amino acid sequences of the SFB alleles were aligned and compared (Figure 5). The F-box motif was conserved at the N-terminal regions of all SFBs. Sequence identities among SFBs were 68.4 to 76.4% (Table 2), which were comparable to those of almond S-RNases (54.2 to 76.2%) (Ushijima et al., 1998; Tamura et al., 2000). Although variable sites are dispersed in the primary structure of the SFB, two regions at the C terminus are quite variable, as they are in the hypervariable region of the S-RNase (Ioerger et al., 1991; Ushijima et al., 1998).
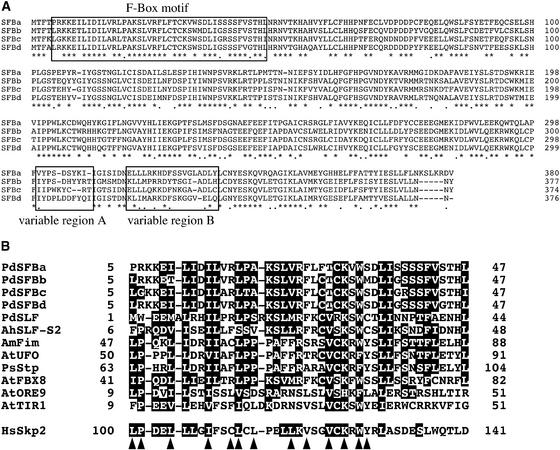
Figure 5.
Alignment of the Amino Acid Sequences of SFBs and the F-Box Motif.
(A) Amino acid sequence alignment of four SFBs. Amino acid sequences of SFBs were aligned using CLUSTAL X (Thompson et al., 1997). The F-box motif and two variable regions are boxed. Sites that are conserved or that have only conservative replacements are marked with asterisks or dots, respectively.
(B) Alignment of amino acid sequences of the F-box motif. Amino acid sequences of F-box motifs from 13 F-box proteins were aligned. Conserved amino acid residues are shown as black boxes. Arrowheads mark the amino acid positions important for the Skp–F-box protein interactions between human Skp1 and Skp2 (Schulman et al., 2000). The species from which each sequence is derived is denoted by the initials of its scientific name (i.e., Pd means Prunus dulcis [almond]). Sequence data included in the alignment are as follows: SLF-S2 Antirrhinum hispanicum; UFO, FBX8, ORE9, and TIR1 of Arabidopsis thaliana; Fim of Antirrhinum majus; Stp of Pisum sativum; and Skp2 of Homo sapiens.
Table 2.
Amino Acid Sequence Identities (%) among SFBs
| SFBb | SFBc | SFBd | |
|---|---|---|---|
| SFBa | 69.0 | 70.1 | 68.4 |
| SFBb | 75.6 | 76.4 | |
| SFBc | 75.8 |
Organ-Specific Expression of the S Locus Genes
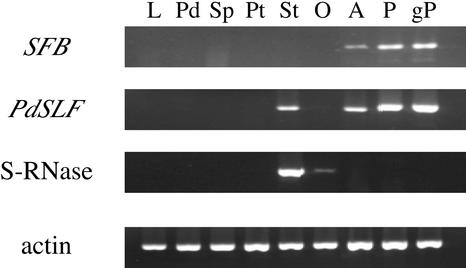
To investigate the expression pattern of SFB and PdSLF, we performed reverse transcriptase–mediated (RT) PCR analyses using RNA from leaf and floral organs of Nonpareil as templates. RT-PCR products for the S-RNase gene and the actin gene, which were used as controls, did not contain genomic DNA–derived PCR products, which are expected to contain introns and thus would be longer than the expected RT-PCR products, suggesting that the templates were not contaminated with genomic DNA. In addition, the forward primer for SFB was designed to include the junction of the two exons to prevent amplification from a genomic DNA template (Figure 3). The cDNA for S-RNase was amplified as expected from the style and at low amounts from the ovary (Figure 6). The cDNA for PdSLF was amplified not only in the male organs (anther, pollen grain, and germinated pollen) but also in the style, where the pollen S gene is not expected to be expressed. On the other hand, SFBc was expressed only in the male organs. Signal intensities of pollen and germinated pollen were higher than that of anther, suggesting that SFB likely is expressed only in pollen.
Figure 6.
Organ-Specific Expression of S Locus Genes.
RNAs were derived from leaf and floral organs of Nonpareil. cDNAs were synthesized from the RNAs and were used for RT-PCR with gene-specific primers. L, leaf; Pd, pedicel; Sp, sepal; Pt, petal; St, style; O, ovary; A, anther; P, pollen; gP, germinated pollen.
Comparison of the Gene Organization of Four S Haplotypes
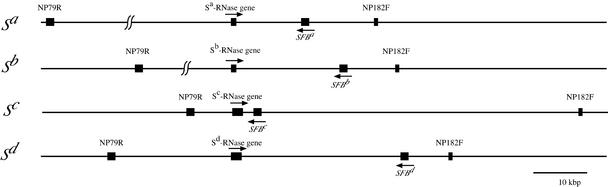
The cosmid or fosmid libraries were constructed from the genomic DNAs of almond cv Nonpareil (ScSd), Sauret No. 1 (SaSc), and Monterey (SbSd). To construct the contigs encompassing the Sa, Sb, and Sd haplotypes, we screened the libraries with probes for SFBs, the S-RNase genes, and the S locus boundary markers NP79R (=PdSLF) and NP182F (Ushijima et al., 2001). The order and orientations of the genes and markers were determined by long PCR. The S-RNase genes and SFBs of all S haplotypes were located in the region between the two boundary markers NP79R and NP182F. The relative order and orientations of the genes and markers were conserved among the four S haplotypes (Figure 7). However, the physical distances between them were significantly different. The physical distance between SFB and the S-RNase gene was ∼1.6 kb in the Sc haplotype but was >10 kb in the other three S haplotypes.
Figure 7.
Gene Organization of Four S Haplotypes of Almond.
Boxes represents exons of the S-RNase genes, SFBs, and the S locus boundary markers NP79R and NP182F (Ushijima et al., 2001). NP79R is part of the PdSLF gene (see text). Arrows denote the orientations of the genes. In the Sa and Sb haplotypes, the physical distance between NP79R and the S-RNase gene was not determined accurately, although it is certain that NP79R links to the S-RNase gene.
DISCUSSION
We previously scanned the S locus region of almond for sequence diversity by DNA gel blot analysis using probes located at varying distances from the S-RNase gene. The results showed that the region between two boundary markers, NP79R and NP182F, is highly diverged among the different S haplotypes, but the regions outside of the two markers are relatively well conserved (Ushijima et al., 2001). In this study, contigs encompassing the two markers were constructed for three additional S haplotypes. Comparative analysis of the contigs showed that the physical distances between genes and markers are quite variable among different S haplotypes, supporting our previous idea that the region is heteromorphic (Ushijima et al., 2001). Sequence analysis of the Sc haplotype region revealed that the region is rich in retrotransposon-like sequences, which may have been causal to the region being heteromorphic. These findings indicate that the almond S locus has been subjected to repeated rearrangements, deletions, and insertions, like the S locus of Brassicaceae (Boyes et al., 1997; Casselman et al., 2000; Kusaba et al., 2001). The structural heteromorphism is a distinctive feature of loci consisting of coadapted gene complexes in different genetic systems (Ferris and Goodenough, 1994; Brown and Casselton, 2001). The heteromorphism of the S locus region is believed to maintain the tight association of the two S specificity genes, the S-RNase gene and the pollen S gene, in the RNase-based GSI and suggests that the pollen S gene is located within the S haplotype–specific region, which is defined by the markers NP79R and NP182F in almond.
Sequence analysis of the Sc haplotype–specific region followed by cDNA cloning revealed that two genes located in the region, PdSLF and SFB, are expressed in pollen. PdSLF corresponded to the S locus boundary marker NP79R and, as expected, showed little sequence polymorphism between alleles. Considering the high degree of sequence diversity of the S-RNases, the pollen S protein is expected to show a high level of sequence diversity for specific interaction with the cognate S-RNase. Therefore, it seems unlikely that PdSLF is the pollen S gene.
In contrast to PdSLF, SFB showed distinctive features of a candidate for the pollen S gene: tight linkage to the S-RNase gene; a high level of sequence polymorphism; and pollen-specific expression. All SFBs were observed to be located at the S haplotype–specific region and to be linked physically (within ∼30 kb) to the S-RNase gene. In addition, heteromorphism at the S haplotype–specific region also could maintain the linkage of the two genes by suppressing recombination at this region, suggesting that SFB and the S-RNase gene are inherited as a unit.
SFB showed a high level of the S haplotype–specific sequence polymorphism, comparable to that observed for the S-RNases of Prunus (Ushijima et al., 1998; Tamura et al., 2000). Amino acid sequence alignment showed that SFB contained two quite variable regions, as did S-RNase (Ioerger et al., 1991; Ushijima et al., 1998). The hypervariable region of S-RNase is exposed on the surface of the folded protein molecule and plays a pivotal role in the recognition of self-pollen (Matton et al., 1997, 1999; Ida et al., 2001; Matsuura et al., 2001). The two variable regions of SFB at the C terminus were found to be hydrophilic enough to be exposed on the surface (data not shown) and may have important recognition functions analogous to those of the S-RNase.
The pollen SI phenotype of Rosaceae and Solanaceae is controlled gametophytically, suggesting that the pollen S gene is expressed only in pollen. RT-PCR analysis revealed that SFB is expressed exclusively in the male reproductive organs and that expression in pollen grains and in germinated pollen is higher than that observed in whole anthers, suggesting that SFB is expressed only in pollen. This result is consistent with our finding that the 1.2-kb promoter region of SFBc directs the expression of a β-glucuronidase reporter gene in pollen but not in anther cells of transgenic tobacco (H. Sassa, K. Ushijima, R. Tao, and H. Hirano, unpublished results). Although we cannot exclude the possibility that the anther cell also expresses a small amount of SFB, it seems unlikely that the sporophytically expressed SFB affects the pollen SI phenotype, because it lacks signal peptide for secretion. In addition, the pollen tube rejection of the RNase-based GSI is a slow reaction that occurs inside the style tissue. This finding is in contrast to the sporophytic SI of Brassica, which is a quick response on the stigma surface, and suggests that exogenous SFB might have little effect on pollen. These findings suggest that SFB functions strictly in a gametophytic manner. All of the features of SFB discussed above are consistent with the hypothesis that SFB controls pollen S specificity in almond, representing the candidate for the pollen S gene of the RNase-based GSI systems. Further analyses for physical interaction between SFB and the S-RNase, as well as functional analyses, will be required to clarify the role of SFB in the RNase-based GSI.
The F-box proteins are known to be a component of a class of ubiquitin ligase, the SCF complex (Deshaies, 1999). In the ubiquitin/26S proteasome pathway of protein degradation, target proteins destined for degradation become modified by covalent attachment of multiple ubiquitins. The polyubiquitinated proteins then are recognized by the 26S proteasome and degraded. The SCF complex catalyzes the final step of ubiquitination of target proteins. The F-box protein serves as a receptor to recruit appropriate target proteins to the core complex for ubiquitination. It associates with an SCF component, Skp1, through the F-box motif at the N terminus, and its C terminus binds specifically to the target (Bai et al., 1996). Our finding of the F-box protein SFB as a candidate for the pollen S gene product of the RNase-based GSI suggests that the likely target of the SCF complex containing the SFB, SCFSFB, is the S-RNase.
The idea that the S-RNase is degraded in pollen tubes through the ubiquitin/26S proteasome pathway seems to be compatible with a proposed model for SI, which hypothesizes that the pollen S gene product is an RNase inhibitor. Despite the apparent difficulty of accommodating the high degrees of structural diversity of the S-RNases, the inhibitor model is supported by recent findings on S haplotype–independent uptake of the S-RNase by the pollen tube (Luu et al., 2000) and the essential role of pollen S products for pollen tube elongation (Golz et al., 2001). As a possible interpretation of the peculiar nature of the pollen S product in the model, it was hypothesized that a pollen S protein contains an RNase inhibitor domain and an S haplotype specificity domain and that it interacts differently with self and nonself S-RNases (McCubbin and Kao, 2000). SFB might function as a component of SCFSFB and ubiquitinate all of the nonself S-RNases for degradation but interact specifically with the self S-RNase to leave it active, leading to the arrest of self pollen tube growth. Recently, it was reported that hnRNP-U functions as a pseudosubstrate that binds to the corresponding F-box protein but is not destined for degradation by the SCF complex (Davis et al., 2002). Based on this example, it is possible that self S-RNase serves as a pseudosubstrate for SCFSFB. The two variable regions located at the C terminus of SFB are implicated in binding with the pseudosubstrate self S-RNase. In plants, the F-box gene family shows a substantial expansion relative to other eukaryotes (Gagne et al., 2002). Identification of almost 700 probable F-box genes in Arabidopsis suggests that the F-box gene family is one of the largest in plants and that the SCF complex–mediated ubiquitin/26S proteasome pathway is a major route for cellular regulation. The RNase-based GSI might represent one of the specialized roles of this highly divergent proteolytic pathway in plants.
METHODS
Plant Materials
Seven cultivars of almond (Prunus dulcis)—Nonpareil (ScSd), Mission (SaSb), Sauret No. 2 (SaSc), Merced (SbSc), Sauret No. 1 (SaSd), Monterey (SbSd), and Jeffries (ScmSd)—were used. Jeffries is a naturally occurring mutant found as a sport on a Nonpareil tree and lacks the Sc haplotype, and the mutated haplotype is designated Scm (Kester et al., 1994; Tao et al., 1997; Ushijima et al., 2001). Pollen was germinated in vitro as described by Lush et al. (1997) with slight modifications.
Shotgun Sequencing
Genomic clones (Figure 1A) were sheared physically by sonication, blunt-ended, and phosphorylated with T4 DNA polymerase and T4 nucleotide kinase (New England Biolabs, Beverly, MA). The resulting fragments were separated on a 1% agarose gel, and the 1.5- to 2.0-kb fragments were cloned into pBluescript II SK− (Stratagene, La Jolla, CA) or pGEM-3Zf(−) (Promega, Madison, WI). The lengths of the inserted fragments were confirmed by PCR. The PCR products were purified by Multiscreen PCR (Millipore, Bedford, MA) and sequenced with an autosequencer (model 4000L; Li-Cor, Lincoln, NE). Sequencing was continued until a minimum of approximately fourfold sequence coverage was achieved for each clone. Sequence alignments were generated using SeqMan II software (DNASTAR, Madison, WI). Gaps between shotgun contigs were amplified and filled by PCR with Pyrobest DNA polymerase (TaKaRa, Otsu, Japan).
Isolation of Nucleic Acids
Freeze-dried leaves were ground into powder using a mortar and pestle. Genomic DNAs were isolated from the ground leaves as described by Ushijima et al. (2001). RNAs were isolated from leaves and floral organs of Nonpareil as described by McClure et al. (1990).
Rapid Amplification of cDNA Ends
Total RNA from the pollen of Nonpareil was used for the synthesis of first-strand cDNA essentially as described for the SMART RACE (rapid amplification of cDNA ends) cDNA Amplification Kit (Clontech, Palo Alto, CA) and by Schmidt and Mueller (1999). The reaction mixture contained 1 × first-strand synthesis buffer (50 mM Tris-HCl, pH 8.3, 75 mM KCl, and 6 mM MgCl2), 1 mM deoxynucleotide triphosphates, 5 mM DTT, 2 mM MnCl2, 1 μM RACE-N (5′-AAGGCTCCGTCGGCATCGATCGCG-CGACTCTTTTTTTTTTTTTTTTT-3′), 1 μM SMART II (5′-AAGCAGTGGTAACAACGCAGAGTACGCGGG-3′), 4 μg of total RNA, and 200 units of SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA) and was incubated at 42°C for 1.5 h. RACE-N and SMART II add the sequences for adapter primers to the cDNA at the 3′ and 5′ ends, respectively. 3′ RACE used the resulting cDNA, ExTaq (TaKaRa), RACE II (5′-AAGGCTCCGTCGGCATCGATC-3′) as the adapter primer, and the gene-specific primers designed from SFBs or the putative open reading frames predicted by the GENSCAN program. 5′ RACE used universal primer mixture (UPM) as the adapter primer instead of RACE II; 1 × UPM contains two primers, 0.1 μM UPM short (5′-CTAATACGACTCACTATA-GGGC-3′) and 0.02 μM UPM long (5′-CTAATACGACTCACTATA-GGGCAAGCAGTGGTAACAACGCAGAGT-3′), and extends the 5′ end of the cDNA for step-out PCR (Matz et al., 1999). If necessary, second-round PCR was conducted with gene-specific primers and adapter primers RACE III (5′-CGGCATCGATCGCGCGACTC-3′) and NUP (5′-AAGCAGTGGTAACAACGCAGAGT-3′) for 3′ and 5′ RACE, respectively.
RACE fragments were subjected to DNA gel blot analyses using the three genomic clones for the Sc haplotype–specific region (Figure 1A) as probes to detect cDNA species with low expression levels. The RACE fragments showing signal(s) were cloned into a plasmid vector. The positive clones for each RACE fragment were selected by colony hybridization with the probes of the genomic clones. For each open reading frame, 6 to 11 independent cDNA clones derived from different primer pairs were subjected to sequence analysis.
Genomic DNA Gel Blot Analysis
Five micrograms of genomic DNAs digested with HindIII was separated and blotted onto a nylon membrane. The membrane was probed with digoxigenin-labeled cDNAs for genes expressed in pollen, washed, and visualized as described by Ushijima et al. (2001).
Reverse Transcriptase–Mediated PCR for Characterization of the Gene Expression Pattern
RNAs from leaves and floral organs of Nonpareil were treated with DNaseI (Invitrogen). Their cDNAs were synthesized by PowerScript (Clontech) with oligo-d(T) primer. The resulting cDNAs were used as templates for PCR amplification with gene-specific primer sets SFBcFmet (5′-CCAACCGCAAAAGATGACATTC-3′) and SFB-C2R (5′-GGG-TTCCATATKTGTATWGG-3′) for SFBc (Figure 3), SCa01F (5′-GATTGG-TGGGGATGTGCTGTAG-3′) and SCa01R (5′-CTTCGGTAACCATAAGAATCTC-3′) for PdSLF, AS1II and Amy-C5R for the S-RNase gene (Tamura et al., 2000), and ActF1 (5′-ATGGTGAGGATATTCAACCC-3′) and ActR1 (5′-CTTCCTGTGGACAATGGATGG-3′) for the actin gene that was used as an internal control. PCR was performed with ExTaq (TaKaRa) using a program of 30 cycles at 94°C for 30 s, 53°C for 30 s, and 72°C for 45 s, an initial denaturing at 94°C for 2.5 min, and a final extension at 72°C for 7 min. PCR products were separated on a 1.5% agarose gel and stained with ethidium bromide.
Construction and Screening of the Cosmid Library
Cosmid or fosmid libraries were constructed from three almond cultivars—Nonpareil (ScSd), Sauret No.1 (SaSc), and Monterey (SbSd)—using the pWEB::TNC Cosmid Cloning Kit or the CopyControl Fosmid Library Production Kit (Epicentre, Madison, WI). The libraries were screened with digoxigenin-labeled probes for the S-RNase gene SFB, NP79R and NP182F, as described by Ushijima et al. (2001). Long PCR with ExTaq or the Expand Long Template PCR System (Roche Diagnostics, Mannheim, Germany) was performed to determine the order and the orientations of the genes on the cosmid/fosmid clones.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Numbers
The accession numbers for the sequences mentioned in this article are as follows: AB081587 (Sc haplotype–specific region), AB092966 (SFBa), AB092967 (SFBb), AB079776 (SFBc), AB081648 (SFBd). Other acces-sion numbers are AJ297974 (SLF-S2 of Antirrhinum hispanicum), NM_102834, AC005825, AF305597, and AF327430 (UFO, FBX8, ORE9, and TIR1 of Arabidopsis thaliana), S71192 (Fim of Antirrhinum majus), AF004843 (Stp of Pisum sativum), and AB050979 (Skp2 of Homo sapiens).
Acknowledgments
We gratefully acknowledge S. Uratsu (University of California, Davis) for collecting and providing plant materials and M. Kusaba (Institute of Radiation Breeding, National Institute of Agrobiological Sciences, Ministry of Agriculture, Forestry, and Fisheries, Japan), for helpful comments. This work was supported by Grant-in-Aid 13660011 for Scientific Research (C) to H.S. and Grant-in-Aid 13460014 for Scientific Research (B) to R.T. from the Japan Society for the Promotion of Science and by grants to A.M.D. from the Almond Board of California.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.009290.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Bai, C., Sen, P., Hofmann, K., Ma, L., Goebl, M., Harper, J.W., and Elledge, S.J. (1996). SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86, 263–274. [DOI] [PubMed] [Google Scholar]
- Bernacchi, D., and Tanksley, S.D. (1997). An interspecific backcross of Lycopersicon esculentum × L. hirsutum: Linkage analysis and a QTL study of sexual compatibility factors and floral traits. Genetics 147, 861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes, D.C., Nasrallah, M.E., Vrebalov, J., and Nasrallah, J.B. (1997). The self-incompatibility (S) haplotypes of Brassica contain highly divergent and rearranged sequences of ancient origin. Plant Cell 9, 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, A.J., and Casselton, L.A. (2001). Mating in mushrooms: Increasing the chances but prolonging the affair. Trends Genet. 17, 393–400. [DOI] [PubMed] [Google Scholar]
- Burge, C., and Karlin, S. (1997). Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268, 78–94. [DOI] [PubMed] [Google Scholar]
- Casselman, A.L., Vrebalov, J., Conner, J.A., Singhal, A., Giovannoni, J., Nasrallah, M.E., and Nasrallah, J.B. (2000). Determining the physical limits of the Brassica S locus by recombinational analysis. Plant Cell 12, 23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman, C.E., and Kao, T.-H. (1992). The flanking regions of two Petunia inflata S alleles are heterogeneous and repetitive sequences. Plant Mol. Biol. 18, 499–511. [DOI] [PubMed] [Google Scholar]
- Davis, M., Hatzubai, A., Andersen, J.S., Ben-Shushan, E., Fisher, G.Z., Yaron, A., Bauskin, A., Mercurio, F., Mann, M., and Ben-Neriah, Y. (2002). Pseudosubstrate regulation of the SCFβ-TrCP ubiquitin ligase by hnRNP-U. Genes Dev. 16, 439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayhoff, M.O., Schwartz, R.M., and Orcutt, B.C. (1979). A model of evolutionary change in proteins. In Atlas of Protein Sequence and Structure, Vol. 5, Suppl. 3, M.O. Dayhoff, ed (Washington, DC: National Biomedical Research Foundation), pp. 345–352.
- de Nettancourt, D. (2001). Incompatibility and Incongruity in Wild and Cultivated Plants. (Berlin: Springer-Verlag).
- Deshaies, R.J. (1999). SCF and cullin/RING H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15, 435–467. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N., Ferguson, C., Clarke, A.E., and Newbigin, E. (1999). Pollen-expressed S-RNases are not involved in self-incompatibility in Lycopersicon peruvianum. Sex. Plant Reprod. 12, 76–87. [Google Scholar]
- Entani, T., Iwano, M., Shiba, H., Takayama, S., Fukui, K., and Isogai, A. (1999). Centromeric localization of an S-RNase gene in Petunia hybrida Vilm. Theor. Appl. Genet. 99, 391–397. [DOI] [PubMed] [Google Scholar]
- Ferris, P.J., and Goodenough, U.W. (1994). The mating-type locus of Chlamydomonas reinhardtii contains highly rearranged DNA se-quences. Cell 76, 1135–1145. [DOI] [PubMed] [Google Scholar]
- Frohman, M.A., Dush, M.K., and Martin, G.R. (1988). Rapid production of full-length cDNAs from rare transcripts: Amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85, 8998–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne, J.M., Downes, B.P., Shiu, S.-H., Durski, A.M., and Vierstra, R.D. (2002). The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 99, 11519–11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golz, J.F., Su, V., Clarke, A.E., and Newbigin, E. (1999). A molecular description of mutations affecting the pollen component of the Nicotiana alata S locus. Genetics 152, 1123–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golz, J.F., Su, V., Oh, H.-Y., Kusaba, M., and Newbigin, E. (2001). Genetic analysis of Nicotiana pollen-part mutants is consistent with the presence of an S-ribonuclease inhibitor at the S locus. Proc. Natl. Acad. Sci. USA 98, 15372–15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S., Lee, H.-S., Karunanandaa, B., and Kao, T.-H. (1994). Ribonuclease activity of Petunia inflata S proteins is essential for rejection of self-pollen. Plant Cell 6, 1021–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ida, K., Norioka, S., Yamamoto, M., Kumasaka, T., Yamashita, E., Newbigin, E., Clarke, A.E., Sakiyama, F., and Sato, M. (2001). The 1.55 Å resolution structure of Nicotiana alata SF11-RNase associated with gametophytic self-incompatibility. J. Mol. Biol. 314, 103–112. [DOI] [PubMed] [Google Scholar]
- Ioerger, T.R., Gohlke, J.R., Xu, B., and Kao, T.-H. (1991). Primary structural features of the self-incompatibility protein in Solanaceae. Sex. Plant Reprod. 4, 81–87. [Google Scholar]
- Kester, D.E., Micke, W.C., and Viveros, M. (1994). A mutation in ‘Nonpareil’ almond conferring unilateral incompatibility. J. Am. Soc. Hortic. Sci. 119, 1289–1292. [Google Scholar]
- Kusaba, M., Dwyer, K., Hendershot, J., Vrebalov, J., Nasrallah, J.B., and Nasrallah, M.E. (2001). Self-Incompatibility in the genus Arabidopsis: Characterization of the S locus in the outcrossing A. lyrata and its autogamous relative A. thaliana. Plant Cell 13, 627–643. [PMC free article] [PubMed] [Google Scholar]
- Lai, Z., Ma, W., Han, B., Liang, L., Zhang, Y., Hong, G., and Xue, Y. (2002). An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Mol. Biol. 50, 29–42. [DOI] [PubMed] [Google Scholar]
- Lee, H.-S., Huang, S., and Kao, T.-H. (1994). S proteins control rejection of incompatible pollen in Petunia inflata. Nature 367, 560–563. [DOI] [PubMed] [Google Scholar]
- Lush, W.M., Opat, A.S., Nie, F., and Clarke, A.E. (1997). An in vitro assay for assessing the effects of growth factors on Nicotiana alata pollen tubes. Sex. Plant Reprod. 10, 351–357. [Google Scholar]
- Luu, D., Qin, X., Morse, D., and Cappadocia, M. (2000). S-RNase uptake by compatible pollen tubes in gametophytic self-incompatibility. Nature 407, 649–651. [DOI] [PubMed] [Google Scholar]
- Matsuura, T., Sakai, H., Unno, M., Ida, K., Sato, M., Sakiyama, F., and Norioka, S. (2001). Crystal structure at 1.5 Å resolution of Pyrus pyrifolia pistil ribonuclease responsible for gametophytic self-incompatibility. J. Biol. Chem. 276, 45261–45269. [DOI] [PubMed] [Google Scholar]
- Matton, D., Maes, O., Laublin, G., Xike, Q., Bertrand, C., Morse, D., and Cappadocia, M. (1997). Hypervariable domains of self-incompatibility RNases mediate allele-specific pollen recognition. Plant Cell 9, 1757–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matton, D.P., Luu, D.T., Xike, Q., Laublin, G., O'Brien, M., Maes, O., Morse, D., and Cappadocia, M. (1999). Production of an S RNase with dual specificity suggests a novel hypothesis for the generation of new S alleles. Plant Cell 11, 2087–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matton, D.P., Mau, S.L., Okamoto, S., Clarke, A.E., and Newbigin, E. (1995). The S-locus of Nicotiana alata: Genomic organization and sequence analysis of two S-RNase alleles. Plant Mol. Biol. 28, 847–858. [DOI] [PubMed] [Google Scholar]
- Matz, M., Shagin, D., Bogdanova, E., Britanova, O., Lukyanov, S., Ditachenco, L., and Chenchik, A. (1999). Amplification of cDNA ends based on template-switching effect and step-out PCR. Nucleic Acids Res. 27, 1558–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure, B.A., Gray, J.E., Anderson, M.A., and Clarke, A.E. (1990). Self-incompatibility in Nicotiana alata involves degradation of pollen rRNA. Nature 347, 757–760. [Google Scholar]
- McClure, B.A., Haring, V., Ebert, P.R., Anderson, M.A., Simpson, R.J., Sakiyama, F., and Clarke, A.E. (1989). Style self-incompatibility gene products of Nicotiana alata are ribonucleases. Nature 342, 955–957. [DOI] [PubMed] [Google Scholar]
- McCubbin, A.G., Chung, Y.-Y., and Kao, T.-H. (1997). A mutant S3 RNase of Petunia inflata lacking RNase activity has an allele-specific dominant negative effect on self-incompatibility interactions. Plant Cell 9, 85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCubbin, A.G., and Kao, T.-H. (2000). Molecular recognition and response in pollen and pistil interactions. Annu. Rev. Cell Dev. Biol. 16, 333–364. [DOI] [PubMed] [Google Scholar]
- McCubbin, A.G., Zuniga, C., and Kao, T.-H. (2000). Construction of a binary bacterial artificial chromosome library of Petunia inflata and the isolation of large genomic fragments linked to the self-incompatibility (S-) locus. Genome 43, 820–826. [PubMed] [Google Scholar]
- Murfett, J., Atherton, T.L., Mou, B., Gasser, C.S., and McClure, B.A. (1994). S-RNase expressed in transgenic Nicotiana causes S-allele-specific pollen rejection. Nature 367, 563–566. [DOI] [PubMed] [Google Scholar]
- Royo, J., Kuntz, C., Kowyama, Y., Anderson, M., and Clarke, A.E. (1994). Loss of a histidine residue at the active site of S-locus ribonuclease is associated with self-compatibility in Lycopersicon peruvianum. Proc. Natl. Acad. Sci. USA 91, 6511–6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata, K., Nagamura, Y., Numa, H., Antonio, B.A., Nagasaki, H., Idonuma, A., Watanabe, W., Shimizu, Y., Horiuchi, I., Matsumoto, T., Sasaki, T., and Higo, K. (2002). RiceGAAS: An automated annotation system and database for rice genome sequence. Nucleic Acids Res. 30, 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassa, H., Hirano, H., Nishino, T., and Koba, T. (1997). Style-specific self-compatible mutation caused by deletion of the S-RNase gene in Japanese pear (Pyrus serotina). Plant J. 12, 223–227. [Google Scholar]
- Schmidt, M.W., and Mueller, M.W. (1999). CapSelect: A highly sensitive method for 5′ CAP-dependent enrichment of full-length cDNA in PCR-mediated analysis of mRNAs. Nucleic Acids Res. 27, e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman, B.A., Carrano, A.C., Jeffrey, P.D., Bowen, Z., Kinnucan, E.R.E., Finnin, M.S., Elledge, S.J., Harper, J.W., Pagano, M., and Pavletich, N.P. (2000). Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408, 381–386. [DOI] [PubMed] [Google Scholar]
- Tamura, M., Ushijima, K., Sassa, H., Hirano, H., Tao, R., Gradziel, T.M., and Dandekar, A.M. (2000). Identification of self-incompatibility genotypes of almond by allele-specific PCR analysis. Theor. Appl. Genet. 101, 344–349. [Google Scholar]
- Tao, R., Yamane, H., Sassa, H., Mori, H., Gradziel, T.M., Dandekar, A.M., and Sugiura, A. (1997). Identification of stylar RNases associated with gametophytic self-incompatibility in almond (Prunus dulcis). Plant Cell Physiol. 38, 304–311. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL X Windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, R.D., and Kirch, H.H. (1992). The S locus of flowering plants: When self-rejection is self-interest. Trends Genet. 8, 381–387. [DOI] [PubMed] [Google Scholar]
- Thompson, R.D., Uhrig, H., Hermsen, J.G.T., Salamini, F., and Kaufmann, H. (1991). Investigation of a self-compatible mutation in Solanum tuberosum clones inhibiting S-allele activity in pollen differentially. Mol. Gen. Genet. 226, 283–288. [DOI] [PubMed] [Google Scholar]
- Ushijima, K., Sassa, H., Kusaba, M., Tao, R., Tamura, M., Gradziel, T.M., Dandekar, A.M., and Hirano, H. (2001). Characterization of the S-locus region of almond (Prunus dulcis): Analysis of a somaclonal mutant and a cosmid contig for an S haplotype. Genetics 158, 379–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima, K., Sassa, H., Tao, R., Yamane, H., Dandekar, A.M., Gradziel, T.M., and Hirano, H. (1998). Cloning and characterization of cDNAs encoding S-RNases from almond (Prunus dulcis): Primary structural features and sequence diversity of the S-RNases in Rosaceae. Mol. Gen. Genet. 260, 261–268. [DOI] [PubMed] [Google Scholar]