Abstract
Urea is the major nitrogen form supplied as fertilizer in agricultural plant production but also an important nitrogen metabolite in plants. We report the cloning and functional characterization of AtDUR3, a high-affinity urea transporter in plants. AtDUR3 contains 14 putative transmembrane-spanning domains and represents an individual member in Arabidopsis that belongs to a superfamily of sodium-solute symporters. Heterologous expression in urea uptake–defective yeast as well as two-electrode voltage clamp and uptake studies using 14C-labeled urea in AtDUR3-expressing oocytes demonstrated that AtDUR3 mediates urea transport. In both heterologous systems, urea transport was stimulated at low pH. In oocytes, inward currents indicated that urea is cotransported with protons. By contrast, a supply of Na+ ions could not stimulate urea transport. Transport of 14C-labeled urea by AtDUR3 in oocytes exhibited saturation kinetics with a Km of ∼3 μM. AtDUR3 was expressed in shoots and roots and upregulated during early germination and under nitrogen deficiency in roots. We propose a role of AtDUR3 in urea uptake by plant cells at low external urea concentrations.
INTRODUCTION
Natural sources of urea arise mostly from the detoxification of nitrogenous compounds by humans and animals, resulting in the synthesis and excretion of urea into the environment. Another main input of urea to the environment derives from the increasing production and application of urea fertilizers. With >50% of the total nitrogen fertilization worldwide, urea has become the dominant nitrogen form for fertilizer application to crop plants (http://www.fertilizer.org/ifa/statistics/STATSIND/). These sources have led to a urea accumulation of ∼10 million tons in the biosphere, which equals ∼1% of the biosphere's protein content.
Despite this important amount in terms of absolute quantity, urea concentrations in soils are rather low as a result of the short half-life of urea, which is degraded rapidly by soil microorganisms (Watson et al., 1994). The hydrolysis of urea to ammonia and carbon dioxide is catalyzed by urease, a Ni-dependent enzyme that occurs in almost all organisms (Gerendás et al., 1999). Because urease is highly stable, the enzyme persists even when microorganisms decay, which leads to a relatively high urea-hydrolyzing activity in most arable soils (Watson et al., 1994). Therefore, most urea-derived nitrogen is believed to enter plant roots in the form of ammonium (Marschner, 1995).
However, physiological evidence for the root uptake of urea before degradation into carbon dioxide and ammonium has been obtained in several independent experiments. For example, when urea was supplied as a nitrogen source to hydroponically grown plants, urea was found in the xylem sap (Hine and Sprent, 1988) or accumulated in leaves of Ni-deficient plants with repressed urease activity (Gerendás et al., 1998). Applying urea as a nitrogen fertilizer directly to leaves caused urea accumulation in leaves and severe leaf tip necrosis, particularly in the presence of a urease inhibitor (Krogmeier et al., 1989), suggesting that urea might enter leaf cells in the intact form. Urea uptake by roots seems to depend on the nitrogen status of the plant, because urea depletion in nutrient solution by nitrogen-sufficient wheat seedlings was lower compared with that by nitrogen-deficient seedlings (Bradley et al., 1989). Although these and related studies presented evidence for urea transport into plant cells, they neither defined the nitrogen form actually passing the membrane nor differentiated in their analyses between urea synthesized internally and that absorbed from the medium.
Wilson and Walker (1988) used independent methods to monitor urea uptake in their model system Chara. Controlling the fate of 14C-labeled urea in short-term uptake studies, they showed that urea uptake was concentration dependent and followed biphasic kinetics, indicating the existence of high- and low-affinity transport systems. In parallel, biphasic uptake kinetics in Chara were confirmed by electrophysiological measurements, in which a sodium-dependent inward current appeared in response to urea (Wilson et al., 1988; Walker et al., 1993). Finally, Wilson et al. (1988) concluded that high-affinity urea uptake in Chara is electrogenic, active, and, because of repressed uptake rates under high nitrogen levels, also under metabolic control.
Using a urea uptake–defective yeast mutant, ElBerry et al. (1993) showed that the uptake of 14C-labeled urea in yeast was restored in the presence of a functional ScDUR3 gene. ScDUR3 expression was regulated in a manner similar to that of other genes in the allantoin pathway (e.g., it was repressed under high nitrogen nutrition). ScDUR3 encodes a hydrophobic protein with 15 putative transmembrane-spanning domains and belongs to a superfamily of sodium-solute symporters (Turk and Wright, 1997; Saier, 2000).
Besides acting as an external nitrogen source, urea is synthesized inside plant cells during Arg degradation via arginase in the Orn (urea) cycle (Polacco and Holland, 1993). Urea accumulates particularly in source leaves of older plants, most likely as a consequence of protein and Arg breakdown for the retranslocation of amino nitrogen (Gerendás et al., 1999). This enhanced nitrogen recycling in senescing tissues also is reflected by an increase in urease and cytosolic Gln synthetase activities, probably as a prerequisite for the generation and refixation of ammonium (Masclaux et al., 2000; Witte et al., 2002). Urea accumulation as a consequence of increased arginase activity also occurs during germination and seedling development, when seed storage proteins are mobilized (Zonia et al., 1995). Because arginases are intramitochondrial, arginase-derived urea cannot diffuse freely to cytoplasmic urease (Polacco and Holland, 1993). Thus, mitochondrial export of urea might be facilitated by transporters that reside in the mitochondrial membrane.
Despite physiological evidence for urea uptake by root and leaf cells and for the intracellular membrane transport of urea, transport pathways for urea are poorly understood at the molecular level. Recently, water channels of the plasma membrane intrinsic protein and tonoplast intrinsic protein subfamilies of aquaporins were shown to permeate small neutral solutes, including urea (Eckert et al., 1999; Gerbeau et al., 1999). Similarly, in mammalian cells, some aquaporins also permeate urea (Echevarria and Ilundain, 1998). However, the physiological significance of urea transport by plant aquaporins remains to be elucidated.
To understand the molecular basis for urea uptake in plants, we searched for genes that mediate urea uptake in other systems. We identified a member of the sodium-solute symporter family, AtDUR3, that complements a yeast Δdur3 deletion mutant. We further expressed AtDUR3 in Xenopus laevis oocytes to characterize the mechanism of urea transport and found that AtDUR3 mediates the proton-dependent, high-affinity transport of urea.
RESULTS
Protonophores Decrease Urea Uptake in Arabidopsis Cell Cultures
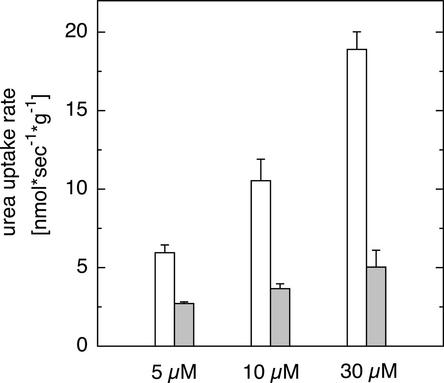
Previous physiological observations indicated that urea might be transported directly across plasma membranes in plant cells. Because the concentration of urea in natural and agriculturally used soils is in the micromolar range, generally <70 μM (Gaudin et al., 1987), urea uptake in Arabidopsis was assayed at 5, 10, and 30 μM urea using 5-day-old Arabidopsis suspension cells (May and Leaver, 1993). Uptake of 14C-labeled urea increased linearly between 0.5 and 5 min at micromolar urea concentrations (data not shown). At pH 5, radioactive tracer accumulated with external application, and uptake was partially blocked by the protonophore carbonyl cyanide m-chlorophenylhydrazone (CCCP) (Figure 1), suggesting that at least one component for urea uptake at micromolar concentrations depends on the proton gradient across the plasma membrane of Arabidopsis cells.
Figure 1.
Inhibition of Urea Uptake by CCCP in Arabidopsis Suspension Cells.
14C-labeled urea was supplied at concentrations of 5, 10, and 30 μM in the absence (open bars) or presence (closed bars) of 100 μM CCCP, and incorporated tracer was assayed after 5 min. Data represent means ± sd, n = 4.
Identification of AtDUR3 as a Urea Transporter in Plants
Urea transporter genes from different unicellular organisms, such as cyanobacteria and yeast, have been identified on the basis of screening and recomplementation of urea uptake–defective mutants (ElBerry et al., 1993; Valladares et al., 2002). Urea transporters from higher eukaryotes have been identified mainly by genetic approaches and heterologous expression in oocytes (Leung et al., 2000; Smith and Rousselet, 2001). Using the BLAST (Basic Local Alignment Search Tool) search algorithm, we searched for ESTs from higher plants that encode proteins homologous with microbial or mammalian urea transporters. We identified a group of plant ESTs (Arabidopsis AV524336, maize BQ164112, rice AU056606, soybean AW164789, barley BF256473, wheat BF202504, and oilseed rape H07563) that showed weak homology with the ScDUR3 gene from yeast (ElBerry et al., 1993). ScDUR3 is a member of the sodium-solute symporter (SSS) gene family, which is widespread in microorganisms, animals, and humans. Members of the SSS family have been described to transport sugars, amino acids, nucleosides, inositols, vitamins, anions, and urea (Reizer et al., 1994; Turk and Wright, 1997; Saier, 2000).
A putative full-length coding sequence of AV524336 from Arabidopsis was deduced from a genomic clone (At5g45380), isolated from a cDNA library (Minet et al., 1992) using gene-specific primers, and cloned into the yeast expression vector pHXT426 carrying a hexose transporter promoter (Wieczorke et al., 1999). The sequence-verified 2428-bp cDNA contained an open reading frame of 2085 bp with an ATG start codon at position 214 and a termination codon at position 2298. An in-frame termination codon 18 bp upstream of the translational initiation codon indicated that the cDNA represented the complete coding region of the respective protein, which was named AtDUR3.
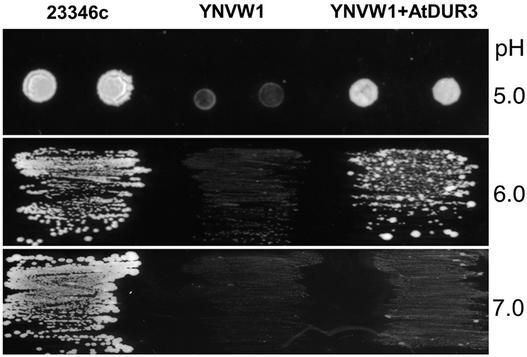
A yeast complementation assay was set up by substituting the ScDUR3 gene in Saccharomyces cerevisiae strain 23346c (Δura3) for a marker gene by PCR-based gene disruption. The resulting mutant, YNVW1 (Δdur3, Δura3), was defective in urea uptake and growth on <5 mM urea as a sole nitrogen source when transformed with the empty vector pHXT426 (Figure 2). Heterologous expression of AtDUR3 enabled the yeast mutant to grow efficiently on 2 mM urea (Figure 2). Growth complementation of the yeast mutant by AtDUR3 depended on medium pH. pHXT426-AtDUR3 transformants attained better growth than the control transformants on 2 mM urea at pH 6.0 and below but showed weak complementation at pH 7.0 (Figure 2). By contrast, wild-type yeast (23346c) also grew better at higher pH, most likely attributable to a different regulation of ScDUR3 gene expression and/or different biochemical properties of the ScDUR3 protein. The pH-dependent growth complementation by AtDUR3 suggested that urea transport is stimulated by external protons.
Figure 2.
Growth Complementation of Yeast by Heterologous Expression of AtDUR3.
The yeast strain YNVW1 (Δdur3, Δura3) was unable to grow on 2 mM urea as a sole nitrogen source. YNVW1 was transformed with the yeast expression vector pHXT426 (Wieczorke et al., 1999) alone or harboring the open reading frame of AtDUR3 and plated on yeast nitrogen base without ammonium and amino acids agar (Difco) adjusted to pH 5, 6, or 7 (because of the softer agar at pH 5, yeast cells were spotted).
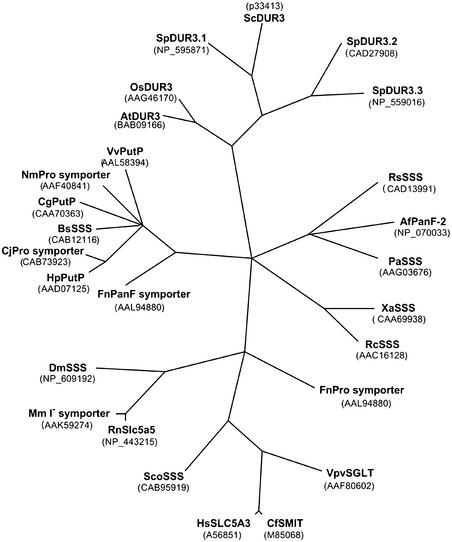
Phylogenetic analysis indicated the closest relationship between DUR3 proteins from plants and yeast (S. cerevisiae and Schizosaccharomyces pombe). These DUR3 proteins form a subfamily that has not been functionally characterized (Figure 3). Among the SSS members from other organisms, urea transport activity has been demonstrated for the low-affinity Na+-Glc cotransporter rbSGLT1, the Na+-iodide cotransporter rNIS, and the Na+-Cl−-γ-aminobutyric acid cotransporter hGAT1 (Leung et al., 2000).
Figure 3.
Phylogenetic Tree of DUR3 and Selected SSS Proteins.
Unrooted, parsimony-based phylogram of AtDUR3 and the 20 most homologous SSS proteins as obtained by BLAST search. The tree was generated using PAUP version 4.0 b5 (D. Swofford, Smithsonian Institution, Washington, DC). Bootstrap values of >50 are indicated (1000 replicates, full heuristic search option, tree bisection reconnection branch swapping with random addition). RbSGLT1, rNIS, and hGAT1, for which urea transport activity has been reported (see Results), are not depicted because of their weak homology with AtDUR3.
Proteins from the SSS family possess between 400 and 700 amino acid residues and are predicted to form 12 to 15 transmembrane-spanning domains (Reizer et al., 1994). Computer-based analysis of the hydrophobicity profile indicated that the AtDUR3 polypeptide represents an integral membrane protein with 14 transmembrane-spanning domains (www.cbs.dtu.dk; Sonnhammer et al., 1998), which are arranged in two stretches of six and an additional stretch of two transmembrane-spanning domains. The N and C termini were predicted to protrude into the extracellular space (Figure 4). No transit peptides for chloroplasts or mitochondrial targeting were indicated. An outside-oriented domain between transmembrane helices 8 and 9 contains a consensus sequence for a P-loop, also designated a “Walker A” motif (Saraste, 1990).
Figure 4.
Amino Acid Sequence Alignment of Members of the DUR3 Subfamily.
Amino acids are given with standard single-letter designations, and dashes indicate gaps. Residues are shown in white letters on black if two or more sequences have identical residues at the aligned positions. Gray boxes above the sequences indicate the positions of 14 potential transmembrane-spanning domains, and the connecting lines indicate potential outside and inside domains as predicted by TMHMM version 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). The indication of a P-loop relates to the consensus sequence A-x(4)-G-K-S designating a Walker A motif. At, Arabidopsis thaliana; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe DUR3.1; Os, Oryza sativa.
AtDUR3 Mediates Urea-Induced Currents
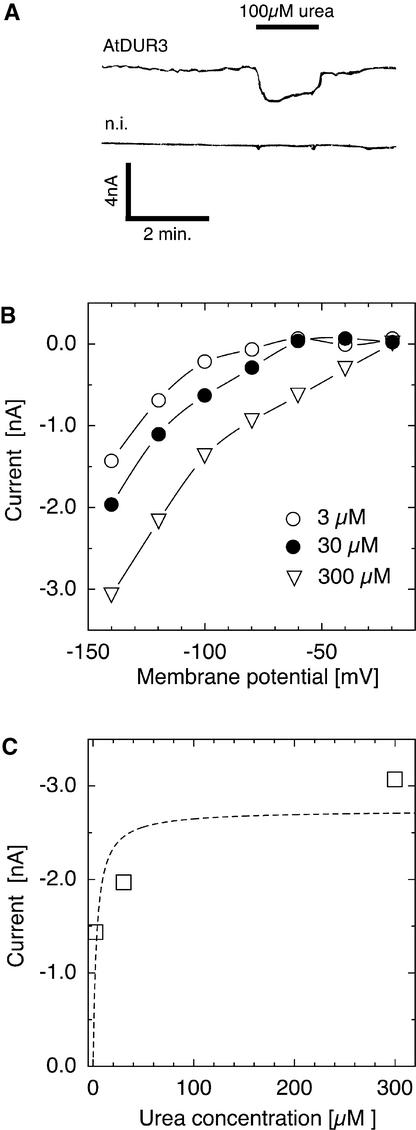
To study the transport mechanism of AtDUR3 in more detail, the protein was expressed heterologously in X. laevis oocytes. Oocytes injected with cRNA encoding AtDUR3 were voltage-clamped to −70 mV in choline-based buffer solution at pH 5.0 and then superfused with buffer containing 100 μM urea. Upon urea addition, small and reversible currents were detected that were absent in noninjected or water-injected control oocytes (Figure 5A). Currents induced by urea were time independent and corresponded to the net influx of positive charge. Weaker currents were observed by superfusion with the structural urea analog thiourea (data not shown). Because urea and thiourea both were uncharged, the current had to be ascribed to another, most likely cotransported ion. No currents were detectable when the following substrates were tested, which have been described as substrates for SSS transporters (Turk and Wright, 1997): Glc, Gal, myo-inositol, and Pro. Urea-induced currents were voltage dependent. The current-voltage relationship was nonlinear, with larger currents at more negative membrane potentials (Figure 5B). Urea-induced currents in AtDUR3-expressing oocytes also were investigated for their dependence on external urea application. Currents increased steeply at lower micromolar concentrations of urea and saturated at >50 μM (Figure 5C).
Figure 5.
Urea-Mediated Currents in the Presence of AtDUR3.
(A) Oocytes were voltage-clamped at −70 mV and bathed in standard solution. The addition of urea (100 μM, indicated by the black bar) reversibly induced small inward currents of similar size in oocytes injected with AtDUR3 cRNA. In water-injected or noninjected (n.i.) control oocytes, currents were unaffected by superfusing oocytes with the same solutions.
(B) Urea (100 μM) induced time-independent but voltage-dependent inward currents in AtDUR3-injected oocytes. Currents increased with increasing negative voltage.
(C) When AtDUR3-injected oocytes were clamped at −140 mV, urea-induced currents saturated at >50 μM external urea.
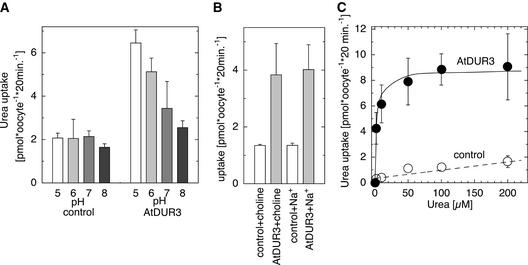
Urea Transport by AtDur3 Is pH and Concentration Dependent
14C-labeled urea uptake into AtDUR3-injected oocytes was investigated at different external pH values. 14C-urea uptake into noninjected oocytes showed no difference from that into water-injected oocytes and served as a control. At pH 5.0, urea uptake was stimulated approximately threefold in AtDUR3-expressing oocytes but not in control oocytes (Figure 6A). The stimulation of 14C-urea uptake by AtDUR3 became less prominent at higher pH, in agreement with the weak growth complementation of yeast by AtDUR3 on 2 mM urea at pH > 6 (Figure 2). This finding indicated that AtDUR3 transports urea in a H+-dependent manner. Because some SSS family members from bacteria and animals were described as Na+-coupled cotransporters, we additionally tested if Na+ influences urea transport by AtDUR3. However, in choline-based solutions at pH 5, the presence of 3 mM Na+ did not affect 14C-urea uptake into oocytes (Figure 6B).
Figure 6.
Characterization of the Uptake of 14C-Labeled Urea in the Presence of AtDUR3.
(A) Uptake of 14C-urea (100 μM) at different pH values in uninjected control and AtDUR3-expressing X. laevis oocytes.
(B) Urea uptake was independent of external sodium (3 mM). Standard choline solutions at pH 5.0 were used as a control.
(C) Concentration-dependent uptake of 14C-urea was saturable and displayed Michaelis-Menten kinetics with a half-maximal saturation at 3 μM urea. Standard choline solutions at pH 5.0 were used.
14C-labeled urea uptake by oocytes was used further to determine concentration-dependent transport by AtDUR3. Uptake periods of 20 min allowed tracer accumulation in oocytes that was sufficient for reliable determination (data not shown) (Leung et al., 2000). 14C-urea transport was saturable at low concentrations of urea, and the urea concentration that permitted half-maximal uptake was ∼3 μM (Figure 6C). Extended uptake periods of up to 2 h yielded similar Km values. Both concentration-dependent transport of 14C-labeled urea and urea-induced currents in AtDUR3-expressing oocytes saturated similarly at ∼50 μM (Figures 5C and 6C).
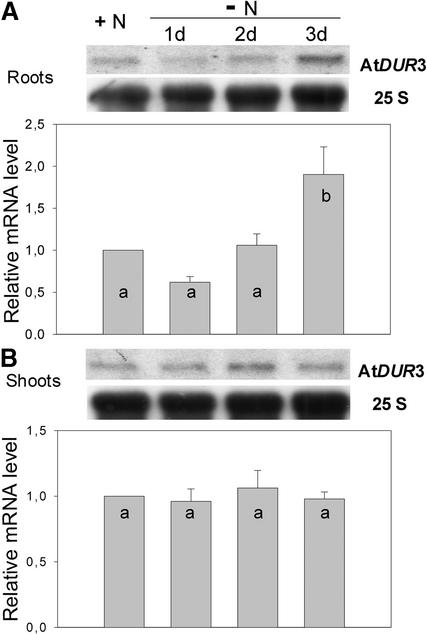
Transcriptional Regulation of AtDUR3 by Nitrogen
Nitrogen-dependent regulation of AtDUR3 gene expression was studied in Arabidopsis plants cultured hydroponically in Magenta boxes for 29 days and then subjected to nitrogen deficiency for 3 days. AtDUR3 was expressed in both shoots and roots under all nitrogen treatments tested (Figure 7). Transcript levels in shoots were not altered significantly under nitrogen starvation. In roots, however, AtDUR3 transcript levels were low under adequate nitrogen supply and increased continuously during 3 days of nitrogen starvation. The same observation was made when roots were analyzed from plants grown in nonaxenic nutrient solution (data not shown).
Figure 7.
Nitrogen-Dependent Expression of AtDUR3.
RNA gel blot analysis performed on hydroponically grown plants that were precultured for 4 weeks in the presence of 2 mM NH4NO3 and starved of nitrogen for 1, 2, or 3 days. Total RNA from roots (A) or shoots (B) was used for hybridization to the complete open reading frame of AtDUR3 and to 25S rRNA (panels show RNA gel blots from one representative experiment). The graphs represent mean values of relative signal intensities from three independent experiments. Mean values were compared pair-wise using Student's t test. Error bars indicate sd, and different letters indicate statistically significant differences at P < 0.05.
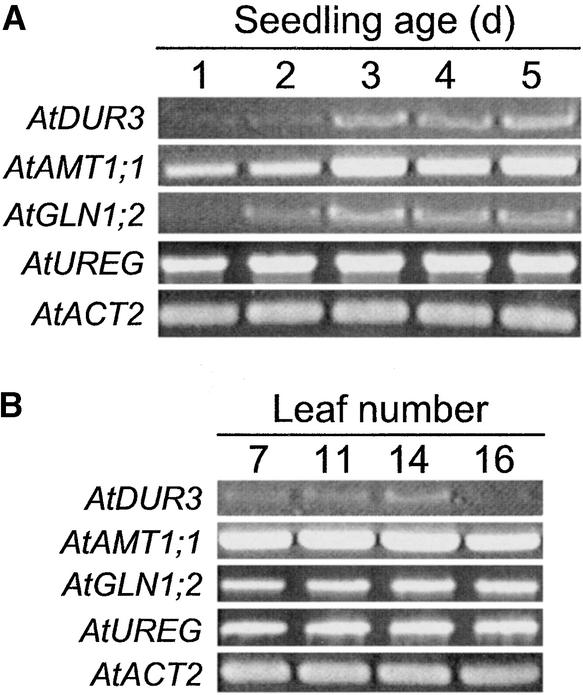
Because urea represents not only a nitrogen form available in the environment but also an important intermediate from the secondary nitrogen metabolism, AtDUR3 expression was monitored during germination and leaf aging, when nitrogen remobilization processes become prominent. Under these conditions, AtDUR3 was only weakly detectable by RNA gel blot analysis; therefore, reverse transcriptase-mediated (RT) PCR was performed. Using RNA from germinating seeds over a 5-day period allowed us to observe an increase in AtDUR3 expression levels between days 2 and 3 (Figure 8A). From day 3 onward, AtDUR3 transcripts were maintained at a higher level, similar to transcript levels of the ammonium transporter gene AtAMT1;1 (Gazzarrini et al., 1999) and cytosolic Gln synthetase (Oliveira and Coruzzi, 1999). In germinating seedlings, AtDUR3 gene expression apparently was not coregulated with the expression of UREG, which encodes a urease accessory protein (Freyermuth et al., 1999). On the other hand, AtDUR3 mRNA was at a similar level in Arabidopsis leaves of different ages, except in the youngest leaves (Figure 8B, leaf 16). Thus, AtDUR3 expression levels also were altered under conditions in which nitrogen remobilization processes were prominent.
Figure 8.
Expression of AtDUR3 in Germinating Seedlings and Leaves of Different Ages.
Gene-specific RT-PCR performed on total RNA isolated from Arabidopsis seedlings that were germinated in Petri dishes prewetted with distilled water and incubated for 1 to 5 days in a growth chamber (see Methods) (A) or from Arabidopsis rosette leaves that were numbered according to their time of formation and harvested from 6-week-old plants (B) (low numbers represent mature leaves; Hinderhofer and Zentgraf, 2001). Reference genes were the ammonium transporter AMT1;1, the cytosolic Gln synthetase GLN1;2, the urease accessory protein UREG, and ACTIN2 (ACT2).
DISCUSSION
AtDUR3 Is Homologous with the Animal and Microbial SSS Family
Our study demonstrated urea transport by AtDUR3 using three independent approaches. First, construction of a yeast dur3 deletion strain and subsequent growth complementation by AtDUR3 on urea as a sole nitrogen source (Figure 2) showed that AtDUR3 mediates urea uptake. Second, heterologous expression of AtDUR3 in oocytes and electrophysiological measurements by two-electrode voltage clamp allowed us to determine small inward-directed currents when urea was present in the medium (Figure 5). Third, heterologous expression of AtDUR3 in oocytes and uptake studies using 14C-labeled urea showed that concentration-dependent substrate accumulation was far greater than in uninjected control oocytes and saturable (Figures 5C and 6C). These observations are in accordance with the characterization of AtDUR3 as a urea transporter. The functional characterization of the AtDUR3 homolog from yeast, ScDUR3, relied on a lower level of uptake of 14C-labeled urea in a dur3 mutant compared with the recomplemented strain (ElBerry et al., 1993), without further investigating the transport mechanism or the biochemical properties of ScDUR3-mediated substrate transport.
AtDUR3 showed no significant homology with any other Arabidopsis protein; thus, it represents the only member of an unidentified gene family in plants. Similarly, in the rice genome, OsDUR3 represents the only sequence that has significant homology with AtDUR3, suggesting that plant DUR3 proteins might represent an individual transporter subfamily consisting of only one member. AtDUR3 is predicted to contain 14 transmembrane-spanning domains, and phylogenetic analysis revealed a close relationship to DUR3 proteins from S. cerevisiae and S. pombe within the large superfamily of SSS from bacteria (BLAST E value of 10−10 to 10−4) and animals (BLAST E value of <10−4). SSS transporters have been described to mediate the transport of various solutes, including Glc and urea in cotransport with Na+. In most cases, SSS transporters seem to preferentially transport one substrate or substrate group (Turk and Wright, 1997), whereas SSS transporters with a broad range of different substrates seem to be the exception (Leung et al., 2000). Among typical substrates for SSS transporters, AtDUR3 transported only urea and its structural homolog thiourea at detectable rates (Figures 2 and 5). Although additional substrates need to be tested in the future, these observations prompted us to classify AtDUR3 as a urea transporter.
To date, the only transporter class from plants shown to permeate urea were aquaporins of the tonoplast intrinsic protein and plasma membrane intrinsic protein subfamilies from tobacco. Oocyte expression and 14C-urea uptake studies have shown that NtTIPa and NtAQP1 permeate 14C-labeled urea (Eckert et al., 1999; Gerbeau et al., 1999). In plants, aquaporin-mediated urea transport potentially can occur across the tonoplast of plants (e.g., via NtTIPa) as well as across the plasma membrane (e.g., via NtAQP1). In mammals, aquaporin-mediated urea transport has been demonstrated for 3 of 10 AQP homologs and seems to occur across different membranes from a large number of tissues or organelles (Echevarria and Ilundain, 1998). In both kingdoms, the physiological function of aquaporin-mediated urea transport remains rather unclear. Like other prominent urea transporters, such as the mammalian UT-type or the Helicobacter pilori Ure1 transporters (Weeks et al., 2000; Smith and Rousselet, 2001), aquaporins represent facilitative membrane transporters for urea that only allow an equilibration of urea gradients across membranes. However, the structure and transport mechanism of AtDUR3 are completely different.
AtDUR3 Mediates H+-Urea Symport at High Affinity
We found that the transport of urea by AtDUR3 depends on urea cotransport with protons: (1) AtDUR3 complemented the growth of the Δdur3 mutant only at pH 6 or lower (Figure 2); (2) AtDUR3-expressing oocytes exhibited a positive inwardly rectifying and voltage-dependent current in response to superfusion with urea (Figure 5); and (3) accumulation of 14C-urea in AtDUR3-expressing oocytes decreased step-wise from pH 5 to 8 (Figure 6A). In our experiments, Na+ could not substitute the effect of protons or stimulate urea uptake in oocytes either when urea-dependent currents were monitored (data not shown) or when uptake of 14C-urea was measured (Figure 6B). These observations led us to conclude that AtDUR3 represents a H+-urea cotransporter.
Unlike other SSS members, DUR3 proteins contain a P-loop, which is a structural domain of ATP-driven pumps. Most recently, ATP-driven urea transport has been indicated by the genetic and physiological characterization of a ureG mutant of Anabaena in which active and nitrogen-controlled urea uptake was decreased strongly. UreG is part of an urt gene cluster that encodes all elements of an ABC-type urea permease in cyanobacteria (Valladares et al., 2002). However, because the P-loop of AtDUR3 is predicted to be located outside of the cytoplasm and urea transport depends on low pH, AtDUR3 more likely represents a secondary active transporter that depends on a H+ gradient rather than ATP hydrolysis as a driving force. Likewise, AtUPS1, an allantoin transporter, also contains a P-loop but energizes allantoin by cotransport with protons (Desimone et al., 2002). Thus, the biochemical significance of the P-loop in AtDUR3 remains to be elucidated.
To characterize the concentration-dependent transport of urea by AtDUR3, we first performed short-term uptake studies in AtDUR3-transformed yeast cells using 14C-labeled urea as a substrate. However, we failed to measure 14C accumulation even after several minutes of incubating yeast cells with the substrate (data not shown), most likely because of the high urease activity in yeast that leads to the generation of volatilizing 14CO2. Probably as a result of this internal urea degradation, ElBerry et al. (1993) determined 14CO2 that had been released from a liquid yeast culture and accumulated in a CO2 trap inside the vessel of a liquid yeast culture. However, this approach required long incubation periods with the tracer and made it impossible to determine the substrate affinity for the ScDUR3 gene product. X. laevis oocytes express neither endogenous urea transporters nor urease activity (Shayakul et al., 1996). As a consequence, the expression of AtDUR3 in oocytes allowed us to determine a sharp Michaelis-Menten–like accumulation of urea (Figure 6C), indicating that mainly one kinetic process was reflected (i.e., urea transport across the oocyte membrane).
14C-urea uptake by AtDUR3 exhibited an affinity constant of ∼3 μM, indicating that AtDUR3 mediates the high-affinity transport of urea. Thus, AtDUR3 clearly differs from most of the urea transporters that have been isolated to date and that saturate at millimolar concentrations or for which affinity constants are not available (Leung et al., 2000; Smith and Rousselet, 2001). The exception is the ABC-type urea transporter UreG from Synechocystis with a substrate affinity of <1 μM, which allows it to transport urea against a concentration gradient (Valladares et al., 2002). High-affinity transport of urea also has been reported for Chara cells that show a transient decrease in membrane potential as a result of the generation of a positive inward current in response to urea. The size of this current was concentration dependent, with a Km of ∼0.3 μM, showing that urea transport was electrogenic. Furthermore, uptake of 14C-labeled urea was inhibited by CCCP and N,N′-dicyclo-hexylcarbodiimide, indicating that high-affinity urea uptake depended on the proton gradient across the plasma membrane (Wilson et al., 1988). This finding is in agreement with urea uptake by suspension cultures from Arabidopsis, which also decreased in the presence of CCCP (Figure 1). When expressed in oocytes, AtDUR3-mediated urea transport depended not only on the availability of protons but also on the membrane potential (Figure 5B). Thus, AtDUR3 most likely uses the electrochemical gradient across plant membranes to energize urea transport.
The Physiological Function of AtDUR3 Is Linked to the Nitrogen Status in Arabidopsis
In yeast and oocytes, AtDUR3 mediated urea import, indicating that AtDUR3 is likely to be involved in urea uptake into plant cells and that plant cells might transport urea directly as a nitrogen-containing metabolite or use it as a nitrogen source. The expression pattern of AtDUR3 in Arabidopsis plants is consistent with this interpretation. RNA gel blot analysis showed that AtDUR3 was expressed in the roots (Figure 6A). Moreover, AtDUR3 gene expression was upregulated under nitrogen deficiency similar to ScDUR3 expression, which is highly sensitive to nitrogen catabolite repression (ElBerry et al., 1993). A nitrogen-repressed transcriptional regulation of AtDUR3 would be similar to that of ammonium (AMT1) and nitrate (NRT2) transporters in Arabidopsis (Gazzarrini et al., 1999; Lejay et al., 1999), further supporting the idea that AtDUR3 is involved in the utilization of urea as an external nitrogen source. Despite a constitutive urease activity in roots, other studies reported urea accumulation in the xylem sap or in leaves when urea was used as a nitrogen source to roots (Hine and Sprent, 1988; Gerendás et al., 1998), indicating that at least part of the root-absorbed urea is translocated to aerial plant organs before urea degradation. Under these conditions or when urea is applied as a nitrogen fertilizer directly to leaves, a low expression level of AtDUR3 in leaves (Figures 7 and 8) might reflect the uptake and utilization of xylem-derived or leaf-absorbed urea by leaf cells. This notion is in agreement with physiological studies showing efficient leaf uptake and retranslocation of urea when supplied to leaves (Shelp and Shattuck, 1986; Nicoulaud and Bloom, 1996).
On the other hand, urea also represents a metabolite from cellular protein catabolism. Arg, which is an important nitrogen transport and storage metabolite, is the immediate precursor of urea in the Orn (urea) cycle of plant cells. Because plant arginases have a mitochondrial location and plant ureases appear to be cytoplasmic, the use of urea requires that it leaves the mitochondrion (Polacco and Holland, 1993). Although in this case, urea can follow the concentration gradient from mitochondria to the cytoplasm, urea transporters might be involved. Therefore, we investigated AtDUR3 expression in tissues with a high turnover of proteins and Arg, because they are represented in germinating seedlings and senescing leaves. In these tissues, AtDUR3 expression was not detectable by RNA gel blot analysis, but gene-specific RT-PCR allowed us to detect an increase in AtDUR3 expression after 3 days of germination under nutrient-free conditions (Figure 8A). In Arabidopsis, nitrogen remobilization during early germination is reflected by parallel increases in arginase and urease activities as well as in urea levels, and the urease-mediated recycling of urea has been proposed as an important process for the utilization of nitrogenous reserves (Zonia et al., 1995). Changes in mRNA levels of arginase (data not shown) or a urease accessory protein during early germination did not coincide with the increase in AtDUR3 expression between days 2 and 3 after germination (Figure 8A). Because there is no physiological evidence for the additional intracellular transport or compartmentation of cytoplasmic urea, and because mitochondrial urea export is extremely unlikely to be driven by proton cotransport, upregulation of AtDUR3 under our conditions most likely reflected the onset of nitrogen deficiency in germinating seedlings, as has been reported for the ammonium transporter gene AtAMT1;1, which is upregulated strongly under N deficiency (Gazzarrini et al., 1999; Rawat et al., 1999).
In general, old leaves remobilize nitrogen partially via the Orn cycle to sustain growth in developing and growing metabolic sinks (Feller and Fischer, 1994) and exhibit far higher urease activities per unit of protein compared with young leaves (Witte et al., 2002). Thus, AtDUR3 might be upregulated by the onset of senescence if it is required for the membrane transport and utilization of urea derived from protein degradation. Although AtDUR3 transcript levels tended to be lowest in the youngest leaves (Figure 8B), a physiological link between protein degradation in senescing leaves and a requirement for AtDUR3-mediated urea transport to support nitrogen remobilization from proteins could not be established and requires further investigation.
In conclusion, the data presented here demonstrate the existence of an active urea transporter in plants, because AtDUR3- mediated membrane transport of urea depends on the availability of protons and an electrical potential gradient. Under nitrogen deficiency, AtDUR3 gene expression was upregulated in roots. Thus, AtDUR3 could mediate urea uptake from the external growth medium and thereby provide a new pathway for the direct utilization of urea nitrogen from the environment. Ongoing studies focus on the membrane localization of AtDUR3 and urea uptake capacities in transgenic lines with repressed AtDUR3 gene expression.
METHODS
Gene Isolation, Generation of a Yeast Mutant, and Functional Complementation
The AtDUR3 open reading frame was amplified from a cDNA library (Minet et al., 1992) by Pfu polymerase (Stratagene) using PCR and AtDUR3-specific primers containing a restriction site for BglII (5′-GAAGAT-CTATGGCTACATGTCCTCCTTTCG-3′ and 5′-GAAGATCTTCAACCTTCTTCATCATTTTTCTTG-3′). The resulting PCR product was cloned into the pGEM-T Easy vector (Promega). The open reading frame of AtDUR3 was sequenced and verified by the sequence in the database. Then, the open reading frame of AtDUR3 with BglII overhangs was ligated into the yeast expression vector pHXT426 (Wieczorke et al., 1999) and into the oocyte expression vector pOO2 (Ludewig et al., 2002) after linearization with BamHI.
The yeast (Saccharomyces cerevisiae) strain Σ 23346c (Mat a, ura3) (Grenson, 1969) was used to generate a mutant defective in urea uptake. The DUR3 gene was deleted by homologous integration of a disruption cassette containing loxP sites flanking the marker gene kan (Güldener et al., 1999). Selection was performed on solid medium containing 100 mg/L G418, and gene deletion was confirmed by PCR. The deficiency of the mutant in urea uptake was determined on agar plates containing different concentrations of urea. The dur3 mutant YNVW1 was transformed by pHXT426 harboring AtDUR3 or by the empty vector alone (Dohmen et al., 1991), and transformants were selected on uracil-deficient yeast nitrogen base without ammonium and amino acid medium (Difco) containing 20 mM NH4+ before a single colony was chosen for growth complementation tests on urea as a sole nitrogen source.
Electrophysiological Studies in Xenopus laevis Oocytes
Capped cRNA was transcribed from pOO2-AtDUR3 in vitro using the mMessage mMachine kit (Ambion, Austin, TX) after linearization of the plasmid with MluI. X. laevis oocytes were removed from adult female frogs by surgery and dissected manually. Oocytes (Dumont stage V or VI) were defolliculated using 10 mg/mL collagenase (Boehringer Mannheim) and trypsin inhibitor (Sigma) for 1 h and injected with 5 to 50 nL of cRNA (15 to 50 ng per oocyte). Each coinjection experiment was repeated multiple times. Oocytes were kept after injection for 2 to 5 days at 16°C in ND96 solution with the following composition: 96 mM NaCl, 2 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 5 mM Hepes, pH 7.4, and gentamycin (20 μg/mL). Data were collected from five batches of oocytes from different frogs. Currents were measured by two-electrode voltage clamp analysis at 3 days after injection using a Dagan CA1 amplifier (Minneapolis, MN) and pClamp 6.0 software (Axon Instruments, Union City, CA). Standard bath solutions contained 100 mM choline-Cl, 2 mM CaCl2, 2 mM MgCl2, and 4 mM Tris, pH adjusted to 5, 6, 7, or 8 with Mes. For the recordings shown in Figure 5A, oocytes were voltage-clamped at a constant negative potential and currents were recorded continuously by a chart recorder. Voltage pulses of 200 ms were applied between 0 and −140 mV for the recordings shown in Figure 5B. For current-voltage analysis, background currents (recorded before and after the addition of the substrate) were subtracted.
Radiotracer Uptake Studies in X. laevis Oocytes
Standard bath solutions were used for uptake experiments. For uptake experiments, oocytes were pooled in groups of three and incubated for 20 min at room temperature in 500 μL of the appropriate buffer containing 10% 14C-labeled urea (specific activity of 57 mCi/mmol; Amersham). Then, oocytes were washed carefully five times in 1 mL of ice-cold buffer with 100-fold excess urea and solubilized in 5% SDS. After the addition of 5 mL of scintillation cocktail (Ultima Gold; Zinsser Analytic, Frankfurt, Germany), washed oocytes were measured in a scintillation counter (Perkin-Elmer, Boston, MA).
Data were processed using SigmaPlot (SPSS, Chicago, IL). All values are given as means ± sd. The concentration dependence of urea uptake was fitted using the equation I = Imax/(1 + Km/c), where Imax is maximal uptake at saturating urea concentration, Km is substrate concentration permitting half-maximal currents, and c is the concentration used in the experiment.
Plant Culture and Gene Expression Analysis
Arabidopsis thaliana seeds (ecotype Columbia 0) were germinated and precultured axenically in Magenta boxes containing 50 mL of nutrient solution of the following composition: 82.5 mg/L NH4NO3, 95 mg/L KNO3, 220 mg/L CaCl2·2H2O, 185 mg/L MgSO4·7H2O, 85 mg/L KH2PO4, 3.1 mg/L H3BO3, 8.45 mg/L MnSO4·H2O, 4.3 mg/L ZnSO4·7H2O, 0.415 mg/L KI, 0.125 mg/L Na2MoO4·2H2O, 0.0125 mg/L CuSO4·5H2O, 0.0125 mg/L CoCl2·6H2O, 18.65 mg/L Na2EDTA·2H2O, 13.93 mg/L FeSO4· 7H2O, 0.25 mg/L nicotinic acid, 0.25 mg/L pyridoxine HCl, 0.05 mg/L thiamine HCl, 1 mg/L Gly, 50 mg/L myo-inositol, 1% Suc, and 10 mM Mes. KOH (1 M) was used to adjust the nutrient solution to pH 5.7. The nutrient solution was changed every 2 days, and plants were cultured for 3 weeks. Magenta boxes were agitated gently on a shaker in a growth chamber at 21°C using a 14-h-light/10-h-dark cycle and light intensity of 250 μmol·m−2·s−1 at plant height. In the 4th week, Suc and vitamins were omitted from the nutrient solution and nutrient solutions were changed every day. Except for the control treatment, with 1 mM NH4NO3, plants were subjected to nitrogen deficiency for 1, 2, and 3 days before harvest. Plant roots and shoots were harvested separately for total RNA extraction. To obtain plant material from germinating seedlings, surface-sterilized seeds were placed in Petri dishes, germinated in autoclaved distilled water, and harvested every day for 5 days. Leaf material of different ages was obtained from Ulrike Zentgraf (Zentrum für Molekularbiologie der Pflanzen) as described by Hinderhofer and Zentgraf (2001).
Isolation of total RNA and RNA gel blot analysis were conducted as described by Gazzarrini et al. (1999). The complete open reading frame of AtDUR3 was used as a probe for hybridization to total RNA. A 25S rRNA probe was used as a reference for the quantifications achieved using a PhosphorImager (Storm; Molecular Dynamics, Sunnyvale, CA). The ratio of AtDUR3 to 25S from the plus-nitrogen treatment was set to 1, and mean values of relative signal intensities from three independent experiments were compared between treatments using Student's t test (SigmaStat 2.03). For reverse transcriptase–mediated PCR analysis, RNAs were converted to cDNAs by reverse transcriptase according to the manufacturer's protocol (MBI Fermentas), and cDNA fragments of AtDUR3 (2085 bp), AtUREG (820 bp), AtGLN1;2 (1073 bp), and AtAMT1;1 (1296 bp) were amplified by 20 PCR cycles using the corresponding gene-specific primers (forward primers: 5′-ATGGCTACATGT-CCTCCTTTCG-3′, 5′-GAAGGCGTCGTGGGTGGG-3′, 5′-ATGAGTCTT-CTTGCAGATCTT-3′, and 5′-ATGCAGCTTGGCTTCGCTAT-3′; reverse primers: 5′-TCAACCTTCTTCATCATTTTTCTTG-3′, 5′-AAGTATTGAAAG-AGTTCCATTCA-3′, 5′-TCAAGGGTTCCAGAGGAGT-3′, and 5′-CGTGGC-TCAACTCTCCTAAG-3′ for AtDUR3, AtUREG, AtGLN1;2, AtAMT1;1, respectively). To ensure that equal amounts of cRNA were used in each PCR procedure, a cDNA fragment of the constitutively expressed ACTIN2 gene was amplified simultaneously by PCR (An et al., 1996). Verification of the amplicons was confirmed further by sequencing.
14C-Urea Uptake Studies in Arabidopsis Suspension Cells
Ten milliliters of 1-week-old Arabidopsis (cv Landsberg) suspension cells was transferred to 90 mL of growth medium (4.3 g/L Murashige and Skoog [1962] medium; Duchefa, Haarlem, The Netherlands), 30 g of Suc, 0.5 mg of yeast nitrogen base without amino acids and ammonium sulfate (NAA; Difco), 0.05 mg of kinetin (Sigma), and 10 mM Mes, adjusted to pH 5.8 (using 1 N KOH), and incubated further for 5 days. Cells were harvested by centrifugation at 3000 rpm for 5 min and resuspended in the same volume of growth medium without NAA and kinetin (buffer solution). Two hundred microliters of cell suspension was used for uptake assays with 14C-labeled urea (specific activity of 57.0 mCi/mmol; Amersham) at 21°C. For the inhibition of urea uptake, cells were preincubated with 100 μM carbonyl cyanide m-chlorophenylhydrazone (Sigma) for 5 min. At the end of the uptake period, cells were washed three times with 2 mL of ice-cold buffer solution containing 10 mM cold urea. After the addition of 5 mL of scintillation cocktail (Ultima Gold; Zinsser), cells were analyzed in a liquid scintillation counter (Wallac).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes.
Accession Number
The GenBank accession number for the cDNA clone from the AtDUR3 gene is AV524336.
Acknowledgments
We thank Doris Rentsch (University of Bern) for her support in generating the yeast mutant, Brigitte Gassert, Dominique Loqué, and Pia Walch-Liu (Hohenheim University) for technical assistance, E. Boles (Institut für Mikrobiologie, Universität Frankfurt) for supplying pHXT426, Ulrike Zentgraf (Zentrum für Molekularbiologie der Pflanzen) for supplying leaf material, the Kazusa DNA Research Institute (Japan) and the Japanese Rice Genome Research Program of the National Institute of Agrobiological Sciences for supplying the rice EST clones, and the Institute of the Society of Techno-Innovation in Agriculture, Forestry and Fisheries for supplying the Arabidopsis EST clones. This study was supported financially by the Deutsche Forschungsgemeinschaft (Bonn, Germany) with a grant to N.v.W. (Wi 1728/2).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.007120.
References
- An, Y.-Q., McDowell, J.M., Huang, S., McKinney, E.C., Chambliss, S., and Meagher, R.B. (1996). Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 10, 107–121. [DOI] [PubMed] [Google Scholar]
- Bradley, D.P., Morgan, M.A., and O'Toole, P. (1989). Uptake and apparent utilization of urea and ammonium nitrate in wheat seedlings. Fertil. Res. 20, 41–49. [Google Scholar]
- Desimone, M., Catoni, E., Ludewig, U., Hilpert, M., Schneider, A., Kunze, R., Tegeder, M., Frommer, W.B., and Schumacher, K. (2002). A novel family of transporters for allantoin and other oxo derivatives of nitrogen heterocyclic compounds in Arabidopsis. Plant Cell 14, 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen, R.J., Strasser, A.W., Honer, C.B., and Hollenberg, C.P. (1991). An efficient transformation procedure enabling long-term storage of competent cells of various yeast genera. Yeast 7, 691–692. [DOI] [PubMed] [Google Scholar]
- Echevarria, M., and Ilundain, A.A. (1998). Aquaporins. J. Physiol. Biochem. 54, 107–118. [PubMed] [Google Scholar]
- Eckert, M., Biela, A., Siefritz, F., and Kaldenhoff, R. (1999). New aspects of plant aquaporin regulation and specificity. J. Exp. Bot. 50, 1541–1545. [Google Scholar]
- ElBerry, H.M., Majumdar, M.L., Cunningham, T.S., Sumrada, R.A., and Cooper, T.G. (1993). Regulation of the urea active transporter gene (DUR3) in Saccharomyces cerevisiae. J. Bacteriol. 175, 4688–4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller, U., and Fischer, A. (1994). Nitrogen metabolism in senescing leaves. Crit. Rev. Plant Sci. 13, 241–273. [Google Scholar]
- Freyermuth, S.K., Forde, B.G., and Polacco, J.C. (1999). Nucleotide sequence of cDNA encoding an Arabidopsis urease accessory protein (accession No. AF 109374) (PGR99-012). Plant Physiol. 119, 364. [Google Scholar]
- Gaudin, R., Dupuy, J., and Bournat, P. (1987). Suivi du contenue en azote de la solution du sol d'une rizière après placement d'urée. Agron. Trop. 42, 13–19. [Google Scholar]
- Gazzarrini, S., Lejay, L., Gojon, A., Ninnemann, O., Frommer, W.B., and von Wirén, N. (1999). Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into Arabidopsis roots. Plant Cell 11, 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbeau, P., Guclu, J., Ripoche, P., and Maurel, C. (1999). Aquaporin Nt-TIPa can account for the high permeability of tobacco cell vacuolar membrane to small neutral solutes. Plant J. 18, 577–587. [DOI] [PubMed] [Google Scholar]
- Gerendás, J., Polacco, J.C., Freyermuth, S.K., and Sattelmacher, B. (1999). Significance of nickel for plant growth and metabolism. J. Plant Nutr. Soil Sci. 162, 241–256. [Google Scholar]
- Gerendás, J., Zhu, Z., and Sattelmacher, B. (1998). Influence of N and Ni supply on urease activity in rice. J. Exp. Bot. 49, 1545–1554. [Google Scholar]
- Grenson, M. (1969). The utilization of exogenous pyrimidines and the recycling of uridine-5′-phosphate derivatives in Saccharomyces cerevisiae, as studied by means of mutants affected in pyrimidine uptake and metabolism. Eur. J. Biochem. 11, 249–260. [DOI] [PubMed] [Google Scholar]
- Güldener, U., Heck, S., Fiedler, T., Beinhauer, J., and Hegemann, J.H. (1999). A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24, 2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinderhofer, K., and Zentgraf, U. (2001). Identification of a transcription factor specifically expressed at the onset of leaf senescence. Planta 213, 469–473. [DOI] [PubMed] [Google Scholar]
- Hine, J.C., and Sprent, J.I. (1988). Growth of Phaseolus vulgaris on various nitrogen sources: The importance of urease. J. Exp. Bot. 39, 1505–1512. [Google Scholar]
- Krogmeier, M.J., McCarty, G.W., and Bremner, J.M. (1989). Phytotoxicity of foliar-applied urea. Proc. Natl. Acad. Sci. USA 86, 8189–8191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejay, L., Tillard, P., Lepetit, M., Olive, F., Filleur, S., Daniel-Vedele, F., and Gojon, A. (1999). Molecular and functional regulation of two NO3− uptake systems by N- and C-status of Arabidopsis plants. Plant J. 18, 509–519. [DOI] [PubMed] [Google Scholar]
- Leung, D.W., Loo, D.D.F., Hirayama, B.A., Zeuthen, T., and Wright, E.M. (2000). Urea transport by cotransporters. J. Physiol. 528, 251–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludewig, U., von Wirén, N., and Frommer, W.B. (2002). Uniport of ammonium by the root hair plasma membrane ammonium transporter LeAMT1;1. J. Biol. Chem. 277, 13548–13555. [DOI] [PubMed] [Google Scholar]
- Marschner, H. (1995). Mineral Nutrition of Higher Plants. (London: Academic Press).
- Masclaux, C., Valadier, M.H., Brugiere, N., Morot-Gaudry, J.F., and Hirel, B. (2000). Characterization of the sink/source transition in tobacco (Nicotiana tabacum L.) shoots in relation to nitrogen management and leaf senescence. Planta 211, 510–518. [DOI] [PubMed] [Google Scholar]
- May, M., and Leaver, C. (1993). Oxidative stimulation of glutathione synthesis in Arabidopsis thaliana suspension cultures. Plant Physiol. 103, 621–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minet, M., Dufour, M.E., and Lacroute, F. (1992). Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNA. Plant J. 2, 417–422. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nicoulaud, B.A.L., and Bloom, A.J. (1996). Absorption and assimilation of foliarly applied urea in tomato. J. Am. Soc. Hortic. Sci. 121, 1117–1121. [Google Scholar]
- Oliveira, I.C., and Coruzzi, G.M. (1999). Carbon and amino acids reciprocally modulate the expression of glutamine synthetase in Arabidopsis. Plant Physiol. 121, 301–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polacco, J.C., and Holland, M.A. (1993). Roles of urease in plant cells. Int. Rev. Cytol. 145, 65–103. [Google Scholar]
- Rawat, S.R., Silim, S.M., Kronzucker, H.J., Siddiqi, M.Y., and Glass, A.D.M. (1999). AtAMT1 gene expression and NH4+ uptake in roots of Arabidopsis thaliana: Evidence for regulation by root glutamine levels. Plant J. 19, 143–152. [DOI] [PubMed] [Google Scholar]
- Reizer, J., Reizer, A., and Saier, M.H., Jr. (1994). A functional superfamily of sodium/solute symporters. Biochim. Biophys. Acta 1197, 133–166. [DOI] [PubMed] [Google Scholar]
- Saier, M.H., Jr. (2000). A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64, 354–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste, M. (1990). Structural features of cytochrome oxidase. Q. Rev. Biophys. 23, 331–366. [DOI] [PubMed] [Google Scholar]
- Shayakul, C., Steel, A., and Hediger, M.A. (1996). Molecular cloning and characterization of the vasopressin-regulated urea transporter of rat kidney collecting ducts. J. Clin. Invest. 98, 2580–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelp, B.J., and Shattuck, V.I. (1986). Accumulation of 14C from foliar applied [14C]urea by developing tomato fruits. Hortic. Sci. 21, 1178–1179. [Google Scholar]
- Smith, C.P., and Rousselet, G. (2001). Facilitative urea transporters. J. Membr. Biol. 183, 1–14. [DOI] [PubMed] [Google Scholar]
- Sonnhammer, E.L., von Heijne, G., and Krogh, A. (1998). A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6, 175–182. [PubMed] [Google Scholar]
- Turk, E., and Wright, E.M. (1997). Membrane topology motifs in the SGLT cotransporter family. J. Membr. Biol. 159, 1–20. [DOI] [PubMed] [Google Scholar]
- Valladares, A., Montesinos, M.L., Herrero, A., and Flores, E. (2002). An ABC-type, high-affinity urea permease identified in cyanobacteria. Mol. Microbiol. 43, 703–715. [DOI] [PubMed] [Google Scholar]
- Walker, N.A., Reid, R.J., and Smith, F.A. (1993). The uptake and metabolism of urea by Chara australis. IV. Symport with sodium: A slip model for the high and low affinity systems. J. Membr. Biol. 136, 263–271. [DOI] [PubMed] [Google Scholar]
- Watson, C.J., Miller, H., Poland, P., Kilpatrick, D.J., Allen, M.B.D., Garret, M.K., and Christianson, C.B. (1994). Soil properties and the ability of the urease inhibitor N-(n-butyl) thiophosphoric triamide (nBTPT) to reduce ammonia volatilization from surface-applied urea. Soil Biol. Biochem. 26, 1165–1171. [Google Scholar]
- Weeks, D.L., Eskandari, S., Scott, D.R., and Sachs, G. (2000). A H+-gated urea channel: The link between Helicobacter pilori urease and gastric colonization. Science 287, 482–485. [DOI] [PubMed] [Google Scholar]
- Wieczorke, R., Krampe, S., Weierstall, T., Freidel, K., Hollenberg, C.P., and Boles, E. (1999). Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 464, 123–128. [DOI] [PubMed] [Google Scholar]
- Wilson, M.R., O'Donoghue, S.I., and Walker, N.A. (1988). The transport and metabolism of urea in Chara australis. III. Two specific transport systems. J. Exp. Bot. 39, 763–774. [Google Scholar]
- Wilson, M.R., and Walker, N.A. (1988). The transport and metabolism of urea in Chara australis. I. Passive diffusion, specific transport and metabolism of urea and methylurea. J. Exp. Bot. 39, 739–751. [Google Scholar]
- Witte, C.P., Tiller, S.A., Taylor, M.A., and Davies, H.V. (2002). Leaf urea metabolism in potato: Urease activity profile and patterns of recovery and distribution of 15N after foliar urea application in wild-type and urease-antisense transgenics. Plant Physiol. 129, 1129–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonia, L.E., Stebbins, N.E., and Polacco, J.C. (1995). Essential role of urease in germination of nitrogen-limited Arabidopsis thaliana seeds. Plant Physiol. 107, 1097–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]