Abstract
Ubiquitin-dependent proteolysis is catalyzed by the 26S proteasome, a dynamic complex of 32 different proteins whose mode of assembly and mechanism of action are poorly understood, in part due to the difficulties encountered in purifying the intact complex. Here we describe a one-step affinity method for purifying intact 26S proteasomes, 19S regulatory caps, and 20S core particles from budding yeast cells. Affinity-purified 26S proteasomes hydrolyze both model peptides and the ubiquitinated Cdk inhibitor Sic1. Affinity purifications performed in the absence of ATP or presence of the poorly hydrolyzable analog ATP-γ-S unexpectedly revealed that a large number of proteins, including subunits of the skp1-cullin-F-box protein ligase (SCF) and anaphase-promoting complex (APC) ubiquitin ligases, copurify with the 19S cap. To identify these proteasome-interacting proteins, we used a recently developed method that enables the direct analysis of the composition of large protein complexes (DALPC) by mass spectrometry. Using DALPC, we identified more than 24 putative proteasome-interacting proteins, including Ylr421c (Daq1), which we demonstrate to be a new subunit of the budding yeast 19S cap, and Ygr232w (Nas6), which is homologous to a subunit of the mammalian 19S cap (PA700 complex). Additional PIPs include the heat shock proteins Hsp70 and Hsp82, the deubiquitinating enzyme Ubp6, and proteins involved in transcriptional control, mitosis, tubulin assembly, RNA metabolism, and signal transduction. Our data demonstrate that nucleotide hydrolysis modulates the association of many proteins with the 26S proteasome, and validate DALPC as a powerful tool for rapidly identifying stoichiometric and substoichiometric components of large protein assemblies.
INTRODUCTION
The 26S proteasome consists of a self-compartmentalized 20S protease core that is capped at one or both ends by the 19S regulatory particle, or cap (also known as PA700 in animal cells). The 20S core particle is made up of two copies each of seven different α and seven different β subunits arranged into four stacked rings (α7β7β7α7). The two outer α rings are catalytically inactive, whereas three of the seven inner β subunits are catalytically active (Voges et al., 1999).
Although the 20S core can degrade fully unfolded proteins in the absence of ATP and ubiquitin (De Mot et al., 1999), protein degradation by 26S proteasomes is strictly ATP dependent, and in most cases requires the presence of a ubiquitin chain on the substrate protein (Verma and Deshaies, 2000). Of the 18 proteins that make up the 19S cap, one, Rpn10, has been demonstrated to bind multiubiquitin chains (Deveraux et al., 1994). However, Rpn10 is dispensable for growth of yeast (van Nocker et al., 1996), implying the existence of other ubiquitin-binding proteins in the 19S cap. Once the ubiquitinated substrate protein is recruited to the 19S cap, it is thought to be unfolded and translocated into the lumen of the 20S core particle where it is degraded. It has been proposed that both unfolding and translocation of substrate are mediated by the six “AAA” ATPases present in the 19S cap (Larsen and Finley, 1997). By analogy to proteasome-like complexes in prokaryotes, these ATPases are presumed to assemble into a six-membered ring that directly abuts the 20S core particle. Because structural studies indicate that the ends of the 20S core are completely closed (Groll et al., 1997), it has been postulated that the ATPases open a channel leading to the proteolytic chamber within the 20S core (Larsen and Finley, 1997).
The composition of eukaryotic 26S proteasomes has been analyzed in several studies. Most of the studies have relied on conventional chromatographic fractionation steps and one- or two-dimensional SDS-PAGE, followed by sequence analysis of individual polypeptides extracted from gel slices. These analyses have defined a core set of subunits that comprise the 26S holoenzyme. Most of these subunits are conserved from yeast to humans (Voges et al., 1999). Investigators using genetic, yeast two-hybrid, and coimmunoprecipitation analyses have identified additional substoichiometric components of 26S proteasome preparations. Examples include the cyclin-dependent kinase Cdc28 and its regulator Cks1 (Kaiser et al., 1999), the DNA repair protein Rad23 (Schauber et al., 1998), and transcription factors such as c-Fos (Wang et al., 1996). It is unclear whether these latter proteins are targets or regulators of the 26S proteasome, and whether these interactions are related to proteolysis or to some other function of the 26S proteasome.
To investigate the composition, function, and regulation of the 26S proteasome in budding yeast, we developed a rapid and reproducible method for isolating highly purified and fully functional 26S complexes, as well as 19S regulatory caps and 20S cores. Using this method, we demonstrate that nucleotide regulates the stable association of a large number of proteins with the 26S proteasome, including subunits of the ubiquitin ligases skp1-cullin-F-box protein ligase (SCF) and anaphase-promoting complex (APC). Electrospray tandem mass spectrometry of a protease digest of the entire proteasome revealed the identities of approximately 24 additional proteasome-interacting proteins (PIPs), which include a new proteasome subunit, ubiquitin pathway components, and transcriptional regulatory proteins. We propose that the PIPs represent potential substrates or regulators of the 26S proteasome, or subunits of multimeric complexes whose dynamic properties are modulated by the 26S proteasome.
MATERIALS AND METHODS
Yeast Strains
RJD 1144 and RJD 1171 (Table 1) were derived from strain JD47-13C by integration of the YIplac211-based plasmids pJD416 and pJD522, respectively. pJD416 (PRE1) and pJD522 (RPT1) contain 3′ segments from the indicated open reading frames, fused in frame to sequences that encode the FlagHis6 tag (FH) followed by the transcriptional terminator from the CYC1 gene. Upon targeted integration of these plasmids into the yeast genome, the corresponding genes were disrupted, such that only the tagged proteins were expressed.
Table 1.
S. cerevisiae strains used in this study
| Strain | Genotype |
|---|---|
| RJD 487 | MATα leu2 ura3 trp1 GAL |
| RJD 497 | MATa leu2 ura3 trp1 bar1∷LEU2 pep4∷TRP1 |
| RJD 1144/JD 122 | MATa his3Δ200 leu2-3,112 lys2-801 trp1Δ63 ura3-52 PRE1FH∷Ylplac211 (URA3) |
| RJD 1171/JD 165 | MATa his3Δ200 leu2-3,112 lys2-801 trp1Δ63 ura3-52RPT1FH∷Ylplac211 (URA3) |
| RJD 1281 | MATa ura3 leu2 trp1 cdc34-2 RPT1FH∷Ylplac211 (URA3) |
| RJD 1294 | MATa RPT1FH∷Ylplac211 (URA3) leu2 cdc34-2 SIC1∷SIC1HAHIS6 (TRP) |
| RJD 1379 | MATα his3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 rub1Δ∷HIS3MX RPT1FH∷Ylplac211 (URA3) |
| RJD 1484 | MATa can1-100 leu2-3,112 his3 trp1-1 ura3-1 ade2-1 pep4∷TRP1 bar1∷LEU2 LEO1∷LEO1TEV2myc9(SpHIS5) |
| RJD 1485 | MATa can1-100 leu2-3,112 his3 trp1-1 ura3-1 ade2-1 pep4∷TRP1 bar1∷LEU2 CTR9∷CTR9TEV2myc9(SpHIS5) |
| RJD 1486 | MATa can1-100 leu2-3,112 his3 trp1-1 ura3-1 ade2-1 pep4∷TRP1 bar1∷LEU2 PAF1∷PAF1TEV2myc9(SpHIS5) |
| RJD 1487 | MATa can1-100 leu2-3,112 his3 trp1-1 ura3-1 ade2-1 pep4∷TRP1 bar1∷LEU2 YLR421C∷YLR421CTEV2myc9(SpHIS5) |
| RJD 1494 | MATa can1-100 leu2-3,112 his3 trp1-1 ura3-1 ade2-1 pep4∷TRP1 bar1∷LEU2 YLR421C∷YLR421CTEV2myc9(SpHIS5) RPT1FH∷Ylplac211 (URA3) |
| RJD 1496 | MATa can1-100 leu2-3,112 his3 trp1-1 ura3-1 ade2-1 pep4∷TRP1 bar1∷LEU2 PAF1∷PAF1TEV2myc9(SpHIS5) RPT1FH∷Ylplac211 (URA3) |
| RJD 1497 | MATa can1-100 leu2-3,112 his3 trp1-1 ura3-1 ade2-1 pep4∷TRP1 bar1∷LEU2 LEO1∷LEO1TEV2myc9(SpHIS5) RPT1FH∷Ylplac211 (URA3) |
| RJD 1498 | MATa can1-100 leu2-3,112 his3 trp1-1 ura3-1 ade2-1 pep4∷TRP1 bar1∷LEU2 CTR9∷CTR9TEV2myc9(SpHIS5) RPT1FH∷Ylplac211 (URA3) |
| RJD 1499 | MATa can1-100 leu2-3,112 his3 trp1-1 ura3-1 ade2-1 pep4∷TRP1 bar1∷LEU2 UBP6∷UBP6TEV2myc9(SpHIS5) |
| RJD 1500 | MATa can1-100 leu2-3,112 his3 trp1-1 ura3-1 ade2-1 pep4∷TRP1 bar1∷LEU2 YGR232W∷YGR232WTEV2myc9(SpHIS5) |
| RJD 1501 | MATa can1-100 leu2-3,112 his3 trp1-1 ura3-1 ade2-1 pep4∷TRP1 bar1∷LEU2 CDC16∷CDC16TEV2HA3(SpHIS5) |
| RJD 1502 | MATa his3Δ200 leu2-3,112 lys2-801 trp1Δ63 ura3-52 RPT1FH∷Ylplac211 (URA3) CDC16∷CDC16TEV2HA3(SpHIS5) |
| RJD 1503 | MATa his3Δ200 leu2-3,112 lys2-801 trp1Δ63 ura3-52 RPT1FH∷Ylplac211 (URA3) CDC23∷CDC23TEV2myc9(SpHIS5) |
| RJD 1504 | MATa can1-100 leu2-3,112 his3 trp1-1 ura3-1 ade2-1 pep4∷TRP1 bar1∷LEU2 CDC23∷CDC23TEV2myc9(SpHIS5) |
| RJD 1521 | MATα his3-1 leu2Δ met15Δ [pUbV76-Val-eΔK-βGal/URA3] |
| RJD 1523 | MATα his3-1 leu2Δ met15Δ daq1∷kanR [pUbV76-Val-eΔK-βGal/URA3] |
Epitope tagging of all other yeast chromosomal genes with either the Myc9 or hemagglutinin (HA)3 epitopes (Table 1) was carried out as described previously (Seol et al., 1999; Seol and Deshaies, unpublished data). Briefly, the tagging cassette encoded two consecutive recognition sites for the tobacco etch virus protease (TEV), fused in frame to either nine copies of the Myc or three copies of the HA epitope, followed by a stop codon and the 3′ untranslated region from the CDC53 locus. The Schizosaccharomyces pombe HIS5 gene, located at the 3′ end of the tagging cassette, served as a selectable marker. The cassette was amplified by polymerase chain reaction (PCR) by using a 5′ oligo homologous to the last 14 codons of the gene of interest and a 3′ oligo homologous to the 45 bp immediately downstream of the stop codon. The PCR product was directly transformed into yeast, yielding an allele of the gene of interest tagged at its 3′ end. Oligodeoxynucleotide sequences are available upon request.
Preparation of Extracts for Immunoprecipitation and Western Blotting
Log phase cultures of yeast (typically 100 ml) were grown in YPD at 30°C to an OD600 of 1.0. Cells were pelleted by centrifugation and washed with 50 mM Tris, pH 7.5, 50 mM sodium fluoride. The pellet was freeze thawed once in liquid N2 and suspended in 1 ml of lysis buffer containing 25 mM Tris, pH 7.5, 200 mM NaCl, 5 mM EDTA, 2.5 mM EGTA, 50 mM NaF, 60 mM β-glycerophosphate, pH 7.5, 0.2% NP-40, 2 mM dithiothreitol, and a protease inhibitor cocktail containing 1 mM phenylmethylsulfonyl fluoride, 0.5 mM 4-(2-aminoethyl)-benzene-sulfonyl fluoride, and 5 μg/ml each of aprotinin, pepstatin, and leupeptin. One milliliter of acid-washed glass beads was added, and tubes were vortexed for 4 min with intermittent cooling. Lysates were clarified by centrifugation at 14,000 rpm in a microfuge at 4°C. Protein concentrations were determined and equal amounts of lysates were used for immunoprecipitation. Primary antibody was used either as is, or after covalent coupling to protein A-Sepharose with dimethylpimelimidate (Harlow and Lane, 1988). After binding for 2 h at 4°C, antibody-coated beads (25 μl) were pelleted in a microfuge and washed three times with a buffer containing 25 mM Tris, pH 7.5, 150 mM NaCl, 0.2% Triton and twice with 25 mM Tris, pH 7.5. Beads were suspended in an equal volume of 2× SDS Laemmli buffer, boiled, and aliquots were resolved on SDS-polyacrylamide gels. Protein was transferred to nitrocellulose membranes and the blot was developed with the appropriate primary antibody, horseradish-peroxidase (HRP)-conjugated secondary antibodies, and chemiluminescent substrate (ECL; Amersham Pharmacia Biotech, Piscataway, NJ). In some instances, anti-HA-biotin/streptavidin-HRP (Boehringer Mannheim, Indianapolis, IN) or HRP-conjugated anti-myc (Santa Cruz Biotechnology, Santa Cruz, CA) were used to detect antigen.
Pulse-Chase Analysis
Wild-type (RJD 1521) or daq1Δ (RJD 1523) yeast cells harboring the UbV76-V-eΔK-βgal reporter plasmid (Johnson et al., 1992) were grown in synthetic selective medium containing 2% galactose and 2% raffinose to an OD600 of 1.0. Cultures were spun down and resuspended in methionine-free medium (10% of original volume) and pulse-labeled with 500 μCi of tran35S-label (ICN Pharmaceuticals, Costa Mesa, CA) for 5 min. Radiolabeled cells were pelleted and resuspended in fresh chase medium containing 1 mg/ml methionine and 0.5 mg/ml cycloheximide. Samples were withdrawn at the indicated time points, and immunoprecipitated with anti-βGal monoclonal antibody (Promega, Madison, WI) as described (Finley et al., 1994).
Purification of 26S, 20S, and 19S Proteasome Complexes
Tagged or untagged strains were grown to an optical density of 2.0, typically in 9 liters of synthetic medium containing 0.67% yeast nitrogen base minus amino acids, 2% dextrose, 0.5% casamino acids, and 20 mg/l adenine and tryptophan. Cells were harvested and washed once with ice cold water. The cell pellet was drop frozen in liquid N2, placed inside a mortar (which in turn was nestled inside an ice bucket filled with dry ice), and manually ground with the pestle to a fine powder (typically 15–30 min, depending on the amount being ground). The pellet being ground was kept frozen by scooping liquid N2 into the mortar every 2 min. The ground powder was collected in a 50-ml screw-cap tube and drop frozen in liquid N2.
The powder was thawed in one pellet volume of 50 mM Tris, pH 7.5, 150 mM NaCl, 10% glycerol, 5 mM MgCl2 (buffer A). ATP and 10× ATP-regenerating mix (ARS) (Verma et al., 1997) were added to final concentrations of 5 mM and 1×, respectively. Where indicated, ATP and ARS were substituted with 5 mM ATP-γ-S. The thawed cell lysate was centrifuged in an SS34 rotor (Sorvall, Newtown, CT) for 20 min at 17,000 rpm, and the pellet was discarded. A 13-ml aliquot of the supernatant (∼130 mg of protein), was supplemented again with 5 mM ATP (or ATP-γ-S) and 1× ARS and was mixed with 300 μl of anti-Flag M2 agarose beads (Sigma, St. Louis, MO) for 90 min on a rotating wheel at 5°C. The beads were then collected, transferred to 2-ml microfuge tubes, and washed with 50 volumes total of buffer A containing 2 mM ATP plus 0.2% Triton. The beads were next washed twice with buffer A containing 2 mM ATP, and specifically bound proteins were eluted for 3 h at 5°C with three bead volumes of elution buffer containing 25 mM Tris, pH 7.5, 150 mM NaCl, 15% glycerol, 5 mM MgCl2, 2 mM ATP, and 100 μg/ml Flag peptide. Typically, the yield was ∼250 μg of purified 26S from 130 mg of lysate.
To purify 20S proteasomes, the entire procedure described above was carried out in the absence of ATP and 1× ARS. To purify 19S caps, the entire protocol described above was carried out using the RPT1FH strain. Purification of the 19S cap was carried out in the absence or presence of ATP and 1× ARS, or in the presence of ATP-γ-S.
Mass Spectrometry
Proteasome samples (20 μg at ∼375 μg/ml) were exchanged into 8 M urea/50 mM Tris, pH 8.5 (by dialysis or gel filtration), and digested over a period of 4 h at 37°C by using 0.3 μg of Endoproteinase Lys-C (Boehringer Mannheim). After the initial digestion with Lys-C, the sample was diluted 4-fold with 50 mM Tris, pH 8.5, and then digested again at 37°C with 0.2 μg of trypsin (Boehringer Mannheim) overnight.
The entire digested proteasome complex was loaded onto a nano-LC ion source as described (Gatlin et al., 1998), which was attached to a Finnigan LCQ ion trap mass spectrometer. An HP-1100 binary pump (Hewlett-Packard, Palo Alto, CA) was programmed to elute the peptides by ramping a linear gradient from 2 to 60% solvent B in 90 min. Solvent A consisted of 0.5% acetic acid and solvent B consisted of 80:20 acetonitrile/water containing 0.5% acetic acid. The flow rate at the tip of the needle was set to 150 nl/min by programming the pump and use of a split line. The mass spectrometer cycled through four scans as the gradient progressed. The first was a full mass scan followed by three tandem mass scans of the three most intense ions. A dynamic exclusion list was used to limit collection of tandem mass spectra for peptides that eluted over a long period of time. All tandem mass spectra were searched by using the SEQUEST program against the yeast ORF database obtained from Stanford University. Each high-scoring peptide sequence was manually compared with the corresponding tandem mass spectrum to ensure the match was correct.
Native Gel Electrophoresis
Proteasome samples were resolved by nondenaturing PAGE as described in Glickman et al. (1998b). Briefly, 4% polyacrylamide gels were run in the cold until the sample dye (xylene cyanol) ran off. The gel was then incubated with the fluorescent peptidase substrate (Suc-LLVY-AMC) for 10 min at 30°C in a sealed bag. Proteasome bands were visualized upon exposure to a UV transilluminator and photographed with a Polaroid camera.
RESULTS
Affinity Purification of 26S Proteasomes, Plus 19S and 20S Subcomplexes
The chromosomal loci encoding several 26S proteasome subunits in budding yeast were modified to encode proteins tagged with an FH epitope. The goal of this effort was to identify subunits that would tolerate the addition of a carboxy-terminal peptide tag that could be used to affinity purify active proteasomes. Strains that expressed FH-tagged Pre1 or FH-tagged Rpt1 exhibited doubling times that were similar to that of the untagged parent strain. Thus, these strains were selected for further analysis. Pre1 is an α subunit of the 20S core, whereas Rpt1 is an ATPase subunit of the 19S regulatory particle.
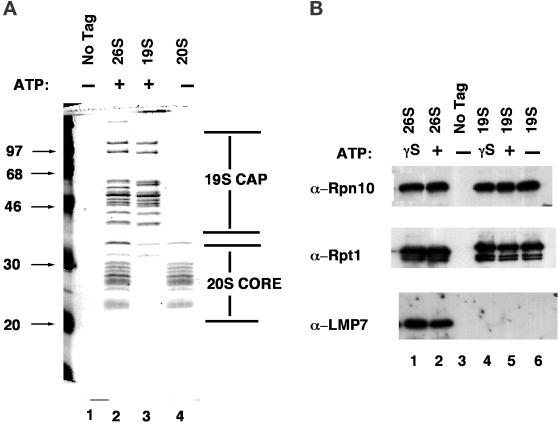
To test whether the FH epitope could be used as an affinity handle to purify proteasomes in a single step, ATP-supplemented lysates of PRE1FH cells were incubated with an anti-Flag antibody affinity resin, unbound proteins were washed away, and specifically bound proteins were eluted with Flag peptide. The results of one such purification are shown in Figure 1A, lane 2. By a variety of criteria, the single-step affinity method yielded highly purified 26S proteasomes. First, no polypeptides were detected in a parallel affinity purification from an untagged strain (lane 1). Second, the polypeptide pattern of the Flag eluate was reminiscent of the pattern observed for budding yeast 26S proteasomes purified by multistep conventional chromatography (Glickman et al., 1998b). Third, mass spectrometry-based sequencing of tryptic peptides derived from the Flag eluate revealed that the sample was comprised almost entirely of known 26S subunits (see below and Table 2). Fourth, the polypeptide pattern observed for 26S proteasomes was a close summation of that observed for 20S (lane 4) and 19S (lane 3) subcomplexes.
Figure 1.
Affinity purification of 26S proteasomes, plus 19S and 20S subcomplexes. (A) Extracts of yeast strains expressing Pre1FH (RJD 1144, lanes 2 and 4) or Rpt1FH (RJD 1171, lane 3) were prepared and bound to anti-Flag M2 resin as described in MATERIALS AND METHODS. Bound proteins were eluted with 100 μg/ml Flag peptide and analyzed by SDS-PAGE and Coomassie blue staining. The entire purification from the PRE1FH strain was carried out either in the presence (lane 2) or absence (lane 4) of ATP. The purification from the RPT1FH strain was carried out in the presence of ATP (lane 3). Lane 1 depicts a control purification performed with extract from an untagged strain (RJD 487) in the absence of ATP. (B) 26S proteasomes, 19S caps, and mock samples were prepared as described in A from RJD 1144, RJD 1171, and RJD 487, respectively, either in the absence of ATP or in the presence of ATP or ATP-γ-S, as indicated in the figure. Purified samples were resolved by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with polyclonal antisera against the mammalian 20S subunit LMP7, which cross-reacts with budding yeast Pre2/Doa3, and the budding yeast 19S subunits Rpn10 and Rpt1.
Table 2.
Mass spectrometric analysis of 26S proteasome prepared in the presence of ATP
| Chromosomal locus | Gene name | Peptides |
|---|---|---|
| 20S core subunits | ||
| YER012W | PRE1 | 1 |
| YPR103W | PRE2 | 1 |
| YJL001W | PRE3 | 3 |
| YFR050C | PRE4 | 3 |
| YMR314W | PRE5 | 6 |
| YOL038W | PRE6 | 6 |
| YBL041W | PRE7 | 2 |
| YGR135W | PRE9 | 5 |
| YOR362C | PRE10 | 3 |
| YGR253 | PUP2 | 2 |
| YER094C | PUP3 | 2 |
| YGL011C | SCL1 | 5 |
| 19S cap subunits | ||
| YHR027C | RPN1; HRD2; NAS1 | 10 |
| YIL075C | RPN2; SEN3 | 3 |
| YER021W | RPN3; SUN2 | 2 |
| YDL147W | RPN5; NAS5 | 3 |
| YDL097C | RPN6; NAS4 | 2 |
| YPR108W | RPN7 | 4 |
| YOR261C | RPN8 | 2 |
| YDR427W | RPN9 | 4 |
| YFR004W | RPN11 | 2 |
| YFR052W | RPN12; NIN1 | 1 |
| YKL145W | RPT1; CIM5 | 3 |
| YDL007W | RPT2; YTA5 | 2 |
| YDR394W | RPT3, YTA2 | 2 |
| YOR117W | RPT5; YTA1 | 2 |
| YGL048C | RPT6; SUG1; CIM3 | 2 |
| Other proteins | ||
| YBR193 | MED8 | 1 |
| YLR421C | DAQ1 | 1 |
Based on the second and fourth criteria mentioned above, the proteins migrating in the 20–30-kDa range comprise the 20S core, whereas the higher-molecular-weight proteins comprise the 19S regulatory particle. The intensities of the Coomassie-stained bands in 26S preparations suggest that the two subparticles were recovered in approximately equal amounts. Native gel electrophoresis (see below) revealed that both singly and doubly capped proteasome complexes were obtained. Immunostaining with antisera to 19S and 20S components confirmed that both subcomplexes were present in the 26S preparation (Figure 1B, lanes 1 and 2), which was further confirmed by mass spectrometry (Table 2).
In contrast to the results obtained with the PRE1FH strain, we were unable to recover intact 26S proteasomes from RPT1FHcells, even though the strain was viable. No 20S core subunits were detected in the Rpt1FH preparation by either Coomassie blue staining (Figure 1A, lane 3), immunoblotting with anti-LMP7 (Doa3) antibody (Figure 1B, bottom, lanes 4–6), or mass spectrometry-based peptide sequencing (Table 3). Thus, the tag on Rpt1FH destabilized the association of the 19S and 20S subcomplexes.
Table 3.
Mass spectrometric analysis of 19S cap prepared in the absence of ATP
| Chromosomal locus | Gene name | Peptides | MW | Codon bias | Comments |
|---|---|---|---|---|---|
| Additional 19S cap subunitsa | |||||
| YHR200W | RPN10; MCB1; | 3 | 29.7 | 0.07 | Δ: Vb |
| YOR259C | RPT4; SUG2 | 4 | 49.4 | 0.26 | Δ: Lb |
| YLR421C | DAQ1; RPN13 | 2 | 17.9 | 0.15 | Δ: V |
| 19S-interacting proteins (PIPs) identified in two of two analyses | |||||
| YGR232W | NAS6 | 4 | 25.6 | 0.13 | Δ = V; Putative cap subunit |
| YNL209W | SSB2 | 7 | 69.6 | 0.88 | Chaperone; Δ: V |
| YLL024C | SSA2; HSP70 | 7 | 69.6 | 0.89 | Chaperone; Δ: V |
| YAL005C | SSA1; HSP70 | 5 | 69.6 | 0.83 | Chaperone; Δ: L |
| YER103W | SSA4 | 3 | 69.6 | 0.24 | Chaperone; Δ: V |
| YMR186W | HSC82 | 3 | 81.2 | 0.69 | Chaperone; Δ: V |
| YFR010W | UBP6 | 6 | 57.1 | 0.19 | Deubiquitinating enzyme; Δ: V |
| YHR174W | ENO1/2 | 4 | 46.8 | 0.93 | Enolase; Δ: V |
| YBR279W | PAF1 | 2 | 51.8 | 0.15 | Transcription; Δ: V |
| YBR118W | TEF1/2 | 2 | 49.9 | 0.92 | Translation elongation factor E1α; Δ: L |
| YGR193C | TDH2/3 | 2 | 35.6 | 0.98 | Glyceraldehyde 3-phosphate dehydrogenase; Δ: V |
| YMR116C | BEL1 | 2 | 34.8 | 0.87 | Protein synthesis; Δ: V |
| YNR016C | ACC1 | 1 | 250.3 | 0.44 | |
| YOR123C | LEO1 | 1 | 53.8 | 0.08 | Unknown; Δ: V |
| YDL082W | RPL13A | 1 | 22.6 | 0.76 | Ribosomal protein L13 Δ: V |
| YGL224W | RTF1 | 8 | 65.8 | 0.16 | Transcription; Δ: V |
| YOL145C | CTR9 | 5 | 124.6 | 0.16 | Transcription; Δ: V |
| YPL110C | ORF | 3 | 138 | 0.03 | PHO81-like |
| YLR058C | SHM2 | 2 | 52.2 | 0.7 | Serine hydroxymethyl transferase; Δ: V |
| YGL026C | TRP5 | 2 | 76.6 | 0.45 | Tryp synthase, last step; Δ: V |
| YJL130C | URA2 | 2 | 254 | 0.36 | Pyrimidine biosynthesis; Δ: V |
| YDL055C | PSA1 | 2 | 39.5 | 0.7 | Mannose-1-phosphate guanyltransferase; periodic mRNA-like CLN1,2; Δ: L |
| YBR031W/YDR012W | RPL4A/4B | 2 | 39 | 0.89 | Ribosomal subunit |
| YIL018W | RPL2B | 2 | 27.4 | 0.84 | Ribosomal subunit; Δ: V |
| YGL147C | RPL9A | 2 | 21.5 | 0.8 | Ribosomal subunit |
| YEL054C | RPL12A/12B | 2 | 15.8 | 0.73/0.85 | |
| YDR224C | HTB1/2 | 2 | 14.2 | 0.7 | Histone; Δ: V |
| YMR247C | ORF | 4 | 180 | ? | Unknown function |
| YPL131W | RPL5 | 3 | 33.5 | 0.9 | Ribosomal subunit; Δ: L |
| YLL054C | ORF | 1 | 89 | 0.04 | Zinc finger transcription factor |
| YER007W | PAC2 | 1 | 59 | 0.04 | Tubulin-specific chaperone; Δ: V |
| YMR076C | PDS5 | 1 | 147 | 0.09 | Mitosis |
| YBR156C | SLI15 | 1 | 79 | 0.02 | Mitotic spindle protein; Δ: L |
| YLR276C | DBP9 | 1 | 68 | 0.13 | DEAD-box RNA helicase |
| YCL009C | ILV6 | 1 | 31 | 0.4 | Amino acid metabolism; mitochondrial; Δ: V |
| YDL126C | CDC48 | 1 | 92 | 0.4 | |
| YDR099W/YER177W | BMH2/BMH1 | 1 | 31 | 0.47 | Mammalian 14-3-3 homolog; Δ: V |
| YNL016W | PUB1 | 1 | 51 | 0.38 | PolyA-binding; Δ: V |
All 19S cap subunits listed in Table 2 were recovered. No 20S subunits were recovered.
Δ: L, Null mutant is lethal; Δ: V, Null mutant is viable.
Proteins that yielded a single peptide in one of two mass spectrometric analyses were divided into two categories. 1) low codon bias (≤0.5) proteins that are listed in the Table and 2) high codon bias (≥0.5) proteins that include the following: YKL152C, YLR044C, YOL086C, YGR209C, YLR150W, YJL138C, YCR031W, YDL075W, YDL081C, YDL082W, YDR418W, YHL001W, YHR203C, YJR123W, YLR441C, YMR230W, and YNL178W.
Stable interaction between the 19S and 20S subcomplexes of the 26S proteasome is dependent upon ATP. Accordingly, anti-Flag affinity purifications conducted with PRE1FH lysate in the absence of ATP yielded only 20S core subunits (Figure 1A, lane 4). Thus, by manipulating ATP levels, the PRE1FH and RPTFH strains could be used to isolate intact 26S proteasomes, as well as individual 19S and 20S subcomplexes.
Functional Characterization of Affinity-purified 26S Proteasomes, 19S Caps, and 20S Cores
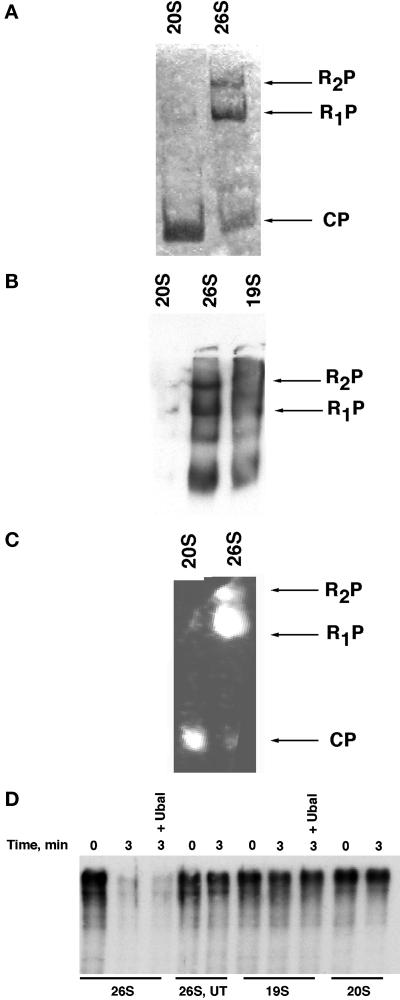
To evaluate the assembly state of the purified proteasome complexes and subcomplexes, we subjected the 26S, 19S, and 20S preparations to native PAGE followed by staining with Coomassie blue (Figure 2A) and immunoblotting with anti-Rpt1 antiserum (Figure 2B). Both methods revealed singly (R1P) and doubly (R2P) capped proteasomes in the 26S preparation, as reported in a prior study (Glickman et al., 1998b). In contrast to 20S and 26S proteasomes, affinity-purified 19S regulatory particles did not resolve as a discrete species on native polyacrylamide gels (Figure 2B; our unpublished data).
Figure 2.
Functional analysis of affinity-purified 26S proteasomes, plus 19S and 20S subcomplexes (A and B) 26S, 19S, and 20S preparations were electrophoresed on nondenaturing polyacrylamide gels and analyzed by Coomassie blue staining directly (A), or transferred to nitrocellulose and immunoblotted with anti-Rpt1 serum (B). CP refers to the 20S core particle, whereas R1P and R2P refer to core particles decorated with either one or two regulatory caps, respectively. Note that no 20S could be detected by Ponceau S staining after transfer to nitrocellulose membranes. (C) Peptidase activity of proteasomal preparations toward the fluorogenic peptide N-succinyl-Leu-Leu-val-Tyr 7-amido-4-methylcoumarin was evaluated by incubating a native gel containing fractionated samples for 10 min at 30°C in the presence of 100 μM substrate and 1 mM ATP. The fluorescent bands were visualized by exposure to UV light (360 nm). (D) Trimeric complex of Sic1/Cdc28/Clb5 purified from insect cells was phosphorylated by immobilized G1 Cdk complexes and subsequently ubiquitinated by immobilized tetrameric SCFCdc4 ubiquitin ligase in the presence of E1, E2 (Cdc34), ATP, and ubiquitin (Seol et al., 1999). The soluble fraction containing ubiquitinated Sic1 (500 nM) was supplemented with 100 nM 26S, 19S, or 20S complexes and incubated at 30°C for 0–3 min in the presence of an ATP-regenerating system. Reactions were terminated by the addition of SDS Laemmli buffer, resolved by SDS-PAGE, and evaluated by immunoblotting with anti-Sic1 polyclonal antibodies. UT refers to Flag eluate from an untagged strain; reactions marked + Ubal contained 2 μM ubiquitin aldehyde (Calbiochem, La Jolla, CA).
To measure the activity of the affinity-purified complexes, two different assays were carried out. First, incubation of native PAGE-fractionated complexes with a fluorogenic peptide reporter for the chymotryptic activity of the proteasome (Glickman et al., 1998b) revealed that both the 20S and 26S preparations contained peptidase activity (Figure 2C). Next, we tested the ability of 26S proteasomes, 19S caps, and 20S cores to degrade a physiological, ubiquitinated substrate, the Cdk inhibitor Sic1 (Seol et al., 1999). Although intact 26S proteasomes rapidly degraded ubiquitinated Sic1, the 19S and 20S subcomplexes were inactive (Figure 2D). The loss of ubiquitinated Sic1 was not due to deubiquitinating (DUB) enzyme activity because it was not prevented by the DUB inhibitor ubiquitin aldehyde (Wilkinson and Hochstrasser, 1998). In addition, no Sic1 was regenerated (our unpublished data). Degradation of ubiquitinated Sic1 by intact 26S proteasomes was specific, in that it depended on ATP, and unmodified Sic1 was not degraded (Verma and Deshaies, unpublished data). Taken together, these observations indicate that the affinity-purified 26S proteasomes had potent proteolytic activity toward a physiological substrate, and that the 20S and 19S subcomplexes were devoid of contaminating 26S proteasome activity.
Nucleotide Hydrolysis Regulates the Association of Multiple Proteins with the 19S Cap
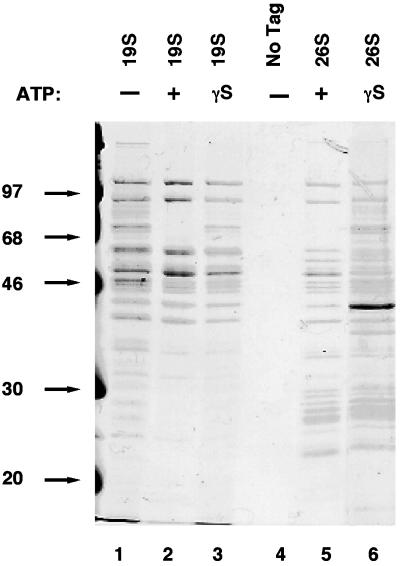
Interestingly, 19S caps prepared from the RPT1FH strain in the absence of ATP yielded a more complicated protein profile than those purified in the presence of ATP (Figure 3, cf. lanes 1 and 2; this effect is illustrated most clearly by the doublet of bands at 70 kDa). Although many of these additional proteins stained more weakly than the bona fide 19S subunits, and are difficult to see in Figure 3, the nucleotide effect was very reproducible. We refer to the ATP-sensitive 19S-associated proteins as PIPs. To distinguish whether nucleotide binding or hydrolysis blocked the coprecipitation of PIPs with 19S complexes, we affinity purified 19S regulatory particles from RPT1FH cells in the presence of the poorly hydrolyzed ATP-γ-S (lane 3). The 19S (ATP-γ-S) and 19S (−ATP) preparations exhibited similar protein profiles (e.g., the doublet of bands at 70 kDa).
Figure 3.
Effect of ATP and ATP-γ-S on composition of proteasome preparations. The entire purification of 19S caps from the RPT1FH strain (RJD 1171) was carried out as described in the legend to Figure 1A in the absence of ATP (lane 1) or in the presence of either ATP (lane 2), or ATP-γ-S (lane 3). Similarly, purification of 26S proteasomes from the PRE1FH strain (RJD 1144) was performed in the presence of ATP (lane 5) or in the presence of ATP-γ-S (lane 6). As a mock control, purification was carried out with extracts prepared from untagged RJD 487 cells (lane 4).
The ATP-modulated detection of PIPs was not restricted to 19S complexes because similar results were obtained when 26S proteasomes were purified in the presence of ATP (lane 5, PIPs absent) or ATP-γ-S (lane 6, PIPs present). Interestingly, the recovery of intact 26S proteasomes in lane 6 indicates that stable docking of 19S caps on the 20S core did not require ATP hydrolysis. However, in the presence of the nonhydrolyzable nucleotide analog 5′-adenylyl imidodiphosphate, the 26S holoenzyme was unstable and Rpn1 and Rpn2 were not recovered in stoichiometric amounts (our unpublished data).
Ubiquitin Ligases SCF and APC Are PIPs
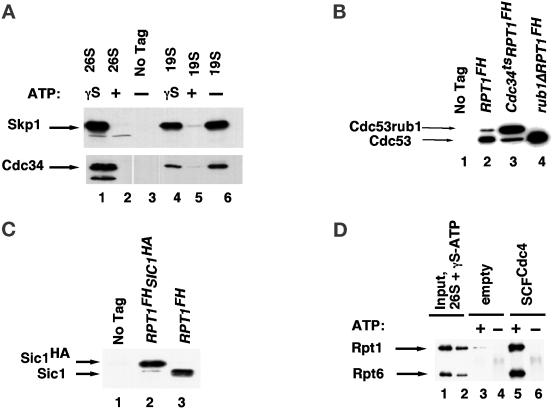
In our initial attempts to identify the PIPs whose binding to the 19S cap was modulated by nucleotide hydrolysis, we took a “candidate protein” approach. In the course of studying the tetrameric E3 ubiquitin ligase SCFCdc4 (Deshaies, 1999), we noticed that it remained tightly bound to its ubiquitinated substrate, Sic1 (our unpublished observations). This observation suggested that the proteasome may recruit ubiquitinated Sic1 that remains bound to SCF, and thus the subunits of SCF might be PIPs. To address this possibility, we evaluated whether components of the SCF pathway could be coimmunoprecipitated with proteasomes. Although the ubiquitin-conjugating enzyme Cdc34 (E2) and the Skp1 and Cdc53 subunits of SCF were bound to 19S regulatory particles prepared in the absence of ATP or presence of ATP-γ-S, these proteins did not coprecipitate with 19S particles in the presence of ATP (Figure 4, A and B; unpublished data). Similar results were observed with 26S proteasomes (Figure 4A).
Figure 4.
SCFCdc4 ubiquitin ligase interacts with the proteasome in a nucleotide-modulated manner. (A) 26S proteasomes (from RJD 1144) and 19S caps (from RJD 1171), prepared in the absence of nucleotide or presence of ATP or ATP-γ-S, as indicated in the figure, were resolved by SDS-PAGE, transferred to nitrocellulose, and immunoblotted for the SCF subunit Skp1 and the cognate E2 Cdc34 by using affinity-purified polyclonal antibodies. As a mock control, purification was carried out with extracts from untagged RJD 487 cells (no tag). (B) 19S caps (−ATP) were prepared from untagged (RJD 487, lane 1), RPT1FH (RJD 1171, lane 2), RPT1FHcdc34ts arrested at 37°C for 6 h (RJD 1281, lane 3), and RPT1FHrub1Δ (RJD 1379, lane 4) cells. Aliquots were resolved by SDS-PAGE, transferred to nitrocellulose, and probed with a polyclonal antibody to the SCF subunit Cdc53. Cdc53rub1 refers to Cdc53 conjugated with the ubiquitin-like Rub1 protein. (C) 19S caps (−ATP) were prepared from untagged (RJD 487, lane 1), RPT1FH SIC1HAcdc34-2 (RJD 1294, lane 2), or RPT1FH (RJD 1171, lane 3) cells. Aliquots were resolved by SDS-PAGE and immunoblotted with polyclonal antibody to Sic1. (D) 26S Proteasomes purified on anti-Flag resin in the presence of ATP were washed free of ATP before being eluted with Flag peptide in the presence of 2 mM ATP-γ-S. Tetrameric SCFCdc4 tagged with a polyoma epitope on the Cdc4 subunit was produced in baculovirus cells and retrieved on antipolyoma resin as described in Seol et al. (1999). Eluted 26S proteasomes (10 μg) were mixed with naked (lanes 3 and 4) or ∼0.5 μg of SCF-coated (lanes 5 and 6) antipolyoma beads in the presence or absence of 2 mM ATP, as indicated. After incubation, polyoma beads were washed and evaluated for their content of 26S proteasome by immunoblotting with anti-Rpt1 and anti-Rpt6 sera. Input lanes 1 and 2 contain 14 and 5% respectively, and bound lanes contain 33% of the material from a single binding reaction.
In addition to SCF subunits, the SCF substrate Sic1 was also coimmunoprecipitated with 19S caps (Figure 4C). Intriguingly, the Sic1 detected in 19S immunoprecipitates was not ubiquitinated, but it remains unclear whether its association with the proteasome was mediated by SCF or by other proteins. To address the converse question of whether the interaction of SCF with proteasomes was mediated by multiubiquitinated substrates bound to SCF, we prepared proteasomes from cdc34ts mutant cells. Cdc34 is the cognate E2 for SCF, and at the nonpermissive temperature for cdc34ts, SCF substrates are not ubiquitinated (Willems et al., 1996). To our surprise, the association of Cdc53 with 19S caps was not diminished in cdc34ts cells held at the restrictive temperature (Figure 4B, lane 3; note that the Rub1-modified form of Cdc53 accumulates preferentially in cdc34ts cells [Lammer et al., 1998]).
If the interaction of SCF with the proteasome was not bridged by ubiquitinated substrates, perhaps SCF was held to the proteasome by the ubiquitin-like Rub1 protein that is attached to Cdc53 (Lammer et al., 1998), much as the ubiquitin-like N terminus of Rad23 mediates association of Rad23 with the proteasome (Schauber et al., 1998). To test this possibility, we performed affinity purifications with strains lacking RUB1. As shown in Figure 4B (lane 4), unmodified Cdc53 could still be immunoprecipitated with the 19S cap. Taken together, these data suggest that SCF binds to the proteasome independently of either Rub1 or the ubiquitination state of SCF substrates.
If 26S proteasomes contain a docking site for SCF, it should be possible to reconstitute SCF–proteasome interaction with purified proteasomes and recombinant SCF. To test this possibility, heterotetrameric SCFCdc4 complex was expressed in insect cells and immunoaffinity purified by using a polyoma epitope tag on Cdc4 (Seol et al., 1999). SCF immobilized on antipolyoma resin was incubated with purified 26S proteasomes prepared in the presence of ATP, but eluted in 2 mM ATP-γ-S. Binding was done in the presence or absence of an additional 2 mM ATP. Association of 26S proteasomes with immobilized SCF was evaluated by immunoblotting the washed beads with α-Rpt1 and α-Rpt6 antibodies. As shown in Figure 4D, 26S proteasomes bound immobilized SCF in the presence but not in the absence of added ATP. This binding was not competed by the addition of 5 μM free tetraubiquitin chains (our unpublished data). In contrast to our finding that SCF was coimmunoprecipitated with proteasomes from cell extracts supplemented with ATP-γ-S, reconstitution of SCF–26S proteasome interaction in vitro required ATP in addition to ATP-γ-S. The significance of these observations is considered further in the DISCUSSION.
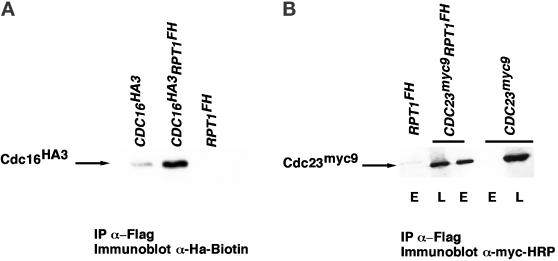
To determine whether other ubiquitin ligases besides SCF also bind to the 26S proteasome, we tested whether epitope-tagged subunits of the APC (Zachariae and Nasmyth, 1999) could be coimmunoprecipitated with Rpt1FH. As shown in Figure 5, both Cdc16HA3 and Cdc23myc9 were specifically recovered in Rpt1FH immunoprecipitates prepared in the absence of ATP.
Figure 5.
Association of subunits of the APC ubiquitin ligase with the proteasome. Extracts from strains with the indicated genotypes were immunoprecipitated with anti-Flag resin in the absence of ATP. Bound proteins were eluted from the resin with either SDS (A) or flag peptide (B). Eluates were resolved by SDS-PAGE and immunoblottted with biotinylated monoclonal antibody to the HA epitope (A) or HRP-conjugated monoclonal antibody against the myc epitope. In B, L refers to crude extract used for immunoprecipitation (“load,” 0.2% of the bound), and E refers to the eluate (4% of total).
Characterization of Purified Proteasomes by Mass Spectrometry
The preceding experiments confirmed that nucleotide hydrolysis regulates the association of multiple proteins with the 19S cap, including subunits of the SCF ubiquitin ligase. These observations suggested that systematic identification of PIPs might reveal novel ubiquitin pathway components, as well as substrates and regulators of the 26S proteasome. To characterize rapidly the polypeptide composition of proteasomes purified in the presence or absence of ATP and various nucleotide analogs, we sought a method that would allow us to query the protein composition of an entire preparation in a single step. Direct analysis of large protein complexes by mass spectrometry (DALPC) (Link et al., 1999) has been successfully used to identify the subunits of purified ribosomes. In this approach, a purified, intact protein complex is directly digested with proteases (without prior fractionation into individual polypeptides by SDS-PAGE) to yield peptides, and the peptides are fractionated by high-pressure liquid chromatography before being introduced by an electrospray interface into an on-line triple-quadrupole mass spectrometer. This procedure allows mass spectra to be collected from a large number of high-pressure liquid chromatography–fractionated peptides in a short period of time.
To evaluate the potential of DALPC, we first applied this method to 26S proteasomes prepared in the presence of ATP. Remarkably, in a single round of analysis, we recovered 85 peptides that identified all known 26S subunits except for Rpn10, Rpt4, Pre8, and Pup1 (Table 2). Moreover, all but two peptides were derived from known subunits of the proteasome, which implies that the affinity-purified 26S proteasomes were ∼97% pure. Of the two nonproteasomal proteins identified, one (Ylr421c) is a 17.9-kDa protein of unknown function that we demonstrate to be a new subunit of budding yeast proteasomes (see below). Thus, this experiment convincingly validated the specificity and reliability of DALPC as a method for characterizing the composition of a complex mixture of proteins.
In contrast to the results obtained by applying DALPC to 26S proteasomes prepared in the presence of ATP, the protein composition of the 19S cap prepared in the absence of ATP was much more complex (Table 3), as expected from the Coomassie blue-staining profile (Figure 3, lane 1). As can be seen in Table 3, which summarizes data from two independent analyses, all of the 19S subunits identified by Glickman et al. (1998a) by using conventional purification followed by SDS-PAGE fractionation and sequence analysis of excised gel bands were also identified by the DALPC approach. In accordance with Glickman and coworkers, we also identified Rpn9. Although this protein has not been found in the mammalian proteasome, clear orthologs exist in sequence databases. In addition, we also detected Ylr421c (which was also found in the 26S preparation; Table 2). Coimmunoprecipitation experiments (see below) revealed that Ylr421c (here referred to as Daq1) is a stoichiometric component of the 19S cap.
Taken together, the two DALPC analyses conducted with 19S caps purified in the absence of ATP revealed 71 unique proteins (Table 3). Of these, 19 corresponded to known 19S subunits, yielding 52 putative PIPs. Twenty-five of these remaining PIPs were excluded from further consideration because they were either ribosomal proteins or abundant glycolytic enzymes, which we presume to be nonspecific contaminants, leaving 27 culled PIPs. Eighteen members of the final group were identified in only one of the two analyses, whereas the remaining nine PIPs were identified in both. Although many PIPs were identified by only a single peptide, note that even in the analysis of 26S proteasomes purified in the presence of ATP, 15 of the 28 subunits that were identified yielded only one or two peptide sequences, whereas five subunits yielded 5–10 peptide sequences (Table 2). Thus, whereas proteins identified by multiple peptides are likely to be abundant components of the sample, the converse is not necessarily true. By analogy to genetic screens, the distribution of peptides recovered per protein identified suggests that our biochemical screen for PIPs is far from saturated, and that more PIPs can be identified by a larger-scale DALPC analysis of 19S (−ATP) caps.
Validation of Proteasome Binding by a Subset of the PIPs Identified by the DALPC Method
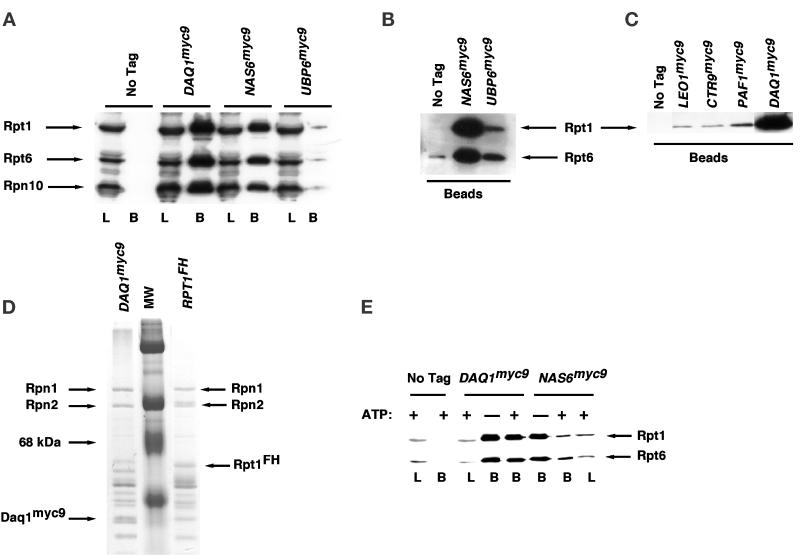
To validate the specificity of the DALPC approach, we sought to confirm the interaction of a subset of the PIPs with the 19S regulatory particle by coimmunoprecipitation experiments. A sample set of proteins from Table 3 was selected for further analysis. PIPs (excluding the abundant heat shock proteins) that were identified in both (2 of 2) mass spectrometric analyses were assigned the highest priority. Among the proteins observed in only one analysis, we focused on PIPs that most closely matched the following three criteria. First, we sought proteins encoded by mRNAs with a codon bias of ≤0.5 because this property is characteristic of relatively inabundant proteins. Such proteins are less likely to arise as spurious contaminants in biochemical purifications. Second, we sought proteins with high PEST scores because many natural substrates of the proteasome contain high-scoring PEST regions (Rechsteiner and Rogers, 1996). Third, we only considered proteins that were represented by at least two or more tryptic peptides. Based on these criteria, we selected Acc1, Nas6, Daq1, Leo1, Rtf1, Ctr9, Paf1, and Ubp6 for further analysis.
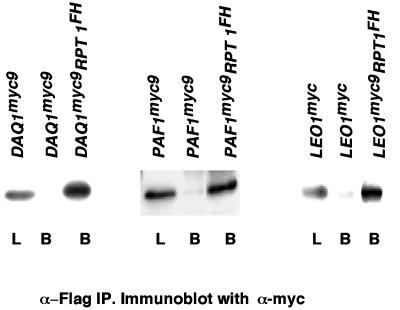
To investigate the chosen subset of proteins, a one-step method of tagging chromosomal loci at their 3′ ends with either nine copies of the myc epitope (myc9) or three copies of the HA epitope (HA3) was used (Seol et al., 1999; Seol and Deshaies, unpublished data). In all cases except for AccI, which was excluded from further analysis, epitope-tagged transformants were recovered that exhibited growth rates identical to the untagged wild-type parent strain. To evaluate the interaction of epitope-tagged PIPs with the 19S cap, cell lysates prepared from tagged transformants were immunoprecipitated in the absence of added ATP with anti-myc antibody resin. The immunoprecipitates were analyzed for 19S subunits by immunoblotting with antibodies against the 19S proteins Rpt1, Rpt6, and Rpn10. All of the proteins tested except for Rtf1myc9 (our unpublished data) specifically coimmunoprecipitated varying amounts of 19S complex (Figure 6, A–C). Because the levels of coprecipitated Rpt1 were extremely low in Leo1myc9, Paf1myc9, and Ctr9myc9 immunoprecipitates, the first two interactions were reexamined by immunoprecipitating Rpt1FH from doubly tagged strains and immunoblotting for the myc9-tagged PIPs. The results in Figure 7 confirm that Leo1myc9 and Paf1myc9 interacted specifically with the 19S regulatory particle. In summary, of seven PIPs selected for analysis, six were confirmed to coimmunoprecipitate specifically with 19S caps (Table 4). Thus, many of the 27 putative PIPs reported in Table 3 may prove to be authentic proteasome-interacting proteins.
Figure 6.
Coimmunoprecipitation of PIPs with the 19S regulatory particle. (A–C) A subset of the genes listed in Table 3 was modified to encode proteins tagged with the TEV2myc9 epitope, and extracts from the resulting tagged strains were prepared as described in MATERIALS AND METHODS. Tagged antigens were immunoprecipitated with anti-myc antibody covalently coupled to protein A, and immunoprecipitates were evaluated for their content of 19S subunits by SDS-PAGE followed by immunoblotting with polyclonal antibodies to Rpt1, Rpt6, and Rpn10, as indicated. In A, L refers to extract used for immunoprecipitation (3% of input) and B refers to the washed and SDS-eluted immunoprecipitates (Beads, 25% of bound). In B and C, only the washed immunoprecipitates (Beads) were evaluated. The experiment shown in B is similar to that shown in A, except that the blot was allowed to develop longer to reveal the specific coimmunoprecipitation of Rpt1 and Rpt6 with Ubp6myc9. (D) Equivalent amounts of extracts from DAQ1myc9 (RJD 1487) and RPT1FH (RJD 1171) were immunoprecipitated in the absence of ATP by using α-myc and α-flag beads, respectively. Equal aliquots of each immunoprecipitate were visualized by Coomassie blue staining. Rpn1 and Rpn2 are highlighted to indicate that equivalent amounts of bona fide 19S subunits are recovered in Daq1myc9 and Rpt1FH immunoprecipitates. (E) Extracts from DAQ1myc9, NAS6myc9, and cells containing no tagged proteins were immunoprecipitated by using α-myc–coated beads in the presence and absence of ATP, as indicated in the figure. Aliquots were resolved on SDS-gels and immunoblotted with anti-Rpt1/Rpt6 polyclonal antibodies. L refers to 2% of the extract loaded for immunoprecipitation, and B refers to the washed and SDS-eluted immunoprecipitates (Beads, 33% of bound).
Figure 7.
Paf1 and Leo1 coimmunoprecipitate with 19S regulatory particles. Extracts were prepared from yeast strains that expressed Daq1myc9 (RJD 1487), Paf1myc9 (RJD 1486), and Leo1myc9 (RJD 1484) or from strains that coexpressed these tagged proteins along with Rpt1FH (RJD 1494, RJD1496, and RJD 1497, respectively). Extracts were immunoprecipitated with anti-Flag resin in the absence of ATP and immunoprecipitates were analyzed by immunoblotting with anti-myc monoclonal antibody. L refers to extract loaded for immunoprecipitation (3% of input), and B refers to the washed and SDS-eluted immunoprecipitates (Beads, 25% of bound).
Table 4.
Summary of analysis of PIPs
| Number of PIPs identified | 52 | See Table 3 |
| Number of PIPs epitope tagged | 8 | ACC1, UBP6, PAF1, LEO1, RTF1, CTR9 YGR232W, YLR421C |
| Number of tagged PIPs that were analyzed | 7a | UBP6, PAF1, LEO1, RTF1, CTR9 YGR232W, YLR421C |
| Number of functional, tagged PIPs that coimmunoprecipitate with the proteasome | 6b | UBP6, PAF1, LEO1, CTR9 YGR232W, YLR421C |
ACC1TEV2myc9 was nonfunctional.
We were unable to confirm specific interaction of RTF1TEV2myc9 with proteasome.
Among the PIPs, Daq1 was unique in that 19S subunits were recovered in equivalent amounts in α-Flag immunoprecipitates from RPT1FH strains and α-myc immunoprecipitates from DAQ1myc9 strains (Figure 6D). However, as was observed for Rpt1FH, no 20S subunits were recovered in Daq1myc9 immunoprecipitates prepared in the presence of ATP (our unpublished data). Perhaps the myc9 tag on Daq1 interfered with binding of 20S to 19S because untagged Daq1 was identified by mass spectroscopy of 26S preparations (Table 2). Taken together, our data suggest that Daq1 is a novel, heretofore undetected subunit of the 19S regulatory particle. Daq1 is a 17.9-kDa protein with higher homologs in sequence databases. Global gene deletion analysis indicates that Daq1 is nonessential (Winzeler et al., 1999). Based on the data presented in Figures 6D and 8 (see below), we propose to rename Daq1 as Rpn13 to reflect its identity as a new subunit of the 19S cap.
Figure 8.
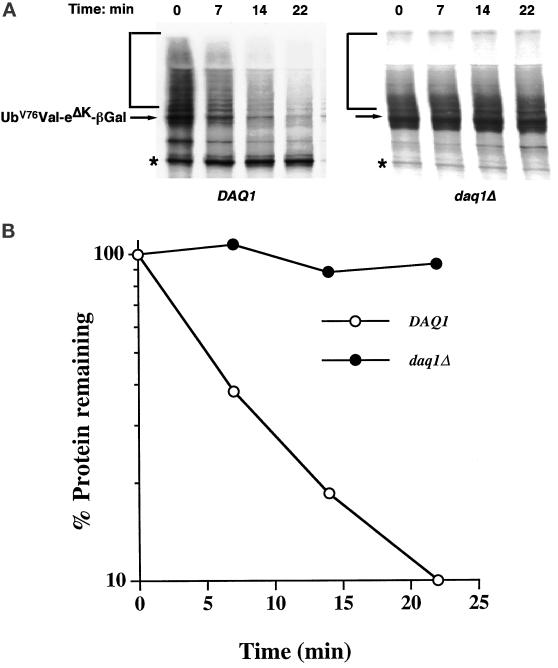
Stabilization of a ubiquitin fusion degradation pathway substrate in daq1Δ cells. Wild-type DAQ1 and daq1Δ mutant cells were transformed with a reporter plasmid expressing the UbV76-Val-eΔK-βgal fusion protein. Metabolic stability of the proteasome reporter substrate in the two strains was monitored by pulse-chase analysis as described in MATERIALS AND METHODS. (A) SDS-PAGE gel analysis of the 35S-labeled substrate protein (arrow). The bracket indicates the ubiquitinated forms of βgal reporter protein and the asterisk the position of the 90-kDa stable breakdown product of the reporter protein. (B). Quantitative analysis. The intensities of the βgal reporter protein bands in A were determined by PhosphorImager analysis and plotted as a funtion of time.
In contrast to Daq1, Ygr232w, which is homologous to the Nas6 subunit of the mammalian PA700 cap complex (Hori et al., 1998), was designated as a PIP for the following reasons. First, the recovery of 19S subunits in Nas6myc9 immunoprecipitates was much lower than the amounts recovered in α-Flag immunoprecipitates from RPT1FH extracts (our unpublished data). Second, unlike Daq1, Nas6 was not identified in mass spectrometric analysis of 26S proteasomes (Table 2). Finally, the association between Nas6 and Rpt1/Rpt6 was modulated by ATP (Figure 6E), as observed for the other PIPs such as SCF (Figure 4A), whereas the association between Daq1 and Rpt1/Rpt6 was not modulated by ATP (Figure 6E). As shown in Tables 2 and 3 and Figures 1 and 3, retention of bona fide subunits within the 19S cap was not influenced by ATP.
Impaired Protein Degradation in daq1Δ Mutants
Cells with mutations in 26S proteasome subunits typically exhibit defects in the degradation of specific ubiquitin pathway substrates. For example, even though Rpn10 is not essential, the rpn10Δ mutant is defective in the turnover of a ubiquitin fusion degradation (Ufd) pathway substrate (van Nocker et al., 1996). Because we have assigned Daq1 as a novel, bona-fide 19S subunit (see above), we explored the degradation of the Ufd pathway substrate UbV76-Val-eΔKβgal (Johnson et al., 1992) in daq1Δ cells. Pulse-chase analyses indicated that this substrate was degraded with a half-life of 7 min in wild-type cells, but was stable in the daq1Δ null mutant (Figure 8).
DISCUSSION
We have developed a simple one-step method to isolate from budding yeast cells highly purified, active 26S proteasomes, as well as 20S cores and 19S caps. The purity of the 26S proteasomes was confirmed by mass spectrometry and the functionality by monitoring the degradation of a physiological substrate: the ubiquitinated Cdk inhibitor Sic1. Because our method enabled purification of proteasomes in a single step, we were able to show that manipulation of nucleotide during the purification procedure influenced the association of a large number of proteins with the 19S regulatory particle, including the ubiquitin ligase SCF.
To identify novel PIPs that coimmunoprecipitated with the proteasome, we used a method, DALPC, that enables rapid compositional analysis of complex protein mixtures (Link et al., 1999). Besides yielding all of the known components of the 19S cap, the DALPC method revealed ∼24 putative PIPs that fall into four classes: 1) new proteasome subunits or proteins implicated in the ubiquitin pathway (such as SCF) or proteasome function; 2) chaperones; 3) regulatory proteins, including transcriptional regulators, that were not previously implicated as either targets or regulators of the proteasome; and 4) abundant proteins, including ribosomal subunits and glycolytic enzymes. Each class of PIPs is discussed in more detail below.
Class I PIPs: New Proteasome Subunits, Ubiquitin Ligases, and Other Proteins Implicated in the Ubiquitin Pathway or Proteasome Function
The first PIPs that were characterized, the ubiquitin ligases SCF and APC, were not detected by mass spectrometry, but were identified as authentic 19S-associated proteins by immunoprecipitation/Western blotting experiments. Interestingly, SCF remained bound to proteasomes in cdc34ts cells at the nonpermissive temperature. Under these conditions, ubiquitination of SCF-bound substrates is greatly diminished, suggesting that SCF was not held to the proteasome indirectly by tightly bound ubiquitinated substrates. Ubiquitin ligases may have other functions besides the recognition of substrate proteins and the catalysis of ubiquitin transfer from E2s to substrate. SCFCdc4 remains tightly bound to ubiquitinated Sic1 in vitro, which raises the possibility that SCFCdc4 contributes to the targeting of its substrates to the 26S proteasome. To address further the role of SCF in targeting ubiquitinated proteins to the 26S proteasome will require the generation of mutant SCF complexes that fail to bind 26S. The ubiquitin ligases Ubr1 and Ufd4 have also shown to be associated with the proteasome, although the effect of nucleotide on this association was not reported (Xie and Varshavsky, 2000).
Of the class I PIPs that were revealed by the DALPC method, our data suggest that Daq1 (Ylr421c) represents a novel subunit of the 19S cap. We have renamed Daq1 as Rpn13 to reflect its identity as a new 19S subunit. Four other class I PIPs, Ygr232w (Nas6), Rpn9, Ubp6, and Cdc48, were previously identified as components of the ubiquitin/proteasome system. Of these four, we have retested (and confirmed) only Nas6 and Ubp6 by immunoprecipitation/Western blotting. Nevertheless, given prior data (see below), all four proteins are likely to be authentic PIPs.
Nas6, which is found in both humans and yeast, contains five copies of the ankyrin repeat. Human Nas6 was identified as a subunit of the human 19S regulatory cap, also known as PA700 (Hori et al., 1998). The yeast Nas6 homolog (Ygr232w), which is dispensible for viability (Hori et al. 1998), was identified as an Rpt3-interacting protein in a two-hybrid screen inserted in list (Uetz et al., 2000). Rpn9 was identified as a yeast proteasomal subunit by Glickman et al. (1998a), but a putative mammalian ortholog has not been shown to be a subunit of the proteasome. Ubp6 is a DUB enzyme that has a ubiquitin-like N terminus. The expression of Ubp6 is increased 5-fold upon treatment of cells with DNA-damaging reagents (Jelinsky and Samson, 1999). It may therefore regulate protein turnover in response to physiological stress. Cdc48 interacts with the proteasome and is linked to the turnover of the immune response inhibitor IκB in mammalian cells (Dai et al., 1998) and is required for degradation of substrates of the Ufd pathway in yeast (Ghislain et al., 1996).
Class II PIPs: Chaperone Proteins
The DALPC analysis uncovered three distinct classes of chaperones: Ssa and Ssb members of the Hsc70 family, Hsc82 (the budding yeast homolog of Hsc90), and Pac2. Hsc70 has been previously implicated in protein turnover by the ubiquitin/proteasome pathway in several studies in vitro, but its detailed role in this process has not been resolved in vivo. Hsc70 interacts with the Escherichia coli DnaJ homolog Ydj1, which is required for the degradation of short-lived proteins (Lee et al., 1996; Yaglom et al., 1996). Because Hsc70 and Hsc90 suppress aggregation during protein folding by binding to segments of nonnative polypeptide, their association with the 26S proteasome may arise from kinetic partitioning of unfolded or misfolded target proteins to a proteolytic fate (Schneider et al., 1996). An alternative possibility is that Hsc70 and Hsc90 are recruited to the proteasome to help disassemble tightly folded ubiquitinated substrates before their destruction (Thrower et al., 2000). In animal cells, the ubiquitin-like Bag1 protein links Hsc70 to the proteasome (Luders et al., 2000). It will be interesting to see whether yeast cells possess a functionally equivalent bridging factor.
Class III PIPs: Miscellaneous Proteins with No Prior Link to the 26S Proteasome
The third category of PIPs detected by mass spectrometry included proteins implicated in transcriptional regulation (Ctr9, Paf1, Yll054, Med8, Rtf1), translation (Tef1, Bel1), cytoskeletal function (Sli5, Pac2), RNA metabolism (Dbp9, Pub1), cell division (Pds5), signal transduction (Bmh1), and metabolism (Shm2, Trp5, Ura2, Psa1, Ilv6, AccI). Of the three members of this class whose proteasomal association was evaluated by coimmunoprecipitation analysis, the transcriptional regulatory factors Paf1 and Ctr9 were shown to be authentic 19S-associated proteins, whereas the third, Rtf9, gave too high a background for a definite conclusion to be reached. Paf1 and Ctr9 are components of an RNA–polymerase II-associated complex that is thought to link protein kinase C to the transcription apparatus (Shi et al., 1997; Chang et al., 1999).
Class IV PIPs: Ribosomal Proteins and Glycolytic Enzymes
Our bias has been to exclude abundant proteins from further consideration on the principle that such proteins are more likely to be nonspecific contaminants. However, a number of observations point to a physiologically relevant interaction between proteasomes and ribosomes (and/or ribosome-associated proteins). Proteasome-mediated processing of p105 to mature nuclear factor-κB p50 occurs cotranslationally (Lin et al., 1998). In mammalian cells as much as 30% of newly synthesized proteins are degraded by the proteasome (Schubert et al., 2000). Some of these substrate proteins are relatively long-lived proteins that are improperly folded. Thus, ribosomal proteins and translation factors identified by DALPC may prove to be authentic PIPs.
Regulation of Proteasome–PIP Interactions by Nucleotide
SCF was stably coimmunoprecipitated with 19S regulatory caps either in the absence of added nucleotide or in the presence of ATP-γ-S but not in the presence of ATP. By analogy to the AAA ATPase katanin (Hartman and Vale, 1999), continuous ATP hydrolysis is predicted to drive cycles of binding and dissociation of target proteins, such that bound proteins are eventually lost during immunoprecipitation. In contrast to the results obtained in coimmunoprecipitation experiments with yeast extracts, ATP promoted efficient binding of immunopurified SCF to purified 26S proteasomes loaded with ATP-γ-S. Similar observations have been reported for BAG-1 (Luders et al., 2000). BAG-1 coimmunoprecipitates proteasomes from HeLa cells in the absence of nucleotide. Paradoxically, although immunopurified BAG-1–proteasome complexes are disassembled by ATP, complex formation between purified BAG-1 and proteasomes in vitro in HeLa cell extract requires ATP.
Why does ATP block the coprecipitation of SCF with 26S proteasomes from yeast extracts, but stimulate the association of SCF with ATP-γ-S–loaded 26S proteasomes in a purified system? We suggest the following speculative model to explain this puzzling result. The 19S ATPases (Rpt proteins) that are nucleotide-free or bound to nonhydrolyzable nucleotide may exist in distinct conformational states, as is the case for the AAA ATPase HslU (Bochtler et al., 2000). Nucleotide-free Rpt proteins might be analogous to the ADP-bound Hsp 70 (Bukau and Horwich, 1998), in that they bind target proteins stably. Upon exchanging ADP for ATP, the Rpt proteins are predicted to undergo a conformational change that prevents further binding of substrates. By analogy to Hsp70, the Rpt proteins loaded with ATP may exist in two conformational states: a weak ATP–Rpt complex that remains bound to target protein, which is rearranged to a tight ATP–Rpt complex with reduced affinity for target protein (Bukau and Horwich, 1998). Perhaps the γ-thio moiety of ATP-γ-S prevents the conversion from the “weakly bound ATP” state to the “tightly bound ATP” state, resulting in stable trapping of the target on 19S. In yeast extract, ATP is rapidly hydrolyzed and the 19S cap presumably exists in the ADP state, which results in the stable binding of targets. In contrast, purified proteasomes loaded with ATP-γ-S can retain previously bound proteins, but are unable to recruit additional proteins unless they cycle through a round of ATP hydrolysis.
Further genetic and biochemical analysis of the proteasome-interacting proteins reported here should shed more light on how the 26S proteasome is integrated into cellular physiology, and may provide insight into the role of the 19S cap in processes other than ubiquitin-dependent proteolysis, including DNA repair (Russell et al., 1999). The association of the 19S cap with the large number of proteins involved in transcription/translation reported here perhaps may relate to its homology to eIF3 and the signalosome in higher eukaryotes (Glickman et al., 1998a).
ACKNOWLEDGMENTS
We thank Craig Correll for antibodies to Cdc53 and Skp1, Kiran Madura for the rub1Δ yeast strain, Wade Harper for Sic1 baculovirus, Carl Mann for antibodies to Rpt1 and Rpt6, John Monaco for antibody to LMP7, Alex Varshavsky laboratory members and Cecile Pickart for tetraubiquitin chains, the Varshavsky laboratory for the ubiquitin fusion reporter plasmid, Zack Pitluck and David Gonda for antibody to Cdc34, Jae Hong Seol for the TEV2myc9-tagging cassette, and Joseph Walker and Richard Vierstra for antibody to Rpn10. We thank members of the laboratory, especially Wenying Shou, for their helpful comments. This work was supported by National Institutes of Health grant GM-52466 to R.J.D. R.J.D. was supported by grant DO-649/1-1 from the Deutsche Forschungsgemeinschaft. J.Y. was supported by the National Center for Research Resources. National Institutes of Health Yeast Biotechnology Resource Center grant RR-11823-03.
Footnotes
Corresponding author. E-mail address: deshaies@its.caltech.edu.
REFERENCES
- Bochtler M, Hartmann C, Song HK, Bourenkov GP, Bartunik HD, Huber R. The structures of HsIU and the ATP-dependent protease HsIU-HsIV. Nature. 2000;403:800–805. doi: 10.1038/35001629. [DOI] [PubMed] [Google Scholar]
- Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- Chang M, French-Cornay D, Fan HY, Klein H, Denis CL, Jaehning JA. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol Cell Biol. 1999;19:1056–1067. doi: 10.1128/mcb.19.2.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai RM, Chen E, Longo DL, Gorbea CM, Li CC. Involvement of valosin-containing protein, an ATPase co-purified with IκBα and 26 S proteasome, in ubiquitin-proteasome-mediated degradation of IκBα. J Biol Chem. 1998;273:3562–3573. doi: 10.1074/jbc.273.6.3562. [DOI] [PubMed] [Google Scholar]
- De Mot R, Nagy I, Walz J, Baumeister W. Proteasomes and other self-compartmentalizing proteases in prokaryotes. Trends Microbiol. 1999;7:88–92. doi: 10.1016/s0966-842x(98)01432-2. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ. SCF and RING H2/Cullin-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. doi: 10.1146/annurev.cellbio.15.1.435. [DOI] [PubMed] [Google Scholar]
- Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26S protease subunit that binds ubiquitin conjugates. J Biol Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- Finley D, Sadis S, Monia BP, Boucher P, Ecker DJ, Crooke ST, Chau V. Inhibition of proteolysis and cell-cycle progression in a multiubiquitination-deficient yeast mutant. Mol Cell Biol. 1994;14:5501–5509. doi: 10.1128/mcb.14.8.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatlin CL, Kleemann GR, Hays LG, Link AJ, Yates JR., III Protein identification at the low femtomole level from silver-stained gels using a new fritless electrospray interface for liquid chromatography-microspray and nanospray mass spectrometry. Anal Biochem. 1998;263:93–101. doi: 10.1006/abio.1998.2809. [DOI] [PubMed] [Google Scholar]
- Ghislain M, Dohmen RJ, Levy F, Varshavsky A. Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J. 1996;15:4884–4899. [PMC free article] [PubMed] [Google Scholar]
- Glickman M, Rubin D, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried V, Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998a;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Fried VA, Finley D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol Cell Biol. 1998b;18:3149–3162. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Hartman JJ, Vale RD. Microtubule disassembly by ATP-dependent oligomerization of the AAA enzyme katanin. Science. 1999;286:782–785. doi: 10.1126/science.286.5440.782. [DOI] [PubMed] [Google Scholar]
- Hori T, Kato S, Saeki M, DeMartino GN, Slaughter CA, Takeuchi J, Toh-e A, Tanaka K. cDNA cloning and functional analysis of p28 (Nas6p) and p40.5 (Nas7p), two novel regulatory subunits of the 26S proteasome. Gene. 1998;216:113–122. doi: 10.1016/s0378-1119(98)00309-6. [DOI] [PubMed] [Google Scholar]
- Jelinsky SA, Samson LD. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc Natl Acad Sci USA. 1999;96:1486–1491. doi: 10.1073/pnas.96.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES, Bartel B, Seufert W, Varshavsky A. Ubiquitin as a degradation signal. EMBO J. 1992;11:497–505. doi: 10.1002/j.1460-2075.1992.tb05080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser P, Moncollin V, Clarke DJ, Watson MH, Bertolaet BL, Reed SI, Bailly E. Cyclin-dependent kinase and Cks/Suc1 interact with the proteasome in yeast to control proteolysis of M-phase targets. Genes Dev. 1999;13:1190–1202. doi: 10.1101/gad.13.9.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammer D, Mathias N, Laplaza JM, Jiang W, Liu Y, Callis J, Goebl M, Estelle M. Modification of yeast Cdc53p by the ubiquitin-related protein Rub1p affects function of the SCFCdc4 complex. Genes Dev. 1998;12:914–926. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen CN, Finley D. Protein translocation channels in the proteasome and other proteases. Cell. 1997;91:431–434. doi: 10.1016/s0092-8674(00)80427-4. [DOI] [PubMed] [Google Scholar]
- Lee DH, Sherman MY, Goldberg AL. Involvement of the molecular chaperone Ydj1 in the ubiquitin-dependent degradation of short-lived and abnormal proteins in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4773–4781. doi: 10.1128/mcb.16.9.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, DeMartino GN, Greene WC. Cotranslational biogenesis of NF-kappaB p50 by the 26S proteasome. Cell. 1998;92:819–828. doi: 10.1016/s0092-8674(00)81409-9. [DOI] [PubMed] [Google Scholar]
- Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., III Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- Luders J, Demand J, Hohfeld J. The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J Biol Chem. 2000;275:4613–4617. doi: 10.1074/jbc.275.7.4613. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- Russell SJ, Reed SH, Huang W, Friedberg EC, Johnston SA. The 19S regulatory complex of the proteasome functions independently of proteolysis in nucleotide excision repair. Mol Cell. 1999;3:687–695. doi: 10.1016/s1097-2765(01)80001-0. [DOI] [PubMed] [Google Scholar]
- Schauber C, Chen L, Tongaonkar P, Vega I, Lambertson D, Potts W, Madura K. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature. 1998;391:715–718. doi: 10.1038/35661. [DOI] [PubMed] [Google Scholar]
- Schneider C, Sepp-Lorenzino L, Nimmesgern E, Ouerfelli O, Danishefsky S, Rosen N, Hartl FU. Pharmacologic shifting of a balance between protein refolding and degradation mediated by Hsp90. Proc Natl Acad Sci USA. 1996;93:14536–14541. doi: 10.1073/pnas.93.25.14536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- Seol J, Feldman R, Zachariae W, Shevchenko A, Correll C, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, Shevchenko A, Nasmyth K, Deshaies R. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Chang M, Wolf AJ, Chang CH, Frazer-Abel AA, Wade PA, Burton ZF, Jaehning JA. Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol Cell Biol. 1997;17:1160–1169. doi: 10.1128/mcb.17.3.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower J, Hoffman L, Rechsteiner M, Pickart C. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- van Nocker S, Sadis S, Rubin DM, Glickman M, Fu H, Coux O, Wefes I, Finley D, Vierstra RD. The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol Cell Biol. 1996;16:6020–6028. doi: 10.1128/mcb.16.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Chi Y, Deshaies RJ. Ubiquitination of cell cycle regulatory proteins in yeast extract. Methods Enzymol. 1997;283:366–376. doi: 10.1016/s0076-6879(97)83030-3. [DOI] [PubMed] [Google Scholar]
- Verma R, Deshaies RJ. A proteasome howdunit: the case of the missing signal. Cell. 2000;101:341–344. doi: 10.1016/s0092-8674(00)80843-0. [DOI] [PubMed] [Google Scholar]
- Voges D, Zwickl P, Baumeister W. The 26S proteasome, a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- Wang W, Chevray PM, Nathans D. Mammalian Sug1 and c-Fos in the nuclear 26S proteasome. Proc Natl Acad Sci USA. 1996;93:8236–8240. doi: 10.1073/pnas.93.16.8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KD, Hochstrasser M. The deubiquitinating enzymes. In: Peters J, Harris J, Finley D, editors. Ubiquitin and the Biology of the Cell. New York: Plenum Press; 1998. pp. 99–120. [Google Scholar]
- Willems AR, Lanker S, Patton EE, Craig KL, Nason TF, Kobayashi R, Wittenberg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M'Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Renuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Veronneau S, Voet M, Volckaert G, Ward TR, Wysocki W, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Xie Y, Varshavsky A. Physical association of ubiquitin ligases and the 26S proteasome [In Process Citation] Proc Natl Acad Sci USA. 2000;97:2497–2502. doi: 10.1073/pnas.060025497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaglom JA, Goldberg AL, Finley D, Sherman MY. The molecular chaperone Ydj1 is required for the p34CDC28-dependent phosphorylation of the cyclin Cln3 that signals its degradation. Mol Cell Biol. 1996;16:3679–3684. doi: 10.1128/mcb.16.7.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W, Nasmyth K. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]