Abstract
In time course experiments, bacterial community compositions were compared between a sulfidogenic and two nonsulfidogenic Cr(VI)-reducing consortia enriched from metal-contaminated sediments. The consortia were subjected to 0 and 0.85 mM or 1.35 mM Cr(VI), and Cr(VI) reduction, growth, and denaturing gradient gel electrophoresis profiles of PCR products of small-subunit (16S) ribosomal genes were compared. Results showed that although Cr(VI) was completely reduced by the three consortia, Cr(VI) inhibited cell growth, with sulfate-reducing bacteria being particularly sensitive to Cr(VI) toxicity relative to other bacteria in the consortia.
Chromium is a metal contaminant that, in nature, exists primarily as the soluble, highly toxic Cr(VI) anion and the less soluble, less toxic Cr(III) species. Many facultative and strictly anaerobic bacteria commonly found in soils and marine sediments are capable of reducing Cr(VI) to Cr(III) (10, 14, 20, 34, 35). Sulfate- and iron-reducing bacteria can indirectly reduce Cr(VI) via their anaerobic metabolic end products, hydrogen sulfide (HS−) and Fe(II), respectively (22, 25, 26, 29, 32). Some bacteria, such as Desulfotomaculum reducens and Pantoea (formerly Enterobacter) agglomerans strain SP-1, may use Cr(VI) as an alternate electron acceptor for anaerobic growth (10, 34), while others have cytochromes (15) (e.g., Desulfovibrio vulgaris) or specific enzymes (3, 17, 21, 33) (e.g., several Pseudomonas species, Escherichia coli, and Shewanella oneidensis strain MR-1) that reduce Cr(VI).
Despite the knowledge regarding Cr(VI) reduction mechanisms that pure cultures have provided, we still know relatively little about Cr(VI) reduction by natural microbial communities. Sulfate-reducing consortia have been studied previously and found to reduce up to 20 mM Cr(VI) (11). However, these samples were not degassed to remove hydrogen sulfide present in the medium prior to the addition of the Cr(VI), as 15% of the Cr(VI) was removed within the first 30 min and the remaining Cr(VI) was removed within 1 h. More recently, Marsh et al. (16) compared Cr(VI) reduction in aquifer sediment slurries amended with different electron acceptors. Results indicated that the reduction of 0.5 mM Cr(VI) in sandy sediment samples amended with sulfate or ferric iron preceded sulfidogenesis or ferric iron reduction and appeared to be mediated by microorganisms. However, more information regarding Cr(VI) reduction by bacterial communities is essential, since pollution levels and other environmental factors influence the development, diversity, and activities of natural microbial communities in different environments.
One powerful molecular biological tool for assessing a microbial population response to a toxic substance is denaturing gradient gel electrophoresis of PCR products of ribosomal small-subunit (16S) genes (PCR-DGGE). PCR-DGGE assesses the diversity and relative dominance of community bacterial ribosomal DNA (rDNA) by usage of 16S rRNA gene banding patterns independent of culturing techniques. Further, DNA sequencing of DGGE bands allows identification of the dominant organisms present (8, 9, 13, 18).
In this study, the changing microbial diversities of one sulfidogenic and two nonsulfidogenic consortia over time during Cr(VI) exposure were compared. Growth, Cr(VI) reduction, and PCR-DGGE of the consortia in combination with DNA sequencing aided in identifying the predominant bacterial populations and those which might be important for Cr(VI) reduction in the environment.
Sediment samples from two sites served as inocula for the consortia. One sediment sample was taken from Green Sands Beach at Mare Island Naval Shipyard (MINSY), San Francisco, Calif., in September 1996, where total Cr concentrations of ∼4.8 mM have been reported (1). By using Widdel's marine medium (39) with lactate (20 mM) as the carbon source, two enrichment consortia, MI-S and MI-A, were obtained. In spite of the presence of sulfate (28 mM) in the medium, only consortium MI-S produced hydrogen sulfide, indicating the presence of sulfate-reducing bacteria (SRB); MI-A did not. Another sample was taken from surficial sediments in June 1999 from Point Mugu Naval Air Weapons Station, Los Angeles, Calif., where total Cr concentrations of 380 mM have been reported (37). The consortium PACC was enriched from this sample by using brackish lactate medium (39) but without sulfate. All cultures were grown in serum bottles with nitrogen gas in the headspace; reductant was excluded to avoid the chemical reduction of Cr(VI). To ensure stable consortia with reproducible growth rates and Cr(VI)-reducing capacities, all three consortia were grown with Cr(VI) concentrations at the upper end of their Cr(VI) tolerance range and transferred often (approximately 10 times).
Time course measurements of Cr(VI) reduction and microbial community composition via PCR-DGGE with each enrichment condition were performed in duplicate experiments to assess reproducibility. To prevent the abiotic reduction and precipitation of chromium, the sulfidogenic enrichment consortium, MI-S, was aseptically degassed with N2 for 25 min prior to inoculation to remove HS−. HS− was not detectable in the experimental bottles immediately after inoculation. For the experiments amended with Cr(VI), 0.85 mM Cr(VI) was added to MI-S and MI-A and 1.35 mM Cr(VI) was added to PACC just prior to inoculation. Consortia without Cr(VI) and uninoculated Cr(VI)-containing media served as controls.
Hydrogen sulfide concentrations were measured colorimetrically at 480 nm with copper sulfate (6). Cr(VI) was measured colorimetrically at 540 nm with diphenylcarbazide (36). The amount of protein was determined as a proxy for microbial biomass with a bicinchoninic acid kit (Pierce) after cells were digested in 1 N NaOH for 5 min at 100°C. Water blanks with 0.85 or 1.35 mM Cr(VI), corresponding to the growth conditions when Cr(VI) was present, were used because there was a slight interference with the bicinchoninic acid assay.
At each time point, high-molecular-weight DNA was isolated. To ensure cell disruption, 1/3 volume of 0.22-μm acid-washed beads was added during the lysis stage and cells were homogenized (Mini Biospec Beadbeater) for 2.5 min. After bead beating and centrifugation, DNA was extracted and purified with a QIAamp tissue kit protocol (Qiagen Inc., Chatsworth, Calif.). DNA was amplified with bacterial primer 1055 and the universal small-subunit rRNA primer 1392, which incorporates a 5′ GC clamp (9, 28). Sequence data were obtained by using these primers sans GC clamp. Standard PCRs were performed by using a single annealing temperature of 50°C. We avoided touchdown PCR protocols to prevent selective amplification of certain templates (7) at the expense of others that may be relevant to Cr(VI) reduction. Three different concentrations of DNA template (approximately 25, 50, and 100 ng/μl) were used to obtain triplicate PCR samples. The PCR products were pooled and purified with a QIAquick PCR purification kit (Qiagen Inc.).
DGGE was carried out with a CBS Scientific Co. (Del Mar, Calif.) system (DGGE-2000). Equal amounts (∼300 ng) of pooled triplicate PCR products were loaded (except where otherwise noted) on 35 to 70% denaturant gels and run at 60°C for either 6 h at 200 V or 16 h at 80 V. Gels were stained with Sybr Gold (Molecular Probes, Eugene, Oreg.), visualized under UV light, and analyzed with a NucleoVision gel documentation system and Gel Expert software (NucleoTech Corp., San Mateo, Calif.). DNA extracted from gel bands by recommended protocols (30, 31) served as templates for PCR reamplification following the same protocol as above and utilizing primers 1055F and 1392R, without GC clamps. PCR products were sequenced according to the manufacturer's instructions (Big Dye v. 2.0) by using an Applied Biosystems 373A automated sequencer. Sequence comparisons to known sequences were performed by using the National Center for Biotechnology Information BLAST website (2). PCR products that did not produce coherent sequences were cloned into the TA cloning vector pCR2.1-TOPO (Invitrogen, San Diego, Calif.), and the clones were sequenced.
The sulfidogenic consortium, MI-S.
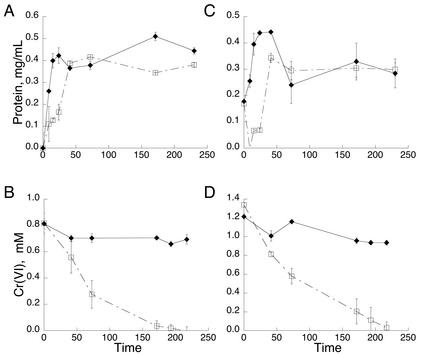
In the absence of Cr(VI), the protein concentration reached its maximum (1.8 mg/ml) by 24 h (Fig. 1A) and the greatest amount of HS− (∼10 mM) was formed by ∼48 h (Fig. 1B). When MI-S was grown with Cr(VI), biomass was depressed until after the complete reduction of Cr(VI), which occurred by 24 h (Fig. 1A and C). HS− was below the detection limit, indicating the absence of sulfate reduction until 100 h (Fig. 1B). The highest levels of protein (1.3 mg/ml) and hydrogen sulfide (6.3 mM) were reached at the end of the experiment (220 h). These findings suggest that Cr(VI) strongly inhibited the activity and growth of the SRB.
FIG. 1.
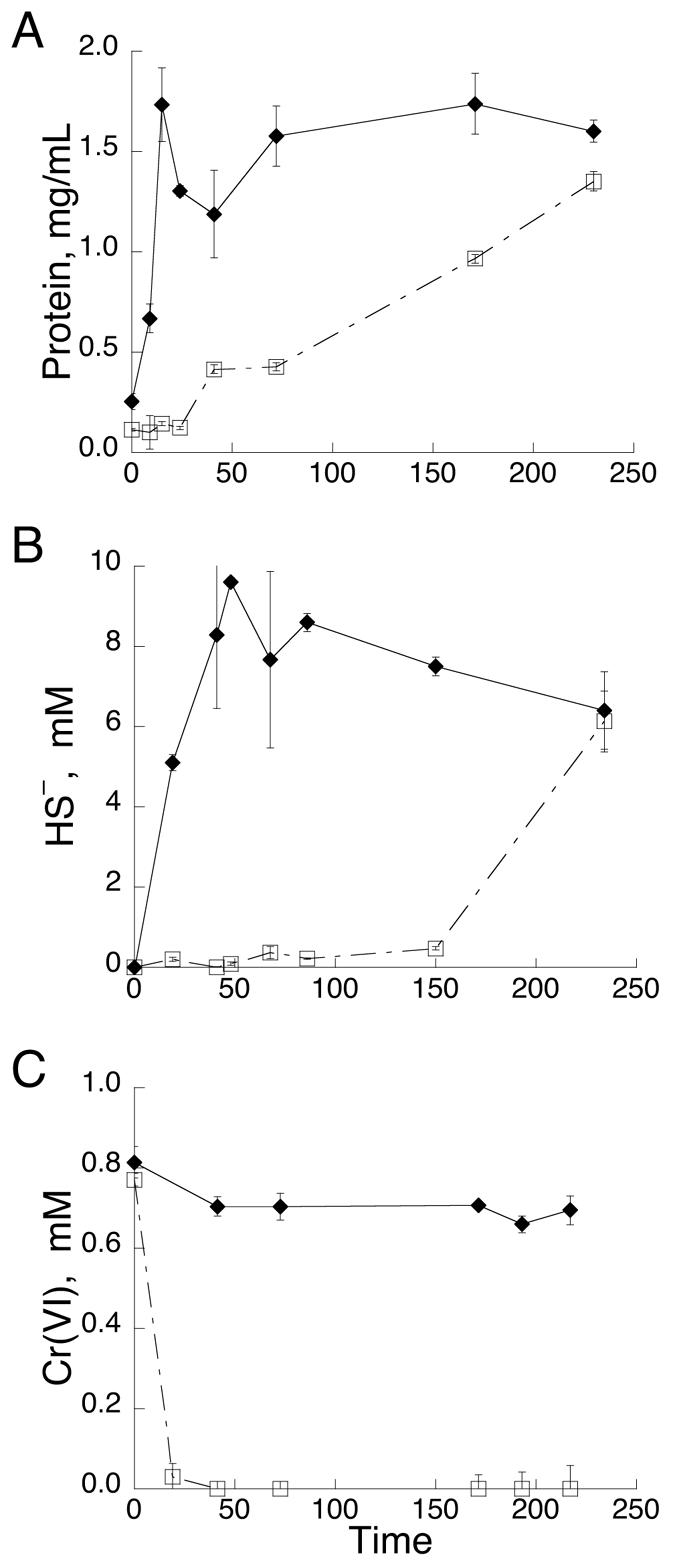
Effect of 0.85 mM Cr(VI) on protein concentration, HS− production, and Cr(VI) reduction by the sulfidogenic enrichment consortium, MI-S, from MINSY. (A) Protein concentration of the control enrichment without Cr(VI) (⧫) and the experimental enrichment subjected to 0.85 mM Cr(VI) (□). (B) HS− production by the control enrichment (⧫) and the experimental enrichment subjected to 0.85 mM Cr(VI) (□). (C) Reduction of 0.85 mM Cr(VI) by medium alone (⧫) and by MI-S (□). Error bars are standard deviations for duplicate experiments.
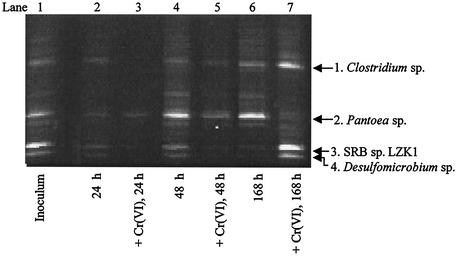
Temporal changes in the microbial composition of MI-S in the presence and absence of Cr(VI) were visualized by PCR-DGGE (Fig. 2), and the relative band intensities were analyzed. In the absence of Cr(VI), MI-S had similar profiles at 24 and 48 h (Fig. 2, lanes 2 and 4). DNA sequencing of the bands allowed tentative identification of the dominant members of the consortium (Table 1). Bacteria most closely related to SRB comprised 40 to 50% of the bacterial community, based on band intensity. By 168 h, a Pantoea sp. had become more dominant in the no-Cr(VI) bacterial community, and the SRB had decreased to ∼10% (Fig. 2, lane 6). In the presence of Cr(VI), at 24 h there were considerably fewer cells than at other time points. Cr(VI) had a striking effect on the microbial composition of the culture at 24 and 48 h, as most of the bands were absent or faint (Fig. 2, lanes 3 and 5). In these samples, the band corresponding to the Pantoea sp. was dominant and remained abundant from the beginning to the end of the experiment (∼20 to 80%). By 48 h, the bands corresponding to the Clostridium sp., Desulfovibrio sp., and Desulfomicrobium sp. were barely detectable, indicating that these organisms were greatly reduced in abundance in the Cr(VI) consortium and were inhibited until long after Cr(VI) was completely reduced (Fig. 2). By ∼168 h, however, the PCR-DGGE profile of the consortium once again resembled the original inoculum (Fig. 2, lane 1). These data corroborate the absence of measurable HS−, indicating that SRB were not active at 24 or 48 h, even though Cr(VI) had been completely reduced by this time (Fig. 1B and C).
FIG. 2.
Time course PCR-DGGE analysis of the sulfidogenic enrichment consortium MI-S exposed to 0.85 mM Cr(VI). 16S rDNA fragments were analyzed on a denaturing gel (35 to 70% denaturant; Sybr Gold stained). Each lane represents the MI-S community DNA at a specific time point in the absence or presence of 0.85 mM Cr(VI). Because there were considerably fewer cells in the presence of Cr(VI) at 24 h than at other time points, for comparative purposes, 150 ng of total rDNA was loaded into lanes 2 and 3. All other lanes were loaded with 300 ng of total rDNA.
TABLE 1.
Bacterial sequences from PCR-DGGE of the Cr(VI)-reducing consortia
| Consortium and best organism match | Band no.a | No. of basesb | % Similarity to known bacteria | Presence in:
|
||
|---|---|---|---|---|---|---|
| MS-1 | MA-1 | PACC | ||||
| MI-S (sulfidogenic) | ||||||
| Clostridium sp.c | 1 | 988 | 98 | + | + | |
| Pantoea sp. | 2 | 326 | 100 | + | ||
| Desulfovibrio sp. LZK1 | 3 | 299 | 99 | + | ||
| Desulfomicrobium sp. | 4 | 149 | 88 | + | ||
| MI-A (nonsulfidogenic) | ||||||
| Clostridium sp.c | 3 | 992 | 98 | + | + | |
| Enterococcus sp.c | 4 | 750 | 91 | + | ||
| Pantoea sp. | 5 | 326 | 100 | + | + | |
| Shewanella alga | 6 | 353 | 98 | + | + | |
| Uncultured bacterium DCE-25 (Spirochaeta sp.) | 7 | 354 | 98 | + | + | |
| PACC (nonsulfidogenic) | ||||||
| Tessaracoccus sp. | 1 | 355 | 97 | + | ||
| Ralstonia sp. | 2 | 170 | 97 | |||
| Clostridium sp. | 3 | 305 | 98 | |||
| Pseudomonas sp.c | 4 | 986 | 98 | + | ||
| Shewanella alga | 5 | 350 | 98 | + | + | |
| Uncultured bacterium DCE-25 (Spirochaeta sp.) | 6 | 351 | 98 | + | + | |
| Acetobacterium | 7 | 268 | 98 | + | ||
Numbers correspond to the bands in Fig. 2 (MI-S), 4 (MI-A), and 5 (PACC).
Number of bases sequenced.
Bacterial isolate obtained from consortium.
The initial hypothesis for rapid Cr(VI) reduction was that hydrogen sulfide produced by the enrichment consortium would chemically reduce Cr(VI) to Cr(III) (Fig. 1C). However, there is a 100- to 150-h lag before the onset of HS− production and a lack of an inverse relationship between Cr(VI) reduction and HS− production. In addition, there is an absence of SRB in the PCR-DGGE analyses until Cr(VI) is completely reduced. All of these findings seem to exclude the possibility of HS− reducing Cr(VI). Instead, either the SRB were lysed or growth was inhibited by Cr(VI); only after several days, after complete Cr(VI) reduction, were measurable HS− produced and SRB-like bacteria detected in the consortium.
The bacterial population implicated by PCR-DGGE as being responsible for Cr(VI) reduction in MI-S is a member of the genus Pantoea: a Pantoea sp. sequence was the only abundant bacterial sequence present in the Cr(VI) enrichment at 24 h, and the Pantoea sp. was the dominant organism at 48 h (Fig. 2), a time during which Cr(VI) was being reduced (Fig. 1C). Recently, Francis et al. (10) demonstrated that P. agglomerans strain SP1 had the capacity to couple the oxidation of lactate, acetate, and H2 to the reduction of 0.1 mM Cr(VI), although SP1 was much less tolerant of Cr(VI) than the MI-S consortium. Whether the tolerance of MI-S to high (>0.8 mM) Cr(VI) concentrations is due to the tolerance and Cr(VI) reduction capacity of the Pantoea sp. or whether other organisms in the consortium moderated the toxicity of Cr(VI) requires further investigation.
We cannot exclude the potential role of SRB in the initial rapid reduction of Cr(VI) in this consortium, even though all HS− was removed from the inoculum. Desulfovibrio spp. are well known to have a c3 cytochrome that functions as a Cr(VI) reductase (15, 23). Thus, although SRB and most other bacteria are strongly affected by Cr(VI) and disappear early on in this culture, it is possible that c3 cytochromes released from lysing SRB could contribute to Cr(VI) reduction.
The nonsulfidogenic consortia, MI-A and PACC.
Both nonsulfidogenic consortia grew well in the absence of Cr(VI) as the protein concentration increased from time zero and reached maximum protein levels by 20 to 24 h (Fig. 3A and C). In Cr(VI)-stressed enrichments, growth of both MI-A and PACC was slightly inhibited and biomass did not equal that of the control sample until 48 h (∼0.4 and 0.35 mg of protein/ml, respectively), when approximately 40% of the initial Cr(VI) concentration had been reduced (Fig. 3A and C). However, both consortia were able to completely reduce Cr(VI) within approximately 200 h (Fig. 3B and D). Even though the time for reduction was similar, the PACC consortium could withstand and reduce a greater concentration of Cr(VI): 1.35 versus 0.85 mM.
FIG. 3.
Effect of 0.85 or 1.35 mM Cr(VI) on protein concentration and Cr(VI) reduction by the nonsulfidogenic enrichment consortia MI-A (A and B) and PACC (C and D). Protein concentration of the control enrichment (⧫) and the experimental enrichment subjected to Cr(VI) (□) is shown for MI-A [0.85 mM Cr(VI)] (A) and PACC [1.35 mM Cr(VI)] (C). (B) Reduction of 0.85 mM Cr(VI) by medium alone (⧫) and by enrichment consortium MI-A (□). (D) Reduction of 1.35 mM Cr(VI) by medium alone (⧫) and by enrichment consortium PACC (▪). Error bars are standard deviations for duplicate experiments.
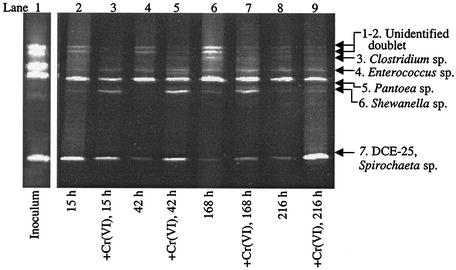
The PCR-DGGE results (Fig. 4) indicate that Cr(VI) did not affect the diversity of the bacterial rDNA of the MI-A consortium as greatly as it affected the sulfidogenic consortium. The Pantoea sp. appears to be the dominant organism. The Pantoea sp. is likely the same organism that was implicated in reducing Cr(VI) in MI-S (Table 1), as the sequences (in base pairs analyzed) were identical. A Spirochaeta sp. is another dominant organism in this consortium. Spirochaeta spp. are known to produce acetate during fermentation (5), which could conceivably be utilized by the Pantoea sp. for the reduction of Cr(VI), similar to P. agglomerans strain SP-1, which reduced Cr(VI) in the presence of acetate (10).
FIG. 4.
Time course PCR-DGGE analysis of the nonsulfidogenic enrichment consortium MI-A exposed to 0.85 mM Cr(VI). 16S rDNA fragments were analyzed on a denaturing gel (35 to 70% denaturant; Sybr Gold stained). Each lane represents the community MI-A DNA at a specific time point in the absence and presence of 0.85 mM Cr(VI); 300 ng was loaded onto each lane. The PCR product used for preparing the inoculum was run on a different DGGE gel and presented alongside the time course experiment. It is apparent that bands in the inoculum correlate reasonably well with the time course experimental profile.
There were two main differences in the PCR-DGGE profiles between Cr(VI)-containing and non-Cr(VI)-containing consortia (Fig. 4). One was that the unidentified top two bands and the Clostridium sp. band were completely absent when Cr(VI) was present and did not reappear until ∼168 h (Fig. 4, lane 7), when Cr(VI) was almost completely reduced (Fig. 3B). Another notable difference in the PCR-DGGE banding profile was the response of Shewanella alga, a metal-reducing bacterium. Without Cr(VI), the S. alga band is a minor member of the consortium at each time point (∼4% of the total community DNA). In the presence of Cr(VI), this band is more abundant, comprising ∼20% of the microbial community. The Shewanella sp. may have contributed to the reduction of Cr(VI) in our consortium, as Shewanella species are known to reduce Cr(VI) enzymatically (19). Alternatively, S. alga may have indirectly reduced Cr(VI) via a catalytic Fe(III)/Fe(II) cycle in which the respiration of Fe(III) produces Fe(II), which in turn reduces Cr(VI) and produces Fe(III) (24, 38, 40). Fe is present as a trace nutrient (7 μM) in Widdel's medium and is a common contaminant in laboratory medium components.
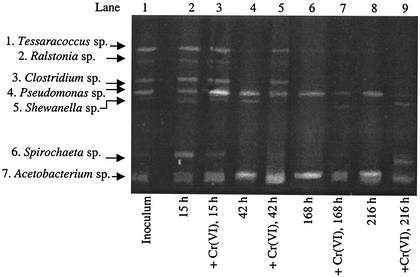
Figure 5 presents microbial community profiles of the PACC consortium. PCR-DGGE analysis indicated that Cr(VI) did not affect the microbial community as greatly as it affected both MINSY sediment consortia. The diversity of organisms in this consortium was similar with and without Cr(VI) at 15 and 42 h (Fig. 5, lanes 3 and 5).
FIG. 5.
Time course PCR-DGGE analysis of the nonsulfidogenic enrichment consortium PACC exposed to 1.35 mM Cr(VI). 16S rDNA fragments were analyzed on a denaturing gel (35 to 70% denaturant; Sybr Gold stained). Each lane represents the PACC community DNA at a specific time point in the absence or presence of 1.35 mM Cr(VI); 300 ng was loaded onto each lane.
Because the Shewanella sp. was implicated in Cr(VI) reduction in the MI-A consortium, we expected it to be active in the PACC consortium as well. However, the Shewanella species is actually less dominant with Cr(VI) at 42 h (Fig. 5, lanes 2 and 5). It is difficult to assess which organisms may be involved in Cr(VI) reduction in the PACC consortium. At this time, the capacity for reduction of Cr(VI) by Spirochaeta spp., Propionibacterium spp., Ralstonia spp., and Acetobacterium spp. has not been established. The Pseudomonas sp. may be contributing to Cr(VI) reduction in this consortium, as reductases capable of reducing Cr(VI) have been found in Pseudomonas ambigua (33) and in several other Pseudomonas spp. (4, 12, 17). Generally, pseudomonads are resistant to extreme Cr(VI) concentrations (10 mM) but do not reduce Cr(VI) (17).
Phylogenetic analysis.
Sequence analyses of the PCR-DGGE bands were used to identify the dominant organisms in the consortia, and in a few cases, bacterial isolates were obtained from the consortia (Table 1). In most cases the sequence similarity to known bacteria was high (>97%) based on the ∼300 bases examined. One SRB-like bacterial sequence was only 88% similar to a Desulfomicrobium sp. sequence. The other SRB was 99% related to the Desulfovibrio strain LZK1, found in naturally occurring radioactive material (27). Some bacterial species were present in two consortia (a Pantoea sp., a Shewanella sp., and the uncultured bacterium DCE-25, a Spirochaeta sp.). These bacterial sequences were aligned with each other and determined to be identical in the sequences examined. Of the isolates obtained in pure culture, none were able to reduce 0.85 mM Cr(VI) in the presence of either acetate or lactate as an electron donor (Table 1).
Conclusions.
This report establishes that the reduction of Cr(VI) at concentrations up to 1.35 mM can occur with microbial consortia enriched from contaminated sites. Although this study examined bacterial consortia, these data also facilitate the understanding of bacterial interactions in natural communities exposed to hexavalent Cr. Since many bacteria are inhibited by Cr(VI), it is inferred that the bacteria resistant to Cr(VI) and/or responsible for its reduction will continue to grow, while other strains in the environment will remain inactive or dormant until Cr(VI) is reduced to noninhibitory levels. Knowing which bacteria are active or thrive during Cr(VI) reduction is useful for site assessment and for designing bioremediation strategies for chromium-contaminated sites.
That SRB are strongly inhibited by Cr(VI) is important because it is often assumed that if SRB are present in Cr(VI)-contaminated environments, then the SRB will be metabolically active and produce HS−, which would abiotically reduce Cr(VI). While this may be true at low Cr(VI) levels, in areas where Cr(VI) levels are high, SRB are probably inhibited relative to other members of the microbial community and, therefore, not the key players in Cr(VI) reduction. This interpretation is consistent with data of Marsh et al. (16), who showed that sulfate reduction and HS− production did not occur when sandy sediments were exposed to 0.5 mM Cr(VI). Similar results have been repeatedly demonstrated in our laboratory in other pure and enrichment cultures of SRB. Our results suggest that facultative anaerobes such as species of Pantoea and Shewanella, at least some of which are capable of Fe(III) reduction, are more likely than SRB to be the key players in Cr(VI) reduction in Cr-contaminated anaerobic environments.
Acknowledgments
We thank Shelly Anghera for providing sediment samples from Point Mugu and Mike Johnson for collecting samples for us at MINSY. We also are grateful to Anna Obraztsova for advice, including critical comments on the manuscript, and to Chris Francis, current Tebo laboratory members, and the reviewers for their helpful suggestions on the manuscript.
Y.M.A. was supported by a MARC Graduate Fellowship through the NIH. This research was supported in part by Office of Naval Research grant N00014-99-1-0107 and the University of California Toxic Substances Research and Training Program.
REFERENCES
- 1.Abu-Saba, K. E. 1998. Spatial and temporal variability in the aquatic cycling of chromium. Ph.D. thesis. University of California, Santa Cruz.
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bae, W., T. Kang, J. Jung, C. Park, S. Choi, and B. Jeong. 2000. Purification and characterization of NADH-dependent Cr(VI) reductase from Escherichia coli ATCC33456. J. Microbiol. Biotechnol. 10:580-586. [Google Scholar]
- 4.Bopp, L. H., and H. L. Ehrlich. 1988. Chromate resistance and reduction in Pseudomonas fluorescens strain LB300. Arch. Microbiol. 150:426-431. [Google Scholar]
- 5.Canale-Parola, E. 1984. The spirochetes, p. 38-70. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams and Wilkins, Baltimore, Md.
- 6.Cord-Ruwish, R. 1985. A quick method for the determination of dissolved and precipitated sulfides in cultures of sulfate-reducing bacteria. J. Microbiol. Methods 4:33-36. [Google Scholar]
- 7.Don, R. H., P. T. Cox, B. J. Wainwright, K. Baker, and J. S. Mattick. 1991. Touchdown PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19:4008.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fandino, L., L. Riemann, G. F. Steward, R. A. Long, and F. Azam. 2001. Variations in bacterial community structure during a dinoflagellate bloom analyzed by DGGE and rDNA sequencing. Aquat. Microb. Ecol. 23:130. [Google Scholar]
- 9.Ferris, M. J., G. Muyzer, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Francis, C. A., A. Y. Obraztsova, and B. M. Tebo. 2000. Dissimilatory metal reduction by the facultative anaerobe Pantoea agglomerans SP1. Appl. Environ. Microbiol. 66:543-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fude, L., B. Harris, M. M. Urrutia, and T. J. Beveridge. 1994. Reduction of Cr(VI) by a consortium of sulfate-reducing bacteria (SRB III). Appl. Environ. Microbiol. 60:1525-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishibashi, Y., C. Cervantes, and S. Silver. 1990. Chromium reduction in Pseudomonas putida. Appl. Environ. Microbiol. 56:2268-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kowalchuk, G. A., J. R. Stephen, W. DeBoer, J. I. Prosser, T. M. Embley, and J. W. Woldendorp. 1997. Analysis of ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl. Environ. Microbiol. 63:1489-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lebedeva, E. V., and N. N. Lyalikova. 1979. Reduction of crocite by Pseudomonas chromatophila sp. nov. Mikrobiologiya 48:517-522. [PubMed] [Google Scholar]
- 15.Lovley, D. R., and E. J. P. Phillips. 1994. Reduction of chromate by Desulfovibrio vulgaris and its C-3 cytochrome. Appl. Environ. Microbiol. 60:726-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marsh, T. L., N. M. Leon, and M. J. McInerney. 2000. Physiochemical factors affecting chromate reduction by aquifer materials. Geomicrobiol. J. 17:291-303. [Google Scholar]
- 17.McLean, J., and T. J. Beveridge. 2001. Chromate reduction by a pseudomonad isolated from a site contaminated with chromated copper arsenate. Appl. Environ. Microbiol. 67:1076-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muyzer, G., E. D. De Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myers, C. R., B. P. Carstens, W. E. Antholine, and J. M. Myers. 2000. Chromium(VI) reductase activity is associated with the cytoplasmic membrane of anaerobically grown Shewanella putrefaciens MR-1. J. Appl. Microbiol. 88:98-106. [DOI] [PubMed] [Google Scholar]
- 20.Ohtake, H., K. Komori, C. Cervantes, and K. Toda. 1990. Chromate-resistance in a chromate-reducing strain of Enterobacter cloacae. FEMS Microbiol. Lett. 67:85-88. [DOI] [PubMed] [Google Scholar]
- 21.Park, C.-H., M. Keyhan, B. Wielinga, S. Fendorf, and A. Matin. 2000. Purification to homogeneity and characterization of a novel Pseudomonas putida chromate reductase. Appl. Environ. Microbiol. 66:1788-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patterson, R. R., S. Fendorf, and M. Fendorf. 1997. Reduction of hexavalent chromium by amorphous iron sulfide. Environ. Sci. Technol. 31:2039-2044. [Google Scholar]
- 23.Peck, H. D. 1959. The ATP-dependent reduction of sulfate with hydrogen in extracts of Desulfovibrio desulfuricans. Proc. Natl. Acad. Sci. USA 45:701-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perry, K. A., J. E. Kostka, G. W. Luther III, and K. H. Nealson. 1993. Mediation of sulfur speciation by a Black Sea facultative anaerobe. Science 259:801-803. [DOI] [PubMed] [Google Scholar]
- 25.Pettine, M., I. Barra, L. Campanella, and F. J. Millero. 1998. Effect of metals on the reduction of chromium (VI) with hydrogen sulfide. Water Res. 32:2807-2813. [Google Scholar]
- 26.Pettine, M., F. J. Millero, and R. Passino. 1994. Reduction of chromium(VI) with hydrogen sulfide in NaCl media. Mar. Chem. 46:335-344. [Google Scholar]
- 27.Phillips, E. J. P., E. R. Landa, T. Kraemer, and R. Zielinski. 2002. Sulfate-reducing bacteria release barium and radium from naturally occurring radioactive material in oil-field barite. Geomicrobiol. J. 18:167-182. [Google Scholar]
- 28.Raskin, L., R. I. Amann, L. K. Poulsen, B. E. Rittmann, and D. A. Stahl. 1995. Use of ribosomal RNA-based molecular probes for characterization of complex microbial communities in anaerobic biofilms. Water Sci. Technol. 31:261-272. [Google Scholar]
- 29.Saleh, F. Y., T. F. Parkerton, R. V. Lewis, J. H. Huang, and K. L. Dickson. 1989. Kinetics of chromium transformations in the environment. Sci. Total Environ. V86:25-41. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., p. E10-E14. Cold Spring Harbor Laboratory Press, New York, N.Y.
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., p. 6.46-6.48. Cold Spring Harbor Laboratory Press, New York, N.Y.
- 32.Sedlak, D. L., and P. G. Chan. 1997. Reduction of hexavalent chromium by ferrous iron. Geochim. Cosmochim. Acta 61:2185-2192. [Google Scholar]
- 33.Suzuki, T., N. Miyata, H. Horitsu, K. Kawai, K. Takamizawa, Y. Tai, and M. Okazaki. 1992. NAD(P)H-dependent chromium(VI) reductase of Pseudomonas ambigua G-1: a chromium(V) intermediate is formed during the reduction of chromium(VI) to chromium(III). J. Bacteriol. 174:5340-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tebo, B. M., and A. Y. Obraztsova. 1998. Sulfate-reducing bacterium grows with Cr(VI), U(VI), Mn(IV), and Fe(III) as electron acceptors. FEMS Microbiol. Lett. 162:193-198. [Google Scholar]
- 35.Turick, C. E., C. Graves, and W. A. Apel. 1998. Bioremediation potential of Cr(VI)-contaminated soil using indigenous microorganisms. Bioremediation J. 2:1-6. [Google Scholar]
- 36.Urone, P. F. 1955. Stability of colorimetric reagent for chromium, S-diphenylcarbazides, in various solvents. Anal. Chem. 27:1354-1355. [Google Scholar]
- 37.U.S. Army Environmental Center. 2000. Final report: in-situ electrokinetic remediation of metal contaminated soils. SFIM-AEC-ET-CR-99022, 12-29. United States Army. [Online.] http://www.estcp.org/documents/techdocs/ISERMS_Report.pdf.
- 38.Venkateswaran, K., D. P. Moser, M. E. Dollhopf, D. P. Lies, D. A. Saffarini, B. J. MacGregor, D. B. Ringelberg, D. C. White, M. Nishijima, H. Sano, J. Burghardt, E. Stackebrandt, and K. H. Nealson. 1999. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int. J. Syst. Bacteriol. 49:705-724. [DOI] [PubMed] [Google Scholar]
- 39.Widdel, F., and F. Bak. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p. 3352-3378. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed. Springer-Verlag, New York, N.Y.
- 40.Wielinga, B., M. M. Mizuba, C. M. Hansel, and S. Fendorf. 2001. Iron promoted reduction of dissimilatory iron-reducing bacteria. Environ. Sci. Technol. 35:522-527. [DOI] [PubMed] [Google Scholar]