Abstract
The Streptomyces sp. strain C5 snp locus is comprised of two divergently oriented genes: snpA, a metalloproteinase gene, and snpR, which encodes a LysR-like activator of snpA transcription. The transcriptional start point of snpR is immediately downstream of a strong T-N11-A inverted repeat motif likely to be the SnpR binding site, while the snpA transcriptional start site overlaps the ATG start codon, generating a leaderless snpA transcript. By using the aphII reporter gene of pIJ486 as a reporter, the plasmid-borne snpR-activated snpA promoter was ca. 60-fold more active than either the nonactivated snpA promoter or the melC1 promoter of pIJ702. The snpR-activated snpA promoter produced reporter protein levels comparable to those of the up-mutated ermE∗ promoter. The SnpR-activated snpA promoter was built into a set of transcriptional and translational fusion expression vectors which have been used for the intracellular expression of numerous daunomycin biosynthesis pathway genes from Streptomyces sp. strain C5 as well as the expression and secretion of soluble recombinant human endostatin.
There has been considerable interest in the use of Streptomyces lividans as a host for the expression of recombinant proteins. This has led to extensive research on the production and secretion of extracellular hydrolases by the streptomycetes, with emphasis on proteinases which may interfere with recombinant protein production. Among the proteinase genes of Streptomyces three similar loci, the snp locus of Streptomyces sp. strain C5 (16), the slp (also known as prt) locus of S. lividans 66 (5, 17), and the mpr locus of Streptomyces coelicolor ‘Müller’ (7) have all been shown to produce similar, small extracellular metalloproteinases. The snp, slp, and mpr loci are composed of two divergently oriented genes, one encoding the proteinase and the other a transcriptional regulator protein belonging to the LysR family (13). The nucleotide sequence of snpA and the biochemical characterization of the unusual proteinase it encodes have been published previously (16). This report describes the nucleotide sequence of snpR as well as the transcriptional properties of the intergenic region. In addition, the development of the locus into a family of expression and secretion vectors and their use for the production of extracellular soluble recombinant human endostatin is described.
MATERIALS AND METHODS
Bacterial strains and plasmids.
S. lividans TK24 and S. lividans 1326 (14) were obtained from D. A. Hopwood, and S. lividans 66 HLP-6 (5), a mutant strain with a deleted slp locus, was obtained from Cangene, Inc. Escherichia coli DH5α (Life Technologies, Inc., Gaithersburg, Md.) and E. coli Top10 (Invitrogen, San Diego, Calif.) were used for propagation of E. coli plasmids. The plasmids used and generated in this study are shown in Table 1.
TABLE 1.
Plasmids used and generated in this study
| Plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| pIJ303 | 10,620 kbp; thiostrepton marker in pIJ101; Thior | D. A. Hopwood |
| pIJ486 | 6.2 kbp; contains aphII gene of Tn5; Thior | D. A. Hopwood |
| pIJ702 | 5.65 kbp; derivative of pIJ350; Thior | D. A. Hopwood |
| pIJ4070 | 2.97 kbp; ermE* promoter fragment in pUC18; Ampr | C. R. Hutchinson |
| pMALc-H#15 | 6.60 kbp; pMALc-H with human endostatin cDNA; Ampr | Merck & Co, Inc. |
| pUC19 | 2.686 kbp; E. coli cloning vector; Ampr | Life Technologies |
| pKC505 | 18.7 kbp; cosmid derivative of SCP*; SP Aprr | 21a |
| pANT42 | 8.76 kbp; Streptomyces sp. strain C5 snp locus in pIJ702; Thior | 16 |
| pANT54 | 3.9 kbp; snpR gene and intergenic region in pUC19; Ampr | 16 |
| pANT806 | 3.937 kbp; aphII gene from pIJ486 in pUC19; Ampr | This study |
| pANT807 | 5.656 kbp; 3.0-kbp BamHI-HindIII fragment of pANT852 containing the snpR-aphII cassette in pUC19; Ampr | This study |
| pANT809 | 4.591 kbp; pANT807 minus SauI-StuI snpR DNA; Ampr | This study |
| pANT824 | 2.949 kbp; pIJ4070 minus EcoRI-AvaI DNA; Ampr | This study |
| pANT825 | 8.220 kbp; 5.293-kbp SphI-PstI fragment of pIJ702 in pIJ4070 with deleted EcoRI, SacI, and KpnI sites; SP Thior Ampr | This study |
| pANT826 | 7.334 kbp; pANT825 with 39-bp synthetic SphI-MluI MCS fragment downstream of ermEp*; SP Thior Ampr | This study |
| pANT827 | 8.617 kbp; 1.3-kbp EcoRI-HindIII fragment of pANT806 in pANT826; SP Thior Ampr | This study |
| pANT841 | 2.746 kbp; pUC19 plus linker containing modified MCS, intact lacZ-α; Ampr | 7a |
| pANT842 | 6.733 kbp; pANT42 minus 1.95-kbp KpnI fragment; Thior | This study |
| pANT849 | 5.343 kbp; pANT842 with 39-bp synthetic SphI-MluI MCS fragment inserted in place of the snpA gene; Thior | This study |
| pANT852 | 6.626 kbp; 1.3-kbp EcoRI-HindIII fragment of pANT806 in pANT849; Thior Neor | This study |
| pANT853 | 5.561 kbp; 1.935-kbp BamHI-HindIII (snpR-aphII) fragment from pANT809 in pANT849; Thior Neor | This study |
| pANT855 | 4.799 kbp; pIJ702 with 39-bp synthetic SphI-MluI MCS fragment inserted in place of the mel genes; Thior | This study |
| pANT856 | 6.082 kbp; 1.3-kbp EcoRI-HindIII fragment of pANT806 in pANT855; Thior Neor | This study |
| pANT857 | 7.97 kbp; 2.364-kbp PvuII fragment of pUC19 containing replicon and bla gene in pANT849; SP Thior Ampr | This study |
| pANT866 | 5.343 kbp; 1.527-kbp KpnI-NotI fragment of pIJ486 with altered rep gene (lacking BamHI site) in pANT849; Thior | This study |
| pANT867 | 9.003 kbp; 1.3-kbp EcoRI-HindIII fragment of pANT806 in pANT857; Thior Neor | This study |
| pANT880 | 4.25 kbp; 1.6-kbp EcoRI-PstI fragment containing acc(3)-IV locus of pKC505 in pUC19; Ampr Aprr | This study |
| pANT881 | 3.498 kbp; 752-bp SstI fragment containing acc(3)-IV gene of pKC505 in pUC19; Ampr | This study |
| pANT883 | 3.638 kbp; 233-bp PstI-SstI fragment containing the bifunctional E. coli-Streptomyces acc(3)-IV promoter in pANT881; Ampr Aprr | This study |
| pANT886 | 3.883 kbp; 1,001-bp SmaI fragment of pANT806 in pANT883, downstream of acc(3)-IV promoter; Ampr Neor | This study |
| pANT890 | 3.891 kbp; pANT886 lacking HindIII and EcoRI sites; Ampr Neor | |
| pANT893 | 3.626 kbp; pANT883 lacking EcoRI site downstream and HindIII, SphI, and PstI sites upstream of the acc(3)-IV gene; Ampr Aprr | This study |
| pANT894 | 6.512 kbp; 3.297-kbp DraI-NdeI fragment of pANT866 in pANT886 variant with deleted EcoRI and HindIII sites; SP Neor | This study |
| pANT895 | 6.247 kbp; 3.297-kbp DraI-NdeI fragment of pANT866 in pANT883 variant with deleted EcoRI, HindIII, SphI, and PstI sites; SP Aprr | This study |
| pANT1200 | 8.052 kbp; pANT857 plus 104-bp synthetic XbaI-HindIII MCS terminator fragment; SP Thior Ampr | This study |
| pANT1201 | 6.601 kbp; pANT894 plus 104-bp synthetic XbaI-HindIII MCS terminator fragment; SP Neor | This study |
| pANT1202 | 6.336 kbp; pANT895 plus 104-bp synthetic XbaI-HindIII MCS terminator fragment; SP Aprr | This study |
| pANT3022 | 6.661 kbp; pANT1201 plus 103-bp synthetic ClaI-HindIII fragment comprising VAA leader peptide; SP Neor | This study |
| pANT3032 | 7.227 kbp; pANT3022 plus cDNA open reading frame from pMalcH#15; SP Neor | This study |
| pANT3052 | 12.582 kbp; VAA-endostatin cassette from pANT3032 in pIJ303; Thior | This study |
Thior, thiostrepton resistance; Neor, neomycin resistance; Aprr, apramycin resistance; Ampr, ampicillin resistance; SP, E. coli-Streptomyces shuttle plasmid.
Media and growth conditions.
E. coli cultures were grown in Luria-Bertani medium as described by Sambrook et al. (23), containing 50 μg of ampicillin/ml, 20 μg of neomycin/ml, or 50 μg of apramycin/ml when necessary. Isopropyl-β-d-thiogalactopyranoside, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside, and allantibiotics used were purchased from Sigma Chemical Company (St. Louis, Mo.). Streptomyces cultures were grown in YEME and R2YE media as described by Hopwood (14). In some cases, streptomycete cultures were also cultivated in TSBP-S (with the following components in grams per liter: tryptic soy broth, 30; yeast extract, 1; dextrose, 5; sucrose, 150) or MSEM (with the following components in grams per liter: maltose, 20; dextrose, 20; yeast extract, 5; MgSO4 · 7H2O, 0.2; K2HPO4 · 3H2O, 0.5; morpholinepropanesulfonic acid, 4; the solution also contained 0.8 ml of 10× trace elements [14]). Streptomyces cultures containing pIJ101-based plasmids were grown on solid or liquid medium containing 50 or 5 μg of thiostrepton/ml, respectively. Recombinant cultures with plasmids based on the aphII or acc(3)-IV genes were grown on medium containing 20 μg of neomycin/ml or 50 μg of apramycin/ml, respectively.
DNA and RNA preparation.
Plasmid DNA was routinely prepared from 5-ml cultures of E. coli or Streptomyces by the method of Carter and Milton (6). Streptomycete RNA was prepared with Trizol (Life Technologies, Inc.) after lysozyme pretreatment of the mycelia for 20 min at 37°C. An additional phenol-chloroform extraction with one volume of buffered phenol, pH 8.0 (Life Technologies), was performed on the RNA preparations if further enzymatic manipulation was planned.
DNA sequencing and analysis.
DNA sequence determination was performed by using the dideoxynucleoside termination procedure of Sanger et al. (24) or an ABI 377 automated sequencing system (Perkin Elmer Applied Biosystems, Foster City, Calif.).
DNA sequence files were managed by using Clone Manager 5 (Scientific & Educational Software, Inc., State Line, Pa.) and BioEdit 4.74 (12), analyzed with software from Genetics Computer Group, University of Wisconsin, Madison, Wis. (8), as well as with the suite of analysis tools available from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) and the Pôle Bio-Informatique Lyonnais Network Protein Sequence Analysis server (http://pbil.ibcp.fr/NPSA; Lyon, France).
Cloning procedures and plasmid construction.
E. coli and Streptomyces cultures were transformed as described by Sambrook et al. (23) and Hopwood (14), respectively. Enzymes were purchased from Life Technologies, Inc., or from New England Biolabs, Inc. (Beverly, Mass.), and were used per the manufacturer's instructions. General cloning methods were conducted as described by Sambrook et al. (23). Double-stranded DNA (dsDNA) linkers for use in plasmid construction were prepared from complementary oligonucleotides (Life Technologies, Inc.) designed to yield dsDNA fragments with the desired 5′ or 3′ overhangs. Equimolar quantities of the separate oligonucleotides were mixed in a buffer containing 10 mM Tris chloride (pH 8.0) and 10 mM MgCl2 that was heated to 99°C for 10 min, allowed to cool, and used for ligations. The oligonucleotides used in this fashion are shown in Table 2.
TABLE 2.
Oligonucleotides used for dsDNA linker preparation
| Designation | Sequence | Cohesive ends | Relevant plasmid(s) |
|---|---|---|---|
| CLDVCT1 | 5′CGCGTTAACTAGTTTAAAGCTTGAGCTCTAGATCTGAATTC GCATG3′ | SphI-MluI | pANT849 |
| CLDVCT2 | 5′CGAATTCAGATCTAGAGCTCAAGCTTTAAACTAGTTAA3′ | ||
| CLDTERM1 | 5′CTAGAGCTCTGCAGGATCCAAGCTTACTAGTGGTACCACG CGTGAATAGATAGAGTGCAGGGGCCCCGACCGTGTCGGGGCCCCTGCACGGGTCCATGG3′ | XbaI-HindIIIa | pANT1200, pANT1201, pANT1202 |
| CLDTERM2 | 5′AGCTCCATGGACCCGTGCAGGGGCCCCGACACGGTCGGG GCCCCTGCACTCTATCTATTCACGCGTGGTACCACTAGTA AGCTTGGATCCTGCAGAGCT3′ | ||
| VAA1 | 5′CGATGGCCAGAAAGACCGTGGCAGCTGCACTCGCCCTTGT GGCGGGAGCCGCTGTGGCCGTCACGGGCAACGCCCCGGCGCAGGCCGTCCCGCCCG3′ | ClaI-BamHI | pANT3022 |
| VAA2 | 5′GATCCGGGCGGGACGGCCTGCGCCGGGGCGTTGCCCGTGA CGGCCACAGCGGCTCCCGCCACAAGGGCGAGTGCAGCTGCCACGGTCTTTCTGGCCAT3′ |
Cohesive end only, does not reform recognition site after ligation with vector.
Reporter protein quantification.
Streptomyces cell lysates were prepared with a Mini-Bead Beater (Biospec Products, Bartlesville, Okla.) per manufacturer instructions and were assessed for reporter protein production by using an NptII enzyme-linked immunosorbent assay kit (5 Prime 3 Prime, Inc., Boulder, Colo.) as described by the manufacturer. The reporter quantification results were assessed for statistical significance with a paired Student's t test.
Primer extension analyses.
Reverse transcription reactions were conducted with the fluoresceinylated primers 5′FGACGACCGGCGCGCCCTCAGCG3′ (snpA) or 5′FGTGTTCGATGCGCCGCAGCTGCGTG3′ (snpR); RNA was prepared from 48-h TSBP-S cultures of S. lividans TK24(pANT842) and rTth reverse transcriptase (Perkin Elmer, Foster City, Calif.) yielding 5′ fluorescein-labeled single-stranded DNA fragments with 3′ termini corresponding to the 5′ termini of the snpA or snpR mRNAs. Both extension reaction mixtures contained 1.0 μg of RNA and were held at 60°C for 70 min and were separated on an ABI 377 along with dideoxy-terminator sequencing reactions generated with the same primer and plasmid template, and the subsequent gel was image cropped and adjusted for contrast by using Adobe Photoshop 5.0 (Adobe Systems Inc., San Jose, Calif.).
Protein electrophoresis, blotting, and immunodetection.
Culture broth samples were clarified by centrifugation at 14,000 × g for 20 min. Equal volumes of cleared broth were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis in a Mini-Protean II apparatus (Bio-Rad) and were transferred to polyvinylidene difluoride membranes with a Mini Trans-Blot cell (Bio-Rad). The blotting procedure was performed as described by Ausubel et al. (1), with polyclonal rabbit anti-human endostatin (1:20,000; Judy Boice at Merck & Co., Inc.) and alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (1:6,000; Bio-Rad) as the primary and secondary antibodies, respectively. Immun-Star substrate (Bio-Rad) was used to indicate bound secondary antibody, and the results were visualized by using X-ray film by standard methods (1). Insoluble recombinant human endostatin produced in E. coli (Judy Boice at Merck & Co., Inc.) was used as a standard.
Nucleotide sequence accession numbers.
The complete Streptomyces sp. strain C5 snpR gene DNA sequence has been deposited in GenBank under the accession number AY072041. Additionally, the DNA sequences for plasmids pANT849, pANT1200, pANT1201, and pANT1202 have been deposited under the accession numbers AY072037, AY072038, AY072039, and AY072040, respectively.
RESULTS
Sequence and analysis of snpR.
The nucleotide sequence of the Streptomyces sp. strain C5 snpR gene was determined from the snpA-snpR subclone plasmid pANT54 by using both manual and automatic methods. The snpR gene is 942 bp in length and has an overall G+C content of 75 mol%. The encoded SnpR protein has 313 residues and a calculated molecular mass of 34,258 Da. The DNA sequence of Streptomyces sp. strain C5 snpR is 80 and 70% identical to those of S. lividans slpR and S. coelicolor mprR, respectively. The amino acid sequence of SnpR was 63, 59, and 17% identical to those of S. lividans SlpR, S. coelicolor MprR, and E. coli LysR, respectively. Aligned as a group, the three streptomycete proteins showed 44% identity.
Analysis of the SnpR, SlpR, and MprR primary sequences by the method of Dodd and Egan (10) indicated that all three were probably strong DNA binding proteins, with helix-turn-helix scores of 5.09, 4.59, and 4.51, respectively. For all three sequences the putative DNA binding region begins at residue 18 (glycine) and continues to residue 39 (arginine/threonine). The strongest conservation between the three proteins was found in the amino-terminal region of the sequences, which contains the helix-turn-helix motif implicated in DNA binding. This is consistent with the observations of Henikoff et al. (13), who found that the original members of the LysR family shared their strongest identity in a region aligning with LysR residues 21 to 40.
Reporter gene constructs.
To study the effect of SnpR on snpA transcription, four reporter gene plasmids were constructed. The reporter gene aphII, encoding neomycin phosphotransferase (NptII), was placed under the control of the snpA promoter with an intact snpR gene in plasmid pANT852. Deletion of the snpR DNA gave pANT853, identical to pANT852 except for the lesion in snpR. To compare snpA promoter activity with another well-characterized streptomycete promoter, the aphII gene was also placed under the transcriptional control of the melC1 promoter in pANT856. In both the snpA and melC1 promoter plasmids the SphI site was used as the 5′ end of the non-snp or non-melC DNA contains an ATG codon encoding the amino-terminal methionine of SnpA or MelC1; thus, the aphII reporter gene was positioned similarly downstream of the two promoters. Additionally, the aphII fragment used in these cloning steps contained its native ribosome binding site.
Comparison of different reporter constructs.
To evaluate the effect of snpR in snpA expression and to compare the snpA and melC1 promoters, MSEM-grown cultures of S. lividans HLP-6, harboring pANT849, pANT852, pANT853, pANT855, or pANT856, were harvested after 48 h of growth, disrupted, and measured for NptII production by enzyme-linked immunosorbent assay (Table 3). Cultures containing pANT849 and pANT855, reporter-negative vector controls, produced only background levels of NptII. The snpR-minus construct and the melC1 promoter construct each produced around 30 pg of NptII per μg of soluble protein, and the snpR-positive construct produced around 1,800 pg of NptII per μg of soluble protein. Thus, the presence of an intact snpR gene conferred a ca. 60-fold increase of NptII production over transcription in its absence or transcription from the melC1 promoter.
TABLE 3.
NptII reporter production by recombinant Streptomyces cultures
| Plasmid | Mean production of NptIIa | SDb |
|---|---|---|
| pANT849 | 7 | 1.8 |
| pANT852 | 1,807*† | 575.0 |
| pANT853 | 30* | 11.2 |
| pANT855 | 5 | 2.9 |
| pANT856 | 29* | 18.7 |
Values are picograms of NptII per microgram of soluble protein. An asterisk indicates significance relative to the pANT849 value, and the dagger indicates significance relative to pANT853 and pANT856 values.
SD, standard deviation.
In a separate experiment with Streptomyces-E.coli shuttle plasmids, SnpR-activated snpAp activity was compared with that of the up-mutated ermE∗ promoter. Plasmid pANT867, which contains an snpR-snpAp-aphII cassette, and pANT827, carrying an ermEp∗-aphII cassette, were generated. Table 4 shows that NptII levels produced by cultures containing the SnpR-activated snpAp construct, although slightly higher on average than those produced by similar cultures containing the ermEp∗ construct, were not significantly different, indicating that the snpR-activated snpA promoter and the mutant ermE∗ promoter were of comparable strength. The negative control produced only background levels of NptII.
TABLE 4.
Comparison of NptII production by the snpA and ermE* promoters
Values are picograms of NptII per microgram of soluble protein. An asterisk indicates significance relative to the pANT857 value.
SD, standard deviation.
Expression of the aphII reporter gene as a function of host growth.
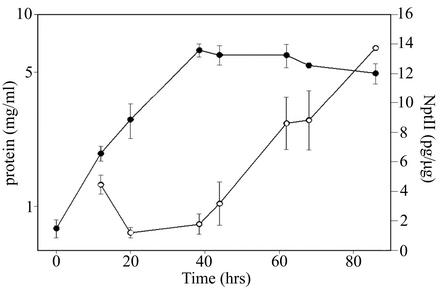
Expression of the pANT852 aphII gene was analyzed in samples taken at various times throughout the incubation of triplicate cultures. Total cellular protein was used as an indicator of culture age and was comparable to growth curves generated by using optical density measurement (data not shown). Accumulation of NptII reached maximal levels during later stationary phase, suggesting that plasmid-borne snpA expression occurs primarily after the exponential growth phase and continues throughout stationary growth phase (Fig. 1).
FIG. 1.
Profile of SnpR-activated snpAp-driven reporter gene expression and culture growth. Samples from triplicate 0.5-liter cultures of S. lividans TK24(pANT852) grown in TSBP-S medium were processed for NptII determination as described in the text. With the mean total cellular protein curve indicating the growth phase of the fermentation (filled circles), NptII accumulation increased with culture age (open circles). This suggests that the snpA promoter is active in stationary phase. Values shown represent means ± standard errors; the two data points without error bars represent the averages of two samples. Streptomycetes growth curves generated from optical density readings and total protein levels were comparable (data not shown).
Mapping of the snpA and snpR transcriptional start sites.
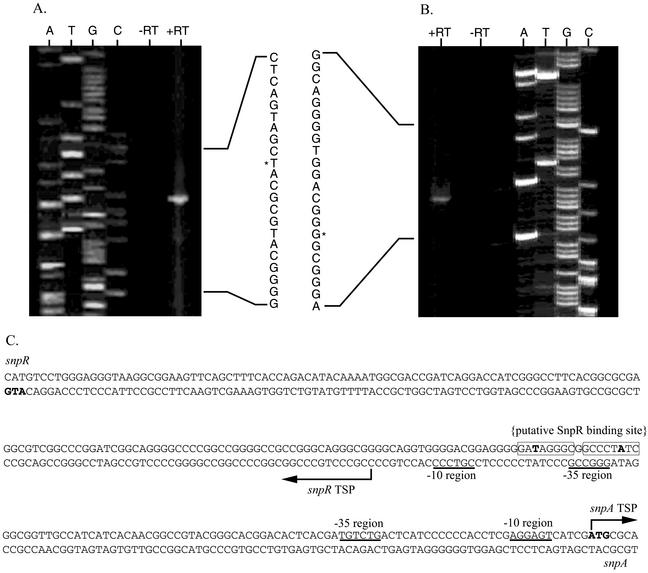
Based on the results of primer extension experiments, the transcriptional start site for snpR is 139 nucleotides upstream of the most likely ATG start codon and is positioned 28 nucleotides upstream of the putative SnpR binding site (Fig. 2). The snpA transcript, on the other hand, starts at the adenine of the ATG start codon most likely to encode the formyl-methionine of SnpA (Fig. 2).
FIG. 2.
Transcriptional mapping, nucleotide sequence, and features of the Streptomyces sp. strain C5 snpA-snpR intergenic region. Primer extension reactions with either an snpA-specific primer (A) or an snpR-specific primer (B) were conducted in the absence and presence of reverse transcriptase (−RT and +RT, respectively) and were run next to dideoxy-terminator sequencing reactions prepared with the same primer. Panel C shows the snpA-snpR intergenic region with the transcriptional start points (TSPs) and optimal −10 and −35 regions for each. Spacing between these putative RNA polymerase binding sites is 19 nucleotides for the snpR TSP and 18 nucleotides for the snpA TSP. The likely SnpR binding site is indicated, with the T-N11-A thymidine and adenine in boldface and the inverted repeat indicated within arrows. The snpA and snpR start codons are also indicated in boldface.
Construction and utilization of Streptomyces expression vectors.
The first snp-based expression vector, pANT849, was generated by replacing the snpA gene with a synthetic multiple cloning site (MCS) such that 10 unique restriction sites were immediately downstream of the SnpR-activated snpA promoter. To improve the utility of this vector, Streptomyces-E. coli shuttle variants were prepared. Plasmid pANT857, which is comprised of the pUC19 replicon and bla gene, is selectable in E. coli with ampicillin and in Streptomyces with thiostrepton.
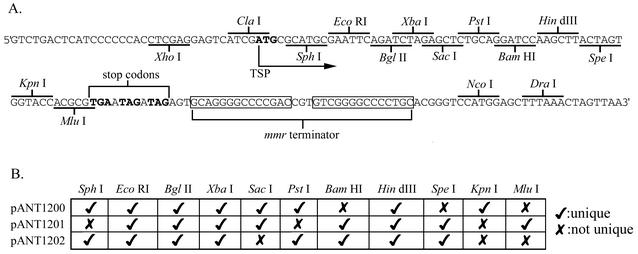
To provide for the use of multiple, compatible, and selectable plasmids, the aphII neomycin phosphotransferase gene and the acc(3)-IV apramycin resistance gene were incorporated, along with an improved MCS, in into new vectors. Because it has been observed that high-copy-number plasmids exceeding approximately 10 kbp in size exhibit instability in both E. coli and Streptomyces (2, 29), the plasmids were reduced in size and unnecessary restriction sites were eliminated. Plasmids pANT894 and pANT895 are neomycin- and apramycin-selectable snp-based expression vectors, respectively, and together with pANT857 they represent the second generation of snp-based expression vectors. These plasmids were further improved with the addition of an enhanced MCS containing additional unique restriction enzyme sites, stop codons in all three frames (for expression of gene fragments lacking their native stop codon), and the strong S. coelicolor mmr transcriptional terminator (18). This third generation of snp-based expression vectors is comprised of three plasmids: pANT1200, which is selectable in E. coli with ampicillin and in Streptomyces with thiostrepton; pANT1201, selectable in both hosts with neomycin; and pANT1202, selectable in both hosts with apramycin. The MCS and unique restriction enzyme sites of these three plasmids are shown in Fig. 3. In support of streptomycete antibiotic biosynthesis studies, snp-based vector systems have been used to express numerous genes from the Streptomyces sp. strain C5 daunomycin biosynthesis gene cluster. The doxA monooxygenase (9), several components of the dps polyketide synthase (20), and the dnm glycosyltransferase (A. J. Woo and W. R. Strohl, unpublished data) all have been successfully expressed by using the snpR-activated snpA promoter.
FIG. 3.
The pANT1200 series MCS. (A) The MCS of the pANT1200 series of vectors contains 11 restriction endonuclease sites, followed by stop codons in three frames (in boldface) and a bidirectional transcriptional terminator. (B) Of the 11 restriction enzyme sites in the MCS, most are unique, with the exceptions shown. The NcoI site downstream of the terminator is not unique in any of the plasmids, while the DraI site is unique in pANT1201 and pANT1202.
Construction and utilization of a Streptomyces secretion vector.
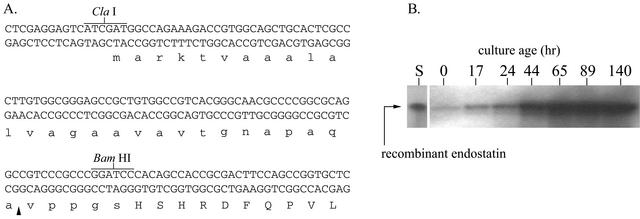
After development of the transcriptional fusion expression vectors, an experiment was undertaken to test the snp system as a translational fusion expression system. A ClaI-BamHI-ended dsDNA linker containing the coding sequence for the Streptomyces venezuale α-amylase (VAA) was inserted into pANT1201 such that the resulting plasmid, pANT3022, contained unique BamHI and HindIII sites. The BamHI site was engineered so that two codons, GGA and TCC, which encode glycine and serine residues, were positioned three codons downstream of the putative signal peptide recognition sites of the VAA signal sequence. A human endostatin cDNA clone modified to contain a 5′ BamHI site encoding an amino-terminal glycine-serine extension was inserted into pANT3022 to generate pANT3032, and the snp-VAA-endostatin cassette was transferred to a pIJ101-based streptomycete vector to make pANT3052. The vector insert joint of the snp-VAA-endostatin cassette is shown in Fig. 4A.
FIG. 4.
Nucleotide sequence of pANT3052 signal peptide-endostatin junction and recombinant endostatin production profile for S. lividans TK24(pANT3052). (A) The important features of the signal peptide-endostatin junction in the VAA-endostatin construct include the ClaI site positioned over the snpA transcription-translation start site; the putative VAA signal peptidase cleavage site, which is indicated by the arrow head; and the BamHI site, which encodes glycine and serine residues. The BamHI site is the point of fusion between the vector DNA and the human endostatin cDNA. The amino acid sequence of the translated product is shown, with the native human endostatin residues capitalized. (B) An anti-human endostatin Western blot of clarified broth samples from an S. lividans TK24(pANT3052) culture showing the accumulation of recombinant endostatin, predominantly after logarithmic growth phase. The lanes are labeled with the time point of the sample in hours. Lane S represents 2.1 ng of recombinant endostatin standard, while the remaining lanes represent 5-μl aliquots of clarified culture broth.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and anti-endostatin Western blot analysis of the culture broths of S. lividans TK24(pANT3052) showed immunoreactive bands migrating identically to pure human endostatin. Whereas the endostatin standard was in the form of denatured inclusion body protein purified from recombinant E. coli, the material produced by the Streptomycetes culture was soluble, as determined by centrifugal clarification before electrophoresis. Figure 4B shows a Western blot of broth samples from a culture of S. lividans(pANT3052). The blot shows that (i) soluble endostatin was produced by the culture, (ii) most of the endostatin production occurred during the stationary growth phase of the culture (after the 44-h time point), and (iii) the protein remained stable (i.e., a single immunoreactive band with no degradation products) out to 140 h of culture time. Densitometric analysis of the Western blot shown in Fig. 4B, which used a range of E. coli-produced endostatin samples as standard (cropped in Fig. 4B), suggested that the level of endostatin produced by S. lividans(pANT3052) was approximately 2 mg per liter of fermentation broth at the 65-h time point (data not shown).
DISCUSSION
The snpR gene encodes a LysR-like protein with a strong amino-terminal helix-turn-helix motif and with significant homology to the two other SnpR-like proteins described from homologous streptomycete loci, SlpR (5, 17) and MprR (7). Reporter gene experiments conducted in this study indicate that SnpR serves as an activator of snpA transcription. On average, SnpR-activated levels of snpA promoter activity exceeded unactivated levels by 60-fold. The function of SnpR as an activator is consistent with the generally observed role of LysR-like transcriptional regulators in the loci they affect. Most serve to stimulate transcription of their target gene(s) at levels usually between 6- and 200-fold (25).
Manual and computer-assisted evaluation of the snpA-snpR intergenic sequence for recognizable DNA motifs suggested a number of potentially important features. The most pronounced was an inverted repeat, centered on a T-N11-A sequence, within 100 nucleotides of the snpA start codon (see Fig. 2C). This motif, originally described by Henikoff et al. (13), is typical of many LysR-like protein binding sites. Thus, this snp intergenic feature is likely to be the SnpR binding site. Searches for sequences resembling promoters, however, were unsuccessful. Neither computer-assisted (21) nor manual screening for sequences resembling consensus streptomycete promoters (4, 26) revealed any candidate sequences.
Primer extension mapping of the snpR transcriptional start point to a position 138 nucleotides upstream of the predicted snpR start codon positions the −35 element within the likely SnpR binding site of the snpA-snpR intergenic region. The overlapped orientation of snpRp and the likely SnpR binding site suggests a potential negative autoregulatory role for the snpR gene product in snpR transcription. Such negative autoregulation is a feature of many LysR-like systems (25). In contrast, the transcriptional start site of snpA mapped to the adenine of the snpA ATG start codon, indicating the proteinase transcript is a leaderless mRNA species. Thus, the AGGA sequence immediately upstream of the snpA ATG, although optimally positioned and originally speculated to function as a Shine-Dalgarno sequence (16), is not involved in translation initiation.
This promoter structure has significant functional implications for the various expression plasmids based on the snpA promoter. Since the snp locus has been successfully used for expression of both streptomycete and non-streptomycete genes, several observations can be made about the translational events possibly occurring. When the DNA inserted into the expression system contains both the gene of interest and its native Shine-Dalgarno sequence, such as with the aphII reporter gene or doxA gene (9), conventional translation initiation may occur. These situations may include the synthesis of a short polypeptide from the 5′ AUG of the chimeric transcript, however. Inspection of the mRNA encoded by the aphII reporter constructs shows that a 21-amino-acid polypeptide would be synthesized by translation of the open reading frame starting with the 5′ AUG before terminating with a UGA codon just 5′ of the aphII AUG. Similarly, the doxA expression cassette of pANT195 encodes an RNA transcript containing an open reading frame with eight codons, starting with the 5′ AUG and terminating at a UGA codon overlapping the doxA AUG (27).
For expression and secretion of endostatin by the snp system, however, the ATG start codon of the new open reading frame (within the ClaI site) is superimposed on the transcriptional start point of the chimeric transcript. Although this translational context is similar to that of wild-type snpA in replacing the native coding sequence with the chimeric open reading frame, any native translational signals downstream of the ATG are lost and are not suitably replaced, as with the aphII and doxA fusions. Thus, the observed production of recombinant endostatin by this construct indicates that the leaderless endostatin expression cassette is recognized by the host ribosomes and is translated properly.
The fact that many leaderless mRNAs are translated with high efficiency despite the absence of Shine-Dalgarno sequences (15) indicates that an alternative method of translation initiation is occurring in these cases. Tedin et al. showed that in E. coli translation initiation factor 3 (IF3) antagonizes translation initiation from leaderless transcripts, and in vivo IF3 levels are inversely proportional to efficiency of leaderless mRNA translation (28). The model proposed suggests that a subpopulation of IF3-deficient 30S ribosomes form ternary complexes on leaderless transcripts during exponential growth, while 70S ribosomes are responsible for these initiation events in stationary phase (28). Notably, 70S ribosomes, which show a high preference for terminal AUG codons, accumulate after logarithmic growth has ceased (28). This may explain the productivity of the snpA promoter during stationary phase observed with both aphII expression (Fig. 1) and endostatin secretion (Fig. 4B).
The production and secretion of soluble endostatin by using the snp system was significant because this human protein has been difficult to produce adequately by other systems. Endostatin is a novel angiogenesis inhibitor of clinical interest (19, 22). It is notable that the endostatin produced in these fermentations was stable in the culture broth and continued to accumulate without noticeable degradation after 120 h of culture incubation. Most likely this results from a combination of vector stability, properly folded protein, and low water activity in the culture medium (at inoculation, the sucrose concentration was 15% [wt/vol]). By semiquantitative Western blot densitometry the level of endostatin produced by S. lividans(pANT3052) was estimated to be 2 mg per liter of fermentation broth at the 65-h time point, well into stationary phase. Even without the extensive optimization that is often applied in such projects (for examples see reference 11), this level of production is similar to those of several other systems designed to produce and secrete mammalian proteins with Streptomyces (3).
Acknowledgments
This work was supported by Merck Research Labs and by the National Science Foundation under grant no. MCB-94-05730.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2000. Current protocols in molecular biology. J. Wiley & Sons, New York, N.Y.
- 2.Balbas, P., X. Soberon, E. Merino, M. Zurita, H. Lomeli, F. Valle, N. Flores, and F. Bolivar. 1986. Plasmid vector pBR322 and its special-purpose derivatives-a review. Gene 50:3-40. [DOI] [PubMed] [Google Scholar]
- 3.Binnie, C., J. D. Cossar, and D. I. Stewart. 1997. Heterologous biopharmaceutical protein expression in Streptomyces. Trends Biotechnol. 15:315-320. [DOI] [PubMed] [Google Scholar]
- 4.Bourn, W. R., and B. Babb. 1995. Computer assisted identification and classification of streptomycete promoters. Nucleic Acids Res. 23:3696-3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butler, M. J., C. C. Davey, P. Krygsman, E. Walczyk, and L. T. Malek. 1992. Cloning of genetic loci involved in endoprotease activity in Streptomyces lividans 66: a novel neutral protease gene with an adjacent divergent putative regulatory gene. Can. J. Microbiol. 38:912-920. [DOI] [PubMed] [Google Scholar]
- 6.Carter, M. J., and I. D. Milton. 1993. An inexpensive and simple method for DNA purifications on silica particles. Nucleic Acids Res. 21:1044.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dammann, T., and W. Wohlleben. 1992. A metalloprotease gene from Streptomyces coelicolor 'Muller' and its transcriptional activator, a member of the LysR family. Mol. Microbiol. 6:2267-2278. [DOI] [PubMed] [Google Scholar]
- 7a.DeSanti, C. L. 2000. Ph.D. thesis. The Ohio State University, Columbus.
- 8.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickens, M. L., and W. R. Strohl. 1996. Isolation and characterization of a gene from Streptomyces sp. strain C5 that confers the ability to convert daunomycin to doxorubicin on Streptomyces lividans TK24. J. Bacteriol. 178:3389-3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dodd, I. B., and J. B. Egan. 1990. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 18:5019-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fornwald, J. A., M. J. Donovan, R. Gerber, J. Keller, D. P. Taylor, E. J. Arcuri, and M. E. Brawner. 1993. Soluble forms of the human T cell receptor CD4 are efficiently expressed by Streptomyces lividans. Bio/Technology 11:1031-1036. [DOI] [PubMed] [Google Scholar]
- 12.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 13.Henikoff, S., G. W. Haughn, J. M. Calvo, and J. C. Wallace. 1988. A large family of bacterial activator proteins. Proc. Natl. Acad. Sci. USA 85:6602-6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hopwood, D. A. 1985. Genetic manipulation of Streptomyces: a laboratory manual. The John Innes Foundation, Norwich, England.
- 15.Janssen, G. R. 1993. Eubacterial, archaebacterial, and eukaryotic genes that encode leaderless mRNA, p.59-67. In R. H. Baltz, G. D. Hegeman, and P. L. Skatrud (ed.), Industrial microorganisms: basic and applied molecular genetics. American Society for Microbiology, Washington, D.C.
- 16.Lampel, J. S., J. S. Aphale, K. A. Lampel, and W. R. Strohl. 1992. Cloning and sequencing of a gene encoding a novel extracellular neutral proteinase from Streptomyces sp. strain C5 and expression of the gene in Streptomyces lividans 1326. J. Bacteriol. 174:2797-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lichenstein, H. S., L. A. Busse, G. A. Smith, L. O. Narhi, M. O. McGinley, M. F. Rohde, J. L. Katzowitz, and M. M. Zukowski. 1992. Cloning and characterization of a gene encoding extracellular metalloprotease from Streptomyces lividans. Gene 111:125-130. [DOI] [PubMed] [Google Scholar]
- 18.Neal, R. J., and K. F. Chater. 1991. Bidirectional promoter and terminator regions bracket mmr, a resistance gene embedded in the Streptomyces coelicolor A3(2) gene cluster encoding methylenomycin production. Gene 100:75-83. [DOI] [PubMed] [Google Scholar]
- 19.O'Reilly, M. S., T. Boehm, Y. Shing, N. Fukai, G. Vasios, W. S. Lane, E. Flynn, J. R. Birkhead, B. R. Olsen, and J. Folkman. 1997. Endostatin: an endogenous inhibitor of angiogenesis and tumor growth. Cell 88:277-285. [DOI] [PubMed] [Google Scholar]
- 20.Rajgarhia, V. B., and W. R. Strohl. 1997. Minimal Streptomyces sp. strain C5 daunorubicin polyketide biosynthesis genes required for aklanonic acid biosynthesis. J. Bacteriol. 179:2690-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reese, M. G., N. L. Harris, and F. H. Eeckman. 1996. Large scale sequencing specific neural networks for promoter and splice site recognition. In L. H. a. T. E. Klein (ed.), Biocomputing: proceedings of the 1996 pacific symposium. World Scientific Publishing Company, Singapore, Republic of Singapore.
- 21a.Richardson, M. A., S. Kuhstoss, P. Solenberg, N. A. Schaus, and R. N. Rao. 1987. A new shuttle cosmid vector, pKC5050, for streptomycetes: its use in the cloning of three different spiramycin-resistance genes from a Streptomyces ambofaciens library. Gene 61:231-241 [DOI] [PubMed] [Google Scholar]
- 22.Saarela, J., R. Ylikarppa, M. Rehn, S. Purmonen, and T. Pihlajaniemi. 1998. Complete primary structure of two variant forms of human type XVIII collagen and tissue-specific differences in the expression of the corresponding transcripts. Matrix Biol. 16:319-328. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook, J., T. Maniatis, and E. F. Fritsch. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 26.Strohl, W. R. 1992. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 20:961-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strohl, W. R., M. L. Dickens, and C. L. DeSanti. October 1999. U.S. patent no. 5,962,293.
- 28.Tedin, K., I. Moll, S. Grill, A. Resch, A. Graschopf, C. O. Gualerzi, and U. Blasi. 1999. Translation initiation factor 3 antagonizes authentic start codon selection on leaderless mRNAs. Mol. Microbiol. 31:67-77. [DOI] [PubMed] [Google Scholar]
- 29.Warnes, A., and J. R. Stephenson. 1986. The insertion of large pieces of foreign genetic material reduces the stability of bacterial plasmids. Plasmid 16:116-123. [DOI] [PubMed] [Google Scholar]