Abstract
PCR-based assays were developed for the detection of plasmid- and chromosome-borne virulence genes in Yersinia enterocolitica and Yersinia pseudotuberculosis, to investigate the distribution of these genes in isolates from various sources. The results of PCR genotyping, based on 5 virulence-associated genes of 140 strains of Y. enterocolitica, were compared to phenotypic tests, such as biotyping and serotyping, and to virulence plasmid-associated properties such as calcium-dependent growth at 37°C and Congo red uptake. The specificity of the PCR results was validated by hybridization. Genotyping data correlated well with biotype data, and most biotypes resulted in (nearly) homogeneous genotypes for the chromosomal virulence genes (ystA, ystB, and ail); however, plasmid-borne genes (yadA and virF) were detected with variable efficiency, due to heterogeneity within the bacterial population for the presence of the virulence plasmid. Of the virulence genes, only ystB was present in biotype 1A; however, within this biotype, pathogenic and apathogenic isolates could not be distinguished based on the detection of virulence genes. Forty Y. pseudotuberculosis isolates were tested by PCR for the presence of inv, yadA, and lcrF. All isolates were inv positive, and 88% of the isolates contained the virulence plasmid genes yadA and lcrF. In conclusion, this study shows that genotyping of Yersinia spp., based on both chromosome- and plasmid-borne virulence genes, is feasible and informative and can provide a rapid and reliable genotypic characterization of field isolates.
Yersinia enterocolitica and Y. pseudotuberculosis, both members of the family Enterobacteriaceae, are comprised of strains with different degrees of pathogenicity. Both pathogenic and nonpathogenic strains are frequently isolated from various animals (birds, mammals, and reptiles) as well as from the environment (water and soil). Rodents (mice and rats), hares, rabbits, and birds serve as reservoirs for Y. pseudotuberculosis (1). Pathogenic strains of Y. enterocolitica and Y. pseudotuberculosis are frequently present in pigs without normally causing disease in these animals. Other food-producing animals, such as cattle, harbor mostly nonpathogenic strains of Y. enterocolitica.
In humans, Y. enterocolitica and Y. pseudotuberculosis are well-known food-borne pathogens and are mainly transmitted through ingestion of contaminated pork, milk, or water. Yersiniosis frequently occurs in young children as enterocolitis with fever, diarrhea, and abdominal cramps. Although the disease is usually self-limiting, complications (e.g., septicemia) are not uncommon in immunocompromised hosts. Furthermore, sequelae, such as reactive arthritis, have been reported (21).
The identification and further typing of subspecies, aiming at recognition of pathogenic strains of Yersinia spp., are traditionally based on phenotypic tests. Y. enterocolitica can be classified into biotype 1A, generally regarded as nonpathogenic (9), and the pathogenic biotypes 1B, 2, 3, 4, and 5. Both species can also be divided into serotypes with predictive values for pathogenicity. Serological and biochemical classification, however, are time consuming and are not generally available in routine laboratories. Alternative phenotypical tests, such as calcium-dependent growth at 37°C, Congo red binding (26), pyrazinamidase testing (16), autoagglutination testing, and serum resistance testing (2, 4, 5, 6, 13, 28) all have limited predictive value for the pathogenicity of Y. enterocolitica and Y. pseudotuberculosis. The tests are frequently ambiguous to read, and their outcome may be unreliable, since they depend on the presence and expression of (plasmid-borne) virulence genes and the virulence plasmid pYV can easily be lost depending on the culture conditions. Therefore, differentiation of pathogenic strains should not rely solely on the expression or detection of the virulence plasmid but also on the detection of chromosomal virulence factors.
The aim of this study was to develop PCR assays for the detection of plasmid- and chromosome-borne virulence genes of Y. pseudotuberculosis and Y. enterocolitica. The obtained results were compared to classical phenotypic subtyping methods. The presence or absence of various virulence genes was compared in strains isolated from human patients, food, (food-producing) animals, and the environment. The following chromosomal virulence genes were included in the analysis: ail, the Y. enterocolitica attachment invasion locus gene, reported to be present in pathogenic strains only (22, 23); ystA, which is responsible for the production of a heat-stable enterotoxin in Y. enterocolitica (12); ystB, which has been observed to encode an enterotoxin present mainly in biotype 1A strains of Y. enterocolitica (27, 29, 33); and inv, which is present in pathogenic Y. pseudotuberculosis (15). The plasmid-borne virulence genes analyzed are yadA, whose product is involved in autoagglutination, serum resistance, and adhesion (30), and virF or lcrF (for Y. enterocolitica and Y. pseudotuberculosis, respectively), which encodes transcriptional activators of the yop regulon (8, 11, 31). The results were compared to phenotypic tests with predictive values for pathogenicity.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
In this study, Y. enterocolitica (140 strains), Y. pseudotuberculosis (40 strains), Yersinia kristensenii (1 strain), Yersinia frederiksenii (4 strains), and Yersinia intermedia (8 strains) were analyzed. These included reference strains, human clinical isolates, animal strains, food strains, and environmental strains (Tables 1 and 2). In addition, the following species were used to test the specificity of the PCR assays: 15 strains comprising 11 Aeromonas spp., 3 Vibrio spp., 2 Campylobacter spp., 2 Staphylococcus spp., 3 type strains of Clostridium perfringens, 3 type strains of Escherichia coli, and type strains of Bacillus cereus, Bacteroides coagulans, Candida albicans, Citrobacter freundii, Enterococcus faecalis, Lactobacillus acidophilus, Listeria monocytogenes, Micrococcus luteus, Proteus vulgaris, Pseudomonas aeruginosa, Salmonella enterica serovar Typhimurium, and Streptococcus agalactiae.
TABLE 1.
Field bacterial strains used in this study
| Species | Source | No. of isolates | No. of isolates of serotype:
|
||
|---|---|---|---|---|---|
| O:3 | O:9 | Other | |||
| Y. enterocolitica | Animal | 53 | 8 | 33 | 12 |
| Y. enterocolitica | Human | 42 | 16 | 11 | 15 |
| Y. enterocolitica | Food | 25 | 3 | 2 | 20 |
| Y. enterocolitica | Environment | 8 | 1 | 0 | 7 |
| Y. pseudotuberculosis | Animal | 34 | |||
| Y. pseudotuberculosis | Human | 5 | |||
| Y. intermedia | Animal | 1 | |||
| Y. intermedia | Environment | 7 | |||
| Y. frederiksenii | Environment | 3 | |||
| Y. kristensenii | Environment | 1 | |||
TABLE 2.
Reference bacterial strains used in this study
| Species | Strain designation(s)a | Biotypeb | Serotype(s) | Source (country) |
|---|---|---|---|---|
| Y. enterocolitica | CIP 80.27 and ATCC 9610 | 1 | O:8 | Face (glanders-like infection) (France) |
| Y. enterocolitica | ATCC 23715 | 1 | O:8 | Blood (not specified) |
| Y. enterocolitica | IP Ye 1105 | (4) | O:8 | Not specified (not specified) |
| Y. enterocolitica | CCTM 3247 | (3) | O:5, O:27 | Not specified (not specified) |
| Y. enterocolitica | LMG 15558 and CCUG 8233 | 4 | O:3 | Mesenteric lymph node of a human with acute terminal ileitis (Sweden) |
| Y. enterocolitica | IP Ye 134 | (4) | O:3 | Not specified (not specified) |
| Y. enterocolitica | CIP 81.42 | 2 | O:9 | Human feces (Belgium) |
| Y. enterocolitica | ATCC 55075 | (2) | O:9 | Not specified (Canada) |
| Y. enterocolitica | IP Ye 21991 | (1A) | O:5 | Not specified (not specified) |
| Y. enterocolitica | IP Ye 102 | (1A) | O:6, O:30 | Not specified (not specified) |
| Y. enterocolitica | IP Ye 106 | (1A) | O:7, O:8 | Not specified (not specified) |
| Y. enterocolitica | IP Ye 1501 | (1A) | O:34 | Not specified (not specified) |
| Y. pseudotuberculosis | CIP 55.85 | Turkey (Sweden) | ||
| Y. frederiksenii | ATCC 33641 | Sewage (Denmark) |
Culture collection designations: ATCC, American Type Culture Collection; CIP, Collection de l'Institut Pasteur; IP, Institut Pasteur; CCUG, Culture Collection, University of Göteborg, Göteborg, Sweden; CCTM, Centre de Collection de Types Microbiens, Université de Lausanne, Lausanne, Switzerland.
The biotypes in parentheses were typed in this study. The biotypes not in parentheses were obtained from the literature or strain collection data.
Phenotyping.
The isolates of Y. enterocolitica were grouped by biotyping with discriminatory tests (lipase, esculin, salicin, indole, xylose, and trehalose) described previously (32) and serotyped by using commercial serum agglutinant anti-Y. enterocolitica O:3 and anti-Y. enterocolitica O:9 (Bio-Rad, Marnes-la-Coquette, France). The calcium dependency of all Y. enterocolitica and Y. pseudotuberculosis strains was tested with magnesium oxalate (MOX) agar as described by Prpic et al. (26) in order to differentiate between plasmid-bearing (resulting in typical pinpoint growth at 37°C) and plasmidless strains. In addition, the ability of Y. enterocolitica and Y. pseudotuberculosis strains to bind Congo red was used to distinguish plasmid-bearing strains (forming small red colonies) from plasmidless strains. The test was performed according to the method described in reference 26 by using CRAMP agar (Sigma, St. Louis, Mo.) incubated at 32°C for 72 h. Moreover, the pyrazinamidase test was used to distinguish potential pathogenic strains from nonpathogenic strains of Y. enterocolitica and performed according to the method described in reference 16.
DNA isolation.
DNA used for PCR was isolated by using the InstaGene matrix (Bio-Rad Laboratories AG). Briefly, 100 μl of cultivated tryptone soy broth was added to 1 ml of sterile double-distilled water and centrifuged for 2 min at 15,300 × g at 4°C. Two hundred microliters of the InstaGene matrix was added to the cell pellet. The mixture was incubated at 56°C for 30 min and then vortexed at high speed for 10 s. Following a boiling water bath for 8 min, lysed cells were mixed and spun at 15,300 × g for 3 min. Five microliters of the resulting supernatant was used as a template for each 50-μl PCR mixture. The remaining supernatant was stored at −20°C for future use.
DNA used for hybridization (dot blot and Southern blot analysis) was isolated by the guanidium thiocyanate extraction method for genomic DNA (25). DNA of gram-positive bacteria was isolated by using the E.Z.N.A. bacterial DNA kit (peqlab Biotechnologie GmbH, Erlangen, Germany) and following the instructions of the manufacturer. If necessary, the DNA was concentrated by using the protocol of the manufacturer to concentrations of at least 50 ng of DNA/μl.
Primers and PCR conditions.
Primers specific for the ail, ystA, and ystB genes of Y. enterocolitica, the yadA and virF/lcrF genes of Y. enterocolitica and Y. pseudotuberculosis, and the inv gene of Y. pseudotuberculosis were designed with Primer Designer software, versions 2.01 and 3.0 (Scientific & Educational Software, Durham, N.C.), and are listed in Table 3. PCRs were performed in 50-μl volumes containing 5 μl of DNA template, 0.2 mM concentrations of deoxynucleoside triphosphates, 5 μl of 10× PCR buffer II (Perkin Elmer, Rotkreuz, Switzerland), 3 mM MgCl2, 1 μM concentrations of each forward and reverse primer, 1.25 U of AmpliTaq Gold (Perkin Elmer), and 2% Tween 20. The thermal cycling conditions performed with a GeneAmp 9600 from Perkin Elmer were as follows: 1 cycle of denaturation at 95°C for 10 min; 25 cycles of melting at 95°C for 15 s, annealing at various temperatures depending on the primer pair used (Table 3) for 30 s, and elongation at 72°C for 30 s; and a final extension at 72°C for 10 min. Afterwards, 10 μl of amplicon was analyzed by electrophoresis on a 2.5% agarose gel. For the production of probes, the PCR was performed as described above with the addition of 40 μM Dig-11-dUTP (Roche Diagnostics, Mannheim, Germany). Labeled PCR amplicons were used as probes for hybridization.
TABLE 3.
Primers used in this study
| Target gene and primer direction | Sequence (5′→3′) | GenBank accession no. | Location (nucleotide) | Amplicon length (bp) | Annealing temp (°C) |
|---|---|---|---|---|---|
| ail | |||||
| Forward | TAATGTGTACGCTGCGAG | M29945 | 00544-00894 | 351 | 57 |
| Reverse | GACGTCTTACTTGCACTG | ||||
| ystA | |||||
| Forward | ATCGACACCAATAACCGCTGAG | X65999 | 00093-00171 | 79 | 61 |
| Reverse | CCAATCACTACTGACTTCGGCT | U09235 | 01181-01259 | 79 | |
| ystB | |||||
| Forward | GTACATTAGGCCAAGAGACG | D88145 | 00143-00288 | 146 | 61 |
| Reversea | GCAACATACCTCACAACACC | ||||
| inv | |||||
| Forward | CGGTACGGCTCAAGTTAATCTG | M17448 | 00923-01105 | 183 | 61 |
| Reverse | CCGTTCTCCAATGTACGTATCC | ||||
| yadA | X13882 | 01024-01872 | 849 | ||
| Forward | CTTCAGATACTGGTGTCGCTGT | AF102990 | 00465-01313 | 849 | |
| Reverse | ATGCCTGACTAGAGCGATATCC | AF056092 | 56159-57007 | 849 | 60 |
| X13881b | 00749-01507 | 759 | |||
| X13883c | 00868-01548 | 681 | |||
| virF/lcrF | |||||
| Forward | GGCAGAACAGCAGTCAGACATA | AF102990 | 32162-32722 | 561 | 63 |
| Reverse | GGTGAGCATAGAGAATACGTCG | M86690d | 00874-00314 | 561 |
From Ramamurthy et al. (27).
Y. enterocolitica serotype O:8.
Y. pseudotuberculosis.
Yersinia pestis lcrF gene.
Hybridization techniques.
Hybridization experiments were performed by dot blot and Southern blot analyses. For dot blots, the template DNA isolated by the miniprep method for genomic DNA was quantified by visual comparison after gel electrophoresis with a DNA standard (100 ng/μl). DNA was denatured in 0.4 M NaOH-10 mM EDTA, and 100 ng of denatured DNA was spotted onto a membrane (Zeta-Probe GT genomic tested blotting membranes; Bio-Rad Laboratories AG). After 30 min of air drying, the membrane was rinsed in 1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). The damp membrane was cross-linked in the GS gene linker UV chamber (Bio-Rad Laboratories AG). Hybridization was performed as described previously (18) at a temperature of 68°C, and washing steps were two times for 5 min in 2× SSC-0.1% sodium dodecyl sulfate at room temperature and two times for 15 min in 0.2× SSC-0.1% sodium dodecyl sulfate at 68°C. Detection of the signals was done by using chromogenic substrates (nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate; Roche Diagnostics) according to the manufacturer’s instructions.
For Southern blots, approximately 100 ng of template DNA was digested with EcoRV at 37°C overnight and separated on a 0.7% agarose gel. Southern blotting was done by alkaline transfer of DNA onto positively charged nylon membranes (Zeta-Probe GT genomic tested blotting membranes; Bio-Rad Laboratories AG) with an LCB 2016 VacuGene vacuum blotting pump (Amersham Pharmacia Biotech, Uppsala, Sweden). Gels were treated according to standard protocols (3). Hybridization was carried out as described above.
Statistical calculations.
The statistical significance of the correlation of Y. enterocolitica biotypes and their origins was calculated by using two-by-two tables and evaluated by Fisher's exact test by using the software NCSS 2000.
RESULTS AND DISCUSSION
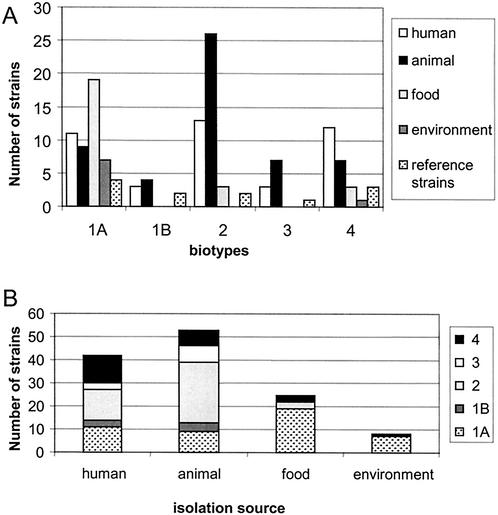
The biotypes of 140 Y. enterocolitica strains included in this study were determined; the distribution of the obtained biotypes for different origins of isolation is shown in Fig. 1. Strains of the pathogenic biotypes 1B, 2, 3, and 4 were significantly more-frequently isolated from humans and animals than from food and environmental sources in comparison to the biotype 1A strains (P < 0.001), indicative of the less-pathogenic nature of biotype 1A. Of 53 animal isolates of Y. enterocolitica, 46 strains (87%) were isolated from pigs, which mainly belonged to the pathogenic biotypes. The other animal isolates were a biotype 2 from a goat, a biotype 4 from a dog, and 5 biotype 1A's from 3 birds, 1 cow, and 1 dog. All field strains were further serotyped for O:3 and O:9, which were shown to be the most important pathogenic serotypes in Switzerland (Tables 1 and 2). Of isolates from human and animal sources, 68 out of 95 (71%) belonged to these serogroups, compared to 6 out of 33 (18%) isolates from food and the environment.
FIG. 1.
Diagram of the distribution of Y. enterocolitica strains included in this study. (A) Strains sorted by biotype and source. (B) Strains stacked by genotype for each isolation source. Reference strains are not included.
Gene-specific PCR tests were developed for 5 well-characterized virulence genes of Y. enterocolitica: the plasmid-borne genes virF and yadA and the chromosomal genes ystA, ystB, and ail. For Y. pseudotuberculosis, yadA and lcrF represented plasmid-borne genes and inv represented a chromosomal gene. The developed PCR tests did not result in a detectable product with any of the control species (results not shown). The PCR results for Y. enterocolitica (grouped with regard to biotype) and Y. pseudotuberculosis are summarized in Table 4. The predominant genotype (40 out of 50, 80%) of biotype 1A strains was ystB+ (lacking ystA, ail, yadA, and virF), showing that these strains are devoid of the virulence plasmid. Six biotype 1A strains isolated from food (3 isolates), 1 reference strain, 1 environmental isolate, and 1 pig isolate lacked both ystA and ystB. Additionally 1 reference strain had a negative result for ystB when analyzed by PCR but a positive result when analyzed by hybridization (Table 5). Eleven human clinical isolates belonged to biotype 1A, suggesting that this biotype is not completely nonpathogenic. All human isolates of biotype 1A contained ystB, and although 3 of the 19 food isolates, 1 of the 9 pig strains, and 1 of the 7 environmental strains lacked ystB, this difference is not statistically significant. The predominant genotype for 1B strains was ystA+ ail+ (lacking ystB, yadA, and virF) (8 out of 9, 89%, the exception was an ail-negative strain). Thus, all biotype 1B strains lacked plasmid-borne virulence genes.
TABLE 4.
Genotyping of Y. enterocolitica and Y. pseudotuberculosis by PCR detection of virulence genes
| Species and biotype (n) | No. of strains with chromosome-borne gene:
|
No. of strains with plasmid-borne gene:
|
|||||
|---|---|---|---|---|---|---|---|
| ystA | ystB | ail | inv | yadA | virF | lcrF | |
| Y. enterocolitica | |||||||
| 1A (50) | 2 | 43 | 2 | 1 | 3 | ||
| 1B (9) | 9 | 0 | 8 | 0 | 0 | ||
| 2 (44) | 44 | 0 | 44 | 29 | 29 | ||
| 3 (11) | 11 | 0 | 11 | 8 | 9 | ||
| 4 (26) | 26 | 3a | 26 | 11 | 12 | ||
| Y. pseudotuberculosis (40) | 40 | 35 | 35 | ||||
Weak signal only.
TABLE 5.
Y. enterocolitica strains with conflicting data from PCR and blotting
| Our strain collection no. | Serotype(s) | Biotype | Resulta for gene:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
yadA
|
virF
|
ail
|
ystA
|
ystB
|
||||||||
| PCR | Blotting | PCR | Blotting | PCR | Blotting | PCR | Blotting | PCR | Blotting | |||
| 22c | O:5 | 1A | − | − | + | − | (+) | − | (+) | − | − | + |
| 38 | NDb | 1A | −e | − | + | − | − | − | (+) | − | + | + |
| 122 | ND | 1A | + | + | + | + | − | − | − | − | + | + |
| 20d | O:5, O:27 | 3 | − | − | (+) | − | + | + | + | + | − | − |
| 32 | O:3 | 4 | −e | − | + | − | + | + | + | + | − | − |
| 54 | O:3 | 4 | − | − | − | − | + | + | + | + | (+) | − |
| 69 | O:3 | 4 | + | + | + | + | + | + | + | + | (+) | − |
| 86 | O:3 | 4 | − | − | − | − | + | + | + | + | (+) | − |
| 75 | ND | 1A | − | − | − | − | − | − | − | − | + | − |
−, negative; +, positive; (+), weak positive.
ND, tested as non-O:3 and non-O:9.
Reference strain IP Ye 21991.
Reference strain CCTM 3247.
Red colonies from CRAMP agar were yadA+ by PCR.
The predominant genotype for biotype 2 strains was ystA+ ail+ yadA+ virF+ (lacking ystB) (29 out of 44, 66%). The same genotype was also found in biotype 3 (8 out of 11, 73%). In addition, one biotype 3 strain tested positive for virF but not for yadA. Variation in plasmid content was also found in biotype 4, with a predominant genotype of ystA+ ail+ (lacking ystB, yadA, and virF) (12 out of 26, 46%). Three strains of biotype 4 were weakly positive for ystB (Table 5).
Our findings indicate that isolates should always be screened for the presence of the virulence plasmid (by PCR detection of yadA) as well as for at least one virulence gene located on the chromosome, in order to avoid the possibility that potentially pathogenic strains will be classified as apathogenic as a result of plasmid loss. We have observed that the virulence plasmid can easily be lost when the strains are subcultivated at temperatures higher than 30°C, if they are repeatedly subcultivated, or if they are stored over time. Even reference strains which are cultivated only at 30°C may lose the plasmid.
The predominant genotype for Y. pseudotuberculosis was inv+ yadA+ lcrF+ (35 out of 40 strains, 88%). It should be noted that 87.5% of the examined Y. pseudotuberculosis strains were plasmid-harboring strains. Absence of the virulence plasmid was less-frequently observed than with Y. enterocolitica, where the percentage of plasmid-harboring strains ranged from 0 to 72.73%, depending on the biotype. Furthermore, phenotypic tests screening for the presence of virulence plasmid, such as calcium-dependent growth at 37°C and Congo red uptake, were easier to read when screening Y. pseudotuberculosis than Y. enterocolitica. These results indicate that the plasmid is less-frequently lost in Y. pseudotuberculosis under the given culture conditions.
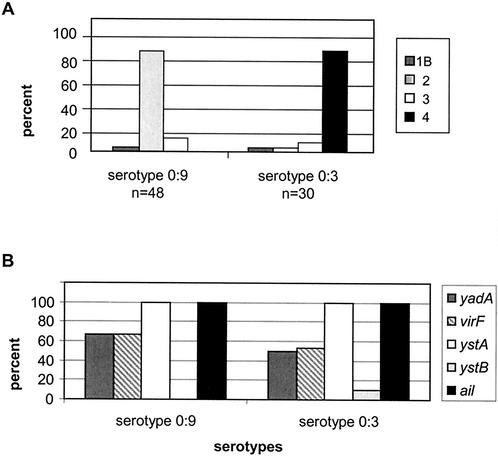
Y. enterocolitica strains of serotypes O:3 (n = 30) and O:9 (n = 48) were next compared for genotype and biotype (Fig. 2). The predominant genotype was ystA+ ail+ yadA+ virF+ (lacking ystB) for serotype O:9 (32 out of 48, 67%) as well as for serotype O:3 (14 out of 30, 47%). Three strains of serotype O:3 had weakly positive results for ystB (Table 5), and one strain of serotype O:3 was positive for virF but not for yadA. Thus, strains of the most common pathotypes in Switzerland, serotypes O:3 and O:9, did not differ in the distribution of their virulence factors.
FIG. 2.
Distribution of virulence genes and biotypes in serotypes O:3 and O:9 of Y. enterocolitica. (A) Distribution of the virulence genes in both serotypes. (B) Distribution of biotypes for the two serotypes
For each gene-specific PCR test, one PCR product was sequenced for confirmation and these products were used as probes in hybridization experiments in which ambiguous strains were further analyzed (Table 5). As a negative control, nonyersiniae and Yersinia spp. other than Y. pseudotuberculosis or Y. enterocolitica were included in the Southern blots. All negative-control lanes were negative (results not shown). PCR testing for ystA demonstrates an agreement of PCR results and hybridization of 97.1%. The presence of ystA or ystB was found to be mutually exclusive in most cases. In four cases, PCR results for ystB were positive but hybridization results were not, and the results were reversed in one case. Table 5 lists some Y. enterocolitica strains that were checked by hybridization because their by PCR results did not fit a typically expected genotype or because the obtained PCR products were weak. For some of these genes, no hybridization signal was obtained, suggesting that the PCR product was nonspecific. In other cases, ambiguous PCR results were obtained due to differences in the sensitivity of PCRs for presumably heterogeneous populations, for instance, in two cases where a virF PCR product but not a yadA product was obtained (both genes are plasmid borne). One such example was biotype 4 strain 32; it was further examined since it was heterogeneous on agar plates, showing a minority of red colonies on CRAMP agar. Such red colonies were genotyped again and were found to have yadA and virF by PCR. This suggests that the PCR for virF displayed a higher sensitivity than that for yadA, since in the heterogeneous population, the latter could not be detected by PCR. In general, strains giving weak PCR signals for virF, ail, ystA, and ystB resulted in negative hybridization, suggesting that such weak PCR bands can be ignored; however, biotype 1A strain 75 was convincingly PCR positive for ystB but negative by hybridization. Unexpectedly, reference strain ATCC 9610 (equivalent to CIP 80.27) was devoid of the ail gene, as shown by PCR and hybridization. In accordance with this result, Blais and Phillippe (7) classify this strain as avirulent because they also found the strain to lack yadA and ail. All other strains had matching data for PCR and hybridization, so that the correlation of PCR and hybridization was 97.1% for yadA, 100% for inv, 94.5% for virF/lcrF, 98.6% for ail, 97.1% for ystA, and 92.9% for ystB.
We next compared the obtained PCR results with phenotypic analysis. Out of 140 strains of Y. enterocolitica, 134 were tested for pyrazinamidase activity (Table 6). Three strains of biotype 1A were incorrectly typed as pyrazinamidase negative (all biotype 1A strains should be positive), and two biotype 2 strains were incorrectly found positive. Seven strains showed an intermediate reaction; 4 of them were biotype 1A strains, which were expected to give a positive result, and 3 of them were of biotype 2, expected to react negatively for pyrazinamidase activity. The pyrazinamidase test was thus found to be less conclusive and more ambiguous than PCR.
TABLE 6.
Correlation of Y. enterocolitica biotypes and results of pyrazinamidase testing
| Y. enterocolitica biotype (n) | No. of strains with pyrazinamidase result
|
No. of strains not tested | ||
|---|---|---|---|---|
| Positive | Negative | Intermediate | ||
| 1A (50) | 42 | 3a | 4b | 1 |
| 1B (9) | 0 | 7 | 0 | 2 |
| 2 (44) | 2c | 39 | 3d | 0 |
| 3 (11) | 0 | 11 | 0 | 0 |
| 4 (26) | 0 | 23 | 0 | 3 |
ystB positive.
All but one strain was ystB positive.
One strain was ystA+ ail+ yadA+ virF+ (lacking ystB), and one strain was ystA+ ail+ (lacking ystB, yadA, and virF).
Two strains were ystA+ ail+ yadA+ virF+ (lacking ystB), and one strain was ystA+ ail+ (lacking ystB, yadA, and virF).
The correlation of MOX agar with the plasmid-specific PCR for yadA and lcrF was 100% in the case of the 40 Y. pseudotuberculosis strains; however, the two assays correlated less well for Y. enterocolitica. Seven Y. enterocolitica strains were read as MOX positive but were negative in both plasmid-specific PCR assays. Ten strains of Y. enterocolitica did not show pinpoint colonies or had only very few pinpoint colonies but were positive in both plasmid-specific PCR assays, and 4 more strains were negative on MOX agar but reacted in PCR (confirmed by Southern blot) as yadA-lacking and virF+. Strains reacting positively on MOX agar were often a mixture of plasmid-positive and plasmid-negative colonies, and the number of pinpoint colonies varied between strains.
As with MOX agar, the correlation of CRAMP agar with the plasmid-specific PCR assays was good for Y. pseudotuberculosis. Only one Y. pseudotuberculosis strain (lacking yadA and lcrF) gave rise to a few positive colonies on CRAMP agar. With regard to Y. enterocolitica strains, no false-negative reactions were seen; however, 19 strains positive on CRAMP agar had a genotype lacking yadA and virF. These consisted of 12 biotype 1A strains and 7 strains of biotype 2 (n = 3), biotype 3 (n = 1), biotype 4 (n = 2,) and biotype 1B (n = 1). Positive CRAMP reactions were hard to read, since intermediate results such as rosa colonies or only a few red colonies were commonly formed. Variation in plasmid carriage is a known problem with Yersinia spp. Robins-Browne et al. (29) found that calcium dependence was the least-sensitive and -specific of the assays for plasmid carriage. Koeppel et al. (17) report that the proportion of microcolonies (plasmid-bearing cells) can range from 5 to over 95%. Plasmid-harboring strains on CRAMP agar in our hands frequently gave rise to an intermediate reaction, probably also due to the loss of a plasmid. Twelve Y. enterocolitica strains of biotype 1A were falsely positive by testing the Congo red uptake. Kwaga et al. (19) and Lewin et al. (20) found Y. enterocolitica strains of biotype 1A with plasmids of various sizes. Some of those strains showed positive reactions in the virulence assay. It was reported (20) that after 16 h all strains had at least a weak-positive reaction for Congo red uptake, but none were positive for low calcium response. Although genotyping is not completely devoid of false-positive or false-negative reactions, it performs at least as well as, and frequently better than, phenotyping. Moreover, the outcome is less subjective than that of phenotyping.
The predictive value of the obtained genotyping data for virulence was assessed. For all strains investigated, Y. enterocolitica strains of pathogenic biotypes (1B and 2 to 4) harbor the chromosome-borne virulence genes ystA and ail independent of their origin of isolation. Thus, a clear correlation exists between chromosome-borne virulence genes and biotype for these biotypes. As expected, the findings for plasmid-borne genes are less consistent due to the possible loss of a plasmid. Gene ystB is exclusively found in biotype 1A strains, independent of the source of isolates. This genotyping test thus performs at least as well as the pyrazinamidase testing, which allows differentiation between the pathogenic biotypes and biotype 1A strains but is difficult to read (Table 6). It is important to note, however, that biotype 1A strains isolated from clinical cases did not differ significantly in their virulence gene content (as detected by PCR) compared to 1A strains isolated from other sources. The genotypic characterization, therefore, has less predictive value for virulence in this group of strains. All Y. pseudotuberculosis isolates harbored the inv gene, and most isolates contained plasmid-borne genes (Table 4). However, lack of discrimination by genotyping based on yadA, lcrF, and inv resulted in little predictive value for the pathogenicity of Y. pseudotuberculosis.
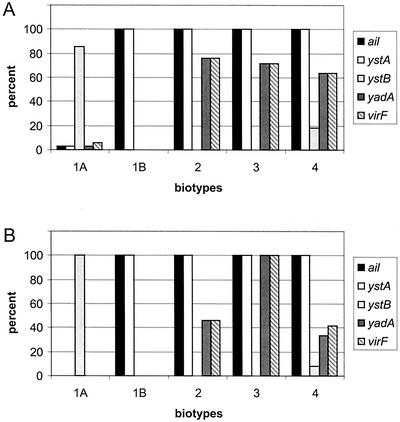
The results of genotyping disseminated to human and nonhuman isolates are presented in Fig. 3. Although there is no striking difference between human and nonhuman isolates, the most remarkable observation is the absence of ystB and the presence of ystA in strains of serotypes 1B and 2 to 4. Human and nonhuman isolates of serotype 1A did not differ in genotype.
FIG. 3.
Diagram of the distribution of Y. enterocolitica genotypes, as determined by PCR, in different biotypes. (A) Distribution of genotypes in biotypes isolated from human sources. (B) Distribution of genotypes in biotypes isolated from nonhuman sources.
The genotyping scheme used in this study clearly separated biotype 1A from the others. However, it did not differentiate between pathogenic and apathogenic strains within this serotype. Our results indicate that the best predictive value for the pathogenicity of the classical pathogenic serotypes of Y. enterocolitica is the genotype ystA+ (lacking ystB). If only these genes were tested, 96% of the strains belonging to biotypes 1B and 2 to 4 would be characterized correctly. However, 26% of human isolates (all biotype 1A) were ystB+ (lacking ystA), which is the predominant genotype for that serotype in our findings. One study reported a frequency of ystB presence in biotype 1A strains of 85% (14); another study (27) described 100% of biotype 1A strains as ystB positive. From our data, the PCR results for 6 strains of biotype 1A (out of 50) were negative for ystA and ystB; none of these strains were of human origin. It is possible that these isolates do not have a gene for enterotoxin or that these genes differ substantially in nucleotide sequence. Ramamurthy et al. (27) described an additional, rare subtype gene, ystC; 16.5% of their strains remained negative with all three probes. Similarly, Grant et al. (14) found 6 strains of Y. enterocolitica biotype 1A which were enterotoxin-producing strains but did not anneal to probes for ystA, ystB, and ystC. In our study, all human biotype 1A strains were positive for ystB, but 80% of the animal, food, and environmental strains were also positive and biotype 1A strains are overrepresented in food and environmental strains. The absence of a striking marker for virulence in biotype 1A strains isolated from human clinical cases may be illustrative of the intrinsic weak pathogenicity of this biotype. The clinical outcome is more likely determined by host factors than by bacterial virulence factors in this case. It has been said that biotype 1A strains, classified as avirulent Y. enterocolitica, are able to evoke clinical disease symptoms similar to those strains belonging to classical pathogenic bioserotypes (10, 24).
In conclusion, PCR-dependent detection of virulence genes results in a rapid characterization of Y. enterocolitica and Y. pseudotuberculosis isolates. These tests can now be further evaluated and refined for their potential to predict pathogenicity, even after loss of the virulent plasmid.
Acknowledgments
We thank Elizabeth Mumford, Colorado State University, for stimulating discussions. Denise Howald and Elisabeth Lüthi of the Swiss Federal Veterinary Office are acknowledged for their exceptional technical support.
REFERENCES
- 1.Aleksic, S., and J. Bockemühl. 1999. Yersinia and other enterobacteriaceae, p. 483-496. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 2.Aulisio, C. C., W. E. Hill, J. T. Stanfield, and R. L. Sellers, Jr. 1983. Evaluation of virulence factor testing and characteristics of pathogenicity in Yersinia enterocolitica. Infect. Immun. 40:330-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1999. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 4.Bhaduri, S., C. Turner-Jones, M. M. Taylor, and R. V. Lachica. 1990. Simple assay of calcium dependency for virulent plasmid-bearing clones of Yersinia enterocolitica. J. Clin. Microbiol. 28:798-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhaduri, S., C. Turner-Jones, and R. V. Lachica. 1991. Convenient agarose medium for simultaneous determination of the low-calcium response and Congo red binding by virulent strains of Yersinia enterocolitica. J. Clin. Microbiol. 29:2341-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhaduri, S., and B. Cottrell. 1997. Direct detection and isolation of plasmid-bearing virulent serotypes of Yersinia enterocolitica from various foods. Appl. Environ. Microbiol. 63:4952-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blais, B. W., and L. M. Phillippe. 1995. Comparative analysis of yadA and ail polymerase chain reaction methods for virulent Yersinia enterocolitica. Food Control 6:211-214. [Google Scholar]
- 8.Bölin, I., A. Forsberg, L. Norlander, M. Skurnik, and H. Wolf-Watz. 1988. Identification and mapping of the temperature-inducible plasmid-encoded proteins of Yersinia spp. Infect. Immun. 56:343-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bottone, E. J. 1997. Yersinia enterocolitica: the charisma continues. Clin. Microbiol. Rev. 10:257-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burnens, A. P., A. Frey, and J. Nicolet. 1996. Association between clinical presentation, biogroups and virulence attributes of Yersinia enterocolitica strains in human diarrhoeal disease. Epidemiol. Infect. 116:27-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornelis, G., C. Sluiters, C. L. de Rouvroit, and T. Michiels. 1989. Homology between VirF, the transcriptional activator of the Yersinia virulence regulon, and AraC, the Escherichia coli arabinose operon regulator. J. Bacteriol. 171:254-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delor, I., A. Kaeckenbeeck, G. Wauters, and G. R. Cornelis. 1990. Nucleotide sequence of yst, the Yersinia enterocolitica gene encoding the heat-stable enterotoxin, and prevalence of the gene among pathogenic and nonpathogenic yersiniae. Infect. Immun. 58:2983-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farmer, J. J., G. P. Carter, V. L. Miller, S. Falkow, and I. K. Wachsmuth. 1992. Pyrazinamidase, CR-MOX agar, salicin fermentation-esculin hydrolysis, and d-xylose fermentation for identifying pathogenic serotypes of Yersinia enterocolitica. J. Clin. Microbiol. 30:2589-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grant, T., V. Bennett-Wood, and R. M. Robins-Browne. 1998. Identification of virulence-associated characteristics in clinical isolates of Yersinia enterocolitica lacking classical virulence markers. Infect. Immun. 66:1113-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isberg, R. R., D. L. Voorhis, and S. Falkow. 1987. Identification of invasin: a protein that allows enteric bacteria to penetrate cultured mammalian cells. Cell 50:769-778. [DOI] [PubMed] [Google Scholar]
- 16.Kandolo, K., and G. Wauters. 1985. Pyrazinamidase activity in Yersinia enterocolitica and related organisms. J. Clin. Microbiol. 21:980-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koeppel, E., R. Meyer, J. Luethy, and U. Candrian. 1993. Recognition of pathogenic Yersinia enterocolitica by crystal violet binding and polymerase chain reaction. Lett. Appl. Microbiol. 17:231-234. [DOI] [PubMed] [Google Scholar]
- 18.Kuhnert, P., J. Hacker, I. Mühldorfer, A. P. Burnens, J. Nicolet, and J. Frey. 1997. Detection system for Escherichia coli-specific virulence genes: absence of virulence determinants in B and C strains. Appl. Environ. Microbiol. 63:703-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwaga, J., J. O. Iversen, and V. Misra. 1992. Detection of pathogenic Yersinia enterocolitica by polymerase chain reaction and digoxigenin-labeled polynucleotide probes. J. Clin. Microbiol. 30:2668-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewin, A., E. Strauch, S. Hertwig, B. Hoffmann, H. Nattermann, and B. Appel. 1996. Comparison of plasmids of strains of Yersinia enterocolitica biovar 1A with the virulence plasmid of a pathogenic Y. enterocolitica strain. Zentbl. Bakteriol. 285:52-63. [DOI] [PubMed] [Google Scholar]
- 21.Lindsay, J. A. 1997. Chronic sequelae of foodborne disease. Emerg. Infect. Dis. 3:443-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, V. L., J. J. Farmer III, W. E. Hill, and S. Falkow. 1989. The ail locus is found uniquely in Yersinia enterocolitica serotypes commonly associated with disease. Infect. Immun. 57:121-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, V. L., J. B. Bliska, and S. Falkow. 1990. Nucleotide sequence of the Yersinia enterocolitica ail gene and characterization of the Ail protein product. J. Bacteriol. 172:1062-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris, J. G., Jr., V. Prado, C. Ferreccio, R. M. Robins-Browne, A.-M. Bordun, M. Cayazzo, B. A. Kay, and M. M. Levine. 1991. Yersinia enterocolitica isolated from two cohorts of young children in Santiago, Chile: incidence of and lack of correlation between illness and proposed virulence factors. J. Clin. Microbiol. 29:2784-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pitcher, D., N. Saunders, and R. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 26.Prpic, J. K., R. M. Robins-Browne, and R. B. Davey. 1983. Differentiation between virulent and avirulent Yersinia enterocolitica isolates by using Congo red agar. J. Clin. Microbiol. 18:486-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramamurthy, T., K. I. Yoshino, X. Huang, G. Balakrish Nair, E. Carniel, T. Maruyama, H. Fukushima, and T. Takeda. 1997. The novel heat-stable enterotoxin subtype gene (ystB) of Yersinia enterocolitica: nucleotide sequence and distribution of the yst genes. Microb. Pathog. 23:189-200. [DOI] [PubMed] [Google Scholar]
- 28.Riley, G., and S. Toma. 1989. Detection of pathogenic Yersinia enterocolitica by using Congo red-magnesium oxalate agar medium. J. Clin. Microbiol. 27:213-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robins-Browne, R. M., T. Takeda, A. Fasano, A. M. Bordun, S. Dohi, H. Kasuga, G. Fang, V. Prado, R. L. Guerrant, et al. 1993. Assessment of enterotoxin production by Yersinia enterocolitica and identification of a novel heat-stable enterotoxin produced by a noninvasive Y. enterocolitica strain isolated from clinical material. Infect. Immun. 61:764-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skurnik, M., and H. Wolf-Watz. 1989. Analysis of the yopA gene encoding the Yop1 virulence determinants of Yersinia spp. Mol. Microbiol. 3:517-529. [DOI] [PubMed] [Google Scholar]
- 31.Straley, S. C. 1991. The low-Ca2+ response virulence regulon of human-pathogenic yersiniae. Microb. Pathog. 10:87-91. [DOI] [PubMed] [Google Scholar]
- 32.Wauters, G., K. Kandolo, and M. Janssens. 1987. Revised biogrouping scheme of Yersinia enterocolitica. Contrib. Microbiol. Immunol. 9:14-21. [PubMed] [Google Scholar]
- 33.Yoshino, K., H. Xiaozhe, M. Miyachi, Y. Hong, T. Takao, H. Nakao, T. Takeda, and Y. Shimonishi. 1994. Amino acid sequence of a novel heat-stable enterotoxin produced by a yst gene-negative strain of Yersinia enterocolitica. Lett. Pept. Sci. 1:95-105. [Google Scholar]