Abstract
Uranium-contaminated sediment and water collected from an inactive uranium mine were incubated anaerobically with organic substrates. Stimulated microbial populations removed U almost entirely from solution within 1 month. X-ray absorption near-edge structure analysis showed that U(VI) was reduced to U(IV) during the incubation. Observations by transmission electron microscopy, selected area diffraction pattern analysis, and energy-dispersive X-ray spectroscopic analysis showed two distinct types of prokaryotic cells that precipitated only a U(IV) mineral uraninite (UO2) or both uraninite and metal sulfides. Prokaryotic cells associated with uraninite and metal sulfides were inferred to be sulfate-reducing bacteria. Phylogenetic analysis of 16S ribosomal DNA obtained from the original and incubated sediments revealed that microbial populations were changed from microaerophilic Proteobacteria to anaerobic low-G+C gram-positive sporeforming bacteria by the incubation. Forty-two out of 94 clones from the incubated sediment were related to sulfate-reducing Desulfosporosinus spp., and 23 were related to fermentative Clostridium spp. The results suggest that, if in situ bioremediation were attempted in the uranium mine ponds, Desulfosporosinus spp. would be a major contributor to U(VI) and sulfate reduction and Clostridium spp. to U(VI) reduction.
Uranium-bearing wastes from nuclear weapons production (22) and mining (31) have generally been disposed of in near-surface environments. Dispersion of toxic aqueous uranium species through groundwater is of great environmental concern (30). In situ stimulation of the growth of microorganisms capable of immobilizing dissolved uranium has been proposed as a potentially cost-effective remediation method (23, 24).
In the laboratory, it has been demonstrated that microorganisms can reduce hexavalent uranium [U(VI)] to tetravalent uranium [U(IV)] and precipitate a U(IV) mineral called uraninite (UO2) (27, 40). Microorganisms that reduce U(VI) in pure culture include a hyperthermophilic archaeon (15), a thermophilic bacterium (19), mesophilic Fe(III)- and sulfate-reducing bacteria (4, 5, 34, 25, 27, 28), and fermentative bacteria (9). Thus, the ability to reduce U(VI) occurs in phylogenetically diverse organisms. In laboratory studies, U(VI) is also reduced by microbes in solutions that contain organic or inorganic ligands or other cations (13, 26, 33) or that contain other electron acceptors such as Fe(III) oxides, sulfate, or selenate (12, 24, 40, 45).
Microbial U(VI) reduction in uranium-contaminated settings has been studied by incubating field-collected sediment and water with organic substrates to stimulate the growth of indigenous microorganisms in the laboratory (1, 2, 15). Although previous studies showed uranium removal from solution during laboratory incubation, the mechanisms by which uranium was removed from solution and the microbial species responsible remain unclear.
In this study, we attempted to better understand the bioremediation process through integration of results obtained from molecular biological, geochemical, and mineralogical studies. Field-collected uranium-contaminated sediment and water samples were incubated anaerobically with organic substrates, which resulted in removal of uranium from solution. Here we studied the mechanisms of uranium removal in detail by analyzing solution chemistry and characterizing solid phases, including minerals and microbial cells. Microbial communities before and after the incubation were also studied by culture-independent molecular biological techniques.
MATERIALS AND METHODS
Sampling site.
The Midnite mine is an inactive open-pit uranium mine located in Stevens County in eastern Washington. Most of the pits were backfilled with waste rock during mining operations. Pits 3 and 4 are open and are partially filled with water. The water in pit 3 comes from various sources, including infiltration, precipitation, and a seep collection system. Groundwater emerging from seeps at the base of a large waste rock pile is collected and pumped to pit 3 to prevent contaminant release to the mine drainage and downstream water bodies. Water from pit 3 is contaminated with uranium, manganese, sulfate, nitrate, and other toxic metals (42). To meet permit limits prior to discharge into one of the mine drainage systems, water from pit 3 is combined with less contaminated water from pit 4 and passed through a lime precipitation treatment plant.
Sample collection.
In July of 2000, sediment was collected from 50 cm below the surface near the water edge of pit 3 at the Midnite mine. The pit water was collected near this sampling site. The pit sediment was transferred into an anaerobic jar (Difco, Detroit, Mich.) with a GasPak Plus (H2 + CO2) (BBL, Cockeysville, Md.) immediately after collection. The pit water sample to be used for chemical analysis was filtered through a 0.2-μm nylon filter with polypropylene housing at the site. Unfiltered pit water was stored aerobically for experiments at 4°C. The sediment and water samples were kept on ice during the 2 days required for shipment to the laboratory and stored at 4°C before the experiments. Subsamples of the pit sediment were stored at −20°C for the molecular analysis described below. The pH, Eh, and conductivity of the pit water were measured on site.
Anaerobic incubation of pit sediment and water with organic substrates.
The pit sediment (5.0 g) and 50 ml of deoxygenated pit water in a serum bottle (100 ml) sealed with a rubber stopper and an aluminum cover with the headspace filled with N2 were autoclaved twice at 120°C for 20 min. Organic substrates (0.01 g each of lactate, acetate, ethanol, benzoic acids, and glucose per liter and 0.02 g each of yeast extract and peptone per liter) were added from anaerobic stocks in an anaerobic chamber (Coy, Grass Lake, Mich.) with an anaerobic gas mixture containing N2, CO2, and H2 (90:5:5). Hereafter, the mixture of the pit sediment and water with the organic substrates is called pit 3 medium. Unautoclaved pit sediment (0.5 g) was inoculated into pit 3 medium. The pit 3 medium was incubated under the anaerobic gas mixture without the addition of unautoclaved sediment as the abiotic control. During incubation, the supernatants from pit 3 medium and the abiotic control were sampled over time, and the supernatants that had been filtered through a 0.2-μm filter were analyzed for manganese, sulfate, sulfide, and uranium concentrations. After 1 month, subsamples of pit 3 medium incubated with the inoculum were stored at −20°C for subsequent molecular biological analysis. Filtered supernatants from pit 3 medium before and after 1 month of incubation were analyzed for their chemical compositions. All experiments were done in triplicate.
Analytical techniques.
Chemical analyses of the pit water, filtered on site, and the filtered supernatants from pit 3 medium were conducted by inductively coupled plasma optical emission spectrometry (ICP-OES) (Jarrell ash IRIS high resolution) and ion chromatography (Dionex DX-500). Mn, sulfate, and sulfide concentrations were measured in the anaerobic chamber by a spectrophotometer (Hach DE/2010) using the methods of periodate oxidation, sulfaVer 4, and methylene blue, respectively (Hach Water Analysis Handbook; Hach, Loveland, Colo.). For uranium, the filtered supernatants were acidified with 1 N HCl in the anaerobic chamber and analyzed by ICP-OES. All of the measurements were carried out at least in duplicate for each sample.
XANES analysis.
To prepare samples for X-ray absorption near-edge structure (XANES) analysis, the samples of incubated pit 3 medium with the inoculum were shaken vigorously for 5 min by hand and then left to sit on a laboratory bench. Large sediment particles settled out within 5 min. Supernatant samples containing finer sediment particles and microbial cells were withdrawn and spun down at 10,000 × g for 5 min. Each of three portions, consisting of pellets, larger sediment particles, and supernatant, were mounted separately on a Plexiglas sample holder, doubly sealed with kapton and a polypropylene bag. All sample preparations were performed in an anaerobic chamber to maintain the anaerobic conditions of the samples.
U L3-edge X-ray absorption fine structure (XAFS) measurements of these samples were performed at the Material Research Collaborative Access Team beamline (38) at the Advanced Photon Source. The data were collected in fluorescence mode due to the small concentrations of U in the samples. The undulator gap was tapered 4 mm to achieve less than 15% change in X-ray energy. An Rh mirror was used to remove high-order harmonics. Slits were used to define the incident X-ray profile of 0.7 mm by 1.0 mm. The incident and transmitted ion chambers were filled with nitrogen gas, and the fluorescence ion chamber was filled with Kr gas. Linearity tests of the experimental setup (18) indicated less than 0.1% nonlinearity for a 50% attenuation of the incident X-ray intensity. The fluorescence measurements were made in slew scanning mode with 0.1-s integration time, resulting in about 2 min per XAFS scan. Five XANES measurements were made on each sample. No time-dependent change was found in the XANES measurements for any of the samples on the 2-min time scale, indicating that the valence state of the uranium in the samples was not affected by the radiation dose on the sample at these time scales.
The normalization was performed with ifeffit (32) and a Cromer-Libermann normalization algorithm. A Y-foil was monitored during the data collection in the manner previously described (6) to align the XANES data to the U(IV) and U(VI) standards, which were taken from UO2 and UO3 powder samples, respectively. The standards, which were diluted in SiO2, were spread on Kapton tape and measured in fluorescence mode. The U(IV) standard used in these experiments may not be considered ideal, as it has been shown in the past to contain up to 25% U(VI). However, it can be used to give an approximation of U(IV) edge position.
TEM.
The supernatant of the incubated pit 3 medium that contained fine sediment and microbial cells was prepared as described above, and 10 μl of the supernatant was mounted onto Formvar-coated 200-nm-mesh Cu or Ni grids in the anaerobic chamber and observed by transmission electron microscopy (TEM) (Philips CM 200). Chemical compositions of materials in the fraction containing finer sediment particles and microbial cells were analyzed by energy-dispersive X-ray spectroscopy. Mineral phases present in the fractions were characterized by selected area electron diffraction (SAED) analysis.
DNA extraction.
With the UltraClean Mega Prep soil DNA kit (Mo Bio Laboratories, Solana Beach, Calif.), community nucleic acids were extracted from the original pit sediment stored at −20°C, and the sediment was incubated anaerobically for 1 month, as described above.
PCR and cloning.
Community 16S rRNA genes were amplified by PCR in mixtures containing 1 to 50 ng of DNA per μl, 1× PCR buffer (Perkin Elmer, Norwalk, Conn.), a 200 μM concentration of each of the four deoxynucleoside triphosphates, 2.5 mM MgCl2, 350 mM each of the forward and reverse primers, and 0.025 U of AmpliTaq Gold (Perkin Elmer) per μl. In reactions, the reverse primer was the universal 1492R (5′-GGTTACCTTGTTACGACTT-3′) (21), and the forward primer was the Bacteria-specific 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) (21) or universal 533F (5′-GTGCCAGCMGCCGCGGTAA-3′, where M is A or C) (21). A Gene Amp 2400 (Perkin Elmer) was used to incubate mixtures through an initial denaturation at 94°C for 5 min, followed by 30 cycles of 94°C for 1 min, 45°C for 45 s, and 72°C for 1.5 min, and completed with an extension period of 20 min at 72°C. The products were purified with QIAquick PCR purification columns (Qiagen, Valencia, Calif.) and quantified by ethidium bromide-UV detection on a 1% agarose gel. The quantified products (an insert-vector ratio of 3:1) were ligated into the vector pGEM-T (Promega, Madison, Wis.), and the inserted vectors were transformed into competent host cells following the manufacturer's instructions.
RFLP screening and sequencing.
For restriction fragment length polymorphism (RFLP) screening and sequencing, the inserted 16S ribosomal DNA (rDNA) was amplified by PCR as described above except that cloned host cells were added directly in the PCR mixture. The vector-specific T7 and SP6 primers were used for PCR. Aliquots of amplified rDNA PCR products were digested with 1 U each of the 4-base-specific restriction endonucleases HinPI and MspI in 1× NEB buffer 2 (New England Biolabs, Beverly, Mass.) and 0.01% Triton X-100 overnight at 37°C. Digested products were separated by agarose (3%) gel electrophoresis. Bands were visualized by staining with ethidium bromide and UV illumination. RFLP patterns were grouped, and representative patterns were selected for sequencing.
Purified PCR products (QIAquick column; see above) were sequenced with the Prism Big Dye terminator sequencing kit (Applied Biosystems, Foster City, Calif.) with 50 to 100 ng of template DNA by following the manufacturer's instructions. Initially, partial sequences were obtained with the universal primer 533F in sequencing reactions. Extended sequences were obtained with primers 27F and 1492R in separate sequencing reactions. DNA sequences were determined on an automated sequencer (ABI 377XL) at the University of Wisconsin Biotechnology Center.
Phylogenetic analysis.
Phylogenetic affiliations of the partial sequences were estimated with the program Blast (Basic Local Alignment Search Tool) (3) and available nucleotide databases. Single primer sequences were aligned with the GDE (Genetic Data Environment) multiple sequence editor against close relatives in ARB (a software environment for sequence data) (41). Similarity of partial sequences was determined with ARB, and those with more than 98% similarity were grouped. Extended sequences of representatives from groups were compiled with SeqEd (Applied Biosystems). Chimeric sequences were checked by the program Check_Chimera (29). Chimeric sequences were also checked by comparing phylogenetic affiliations of the 5′ and 3′ halves of each sequence. Sequences were managed in ARB and reduced to unambiguously alignable positions with the mask (21). Evolutionary analysis of alignments was performed by distance methods, parsimony, and maximum likelihood with PAUP (44).
RESULTS
Chemical characteristics of original pit 3 water.
The temperature, pH, Eh, and conductivity of the surface pit water, measured on site in July 2000, were 22.8°C, 4.12, 222 mV, and 3.1 mS/cm, respectively. The chemical composition of the pit water analyzed by ICP-OES and ion chromatography is shown in Table 1. As described previously (42), the pit water contained high concentrations of U, Mn, and sulfate at 23.87, 143, and 3014 ppm, respectively. The water also contained relatively high concentrations of nitrate and toxic metals such as Co, Ni, and Zn.
TABLE 1.
Chemical compositions of the original water samples from pit 3 (P30), the supernatant (sup) from the pit3 medium before incubation with the sediment inoculum, the supernatant after 1 month of incubation with the inoculum, and the supernatant after 1 month incubation without the inoculum (the abiotic control)a
| pH or chemical | Concn (mg/liter)
|
|||
|---|---|---|---|---|
| P30 | Sup | Incubated sup
|
||
| Inoculated | Uninoculated | |||
| pH | 4.1 | 6.1 | 6.9 | 6.9 |
| K | 5.1 | 18 | 17 | 17 |
| Na | 33 | 102 | 163 | 106 |
| Li | 0.45 | 0.10 | 0.04 | 0.05 |
| Mg | 309 | 218 | 214 | 223 |
| Ca | 618 | 501 | 541 | 507 |
| Mn | 143 | 81 | 79 ± 11b | 85 |
| Fe | 0.18 | ND | ND | 0.02 |
| Co | 1.6 | 0.04 | ND | 0.05 |
| Ni | 2.7 | 0.11 | ND | 0.08 |
| Cu | 0.18 | ND | ND | 0.03 |
| Zn | 5.6 | 0.30 | ND | 0.35 |
| Cd | 0.05 | 0.02 | ND | 0.02 |
| Pb | 0.04 | 0.05 | 0.05 | 0.04 |
| U | 24 | 13 | 0.30 | 16 |
| F− | 3.8 | 1.7 | 1.3 | 1.7 |
| Cl− | 38 | 70 | 139 | 73 |
| NO3− | 3.0 | 5.5 | ND | 5.5 |
| PO43− | NDc | ND | ND | ND |
| SO42− | 3,010 | 2,710 | 2,320 | 2,730 |
| S2− | ND | ND | 13 ± 3.5 | ND |
All values are the means of the triplicate measurements and range within less than 10% errors, otherwise indicated.
Average ± SD.
ND, not detected by ICP-OES or ion chromatograply.
Anaerobic incubation of pit 3 medium.
The chemical composition and pH of the filtered supernatants from pit 3 medium at the beginning and the end of the incubation are shown in Table 1. The pH of the original pit 3 water increased from 4.12 to 6.14 after autoclaving with the pit 3 sediment and the organic substrates, probably due to the dissolution of carbonate minerals present in the pit sediment during autoclaving. The concentrations of the major elements, toxic metals, and anions (except some monovalent cations and anions such as K+, Na+, Cl−, and NO3−) decreased notably after autoclaving, likely due to precipitation or adsorption. After 1 week, pit 3 medium incubated with the inoculum became turbid, and microbial cells were observed by light microscopy. After 3 weeks of incubation, the medium turned black. In contrast, no change in color was observed for the abiotic controls.
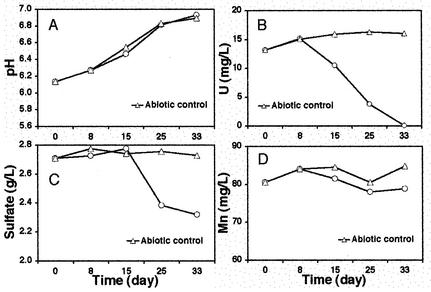
During incubation, the changes in the pH and the concentrations of U, Mn, and SO42− were monitored. Results are shown in Fig. 1. The pH of the pit 3 medium and the abiotic control increased from 6.14 to around 6.9. The nitrate concentration in pit 3 medium decreased from 5 ppm to under the detection limit (0.1 ppm) at the end of the incubation. In contrast, no change was observed for the abiotic control. The concentration of U in pit 3 medium started decreasing after 8 days of incubation. After 1 month, the U concentration decreased down to 0.3 ppm. The U concentration in the abiotic control increased slightly, probably because U adsorbed or precipitated during autoclaving was slowly released into solution during incubation at room temperature. The concentration of sulfate in pit 3 medium began to decrease after 15 days of incubation and reached 2.3 g per liter after 1 month. We also detected ∼13 mg of sulfide per liter in solution after 1 month of incubation. No changes in the sulfate and sulfide concentrations were observed for the abiotic control. In contrast to nitrate, U, and sulfate, the concentration of Mn in solution was constant at around 80 ppm throughout the incubation.
FIG. 1.
Changes in the pH (A) and in the concentrations of uranium (B), sulfate (C), and manganese (D) in pit 3 medium during anaerobic incubation. All data points are the means of triplicate measurements.
XANES.
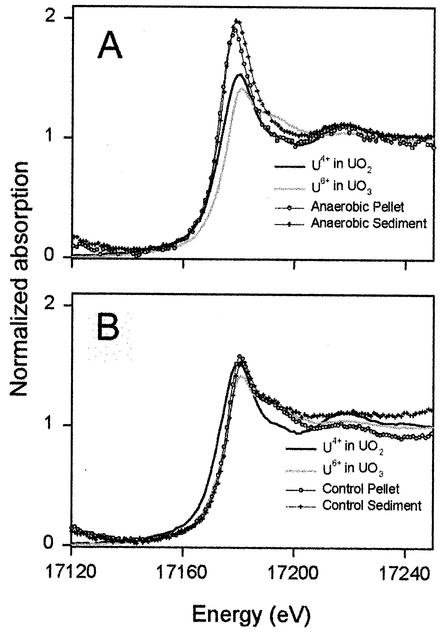
Naturally occurring samples typically contain yttrium (Y) concentrations similar to uranium concentrations. This is problematic because the Y L3-edge energy is just 100 eV below that of uranium and affects the normalization factor by approximately 10%. The Y L3-edge also causes the nonlinear pre-edge adsorption slopes shown in Fig. 2. Therefore, our ability to resolve the U(VI) and U(IV) edge positions was limited to about 10%.
FIG. 2.
Averaged normalized U L3-edge absorption data for the anaerobic pit 3 (A) and abiotic control (B) samples and for the U(IV) and U(VI) standards from UO2 and UO3, respectively.
Figures 2A and B show the averaged normalized XANES spectra of the U(IV) and U(VI) standards and samples from the incubated pit 3 medium and the abiotic control, respectively. The uranium L3-edge positions from the pellet and the larger sediment particles in pit 3 medium (referred to as the anaerobic pellet and anaerobic sediment, respectively, in Fig. 2A) were consistent with the edge position of the U(IV) standard, indicating that the vast majority (>90%) of the uranium in these samples has been reduced to U(IV). The uranium L3-edge positions from the pellet and the large sediment particles in the abiotic control (referred to as the control pellet and control sediment, respectively, in Fig. 2B) were the same as the edge position of the U(VI), indicating that the majority (>90%) of the uranium is U(VI) in these samples. The uranium L3-edge from the supernatant from pit 3 medium and the control were not detected due to their low concentrations.
TEM.
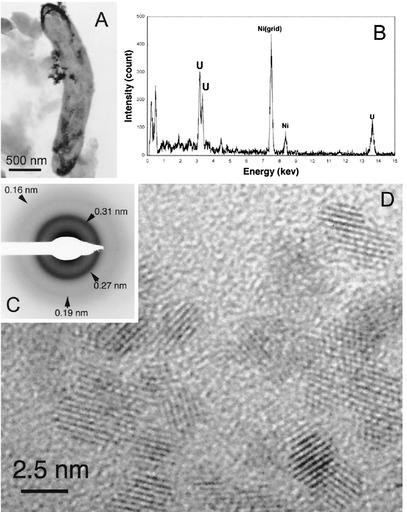
TEM observations of the fraction analyzed by XANES that contained microbial cells and small particles revealed that microbial cells were relatively abundant. Energy-dispersive X-ray spectroscopy analyses (∼50-nm resolution in diameter) of many bacterial cells showed that the cells were significantly enriched with uranium (Fig. 3A and B). SAED pattern analysis of the microbial cells enriched with uranium showed that these cells were associated with crystalline precipitates, which had interplanar spacings matching those of uraninite (UO2, Fig. 3C). High-resolution TEM observations of the microbial cells enriched with uranium revealed that the cell surfaces were entirely coated with nanometer-scale crystalline particles of uraninite (Fig. 3D).
FIG. 3.
(A) TEM images showing that microbial cells are entirely coated with extracellular precipitates. (B) Energy-dispersive X-ray spectrum obtained from the extracellular precipitates. (C) SAED pattern of the extracellular UO2 with interplanar spacings. (D) High-resolution TEM image of nanocrystalline UO2 in the precipitates.
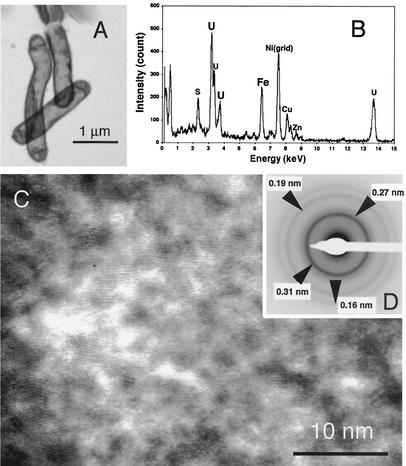
TEM observations and energy-dispersive X-ray spectroscopy analyses showed that some microbial cells were enriched with Cu, Fe, Zn, and S as well as U (Fig. 4A and B). High-resolution TEM observations showed that the microbial cells were entirely coated with nanometer-scale crystalline particles (Fig. 4C). The analysis of the SAED pattern obtained from the particles had only interplanar spacings matching those of uraninite, although the particles contained S, Fe, Cu. and Zn besides U (Fig. 4D).
FIG. 4.
(A) TEM image showing that microbial cells are associated with extracellular precipitates of UO2 and metal sulfides. (B) Energy-dispersive X-ray spectrum obtained from the UO2 and metal sulfides. (C) SAED pattern of the UO2 and metal sulfides. (D) High-resolution TEM image of UO2 and metal sulfides.
Analysis of rRNA-based libraries from original and incubated pit 3 sediments.
PCR-based methods were used to study how microbial populations in the original pit 3 sediment were changed by anaerobic incubation with organic substrates. 16S rRNA genes (rDNA genes) were amplified from the original and the inoculated, incubated sediments and sequenced. Amplification of 16S rDNA with an Archaea-specific primer set (7) was unsuccessful. Clones from the original sediment, designated P3OB or P3OU, and from the incubated sediment, designated P3IB or P3IU, were analyzed further.
Thirty-two RFLP groups were obtained from the original pit 3 sediment (P3O) and 24 from the incubated pit 3 sediment (P3I). Representatives from the RFLP groups were sequenced with the single universal primer (533F), and sequences with more than 98% similarity were grouped. The RFLP groups were reduced to 15 and 10 representatives of P3O and P3I, respectively. Efficacy of the grouping was confirmed by partial sequencing of multiple representatives from the larger RFLP groups. The representative sequences were extended with the primers 1492R and 27F. The collection of pit 3 sequences was inspected for the occurrence of chimeric sequences, as described above. Three clones were found to be strongly chimeric and excluded from further analysis.
Phylogenetic distribution of pit 3 sequences.
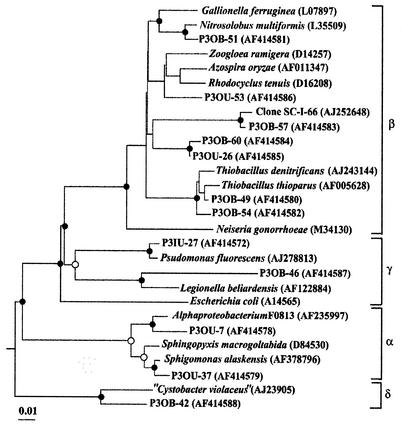
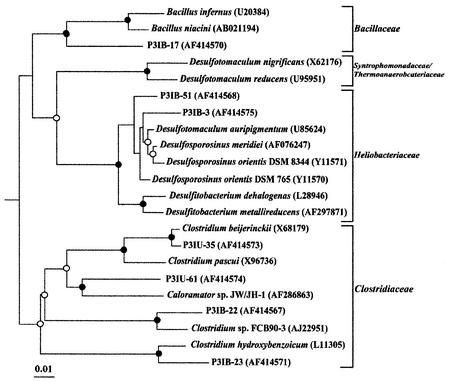
Evolutionary distance dendrograms of pit 3 type sequences and sequences available from the databases are shown in Fig. 5 and 6. In the original pit 3 sediment, most of the sequences were clustered within the taxonomic division of Proteobacteria (Fig. 5). In sharp contrast, most of the sequences obtained from the incubated sediment were positioned within low-G+C gram-positive bacteria (Fig. 6). The type sequences of P3OU-1 and P3IB-39, representing 6 of 66 P3O clones and 16 of 94 P3I clones, respectively, were closely related to the same clone sequence in the genus Cytophaga. Type sequence P3IU-27 was the only sequence in the P3I libraries belonging to the γ-Proteobacteria and was closely related to Pseudomonas fluorescens (>98% sequence similarity) (Fig. 5).
FIG. 5.
Evolutionary distance dendrogram of pit 3 clones with sequences within the Proteobacteria division based on 1,220 nucleotides of 16S rDNA. Classes are bracketed at the right of the figure. Bacillus subtilis was used as the outgroup (not shown). Branch points supported by distance, maximum-likelihood, and parsimony estimations (bootstrap values ≥75%) are indicated by solid circles. Marginally supported branch points (supported by most phylogenetic analyses with bootstrap values of 50 to 74%) are indicated by open circles. Branch points without circles are not supported by the majority of analyses. Evolutionary distances are indicated by the sum of horizontal branch lengths. The scale bar represents changes per nucleotide.
FIG. 6.
Evolutionary distance dendrogram of pit 3 clones with low-G+C gram-positive bacteria sequences based on 1,220 nucleotides of 16S rDNA. Families are bracketed at the right of the figure. Escherichia coli was used as the outgroup (not shown). Indication of branch point support and evolutionary distance are as described for Fig. 5.
The type sequences in P3O libraries positioned within the Proteobacteria were distributed over the α, β, γ, and δ classes. Forty-one of 66 clones were clustered within the β class (Fig. 5). The most abundant type sequence in the P3O libraries, representing 14 of 66 P3O clones, was P3OB-49. This sequence was closely related to Thiobacillus thioparus and Thiobacillus denitrificans (>98% sequence similarity, as shown in Fig. 5). Initially, type sequences of P3OB-60 and P3OU-26, which comprised 14 of 66 clones, had database matches with Zoogloea ramigera (93% database match) and Azospira oryzae (93% database match), respectively, in the Rhodocyclus-Azoarcus subgroup of the β-Proteobacteria. However, after further phylogenetic analysis, these type sequences clustered separately from these organisms. P3OB-51, representing 5 of 66 P3O clones, was closely related to Nitrosolobus multiformis, having 98% similarity (Fig. 5). We obtained two type sequences that belong to α-Proteobacteria and were related to Sphingomonas spp. (98% sequence similarity). In the P3O libraries, the type sequence of P3OB-46 had the lowest similarity to known sequences (<90% sequence similarity).
All type sequences in the P3I libraries except P3IB-39 and P3IU-27 were clustered within the low-G+C gram-positive bacteria. The most abundant sequence type in the P3I libraries was P3IB-3 (34 of 94 P3I clones). This sequence was strongly clustered with gram-positive sulfate-reducing bacteria, Desulfosporosinus spp. The sequence type P3IB-51, representing eight clones, was also related to Desulfosporosinus spp. Four type sequences comprising P3IU-35, P3IU-61, P3IB-22, and P3IB-23 were positioned within the family Clostridiaceae and accounted for 23 of 94 P3I clones. The type sequence of P3IU-35, representing eight clones, was closely related to Clostridium beijerinckii (>98% sequence similarity).
DISCUSSION
Inference of physiology from phylogeny of organisms in P3O libraries.
The physiological properties of organisms represented by type sequences found in the P3O libraries were inferred from the properties of their characterized relatives. Approximately 24% of the clones analyzed in the P3O libraries were associated with Thiobacillus denitrificans. T. denitrificans is autotrophic, capable of facultative anaerobic growth by coupling nitrate reduction to oxidation of S compounds such as sulfur, sulfide, thiosulfate, tetrathionate and thiocyanate (17). Optimum temperature and pH for growth are 28 to 32°C and 6.8 to 7.4, respectively. Nitrosolobus multiformis oxidizes reduced N compounds such as ammonia and nitrite (47). P3OB-60 and P3OU-26 and P3OU-53 made up about 24% of the clones analyzed in the P3I libraries and were to some extent related to the Rhodocyclus-Azoarcus subgroup of the β-Proteobacteria (Fig. 5). Many species in this group are capable of facultative anaerobic growth, with denitrification coupled to degradation of aromatic compounds (35, 39). In addition, Sphingomonas spp. have been reported to degrade aromatic compounds (10, 11).
The physiological inferences described above suggest an abundance of denitrifying organisms in the pit 3 sediment that was collected from 50 cm below the sediment-air interface at the pond edge. Thus, we suggest that this habitat was probably microaerophilic. As we found some organisms that are capable of degrading aromatic compounds in the libraries, naturally occurring aromatic compounds, such as lignin, may be a major organic carbon source available to organisms in the sediment. We also found organisms related to lithotrophs that use S or N compounds for energy sources. Thus, the original natural pit 3 sediment (prior to incubation with organic compounds) was suggested to be oligotrophic and to sustain organisms that can degrade aromatic compounds and/or grow chemolithotrophically.
Biogeochemical processes occurring during anaerobic incubation with organic substrates.
As a result of the change from the microaerophilic and oligotrophic conditions of the original pit 3 sediment to anaerobic and nutrient-rich conditions, microbial populations present in the original sediment changed significantly. Although clone libraries may not be fully representative of the assemblage present due to PCR amplification bias, our results suggest that sporeforming anaerobic bacteria, including Clostridium and Desulfosporosinus spp., are abundant in the incubated sediments. As the original sediment was inferred to be microaerophilic, it is likely that these organisms were dormant in the original sediment and not detected by the RNA-based molecular biological techniques. C. beijerinckii, C. hydroxybenzoicum, and Desulfosporosinus spp. are capable of degrading aromatic compounds (14, 37, 48). Under natural conditions, seasonal fluctuations may cause the environment in the sediment to become anaerobic, stimulating the metabolism of naturally occurring aromatic compounds by these organisms.
Nitrate reduction.
For anaerobic respiration, microorganisms utilize electron acceptors other than oxygen. Coupling oxidation of organic compounds with nitrate reduction is more thermodynamically favorable than reduction of other electron acceptors abundantly present in the medium [e.g., Mn(IV), U(VI), and sulfate]. Thus, nitrate was probably reduced before Mn(IV), U(VI), and sulfate. Type sequence P3IU-27 was closely related to sequences from denitrifying bacteria such as Pseudomonas fluorescens. This organism may be responsible for removal of nitrate prior to the onset of Mn, U, and sulfate reduction.
Fe(III) and Mn(IV) reduction.
Although the total Fe concentrations in the supernatants were under our detection limit (0.01 ppm) throughout the incubation, Fe(II) was precipitated with sulfide produced by sulfate-reducing bacteria (Fig. 4). C. bijerinckii, Desulfosporosinus orientis DSM 765, and Desulfosporosinus meridiei have been described to reduce Fe(III) (8, 37). Thus, organisms in pit 3 sediments that are related to these organisms may reduce Fe(III) to Fe(II). Mn(IV) is well known to serve as an electron acceptor in a variety of microbial metabolisms. The pit 3 water originally contained 143 ppm of Mn, probably as Mn(II). Autoclaving the pit water with the sediment resulted in removal of Mn from solution (down to ∼80 ppm), most likely due to oxidative precipitation of Mn(IV) solid phases. This indicates that Mn(IV) was in a form that could be utilized by microorganisms. However, the Mn concentrations in the supernatants were constant throughout the incubation, regardless of whether microbes were active. This indicates that microbes did not catalyze reductive dissolution of Mn(IV) solid phases and that Mn(IV) may not be a preferred electron acceptor for organisms in the pit 3 sediment.
U(VI) reduction.
In previous studies with field-collected uranium-contaminated samples, Abdelouas et al. (1, 2), Kauffman et al. (16), and Lovley and Phillips (26) demonstrated uranium removal from solution by enhanced microbial activities when organic substrates were added to the samples.
Kauffman et al. (16) showed that uranium is removed from uranium mine discharge water when the discharge water was percolated through a column packed with soil collected from the uranium mine with the addition of sucrose. As the uranium removal occurred after hydrogen sulfide was produced, Kauffman et al. (16) inferred that hydrogen sulfide, which is produced by native sulfate-reducing bacteria in the soil, reduces U(VI) indirectly. Lovley and Phillips (26) showed that a pure culture of Desulfovibrio desulfuricans removes U(VI) from various uranium-contaminated waters collected from a uranium mine and the Department of Energy site in Hanford, Wash. Abdelouas et al. (1, 2) showed microbial cells precipitating UO2 from a solution to which about 250 ppm of uranium was added in the laboratory. These investigations suggested that removal of uranium in other samples to which no uranium was added also resulted from reductive microbial precipitation of UO2 and that other possible mechanisms, such as adsorption to solid surfaces and coprecipitation, were unlikely.
In this study, XANES analysis confirmed that most of the uranium removed from solution was U(IV). Without microbial activity, no U(IV) was detected by XANES, indicating that U(VI) reduction requires microbial activity. In this study, no uranium was added in the laboratory. Thus, microorganisms can induce precipitation of UO2 from relatively dilute solutions with initial concentrations of U(VI) at ∼20 ppm. TEM observations and energy-dispersive X-ray spectroscopy data show that UO2 was directly associated with microbial cells. These observations, taken alone, do not rule out indirect reduction by sulfide produced by sulfate-reducing bacteria. However, data showing that U(VI) reduction was initiated before sulfate reduction started (Fig. 1) support the conclusion that uranium reduction occurred via a direct enzymatic pathway.
Francis et al. (9) showed U(VI) reduction by a Clostridium sp. Thus, it is inferred that organisms related to Clostridium spp. are partially responsible for U(VI) reduction and removal of uranium from solution. This result implies that U(VI) can be reduced in natural and contaminated anaerobic sediments during fermentation, before the onset of dissimilatory Fe(III) or sulfate reduction with H2 and short-chain fatty acids produced by fermentation.
Sulfate and U(VI) reduction.
The sulfate concentration decreased after U(VI) reduction started (Fig. 1). After 1 month of incubation, sulfide (∼13 ppm) was detected (Table 1). We found microbial cells precipitating solid phases containing Fe, Cu, Zn, S, and U, as shown in Fig. 4B. The patterns from sphalerite (ZnS) and chalcopyrite (CuFeS2) were indistinguishable from that of uraninite due to their almost identical lattice parameters. Thus, we infer that the SAED pattern of the precipitates (Fig. 4C) includes those from the three different mineral phases and that sulfate-reducing bacteria precipitated nanocrystalline uraninite, sphalerite and chalcopyrite.
In the P3I clone libraries, all sequence types that were related to sulfate-reducing bacteria clustered with Desulfosporosinus spp. In a separate study, we isolated an organism related to Desulfosporosinus spp. from the pit 3 sediment and showed that this, as well as a type species, enzymatically reduced U(VI) (Y. Suzuki, S. D. Kelly, K. M. Kemner, and J. F. Banfield, submitted for publication). Thus, we conclude that organisms closely related to Desulfosporosinus spp. will reduce U(VI) and sulfate in the natural sediment when it becomes anaerobic and during bioremediation.
Considering the biases associated with analysis of microbial communities based on the PCR methods used here (36, 43), quantification of organism populations based on the numbers of clones obtained in the libraries is impossible. Although other methods such as in situ hybridization are needed to confirm the abundance of Desulfosporosinus spp., the clone library provides an initial indication of microbial diversity and confirms the presence of likely U(VI)-reducer organisms (Desulfosporosinus and Clostridium spp.). Organisms other than those found in the P3I libraries may have played roles in U(VI) reduction during the incubation. Isolation of these organisms and testing for U(VI) reduction by the isolates are necessary to determine the full range of organisms that may contribute to U(VI) reduction.
Implications for in situ bioremediation of groundwater at Midnite mine and other uranium-contaminated sites.
We showed that the uranium content in solution was reduced from ∼20 to ∼0.3 ppm by the enhanced activities of indigenous anaerobic microorganisms. As groundwater at the Midnite mine is contaminated with uranium and sulfate, addition of adequate nutrients may stimulate anaerobic sporeforming gram-positive bacteria, and the activities of these microorganisms may result in removal of uranium from solution. As dormant, sporeforming Desulfosporosinus and Clostridium spp. are ubiquitous even in oxic sediments and soils (46), these organisms may be stimulated with addition of sulfate and nutrients to remove uranium from ground water at contaminated sites.
Acknowledgments
This research was supported by a grant from the U.S. Department of Energy, Basic Energy Science Program, and the U.S. Department of Energy, Biological and Environmental Research (NABIR) Program. Y.S. is grateful to the providers of the Yoshida Scholarship for additional financial support. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract W-31-109-Eng-38. Work performed at the MR-CAT beamline is supported, in part, by funding from the Department of Energy under grant number DEFG2000ER45811.
We thank Donna Bruce and Ellie Hale for assistance and supervising sample collection. Laboratory assistance provided by Matthias Labrenz and Philip Bond is greatly appreciated. We acknowledge Edward O'Loughlin for assistance in collecting the XANES data. Comments from Philip Hugenholz were helpful.
REFERENCES
- 1.Abdelouas, A., W. Lutze, and H. E. Nuttal. 1999. Oxidative dissolution of uraninite precipitated on Navajo sandstone. J. Contam. Hydrol. 36:353-375. [Google Scholar]
- 2.Abdelouas, A., W. Lutze, W. Gong, E. H. Nuttall, B. A. Strietelmeier, and B. J. Travis. 2000. Biological reduction of uranium in groundwater and subsurface soil. Sci. Total Environ. 250:21-35. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Coates, J. D., T. Councell, D. J. Ellis, and D. R. Lovley. 1998. Carbohydrate oxidation coupled to Fe(III) reduction, a novel form of anaerobic metabolism. Anaerobe 4:277-282. [DOI] [PubMed] [Google Scholar]
- 5.Coates, J. D., V. K. Bhupathiraju, L. A. Achenbach, M. J. Mclnerney, and D. R. Lovley. 2001. Geobacter hydrogenophilus, Geobacter chapellei and Geobacer grbiciae, three new, strictly anaerobic, dissimilatory Fe(III)-reducers. Int. J. Syst. Evol. Microbiol. 51:581-588. [DOI] [PubMed] [Google Scholar]
- 6.Cross, J. L., and A. I. Frenkel. 1998. Use of scattered radiation for absolute energy calibration. Rev. Sci. Instrum. 70:38-40. [Google Scholar]
- 7.Delong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dobbin, P. S., J. P. Carter, C. G. Sanjuan, M. V. Hobe, A. K. Powell, and D. J. Richardson. 1999. Dissmilatory Fe(III) reduction by Clostridium beijerinckii isolated from freshwater sediment with Fe(III) maltol enrichment. FEMS Microbiol. Lett. 176:131-138. [DOI] [PubMed] [Google Scholar]
- 9.Francis, A. J., C. J. Dodge, F. Lu, G. P. Halada, and C. R. Clayton. 1994. XPS and XANES studies of uranium reduction by Clostridium sp. Environ. Sci. Technol. 28:636-639. [DOI] [PubMed] [Google Scholar]
- 10.Fredrickson, J. M., D. L. Brockman, D. J. Workman, S. W. Li, and T. O. Stevenson. 1991. Isolation and characterization of a subsurface bacterium capable of growth on toluene, naphthalene, and other aromatic compounds. Appl. Environ. Microbiol. 57:796-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fredrickson, J. M., D. L. Brockman, G. R. Drake, M. F. Romine, D. B. Ringelberg, and D. C. White. 1995. Aromatic-degrading Shingomonas isolates from the deep subsurface. Appl. Environ. Microbiol. 61:1917-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fredrickson, J. M., J. M. Zachara, D. W. Kennedy, M. C. Duff, Y. A. Gorby, S. W. Li, and K. M. Krupka. 2000. Reduction of U(VI) in goethite (α-FeOOH) suspensions by a dissimilatory metal-reducing bacterium. Geochim. Cosmochim. Acta 64:3085-3098. [Google Scholar]
- 13.Ganesh, R., K. G. Robinson, G. D. Reed, and G. S. Sayler. 1997. Reduction of hexavalent uranium from organic complexes by sulfate- and iron-reducing bacteria. Appl. Environ. Microbiol. 63:4385-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou, L., and S. K. Dutta. 2000. Phylogenetic characterization of several para- and meta-PCB dechlorinating Clostridium species: 16s rDNA sequence analyses. Lett. Appl. Microbiol. 30:238-243. [DOI] [PubMed] [Google Scholar]
- 15.Kashefi, K., and D. R. Lovley. 2000. Reduction of Fe(III), Mn(IV), and toxic metals at 100 by Pyrobaculum islandicum. Appl. Environ. Microbiol. 66:1050-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kauffman, J. W., W. C. Laughlin, and R. A. Baldwin. 1986. Microbiological treatment of uranium mine waters. Environ. Sci. Technol. 20:243-248. [DOI] [PubMed] [Google Scholar]
- 17.Kelly, D. P., and A. P. Wood. 2000. Confirmation of Thiobacillus denitrificans as a species of the genus Thiobacillus, in the β-subclass of the Proteobacteria, with strain NCIMB 9548 as the type strain. Int. J. Syst. Evol. Microbiol. 50:511-516. [DOI] [PubMed] [Google Scholar]
- 18.Kemner, K. M., A. J. Kropf, and B. A. Bunker. 1994. A low-temperature total electron yield detector for X-ray absorption fine structure spectra. Rev. Sci. Instrum. 65:3667-3669. [Google Scholar]
- 19.Kieft, T. L., J. K. Fredrickson, T. C. Onstott, Y. A. Gorby, H. M. Kostandarithes, T. J. Bailey, D. W. Kennedy, S. W. Li, A. E. Plymale, C. M. Spadoni, and M. S. Gray. 1999. Dissimilatory reduction of Fe(III) and other electron acceptors by a Thermus isolate. Appl. Environ. Microbiol. 65:1214-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Labrenz, M., G. K. Druschel, T. Thomsen-Ebert, B. Gilbert, S. A. Welch, K. M. Kemner, G. A. Logan, R. E. Summons, G. D. Stasio, P. L. Bond, C. Lai, S. D. Kelly, and J. F. Banfield. 2000. Formation of sphalerite (ZnS) deposits in natural biofilms of sulfate-reducing bacteria. Science 290:1744-1747. [DOI] [PubMed] [Google Scholar]
- 21.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, N.Y.
- 22.Linking Legacies. 1997. Connecting the Cold War Nuclear Weapons Production Processes to Their Environmental Consequences. U.S. Department of Energy, Office of Environmental Management, January 1997. DOE/EM-0319. Office of Environmental Management, Department of Energy, Washington. D.C.
- 23.Lovley, D. R. 1995. Bioremediation of organic and metal contaminants with dissimilatory metal reduction. J. Ind. Microbiol. 14:85-93. [DOI] [PubMed] [Google Scholar]
- 24.Lovley, D. R. E. 2001. Anaerobes to the rescue. Science 293:1444-1446. [DOI] [PubMed] [Google Scholar]
- 25.Lovley, D. R., and E. J. P. Phillips. 1992. Reduction of uranium by Desulfovibrio desulfuricans. Appl. Environ. Microbiol. 58:850-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lovley, D. R., and E. J. P. Phillips. 1992. Bioremediation of uranium contamination with enzymatic uranium reduction. Environ. Sci. Technol. 26:2228-2234. [Google Scholar]
- 27.Lovley, D. R., E. J. P. Phillips, Y. A. Gorby, and E. R. Landa. 1991. Microbial reduction of uranium. Nature 350:413-416. [Google Scholar]
- 28.Lovley, D. R., E. E. Roden, E. J. P. Phillips, and J. C. Woodward. 1993. Enzymatic iron and uranium reduction by sulfate-reducing bacteria. Marine Geol. 113:41-53. [Google Scholar]
- 29.Maidak, B. L., J. R. Cole, C. T. Parker, G. M. Garrity, N. Larsen, B. Li, T. G. Liburn, M. J. McCaughey, G. J. Olsen, R. Overbeek, S. Pramanik, T. M. Schmidt, J. M. Tiedje, and C. R. Woose. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milvy, P., and C. R. Cothern. 1990. Scientific background for the development of regulations for radionuclides in drinking water, p. 1-16. In C. R. Cothern and P. A. Rebers (ed.). Radon, radium and uranium in drinking water. Lewis Publishers, Chelsea, Mich.
- 31.Morrison, S. J., and R. R. Spangler. 1992. Extraction of uranium and molybdenum from aqueous solutions. A survey of industrial materials for use in chemical barriers for uranium mill tailings remediation. Environ. Sci. Technol. 26:1922-1931. [Google Scholar]
- 32.Newville, M. 2001. IFEFFIT: interactive EXAFS analysis and FEFF fitting. J. Synchrotron Rad. 8:322-324. [DOI] [PubMed] [Google Scholar]
- 33.Phillips, E. J. P., E. R. Landa, and D. R. Lovley. 1995. Remediation of uranium contaminated soils with bicarbonate extraction and microbial U(VI) reduction. J. Ind. Microbiol. 14:203-207. [Google Scholar]
- 34.Pietzsch, K., B. C. Hrad, and W. Babel. 1999. A Desulfovibrio sp. capable of growing by reducing U(VI). J. Basic Microbiol. 39:365-372. [Google Scholar]
- 35.Reinhold-Hurek, B., and T. Hurek. 2000. Reassessment of the taxonomic structure of the diazotrophic genus Azoarcus sensu lato and description of three new genera and new species, Azovibrio restrictus gen. nov., sp. nov., Azospira oryzae gen. nov., sp. nov., and Azonexus fungiphilus gen. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 50:649-659. [DOI] [PubMed] [Google Scholar]
- 36.Reysenbach, A.-L., L. J. Giver, G. S. Wickham, and N. R. Pace. 1992. Differential amplification of rRNA genes by polymerase chain reaction. Appl. Environ. Microbiol. 58:3417-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robertson, W. J., J. P. Brownman, P. D. Franzmann, and B. J. Mee. 2001. Desulfosporosinus meridiei nov., a spore-forming sulfate-reducing bacterium isolated from gasolene-contaminated groundwater. Int. J. Syst. Evol. Microbiol. 51:133-140. [DOI] [PubMed] [Google Scholar]
- 38.Segre, C. U., N. E. Leyarovska, L. D. Chapman, W. M. Lanender, P. W. Plag, A. S. King, A. J. Kropf, B. A. Bunker, K. M. Kemner, P. Dutta, R. S. Druan, and J. Kaduk. 2001. The MRCAT insertion device beamline at the Advanced Photon Source. Synch. Rad. Instrum. 11th U.S. Conference DP521 419-422.
- 39.Song, P., N. J. Palleroni, L. J. Kerlhof, and M. M. Hägglom. 2001. Characterization of halobenzoate-degrading, denitrying Azoarcus and Thauera isolates and description of Thauera chlorobenzoica sp. nov. Int. J. Syst. Evol. Microbiol. 51:589-602. [DOI] [PubMed] [Google Scholar]
- 40.Spear, J. R., L. A. Figueroa, and B. D. Honeyman. 2000. Modeling reduction of uranium U(VI) under variable sulfate concentrations by sulfate-reducing bacteria. Appl. Environ. Microbiol. 66:3711-3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strunk, O., W. and Ludwig. 1995. ARB—a software environment for sequence data. Department of Microbiology, Technical University of Munich, Munich, Germany.
- 42.Sumioka, S. S. 1991. Quality of water in an inactive uranium mine and its effects on the quality of water in Blue Creek, Stevens County, Washington. U.S. Geological Survey Water Resources Investigations Report 89-4110. U.S. Geological Survey, Washington, D.C.
- 43.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swofford, D. L. 1999. PAUP*: phylogenetic analysis with parsimony (* and other methods), version 4.02 ed. Sinauer Associates, Sunderland, Mass.
- 45.Uhrie, J. L., J. I. Drever, P. J. S. Colberg, and C. C. Nesbitt. 1996. In situ immobilization of heavy metals associated with uranium leach mines by bacterial sulfate reduction. Hydrometallurgy 43:231-239. [Google Scholar]
- 46.Widdel, F. 1991. The genus Desulfotomaculum. p. 1792-1799. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed., vol. 2. Springer-Verlag, New York, N.Y.
- 47.Woese, C. R., W. G. Weisburg, B. J. Paster, C. M. Hahn, R. S. Tanner, N. R. Krieg, H.-P. Koops, H. Harms, and E. Stackebrandt. 1984. The phylogeny of purple bacteria: the beta subdivision. Syst. Appl. Microbiol. 5:327-336. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, X. L., L. Mandelco, and J. Wiegel. 1994. Clostridium hydroxybenzoicum sp. nov., an amino acid-utilizing, hydroxybenzoate-decarboxylating bacterium isolated from methanogenic freshwater pond sediment. Int. J. Syst. Bacteriol. 44:214-222. [DOI] [PubMed] [Google Scholar]