Abstract
Temporal pigmentation changes resulting from the development of a purple color in anaerobic swine waste lagoons were investigated during a 4-year period. The major purple photosynthetic bacterium responsible for these color changes and the corresponding reductions in odor was isolated from nine photosynthetic lagoons. By using morphological, physiological, and phylogenetic characterization methods we identified the predominant photosynthetic bacterium as a new strain of Rhodobacter, designated Rhodobacter sp. strain PS9. Rhodobacter sp. strain PS9 is capable of photoorganotrophic growth on a variety of organic compounds, including all of the characteristic volatile organic compounds (VOC) responsible for the odor associated with swine production facilities (J. A. Zahn, A. A. DiSpirito, Y. S. Do, B. E. Brooks, E. E. Copper, and J. L. Hatfield, J. Environ. Qual. 30:624-634, 2001). The seasonal variations in airborne VOC emitted from waste lagoons showed that there was a 80 to 93% decrease in the concentration of VOC during a photosynthetic bloom. During the height of a bloom, the Rhodobacter sp. strain PS9 population accounted for 10% of the total community and up to 27% of the eubacterial community based on 16S ribosomal DNA signals. Additional observations based on seasonal variations in meteorological, biological, and chemical parameters suggested that the photosynthetic blooms of Rhodobacter sp. strain PS9 were correlated with lagoon water temperature and with the concentrations of sulfate and phosphate. In addition, the photosynthetic blooms of Rhodobacter sp. strain PS9 were inversely correlated with the concentrations of protein and fluoride.
Improved swine production efficiency over the last few decades has resulted in a trend towards concentrated animal feeding operations (15). This trend has also resulted in increased private and government awareness of the potential effects of high-density production facilities on water and air quality associated with manure storage and treatment systems (3, 32, 43, 45, 46, 52). Swine production facilities generally store manure in earthen, concrete, or steel-lined storage basin, pit, or lagoon systems for up to 14 months prior to land application (14, 16). During storage, the effluent becomes anaerobic, and emission of malodorous compounds often becomes a problem. Several treatment systems have been described to control the release of odorous compounds from livestock wastes (26, 56). Aeration, addition of chemical oxidants, and methanogenic digestors all reduce odors generated during storage and treatment (1, 37). However, the use of these techniques has not received widespread acceptance because of the initial expense, management time, and equipment maintenance required. Other treatments designed to reduce odors from swine production facilities include addition of chemicals, including sodium nitrate and enzymes such as yucca extracts, and addition of mixed microbial populations to storage systems (56). These treatments are expensive and have produced inconsistent results (38).
Because of cost and space limitations, anaerobic lagoons are often used for animal waste management systems. These lagoons serve as storage facilities and support anaerobic degradation of complex organic compounds. However, in the absence of oxygen and most other terminal electron acceptors, the incomplete oxidation of the complex organic compounds in livestock waste produces offensive by-products, such as volatile organic compounds (VOC), hydrogen sulfide, and ammonia (21, 32, 38, 45,46, 54). The concentration of odorous compounds from anaerobic lagoons can be minimized in many cases with good management practices (56). However, regardless of the management practices used, the concentration of odors from lagoons appears to be dependent on the microflora that develops in the system. For example, it is common practice to seed new anaerobic swine waste lagoons with material from working lagoons to accelerate the development of a stable microflora (56).
One microbial event that has been observed by a number of investigators in relation to odors from swine waste storage lagoon systems is the temporal change in the pigmentation of the lagoons from brown to rose to pink (11, 44, 50, 53). In field reports workers have described a relationship between decreased odor intensity and a change in lagoon color to purple. Studies of Zahn et al. (59) confirmed the field reports and showed that the horizontal flux rates of VOC from anaerobic swine waste lagoons that exhibit photosynthetic blooms are lower than the horizontal flux rates of VOC from anaerobic swine waste lagoons that do not exhibit photosynthetic blooms. In this study we focused on characterization of the photosynthetic blooms in swine waste lagoons and on the relationship between the photosynthetic blooms and VOC emissions.
MATERIALS AND METHODS
Description of anaerobic swine waste storage lagoons.
Samples from nine anaerobic swine waste lagoons (five lagoons in central Iowa and four lagoons in northern Missouri) known to turn purple, which are referred to below as photosynthetic lagoons, were used as inocula for enrichment cultures or were plated directly onto solid media. In addition to the nine photosynthetic lagoons, six anaerobic swine waste lagoons (two lagoons in central Iowa and four lagoons in northern Missouri) in which photosynthetic blooms had never been observed, which are referred to below as nonphotosynthetic lagoons, were also monitored periodically for 4 years. Single samples were also taken from photosynthetic lagoons in North Carolina (n = 2) and Minnesota (n = 3) and were assayed for the presence of Rhodobacter sp. strain PS9 by microbial identification system fatty acid methyl ester (MIDI-FAME) analysis as described below. All of the lagoons studied were earthen lined, 64 to 120 by 61 to 120 m, and 4 to 8 m deep. To examine the temporal events that occur during a blooming event, one site in Ogden, Iowa, was monitored weekly for 1 year and periodically (at 1- to 3-month intervals) over a 4-year period. At this site a two-stage anaerobic lagoon system was utilized to process waste from 1,200 sows with piglets and 3,000 feeder animals. All confinement buildings had slotted floors and shallow underfloor pits which were flushed periodically with lagoon surface water. The diluted waste in the pits was pumped into outside storage systems through a pipe. The first-stage lagoon received all diluted waste from the buildings and was connected to the second-stage lagoon with a pipe. The first-stage earthen slurry storage anaerobic lagoon systems were 64 by 64 m and 7 m deep (primary lagoon) and 61 by 95 m and 4 m deep (secondary lagoon).
Weather station.
Meteorological conditions (wind speed, solar radiation, relative humidity, air temperature, and solution temperature for every 10 cm of depth) were monitored continuously with an integrated weather station (42) as previously described (58).
Field monitoring.
Effluent samples were taken at four locations around the perimeter of each lagoon system at a distance of at least 2.0 m from the edge of the storage impoundment and at a depth of 0.2 to 0.3 m by using the method of DiSpirito et al. (7). Lagoon liquid samples were filtered with a single layer of cheesecloth to remove large particulate material directly into 110-ml serum vials that were flushed with argon gas to maintain anaerobic conditions and were sealed with Teflon-lined silicon septa (Tuf-Bond; Pierce Chemical Co., Rockford, Ill.). The samples either were stored on ice if they were processed within 24 h or were frozen and stored at −20°C if they were analyzed later. Samples used for RNA or DNA isolation were centrifuged at 13,000 × g for 15 min at 4°C, and the pellets were washed twice with RNase-free 10 mM phosphate buffer (pH 7.0), centrifuged at 13,000 × g for 15 min, and stored at −80°C until they were processed for nucleic acid recovery.
Chemical characterization of swine waste lagoons.
The organic solid content was estimated by determining the dry weight (20) and by determining the protein concentration (22). Analyses of soluble anions were performed by using a DX 500 chromatography system (Dionex Corp., Sunnyvale, Calif.) equipped with a Dionex CD20 conductivity detector and an IonPac AS9-SC analytical column (4 by 200 mm; Dionex Corp.). Chemical identities and concentrations were determined by using standards obtained from Dionex.
Collection of environmental odor pollutants.
VOC emitted from the lagoons were collected on thermal desorption tubes with a multibed combination of Tenax TA (Alltech Associates Inc., Deerfield, Ill.) and Carbopack C (Supelco Inc., Bellefonte, Pa.) at the ambient temperature as previously described (7, 9, 10). Thermal desorption tube samples were collected on the downwind edge of a lagoon or from the surface of the lagoon by using sampling pumps in the vacuum mode (56). Samples from the surface of the lagoon were collected with dynamic flux chambers as previously described (9, 10). In order to collect sufficient VOC for analysis, 50 liters of air at a flow rate of 500 ml · min−1 was passed through a thermal desorption tube (59). After sampling, the tubes were sealed in glass storage tubes, transported to the laboratory on ice, and stored at −20°C until analysis. All thermal desorption tube samples were processed and analyzed within 7 days (28).
Analysis of environmental odor pollutants in the air.
Chemical analytes concentrated on the thermal desorption tubes were analyzed by using a Dynatherm thermal desorption unit (model 890; Supelco Inc., Bellefonte, Pa.) coupled to a Trimetrics model 9001 gas chromatograph (Finnigan Corp., Austin, Tex.) equipped with a flame ionization detector. Tubes were desorbed at 300°C for 5 min at a carrier flow rate of 2.2 ml · min−1. Analytes were transferred from the desorber oven to an HP-Innowax capillary column (30 m by 0.25 mm by 0.25 μm; Hewlett-Packard, Rolling Meadows, Ill.) through a nickel transfer line heated at 240°C. Analytes were adequately focused on the front of the column by using an initial oven temperature of 50°C for 3 min. The oven temperature was increased to 210°C at a rate of 8°C · min−1, maintained at 210°C for 3 min, and then increased to 260°C at a rate of 10°C · min−1. The identity of each VOC was determined based on the retention times of authentic standards purchased from Aldrich Chemical Co. (Milwaukee, Wis.) and was confirmed by gas chromatography-mass spectrometry. Authentic standards were trapped on thermal desorption tubes by using a model 1000 dynamic thermal stripper (Supelco Inc.) by heating standard solutions of each VOC to 92°C for 30 min. Standard solutions were purged with activated-carbon-filtered nitrogen gas during the heating procedure and then trapped on thermal desorption tubes inside a heated jacket (70°C).
The gas chromatography-mass spectrometry analysis was performed by using a Hewlett-Packard model 6890 gas chromatograph equipped with a mass spectrometer and by using either an HP-Innowax capillary column or a PTE-5 QTM capillary column (30 m by 0.25 mm by 0.25 μm; Supelco Inc.) as previously described (58, 59).
Correlation of a human olfactory response with a VOC concentration was determined by using an equation based on Stevens' law of perception (47-49), as described previously (57).
Isolation of photosynthetic bacteria from lagoon samples by using enrichment cultures.
The following two media were used for isolation and maintenance of photosynthetic bacteria from swine waste lagoons: photosynthetic mineral salts broth (PMSB) (34) and photosynthetic mineral salts medium (PMS) (12). In addition, six reductants (hydrogen sulfide, succinate, valeric acid, butanoic acid, phenol, and 3-methyl indole) were tested with each medium with samples from each site. Samples (10 ml) of lagoon water filtered through cheesecloth were used to inoculate 100 ml of PMS or PMSB in 110-ml glass Hypo vials sealed with Teflon-lined silicon septa. The cultures were incubated under anaerobic conditions in the presence of light (General Electric Bright Stik Gro and Sho bulbs) for 10 days at room temperature (20 to 23°C). When a culture reached the stationary phase, a 10-ml inoculum was transferred into fresh mineral salts medium and incubated as described above; a total of four transfers were made. Cells from the fourth enrichment culture were then spread onto PMS agar plates and incubated in an Oxoid anaerobic jar (Unipath Ltd., Basingstoke, Hampshire, United Kingdom). The plates were incubated under grow lights at room temperature for 10 days, and individual colonies were streaked for single-colony isolation.
Identification of photosynthetic bacteria from lagoon samples on solid media.
Lagoon samples were also plated on solid PMBS or PMS containing 5% filter-sterilized lagoon water. The lagoon water was sterilized by centrifugation at 13,200 × g for 20 min at 4°C and then centrifugation of the supernatant at 13,200 × g for 20 min at 4°C and subsequent filter sterilization with a 0.22-μm-pore-size filter. The substrates and carbon sources described above for liquid enrichment cultures were used for all three solid media. Plates were incubated under anaerobic conditions under grow lights at room temperature for 10 days. The green and purple colonies were picked and streaked for single-colony isolation.
Isolation and identification of bacteriochlorophylls.
Chlorophylls were isolated, characterized, and identified as described by Oelze (29). Spectra were obtained with an Aminco DW-2000 UV-visible spectrophotometer in the split beam mode.
Cellular fatty acid analysis.
Photosynthetic isolates were cultured on Bacto tryptic soy broth agar (Difco) and incubated at 28°C for 48 h, and the cellular fatty acid contents were determined by using the MIDI procedure (MIDI, Inc., Newark, Del.). Identification of isolates was based on a comparison of fatty acid profiles with the fatty acid profiles in tryptic soy broth agar anaerobe databases (MIDI Inc.) (27, 40, 41).
Phylogenetic analysis.
The sequence of the gene coding for the 16S rRNA from nine purple non-sulfur bacterial isolates, one from each photosynthetic lagoon tested, was determined. The 16S rRNA-encoding gene was PCR amplified from genomic DNA by using primers 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (GGTTACCTTGTTACGACTT-3′) as previously described (4). Approximately 500 bases of sequence was determined directly from each amplicon by using primer 519R (5′ GTATTACCGCGGCTGCTGG). The sequences of the isolates were identical. Strain PS9 (8) was selected as a representative strain, and a collection of primers based on known sequences was used to determine the entire 1,419-base sequence of the amplified 16S rRNA gene at an average coverage of 2.5. The consensus 16S ribosomal DNA (rDNA) sequence of strain PS9 was initially aligned with a collection of rDNA sequences in the Ribosomal Database Project (release 8.0) (24) by using the automated alignment tool in ARB (http://www.arb-home.de). The alignment was then adjusted manually to align regions of conserved primary sequence and secondary-structure elements.
A phylogenetic tree was created by using the maximum-likelihood method FastDNAml (31). Bootstrap values were computed with the neighbor-joining program in ARB. The GenBank accession numbers for sequences used in the phylogenetic reconstruction are as follows: Rhodobacter gluconicum, AB077986.1; Rhodobacter blasticus, D16429; Rhodobacter sp. strain TCRI3, AB017796; Rhodobacter apigmentum, AF035433; Rhodobacter azotoformans, D70847; Rhodobacter sphaeroides, X53855; Roseobacter litoralis, X78312; Roseobacter denitrificans, M59063; Roseobacter gallaeciensis, Y13244; Rhodovulum iodosum, Y15011; and Rhodovulum adriaticum, D16418.
Nucleic acid extraction and analysis of lagoon samples.
Sufficient quantities of nucleic acids for filter hybridization experiments were obtained from 0.5-g (wet weight) portions of harvested lagoon pellets by using modifications of the method of Buckley et al. (4). Lagoon pellets were resuspended in 0.7 ml of homogenization buffer (4 M guanidium thiocyanate, 200 mM sodium phosphate [pH 8], 25 mM sodium citrate, 0.5% N-lauryl sarcosine, 1.7% polyvinylpolypyrrolidone) (30) and combined with 0.7 g of 0.1-mm-diameter zirconia-silica beads (Biospec Products, Bartlesville, Okla.). Cells were lysed with a bead beater (Biospec Products) at 0 to 4°C and centrifuged at 3,000 × g for 10 min, and the supernatant was mixed with 0.1 volume of 5 M sodium chloride and 0.5 volume of 50% polyethylene glycol 8000 and incubated on ice for 2 h. Samples were centrifuged at 15,000 × g for 30 min, washed with 70% ethanol, and then resuspended in 2 ml of 120 mM sodium phosphate buffer (pH 7.2). Each extract was combined with 0.1 volume of 10% hexadecyltrimethylammonium bromide and heated at 60°C for 5 to 8 min to remove some humic acid contamination. The nucleic acids were purified further by extraction with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) at pH 4.7 for RNA and at pH 7.0 for DNA. Hydroxyapatite spin columns (Bio-Rad Laboratories, Hercules, Calif.) were used to remove the remaining humic acid contaminants by the method of Purdy et al. (35), desalted with Sephadex G-75 spin columns (Pharmacia Biotech, Uppsala, Sweden) precipitated with sodium acetate-isopropanol (1:6, vol/vol) (35), and resuspended in diethyl pyrocarbonate-treated RNase-free water (39). For extraction of DNA, 100 μg of RNase A (Sigma Chemical Co., St. Louis, Mo.) per ml was added following the hydroxyapatite spin column treatment, and the sample was incubated for 1 h at 37°C.
Oligonucleotide probes and labeling.
Total 16S rDNA and 16S rRNA were quantified with the universal oligonucleotide probe Univ1390 (GAC GGG CGG TGT GTA CAA) (60) as described previously, and the contribution of members of the domain Eubacteria was determined by hybridization with the Eubacteria-specific oligonucleotide probe Eub338 (GCT GCC TCC CGT AGG AGT) as described previously (2). Like the Univ1390 and Eub338 probes, probe Rhodo2 (ACC ATC TCT GGA ACC GCG) complementary to regions of the small-subunit rRNA molecule was designed for detection and monitoring of a taxon, Rhodobacter sp. strain PS9. Rhodo2 was designed by using the Probe Design function in the ARP program (http://www.arb-home.de). Probes Rhodo1 (CTG GAA CCG CGA TCG CCA) and Rhodo2, suggested by the Probe Design program, were tested with a variety of bacterial species, including R. sphaeroides. Rhodo2 was chosen based on its specificity for Rhodobacter sp. strain PS9 under the hybridization conditions described below. Rhodo1 was less specific and showed similar hybridization to R. sphaeroides and Rhodobacter sp. strain PS9. Oligonucleotide probes were 5′ end labeled by using T4 polynucleotide kinase (Promega Corp., Madison, Wis.) with [γ-32P]ATP as previously described (39).
Quantitative filter hybridization.
Quantitative filter hybridization was performed by the method of DeLong (6), with minor modifications. Nucleic acids from lagoon samples and cultures were denatured with 2% glutaraldehyde-50 mM Na2HPO4 and blotted onto Magna Charge nylon membranes (MSI Micron Separations Inc., Westborough, Mass.) with a Bio-Dot microfiltration apparatus (Bio-Rad Laboratories). Replicate membranes were hybridized with 32P-labeled oligonucleotide probes (Univ1390, Eub338, or Rhodo2) for a minimum of 24 h at 40°C and washed twice for 30 min at 45°C in 2× ST solution containing 0.1% sodium dodecyl sulfate (6, 36). The membranes were then dried and cut into separate hybridization spots, and the radioactivity was quantified with a Packard Tri-Carb 2100TR liquid scintillation analyzer.
The relative abundance of 16S rDNA and the relative abundance of 16S rRNA were calculated by determining the ratio of the signal derived from domain- or group-specific probes to the signal derived from the universal probe (6, 13). To calculate the relative abundance of Rhodobacter sp. strain PS9, the slopes of the probe binding curves were determined for serial dilutions of positive controls (Rhodobacter sp. strain PS9), negative controls (Saccharomyces cerevisiae ATCC 32747, Halobacterium sp., Escherichia coli ATCC e11303, Methylococcus capsulatus Bath, and R. sphaeroides ATCC 17023), and lagoon samples. Positive controls were included on each membrane. The relative abundance of 16S rDNA and the relative abundance of 16S rRNA from Rhodobacter sp. strain PS9 in swine lagoon samples (expressed as percentages) were determined by using the following equation:
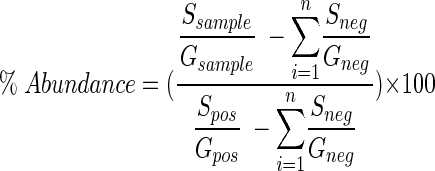 |
To account for the contribution of nonspecific binding, the ratio of specific probe (S) binding to universal or general probe (G) binding for each of the negative controls (Sneg/Gneg) was subtracted from ratio of the signal intensities for each of the samples (Ssample/Gsample). To account for the difference in probe-specific activity, the ratio of specific probe binding to universal probe binding for positive controls (Spos/Gpos) was determined. Because nonspecific binding could occur for positive controls, Sneg/Gneg was subtracted from Spos/Gpos. Under the hybridization conditions described above, Rhod2 showed no cross-reactivity with the negative controls tested. The equation given above was used to correct for the different specific activities of the probes used and to account for nonspecific binding.
Microscopy.
Phase-contrast microscopy and fluorescent microscopy were performed with a Zeiss Axioplan II microscope equipped with an Axio Cam HRC camera. Fluorescent microscopy was carried out in a dark room to minimize background light by using a 4′,6′-diamidino-2-phenylindole (DAPI) filter.
Electron microscopy samples were prepared at the Iowa State University Bessey Microscopy Facility as previously described (51). Images were obtained with a JEOL 1200EX scanning transmission electron microscope (Japan Electron Optic Laboratories, Akishima, Japan).
Statistical analysis.
Pearson correlation coefficients and probabilities were determined by bivariate correlation analysis by using SPSS 10.1.3 (SPSS Inc., Chicago, Ill.).
Nucleotide sequence accession number.
The consensus 16S rDNA sequence of strain PS9 has been deposited in the GenBank database under accession number AF515782.
RESULTS
Field observations.
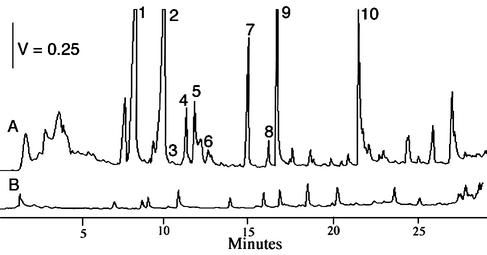
Several investigators have described temporal changes in the pigmentation of swine waste lagoons (50). In field reports workers have also described reductions in odor with coincidental changes in lagoon color to purple. To investigate these temporal events, the emission of VOC prior to and following photosynthetic blooms was monitored in nine lagoons. The concentration of VOC was used to monitor odor levels since it is the only reliable predictor of odor intensity at swine production facilities (19, 55, 57-59). An example of the VOC emissions before and after a photosynthetic bloom in an anaerobic swine waste lagoon in central Iowa is shown in Fig. 1. Reductions in the VOC emissions of 80 to 93% were observed following photosynthetic blooms in all nine test lagoons. Conversely, the VOC emissions from the six nonphotosynthetic lagoons increased 23 to 36% during the same time period. These results are consistent with studies of Zahn et al. (59) that showed that the levels of VOC emissions from photosynthetic lagoons were 10 to 20% of the levels of emissions from nonphotosynthetic lagoons or from other swine waste storage or treatment systems at test sites in Iowa, Oklahoma, Minnesota, and North Carolina (59). On a 1 to 100% human response scale (see Materials and Methods) (57) the odor intensity level decreased from more than 100% where the average character descriptors associated with the odor at or above the 100% level include very bad, strong, powerful, sickening, astringent, etc. to 3 to 15% where the average character descriptors include barely detectable, noticeable, barely present, slightly unpleasant, etc. (57).
FIG. 1.
Chromatographic profiles of airborne VOC emitted from anaerobic swine waste lagoons before (profile A) and after (profile B) photosynthetic blooms. The compounds of interest include acetic acid (peak 1), propionic acid (peak 2), isobutyric acid (peak 3), butyric acid (peak 4), isovaleric acid (peak 5), valeric acid (peak 6), caproic acid (peak 7), phenol (peak 8), para-cresol (peak 9), and 3-methyl indole (peak 10).
Isolation of photosynthetic bacteria.
To determine the type(s) of photosynthetic bacteria involved in the blooms, samples were obtained from nine photosynthetic lagoons (five lagoons in central Iowa and four lagoons in northern Missouri) before and after photosynthetic blooms. The samples were used as inocula for either PMS or PMSB in enrichment cultures or were plated directly on solid media containing 5% filter-sterilized lagoon water in PMS or PMSB and grown either photolithoautotrophically with hydrogen sulfide, hydrogen, or sulfur and carbon dioxide or photooganotrophically with succinate, valeric acid, butanoic acid, or 3-methyl indole as a carbon and reductant source. In addition, prebloom lagoon samples were incubated at room temperature in the dark or under grow lights under anaerobic conditions.
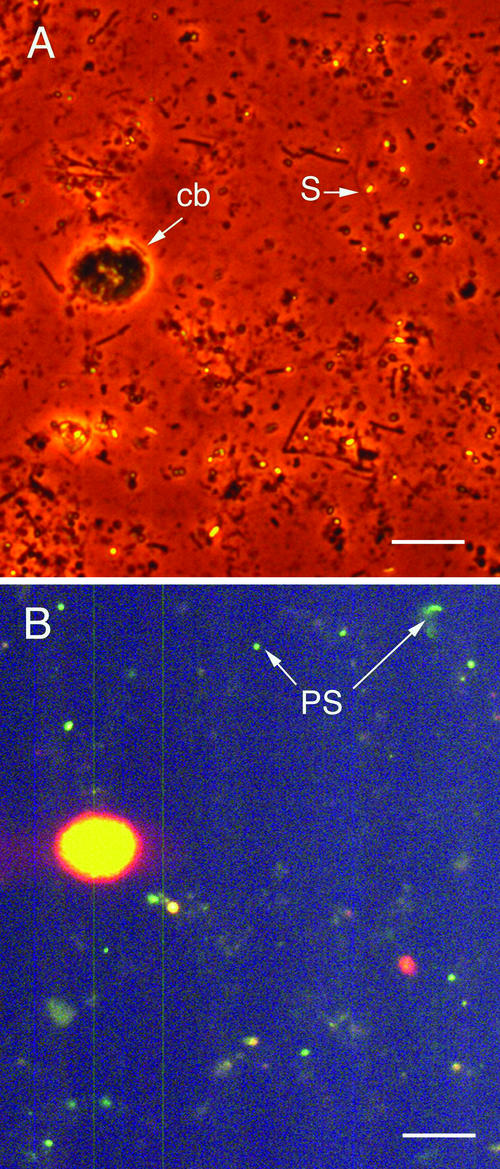
Microscopic examination of lagoon samples indicated that both purple and green photosynthetic bacteria, as well as cyanobacteria, were present in all nine photosynthetic swine waste lagoons tested. When fluorescent microscopy with a DAPI filter was used, pure cultures of Rhodobacter sp. strain PS9 showed weak green fluorescence and cyanobacterial enrichment cultures exhibited bright yellow-orange fluorescence. Based on these observations, the weakly green fluorescent cells in lagoon water samples were assumed to be bacteriochlorophyll a-containing photosynthetic bacteria, and the large (diameter, approximately 5 μm) yellow-orange fluorescent cells were assumed to be cyanobacteria. The appearance of non-cell-associated sulfur granules during a photosynthetic bloom was taken to indicate that green sulfur photosynthetic bacteria and/or Rhodospirillum rubrum was present (Fig. 2A).
FIG. 2.
Phase-contrast (A) and fluorescence (B) micrographs of a lagoon sample taken on 9 June 1997. Refractile sulfur particles (S) are evident in the phase-contrast micrograph, and the yellow-orange fluorescence of chlorophyll a from a cyanobacterium (cb) and the weak green fluorescence of bacteriochorophyll a from photosynthetic bacteria (PB) are evident in the fluorescence micrograph. Bars = 5 μm.
Direct plating of lagoon water samples onto solid media also showed that both green and purple photosynthetic bacteria, as well as cyanobacteria, were present. However, 92% of the colonies were purple non-sulfur photosynthetic bacteria. Thus, microscopic examination and direct plating of lagoon samples on solid media, as well as the results described below, showed that the dominant photosynthetic bacteria in all nine test lagoons were purple non-sulfur bacteria similar to R. sphaeroides. Previous studies of photosynthetic bacteria in a sewage treatment plant in Göttingen, Germany, also showed that cyanobacteria and green and purple photosynthetic bacteria were present and that the dominant group was the purple non-sulfur bacteria similar to R. sphaeroides (44). Below we describe the properties of one of the R. sphaeroides-like isolates, Rhodobacter sp. strain PS9.
Properties of Rhodobacter sp. strain PS9.
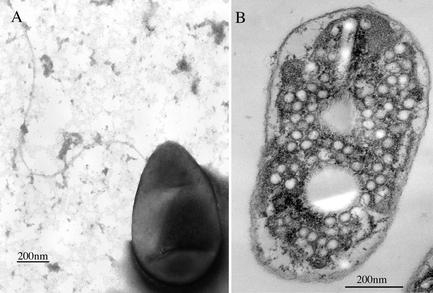
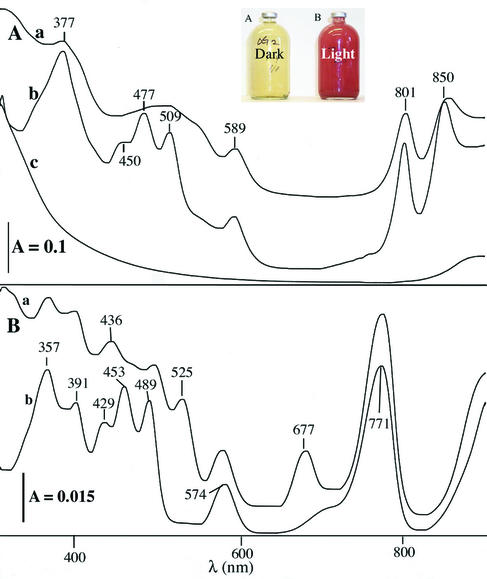
Rhodobacter sp. strain PS9 is a gram-negative organism with ovoid cells that are 0.65 to 0.73 μm wide and 0.95 to 1.4 μm long and have a single subpolar flagellum (Fig. 3A) and an intracytoplasmic membrane arrangement typical of Rhodobacter species (Fig. 3B). The spectral properties of this bacterium cultured under anaerobic conditions in the presence of light were similar to the spectral properties obtained directly for anaerobic swine waste lagoons during photosynthetic blooms in late summer, as well as the spectral properties of prebloom lagoon samples incubated in the presence of light at room temperature (approximately 22°C) (Fig. 4). Whole-cell UV-visible absorption spectra had absorption maxima at 377, 450, 477, 509, 589, 801, 850, and 951 nm. The whole-cell absorption spectrum of Rhodobacter sp. strain PS9 (Fig. 4A) indicated that the cells contained both light-harvesting I and light-harvesting II photocomplexes (23, 61) and the B800-850-type antenna complex, as well as carotenoids, all of which are characteristic of purple non-sulfur photosynthetic bacteria (23, 29, 33, 61).
FIG. 3.
Electron micrographs of negatively stained (A) and ultrathin-sectioned (B) cells of Rhodobacter sp. strain PS9, showing ovoid cells with a subpolar flagellum and a vesicular type of intracytoplasmic membrane arrangement, respectively.
FIG. 4.
UV-visible absorption spectra. (A) Spectra for anaerobic swine waste lagoons before (trace c) and after (trace a) purple photosynthetic blooms, compared with the spectrum for Rhodobacter sp. strain PS9 grown photosynthetically (trace b). (B) Spectra for methanol extracts of bacteriochorophyll from a purple photosynthetic lagoon (trace a) and Rhodobacter sp. strain PS9 (trace b). (Inset) Lagoon liquid samples taken prior to a photosynthetic bloom and incubated for 20 days under anaerobic conditions in the dark (serum vial A) or under grow lights (serum vial B) at room temperature.
Bacteriochlorophyll extracts resuspended in ethyl ether also produced UV-visible absorption maxima characteristic of bacteriochlorophyll a and of carotenoids and the spheroidene series (Fig. 4B) (17, 22, 28). The spectra of bacteriochlorophyll extracts of lagoon samples obtained during the blooms were similar to the spectra of extracts of Rhodobacter sp. strain PS9, and additional absorption maxima at 436, 524, and 677 nm indicated that chlorophyll a was present (Fig. 4B). The high concentrations of chlorophyll a observed in the extracts of lagoon samples (Fig. 4B) were unexpected since macroscopic and microscopic examinations of lagoon water showed that little or no plant material, algae, or cyanobacteria were present. Cyanobacteria could be detected during enrichment in most of the lagoons tested, but few cyanobacteria were detected prior to enrichment. Since there was no other identifiable source of chlorophyll a, the source of chlorophyll a was probably the corn- and soybean-based animal feed. Chlorophyll a is a pigment associated with manure from animal feeds containing corn and soybean products and can be used to detect fecal contamination (5).
By using MIDI-FAME analysis the major purple photosynthetic isolates from all nine test photosynthetic lagoons were identified as R. sphaeroides strains with similarity indices of 0.9 ± 0.05. The predominant fatty acid (84.3% ± 6.8% of the total fatty acids) was 18:1 ω7c, followed by 18:0 (3.6% ± 0.5%), 10:0 3OH (3.3% ± 0.5%), 16:0 (1.1% ± 0.12%), 17:0 (1% ± 0.2%), and 11-methyl 18:1 ω7c (0.9% ± 0.1%). In addition to the samples from the nine photosynthetic lagoons in Iowa and Missouri tested, single samples from five additional photosynthetic swine waste lagoons in Minnesota (n =3) and North Carolina (n = 2) were used for enrichments. The 17 purple non-sulfur photosynthetic isolates obtained from these five photosynthetic lagoons were similar to Rhodobacter sp. strain PS9 and showed MIDI-FAME similarity indices of 0.91 ± 0.07 to R. sphaeroides.
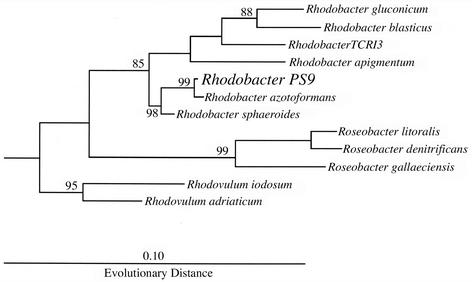
The phylogenetic tree based on the 16S rRNA gene sequence (Fig. 5) also grouped Rhodobacter sp. strain PS9 with other members of the genus Rhodobacter. Rhodobacter sp. strain PS9 grouped closest to R. azotoformans, a denitrifying purple non-sulfur photosynthetic bacteria isolated from an activated sludge pit (17, 18), exhibiting a 2% sequence difference.
FIG. 5.
Phylogenetic analysis of Rhodobacter sp. strain PS9 and select photosynthetic bacteria in the alpha subdivision of the division Proteobacteria. Bootstrap values greater than 80% are indicated at the nodes. The 16S rDNA sequence of Caulobacter vibrioides (data not shown) was used to root the phylogenetic tree. The evolutionary distance is based on the number of fixed point mutations per position.
Four Rhodobacter isolates from swine waste lagoons were tested to determine the range of suitable electron donors and carbon sources during photosynthetic growth. The results were the same for all four isolates, and the results for Rhodobacter sp. strain PS9 are shown in Table 1. Rhodobacter sp. strain PS9 grew photoorganotropically on a wide variety of organic compounds, including short-chain fatty acids, sugars, amino acids, and aromatic carbon compounds, but not on chemolithotrophic substrates. Rhodobacter sp. strain PS9 also did not grow in the dark on any of the growth substrates tested.
TABLE 1.
Utilization of electron donor and carbon sources for phototrophic growth by Rhodobacter sp. strain PS9 and R. azotoformansa
| Substrate | Utilization by:
|
|
|---|---|---|
| Rhodobacter sp. strain PS9 | R. azotoformans | |
| Organic acids | ||
| Formic acid | +b | + |
| Acetic acidc | + | + |
| Propoinic acidc | + | + |
| Butyric acidc | + | + |
| Valeric acidc | + | + |
| Caproic acidc | + | + |
| Heptonic acidc | + | NT |
| Caprylic acid | − | − |
| Pelargonic acid | − | NT |
| Pyruvic acid | + | + |
| Lactic acid | − | + |
| Malic acid | − | + |
| Succinic acid | + | + |
| Fumaric acid | − | + |
| Tartaric acid | + | − |
| Citric acid | − | − |
| Gluconic acid | − | NT |
| Benzoic acidc | + | − |
| Sugars and alcohols | ||
| Glucose | + | + |
| Fructose | + | + |
| Manitol | + | + |
| Sorbitol | + | + |
| Amino acids | ||
| Aspartic acid | − | + |
| Arginine | + | NT |
| Glutamic acid | − | + |
| Aromatic carbon compounds | ||
| Phenolc | + | NT |
| p-Cresolc | + | NT |
| 3-Methyl indolec | + | NT |
| Chemolithoautotrophic substrates | ||
| Hydrogen + CO2d | − | NT |
| Sulfide + CO2d | − | NT |
| Sulfur + CO2d | − | NT |
| Thiosulfate + CO2 | − | − |
Data for R. azotoformans were obtained from reference 15. All cultures were incubated at room temperature in the presence of light for 14 days.
+, growth (optical density at 660 nm, 0.5 to 1.0); −, no growth; NT, not tested.
Volatile substrate commonly detected in anaerobic swine waste lagoons.
Hydrogen, sulfide, and sulfur are commonly detected in anaerobic swine waste lagoons.
Quantification of Rhodobacter sp. strain PS9 in anaerobic swine waste lagoons by oligonucleotide probe hybridization.
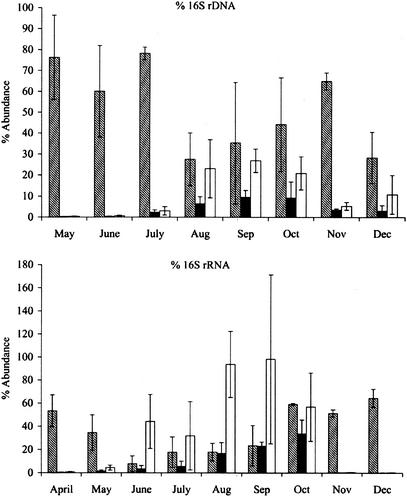
Lagoon samples were taken from four locations around the perimeter of a lagoon at a depth of 0.2 to 0.3 m. Previously obtained depth profiles for seven of the nine test lagoons demonstrated that above the sludge layer, 0.5 to 1 m from the bottom, the concentrations of target microorganisms, bacteriochlorophyll, and total protein were essentially constant with depth (7). This high degree of homogeneity in the water columns of the test lagoons eliminated the need for depth profiling during this phase of the study. Figure 6 shows the relative abundance of the Rhodobacter sp. strain PS9 population in an anaerobic swine waste lagoon during 1997 based on rDNA and rRNA signals. For simplicity, weekly hybridization and VOC measurements were combined to obtain monthly averages. By using the universal oligonucleotide probe (Univ1390) and probes specific for eubacteria (Eub338) and Rhodobacter sp. strain PS9 (Rhodo2), the abundance of 16S rDNA and the abundance of 16S rRNA were determined. The concentration of rDNA was used to estimate population density, whereas the concentration of rRNA was used to estimate the activity level of this bacterium. During the 3-month period from April to June, the contribution of Rhodobacter sp. strain PS9 16S rDNA to the total community 16S rDNA was low (approximately 0.3%), as was the contribution of the 16S rRNA of this organism to the total community 16S rRNA (approximately 0.8%). However, the relative levels of Rhodobacter sp. strain PS9 16S rDNA and 16S rRNA increased rapidly in late June and eventually were 10% ± 3% and 34% ± 12% of the total community levels, respectively, in September. Moreover, the relative levels of Rhodobacter sp. strain PS9 16S rDNA and 16S rRNA increased to 27 and 99% of the eubacterial community levels in September, respectively. The results obtained were consistent with the results of previous studies of Merrill and Halverson (25) for community structure in anaerobic swine wastes lagoons obtained by using MIDI-FAME analysis. Taken together, the results demonstrate that during the photosynthetic blooms, which peak in the late summer to the early fall, Rhodobacter sp. strain PS9 is the major photosynthetic bacterium in the lagoons.
FIG. 6.
Relative abundance of Rhodobacter sp. strain PS9 16S rDNA (top panel) and relative abundance of Rhodobacter sp. strain PS9 16S rRNA (bottom panel) in liquid samples obtained from anaerobic swine waste lagoons during 1997, showing the contribution of eubacteria to the total community (cross-hatched bars), the contribution of Rhodobacter sp. strain PS9 to the total community (solid bars), and the contribution of Rhodobacter sp. strain PS9 to the total eubacterial community (open bars).
Correlation of photosynthetic blooms with environmental conditions.
In an attempt to identify the environmental conditions conducive to photosynthetic blooming, seasonal variations in the meteorological, biological, and chemical factors were measured and related to changes in the microbial populations. To minimize variables due to site, lagoon construction, weather, management practices, animal genetics, and feed variability, eight test lagoons (four photosynthetic lagoons and four nonphotosynthetic lagoons) in northern Missouri at the same production facility were studied. At this facility the same feed (mixed on site), building and lagoon construction, number of animals per building, and animal genetic lines were utilized for all buildings and lagoons. The only discernible difference was the size and age of the animals in the buildings. The lagoons servicing the buildings devoted to finisher pigs had higher organic loading rates and higher residual organic contents (more than 5 mg [dry weight]/ml) than the lagoons servicing buildings devoted to farrowing pigs (usually less than 4 mg [dry weight]/ml). Of the four photosynthetic lagoons examined in northern Missouri, three serviced farrowing pigs, while one serviced a finisher pig site.
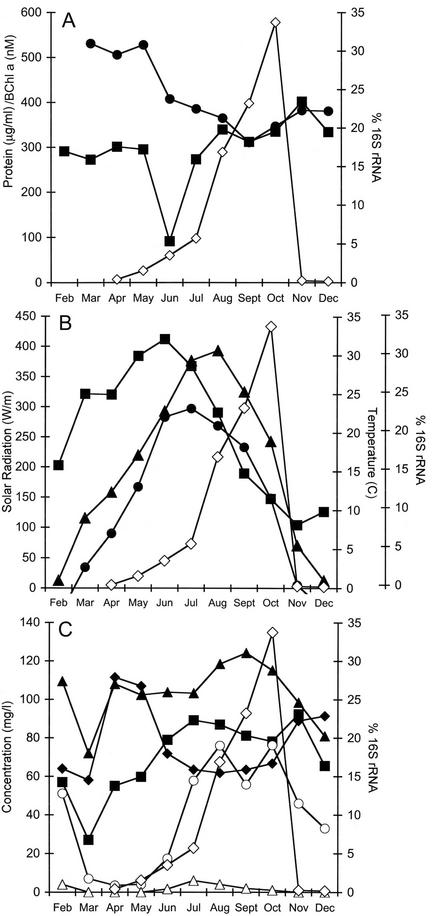
Figure 7 shows the seasonal variations in biomass (protein) and bacteriochlorophyll concentrations (Fig. 7A), air and lagoon water temperatures (Fig. 7B), solar radiation (Fig. 7B), and nitrate, sulfate, fluoride, chloride, and phosphate ion concentrations (Fig. 7C) before and after a photosynthetic bloom. Of the lagoon water parameters monitored, the concentrations of sulfate and phosphate showed the strongest positive correlation with the photosynthetic bloom, and the protein and fluoride concentrations showed the strongest negative correlation (Table 2).
FIG. 7.
Seasonal variations in the meteorological, biological, and chemical factors in two-stage anaerobic swine waste lagoons. (A) Monthly averages for the concentrations of bacteriochlorophyll (▪) and protein (•) and the contribution of Rhodobacter sp. strain PS9 to the total community as measured by the abundance of 16S rRNA (⋄). (B) Monthly averages for solar radiation (▪), water temperature (•), air temperature (▴), and the contribution of Rhodobacter sp. strain PS9 to the total community as measured by the abundance of 16S rRNA (⋄). (C) Monthly averages for the concentrations of phosphate (▴), fluoride (♦), chloride (▪), sulfate (○), and nitrate (▵) and the contribution of Rhodobacter sp. strain PS9 to the total community as measured by the abundance of 16S rRNA (⋄).
TABLE 2.
Bivariate correlations for abundance of the Rhodobacter sp. strain PS9 population.
| Characteristic | Parameter | PS9 abundance as determined by:
|
Bacterio- chlorophyll content | Protein content of lagoon water | Water temp | Air temp | Solar radiation | Concn of:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % of total community 16S rRNA | % of total eubacterial 16S rRNA | % of total community 16S rDNA | % of total eubacterial 16S rDNA | SO42− | NO3− | PO42− | Cl− | F− | Fatty acids | Aromatic compounds | |||||||
| PS9 abundance as determined by: | |||||||||||||||||
| % of total community 16S rRNA | r | 1.000 | |||||||||||||||
| % of total eubacterial | r | 0.771 | 1.000 | ||||||||||||||
| 16S rRNA | P | 0.015 | |||||||||||||||
| % of total community | r | 0.928 | 0.864 | 1.000 | |||||||||||||
| 16S rDNA | P | 0.000 | 0.003 | ||||||||||||||
| % of total eubacterial | r | 0.819 | 0.804 | 0.934 | 1.000 | ||||||||||||
| 16S rDNA | P | 0.007 | 0.009 | 0.000 | |||||||||||||
| Bacteriochlorophyll | r | 0.189 | −0.068 | 0.269 | 0.416 | 1.000 | |||||||||||
| content | P | 0.626 | 0.862 | 0.484 | 0.265 | ||||||||||||
| Protein content of | r | −0.630 | −0.668 | −0.823 | −0.792 | −0.226 | 1.000 | ||||||||||
| lagoon water | P | 0.069 | 0.049 | 0.006 | 0.011 | 0.558 | |||||||||||
| Water temp | r | 0.474 | 0.762 | 0.461 | 0.288 | −0.356 | −0.247 | 1.000 | |||||||||
| P | 0.197 | 0.017 | 0.212 | 0.452 | 0.347 | 0.521 | |||||||||||
| Water temp shiftedb | r | 0.735 | 0.844 | 0.827 | 0.654 | ||||||||||||
| P | 0.038 | 0.005 | 0.011 | 0.078 | |||||||||||||
| Air temp | r | 0.344 | 0.669 | 0.322 | 0.125 | −0.563 | −0.157 | 0.970 | 1.000 | ||||||||
| P | 0.364 | 0.049 | 0.398 | 0.749 | 0.114 | 0.686 | 0.000 | ||||||||||
| Solar radiation | r | −0.311 | 0.003 | −0.434 | −0.554 | −0.750 | 0.562 | 0.558 | 0.686 | 1.000 | |||||||
| P | 0.416 | 0.995 | 0.243 | 0.122 | 0.020 | 0.115 | 0.118 | 0.041 | |||||||||
| Solar radiation shiftedc | r | 0.448 | 0.818 | 0.480 | 0.489 | ||||||||||||
| P | 0.313 | 0.024 | 0.276 | 0.053 | |||||||||||||
| Concn of: | |||||||||||||||||
| SO42− | r | 0.743 | 0.672 | 0.824 | 0.779 | 0.434 | −0.838 | 0.421 | 0.255 | −0.454 | 1.000 | ||||||
| P | 0.022 | 0.047 | 0.006 | 0.013 | 0.243 | 0.005 | 0.259 | 0.507 | 0.219 | ||||||||
| NO3− | r | 0.236 | 0.528 | 0.303 | 0.184 | −0.149 | −0.388 | 0.806 | 0.758 | 0.372 | 0.557 | 1.000 | |||||
| P | 0.542 | 0.144 | 0.427 | 0.635 | 0.703 | 0.302 | 0.009 | 0.018 | 0.324 | 0.120 | |||||||
| PO42− | r | 0.728 | 0.811 | 0.714 | 0.565 | 0.006 | −0.303 | 0.738 | 0.633 | 0.132 | 0.441 | 0.346 | 1.000 | ||||
| P | 0.026 | 0.008 | 0.031 | 0.113 | 0.988 | 0.429 | 0.023 | 0.067 | 0.736 | 0.235 | 0.362 | ||||||
| Cl− | r | 0.310 | 0.478 | 0.510 | 0.384 | 0.146 | −0.730 | 0.410 | 0.353 | −0.224 | 0.745 | 0.599 | 0.272 | 1.000 | |||
| P | 0.417 | 0.193 | 0.160 | 0.307 | 0.707 | 0.025 | 0.273 | 0.351 | 0.563 | 0.021 | 0.088 | 0.480 | |||||
| F− | r | −0.660 | −0.818 | −0.769 | −0.640 | 0.106 | 0.845 | −0.687 | −0.630 | 0.074 | −0.829 | −0.730 | −0.495 | −0.785 | 1.000 | ||
| P | 0.053 | 0.007 | 0.015 | 0.063 | 0.785 | 0.004 | 0.041 | 0.069 | 0.850 | 0.006 | 0.025 | 0.175 | 0.012 | ||||
| Fatty acids | r | −0.763 | −0.767 | −0.904 | −0.910 | −0.308 | 0.822 | −0.266 | −0.148 | 0.548 | −0.771 | −0.122 | −0.528 | −0.591 | 0.693 | 1.000 | |
| P | 0.017 | 0.016 | 0.001 | 0.001 | 0.420 | 0.007 | 0.488 | 0.704 | 0.127 | 0.015 | 0.754 | 0.144 | 0.093 | 0.038 | |||
| Aromatic compounds | r | −0.786 | −0.758 | −0.916 | −0.888 | −0.433 | 0.892 | −0.408 | −0.248 | 0.513 | −0.956 | −0.461 | −0.510 | −0.714 | 0.825 | 0.883 | 1.000 |
| P | 0.012 | 0.018 | 0.001 | 0.001 | 0.244 | 0.001 | 0.275 | 0.519 | 0.158 | 0.000 | 0.212 | 0.161 | 0.031 | 0.006 | 0.002 | ||
As determined by the percentage of total community 16S rRNA or 16S rDNA and the percentage of the total eubacterial 16S rRNA or 16S rDNA, the bacteriochlorophyll content, the protein content of lagoon water, the water temperature, the water temperature data moved forward 14 days, the air temperature, the solar radiation, the solar radiation data moved forward 2 months, and the concentrations of sulfate, nitrate, phosphate, chloride, fluoride, fatty acids, and aromatic compounds in the anaerobic swine waste lagoon at the Ogden, Iowa, site. Correlations that were significant at the ≤0.05 level are indicated by boldface type.
Water temperature values shifted 14 days forward from the sampling date.
Solar radiation values shifted 2 months forward from the sampling date.
On the basis of organic loading, the test lagoon chosen for weekly monitoring appeared to be ideal for culturing photosynthetic bacteria; it had an organic load of approximately 3 mg [dry weight]/ml throughout the year. Previous studies of Seifert et al. (44) showed that the population of photosynthetic bacteria in sewage treatment plants increased with organic loading up to the highest concentration tested, 2.5 mg (dry weight)/ml. The results of this study showed that this trend continued up to a maximum value of 3.0 to 3.5 mg (dry weight)/ml and showed that the trend was the opposite at higher organic loading rates. In the lagoons that were monitored or sampled, photosynthetic blooms were observed only in swine waste lagoons with protein concentrations of less than 4 g (dry weight)/liter or 1 g of protein/liter. Organic loading was not a determining variable in the Ogden, Iowa, lagoon used for weekly monitoring in this study since the concentration in the test lagoon never exceeded 1 g of protein/ml. However, even in this lagoon, a strong negative correlation between the organic load as measured by protein concentration (Fig. 7A) and the photosynthetic bloom was still apparent.
To our surprise, the correlation between temperature and a photosynthetic bloom was significant only when the data were compared to the abundance of Rhodobacter sp. strain PS9 16S rRNA in the total eubacterial population (Table 2). This was surprising considering that in all nine test lagoons, as well as in more than 30 lagoons visited over a 4-year period, initiation of the photosynthetic bloom occurred only when the lagoon water temperature reached 15 to 20°C. A possible explanation for this unexpected result was obtained if the water temperature values were moved forward 14 days. When this shift was made, statistically significant correlations were observed between lagoon water temperature and the photosynthetic blooms (Table 2). The 14-day shift in the temperature data was chosen based on the incubation time needed for a photosynthetic bloom to occur in the laboratory when prebloom lagoon samples are incubated at 23°C (Fig. 4, inset). Thus, a positive correlation between temperature and the photosynthetic bloom was observed when the physiological response time was taken into consideration.
Little correlation was also observed between light intensity and the photosynthetic blooms. As illustrated in Fig. 4, light was necessary for a bloom to occur; however, there was little correlation between the level of solar radiation and the development of a photosynthetic bloom (Table 2; Fig. 7B). In fact, the largest population and highest activity levels of Rhodobacter sp. strain PS9 were observed during periods of low light intensity. However, as with lagoon temperatures, if the solar radiation data and the time of a population bloom were offset by 2 months, then some correlation between solar radiation and the photosynthetic bloom was observed (Table 2). As observed with temperature, there appeared to be a lag period between the exposure to solar radiation and the physiological response leading to higher population levels.
A second unexpected result was the positive correlation between a photosynthetic bloom and the concentration of sulfate in the lagoon water, as well as the presence of nitrate during the photosynthetic bloom (Table 2; Fig. 7C). These results suggest that there may be competition among sulfate-reducing bacteria, denitrifying bacteria, and photosynthetic bacteria, possibly for low-molecular-mass organic substrates. However, this competition is only inferred since the populations of sulfate-reducing and denitrifying bacteria were not monitored in this study.
At this time we have no explanation for the positive correlation between the phosphate concentration and the photosynthetic blooms and the negative correlation between the fluoride concentration and the photosynthetic blooms (Table 2).
Correlation between photosynthetic blooms and rate of emission of VOC.
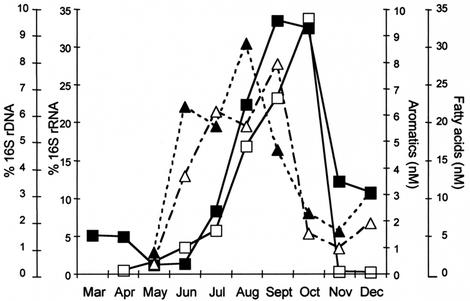
The seasonal rates of emission of VOC, including short-chain fatty acids, such as acetic acid, propionic acid, butyric acid, valeric acid, isovaleric acid, caproic acid, and isocaproic acid, and aromatic carbon compounds, such as phenol, para-cresol, and 3-methyl indole, were measured, and they were inversely correlated with Rhodobacter sp. strain PS9 population and activity levels (Table 2; Fig. 8). For simplicity, the concentrations of individual fatty acids and individual aromatic compounds were combined for Fig. 8, since the compounds in each group exhibited similar temporal trends. The rates of emission of short-chain fatty acids and aromatic carbon compounds were highest during August and September, respectively, and decreased to 80% and 85% of the maximal values, respectively, during the photosynthetic blooms in September and early November. Figure 8 also shows that the concentrations of volatile short-chain fatty acids decreased first and that these decreases were followed by decreases in the concentrations of aromatic compounds.
FIG. 8.
Seasonal fluctuations in the concentrations of volatile fatty acids (acetic acid, propionic acid, butyric acid, isobutyric acid, valeric acid, isovaleric acid, caproic acid, isocaproic acid) (▴) and aromatic carbon compounds (4-methyl phenol, 4-ethyl phenol, para-cresol, indole, 3-methyl indole) (▵) compared with the abundance of Rhodobacter sp. strain PS9 16S rDNA (▪) and the abundance of Rhodobacter sp. strain PS9 16S rRNA (□) in the total community in liquid lagoon samples.
DISCUSSION
Temporal changes in the pigmentation of waste treatment lagoons from pink to rose to brown have previously been observed by a number of researchers (50, 53). The color changes reflect changes in physical and chemical factors that result in the selection of different bacterial groups, and this is especially evident in anaerobic livestock waste lagoons (53). Moreover, field reports indicate that there is a correlation between odor reduction and pigmentation changes in lagoons to purple. However, the organism(s) responsible for odor reduction has never been identified, nor have the population levels of the microorganisms responsible for the change to a purple color been studied in a systematic manner over time.
In this study nine anaerobic swine waste lagoons known to develop photosynthetic blooms were monitored periodically over a 4-year period, and one lagoon was monitored weekly for 1 year. In addition, six anaerobic swine waste lagoons in which photosynthetic blooms had never been observed were also monitored periodically over a 4-year period. In the lagoons known to develop photosynthetic blooms, the organic loading and temperature of the lagoon water appeared to be the main parameters controlling the bloom, although the correlation between temperature and the photosynthetic bloom was not strong unless the lagoon water temperatures were offset by 14 days (Table 2). In the lagoons examined, a photosynthetic bloom was never observed before the lagoon water temperature reached a minimum of 15°C and often required temperatures of 19 to 21°C, and the blooms ended when the lagoon water temperature dropped below 20°C.
Light was also required for a bloom to occur under laboratory conditions. However, no direct correlation was observed between the intensity of solar radiation and the population densities of photosynthetic bacteria. As in the case of water temperature, there were indirect correlations between solar radiation and population densities of photosynthetic bacteria, but only if the results were offset by 2 months. The response time should not be considered unusual considering the time required for solar warming of the lagoon water and the physiological response by the photosynthetic population to light and temperature changes. In addition to temperature and perhaps solar radiation, organic loading appeared to be a major determinant of whether a photosynthetic bloom occurred in an anaerobic swine waste lagoon or outdoor pit storage system. Photosynthetic blooms were never observed in outdoor waste storage systems if the organic load exceeded 1 g of protein/liter.
The microbial community structure in a lagoon before and during a photosynthetic bloom appeared to be complex. Based on chemical analyses of air and effluent samples and on the population levels of Rhodobacter sp. strain PS9, the following yearly sequence of events is postulated to occur. First, as a lagoon thaws and the temperature increases to approximately 10°C, the microbial population responsible for the generation of VOC becomes active, and the levels of VOC increase (Fig. 8). Second, based on hydrogen sulfide and methane emission rates (59), as well as the concentrations of sulfate and nitrate in the lagoon effluent (Fig. 7), sulfate-reducing bacteria, methanogens, and denitrifying bacteria appear to be the main bacterial groups utilizing the VOC. During this period, the levels of sulfate and nitrate in the lagoon water are not detectable, and the rates of emission of VOC, methane, and hydrogen sulfide are high (59; J. A. Zahn, A. A. DiSpirito, and Y. S. Do, unpublished results). As the temperature increases above 20°C, the population levels of photosynthetic bacteria, including Rhodobacter sp. strain PS9, increase rapidly, and these bacteria become the predominant microbial group in the lagoon system (Fig. 6 to 8). According to quantitative oligonucleotide probe hybridization data, the size of the Rhodobacter sp. strain PS9 population increases, as indicated by the relative abundance of its 16S rDNA (up to 10% of the total community 16S rDNA and up to 27% of the eubacterial community 16S rDNA). Estimation of Rhodobacter sp. strain PS9 activity, as indicated by the relative abundance of its 16S rRNA, showed that this activity increased to 34% of the total community activity and a surprising 99% of the eubacterial community activity during the height of a bloom. During a photosynthetic bloom the flux rates of methane and hydrogen sulfide increased 35 and 16%, respectively (59), concomitant with 80 to 85% decreases in the rates of emission of VOC and increases in the concentrations of sulfate and nitrite in the lagoon water. The increases in the sulfate and nitrate concentrations in lagoon water that occur with the increased size of the Rhodobacter sp. strain PS9 population may be interpreted as indicating that the photosynthetic bacteria outcompete sulfate-reducing bacteria, methanogens, and denitrifiers for the uptake and utilization of VOC.
This study was undertaken to improve our understanding of the microbial, chemical, and physical parameters involved in the photosynthetic blooms in anaerobic swine waste lagoons, and the objective is to manipulate microbial populations for odor control. The results indicate that population changes in anaerobic swine waste lagoons can occur naturally and reduce emissions of the VOC associated with odor. The results also indicate the potential of photosynthetic bacteria for controlling the rates of emission of VOC in anaerobic swine waste lagoon systems and provide useful base information for future attempts to manipulate microbial populations in these systems. For example, the results presented here support an initial strategy for initiation or extension of photosynthetic blooms in anaerobic swine waste lagoons which involves maintaining temperatures above 20°C and organic loads of less than 4 g (dry weight) per liter. Although not evident from the results presented here, increasing light intensity and/or duration should also aid in the initiation and maintenance of photosynthetic blooms involving Rhodobacter sp. strain PS9. In addition, the observation that Rhodobacter sp. strain PS9 is the dominate photosynthetic bacterium in all anaerobic swine waste lagoons tested in Minnesota, Iowa, Missouri, and North Carolina suggests that inoculation may not be necessary in initial attempts to induce a photosynthetic bloom involving purple non-sulfur bacteria in an anaerobic lagoon system.
Acknowledgments
Research support from the Iowa Soybean Promotion Board, the Iowa Corn Board, the Iowa Pork Producers Association, and the Iowa State University Office of Biotechnology (to A.A.D.) is gratefully acknowledged.
We thank L. Halverson (Iowa State University) for his assistance with the MIDI-FAME analysis and T. M. Pepper (Iowa State University microscopy facility) for performing electron microscopy.
REFERENCES
- 1.Al-Kanani, T., E. Akochi, A. F. MacKenzie, I. Alli, and S. Barrington. 1992. Odor control in liquid hog manure by added amendments and aeration. J. Environ. Qual. 21:704-708. [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barth, C. L., and S. W. Melvin. 1984. Odor, p. 97-106. In J. M. Sweeten and F. J. Humenlik (ed.), Agriculture and the environment. American Society of Agricultural Engineers, St. Joseph, Mich.
- 4.Buckley, D. H., J. R. Graber, and T. M. Schmidt. 1998. Phylogenetic analysis of nonthermophilic members of the kingdom Crenarchaeota and their diversity and abundance in soils. Appl. Environ. Microbiol. 64:4333-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casey, T. A., R. A. Rasmussen, and J. W. Petrich. June 1999. Method and system for detecting fecal ingesta contamination on carcasses of meat animals. U.S. patent 5,914,247.
- 6.DeLong, R. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiSpirito, A. A., Y. S. Do, C. L. Krema, J. Emerson, and J. A. Zahn. 1995. Methods to monitor microbial populations and odors from livestock wastes, p. 80-85. In H. L. Harris (ed.), International Livestock Odor Conference. Iowa State University, Ames.
- 8.DiSpirito, A. A., Y. S. Do, G. J. Philips, and J. A. Zahn. January 2003. A Rhodobacter strain for odor remediation of anaerobic livestock waste lagoons and biomass production. U.S. patent 6,489,156.
- 9.DiSpirito, A. A., and J. A. Zahn. June 1998. Device for quantification of odors from liquid livestock wastes. U.S. patent 5,766,551.
- 10.DiSpirito, A. A., and J. A. Zahn. April 1999. Methods and means for quantification of odors from livestock wastes. U.S. patent 5,898,003.
- 11.Do, Y. S. 2001. Role of the dominant phototrosynthetic bacterium in anaerobic swine waste lagoons, Rhodobacter sp. PS9, in odor remediation. Ph.D. thesis. Iowa State University, Ames.
- 12.Gemerden, H. V., and H. H. Beeftink. 1983. Ecology of phototrophic bacteria, p. 146-185. In J. G. Ormerod (ed.), The phototrophic bacteria: anaerobic life in light, vol. 4. University of California Press, Los Angeles.
- 13.Giovannoni, S. J., T. B. Britsschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60-63. [DOI] [PubMed] [Google Scholar]
- 14.Grady, L. C. P., T. T. Daigger, and H. C. Lim. 1999. Biological waste water treatment, 2nd ed. Marcel Dekker, New York, N.Y.
- 15.Harkin, T. 1997. Animal waste pollution in America: an emerging national problem. [Online.] The Minority Staff of the United States Senate Committee on Agriculture, Nutrition, and Forestry, Washington, D.C. www.senate.gov/∼agriculture/animalw.htm.
- 16.Harper, L. A., and R. R. Sharpe. 1997. Lagoon nutrient cycling and atmospheric nitrogen losses. USDA Agricultural Research Service, Beltsville, Md.
- 17.Hiraishi, A., K. Muramatsu, and Y. Ueda. 1996. Molecular genetic analysis of Rhodobacter azotoformans sp. nov. and related species of phototrophic bacteria. Syst. Appl. Microbiol. 19:168-177. [Google Scholar]
- 18.Hiraishi, A., K. Muramatsu, and K. Urata. 1995. Characterization of new denitrifying Rhodobacter strains isolated from photosynthetic sludge for wastewater treatment. J. Ferment. Bioeng. 79:39-44. [Google Scholar]
- 19.Hobbs, P. J., T. H. Misselbrook, and B. F. Pain. 1995. Assessment of odors from livestock wastes by a photoionization detector, an electronic nose, olfactometry and gas chromatography-mass spectrometry. J. Agric. Eng. Res. 60:137-144. [Google Scholar]
- 20.Koch, A. L. 1994. Growth measurement, p. 249-277. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular biology. American Society for Microbiology, Washington, D.C.
- 21.Lindley, J. A. 1982. Processing manure for feed components. Agricultural Experimental Station North Central Regional Research publication 284. USDA Agricultural Research Service, Beltsville, Md.
- 22.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 23.Madigan, M. T., D. O. Jung, C. R. Woese, and L. A. Achenbach. 2000. Rhodoferax antarcticus sp. nov., a moderately psychrophilic purple nonsulfur bacterium isolated from an Antarctic microbial mat. Arch. Microbiol. 173:269-277. [DOI] [PubMed] [Google Scholar]
- 24.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, P. R. Saxman, R. J. Ferris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP (Ribosomal Database Project) moves forward. Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merrill, L. S. 1999. Microbial community structure and malodorous compounds within swine waste storage systems. M.S. thesis. Iowa State University, Ames.
- 26.Miner, J. R. 1982. Controlling odors from livestock production facilities. Agricultural Experimental Station North central Regional Research publication 284. USDA Agricultural Research Service, Beltsville, Md.
- 27.Mukwaya, G. M., and D. F. Welch. 1989. Subgrouping of Pseudomonas cepacia by cellular fatty acid composition. J. Clin. Microbiol. 27:2640-2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Odam, E. M., J. Page, M. G. Townsend, and J. P. Wilkins. 1986. Identification of volatile components in headspace from animal slurries, p. 284-295. In V. C. Nielsen, J. H. Voorburg, and P. L. Hermite (ed.), Odor prevention and control of organic sludge and livestock farming. Elsevier Applied Science Publishers, New York, N.Y.
- 29.Oelze, J. 1985. Analysis of bacteriochlorophylls. Methods Microbiol. 18:275-284. [Google Scholar]
- 30.Ogram, A., W. Sun, F. J. Brockman, and J. K. Frederickson. 1995. Isolation and characterization of RNA from low-biomass deep-subsurface sediments. Appl. Environ. Microbiol. 61:763-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olsen, G. J., H. Matsuda, R. Hagstrom, and R. Overbeek. 1994. FasDNAml: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput. Applic. Biosci. 10:41-48. [DOI] [PubMed] [Google Scholar]
- 32.Pain, B. F., C. R. Clarkson, V. R. Phillips, J. V. Klarenbeek, T. H. Misselbrook, and M. Bruins. 1991. Odor emission arising from application of livestock slurries on land: measurements following spreading using a micrometeorological technique and olfactometry. J. Agric. Eng. Res. 48:101-110. [Google Scholar]
- 33.Pfennig, N., and H. G. Trüper. 1992. The family Cromatiaceae, p. 3200-3221. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes. Springer-Verlag, Berlin, Germany.
- 34.Phillips, J. A., and T. D. Brock. 1991. Laboratory manual: biology of microorganisms, 6th ed. Prentice Hall, Englewood Cliffs, N.J.
- 35.Purdy, K. J., T. M. Embley, S. Takii, and D. B. Nedwell. 1996. Rapid extraction of DNA and rRNA from sediments by a novel hydroxyapatite spin-column method. Appl. Environ. Microbiol. 62:3905-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raskin, L., J. M. Stromley, B. E. Rittmann, and D. A. Stahl. 1994. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl. Environ. Microbiol. 60:1232-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ritter, W. F. 1981. Chemical and biochemical odor control of livestock wastes: review. Can. Agric. Eng. 23:1-4. [Google Scholar]
- 38.Ritter, W. F. 1989. Odor control of livestock wastes: state-of-the-art in North America. J. Agric. Eng. Res. 42:51-62. [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Sasser, M. 1990. Identification of bacteria by gas chromatography of cellular fatty acids. MIDI technical note 101. MIDI, Newark, Del.
- 41.Sasser, M. 1990. “Tracking” a strain using the Microbial Identification System. MIDI technical note 102. MIDI, Newark, Del.
- 42.Sauer, P. J., and J. L. Hatfield.1994. Walnut Creek watershed research protocol report. Bulletin 94-1. National Soil Tilth Lab, USDA Agricultural Research Service, Beltsville, Md.
- 43.Schiffman, S. S., E. A. S. Miller, M. S. Suggs, and B. G. Graham. 1995. The effect of environmental odors emanating from commercial swine operations on the mood of nearby residents. Brain Res. Bull. 37:369-375. [DOI] [PubMed] [Google Scholar]
- 44.Siefert, E., R. L. Igrens, and N. Pfennig. 1978. Phototrophic purple and green bacteria in a sewage treatment plant. Appl. Environ. Microbiol. 35:38-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spoelstra, S. F. 1980. Origin of objectionable odorous components in piggery wastes and the possibility of applying indicator components for studying odor development. Agric. Environ. 5:241-260. [Google Scholar]
- 46.Spoelstra, S. F. 1977. Simple phenols and indoles in anaerobically stored piggery wastes. J. Sci. Food Agric. 28:415-423. [Google Scholar]
- 47.Stevens, S. S. 1970. Neural events and the psychological law. Science 170:1043-1050. [DOI] [PubMed] [Google Scholar]
- 48.Stevens, S. S. 1962. The surprising simplicity of sensory metric. Am. Psychol. 17:29-39. [Google Scholar]
- 49.Stevens, S. S. 1961. To honor Fechner and repeal his law. Science 133:80-86. [DOI] [PubMed] [Google Scholar]
- 50.Thauer, R. K. 1989. Energy metabolism of sulfate-reducing bacteria, p. 397-414. In H. G. Schlegel and B. Browien (ed.), Autotrophic bacteria. Springer-Verlag, New York, N.Y.
- 51.Trampel, D. W., T. M. Pepper, and B. L. Blaburn. 2000. Urinary tract cryptosporidiosis in commercial laying hens. Avian Dis. 44:479-484. [PubMed] [Google Scholar]
- 52.Warner, P. O., K. S. Sidhu, and L. Chadzynski. 1990. Measurement and impact of agricultural odors from a large scale swine production farm. Vet. Hum. Toxicol. 32:319-323. [PubMed] [Google Scholar]
- 53.Wenke, T. L., and J. C. Vogt. 1981. Temporal changes in a pink feedlot lagoon. Appl. Environ. Microbiol. 41:381-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams, A. G. 1984. Indicators of piggery slurry odor offensiveness. Agric. Wastes 10:15-36. [Google Scholar]
- 55.Williams, A. G., and M. R. Evans. 1981. Storage of piggery slurry. Agric. Wastes 3:311-322. [Google Scholar]
- 56.Zahn, J. A. 1997. Swine odor and emissions from pork production, p. 20-122. In K. McGuire (ed.), Environmental assurance program. National Pork Producers Council, Clive, Iowa.
- 57.Zahn, J. A., A. A. DiSpirito, Y. S. Do, B. E. Brooks, E. E. Copper, and J. L. Hatfield. 2001. Correlation of human olfactory responses to airborne concentrations of malodorous volatile organic compounds emitted from swine effluent. J. Environ. Qual. 30:624-634. [DOI] [PubMed] [Google Scholar]
- 58.Zahn, J. A., J. L. Hatfield, Y. S. Do, A. A. DiSpirito, D. A. Laird, and R. L. Pfeiffer. 1997. Characterization of volatile organic emissions and wastes from a swine production facility. J. Environ. Qual. 26:1687-1696. [Google Scholar]
- 59.Zahn, J. A., J. L. Hatfield, D. A. Laird, T. T. Hart, Y. S. Do, and A. A. DiSpirito. 2001. Functional classification of swine manure management systems based on effluent and gas emission characteristics. J. Environ. Qual. 30:635-647. [DOI] [PubMed] [Google Scholar]
- 60.Zheng, D., L. Raskin, E. W. Alm, and D. A. Stahl. 1996. Characterization of universal small-subunit rRNA-targeted oligonucleotide hybridization probes. Appl. Environ. Microbiol. 62:4504-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zuber, H., and R. J. Cogdell. 1995. Structure and organization of purple bacterial antenna complexes, p. 315-348. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.