Abstract
Clostridium perfringens is a leading cause of bacterial food-borne illness in countries where consumption of meat and poultry is high. For example, each year in the United States, this organism is the second or third most common cause of confirmed cases of food-borne illness. Surveys of the incidence of this organism in retail foods were done in the 1960s without regard to whether isolates were enterotoxigenic. It is now known that not all strains of this organism possess the enterotoxin gene responsible for illness. We examined the incidence of this organism in 131 food samples from retail food stores in an area of the northeastern United States. Forty isolates were obtained by using the iron milk method at 45°C, with confirmation by use of motility nitrate and lactose gelatin media. The presence of the C. perfringens enterotoxin (cpe) and alpha toxin (cpa) genes was determined by PCR using previously published primer sequences. All isolates possessed cpa. None of the isolates were identified as carrying the cpe gene by this method or by another method using a digoxigenin-labeled gene probe. Consistent with these results, none of the sporulating-cell extracts contained enterotoxin as determined by reverse passive latex hemagglutination. Pulsed-field gel electrophoresis was used to determine the genetic relatedness of the isolates. About 5% of the isolates were considered to be closely related (2- to 3-band difference). The others were considered to be unrelated to one another. The results demonstrate the rarity of cpe+ strains in retail foods and the genetic diversity among nonoutbreak strains.
Food-borne illness caused by Clostridium perfringens is among the most common illnesses resulting from the consumption of contaminated food. In the United States in the period from 1983 to 1997, this organism was the third most common cause of confirmed outbreaks and cases of food-borne illness (4, 5, 6). The illness is caused by an enterotoxin produced during sporulation, and the vehicles of infection are typically meat and poultry products, usually those found in food service settings. Recent reviews have examined in detail food-borne illness caused by C. perfringens (13, 23).
While the presence of C. perfringens enterotoxin (CPE) in patients' stools is a definitive indicator for implicating C. perfringens as the etiological agent in food-borne disease, CPE cannot be routinely detected directly in foods. Thus, it is difficult to link patient isolates with suspect foods. Serotyping of food and stool isolates has been used but has significant drawbacks. For example, in one large-scale study in the United States, only about one-third of the isolates tested were confirmed by serotype results (12). Similarly, using commercially available antisera, Saito reported that only 17% of 131 nonoutbreak strains tested could be serotyped (27).
Among the many genetic methods for subtyping C. perfringens, pulsed-field gel electrophoresis (PFGE) has shown substantial discriminatory power (7, 8, 16, 30) and has been used to subtype food-borne C. perfringens strains associated with outbreaks (22, 26, 28). In the case of at least one outbreak, both cpe-negative and cpe+ strains were isolated from patients' feces and possessed the same PFGE profile (26).
Here we report on the enterotoxigenicity and presence of the cpe gene encoding enterotoxin of 40 isolates of C. perfringens from retail, nonoutbreak foods. PFGE was used to subtype isolates to provide information on the molecular relatedness of the isolates.
MATERIALS AND METHODS
Strains, media, and culture conditions.
A total of 133 food samples were obtained from local retail outlets in western Massachusetts, including open-air markets, neighborhood grocery stores, large supermarkets, and small butcher shops. Samples were transported to the laboratory on ice and processed immediately or held for a maximum of 24 h before processing. Ten-gram portions of samples were diluted in 99 ml of sterile 0.1% peptone water and blended in a stomacher. A three-tube most probable number (MPN) method using iron milk medium (IMM) was used to detect C. perfringens. For this procedure, two consecutive 1:10 dilutions were prepared. One milliliter of each of the three dilutions was added to three separate tubes of IMM. After incubation at 45°C, C. perfringens produces a unique stormy fermentation after 18 h (31). Although others have suggested that the combination of rapid growth in IMM at 45°C and a stormy fermentation within 18 h is adequate for confirmation (1, 2, 31), we chose to confirm all positive isolates by using motility nitrate (MN) and lactose gelatin (LG) agars (11). For this purpose, 0.1 ml from the highest MPN dilution was pour plated by using tryptose sulfite cycloserine agar and the plates were overlayed by using the same medium. After incubations at 37°C for 18 to 20 h, five black colonies were picked, and each was inoculated into 10 ml of fluid thioglycolate broth (FTG). After 16 to 18 h at 37°C, the MN- and LG-containing tubes were stab inoculated and incubated at 35°C, and the results were evaluated as described elsewhere (11).
PCR for cpe and cpa genes.
Template DNA was obtained from cultures of one of the five confirmed isolates obtained from each food containing C. perfringens. Cells were grown for 18 to 20 h at 37°C in FTG. The FTG was prepared by using individual components, except agar. One milliliter of culture was centrifuged at 5,000 × g for 15 min, and the cell pellet was washed twice with sterile saline and resuspended in 200 μl of high-pressure liquid chromatography-grade water and then placed in a boiling water bath for 20 min. After centrifugation, 10 μl of supernatant fluid was used as the template for PCR. The sequences of the primers for the C. perfringens alpha toxin gene (cpa), slightly modified from those previously published (24), were 5′-GCTAATGTTACTGCCGTTGA and 5′-CCTCTGATACATCGTGTAAG. The sequences of the primers for the enterotoxin gene (cpe) were 5′-GGAGATGGTTGGATATTAGG and 5′-GGACCAGCAGTTGTAGATA (24). The 50-μl PCR volume included 5 μl of Taq DNA polymerase assay buffer (Fisher), 10 μl of template DNA, 1 μM concentrations of each primer, 0.2 mM concentrations of deoxynucleoside triphosphates, 1.5 mM MgCl2, and 2 U of Taq DNA polymerase (Fisher). Amplification was carried out in a Temperature-Tronic thermal cycler (Barnstead/Thermolyne Corp.), with 30 cycles of 1 min at 94°C, 2 min at 55°C, 3 min at 72°C, and a final dwell time of 4 min at 72°C. The results were determined by electrophoresis of 20 μl of PCR products in a 1.5% agarose gel for 30 min at 80 V and staining with ethidium bromide. The 324- and 233-bp PCR products of cpa and cpe, respectively, were observed. In the case of negative results for cpe, PCR was repeated by using an internal positive cpe control, C. perfringens NCTC 8239. PCR markers (Promega Corp.) consisting of six DNA fragments with sizes of 50, 150, 300, 500, 750, and 1,000 bp were used as the standards. Amplified bands were visualized by UV illumination and photographed on high-density thermal paper film (Mitsubishi Electronics America, Inc.).
PFGE.
The preparation of DNA (buffers, enzymes, and agarose) was modified based on a CHEF bacterial genomic DNA plug kit (Bio-Rad Laboratories). One milliliter of an overnight culture (10 ml of FTG) of each isolate was centrifuged and washed once with SE wash buffer (75 mM NaCl, 25 mM EDTA [pH 8]). Cells were resuspended to an absorbance at 600 nm of 1.3 by using cell suspension buffer (10 mM Tris [pH 7.2], 20 mM NaCl, 50 mM EDTA). One milliliter of cell suspension was added to a 1.5-ml microcentrifuge tube and centrifuged for 5 min at 13,000 × g to pack cells. The supernatant was carefully removed, and the cells were resuspended in cell suspension buffer to one-half the final volume of the plugs. The suspension was equilibrated to 50°C. The cell suspension was combined with an equal volume (40 μl) of 2% CleanCut agarose (Bio-Rad Laboratories) and mixed gently at 50°C. The mixture was transferred to plug molds by using sterile transfer pipettes, and the agarose was allowed to solidify. Five milliliters of lysozyme buffer (10 mM Tris [pH 7.2], 50 mM NaCl, 0.2% sodium deoxycholate, 0.5% sodium lauryl sarcosine, 1 mg of lysozyme/ml) was added to each milliliter of agarose plugs. The solidified agarose plugs were placed in the centrifuge tubes containing lysozyme buffer and incubated for 16 h at 37°C. After cell lysis, the lysozyme buffer was removed and the plugs were rinsed in sterile distilled water with vigorous agitation. Five milliliters of proteinase K reaction buffer (100 mM EDTA [pH 8.0], 0.2% sodium deoxycholate, 1% sodium lauryl sarcosine, 1 mg of proteinase K/ml) was added for each milliliter of agarose plugs, and the plugs were incubated overnight at 50°C without agitation. Next, the plugs were washed four times in 50 ml of wash buffer (20 mM Tris [pH 8.0], 50 mM EDTA) for 40 min each time at room temperature with gentle agitation, and then the plugs were stored at 4°C. DNA samples in agarose were digested with 20 U of enzyme ApaI (New England Biolabs) for 4 h at 25°C in 200 μl of the buffer as recommended by the manufacturer. After digestion, the buffer was removed and the plugs were incubated in 1 ml of 0.66× TBE buffer (1× TBE is 89 mM Tris-89 mM boric acid-2 mM EDTA, pH 8.3) for approximately 30 min with gentle agitation. Sample plugs containing DNA were placed on a smooth clean surface and cut to size by using a razor blade or spatula. Low-melt preparative-grade agarose (1%, 100 ml) was cast around the comb. The plug remained in place when the comb was removed. PFGE was performed with a Bio-Rad CHEF-DR II apparatus with pulse times ramped from 0.5 to 40 s over 20 h. A lambda standard ladder and Saccharomyces cerevisiae DNA size standards (Bio-Rad) were used as molecular size markers. Analysis of the banding pattern was done by using GelCompar II (Biosystematica). In general, strains were considered clonal if they showed 100% similarity with a 1% tolerance for fragment shifts.
DIG-labeled cpe gene probe protocol.
Isolates were grown in FTG at 37°C for 16 to 18 h. Twenty-five or 50 μl was transferred under vacuum to a dot blot microfiltration apparatus (Bio-Rad Laboratories) containing a 0.45-μm-pore-size nylon membrane. Membranes were left to dry at room temperature. cpe+ (NCTC 8798) and cpe-negative (ATCC 3624) strains were included as controls. A 40-oligomer digoxigenin (DIG)-labeled cpe probe (probe A [36]) was obtained from NBI/Genovus (Plymouth, Minn.). The DNA dot blots were prehybridized and then hybridized with the DIG-labeled cpe probe according to the manufacturer's instructions (Boehringer Mannheim). Anti-DIG-alkaline phosphate and a mixture of the color substrates nitroblue tetrazolium chloride and BCIP (5-bromo-4-chloro-3-indolylphosphate) were used to detect DIG-labeled nucleic acid hybrids according to the manufacturer's instructions (formerly Genius 3 system; Boehringer Mannheim).
Enterotoxin detection.
The presence of the cpe gene does not necessarily mean that it is expressed (18). Indeed, certain C. perfringens type E isolates contain silent cpe sequences (3). We therefore also sought to determine the ability of isolates to produce CPE. CPE is only produced during the sporulation process, so a number of sporulation media were employed (see below). The presence of CPE in sporulating-cell extracts was determined by reversed passive latex agglutination (RPLA) using a commercially available kit (Oxoid, Ogdensburg, N.Y.). For this purpose, 200 ml of sporulation medium was inoculated (1%) with an overnight FTG culture. After incubation at 37°C for 7 h, the percentage of sporulating cells (200 cells were observed) was determined by phase-contrast microscopy. The cells were collected by centrifugation, washed once with 5 ml of 0.1 M phosphate buffer (pH 6.8), and disrupted by sonication while chilled in an ice-water bath (6 to 7 30-s cycles with 30-s cooling periods). Disrupted cells were collected by centrifugation at 10,000 × g for 20 min, and the supernatant fluid was frozen at −20°C for subsequent analysis by RPLA according to the manufacturer's protocol. CPE+ (NCTC 8798) and CPE− (ATCC 3624) strains were included in all assays.
Sporulation media.
Although formulae for sporulation media are included with commercial CPE assay kits, no single medium will support the sporulation of all or most strains of C. perfringens. It has been our experience that nonoutbreak strains are especially reluctant to sporulate (R. Labbe, unpublished data). All strains in this study were grown in Duncan-Strong sporulation medium (10) or a modified version in which raffinose (0.4%) replaced soluble starch (21). Various adjuncts previously shown to enhance sporulation of C. perfringens were also added to each basal medium. The optimal concentration of each adjunct varied with each isolate, ranging from 50 to 200 μg/ml for caffeine, theobromine (20), or papaverine (Labbe, unpublished); 0.12 to 2.0 mg/ml for bovine bile salts or sodium taurocholate (14); and 15 to 150 μl of a 40-fold concentrate of a putative sporulation factor (29, 35). All chemicals were obtained from Sigma. Fresh, filter-sterilized cattle rumen fluid was also added (after repeated centrifugations to remove particulate matter) as an adjunct (5, 10, and 20%) to each basal medium. Thus, 16 variations of sporulation media were employed.
RESULTS
Isolates.
Thirty-nine of the 132 (30%) retail food samples contained C. perfringens at levels between 3 and 292 CFU/g. One additional sample (chicken leg) had >1,100 CFU/g (Table 1). All isolates were confirmed by using MN and LG media. Meat (24 isolates) was the most common food item found to contain C. perfringens, followed by poultry and fish (5 isolates each), soup mixes (2 isolates), pasta sauce, fresh mushrooms, and herbs (1 isolate each) (Table 2). One isolate from each food item was tested by PCR and gene probe assays.
TABLE 1.
Levels and characteristics of C. perfringens isolated from retail foods
| Food product | MPN/g | Presence or absence of:
|
|||
|---|---|---|---|---|---|
| cpa genea |
cpe gene
|
CPEb | |||
| By PCR | By dot blot | ||||
| Mushroom | 3.05 | + | − | − | − |
| Lamb | 7.36 | + | − | − | − |
| Ground beef 1 | 20.5 | + | − | − | − |
| Chicken neck | 9.18 | + | − | − | − |
| Ground veal | 93.3 | + | − | − | − |
| Ground beef 2 | 14.7 | + | − | − | − |
| Ground beef 3 | 9.18 | + | − | − | − |
| Burger meat | 7.36 | + | − | − | − |
| Minestrone | 6.19 | + | − | − | − |
| Sicilian soup mix | 6.11 | + | − | − | − |
| Fine herb | 3.57 | + | − | − | − |
| Pasta sauce | 3.01 | + | − | − | − |
| Gumbo mix | 3.57 | + | − | − | − |
| Haddock | 6.11 | + | − | − | − |
| Shrimp | 93.3 | + | − | − | − |
| Flounder | 11 | + | − | − | − |
| Clam | 23.1 | + | − | − | − |
| Salmon | 11.2 | + | − | − | − |
| Ground beef | 7.23 | + | − | − | − |
| Pork bone | 7.36 | + | − | − | − |
| Beef | 7.23 | + | − | − | − |
| Ground pork | 42.7 | + | − | − | − |
| Beef | 14.7 | + | − | − | − |
| Ground pork | 3.57 | + | − | − | − |
| Beef liver | 14.7 | + | − | − | − |
| Turkey leg | 42.7 | + | − | − | − |
| Ground pork | 3.57 | + | − | − | − |
| Beef | 9.18 | + | − | − | − |
| Ground beef 4 | 9.18 | + | − | − | − |
| Sausage | 7.36 | + | − | − | − |
| Pork | 7.36 | + | − | − | − |
| Ground chuck | 149 | + | − | − | − |
| Spare ribs | 292 | + | − | − | − |
| Chicken leg | >1,100 | + | − | − | − |
| Sausage | 7.36 | + | − | − | − |
| Chicken | 3.05 | + | − | − | − |
| Ground veal | 7.36 | + | − | − | − |
| Ground veal | 3.57 | + | − | − | − |
| Chicken | 11.2 | + | − | − | − |
| Pork | 20.5 | + | − | − | − |
Determined by PCR.
CPE in cell extract, determined by RPLA.
TABLE 2.
Proportion of MPN-positive C. perfringens isolates in retail food samples by category
| Sample | Total no. tested | No. (%) of MPN-positive isolates |
|---|---|---|
| Meat products | 66 | 24 (36) |
| Poultry | 19 | 5 (26) |
| Vegetable | 10 | 1 (10) |
| Fish | 18 | 5 (28) |
| Instant soups and dry seasonings | 20 | 5 (25) |
| Total | 133 | 40 (30) |
cpa and cpe gene detection.
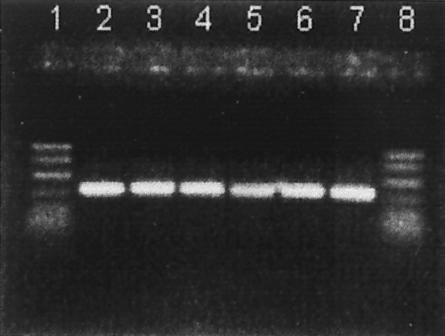
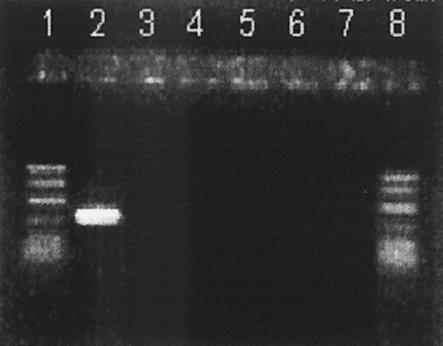
PCR was used to detect the cpa and cpe genes. As expected, all isolates possessed cpa encoding alpha toxin (Fig. 1) (Table 1). None of the isolates possessed the cpe gene (Fig. 2). Unlike what was reported in at least one previous study (18), no assay interference was noted (in either assay) in positive controls when crude culture lysates obtained by boiling were used. In addition, none of the isolates was positive for cpe when a DIG-labeled cpe-specific gene probe was used (dot blot) (Table 1) (Fig. 3).
FIG. 1.
PCR products obtained from amplification of cpa (324 bp). Lanes 1 and 8, PCR markers (50 to 1,000 bp); lane 2, positive control (strain FD-1041); lanes 3 to 7, representative C. perfringens isolates from retail foods.
FIG. 2.
PCR products obtained from amplification of cpe (233 bp). Lanes 1 and 8, PCR markers (50 to 1,000 bp); lane 2, positive control (strain FD-1041); lanes 3 to 7, representative C. perfringens isolates from retail foods.
FIG. 3.
Dot blot for cpe gene by use of DIG-labeled cpe-specific gene probe. Fifty or 25 μl of 18-h cell cultures of each C. perfringens isolate was added to separate grids. B1 and B2, positive control (strain NCTC 8239); D1 and D2, positive control (strain FD 1041); C1 and C2, negative control (strain FD1); E1 and E2, negative control (strain NCTC 3624). Columns 3 to 12, C. perfringens food isolates (50 μl).
In vitro sporulation.
In addition to the presence of cpe, we determined, using RPLA, the ability of isolates to produce CPE. For this purpose, sporulating cells were required. A minimal sporulation level of 10% for each isolate was attempted. The standard basal medium used was Duncan-Strong medium (10) with either 0.4% starch (DS-S) or raffinose (DS-R) as the carbohydrate. Of the 40 isolates, 19 and 25 sporulated at a level of 10% or more in DS-S and DS-R, respectively (Table 3). The highest levels of sporulation were 47 and 55% for DS-S and DS-R, respectively. Cultures of isolates with the highest level of sporulation in DS-S or DS-R were kept for CPE assay. Various adjuncts increased sporulation from <10% to ≥10% (Table 3). Altogether, a total of 31 isolates sporulated at 10% or higher, and the cell extracts of these were assayed for CPE. In the case of four isolates, no sporulation was observed with any medium formulation. Similarly, use of the sporulation medium of Tortora (33) failed to induce the sporulation of these four isolates.
TABLE 3.
Media formulae supporting ≥10% sporulation of retail isolates of C. perfringens
| Medium and adjunct | No. of isolates with ≥10% sporulationa |
|---|---|
| DS-S | 19 (11-47)b |
| Caffeine | 7 (10-42) |
| Theobromine | 5 (10-37) |
| Papaverine | 7 (11-26) |
| Bile salts | 5 (12-33) |
| Sodium taurocholate | 2 (12-31) |
| Rumen fluid | 7 (11-48) |
| Sporulation factor | 5 (11-26) |
| DS-R | 25 (11-55) |
| Caffeine | 6 (11-53) |
| Theobromine | 5 (11-55) |
| Papaverine | 3 (11-25) |
| Bile salts | 4 (11-40) |
| Sodium taurocholate | 2 (11-36) |
| Rumen fluid | 3 (12-44) |
| Sporulation factor | 2 (10-25) |
Values corresponding to DS-S and DS-R are the number of isolates with ≥10% sporulation. Values corresponding to the adjuncts are the number of additional isolates for which each adjunct increased sporulation from <10% to ≥10%. The cultures of each isolate with the highest percentage of sporulation were kept for CPE analysis.
Values in parentheses indicate the range of the percentage of sporulation.
RPLA.
The presence of CPE in sporulating-cell extracts was determined for those strains which sporulated at a level of 10% or more. No CPE was detected in the 31 isolates (Table 1).
PFGE.
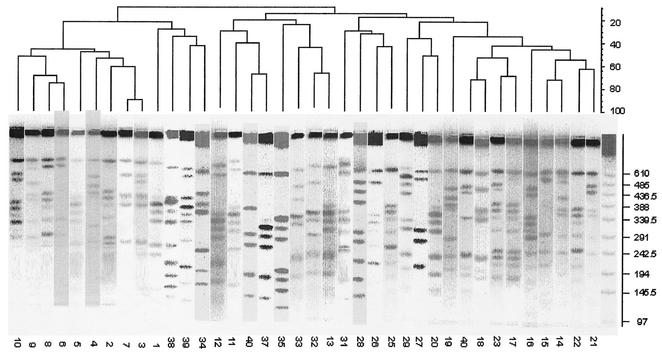
The clonal relationship of the 39 isolates from each C. perfringens-positive food sample was determined by PFGE. Restriction enzyme profiles of the isolates' genomic DNA obtained by using ApaI showed 6 to 10 fragments ranging in size from ∼50 to 610 kb (Fig. 4). According to the criteria of Tenover et al. (32), none of the isolates can be considered genetically indistinguishable. Two isolates were closely related, with 2- to 3-band differences. These isolates were both from beef products taken from the same retail outlet but at different times. The restriction enzyme profiles of the genomic DNA of these isolates were reanalyzed by using MluI. In this case, the two isolates differed by 4 bands, confirming their genetic unrelatedness.
FIG. 4.
Dendrogram of PFGE subtype patterns of C. perfringens isolated from retail food. Fragments ranged in size from 50 to 610 kb. Isolate numbers are shown below each lane.
DISCUSSION
In recent decades, many surveys on the incidence of C. perfringens in raw and processed foods have been conducted (19), and an incidence level of 30 to 80% in raw or frozen meat and poultry items has been found. These early reports did not distinguish between CPE+ and CPE− isolates. In the present study, about 30% of the samples were positive for this organism. The development of immunoassays for CPE has permitted the differentiation of CPE+ and CPE− strains with relative ease. However, since sporulation is required for CPE expression, the well-known reluctance of C. perfringens to sporulate in vitro can result in false negatives.
In recent years, the development of synthetic DNA probes and PCR for detection of the cpe gene has led to surveys of the cpe gene in isolates associated with food-borne outbreaks and from healthy and diseased animals. In one study of isolates from human and nonhuman animals having digestive tract disorders, the frequency of cpe+ isolates was 1% by DNA colony hybridization (9). As part of the same study, none of the 45 isolates from nonoutbreak foodstuffs of animal origin hybridized with the enterotoxin gene probe. An additional study of clostridial enteric diseases in animals (30) indicated that about 8% of isolates tested were PCR positive for cpe. In the case of the intestinal contents of healthy pigs and swine feed, none of the 97 C. perfringens isolates tested in a previous study were positive for the enterotoxin gene (15). Using a cpe DIG probe, Tschirdewahn et al. (34) reported that cpe+ strains were present in 14, 22, 10, and 0% of fecal isolates from horses, cattle, poultry, and pigs, respectively, while Van Damme-Jongsten et al. (37) reported that only 6% of the 98 C. perfringens isolates they obtained from a variety of animals were cpe+.
There are few reports of the prevalence of cpe+ strains in nonoutbreak foods. In a recent study, C. perfringens was present in an average of 37% of meat and poultry samples (25), yet cpe+ strains amounted to only 17% of isolates. Using an RPLA assay, Saito (27) reported that only 2% of C. perfringens strains isolated from meat and fish were CPE+, though whether each isolate sporulated, a requirement for the assay, was not reported.
In this work, we found that of the 40 isolates of C. perfringens obtained from retail outlets, none possessed the cpe gene. Of the 31 strains in which a level of 10% sporulation was achieved, none possessed detectable CPE in their cell extracts. None of the isolates possessed identical PFGE profiles, with only two being closely related. These results demonstrate the wide genetic variation among CPE− strains. Similarly, using ribotyping, Kilic et al. (17) found only four identical ribotype patterns among 111 C. perfringens isolates from ground meat.
Despite the low incidence of cpe+ strains in food products, they are nevertheless able to function as a reservoir for enterotoxin-producing strains. Given the number of outbreaks caused by this organism, our finding that a reasonably high level (30%) of the retail food sampled contained C. perfringens suggests that there may be many instances where this organism may grow to significant levels in temperature-abused foods. The relative rarity of CPE+ strains may be one reason that this organism is not more of a public health problem.
Acknowledgments
This work was supported in part by the U.S. Department of Agriculture International Cooperation and Development Program and by the USDA Massachusetts Agricultural Experiment Station under project no. mas09702553.
REFERENCES
- 1.Abeyta, C., Jr., A. Michalovskis, and M. Wekell. 1985. Differentiation of Clostridium perfringens from related clostridia in iron milk medium. J. Food Prot. 48:130-134. [DOI] [PubMed] [Google Scholar]
- 2.Abeyta, C., Jr. 1983. Comparison of iron milk and official AOAC methods for enumeration of Clostridium perfringens from fresh seafoods. J. Assoc. Off. Anal. Chem. 66:1175-1179. [PubMed] [Google Scholar]
- 3.Billington, S., E. Wieckowski, M. Sarker, D. Bueschel, J. G. Songer, and B. McClane. 1998. Clostridium perfringens type E animal enteritis isolates with highly conserved, silent enterotoxin gene sequences. Infect. Immun. 66:4531-4536. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Centers for Disease Control. 1990. Foodborne disease outbreaks, 1983-1987. Centers for Disease Control, Atlanta, Ga.
- 5.Centers for Disease Control and Prevention. 1996. Surveillance for foodborne-disease outbreaks—U.S. 1998-1992. Centers for Disease Control and Prevention, Atlanta, Ga.
- 6.Centers for Disease Control and Prevention. 2000. Surveillance for foodborne-disease outbreaks—U.S. 1993-1997. Centers for Disease Control and Prevention, Atlanta, Ga.
- 7.Collie, R., and B. McClane. 1998. Evidence that the enterotoxin gene can be episomal in Clostridium perfringens isolates associated with non-food-borne human gastrointestinal diseases. J. Clin. Microbiol. 36:30-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collie, R., J. Kokai-Kun, and B. McClane. 1998. Phenotypic characterization of enterotoxigenic Clostridium perfringens isolates from non-foodborne human gastroenteritis. Anaerobe 4:69-79. [DOI] [PubMed] [Google Scholar]
- 9.Daube, G., P. Simon, B. Limbourg, C. Manteca, J. Mainil, and A. Kaeckenbeeck. 1996. Hybridization of 2659 Clostridium perfringens isolates with gene probes for seven toxins (α, β, ɛ, ι, θ, μ, and enterotoxin) and for sialidase. Am. J. Vet. Res. 57:496-501. [PubMed] [Google Scholar]
- 10.Duncan, C., and D. Strong. 1968. Improved medium for sporulation of Clostridium perfringens. Appl. Microbiol. 16:82-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Food and Drug Administration. 1998. Bacteriological analytical manual, 8th ed. Association of Official Analytical Chemists International, Gaithersburg, Md.
- 12.Hatheway, C., D. Whaley, and V. Dowell, Jr. 1980. Epidemiological aspects of Clostridium perfringens foodborne illness. Food Technol. 34:77-79, 90.
- 13.Heredia, N., and R. Labbe. 2001. Clostridium perfringens, p. 133-141. In R. Labbe and S. Garcia, (ed.), Guide to foodborne disease. John Wiley and Sons, New York, N.Y.
- 14.Heredia, N., R. Labbe, M. A. Rodriguez, and J. S. Garcia-Alvarado. 1991. Growth, sporulation and enterotoxin production by Clostridium perfringens type A in the presence of human bile salts. FEMS Microbiol. Lett. 68:15-21. [DOI] [PubMed] [Google Scholar]
- 15.Kanakaraj, R., D. Harris, J. G. Songer, and B. Bosworth. 1998. Multiplex PCR assay for detection of Clostridium perfringens in feces and intestinal contents of pigs and in swine feed. Vet. Microbiol. 63:29-38. [DOI] [PubMed] [Google Scholar]
- 16.Katayama, S., B. Dupuy, G. Daube, B. China, and S. Cole. 1996. Genome mapping of Clostridium perfringens strains with I-CeuI shows many virulence genes to be plasmid-borne. Mol. Gen. Genet. 251:720-726. [DOI] [PubMed] [Google Scholar]
- 17.Kilic, U., B. Schalch, and A. Stolle. 2002. Ribotyping of Clostridium perfringens from industrially produced ground meat. Lett. Appl. Microbiol. 34:238-243. [DOI] [PubMed] [Google Scholar]
- 18.Kokai-Kun, J. F., J. G. Songer, J. R. Czeczulin, F. Chen, and B. A. McClane. 1994. Comparison of western immunoblots and gene detection assays for identification of potentially enterotoxigenic isolates of Clostridium perfringens. J. Clin. Microbiol. 32:2533-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Labbe, R. 1989. Clostridium perfringens, p. 191-233. In M. Doyle (ed.), Foodborne bacterial pathogens. Marcel Dekker, Inc., New York, N.Y.
- 20.Labbe, R., and L. Nolan. 1981. Stimulation of Clostridium perfringens enterotoxin formation by caffeine and theobromine. Infect. Immun. 34:50-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Labbe, R., and D. Rey. 1979. Raffinose increases sporulation and enterotoxin production by Clostridium perfringens type A. Appl. Environ. Microbiol. 37:1196-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maslanka, S., J. Kerr. G. Williams, J. Barbaree, L. Carson, J. Miller, and B. Swaminathan. 1999. Molecular subtyping of Clostridium perfringens by pulsed-field gel electrophoresis to facilitate food-borne disease outbreak investigations. J. Clin. Microbiol. 37:2209-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McClane, B. 2001. Clostridium perfringens, p. 351-372. In M. Doyle, L. Beuchat, and T. Montville (ed.), Food microbiology: fundamentals and frontiers. American Society for Microbiology, Washington, D.C.
- 24.Meer, R., and J. G. Songer. 1997. Multiplex polymerase chain reaction assay for genotyping Clostridium perfringens. Am. J. Vet. Res. 58:702-705. [PubMed] [Google Scholar]
- 25.Miwa, N., T. Nishina, S. Kubo, M. Atsumi, and H. Honda. 1998. Amount of enterotoxigenic Clostridium perfringens in meat detected by nested PCR. Int. J. Food Microbiol. 42:195-200. [DOI] [PubMed] [Google Scholar]
- 26.Riddell, J., J. Bjorkvoth, H. Eisgruber, B. Schalch, A. Stolle, and H. Korkeala. 1998. Prevalence of the enterotoxin gene and clonality of Clostridium perfringens strains associated with food poisoning outbreaks. J. Food Prot. 61:240-243. [DOI] [PubMed] [Google Scholar]
- 27.Saito, M. 1990. Production of enterotoxin by Clostridium perfringens derived from humans, animals, foods, and the natural environment in Japan. J. Food Prot. 53:115-118. [DOI] [PubMed] [Google Scholar]
- 28.Schalch, B., B. Sperner, H. Eisgruber, and A. Stolle. 1999. Molecular methods for the analysis of Clostridium perfringens relevant to food hygiene. FEMS Immnol. Med. Microbiol. 24:281-286. [DOI] [PubMed] [Google Scholar]
- 29.Shih, N.-J., and R. Labbe. 1996. Sporulation-promoting ability of Clostridium perfringens culture fluids. Appl. Environ. Microbiol. 62:1441-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sparks, S., R. Carman, M. Sarker, and B. McClane. 2001. Genotyping enterotoxigenic Clostridium perfringens fecal isolates associated with antibiotic-associated diarrhea and food poisoning in North America. J. Clin. Microbiol. 39:883-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.St. John, W., J. Matches, and M. Wekell. 1982. Use of iron milk medium for enumeration of Clostridium perfringens. J. Assoc. Off. Anal. Chem. 65:1129-1133. [Google Scholar]
- 32.Tenover, F., R. Arbeit, R. Goering, P. Mickelsen, B. Murray, D. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tortora, J. 1984. Alternative medium for Clostridium perfringens sporulation. Appl. Environ. Microbiol. 47:1172-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tschirdewahn, B., S. Notermans, K. Wernars, and F. Untermann. 1991. The presence of enterotoxigenic Clostridium perfringens strains in feces of various animals. Int. J. Food Microbiol. 14:175-178. [DOI] [PubMed] [Google Scholar]
- 35.Tseng, W. J., and R. Labbe. 2000. Characteristics of a sporulation stimulating factor from Clostridium perfringens type A. Lett. Appl. Microbiol. 30:254-257. [DOI] [PubMed] [Google Scholar]
- 36.Van Damme-Jongsten, M., J. Rodhouse, R. J. Gilbert, and S. Notermans. 1990. Synthetic DNA probes for detection of enterotoxigenic Clostridium perfringens strains isolated from outbreaks of food poisoning. J. Clin. Microbiol. 28:131-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Damme-Jongsten, M., K. Werners, and S. Notermans. 1989. Cloning and sequencing of the Clostridium perfringens enterotoxin gene. Antonie Leeuwenhoek 56:181-190. [DOI] [PubMed] [Google Scholar]