Abstract
The Caenorhabditis elegans UNC-13 protein and its mammalian homologues are important for normal neurotransmitter release. We have identified a set of transcripts from the unc-13 locus in C. elegans resulting from alternative splicing and apparent alternative promoters. These transcripts encode proteins that are identical in their C-terminal regions but that vary in their N-terminal regions. The most abundant protein form is localized to most or all synapses. We have analyzed the sequence alterations, immunostaining patterns, and behavioral phenotypes of 31 independent unc-13 alleles. Many of these mutations are transcript-specific; their phenotypes suggest that the different UNC-13 forms have different cellular functions. We have also isolated a deletion allele that is predicted to disrupt all UNC-13 protein products; animals homozygous for this null allele are able to complete embryogenesis and hatch, but they die as paralyzed first-stage larvae. Transgenic expression of the entire gene rescues the behavior of mutants fully; transgenic overexpression of one of the transcripts can partially compensate for the genetic loss of another. This finding suggests some degree of functional overlap of the different protein products.
INTRODUCTION
Transfer of information between neurons or between neurons and muscles requires the Ca2+-dependent release of neurotransmitters from synaptic vesicles. According to the SNARE hypothesis, specific synaptic vesicle proteins interact with specific plasma membrane proteins at docking sites at the active zone (Söllner et al., 1993a,b). Interacting proteins include synaptobrevin (also known as VAMP) and synaptotagmin in the synaptic vesicle and syntaxin and SNAP-25 in the presynaptic membrane (Hayashi et al., 1993). Soluble factors, including N-ethylmaleimide–sensitive factor and α- and β-soluble N-ethylmaleimide–sensitive factor attachment proteins (Söllner et al., 1993b), are involved in the formation and/or regulation of these proteins. Although many components of synaptic vesicle targeting and neurotransmitter release have been identified, the regulation of these processes cannot be completely explained.
unc-13 was originally identified in Caenorhabditis elegans as a gene important for normal locomotion. Mutations in this gene result in severely uncoordinated movement, increased accumulation of the neurotransmitter acetylcholine, and resistance to acetylcholinesterase inhibitors (Brenner, 1974; Rand and Russell, 1985; Hosono et al., 1989; Nguyen et al., 1995). These phenotypes suggest a decrease in the synaptic release of acetylcholine, and other studies have indicated that unc-13 mutants are deficient in the release of most or all neurotransmitters (Miller et al., 1996).
The unc-13 gene encodes a protein (UNC-13) containing C1 and C2 homology domains (Maruyama and Brenner, 1991). Mammalian homologues of UNC-13, Munc13-1, Munc13-2, and Munc13-3 in rat brain (Brose et al., 1995) and Hmunc13 in human kidney (Song et al., 1999), also contain one C1 and two or three C2 domains. C1 and C2 regulatory domains are found in PKC and other proteins (Coussens et al., 1986); C1 domains bind diacylglycerol (DAG) and phorbol esters (Burns and Bell, 1991), and C2 domains bind Ca2+ and phospholipids (Kaibuchi et al., 1989). C2 domains are also present in synaptotagmin and are necessary for its function (Perin et al., 1990; Davletov and Südhof, 1993; Hata et al., 1993; Zhang et al., 1994; Li et al., 1995). In UNC-13, the C1 and C2 domains enable the protein to bind DAG and phorbol esters and to bind phospholipids in a Ca2+-dependent manner (Maruyama and Brenner, 1991; Ahmed et al., 1992; Kazanietz et al., 1995).
Phorbol esters affect the localization and function of Munc13-1 and Hmunc13 (Betz et al., 1998; Song et al., 1999). When Munc13-1 is overexpressed in Xenopus motor neurons, it is distributed throughout the cytoplasm, but it becomes associated with the plasma membrane in the presence of phorbol esters (Betz et al., 1998). Overexpression of Munc13-1 also increases spontaneous and evoked neurotransmitter release in culture, and this release is enhanced by phorbol esters (Betz et al., 1998). The subcellular localization of Hmunc13 is also dependent on the presence of DAG and phorbol esters. This cytoplasmic protein translocates to the Golgi apparatus in the presence of phorbol esters, and its translocation is associated with the induction of apoptosis (Song et al., 1999). These results suggest that unc-13 homologues may function as DAG-mediated regulators of membrane trafficking and/or secretion.
Several proteins involved in neurotransmitter release have been shown to interact with UNC-13 or Munc13-1 in the yeast two-hybrid system or in vitro. These proteins include the synaptic vesicle protein Doc2 (Orita et al., 1997) and the plasma membrane proteins syntaxin (O'Connor et al., 1997) and UNC-18/nSec-1 (Sassa et al., 1999). Interactions between UNC-13 and UNC-18 have been implicated in regulating UNC-18 interactions with syntaxin (Sassa et al., 1999). These protein–protein interactions support a role for UNC-13 in regulating neurotransmitter release.
We now show that alternative splicing and apparent alternative promoters lead to the expression of two major protein products from the C. elegans unc-13 gene. These products share their C-terminal region; this is the region most highly conserved in the mammalian homologues. The most abundant form localizes to most or all synapses. We have analyzed the sequence alterations, immunostaining patterns, and behavioral phenotypes of 31 independent unc-13 alleles. Many of these mutations are transcript-specific; their phenotypes suggest that the different UNC-13 forms have different cellular functions. A deletion allele that is predicted to disrupt all UNC-13 protein products leads to lethality as paralyzed first-stage larvae.
Preliminary results have been reported in abstract form (Eustance et al., 1999).
MATERIALS AND METHODS
Growth and Culture of C. elegans and Mutants
C. elegans was grown on modified nematode growth medium (Brenner, 1974; Johnson et al., 1988). Wild-type nematodes were N2 Bristol. The unc-13 mutations with md designations (except md2413, md2414, and md2415) were isolated in screens for resistance to the cholinesterase inhibitor aldicarb (Nguyen et al., 1995; Miller et al., 1996). Strains containing the unc-13 alleles e51 and e1091 were obtained from the C. elegans Stock Center (St. Paul, MN), the s69 mutation was from Ann Rose (University of British Columbia, Vancouver, BC), and the n2813 allele was from Erik Jorgensen (University of Utah, Salt Lake City, UT); all alleles were outcrossed at least twice before analysis.
Northern Transfers
Poly(A)+ selected RNA was purified for Northern transfers from a mixed-stage culture of wild-type worms by means of standard procedures (Sambrook et al., 1989). mRNA was separated by SDS-PAGE and transferred to nitrocellulose. Probes recognizing the unc-13L, unc-13M, and unc-13R regions were PCR amplified from a random primed cDNA library, end labeled with 32P, and imaged with a phosphorimager.
cDNA Library Screens
unc-13 cDNAs were isolated from the random primed cDNA library λActRB2. Probes were PCR amplified from plasmids containing either genomic or cDNA inserts. cDNAs were sequenced at the Oklahoma Medical Research Foundation DNA sequencing facility.
Rapid Amplification of cDNA Ends
5′ rapid amplification of cDNA ends (RACE) was performed on mixed-stage wild-type poly(A)+ selected RNA (Sambrook et al., 1989) with the use of a kit from GIBCO-BRL (Gaithersburg, MD). cDNAs were initiated from primers binding in exon 14 (in unc-13M) or in exon 18 or 24 (in unc-13R). After dC (deoxycytidine) tailing, nested PCR reactions with primers binding to exon 14, 18, or 24 were used to amplify the products. cRACE was performed as described previously (Maruyama et al., 1995).
Sequence of unc-13 Mutations
A set of 1- to 2-kilobase (kb) genomic PCR fragments spanning the unc-13 exons were synthesized by direct single-worm PCR from individual mutant animals (Williams et al., 1992). Mutations were then localized to a particular fragment with the use of a modified restriction endonuclease fingerprinting protocol (Liu and Sommer, 1995; K. Grundahl and J.B. Rand, unpublished data), followed by sequencing with internal primers directly from the PCR product.
Primary Antibody Production
The coding sequence for most of UNC-13L (amino acids 106–528 in the longest form of the UNC-13 protein) was amplified from a random primed cDNA library and cloned into pRSETA (Invitrogen, San Diego, CA). Overexpression of the fusion protein in Escherichia coli strain BL21/DE3 was induced by isopropylthio-β-galactoside, and fusion protein was purified with nickel columns (Invitrogen). Serum from immunized rabbits was affinity depleted with BL21/DE3 cells and then was affinity purified with the use of fusion proteins bound to nitrocellulose membranes with methanol (Smith and Fisher, 1984; Harlow and Lane, 1988). Bound antibody was eluted with 5 mM glycine, 0.5 M NaCl, pH 2.3, followed by a high-pH wash (50 mM triethanolamine, pH 11.5). Antibody was exchanged into PBS and concentrated with Centriprep 30 ultrafiltration (Amicon, Beverly, MA). A mAb (mAb 1403) to UNC-17/VAChT (J.S. Duerr, unpublished data) was used for double staining.
Immunocytochemical Staining
Nematodes were freeze-cracked on slides (Duerr et al., 1999) and fixed in methanol for 2 min and in acetone for 4 min. After rinsing and blocking, nematodes were incubated overnight in primary antibodies (1:50). After rinsing, nematodes were incubated for 4 h in secondary antibodies labeled with Cy3 (Jackson ImmunoResearch, West Grove, PA) or Oregon Green 488 (Molecular Probes, Eugene, OR). Slides were rinsed and mounted with anti-bleaching medium. Antibody staining patterns were visualized with the use of a Zeiss (Thornwood, NY) fluorescence microscope or a Leica (Wetzlar, Germany) TCS NT confocal microscope.
Behavioral and Fertility Assays
Body thrashing in liquid medium and pharyngeal pumping in the presence of food were measured as described previously (Miller et al., 1996; Duerr et al., 1999). Each behavior was measured in 10–50 young adults of each strain.
To measure fertility and viability, individual fourth-stage larvae were selected and transferred to new plates covered with bacteria. The size and coordination of unc-13(s69), and to some extent unc-13(e1091), individuals in a population were variable; for these tests, healthier individuals were selected. (The size and coordination of the heterozygous and wild-type individuals were relatively invariant, with virtually all appearing “healthy.”) Five to 20 individual hermaphrodites were transferred one to three times daily to new plates until they ceased laying eggs and, eventually, died. The total number of hatched progeny for each individual was determined. Plates from a representative subset of individuals were observed for at least 10 d after removal of the adult. The percentage of hatched first-stage larvae that progressed to egg-laying adults by the end of that 10-d period was determined. (The normal time from the laying of the egg to adult fertility was ∼3 d under laboratory conditions.) In addition, at least 100 eggs laid by young adults of a given phenotype were monitored for hatching.
Western Transfers
Western transfers were performed on mixed populations of nematodes as described previously (Moerman et al., 1988). Nematodes were rinsed well, and an equal volume of nematode solubilization buffer (0.3% ethanolamine, 2 mM EDTA, 5 mM DTT) was added. The nematodes were microwaved for 25 s. An equal volume of 2× Laemmli sample buffer plus protease inhibitors (0.1 mM PMSF, 1 mg/ml pepstatin A, 1 mg/ml leupeptin, 1 mg/ml chymostatin, 1 mM benzamidine, 1 mg/ml Nα-p-tosyl-L-arginine methyl ester, 0.1 mg/ml aprotinin, 0.1 mg/ml EDTA) was added, and the tube was placed in a boiling-water bath for 7 min. Lysate was passed through a 26-gauge needle, spun, and then used immediately or frozen at −80°C. After incubation at 95°C for 10 min, protein was separated with SDS-PAGE and transferred to nitrocellulose membrane (Sartorius, Göttingen, Germany) (Harlow and Lane, 1988). After blocking, membranes were incubated overnight with primary antibody. After rinsing, membranes were incubated with HRP-labeled secondary antibodies. Protein bands were visualized with tetramethylbenzidine membrane substrate (Kirkegaard & Perry Laboratories, Gaithersburg, MD).
PCR Selection and Balancing of unc-13 Deletion Mutants
C. elegans libraries mutagenized with psoralen were screened for deletions in the unc-13 gene. Worms were pooled, genomic DNA was extracted, and deletions were detected with nested PCR reactions as described previously (Barstead, 1999). To detect deletions in the unc-13M region, we used primers that gave a PCR product of 3.3 kb from wild-type genomic DNA. In md2413, 1041 base pairs (bp) are deleted, including 148 bp of the intron preceding exon 14 and 893 bp of exon 14 (UNC-13M is encoded by exon 14). In md2414, 1478 bp are deleted, including 1171 bp of the intron preceding exon 14 and 307 bp of exon 14. Strains containing the md2413 and md2414 mutations were outcrossed four times.
To detect deletions in the unc-13R region, we used primers that gave a PCR product of 3.0 kb from wild-type genomic DNA. In md2415, 2731 bp are deleted, including 311 bp from the 3′ end of exon 24 through 47 bp from the 5′ end of exon 30. The strain containing the lethal allele md2415 was outcrossed 10 times and was maintained with the balancer hT1(I;V) (McKim et al., 1988).
Transgenic Methods
DNA transformation methods for C. elegans were essentially those of Mello et al. (1991). The genomic cosmid ZK524 (100 μg/ml) or C44E1 (1–70 μg/ml) (obtained from Alan Coulson, Sanger Centre, Hinxton, United Kingdom) was injected along with a plasmid containing the synaptobrevin promoter (Nonet et al., 1998) driving the expression of green fluorescent protein (GFP; 50–100 μg/ml) as a visible marker for the array. Three similar healthy stable lines were isolated for ZK524; one line containing array mdEx42 was used for further analysis. Only two stable expressing arrays were generated after injection of C44E1; the healthier array, mdEx43, was used for behavioral characterization. Arrays were maintained in a wild-type background by selection of GFP-expressing nematodes. The arrays were crossed into unc-13 mutants with the use of standard genetic methods. Because of the occasional loss and variable expression of these transgenic arrays, the behavior and progeny yield of individual nematodes carrying arrays were quite variable. The healthiest individuals from a population carrying the array were selected for behavioral testing. Nematodes carrying transgenic arrays with very high copy numbers of either ZK524 or C44E1 were often highly uncoordinated and semisterile. This might be due to overexpression of the unc-13 gene or other sequences present on these arrays.
RESULTS
Structures of unc-13 Transcripts
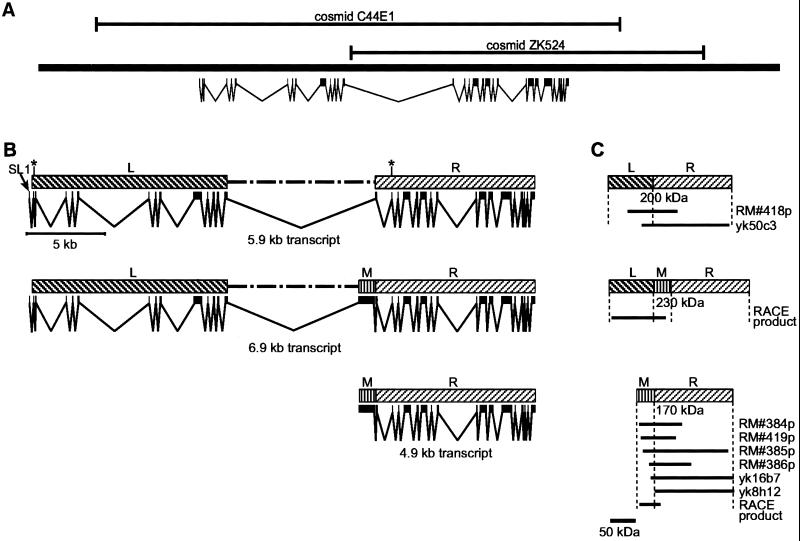
Analysis of the unc-13 genomic region and unc-13 cDNAs indicates three important genomic regions, which we designate the L, M, and R regions (left, middle, and right; Figure 1). We have identified two major transcript classes derived from the unc-13 locus. An ∼5.9-kb transcript including unc-13L and unc-13R has been described previously (Maruyama and Brenner, 1991) (GenBank accession number U50735). Subsequent cDNA analysis has shown that this transcript extends farther in the 5′ direction than reported previously (GenBank accession number M62830). In particular, the additional N-terminal sequence of the predicted protein includes a third (relatively diverged) C2 domain. This structure is quite similar to that reported for the Munc13-1 transcript and protein (Brose et al., 1995) (Figure 2).
Figure 1.
Genomic region around unc-13, with predicted unc-13 transcripts and protein products. (A) Genomic sequence indicating the locations of the major unc-13 transcript and the extents of cosmids C44E1 and ZK524. (B) Genomic sequence with introns and exons indicating the production of three predicted unc-13 transcripts. Region L (left) corresponds to the less conserved 5′ end of unc-13, and region R (right) corresponds to the more highly conserved 3′ end of unc-13. Region M (middle) corresponds to an exon that was not originally identified in the published sequence of unc-13. Asterisks mark positions of alternative splicing. (C) Proteins expressed from the predicted transcripts are shown. cDNA library screens and RACE products indicate the existence of unc-13 products. The cDNAs yk50c3 (GenBank accession number D67349), yk16b7 (GenBank accession numbers D26913 and D35032), and yk8h12 (GenBank accession number D37721) were isolated by Yuji Kohara from a dT-primed C. elegans library. The remaining cDNAs (RM#418p, RM#384p, RM#419p, RM#385p, and RM#386p) were isolated in our laboratory from a random primed C. elegans library. 5′ RACE and cRACE (Maruyama et al., 1995) were used to produce RACE products.
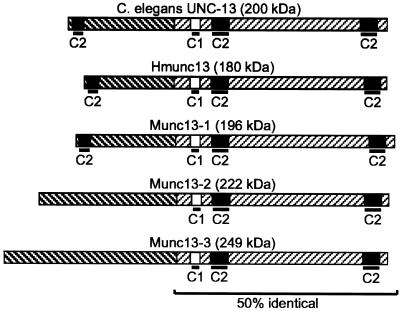
Figure 2.
UNC-13 has several mammalian homologues. The UNC-13L-R structure is shown. This form is similar to the published form of UNC-13 (Maruyama and Brenner, 1991). Additional N-terminal regions, including a third C2 domain, were identified in cDNA library screens (GenBank accession number M62830). C1 and C2 represent PKC homology domains. Munc13-1, Munc13-2, and Munc13-3 (GenBank accession numbers U24070, U24071, and U75261, respectively) are rat homologues of C. elegans UNC-13 (Brose et al., 1995). Hmunc13 (GenBank accession number AF020202) is a human homologue of UNC-13 (Song et al., 1999). The UNC-13L region is 30% identical to the N-terminal region of Munc13-1.
Another major type of unc-13 cDNA has been isolated in our laboratories and also by Yuji Kohara (National Institute of Genetics, Mishima, Japan); these cDNAs include a previously undescribed exon from the unc-13M region as well as sequences from unc-13R (Figure 1). Sequence analysis of the M region reveals an ORF of 1017 nucleotides bounded by splice donor and acceptor sites, suggesting a single exon encoding an ORF of up to 339 amino acids. The predicted UNC-13M amino acid sequence has no homology to known proteins.
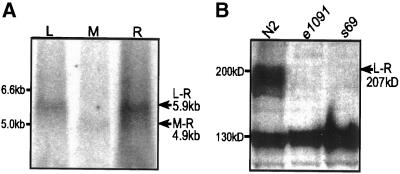
In a random primed library, cDNAs containing the unc-13R region were more abundant than those containing unc-13M (4/106 versus <1/106). All isolated cDNAs that included unc-13M also included a portion of unc-13R, and none of them included unc-13L (Figure 1C), suggesting that the M region is usually present as part of an M-R transcript. Northern analysis with the use of probes specific for either unc-13L or unc-13R reveals an ∼5.9-kb transcript (corresponding in size to unc-13L-R) (Figure 3A). Probes specific for unc-13M label a smaller ∼4.9-kb transcript (the predicted size of an unc-13M-R transcript). The M-containing cDNAs that extend the farthest in the 5′ direction include all but six bases of the proposed M exon. We were not able to find either SL1 or SL2 trans-spliced leader sequences on any M-containing cDNA, nor were we able to PCR amplify any SL1- or SL2-containing M product from cDNA libraries. We tentatively conclude that the M-R transcript is not trans spliced and that it initiates near the beginning of the M exon, presumably being driven from a promoter within the large 7.5-kb intron between the L and M regions. Assuming such a start site, the first in-frame start codon is located at nucleotide 241 of the M exon, so that an M-R protein is predicted to include 259 amino acids from M.
Figure 3.
Northern and Western transfers show products of the unc-13 gene. (A) Northern transfers indicate the presence of unc-13L-R and unc-13M-R transcripts. L, M, and R indicate lanes incubated with 32P-labeled probes that hybridize specifically to the unc-13L, unc-13M, and unc-13R regions, respectively. (B) Western transfers indicate the presence of several high-molecular-mass protein products from the unc-13 gene. Proteins from N2, unc-13(e1091), and unc-13(s69) bound to nitrocellulose membrane were labeled with anti-UNC-13L antibody. The band present at ∼130 kDa is nonspecific.
We have also obtained evidence for another minor transcript type that includes the M region (Figure 1). 5′-RACE and cRACE experiments indicate the presence of transcripts containing the L region connected to the M region (Figure 1C), suggesting the presence of a transcript class with an L-M-R structure (presumably derived by alternative splicing). However, because there is no evidence on Northern transfers for a 6.9-kb L-M-R transcript (Figure 3A), and none of the isolated M-containing cDNAs contained L, the L-M-R transcript must be considerably less abundant than the other transcript forms and may merely result from aberrant splicing.
Within each of the cDNA classes described above, we found evidence of heterogeneity that apparently results from alternative splicing. RACE experiments with the use of L-region primers identified two alternative sites in the first exon used for trans splicing of the SL1 leader sequence. The two sites are 157 nucleotides apart, and they both follow consensus splice acceptor sequences. The longer transcript encodes a protein with 15 additional amino acids at the N terminus; 6 of the 15 are aspartate, suggesting that the longer protein form might contain a functionally important acidic domain. In addition, the very small (nine nucleotides) second exon of the R region was not present in all R-containing cDNAs; this exon encodes a VLK tripeptide upstream of the C1 domain. The biological importance of this variability is not yet known.
Mutations and Mutant Phenotypes
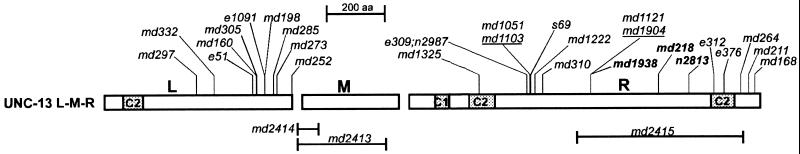
A large number of unc-13 alleles have been isolated previously; most were isolated as homozygotes that were uncoordinated and/or resistant to inhibitors of cholinesterase (Brenner, 1974; Miller et al., 1996). Two of these alleles, e51 and e1091, were shown to be caused by amber mutations in the unc-13L region (Maruyama and Brenner, 1991). We have sequenced 29 additional unc-13 mutant alleles (Figure 4), and some of their properties are presented in Table 1.
Figure 4.
Sequenced unc-13 alleles. The positions of mutations are indicated on a diagram of an UNC-13L-M-R protein. Alleles with names shown in bold encode in-frame (i.e., missense) mutations. The unc-13L region (L) includes exons 1–13; unc-13M (M) consists of exon 14; and unc-13R (R) includes exons 15–31.
Table 1.
unc-13 allele mutations, phenotypes, and staining patterns
| Allele | Molecular defect | Region | Staininga | Thrashingb | Pumpingb |
|---|---|---|---|---|---|
| e51 | Stop exon 11 | L | + | 2 ± 2 | 52 ± 10 |
| e1091 | Stop exon 12 | L | − | 1 ± 1 | 52 ± 8 |
| md160 | 1-bp insertion exon 11 | L | − | 31 ± 9 | 67 ± 20 |
| md198 | 2-bp deletion exon 12 | L | − | 78 ± 9 | 99 ± 11 |
| md252 | 1-bp deletion exon 13 | L | + | 8 ± 4 | 56 ± 15 |
| md273 | Stop exon 13 | L | − | 2 ± 1 | 49 ± 14 |
| md285 | 32-bp deletion exon 13 | L | − | 12 ± 4 | 56 ± 11 |
| md297 | 1-bp deletion exon 9 | L | − | 79 ± 8 | 67 ± 18 |
| md305 | 2-bp insertion exon 11 | L | − | 2 ± 2 | 34 ± 6 |
| md332 | 34-bp deletion exon 9 | L | − | 5 ± 3 | 58 ± 12 |
| md1072 | Deletion exons 1–13 | L | − | 2 ± 2 | 53 ± 14 |
| md2413 | 1.5-kb deletion exon 14 | M | + | 92 ± 5 | 96 ± 5 |
| md2414 | 1-kb deletion exon 14 | M | + | 88 ± 3 | 96 ± 6 |
| e309 | 1-bp change in splice site after exon 20 | R | − | ND | ND |
| e312 | Stop exon 28 | R | ND | 13 ± 7 | 95 ± 6 |
| e376 | Stop exon 28 | R | ND | 0.9 ± 0.4 | 60 ± 10 |
| md168 | 1-bp deletion exon 31 | R | − | 40 ± 27 | 89 ± 6 |
| md211 | Splice site change after exon 30 | R | + | 37 ± 12 | 65 ± 12 |
| md218 | 3-bp deletion exon 26 | R | + | 47 ± 11 | 64 ± 9 |
| md264 | Stop exon 30 | R | + | 3 ± 1 | 61 ± 8 |
| md310 | Stop exon 22 | R | − | 0.3 ± 0.6 | 26 ± 7 |
| md1051 | Tc1 exon 21 | R | ND | 74 ± 12 | 74 ± 17 |
| md1103 | Tc1 exon 21 | R | ND | 87 ± 7 | 68 ± 21 |
| md1121 | Tc1 exon 24 | R | + | 0.1 ± 0.2 | 31 ± 8 |
| md1222 | Tc1 exon 21 | R | − | 30 ± 21 | 76 ± 16 |
| md1325 | 4-bp insertion exon 19 | R | + | 0.1 ± 0.2 | 50 ± 16 |
| md1904 | Tc1 exon 24 | R | + | ND | ND |
| md1938 | Imprecise Tc1 excision from exon 24; Glu inserted | R | + | ND | ND |
| md2415 | 2.7-kb deletion exons 24–30 | R | + | ND | ND |
| n2813 | Transversion exon 26 | R | ND | 78 ± 8 | 90 ± 8 |
| s69 | 5-bp deletion exon 21 | R | − | 0 | 41 ± 13 |
Mutant alleles of unc-13 are listed alphabetically within region, followed by descriptions of their molecular defects and the region in which the defects occur. Staining indicates the level of immunoreactivity in the nervous system with an antibody to the L region of UNC-13. Staining is reduced or absent when the antigenic portion of the L region of UNC-13 is not present in its proper concentration and/or location. Pumping and thrashing behavioral assays were performed as described on 10–50 individuals of each genotype, and are presented as the percent of the wild-type value.
Level of staining in the nervous system: +, approximately normal; −, decreased or undetectable. ND indicates not done.
Values presented are means ± SD.
Ten mutations (including e51 and e1091) in the unc-13L region cause either frame shifts or stop codons (Figure 4). These mutations are predicted to disrupt the unc-13L-R and unc-13L-M-R transcripts. Although these animals can barely move, they all grow and reproduce relatively well (Table 2). The mutation md1072 is associated with a 26- to 38-kb deletion that eliminates all of the L coding region. The left end point of the deletion is near (or perhaps within) the next upstream predicted ORF, C15A11.7, so it is likely to delete all or most of the L-region promoter. The right end point removes ∼3 kb of the 8-kb intron between L and M. The phenotype of this mutant is similar to that of a number of other L alleles, including e51 and e1091; these mutations likely represent null alleles for the L region.
Table 2.
Growth and fertility of unc-13 mutants
| Strain | Total progeny per adult | Percent of eggs hatching | Percent L > A (in <10 d) |
|---|---|---|---|
| Wild type (+/+) | 294 ± 38 (n = 10) | 99% (n = 102) | 99.8% (n = 1307) |
| e1091/e1091 | 244 ± 18 (n = 5)a | 99% (n = 217) | 98.2% (n = 1218)b |
| s69/s69 | 134 ± 71 (n = 20)c | 99% (n = 135) | 78.2% (n = 650)b |
| e1091/+ | 302 ± 40 (n = 5) | ND | 99.6% (n = 1510) |
| s69/+ | 280 ± 21 (n = 12) | ND | 98.1% (n = 1999)b |
The progeny of individual hermaphrodites was monitored as described in MATERIALS AND METHODS. The total number of progeny per hermaphrodite was determined for a given number of hermaphrodites; numbers given are means ± SD. The fate of a subset of the eggs laid by young adults was monitored for their ability to hatch as stage 1 larvae (percent of eggs hatching). Additionally, the fate of a number (n) of first-stage larvae was monitored; the percentage that reached adulthood within 10 d is given.
Significantly different from wild type (Student's t test; p < 0.02).
Significantly different from wild type (χ2 test; p < 0.001).
Significantly different from wild type (Student's t test; p < 0.001).
Seventeen mutations have been localized to the unc-13R region; these mutations should affect all unc-13 transcripts. They include stop codons, frame shifts, and transposon insertions as well as some mutations that preserve reading frame. These mutations lead to a considerable range in the severity of the deficits in two behaviors, thrashing and pharyngeal pumping (Table 1). There also seemed to be developmental problems associated with severe R-region mutations; we explored this carefully for s69 homozygotes. The brood size of s69 animals is less than half that of either wild-type or e1091 animals; this appears to be a problem with gamete production and/or fertilization rather than a block during embryogenesis, because essentially all of the eggs laid by s69 homozygotes hatch (Table 2). In addition, almost one-quarter of the s69 larvae produced fail to reach adulthood (Table 2), although we were unable to observe any specific developmental stage at which such animals were arrested. The s69 homozygotes that became adults were considerably smaller than their wild-type counterparts (Figure 5). Comparable studies showed that the development and size of the severe L-region mutation e1091 were only slightly (but significantly) different from those of wild type and also that the developmental problems associated with s69 were recessive (Table 2, Figure 5).
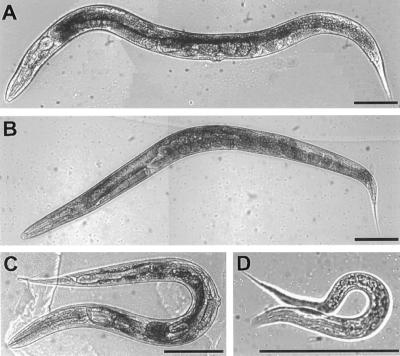
Figure 5.
Appearance of unc-13 mutants. (A) N2 (wild-type) young adult. (B) Young adult homozygous for the severe L-region mutation e1091. (C) Young adult homozygous for the severe R-region mutation s69. (D) Recently hatched larva homozygous for the lethal mutation md2415. These animals do not grow; they barely move, and they die within a few days. Bars, 100 μm.
Isolation of Additional Deletions Within unc-13
The phenotypes of mutants varied significantly for the different regions of unc-13. Numerous mutations in the L region, including the large deletion md1072, suggest that total disruption of the L region, and the presumed L-R and L-M-R transcripts, leads to paralysis and resistance to cholinesterase inhibitors but almost normal viability. We were struck by the fact that no mutations in M were isolated, either in the selections for resistance to cholinesterase inhibitors or in the screens for uncoordinated mutants. Therefore, it appeared likely that a mutation in M would confer neither uncoordinated locomotion nor resistance to cholinesterase inhibitors. Finally, none of the available R mutants were guaranteed to be completely null for that region, and the decreased viability of some of the R mutants suggested that complete disruption of R might lead to severe defects that prevented their isolation in our previous screens for viable mutants.
To address the issue of region-specific deficits, we used PCR selection to isolate two deletions in the unc-13M region, md2413 and md2414, and one deletion in the unc-13R region, md2415 (see MATERIALS AND METHODS) (Figure 4). Both of the deletions in unc-13M remove part of the intron preceding the unc-13M exon and part of the unc-13M exon itself, and they should affect both the unc-13L-M-R and unc-13M-R transcripts. As predicted, animals homozygous for M deletions are not resistant to cholinesterase inhibitors (our unpublished results). In addition, we were unable to find any obvious behavioral defects associated with these mutations (Table 1).
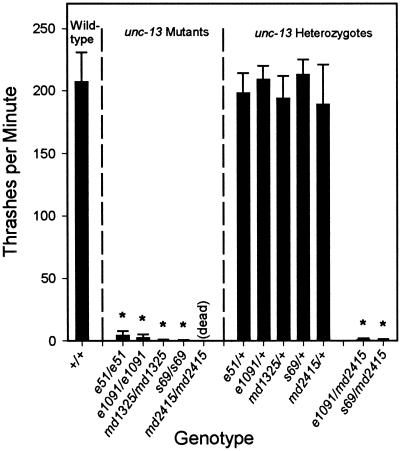
The unc-13 (md2415) allele has a 2.7-kb deletion in unc-13R and is predicted to disrupt all unc-13 transcripts (Figures 1 and 4). This mutation leads to lethality in the first larval stage. These larvae are almost completely paralyzed, maintaining a mostly coily posture with occasional slow movement of their heads. The md2415 mutation fails to complement the behavioral defects of strong L-region (e1091) as well as strong R-region (s69) mutations (Figure 6). We believe that md2415 is a null allele of the unc-13 locus (see below).
Figure 6.
unc-13 behavioral phenotypes are recessive, and L and R mutants fail to complement one another. Thrashing values are body bends per minute measured during a 2-min interval; values are means of 10–20 nematodes ± SDs. The wild-type data are representative of tests on a single day. Probabilities were calculated with the use of Student's t test for each strain versus a matched set of wild-type nematodes. *Significantly different from wild-type (p < 0.001). The homozygous unc-13 alleles (e51, e1091, md1325, and s69) are not significantly different from each other or from e1091/md2415 or s69/md2415.
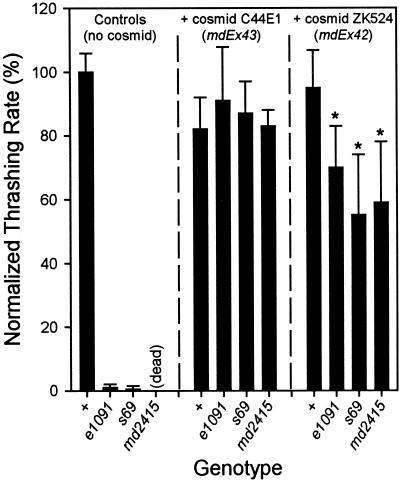
Cosmid Rescue of the unc-13 Mutant
Cosmid C44E1 contains a 44.6-kb genomic insert whose left end extends 8.7 kb upstream of the first unc-13 exon and whose right end extends 4.5 kb downstream from the unc-13 poly(A) addition site (Figure 1). Thus, this cosmid includes all of the unc-13 coding sequences and is likely to include all of its promoter(s). Transgenic arrays carrying this cosmid were generated and crossed into different unc-13 mutants. The presence of one such array, mdEx43, led to slight defects in the behavior of wild-type nematodes (Figure 7). When expressed in a mutant background, the array led to virtually wild-type behavior in the L mutant e1091 and two R mutants, s69 and the deletion mutant md2415.
Figure 7.
Behavioral phenotypes of transgenic strains. Transgenic arrays including the cosmid ZK524 (mdEx42) or the cosmid C44E1 (mdEx43) were generated by injection of wild-type nematodes. The arrays were crossed into the unc-13 mutants e1091, s69, and md2415. All strains were assayed for thrashing in liquid. Values given are normalized to N2 values measured on the same day. *Significantly different from N2 plus ZK524 (p < 0.01).
Cosmid ZK524 contains a 30.2-kb genomic insert. At its left end, the cosmid sequence includes most of the large intron between the unc-13L and unc-13M regions (Figure 1), and its right end lies ∼12 kb downstream of the unc-13 poly(A) addition site. This cosmid, therefore, contains none of the unc-13L coding sequence but all of the M and R sequences, as well as the putative regulatory information governing expression of the M-R (and perhaps other non-L–containing?) transcripts. Transgenic expression of ZK524 would be expected to provide all of the non-L–containing transcripts and thus to rescue the lethality of md2415 homozygotes. However, because none of the L-region coding sequence is present on ZK524, we expected that the phenotype of the rescued md2415 animals would be similar to the phenotype of L-region mutations (e.g., e1091). In addition, we predicted that the uncoordinated behavior caused by L-region mutations would not be rescued by the cosmid.
We generated transgenic arrays containing ZK524, and as predicted, we found that they were able to rescue the lethality of md2415 homozygotes. However, the behavior of the rescued animals was better than we had expected: although the transgenic animals were still uncoordinated, their locomotion was considerably better than that of the severe L-region mutants (Figure 7). These arrays also led to improved behavior of s69 (R) and e1091 (L) mutants (Figure 7). These data indicate that overexpression of non-L–containing transcripts can partially compensate for genetic loss of L-containing transcripts. However, note that this rescued behavior is not as coordinated as that seen in wild-type animals or in mutants carrying an array including the entire unc-13 gene.
unc-13 Protein Products
The UNC-13L-R protein is predicted to be 207 kDa, and Western transfers indicate the presence of several high-molecular-mass proteins (190–210 kDa) recognized by anti-UNC-13L antibodies (Figure 3B). The different sized proteins seen on Western transfers may result from either alternative splicing or posttranslational modification. The high-molecular-mass bands all appear to represent L-containing UNC-13, because all are absent in animals homozygous for either e1091 or s69 (Figure 3B). UNC-13L-M-R protein is predicted to be 245 kDa, and we have found no evidence for an immunoreactive protein of this size on Western transfers. Furthermore, we detected no difference in the observed protein bands in the M-deletion mutants (our unpublished results). We conclude that the L-M-R protein, if made at all, is present at too low a level to be detected.
UNC-13 Protein Localization
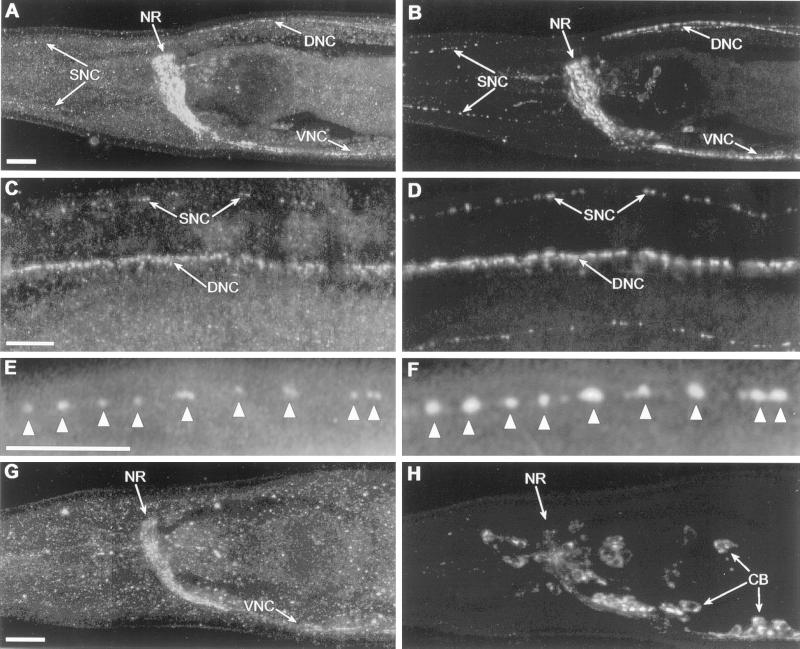
Antibodies produced against the UNC-13L region labeled most or all neurons (Figure 8) as well as nuclei in the gut and gonad (our unpublished results). The staining in the nervous system appeared to be specific for UNC-13, because it was decreased in many unc-13 mutants (Table 1). The nuclear staining in the gut and gonad was not altered in any of the unc-13 mutants and therefore is likely to be nonspecific. Antibody staining in the nervous system is punctate and appears to be synaptic, because in cholinergic neurons it colocalized with antibodies to the synaptic vesicle protein UNC-17/VAChT (Figure 8). However, analysis of unc-104 mutants suggested that the UNC-13 immunoreactivity was not associated with synaptic vesicles. The unc-104 gene encodes a kinesin-related protein required for the transport of synaptic vesicles from neuronal cell bodies along the axons to synapses (Hall and Hedgecock, 1991; Otsuka et al., 1991). In unc-104 mutants, most synaptic vesicles are not found at synapses but rather are found in large clusters in cell bodies (Hall and Hedgecock, 1991). In addition, a number of synaptic vesicle–associated proteins, such as synaptotagmin, synaptobrevin, and the vesicular neurotransmitter transporters, are mislocalized to neuronal cell bodies (Alfonso et al., 1993; Nonet et al., 1993, 1998; McIntire et al., 1997; Duerr et al., 1999). We find that UNC-13L staining remains at synapses in unc-104 mutants (Figure 8), suggesting that it is not transported to synapses by the UNC-104 protein, and therefore is unlikely to be on synaptic vesicles.
Figure 8.
Anti-UNC-13L antibody stains a form of UNC-13 protein that is synaptic but is unlikely to be on synaptic vesicles. C. elegans were double stained with anti-UNC-13L (left) and anti-UNC-17/VAChT (right). (A) Anti-UNC-13L in an adult wild-type head stains the major synaptic regions, including the nerve ring (NR), ventral nerve cord (VNC), dorsal nerve cord (DNC), and sublateral nerve cords (SNC). (B) Anti-UNC-17/VAChT stains a subset of the region stained by anti-UNC-13L. (C and D) A slightly higher magnification shows punctate staining with both antibodies in the dorsal nerve cord and sublateral nerve cords in the body. (E and F) A higher magnification shows the colocalization of anti-UNC-13L (E) and anti-UNC-17/VAChT (F) in synaptic regions (arrowheads) in one of the sublateral nerve cords in the head. (G and H) In an adult unc-104 mutant, staining of anti-UNC-13L is somewhat less intense but remains synaptic, whereas anti-UNC-17/VAChT is now localized in neuronal cell bodies (CB). Anterior is left; bars, 10 μ m.
In most of the mutants with defects in unc-13L or unc-13R, synaptic staining with anti-UNC-13L is partially or completely deficient (Table 1). Two unc-13 mutants with residual staining had less severe behavioral phenotypes (md211 and md218), but several mutants with normal immunoreactivity were strongly paralyzed (e51, md252, md1325, md2415, and md1121). Mutants with deletions in the unc-13M region did not show any alterations in UNC-13L immunostaining. The presence of antibody staining in strongly paralyzed mutants presumably results from the presence of stable abnormal protein products. If the UNC-13 protein functioned as part of a multimeric assembly, an abnormal protein might have a dominant negative effect, i.e., it might interfere with the assembly and/or function of other synaptic molecules. However, careful behavioral measurements of animals heterozygous for a number of such alleles (Figure 6) indicate that such mutations appear to be recessive. Although we have not performed such quantitative measurements on heterozygotes of all of the unc-13 alleles, it seems likely that the residual immunoreactivity in some of the mutants is not associated with any dominant negative effects.
DISCUSSION
Multiple unc-13 Transcripts
We have shown that several transcripts are expressed from the unc-13 gene in C. elegans and that the protein encoded by the most abundant transcript localizes primarily to synapses in the nervous system. The protein appears to be presynaptic and is unlikely to be located on synaptic vesicles. The mammalian homologues of UNC-13, Munc13-1 from rats and Hmunc13 from human, are most like C. elegans UNC-13L-R. Munc13-1 is expressed in brain and is localized to synapses (Brose et al., 1995). In addition, Munc13-1 and Hmunc13 are expressed in kidney (Song et al., 1999).
The unc-13 Null Phenotype Is Lethal
The md2415 deletion removes 2.7 kb from the unc-13R region and is predicted to affect all unc-13 transcripts. In genetic tests, the md2415 mutation is recessive and does not seem to be associated with a dominant negative phenotype. We believe that the lethality associated with md2415 homozygotes is due to the deletion, rather than a second-site mutation, because the lethal phenotype is rescued by the cosmids C44E1 and ZK524. We conclude, therefore, that md2415 is a null allele and that posthatching lethality represents the unc-13 null phenotype. This phenotype is consistent with a profound defect in neurotransmitter release (Richmond et al., 1999) and is similar to that of a synaptobrevin null mutation (Nonet et al., 1998).
unc-13 Mutants
Most mutations in the unc-13L region, including the md1072 deletion, result in a paralyzed but viable phenotype, the “traditional” unc-13 phenotype (Brenner, 1974; Rand and Russell, 1985; Hosono et al., 1989; Nguyen et al., 1995; Miller et al., 1996). The phenotypic similarities of several of the sequenced L-region mutants, including e1091, to md1072 indicate that this phenotype represents the total loss of L-encoded function(s). Therefore, we suggest that the e1091 allele, a premature stop, be used as the reference allele for the “L-null” type of unc-13 mutation.
Viable mutations specific to the unc-13R region result in slightly to severely paralyzed phenotypes, depending on the exact location and type of mutation. Frame shifts, stops, and Tc1 transposon insertions in different exons lead to different severity of defects; these differences may result from alternative splicing. The more severe viable unc-13R–region mutations also lead to developmental problems and a decreased progeny yield, phenotypes not found in the UNC-13L mutants. The R-region null mutant, md2415, is severely paralyzed and dies as a first-stage larva.
The difference in phenotypes between L and R mutants is presumably due to the presence of M-R transcripts. Therefore, we expected M mutants to have a noticeable effect on both behavior and viability, so that a combination of L deficits and M deficits would lead to the severe defects in coordination and the lethality seen in R mutants. However, mutants containing a deletion disrupting the unc-13M region appear similar to wild type. This observation suggests that alternative splicing or an additional promoter (perhaps within the R region) allows truncated yet partially functional forms of UNC-13 protein to be expressed in the M deletion mutants. Further behavioral analysis may identify a specific role for protein products from the unc-13L-M-R and unc-13M-R transcripts. However, at present, the function(s) of the M region and of M-containing transcripts remains unclear.
Functional Overlap of Different UNC-13 Proteins
Most of the known functionality of UNC-13 (the phorbol ester- and DAG-binding C1 domain, two calcium- and phospholipid-binding C2 domains, and the syntaxin-binding region) is associated with the R region, which we believe is present in all unc-13 transcripts. However, the different forms of unc-13 are not functionally equivalent. Recent electrophysiological data indicate that unc-13 mutants have significant defects in neurotransmitter release at the neuromuscular junction (Richmond et al., 1999). In a mutant in the unc-13L region (e1091), there are severe decreases in evoked release without similar decreases in spontaneous release. In a mutant with a defect in unc-13R (s69), both evoked and spontaneous neurotransmitter releases are almost completely absent. These data support the hypothesis that different forms of UNC-13 may play different roles in neurotransmitter release in C. elegans.
There is also evidence that the different forms can partially substitute for each other. The partial rescue of e1091 homozygotes by the ZK524 cosmid suggests that an excess of M-R transcripts can partially substitute for L-R transcripts. However, the lack of complete rescue argues against full equivalence of the two transcripts. In addition, the partial phenotypic rescue is seen only when the M-R transcript is overexpressed; it is likely that mutants with disruptions in the L region are already making wild-type levels of the M-R product. In contrast, transgenic expression of the C44E1 cosmid, which includes all of unc-13, provides virtually complete phenotypic rescue of L (e1091) or R (md2415, s69) mutants.
Localized versus Recruitable UNC-13 Molecules
It is possible that one of the major differences between the UNC-13 isoforms (in particular, between the UNC-13L-R protein and the UNC-13M-R protein) is in subcellular localization. Although our immunocytochemical results indicate that the UNC-13L-R protein is localized to synapses (as is the Munc13-1 protein; Betz et al., 1998), there is evidence that the M-R protein has a more diffuse distribution in neurons. Two recent studies (Lackner et al., 1999; Nurrish et al., 1999) used a transgene derived from cosmid ZK524 that resulted in overexpression of a GFP-tagged functional UNC-13 protein (presumably UNC-13M-R). The GFP-tagged protein was distributed throughout the axon cytoplasm and was reported to rescue the uncoordinated phenotype of an L-region mutant, unc-13(e51) (Nurrish et al., 1999).
In the goa-1 (Gαo) mutant, which is hypothesized to have increased levels of DAG, the GFP-tagged UNC-13 protein was distributed more synaptically (Nurrish et al., 1999). This change is reminiscent of the phorbol ester–induced changes in the subcellular localization of Munc13-1 and Hmunc13 (Betz et al., 1998; Song et al., 1999), in which phorbol esters stimulate association of the protein with membranes. Furthermore, when cultured neurons that overexpress Munc13-1 are treated with phorbol esters, neurotransmitter release is enhanced (Betz et al., 1998). This increased release might be due in part to the translocation of the protein to the cell membrane and the resultant increased local concentrations of Munc13. However, in C. elegans, phorbol esters do not cause relocation of GFP-tagged UNC-13 to synapses (Nurrish et al., 1999). Other induced changes, such as the phorbol ester–dependent binding of Doc2 to Munc13-1 (Orita et al., 1997), might be responsible for the observed increase in neurotransmitter release. The effects of phorbol esters on UNC-13 subcellular localization and/or function might be responsible for our laboratory's previous observation that treatment of wild-type C. elegans with phorbol ester leads to hypersensitivity to cholinesterase inhibitors, presumably resulting from excess release of acetylcholine (Miller et al., 1999).
A plausible model for UNC-13 function suggests that the L-R protein is synaptically localized and is required for normal neurotransmitter release, whereas the M-R protein is distributed diffusely along axons and normally plays a minimal role in synaptic transmission. This might explain the lack of observable phenotype associated with M-region deletions. However, under conditions of high neuronal activity (or perhaps metabolic stress), a signal transduction pathway is activated that operates through Gαq and leads to elevated DAG levels (Lackner et al., 1999; Miller et al., 1999). This in turn helps to activate the M-R protein and perhaps recruit some of it to synapses, where it can supplement the L-R protein, thus leading to increased synaptic release.
This interpretation suggests that the M-R protein is not necessary for stimulated transmitter release (hence, the lack of observable phenotype associated with M-region deletions) but that one of its roles is to form a diffuse reserve pool, which can augment some (but not all) of the synaptic functions of the synaptic L-R protein. Thus, in the L-region mutants, the endogenous diffuse M-R protein is enough to maintain minimal transmitter release and thus permit survival (the traditional unc-13 phenotype). Cosmid overexpression leads to high levels of M-R protein, which would presumably increase the synaptic level of UNC-13 protein and provide some additional neuromuscular function.
ACKNOWLEDGMENTS
We thank Kiely Grundahl for adapting restriction endonuclease fingerprinting methodology to C. elegans. We also thank Alan Coulson for providing cosmids, Ann Rose for providing the s69 allele, Erik Jorgensen for providing the n2813 and n2987 alleles, Ken Miller for providing the neuronal GFP marker plasmid and the md1904 and md1938 alleles, and the Caenorhabditis Genetics Center (funded by the National Institutes of Health National Center for Research Resources) for providing strains containing the e51 and e1091 alleles and the balancer hT1(I;V). This research was funded by grants from the National Institute of Neurological Disorders and Stroke to J.B.R. (NS33187) and I.N.M (NS31439), a National Research Service Award from the National Institute of Neurological Disorders and Stroke to R.E.K., and a grant from the Oklahoma Center for the Advancement of Science and Technology to J.S.D.
REFERENCES
- Ahmed S, Maruyama IN, Kozma R, Lee J, Brenner S, Lim L. The Caenorhabditis elegans unc-13 gene product is a phospholipid-dependent high-affinity phorbol ester receptor. Biochem J. 1992;287:995–999. doi: 10.1042/bj2870995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfonso A, Grundahl K, Duerr JS, Han H-P, Rand JB. The Caenorhabditis elegans unc-17 gene: a putative vesicular acetylcholine transporter. Science. 1993;261:617–619. doi: 10.1126/science.8342028. [DOI] [PubMed] [Google Scholar]
- Barstead RJ. Reverse genetics. In: Hope IA, editor. C. elegans: A Practical Approach. Oxford, UK: Oxford University Press; 1999. pp. 97–118. [Google Scholar]
- Betz A, Ashery U, Rickmann M, Augustin I, Neher E, Südhof TC, Rettig J, Brose N. Munc13-1 is a presynaptic phorbol ester receptor that enhances neurotransmitter release. Neuron. 1998;21:123–136. doi: 10.1016/s0896-6273(00)80520-6. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brose N, Hofmann K, Hata Y, Südhof TC. Mammalian homologues of Caenorhabditis elegans unc-13 gene define novel family of C2-domain proteins. J Biol Chem. 1995;270:25273–25280. doi: 10.1074/jbc.270.42.25273. [DOI] [PubMed] [Google Scholar]
- Burns DJ, Bell RM. Protein kinase C contains two phorbol ester binding domains. J Biol Chem. 1991;266:18330–18338. [PubMed] [Google Scholar]
- Coussens L, Parker PJ, Rhee L, Yang-Feng TL, Chen E, Waterfield MD, Franke U, Ullrich A. Multiple, distinct forms of bovine and human protein kinase C suggest diversity in cellular signaling pathways. Science. 1986;233:859–866. doi: 10.1126/science.3755548. [DOI] [PubMed] [Google Scholar]
- Davletov BA, Südhof TC. A single C2 domain from synaptotagmin I is sufficient for high affinity Ca2+/phospholipid binding. J Biol Chem. 1993;268:26386–26390. [PubMed] [Google Scholar]
- Duerr JS, Frisby DL, Gaskin J, Duke A, Asermely K, Huddleston D, Eiden LE, Rand JB. The cat-1 gene of C. elegans encodes a vesicular monoamine transporter required for specific monoamine-dependent behaviors. J Neurosci. 1999;19:72–84. doi: 10.1523/JNEUROSCI.19-01-00072.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustance RJ, Duerr JS, McManus J, Duke A, Molder G, Maruyama H, Maruyama IN, Barstead R, Rand JB. UNC-13 in the C. elegans nervous system. Mol Biol Cell. 1999;10:216a. doi: 10.1091/mbc.11.10.3441. (Abstract). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DH, Hedgecock EM. Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell. 1991;65:837–847. doi: 10.1016/0092-8674(91)90391-b. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Hata Y, Davletov B, Petrenko AG, Jahn R, Südhof TC. Interaction of synaptotagmin with the cytoplasmic domains of neurexins. Neuron. 1993;10:307–315. doi: 10.1016/0896-6273(93)90320-q. [DOI] [PubMed] [Google Scholar]
- Hayashi T, McMahon H, Yamasaki S, Binz T, Hata Y, Südhof TC. Synaptic vesicle membrane fusion complex: action of clostridial neurotxin on assembly. EMBO J. 1993;13:5051–5061. doi: 10.1002/j.1460-2075.1994.tb06834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosono R, Sassa T, Kuno S. Spontaneous mutations of trichlorfon resistance in the nematode Caenorhabditis elegans. Zool Sci. 1989;6:697–708. [Google Scholar]
- Johnson CD, Rand JB, Herman RK, Stern BD, Russell RL. The acetylcholinesterase genes of C. elegans: identification of a third gene (ace-3) and mosaic mapping of a synthetic lethal phenotype. Neuron. 1988;1:165–173. doi: 10.1016/0896-6273(88)90201-2. [DOI] [PubMed] [Google Scholar]
- Kaibuchi K, Fukumoto Y, Oku N, Takai Y, Arai KI, Muramatsu M. Molecular genetic analysis of the regulatory and catalytic domains of protein kinase C. J Biol Chem. 1989;264:13489–13496. [PubMed] [Google Scholar]
- Kazanietz MG, Lewin NE, Bruns JD, Blumberg PM. Characterization of the cysteine-rich region of the Caenorhabditis elegans protein Unc-13 as a high affinity phorbol ester receptor: analysis of ligand-binding interactions, lipid cofactor requirements, and inhibitor sensitivity. J Biol Chem. 1995;270:10777–10783. doi: 10.1074/jbc.270.18.10777. [DOI] [PubMed] [Google Scholar]
- Lackner MR, Nurrish SJ, Kaplan JM. Facilitation of synaptic transmission by EGL-30 Gqα and EGL-8 PLCβ: DAG binding to UNC-13 is required to stimulate acetylcholine release. Neuron. 1999;24:335–346. doi: 10.1016/s0896-6273(00)80848-x. [DOI] [PubMed] [Google Scholar]
- Li C, Davletov BA, Südhof TC. Distinct Ca2+ and Sr2+ binding properties of synaptotagmins: definition of candidate Ca2+ sensors for the fast and slow components of neurotransmitter release. J Biol Chem. 1995;270:24898–24902. doi: 10.1074/jbc.270.42.24898. [DOI] [PubMed] [Google Scholar]
- Liu Q, Sommer SS. Restriction endonuclease fingerprinting (REF): a sensitive method for screening mutations in long, contiguous segments of DNA. BioTechniques. 1995;18:470–477. [PubMed] [Google Scholar]
- Maruyama IN, Brenner S. A phorbol ester/diacylglycerol-binding protein encoded by the unc-13 gene of Caenorhabditis elegans. Proc Natl Acad Sci USA. 1991;88:5729–5733. doi: 10.1073/pnas.88.13.5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama IN, Rakow TL, Maruyama HI. cRACE: a simple method for identification of the 5′ end of mRNAs. Nucleic Acids Res. 1995;23:3796–3797. doi: 10.1093/nar/23.18.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntire SL, Reimer RJ, Schuske K, Edwards RH, Jorgensen EM. Identification and characterization of the vesicular GABA transporter. Nature. 1997;389:870–876. doi: 10.1038/39908. [DOI] [PubMed] [Google Scholar]
- McKim KS, Howell AM, Rose AM. The effects of translocations on recombination frequency in Caenorhabditis elegans. Genetics. 1988;120:987–1001. doi: 10.1093/genetics/120.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KG, Alfonso A, Nguyen M, Crowell JA, Johnson CD, Rand JB. A genetic selection for Caenorhabditis elegans synaptic transmission mutants. Proc Natl Acad Sci USA. 1996;93:12593–12598. doi: 10.1073/pnas.93.22.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KG, Emerson MD, Rand JB. Goα and diacylglycerol kinase negatively regulate the Gqα pathway in C. elegans. Neuron. 1999;24:323–333. doi: 10.1016/s0896-6273(00)80847-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman DG, Benian GM, Barstead RJ, Schriefer LA, Waterston RH. Identification and intracellular localization of the unc-22 gene product of Caenorhabditis elegans. Genes Dev. 1988;2:93–105. doi: 10.1101/gad.2.1.93. [DOI] [PubMed] [Google Scholar]
- Nguyen M, Alfonso A, Johnson CD, Rand JB. Caenorhabditis elegans mutants resistant to inhibitors of acetylcholinesterase. Genetics. 1995;140:527–535. doi: 10.1093/genetics/140.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet ML, Grundahl K, Meyer BJ, Rand JB. Synaptic function is impaired but not eliminated in C. elegans mutants lacking synaptotagmin. Cell. 1993;73:1291–1305. doi: 10.1016/0092-8674(93)90357-v. [DOI] [PubMed] [Google Scholar]
- Nonet ML, Saifee O, Zhao H, Rand JB, Wei L. Synaptic transmission deficits in Caenorhabditis elegans synaptobrevin mutants. J Neurosci. 1998;18:70–80. doi: 10.1523/JNEUROSCI.18-01-00070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurrish S, Ségalat L, Kaplan JM. Serotonin inhibition of synaptic transmission: Gαo decreases the abundance of UNC-13 at release sites. Neuron. 1999;24:231–242. doi: 10.1016/s0896-6273(00)80835-1. [DOI] [PubMed] [Google Scholar]
- O'Connor V, et al. Disruption of syntaxin-mediated protein interactions blocks neurotransmitter secretion. Proc Natl Acad Sci USA. 1997;94:12186–12191. doi: 10.1073/pnas.94.22.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orita S, Naito A, Sakaguchi G, Maeda M, Igarashi H, Sasaki T, Takai Y. Physical and functional interactions of Doc2 and Munc13 in Ca2+-dependent exocytotic machinery. J Biol Chem. 1997;272:16081–16084. doi: 10.1074/jbc.272.26.16081. [DOI] [PubMed] [Google Scholar]
- Otsuka AJ, Jeyaprakash A, Garcia-Añoveros J, Tang LZ, Fisk G, Hartshorne T, Franco R, Born T. The C. elegans unc-104 gene encodes a putative kinesin heavy chain-like protein. Neuron. 1991;6:113–122. doi: 10.1016/0896-6273(91)90126-k. [DOI] [PubMed] [Google Scholar]
- Perin MS, Fried VA, Mignery GA, Jahn R, Südhof TC. Phospholipid binding by a synaptic vesicle protein homologous to the regulatory region of protein kinase C. Nature. 1990;345:260–263. doi: 10.1038/345260a0. [DOI] [PubMed] [Google Scholar]
- Rand JB, Russell RL. Molecular basis of drug-resistance mutations in the nematode Caenorhabditis elegans. Psychopharmacol Bull. 1985;21:623–630. [PubMed] [Google Scholar]
- Richmond JE, Davis WS, Jorgensen EM. UNC-13 is required for synaptic vesicle fusion in C. elegans. Nat Neurosci. 1999;2:959–964. doi: 10.1038/14755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Sassa T, Harada S, Ogawa H, Rand JB, Maruyama IN, Hosono R. Regulation of the UNC-18-Caenorhabditis elegans syntaxin complex by UNC-13. J Neurosci. 1999;19:4772–4777. doi: 10.1523/JNEUROSCI.19-12-04772.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DE, Fisher PA. Identification, developmental regulation, and response to heat shock of two antigenically related forms of a major nuclear envelope protein in Drosophila embryos: application of an improved method for affinity purification of antibodies using polypeptides immobilized on nitrocellulose blots. J Cell Biol. 1984;99:20–28. doi: 10.1083/jcb.99.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söllner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993a;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- Söllner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993b;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- Song Y, Ailenberg M, Silverman M. Human munc13 is a diacylglycerol receptor that induces apoptosis and may contribute to renal cell injury in hyperglycemia. Mol Biol Cell. 1999;10:1609–1619. doi: 10.1091/mbc.10.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BD, Schrank B, Huynh C, Shownkeen R, Waterston RH. A genetic mapping system in Caenorhabditis elegans based on polymorphic sequence-tagged sites. Genetics. 1992;131:609–624. doi: 10.1093/genetics/131.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JZ, Davletov BA, Südhof TC, Anderson RGW. Synaptotagmin I is a high affinity receptor for clathrin AP-2: implications for membrane recycling. Cell. 1994;78:751–760. doi: 10.1016/s0092-8674(94)90442-1. [DOI] [PubMed] [Google Scholar]