Abstract
Two major virulence factors are associated with epidemic strains (O1 and O139 serogroups) of Vibrio cholerae: cholera toxin encoded by the ctxAB genes and toxin-coregulated pilus encoded by the tcpA gene. The ctx genes reside in the genome of a filamentous phage (CTXφ), and the tcpA gene resides in a vibrio pathogenicity island (VPI) which has also been proposed to be a filamentous phage designated VPIφ. In order to determine the prevalence of horizontal transfer of VPI and CTXφ among nonepidemic (non-O1 and non-O139 serogroups) V. cholerae, 300 strains of both clinical and environmental origin were screened for the presence of tcpA and ctxAB. In this paper, we present the comparative genetic analyses of 11 nonepidemic serogroup strains which carry the VPI cluster. Seven of the 11 VPI+ strains have also acquired the CTXφ. Multilocus sequence typing and restriction fragment length polymorphism analyses of the VPI and CTXφ prophage regions revealed that the non-O1 and non-O139 strains were genetically diverse and clustered in lineages distinct from that of the epidemic strains. The left end of the VPI in the non-O1 and non-O139 strains exhibited extensive DNA rearrangements. In addition, several CTXφ prophage types characterized by novel repressor (rstR) and ctxAB genes and VPIs with novel tcpA genes were found in these strains. These data suggest that the potentially pathogenic, nonepidemic, non-O1 and non-O139 strains identified in our study most likely evolved by sequential horizontal acquisition of the VPI and CTXφ independently rather than by exchange of O-antigen biosynthesis regions in an existing epidemic strain.
Vibrio cholerae is a serologically diverse, environmental, gram-negative bacterial species. Although V. cholerae comprises more than 200 O-antigen-based serogroups (35), only the O1 and O139 serogroup strains are known to cause epidemics of cholera, a severe diarrheal disease. Ever since cholera epidemics caused by V. cholerae O139 Bengal surfaced in 1992 in the Bay of Bengal region (1, 31), there has been renewed interest in the pathogenesis and evolutionary mechanisms of non-O1 vibrios. Several studies have reported (8, 9, 11, 15, 28-30, 33, 36) the presence of the two virulence genes found in epidemic strains, tcpA and ctxAB, in environmental and clinical strains of serogroups other than O1 and O139. However, the mechanisms of origin of these strains are not completely understood at the present time.
Cholera toxin encoded by ctxAB is responsible for the severe diarrheal symptoms elicited by the bacterium (20), and toxin-coregulated pilus (TCP) encoded by tcpA is responsible for efficient colonization of the human intestinal tract by the bacterium (42, 43). In addition, the O antigen is the bacterium's major protective antigen, and therefore, changes in the O antigen of a preexisting epidemic strain may result in a new pathogen capable of causing disease in populations immune to the original epidemic strain (1, 31). For example, the V. cholerae O139 Bengal strain emerged from an O1 epidemic strain by genetic exchange of O-antigen biosynthesis regions (3, 26, 40), and O139 strains cause disease in persons immune to O1 strains (4, 27).
The ctxAB genes are carried in the genome of a filamentous, single-stranded DNA phage designated CTXφ (44), and their dissemination to nonpathogenic strains may, therefore, occur via phage-mediated horizontal gene transfer. Since the discovery of CTXφ, intra- and interspecies transfers of ctxAB genes in V. cholerae and Vibrio mimicus, via CTXφ-mediated transduction, have been demonstrated under both laboratory and natural conditions (6, 7, 13, 14).
The TCP structural gene, tcpA, has been mapped (21) to a gene cluster designated the vibrio pathogenicity island (VPI), and the VPI has recently been proposed (22) to also be a filamentous phage, VPIφ. Unlike for the CTXφ, convincing data are lacking for the existence or the horizontal transfer of VPIφ. At the present time, none of the 29 genes in the VPI (other than tcpA) has been assigned, based on experimental data, any function in the proposed phage's life cycle or phage transduction. However, TCP has clearly been demonstrated (32, 43, 44) to be the receptor for CTXφ and to be the bacterium's colonization factor.
V. cholerae non-O1 and non-O139 strains have been occasionally isolated from humans with gastroenteritis, extraintestinal infections, and cholera-like illness (20, 34). Strains of serogroup O37 have been implicated in localized outbreaks of cholera in the past (2, 19). In addition, Mukhopadhyay et al. (28) reported the identification and genetic characterization of potentially pathogenic non-O1 and non-O139 V. cholerae strains from environmental samples in the Calcutta region. However, how frequently potentially epidemic non-O1 and non-O139 strains arise, and how widely they are distributed in nature, are not known at the present time.
Thus, the aims of our studies were to identify potentially pathogenic non-O1 and non-O139 strains in a large collection of V. cholerae isolates not examined previously and to determine the identified strains' evolutionary history. Here we report the identification and comparative genetic analyses of diverse non-O1 and non-O139 strains which possess VPI only or VPI and pre-CTXφ (a precursor of CTXφ which lacks ctxAB genes) (7) or CTXφ prophage and have distinct lineages compared to the epidemic strains, thus indicating independent horizontal transfer and acquisition of the virulence regions. While this manuscript was under preparation, Boyd and Waldor (8) also reported the evolutionary and functional analyses of several novel TcpA alleles carried by non-O1 and non-O139 strains.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The V. cholerae strains used in the preliminary screening for the presence of tcpA and ctxAB included 194 serogroup strains from the Shimada type culture collection (35), 36 clinical strains, and 70 clinical and environmental strains from the Smith collection (37). N16961 and 395 represented the O1 El Tor and O1 classical strains, respectively. Other strains characterized in detail are described in Table 1. Bacterial strains were grown in Luria-Bertani medium, and other methods pertaining to bacterial growth and storage conditions have been described previously (38).
TABLE 1.
Characterization of non-O1 and non-O139 V. cholerae strains with pathogenic potentialg
| Strain | History of isolation (country/year/source) | Serogroup | Presence of VPI
|
CTXφ type
|
ctx coreh | |||
|---|---|---|---|---|---|---|---|---|
| Clustera | tcpA type | ctxA | ctxB | rstR | ||||
| 395 | India/1966/diarrhea | O1 Cla | + | Cla | WT | Cla | Cla | + |
| N16961 | Bangladesh/1975/diarrhea | O1 ET | + | ET | WT | ET | ET | + |
| 153-94 | Unknown/1994/CDC | O8 | +▴/▾ | O8 | WT | ET | ET, O8b | + |
| No. 63 | Japan/1991/diarrhea from travel in Thailand | O26 | +▾ | O26 | WT | NT | Cla | + |
| 506-94 | Thailand/1994/diarrhea | O44 | +▾ | ET | WT | O26 | ET, O44NT | + |
| AQ1875 | Japan/1998/tortoise imported from Taiwan | O48 | +▾ | O48 | rstR-4**c, O44NT | − | ||
| 507-94 | Thailand/1994/diarrhea | O49 | +▴ | O49 | WT | ET | ET, O8b | + |
| 8-76 | India/1976/diarrhea | O77 | +▾ | O77 | − | |||
| 1421-77 | India/1977/diarrhea | O80 | +▾ | O77 | − | |||
| 571-88 | China/1988/diarrhea | O105 | +▴/▾ | O27d | NTe | NT | ET | + |
| 523-80 | United States/1980/diarrhea | O115 | +▴ | O115 | ET | − | ||
| 203-93 | India/1993/diarrhea | O141 | +▴/▾ | O53Gf | NTe | Cla | Cla | + |
| 366-96 | Japan/1996/prawn imported from Thailand | O191 | +▴ | O191 | + | Cla | Cla | + |
| AM2 | India/1995/diarrhea | O9 | − | |||||
| AM107 | India/1996/diarrhea | O144 | − | |||||
| NRT36-S | Japan/1990/diarrhea | O31 | − | |||||
| 117-94 | Korea/1994/river water | O35 | − | |||||
| 984-81 | India/1981/diarrhea | O89 | − | |||||
+, presence of the entire VPI cluster based on restriction mapping and hybridization.
rstRO8 is different from rstR-4** at the following three positions: rstR-4**H2RO8, rstR-4**G5DO8, and rstR-4**R7H08.
rstR-4** is SCE223 (28).
Differs from tcpA-env allele of reference 28 at one position, O27V9Denv.
A single amino acid substitution: wtS46NO37,O105,O141.
O53G is SCE5 from reference 29; rstRO44 is a novel allele, provisionally designated rstR6.
Abbreviations: Cla, classical; ET, EI Tor; WT, wild type; NT, novel type; ▾, insertion; ▴, deletion; CDC, Centers for Disease Control and Prevention.
Presence of CTXφ genes other than ctxAB.
PCR.
PCR amplification of various segments of the VPI and CTX regions to be used as probes in dot blot and restriction fragment length polymorphism (RFLP) analyses and PCR amplification of the tcpA, rstR, pgm, recA, and gyrB genes for sequencing were performed by using Taq DNA polymerase (Promega Corp. Madison, Wis). Primers used in PCRs were reported previously (25) and were designed based on the V. cholerae genome sequence (16). Five additional pairs of primers used to prepare PCR probes to characterize the left end of the VPI and the primers used to amplify fragments of pgm (phosphoglucomutase), gyrB, and recA for multilocus sequence typing (MLST) are listed in Table 2. For most PCRs, the amplification conditions were as follows: 92°C for 5 min, followed by 35 amplification cycles, each consisting of sequential incubation at 92°C (30 s), 55°C (1 min), and 72°C (1 to 2 min), and a final extension at 72°C (5 min). In some PCRs, optimal annealing temperatures were determined by gradient PCR analysis with a Robocycler Gradient 96 (Stratagene, Inc.) thermocycler.
TABLE 2.
List of primers
| Primer | Gene, probe | Sequence (5′-3′) | Reference |
|---|---|---|---|
| Sulak 80 | VPI-1, for-a* | GCA ACA GGA TGA GTA ATC GAG | This study |
| Sulak 81 | VPI-1, rev-a* | CGA TTT CAC AAA GTA GCT CAC | This study |
| Sulak 84 | VPI-3, for-c* | TGA GCC TGA AAT AAT CAC AGG | This study |
| Sulak 85 | VPI-3, rev-c* | GAT GAT GAA GTG TAT ATC TAC | This study |
| Sulak 86 | VPI-4, for-c** | TGG GGA AGA CTT TTG CTC AAG | This study |
| Sulak 87 | VPI-4, rev-c** | ATA TCT TGA ATG GGC TTT ACC | This study |
| Sulak 121 | VPI-5, for-b* | ACG GTT AGC TCT TCC ATC GAC | This study |
| Sulak 122 | VPI-5, rev-b* | CAG AGC GTC TCA TCA AGA TTC AC | This study |
| Sulak 123 | VPI-6, for-b** | GTG AAT CTT GAT GAG ACG CTC TG | This study |
| Sulak 124 | VPI-6, rev-b** | GGT GAG CCA GGC TTA TTT GGG | This study |
| Sulak 76 | pgm, for | AAA GAT ACT CAY GCS CTG TC | This study |
| Sulak 77 | pgm, rev | AAC CAG CGT TTT ACC GAC GGC AAC A | This study |
| Sulak 78 | gyrB, for | GAA GGB GGT ATT CAA GC | This study |
| Sulak 79 | gyrB, rev | GAG TCA CCC TCC ACW ATG TA | This study |
| rec, 1 | GAA ACC ATT TCG ACC GGT TC | 39 | |
| rec, 2 | CCG TTA TAG CTG TAC CAA GCG CCC | 39 | |
| M 407 | rstR1 | GAC GTA GCG TGC GGA GTC GCG TTG | 25 |
| M 408 | rstR2 | TGA AGC ATA AGG AAC CGA CCA AGC | 25 |
| M 573 | rstA1 | ACT CGA TAC AAA CGC TTC TC | 25 |
| M 574 | rstA2 | AGA ATC TGG AGG TTG AGT G | 25 |
MLST.
A 900-bp fragment of pgm, a 650-bp fragment of gyrB, a 785-bp fragment of recA, a 1.8-kb fragment encompassing the tcpA gene, and a 1.5-kb fragment of the ctxAB genes were PCR amplified, and the amplified fragments were sequenced (in both directions) by using the BigDye terminator cycle sequencing kit (Applied Biosystems, Inc., Foster City, Calif.) and either an ABI 377 Prism automated sequencer or an ABI 3700 DNA analyzer (Applied Biosystems, Inc.). Sequence alignments and dendrograms were generated as described previously (24) by using the Clustal-X (18) and PAUP (D. Swofford, Sinauer Associates, Sunderland, Mass.) programs, respectively. The rstR gene (750 bp) was PCR amplified with primers M407 (rstR1) and M408 (rstR2) (25), subcloned in the pCR2.1 vector (Invitrogen Life Technologies, Carlsbad, Calif.), and sequenced. When two different-sized fragments were obtained in a PCR of a single strain, the fragments were gel purified, cloned separately, and sequenced.
DNA dot blot and Southern analyses.
Screening for the presence of tcpA and ctxAB was performed by DNA dot blot analysis as described previously (38). The presence of the aldA, toxT, int, rstR, and rstA genes was determined by Southern hybridization of the genomic DNAs with PCR fragments of these genes amplified from an El Tor strain, N16961. The rstA gene was PCR amplified with primers M573 (rstA1) and M574 (rstA2) (Table 2), and all of the other primers were reported earlier (25).
RFLP analysis.
Two ∼50-kb genomic segments encompassing the VPI and CTXφ prophage regions were analyzed by RFLP. Genomic DNAs were restriction digested with XmnI (for analysis of the VPI region) and with EcoRI or SphI (for analysis of the ctx region), electrophoresed in an agarose gel, transferred onto a Zetaprobe membrane (Bio-Rad Laboratories, Hercules, Calif), and simultaneously hybridized with multiple nonradioactive probes prepared with an enhanced chemiluminescence kit (Amersham Biosciences, Piscataway, N.J.). Hybridization conditions, the various probes, the lengths and order of the restriction fragments probed, and the exact endpoints of the two regions analyzed by RFLP were reported previously (25). The identity of the individual bands was confirmed by hybridizing the same blot with individual probes.
Nucleotide sequence accession number.
The nucleotide sequences of the rstR (accession numbers AF452585 to AF452586),tcpA (accession numbers AF452570 to AF452580), ctxA (accession numbers AF452584, AF46340, and AF463401), ctxB (accession numbers AF463402 and AF452581 to AF452583), and gyrB alleles (accession numbers AF501888 to AF501913) of the non-O1 and non-O139 serogroup strains have been deposited in the GenBank database.
RESULTS
Identification of non-O1 and non-O139 strains containing VPI and CTXφ.
To determine the prevalence of tcpA and ctxAB in non-O1 and non-O139 V. cholerae, 300 strains were screened by DNA dot blot analysis for the presence of the genes. Fifteen non-O1 and non-O139 strains carried the tcpA gene, 9 of the 15 strains carried the ctxAB genes, and none of the 300 strains carried ctxAB alone. The 15 tcpA+ strains were further analyzed, by Southern hybridization, for the presence of other genes in the VPI and CTX regions, and the results are summarized in Table 3. All of the tcpA+ strains also carried three other genes, aldA, toxT, and int, from the left, middle, and right ends of the VPI cluster, respectively, which indicated that the entire VPI region might be present in these strains. Thirteen of the 15 tcpA+ strains carried rstR and rstA of the CTXφ genome, and two other strains (serogroups O77 and O80) did not carry any of the CTXφ genes. Nine of the 13 rstA+rstR+ strains also carried ctxAB, one strain (serogroup O115) carried only rstA and rstR, and three strains (serogroups O48, O53, and O65) carried rstR, rstA, and the genes of the core region, except ctxAB. Li et al. (25) previously reported the genetic characterization of four of the 15 strains (serogroups O27, O37, O53, and O65) carrying the entire VPI and a CTXφ or a pre-CTXφ (7), i.e., CTXφ lacking the ctxAB genes and an epidemic genetic backbone. In the present study, we examined the 11 remaining strains which have genetic backbones different from those of the epidemic strains, and we found that seven of the strains carried the CTXφ, one carried the pre-CTXφ, and one only had the repeat sequence element.
TABLE 3.
Classification of screened V. cholerae strainsa
| Genetic background | No. (serogroup[s]) of strains containing:
|
||||
|---|---|---|---|---|---|
| CTXφ only | VPI only | VPI and RS1b | VPI and pre-CTXφc | VPI and CTXφd | |
| Epidemic | 0 | 0 | 0 | 2 (O53, O65) | 2 (O27, O37) |
| Nonepidemic | 0 | 2 (O77, O80) | 1 (O115) | 1 (O48) | 7 (—e) |
A total of 300 strains were screened, and 285 strains lacked VPI and CTXφ.
rstR+ rstA+.
rstR+ rstA+ ctx core+.
rstA+ rstR+ ctxAB+ ctx core+.
O8, O26, O44, O49, O105, O141, and O191.
Genetic relatedness of the non-O1 and non-O139 strains to the epidemic strains.
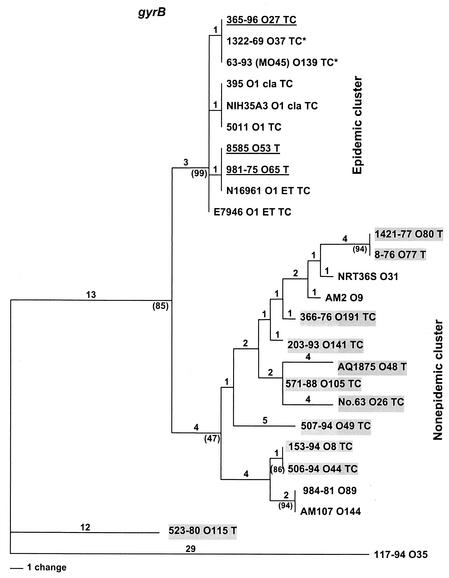
To determine the genetic relatedness of the 11 tcpA+ nonepidemic strains to the tcpA+ epidemic strains, MLST was performed. A 900-bp fragment of pgm (phosphoglucomutase), a 650-bp fragment of gyrB, and a 785-bp fragment of recA were sequenced. The nucleotide sequences were analyzed by using the maximum-parsimony and maximum-likelihood methods reported previously (17) to be well suited for determining phylogenetic relationships among various bacterial strains and species, and both methods gave consistent results for each of the three genes. The epidemic and nonepidemic strains were topologically separated into two major branches of the phylogenetic tree. The phylogram (Fig. 1) generated for one of the genes (gyrB) revealed a tight clustering of the strains of epidemic serogroups (O1 and O139) and strains of four other serogroups (O27, O37, O53, and O65), thus indicating the clonal nature of these strains. The branch node of the epidemic cluster was supported by a value of 99% in a 1,000-replicate bootstrap analysis. Similar clustering of epidemic strains was seen with the pgm and recA gene trees (data not shown), where the number of nucleotide changes per site among the strains was greater than with gyrB. The branch carrying 10 tcpA+ non-O1 and non-O139 strains and several other non-O1 and non-O139 strains had a low bootstrap value (47%) (Fig. 1), which suggests that the strains in this group are nonclonal in nature and, accordingly, mostly formed independent secondary branches. Although a few strains within this group formed clusters with high bootstrap values (serogroups O77 to O80, O8 to O44, and O89 to O144) (Fig. 1), this clustering was not consistent with the pgm- and recA-based trees. The remaining one tcpA+ non-O1 and non-O139 strain (serogroup O115) was a V. mimicus strain and was an outlier in the tree. Apparently, this strain has acquired the VPI region via interspecies horizontal transfer. Based on these data, we conclude that the 11 tcpA+ non-O1 and non-O139 strains have distinct lineages and did not originate directly from epidemic strains by O-antigen switching.
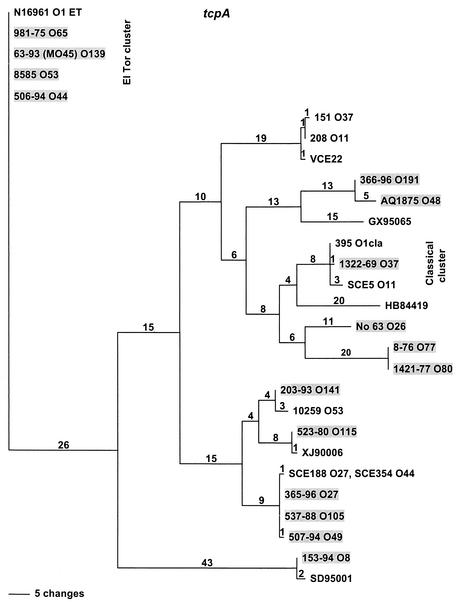
FIG. 1.
Neighbor-joining tree constructed by the maximum-parsimony method with the nucleotide sequences of gyrB gene fragments of V. cholerae strains. Labels at the branch tips represent strain designation, serogroup, and the virulence gene (T for tcpA and C for ctxAB), if present. The horizontal length represents the genetic distance, and the vertical lengths are not meaningful. The numbers above the branch lines represent the number of changes, and the numbers in parentheses below the branch lines at the branch node are the bootstrap values. Strains for which O-antigen switching is the proposed mechanism are underlined, and strains in which O-antigen switching is supported by sequence evidence are indicated by asterisks. Strains which appear to have acquired VPI and CTXφ or pre-CTXφ by independent horizontal gene transfer are highlighted.
Genetic organization of VPIs.
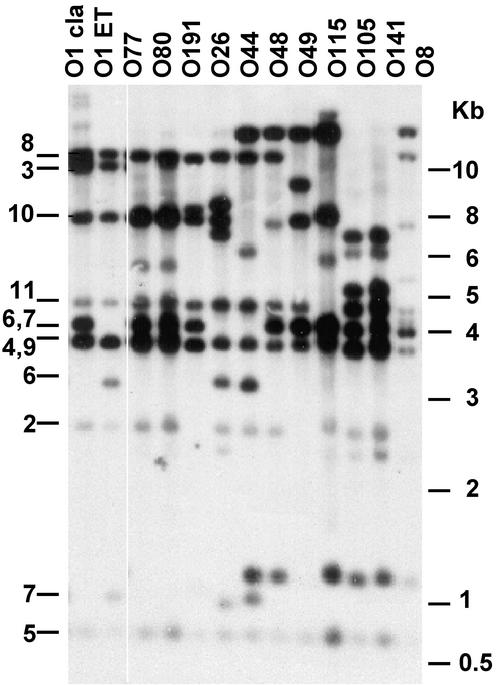
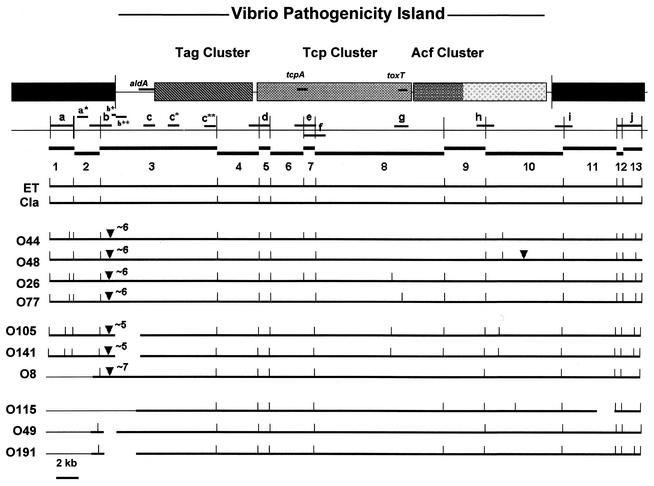
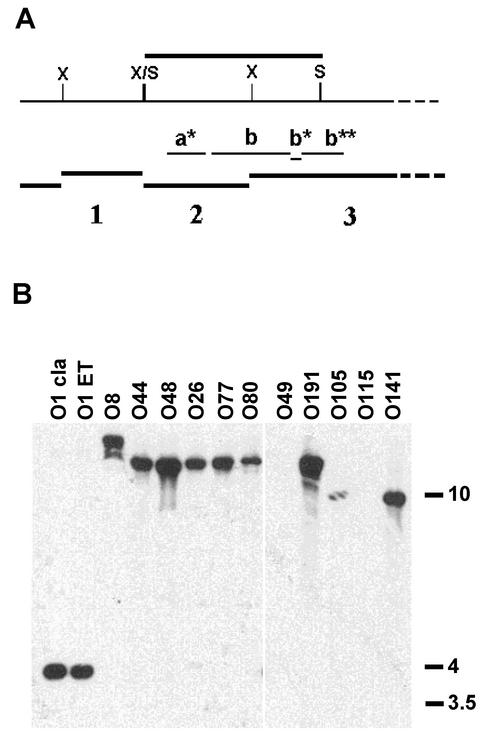
The genetic organization of the VPI region in epidemic and nonepidemic strains was compared by RFLP analysis of genomic DNAs digested with XmnI and hybridized with various probes derived from the VPI region (Fig. 2). The VPIs of the O1 classical and O1 El Tor strains had distinct RFLP patterns (Fig. 2, lanes O1 cla and O1 ET). Ten of the 11 non-O1 and non-O139 strains had distinct RFLP patterns that were different from those of the epidemic strains, with additional XmnI sites and/or genetic rearrangements (insertions and deletions) at the left end of the VPI (Fig. 2, lanes O77 through O8), and O77 and O80 serogroups had identical clusters. The deduced restriction maps of the VPI regions in the 10 strains are shown in Fig. 3. The insertions and deletions at the VPI's left end in the non-O1 and non-O139 strains were confirmed by hybridization of SalI-digested genomic DNAs (Fig. 4). A SalI-generated fragment of the expected size of 4,299 bp in length was seen in the wild-type VPI (Fig. 4b, lanes O1 cla and O1 ET). However, in the non-O1 and non-O139 strains, a fragment of >10 to 12 kb was observed (Fig. 4b, lanes O8 to O80, O191, O105, and O141), thus indicating a DNA insertion (∼5 to 7 kb) in the VPI region. Also, the O49 and O115 strains did not hybridize with this probe, which indicates a deletion in this region. Thus, these non-O1 and non-O139 strains could be divided into three groups with respect to their VPIs' left ends. First, strains of serogroups O26, O44, O48, O77, and O80 contained an insertion of an approximately 6-kb fragment between the VPI left att site and the SalI site within open reading frame (VC0817) at the left end of the VPI. Second, strains of serogroups O8, O105, and O141 carried a deletion and an insertion of an approximately 7-kb fragment (O8) or an approximately 5-kb fragment (O105 and O141) at the left end of the VPI. Third, strains of serogroups O49, O115, and O191 carried deletions of various lengths at the left end of the VPI.
FIG. 2.
RFLP analysis of the VPI region in various V. cholerae strains. Southern analysis of the XmnI-digested genomic DNAs of the indicated strains was done by simultaneous hybridization with multiple probes. The probes used were b, d, e, g, h, and i, as shown in Fig. 3. The corresponding restriction fragments (shown in Fig. 3) of the El Tor (ET) VPI detected by these probes are indicated by the numbers on the left side of the figure. The size markers (in kilobases) are indicated on the right side of the figure. cla, classical.
FIG. 3.
Genetic organization of the VPI cluster. A schematic diagram of the VPI region in an O1 El Tor strain, based on the V. cholerae (16) genome sequence, is shown. The top bar represents the various subclusters within the VPI region, the filled bar indicates the junction segments of the VPI region, and the vertical lines mark the ends of the VPI cluster. The various probes are indicated in the second row (a through j). The predicted XmnI fragments of the VPI region of the El Tor chromosome (numbered 1 to 13) are indicated in the third row. The observed XmnI fragments of the VPI region deduced from hybridization analyses and the resulting genetic maps of the El Tor (ET), classical (Cla), and various non-O1 and non-O139 strains of V. cholerae are shown below. DNA insertions and their sizes in kilobases are indicated by triangles and the numbers above the triangles, broken lines represent deletions, and thin lines indicate the adjoining sequences on the chromosome. The El Tor type included the O53, O65, and O139 serogroups, the classical type included the O37 serogroup, and the O77 type included the O80 serogroup.
FIG. 4.
RFLP analysis of the left end of the VPI region in V. cholerae strains. The schematic diagram of the left end of the VPI region and the expected SalI- and XmnI-generated fragments 1 to 3 as shown in Fig. 3 are indicated at the top (A). SalI-digested genomic DNAs were hybridized with the b* probe (B). Strains of serogroups O49 and O115 have a deletion in the region of the probe. ET, El Tor; cla, classical.
To elucidate further the origin of the VPIs found in the above-described non-O1 and non-O139 strains, the tcpA genes located within the VPIs were sequenced. As observed earlier in other studies (8, 9, 15, 28-30), extensive divergence in the tcpA gene sequences of these strains was observed. The results are summarized in Table 1, and a phylogenetic tree based on the amino acid sequences (12 tcpA sequences from this study and 12 previously published tcpA sequences) is shown in Fig. 5. Four strains (serogroups O44, O53, O65, and O139) carried an El Tor allele, and strain 203-93 (serogroup O141) carried an allele very similar to the one found previously in strain 10259 (serogroup O53), except at three positions (O53A190GO141, O53P191TO141, and O53G193SO141) (15). The tcpA alleles in serogroups O27 and O105 were very similar to the recently described tcpA-env allele (28), except at one position (O27,O105D9VtcpA-env), and the O49 allele differed from the O27 and O105 alleles at one position (O27,O105N56HO49). Seven new alleles in serogroups O8, O26, O48, O49, O77, O141, and O191 were identified in this study. Taken together, these data suggest that the VPIs of a majority of the non-O1 and non-O139 strains differ from those of the epidemic strains (classical and El Tor) as do their tcpA genes.
FIG. 5.
Neighbor-joining tree constructed by the maximum-parsimony method with the amino acid sequences of the tcpA gene fragments of V. cholerae strains. The labels at the branch tips represent the strain designation and serogroup, if known. A total of 28 tcpA sequences, which included alleles described in this study (highlighted) and in previously published studies, were used for the analysis. The strain designations of published sequences are as follows (serogroups, if known, and GenBank accession numbers are in parentheses): N16961-ET (O1, AF325734), 395 classical (O1, AF325733), 151 (O37, AF030546), 208 (O11, AF030309), VCE22 (AF414371), SCE5 (O11, AB012946), 10259 (O53, AF139626), SCE188, SCE354 (O27, O44, AF208385), GX95065 (AY056618), HB84419 (AY052830), XJ90006 (AY056619), SD95001, (AY052831), and 365-96 (O27, AF390571). The strain designations of novel alleles reported in this paper are as follows (serogroups, if known, and GenBank accession numbers are in parentheses): 366-96 (O191, AF452580), AQ1875 (O48, AF452573), 1322-69 (O37), No. 63 (O26, AF452571), 8-76 (O77, AF452575), 203-93 (O141, AF452579), 523-80 (O115, AF452578), 507-94 (O49, AF452574), 153-94 (O8, AF452570), 571-88 (AF452577), 1421-77 (O80, AF452576), and 506-94 (O44, AF452572). ET, El Tor; cla, classical.
Genetic organization of the ctx region.
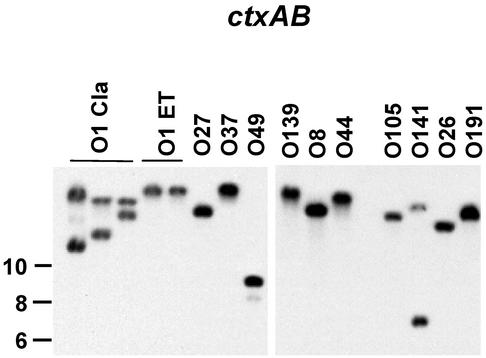
The organization of the ctx region in the seven non-O1 and non-O139 strains that were ctxAB+ (serogroups O8, O26, O44, O49, O105, O141, and O191) was determined by RFLP analysis. EcoRI does not cut within the CTXφ genome; thus, digestion of the genomic DNAs with EcoRI and hybridization with ctxAB probes revealed a single copy of the CTXφ genome in strains of serogroups O8, O26, O44, O49, O105, and O191 and two copies in O141 (Fig. 6). Five other previously described (25) probes, ctxrgn 1 to 5, were used to scan the ctx region, and the genetic organization of that region in the non-O1 and non-O139 strains was found to be quite heterogeneous and distinct from that of the epidemic strains. Also, the non-O1 and non-O139 strains lacked pTLC, an element carried by all of the epidemic strains, and they possessed an El Tor-like RTX cassette (data not shown).
FIG. 6.
Analysis of the CTXφ prophage in V. cholerae strains. Southern blot analysis of EcoRI-digested genomic DNAs probed with the ctxAB gene probes is shown; each band represents one copy of the CTXφ prophage genome. ET, El Tor; Cla, classical.
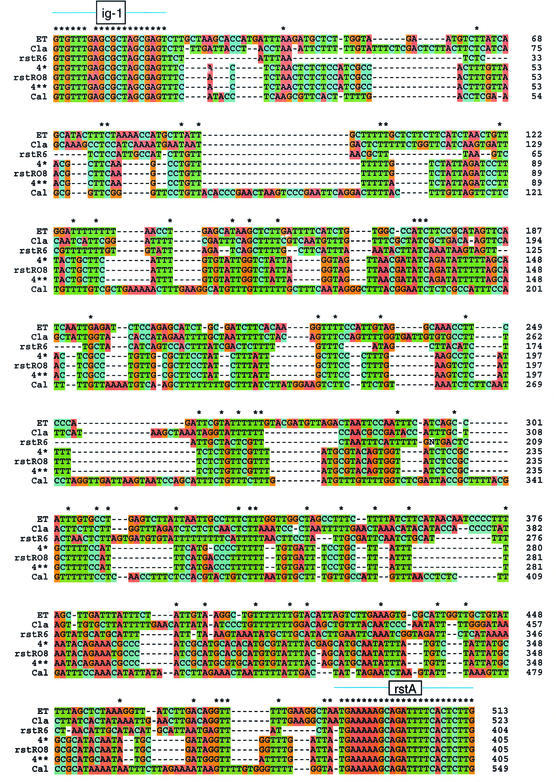
To understand further the origin of the CTXφs present in the non-O1 and non-O139 strains, the nucleotide sequences of the rstR and ctxAB genes were determined (the allelic types based on their sequences are listed in Table 1). The rstR sequences were diverse among the strains examined in this study. Four strains (serogroups O8, O44, O48, and O49) had two alleles (double lysogens), one of which was an El Tor type in the O8, O44, and O49 serogroups, three strains carried the rstRcla allele (serogroups O26, O141, and O191), two strains carried only an rstRET allele (serogroups O105 and O115) and two new alleles were found, rstRO8 and rstRO44 (serogroups O8, O44, and O49). rstRO8 is a variant of the rstR4** type (28), and rstRO44 is a novel type, which we provisionally designate rstR6, and is similar to a sequence already available in the GenBank database (accession number AF302794). A Clustal-X alignment of the nucleotide sequences of the rstR type alleles and two variants is shown in Fig. 7. Twenty-four rstR sequences available in the GenBank database were aligned. They fall into 5 rstR types: El Tor type (5 sequences), classical type (4 sequences), Calcutta type (4 sequences), rstR4** type (9 sequences), and rstR6 (2 sequences). The sequences upstream and downstream of rstR (ig-1 and rstA) are highly conserved in all of the alleles while the rstR region showed extensive variations, as do their respective RstR proteins.
FIG.7.
Clustal-X alignment of rstR sequences. A total of 24 rstR sequences available in the GenBank database, which included alleles described in this study, were used for the analysis. Only the rstR type sequences that encode different RstR proteins (5 type sequences, i.e., El Tor [ET], classical [Cla], Calcutta [Cal], rstR4**, and rstR6) and two variants are shown in the figure. One sequence (accession number AF133308, designated rstR5 from strain SCE264) of the Calcutta type shows extensive variations, its RstR protein exhibited the least similarity to all of the other alleles (28), and hence, it was not included in the alignment. The strain designations of the published sequences are as follows (the serogroup [if known] and the GenBank accession numbers are in parentheses: El Tor (ET) type, E7946 (O1 ET, U83795), E7946 (O1 ET, U83796), SC8511 (O1 ET, AF511000), N16961 (O1 ET, AE004224), and JS9803 (O139, AY101180); classical (Cla) type, O39 (O1 Cla, AF262318), SC9773 (O1 ET, AF510999), 569B (O1 Cla, AF05890), and 86015 (O1 ET, AF220606); Calcutta (Cal) type, AS207 (O139, AF110029), SCE188 (O44, AF133310), FJ98352 (O139, AF511001), and SCE264 (O42, AF133308); rstR4** type, SCE223 (O27, AF133307), 365-96 (O27, AF390570), JX94484 (O139, AF511001), VCE22 (O36, AY145124), VCE228 (O27, AY145125), VCE232 (O4, AY145126), VCE233 (O27, AY145127), 153-94 (O8, AF452585), and SCE263 (O10, AF133309); Novel type, 506-94 (O44, AF452586) and 9803 (O139, AF302794). Nucleotides CT and AG are in green and red, respectively. ig-1, intergenic region 1; rstA, the 5′ end of the rstA gene. Nucleotides that are identical in all sequences are indicated by asterisks.
The CtxA protein was conserved in all seven strains, except at one position in two strains (wtS46NO105,O141). The CtxB protein was of the El Tor type in serogroups O8 and O49 and was of the classical type in serogroups O141 and O191. The classical and El Tor CtxB alleles differed from each other at two positions (ETY39Hcla and ETI68Tcla). In the O26, O44, and O105 serogroups, novel alleles with four amino acid substitutions (wtT36A, wtF46L, wtK55N, and ETI68Tcla,O26,O44,O105) were found.
DISCUSSION
In this study we utilized a panel of 300 clinical and environmental strains of V. cholerae to identify and characterize 15 novel pathogens which belong to serogroups traditionally considered nonpathogenic and nonepidemic and which carry the VPI or the VPI and CTX prophage (regions originally thought to be exclusive to O1 and O139 serogroups). Mukhopadhyay et al. (28) reported the identification and characterization of similarly unusual V. cholerae pathogens (serogroups O8, O10, O11, O27, O35, O42, and O69) isolated from environmental samples from the Calcutta region, and very recently, Boyd and Waldor (8) reported several additional isolates (serogroups O8, O37 and O141). Thus, our study further expands the repertoire of novel V. cholerae serogroups (O26, O37, O44, O48, O49, O53, O65, O77, O80, O105, O115, O141, and O191) found to carry virulence genes.
The 15 non-O1 and non-O139 strains carrying the VPI cluster could be divided into two groups, based on comparative genomic analyses (MLST and RFLP) (Fig. 1 and 2 to 4, respectively). First, strains of serogroups O27, O37, O53, and O65 have very similar genetic backgrounds and cluster with the epidemic strains, and they most likely emerged by horizontal transfer and exchange of O-antigen biosynthesis regions. In that regard, previous studies (5, 26) and the results we obtained during our sequencing of the O-antigen biosynthesis region in an O37 serogroup strain (25) support that emergence mechanism. Second, strains of serogroups O8, O26, O44, O48, O49, O77, O80, O105, O115, O141, and O191 have distinct genetic backgrounds and virulence regions compared to the epidemic strains, and they most likely arose by independent acquisition of the VPI. Also, several strains in the second group subsequently acquired a CTXφ or a pre-CTXφ. None of the 300 strains screened in our study had the CTXφ alone, which supports the two-step sequential model for the acquisition of VPI and CTXφ (7). Also, as previously reported (7), intermediates in the evolution of the CTXφ have also been found, i.e., a pre-CTXφ lacking ctxAB has been found in three different non-O1 serogroups (O48, O53, and O65).
The tcpA, rstR, and ctxAB diversity observed in this study is similar to that reported in previous publications. For example, the two new rstR alleles identified in the present work (rstRO8 and rstRO44) are very similar to the rstR4 allele (28) and to another sequence deposited in the GenBank database (accession number AF302794). In addition, an insertion and deletion observed at the left end of the VPIs in some non-O1 and non-O139 strains has been reported by Mukhopadhyay et al. (28). However, the sizes of the inserted fragments observed by us are much larger than the 1.6-kb segment of chromosome II reported, in the above-referenced study (28), to have been translocated to the left end of the VPI on chromosome I in place of a 300-bp segment. The genetic contents of the inserts and the deletion end-points need to be determined in order to understand whether translocation of the same region has occurred in the non-O1 and non-O139 strains described in this paper.
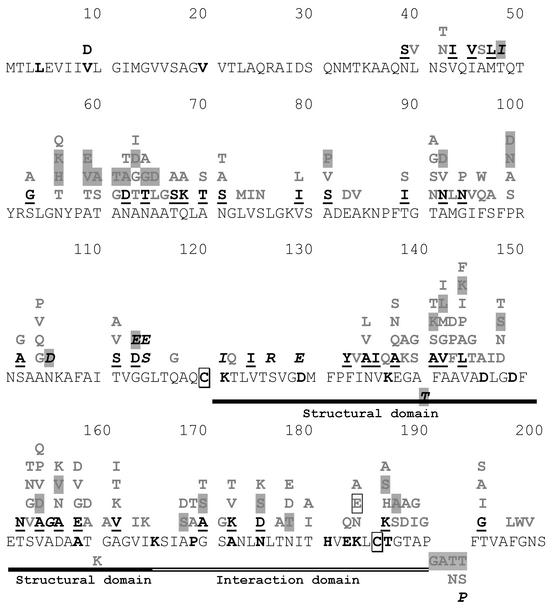
Extensive site-directed mutagenesis analysis of the tcpA gene (10, 23, 41) has yielded useful insights into the structure-function relationship of the TCP. Based on functional analysis of mutant TcpA pilins, the TcpA protein has been proposed to have three distinct domains, N-terminal, C-terminal structural, and C-terminal interaction domains (10, 23, 41). As observed earlier (8), the various tcpA alleles identified in our study have variations in the amino acid residues previously demonstrated (23, 41) to have functional significance, i.e., the majority of the changes are found in the C-terminal structural and interaction domains (Fig. 8). The novel finding, however, is further delineation of the region between amino acid residues 50 and 120, which carries two mutational hotspots, residues 53 to 75 and 90 to 105, where multiple variations are seen (Fig. 8). The role of this region in the structure-function relationship of TcpA pilin remains to be determined. Since there are multiple variations within any given tcpA allele, assessing the effect of these changes may be difficult, especially if the changes in one domain are the result of intragenic suppression of mutations elsewhere in the gene. Site-directed mutagenesis studies of this region (residues 50 to 120) in a wild-type tcpA may reveal the functional significance of this domain.
FIG. 8.
Domain structure of TcpA. The primary amino acid sequence of the El Tor TcpA is shown. The residues shown to have functional significance, based on site-directed mutagenesis (10, 23, 41), are indicated in boldface type, the two cysteine residues predicted to be essential for the structure and function of TcpA pilin are indicated by boxes, and the proposed domains are indicated by bars (10, 23, 41). The variations in the amino acid residues found in various tcpA alleles described in other studies (9, 15, 28-30) and in this work (highlighted) are indicated above the primary sequences. The variations seen in the classical allele are underlined. The changes unique to a single strain are indicated in italics. The O37 serogroup has a classical TcpA with a single change (K184 to E) indicated by a shaded box.
Horizontal gene transfer plays an important role in the evolution of pathogenic bacteria, including V. cholerae, which has become a paradigmatic organism for studying horizontal gene transfer mechanisms. All three of the major known V. cholerae virulence markers (CTXφ, VPI, and O-antigen biosynthesis regions) are believed to have been acquired by horizontal gene transfer. Two of these markers (CTXφ and VPI) have been introduced into the V. cholerae chromosome via phage-mediated transduction (22, 44); the mechanisms responsible for acquiring the O-antigen biosynthesis regions are unknown at the present time. However, acquisition of VPI and CTXφ appears to be more frequent than exchange of O antigens in epidemic serogroups, which may reflect mechanistic differences in their horizontal transfer. Acquisition of VPI and CTXφ involves single-stranded phages and a site-specific recombination process, whereas acquisition of the O-antigen region probably involves generalized transducing phages and a homologous recombination mechanism. Hence, it is tempting to speculate that horizontal transfer of O-antigen regions might be subjected to DNA restriction and recombination barriers.
The present study provides further support to the growing body of evidence that the classical V. cholerae virulence markers, ctxAB and tcpA, are not unique to epidemic strains and that they are found in at least some nonepidemic, non-O1 and non-O139 V. cholerae serogroups. However, the epidemic potential of these strains is not clear at the present time, especially since—and in clear contrast to O1 and O139 isolates—none of the non-O1 and non-O139 strains has been associated with past epidemics and/or pandemics of cholera. However, these observations raise the intriguing possibility that the presence of ctxAB and tcpA in V. cholerae, while critical, is not sufficient for the full-blown epidemics of cholera. In this context, Dziejman et al. (12) recently reported comparative genome analyses of 11 V. cholerae strains of epidemic serogroups, and they identified two clusters of genes differentiating pandemic strains from other strains. Similar comparative genomic analysis of epidemic strains with the nonepidemic, non-O1 and non-O139 strains identified in this study is likely to provide invaluable information regarding the mechanisms responsible for the emergence of the epidemic V. cholerae serogroups and strains.
Acknowledgments
We thank Arnold Kreger, Alexander Sulakvelidze, and Colin Stine for helpful comments and editorial assistance, Judy Johnson for providing access to the Smith strain collection, and Glenn Morris for support and encouragement during the course of this study.
M.K. was supported by an International Training and Research in Emerging Infectious Diseases grant from the Fogarty International Center, National Institutes of Health. Funding for our studies was provided by a University of Maryland intramural grant (to S.S.) and BREF intramural support from the Department of Veterans Affairs (to S.S.).
REFERENCES
- 1.Albert, M. J., M. Ansaruzzaman, P. K. Bardhan, A. S. G. Faruque, S. M. Faruque, M. S. Islam, D. Mahalanabis, R. B. Sack, M. A. Salam, A. K. Siddique, M. D. Yunus, and K. Zaman. 1993. Large epidemic of cholera-like disease in Bangladesh caused by Vibrio cholerae O139 synonym Bengal. Lancet 342:387-390. [PubMed] [Google Scholar]
- 2.Aldova, E., K. Laznickova, E. Stepankova, and J. Lietava. 1968. Isolation of nonagglutinable vibrios from an enteritis outbreak in Czechoslovakia. J. Infect. Dis. 118:25-31. [DOI] [PubMed] [Google Scholar]
- 3.Berche, P., C. Poyart, E. Abachin, H. Lelievre, J. Vandepitte, A. Dodin, and J.-M. Fournier. 1994. The novel epidemic strain O139 is closely related to pandemic strain O1 of Vibrio cholerae. J. Infect. Dis. 170:701-704. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharya, S. K., M. K. Bhattacharya, G. B. Nair, D. Dutta, A. Deb, T. Ramamurthy, S. Garg, P. K. Saha, P. Dutta, A. Moitra, B. K. Mandel, T. Shimada, Y. Takeda, and B. C. Deb. 1993. Clinical profile of acute diarrhoea cases infected with the new epidemic strain of V. cholerae O139: designation of disease as cholera. J. Infect. Dis. 27:11-15. [DOI] [PubMed] [Google Scholar]
- 5.Bik, M., R. D. Gouw, and F. R. Mooi. 1996. DNA fingerprinting of Vibrio cholerae strains with a novel insertion sequence element: a tool to identify epidemic strains. J. Clin. Microbiol. 34:1453-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd, E. F., K. E. Moyer, L. Shi, and M. K. Waldor. 2000. Infectious CTXφ and the vibrio pathogenicity island prophage in Vibrio mimicus: evidence for recent horizontal transfer between V. mimicus and V. cholerae. Infect. Immun. 68:1507-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd, E. F., A. J. Heilpern, and M. K. Waldor. 2000. Molecular analyses of a putative CTXφ precursor and evidence for independent acquisition of distinct CTX φs by toxigenic Vibrio cholerae. J. Bacteriol. 182:5530-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyd, E. F., and M. K. Waldor. 2002. Evolutionary and functional analyses of variants of the toxin-coregulated pilus protein TcpA from toxigenic Vibrio cholerae non-O1/non-O139 serogroup isolates. Microbiology 148:1655-1666. [DOI] [PubMed] [Google Scholar]
- 9.Chakraborty, S., A. K. Mukhopadhyay, R. K. Bhadra, A. N. Ghosh, R. Mitra, T. Shimada, S. Yamasaki, S. M. Faruque, Y. Takeda, R. R. Colwell, and G. B. Nair. 2000. Virulence genes in environmental strains of Vibrio cholerae. Appl. Environ. Microbiol. 66:4022-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiang, S. L., R. K. Taylor, M. Koomey, and J. J. Mekalanos. 1995. Single amino acid substitutions in the N-terminus of Vibrio cholerae TcpA affect colonization, autoagglutination, and serum resistance. Mol. Microbiol. 17:1133-1142. [DOI] [PubMed] [Google Scholar]
- 11.Dalsgaard, A., O. Serichantalergs, A. Forslund, W. Lin, J. Mekalanos, E. Mintz, T. Shimada, and J. G. Wells. 2001. Multiple clinical and environmental isolates of Vibrio cholerae serogroup O141 carry the ctx phage and the genes encoding the toxin coregulated pili. J. Clin. Microbiol. 39:4086-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dziejman, M., E. Balon, D. Boyd, C. M. Fraser, J. F. Heidelberg, and J. J. Mekalanos. 2002. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc. Natl. Acad. Sci. USA 99:1556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faruque, S. M., M. M. Rahman, Asadulghani, K. M. Nasirul Islam, and J. J. Mekalanos. 1999. Lysogenic conversion of environmental Vibrio mimicus strains by CTXφ. Infect. Immun. 67:5723-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faruque, S. M., Asadulghani, M. M. Rahman, M. K. Waldor, and D. A. Sack. 2000. Sunlight-induced propagation of the lysogenic phage encoding cholera toxin. Infect. Immun. 68:4795-4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh, C., R. K. Nandy, S. K. Dasgupta, G. B. Nair, R. H. Hall, and A. C. Ghose. 1997. A search for cholera toxin (CT), toxin coregulated pilus (TCP), the regulatory element ToxR and other virulence factors in non-O1/non-O139 Vibrio cholerae. Microb. Pathog. 22:199-208. [DOI] [PubMed] [Google Scholar]
- 16.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hillis, D. M., M. W. Allard, and M. M. Miyamoto. 1993. Analysis of DNA sequence data: phylogenetic inference. Methods Enzymol. 224:456-487. [DOI] [PubMed] [Google Scholar]
- 18.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 19.Kamal, A. M. 1971. Outbreak of gastro-enteritis by non-agglutinable (NAG) vibrios in the republic of the Sudan. J. Egypt. Public Health Assoc. 46:125-159. [Google Scholar]
- 20.Kaper, J. B., J. G. Morris, Jr., and M. M. Levine. 1995. Cholera. Clin. Microbiol. Rev. 8:48-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karaolis, D. K., J. A. Johnson, C. C. Bailey, E. C. Boedeker, J. B. Kaper, and P. R. Reeves. 1998. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc. Natl. Acad. Sci. USA 95:3134-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karaolis, D. K., S. Somara, D. R. Maneval, Jr., J. A. Johnson, and J. B. Kaper. 1999. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature 399:375-379. [DOI] [PubMed] [Google Scholar]
- 23.Kirn, T. J., M. J. Lafferty, C. M. P. Sandoe, and R. K. Taylor. 2000. Delineation of pilin domains required for bacterial association into microcolonies and intestinal colonization by Vibrio cholerae. Mol. Microbiol. 35:896-910. [DOI] [PubMed] [Google Scholar]
- 24.Kotetishvili, M., O. C. Stine, A. Kreger, J. G. Morris, Jr., and A. Sulakvelidze. 2002. Multilocus sequence typing for characterization of clinical and environmental Salmonella strains. J. Clin. Microbiol. 40:1626-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, M., T. Shimada, J. G. Morris, Jr., A. Sulakvelidze, and S. Sozhamannan. 2002. Evidence for the emergence of non-O1 and non-O139 Vibrio cholerae strains with pathogenic potential by exchange of O-antigen biosynthesis regions. Infect. Immun. 70:2441-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mooi, F. R., and E. M. Bik. 1997. The evolution of epidemic Vibrio cholerae strains. Trends Microbiol. 4:161-165. [DOI] [PubMed] [Google Scholar]
- 27.Morris, J. G., Jr., G. E. Losonsky, J. A. Johnson, C. O. Tacket, J. P. Nataro, P. Panigrahi, and M. M. Levine. 1995. Clinical and immunological characteristics of Vibrio cholerae O139 Bengal infection in North American volunteers. J. Infect. Dis. 171:903-908. [DOI] [PubMed] [Google Scholar]
- 28.Mukhopadhyay, A. K., S. Chakraborty, Y. Takeda, G. B. Nair, and D. E. Berg. 2001. Characterization of VPI pathogenicity island and CTXφ prophage in environmental strains of Vibrio cholerae. J. Bacteriol. 183:4737-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nandi, B., R. K. Nandy, A. C. Vicente, and A. C. Ghose. 2000. Molecular characterization of a new variant of toxin-coregulated pilus protein (TcpA) in a toxigenic non-O1/non-O139 strain of Vibrio cholerae. Infect. Immun. 68:948-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Novais, R. C., A. Coelho, C. A. Salles, and A. C. Vicente. 1999. Toxin co-regulated pilus cluster in non-O1, non-toxigenic Vibrio cholerae: evidence of a third allele of pilin gene. FEMS Microbiol. Lett. 171:49-55. [DOI] [PubMed] [Google Scholar]
- 31.Ramamurthy, T., S. Garg, R. Sharma, S. K. Bhattacharya, G. B. Nair, T. Shimada, T. Takeda, T. Karasawa, H. Kurazano, A. Pal, and Y. Takeda. 1993. Emergence of novel strain of Vibrio cholerae with epidemic potential in southern and eastern India. Lancet 341:703-704. [DOI] [PubMed] [Google Scholar]
- 32.Rhine, J. A., and R. K. Taylor. 1994. TcpA pilin sequences and colonization requirements for O1 and O139 Vibrio cholerae. Mol. Microbiol. 13:1013-1020. [DOI] [PubMed] [Google Scholar]
- 33.Rivera, I. N., J. Chun, A. Huq, R. B. Sack, and R. R. Colwell. 2001. Genotypes associated with virulence in environmental isolates of Vibrio cholerae. Appl. Environ. Microbiol. 67:2421-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma, C., M. Thungapathra, A. Ghosh, A. K. Mukhopadhyay, A. Basu, R. Mitra, I. Basu, S. K. Bhattacharya, T. Shimada, T. Ramamurthy, T. Takeda, S. Yamasaki, Y. Takeda, and G. B. Nair. 1998. Molecular analysis of non-O1, non-O139 Vibrio cholerae associated with an unusual upsurge in the incidence of cholera-like disease in Calcutta, India. J. Clin. Microbiol. 36:756-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimada, T., E. Arakawa, K. Itoh, T. Okitsu, A. Matsushima, Y. Asai, S. Yamai, T. Nakazato, G. B. Nair, M. J. Albert, and Y. Takeda. 1994. Extended serotyping scheme for Vibrio cholerae. Curr. Microbiol. 28:175-178. [Google Scholar]
- 36.Singh, D. V., M. H. Matte, G. R. Matte, S. Jiang, F. Sabeena, B. N. Shukla, S. C. Sanyal, A. Huq, and R. R. Colwell. 2001. Molecular analysis of Vibrio cholerae O1, O139, non-O1, and non-O139 strains: clonal relationship between clinical and environmental isolates. Appl. Environ. Microbiol. 67:910-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith, H. L. 1979. Serotyping of non-cholera vibrios. J. Clin. Microbiol. 10:85-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sozhamannan, S., Y. K. Deng, M. Li, A. Sulakvelidze, J. B. Kaper, J. A. Johnson, G. B. Nair, and J. G. Morris Jr. 1999. Cloning and sequencing of the genes downstream of the wbf gene cluster of Vibrio cholerae serogroup O139 and analysis of the junction genes in other serogroups. Infect. Immun. 67:5033-5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stine, O. C., S. Sozhamannan, Q. Gou, S. Zheng, J. G. Morris, Jr., and J. A. Johnson. 2000. Phylogeny of Vibrio cholerae based on recA sequence. Infect. Immun. 68:7180-7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stroeher, U. H., and P. A. Manning. 1997. Vibrio cholerae serotype O139: swapping genes for surface polysaccharide biosynthesis. Trends Microbiol. 5:178-180. [DOI] [PubMed] [Google Scholar]
- 41.Sun, D., M. J. Lafferty, J. A. Peek, and R. K. Taylor. 1997. Domains within the Vibrio cholerae toxin coregualted pilin subunit that mediate bacterial colonization. Gene 192:79-85. [DOI] [PubMed] [Google Scholar]
- 42.Tacket, C. O., R. K. Taylor, G. Losonsky, Y. Lim, J. P. Nataro, J. B. Kaper, and M. M. Levine. 1998. Investigation of the roles of toxin-coregulated pili and mannose-sensitive hemagglutinin pili in the pathogenesis of Vibrio cholerae O139 infection. Infect. Immun. 66:692-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor, R. K., V. L. Miller, D. B. Furlong, and J. J. Mekalanos. 1987. The use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. USA 84:2833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]