Abstract
Quorum-sensing-controlled processes are considered to be important for the competitiveness of microorganisms in the rhizosphere. They affect cell-cell communication, biofilm formation, and antibiotic production, and the GacS-GacA two-component system plays a role as a key regulator. In spite of the importance of this system for the regulation of various processes, strains with a Gac− phenotype are readily recovered from natural habitats. To analyze the influence of quorum sensing and the influence of the production of the antibiotic phenazine-1-carboxamide on rhizosphere colonization by Pseudomonas chlororaphis, a gnotobiotic system based on Arabidopsis thaliana seedlings in soil was investigated. Transposon insertion mutants of P. chlororaphis isolate SPR044 carrying insertions in different genes required for the production of N-acyl-homoserine lactones and phenazine-1-carboxamide were generated. Analysis of solitary rhizosphere colonization revealed that after prolonged growth, the population of the wild type was significantly larger than that of the homoserine lactone-negative gacS mutant and that of a phenazine-1-carboxamide-overproducing strain. In cocultivation experiments, however, the population size of the gacS mutant was similar to that of the wild type after extended growth in the rhizosphere. A detailed analysis of growth kinetics was performed to explain this phenomenon. After cells grown to the stationary phase were transferred to fresh medium, the gacS mutant had a reduced lag phase, and production of the stationary-phase-specific sigma factor RpoS was strongly reduced. This may provide a relative competitive advantage in cocultures with other bacteria, because it permits faster reinitiation of growth after a change to nutrient-rich conditions. In addition, delayed entry into the stationary phase may allow more efficient nutrient utilization. Thus, GacS-GacA-regulated processes are not absolutely required for efficient rhizosphere colonization in populations containing the wild type and Gac− mutants.
In natural habitats microorganisms compete for nutrients to ensure that they can multiply. Compared to the surrounding soil, the rhizosphere of plants is a comparably nutrient-rich habitat, and various factors have been shown to be required for successful colonization by fluorescent pseudomonads (10, 11, 20, 27, 32, 42). The introduction of Pseudomonas spp. for plant growth promotion or biocontrol is of biotechnological interest. However, the impact of abiotic and biotic factors on the competitiveness of these organisms with the indigenous microflora is largely unknown. Different factors, including the production of antimicrobial agents, contribute to bacterial competitiveness in the rhizosphere (7, 29, 46). Some of these factors are regulated by changes in the cell density, a phenomenon known as quorum sensing (15, 45). This regulation is based on the synthesis of low-molecular-mass signals, including N-acyl-homoserine lactones (acyl-HSLs), which freely diffuse out of the cell. As the cell density increases, acyl-HSLs accumulate in the environment. At intracellular concentrations above a certain threshold concentration, the signal molecules productively interact with transcriptional regulators of the LuxR family, and the expression of quorum-sensing-regulated genes is initiated (13).
In Pseudomonas spp., the production of metabolites like HCN, 2,4-diacetylphloroglucinol, pyrrolnitrin, and extracellular proteases (8, 26), which have been shown to be important for biocontrol activity, is controlled by a conserved two-component system comprising the response regulator GacA and the sensor kinase GacS (31). The GacA-GacS two-component system and the acyl-HSL-mediated regulatory systems do not act individually, but they are part of a complex regulatory cascade (33). gacA and gacS mutants do not produce these secondary metabolites and exoenzymes and are less effective than the wild type in disease suppression in the rhizosphere (26). Whereas antibiosis is generally considered to be a competitiveness factor, spontaneous gacA and gacS mutants are isolated from bacterial cultures, which is a problem for the formulation for biocontrol agents (12). In addition, gacA mutants have been isolated from the rhizosphere, suggesting that the absence of this cascade may confer a competitive advantage under certain conditions (31, 36).
The limited success of biocontrol field experiments has often been shown to correlate with poor colonization of the roots (47). Thus, when the production of antimicrobial metabolites by Pseudomonas fluorescens strain 79-2 was exploited to suppress the fungus Graeumannomyces graminis var. tricitii in the wheat rhizosphere, the functionality of quorum sensing was shown to be positively correlated with the population size (2). However, despite the obvious importance of quorum sensing for cell-cell communication, there is relatively little information concerning the influence of quorum sensing on rhizosphere colonization. To address this issue, we used a microcosm-based closed system with low intrinsic variability, which was suitable for detection of subtle differences in rhizosphere colonization. A natural isolate of Pseudomonas chlororaphis, strain SPR044 (40), was used as a model for studies on the influence of quorum sensing and phenazine-1-carboxamide (PCN) production. Rhizosphere colonization by the wild type and rhizosphere colonization by mutants carrying insertions in genes required for homoserine lactone and/or PCN biosynthesis were compared, and the results showed that these traits per se do not offer competitive advantages. In contrast, defects in the quorum-sensing system may be advantageous under certain conditions.
MATERIALS AND METHODS
Organisms, plasmids, and growth conditions.
Escherichia coli JM109 and HB101 were grown at 37°C in Luria-Bertani (LB) medium, whereas Pseudomonas spp., Bacillus subtilis, Chromobacterium violaceum, and Agrobacterium tumefaciens were grown in LB medium at 26°C. Alternatively, P. chlororaphis was grown at 26°C in liquid M9 minimal medium supplemented with 0.5% glucose or 0.5% myo-inosit (35). For plasmid propagation and selection of transformants media were supplemented with the following antibiotics: carbenicillin (100 μg/ml), chloramphenicol (20 μg/ml), tetracycline (5 μg/ml), kanamycin (50 μg/ml for E. coli and C. violaceum and 100 μg/ml for P. chlororaphis), streptomycin (50 μg/ml for E. coli and 200 μg/ml for P. chlororaphis), and spectinomycin (50 μg/ml for E. coli and 200 μg/ml for P. chlororaphis). The strains and plasmids used in this study are described in Table 1.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| P. chlororaphis SPR044 | Wild type, P. chlororaphis natural isolate | 40 |
| SPR144 | SPR044, TnMod insertion in gene for putative RNA 3′-terminal phosphate cyclase, Kanr | This study |
| SPR244 | SPR044, gacS::TnMod, Kanr | This study |
| SPR344 | SPR044, TnMod insertion in gene for putative glycosyl transferase, Kanr | This study |
| SPR544 | SPR044, TnMod insertion in gene for putative transcriptional regulator, Kanr | This study |
| SPR644 | SPR044, htpX::TnMod, Kanr | This study |
| SPR744 | SPR044, phzF::TnMod, Kanr | This study |
| SPR844 | SPR044, phzE::TnMod, Kanr | This study |
| P. aureofaciens 30-84I | phzl::Km, HSL−, Phz−, Kanr | 49 |
| E. coli HB101 | K12/B hybrid, StrrrecA thi pro leu hsdRM+ | 21 |
| E. coli JM109 | endA1 gyrA96 thi hsdR71 supE44 recA1 relA1 Δ(lac-proAB) (F′ traD36 proAB+ lacIqlacZΔM15) | 50 |
| A. tumefaciens A136 | pCF218 traR, pCF372 traI-lacZ, Tetr, Spcr, acyl-HSL detection strain | 14 |
| B. subtilis | Wild type | ATCC 1498 |
| Plasmids | ||
| pRK600 | ColE1 oriV, RP4 oriT, Camr, helper plasmid in triparental matings | 21 |
| pTrc200 | pVS1 replicon, lacIq Strr Spcr, trc promoter | 39 |
| pTrc300 | pVS1 replicon, lacIq Strr Spcr, trc promoter | This study |
| pTrcGacS | pTrc300, gacS expression plasmid | This study |
| pTnMod-OKm | OriR, Kanr, TnMod delivery plasmid | 9 |
TnMod mutagenesis and mutant analysis.
TnMod was introduced into P. chlororaphis by triparental mating. The recipient, the donor E. coli JM109 harboring pTnMod-OKm, and E. coli HB101 containing the helper plasmid RK600 were propagated in LB medium supplemented with antibiotics to the mid-log phase, sedimented by centrifugation, and resuspended in LB medium. Equal amounts of donor, recipient, and helper (10 μl of each, corresponding to 107 cells) were mixed on prewarmed LB agar plates and incubated for 16 h at 26°C to allow plasmid transfer. The cells were washed from the plates with LB medium and plated on agar supplemented with 100 μg of kanamycin per ml and 100 μg of carbenicillin per ml to select for transposon insertions into the P. chlororaphis chromosome. Transposon-carrying derivatives affected in acyl-HSL production were identified due to their inability to restore orange pigment production to phzI-deficient Pseudomonas aureofaciens 30-84I streaked in close proximity on LB agar (48). Strains affected in PCN production were identified by visual inspection for colonies with altered amounts of green PCN crystals. To determine the chromosomal site of insertion, the replication-competent plasposon and flanking DNA were recloned from the chromosome. DNA was isolated (Quantum Prep AquaPure genomic DNA isolation kit; Bio-Rad) and cleaved with SalI or PstI; this was followed by religation, electroporation into JM109, and selection for kanamycin resistance. The resulting plasmids were sequenced by using primers Ori5seq (5′-GCCTTTTGCTCACATGTTCTTTCC-3′) and Ori3seq (5′-CCCCGAGCTCTTAATTAATTTAAATC-3′), which anneal to the two ends of the TnMod insertion, which allows identification of the insertion site.
TLC analysis of acyl-HSLs released by P. chlororaphis SP044.
Acyl-HSLs produced by SPR044 and its TnMod insertion derivatives were isolated essentially as described previously (52). Briefly, after ethyl acetate extraction of cell-free supernatant and subsequent evaporation to dryness, the samples were dissolved in 50 μl of ethyl acetate. Ten-microliter aliquots were applied to C18 reversed-phase thin-layer chromatography (TLC) plates (RP18 F254; Merck) and developed in 60% methanol-40% water. After chromatography, the plates were each dried and overlaid with 300 ml of LB agar inoculated with 10 ml of an exponentially growing culture of the indicator strain P. aureofaciens 30-84I (phzI negative) (49) or the indicator strain A. tumefaciens A136(pCF218)(pCF372) (14). Acyl-HSL-induced orange phenazine production by P. aureofaciens 30-84I was determined by visual inspection, whereas β-galactosidase activity in the A. tumefaciens indicator strain was monitored in agar supplemented with 40 μg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactoside) per ml. For identification of inducing compounds from strain SPR044, the positions of the compounds after TLC separation were compared to those of the reference compounds butyryl-homoserine lactone (BHSL), hexanoyl-homoserine lactone (HHSL), and octanoyl-homoserine lactone (OHSL) (Sigma).
Assays for production of secondary metabolites.
Assays to examine production of BHSL, antimicrobial agents, and extracellular proteases were performed by the method of Ravn et al. (34), with minor modifications. Briefly, cell-free supernatants from cultures grown for 17 h in M9 minimal medium with glucose were screened in agar well diffusion assays. For quantitation of acyl-HSL production, synthetic BHSL (1 to 100,000 pmol) was analyzed simultaneously. The agar was inoculated with 10 ml of an exponentially growing culture of P. aureofaciens 30-84I or with B. subtilis for monitoring antimicrobial agents. The plates were incubated at 26°C for 48 h before the diameters of the zones induced by acyl-HSLs (color produced by indicators) or antimicrobial metabolites (growth inhibition) were measured. For the analysis of extracellular proteases, an agar containing 0.3% yeast extract, 0.5% K2HPO4, and 1% skim milk powder (pH 7) was used, and lysis zones were monitored as described above. Phenazine production was analyzed after chloroform extraction (1:1, vol/vol) of cell-free supernatants from cultures grown to the stationary phase in LB medium. After evaporation to dryness, samples were each suspended in 1 ml of chloroform, and the production of phenazines was assessed after spectral analyses (200 to 500 nm) (28).
DNA manipulations.
DNA purification, modification, and cloning were performed by using standard protocols (35) with enzymes purchased from MBI Fermentas and New England Biolabs. DNA sequence analyses were performed with an ABI Prism 377 sequencer. The GacS coding region was PCR amplified with Taq polymerase from 1 ng of chromosomal DNA of P. chlororaphis SPR044 by using oligonucleotides GacS5′ (5′-GGCAGAGCTCGTTAGCAGGAGAGTTGCGTGCTTAAG-3′) and GacS3′ (5′-GCGGGTCTAGATCAGGCGTTGATGCGGGCCT-3′) and the following cycling conditions: 2 min at 95°C for one cycle; 44°C for 1 min, 72°C for 1 min, and 95°C for 30 s for 30 cycles; and 44°C for 3 min and 72°C for 7 min for one cycle. The resulting 2.7-kb fragment was cleaved with SacI and XbaI and ligated into SacI- and XbaI-cleaved pUC19, resulting in pUC-GacS. The sequence of the insert was determined with specific primers. For expression of gacS in P. chlororaphis, pTrc200 (39) was cut with NcoI, and the overhanging single-stranded DNA was removed with S1 nuclease and religated, resulting in vector pTrc300. The GacS coding sequence was excised from pUC-GacS by using SacI and HindIII and was ligated into SacI- and HindIII-cleaved pTrc300, resulting in pTrcGacS.
SDS-PAGE and protein analysis.
Protein lysates were analyzed after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in acrylamide-containing gels (23) followed by Western blotting, incubation with specific polyclonal antisera, and detection with anti-rabbit horseradish peroxidase-conjugated secondary antibody (Bio-Rad) by using a chemiluminescence-based detection system (NEN). RpoS-specific antiserum was used at a 1:5,000 dilution.
Microcosm design and sampling procedure.
Seed sterilization, coating with bacteria, and growth of Arabidopsis thaliana were performed as described previously (40). Ten plants were harvested randomly after 14, 28, and 42 days. After removal of unattached soil, the root weight was determined. The roots were transferred into flasks containing 10 ml of phosphate-buffered saline (pH 7.2) and 0.5 g of glass beads (diameter, 0.25 to 0.3 mm; B. Braun Biotech International) and subjected to vigorous shaking for 20 min. Serial dilutions were plated on LB medium supplemented with kanamycin if required for discrimination of SPR044 and the TnMod derivatives. After 36 h of incubation at 26°C colonies were counted, and the number of CFU per gram of root was determined.
Statistical methods.
Data were analyzed for significance by using the software SPSS10 for Macintosh. Normal distribution of the data was analyzed with the Kolmogorov-Smirnov test, and the homogeneity of the variances was analyzed with the Levene test. Since the doubling times and lag phases in the growth experiments showed normal distribution and homogeneity of the variances, they were compared by analysis of variance. Pairwise comparisons between the wild type and mutants were performed with the post hoc test of Bonferroni. The values for log CFU per gram of root obtained in the plant colonization assays did not show a normal distribution and therefore were compared by using the Kruskal-Wallis test. With this nonparametric test we compared the medians of multiple samples for each derivative and each time point. Pairwise comparisons between the wild type and mutants were performed with the Mann-Whitney U test.
Nucleotide sequence accession numbers.
The nucleotide sequences of complete genes have been deposited in the GenBank database under accession numbers AF517687 (gacS) and AF517688 (putative glycosyl transferase gene).
RESULTS
Isolation and characterization of mutants affected in acyl-HSL and/or PCN biosynthesis.
P. chlororaphis SPR044 was mutagenized by triparental mating (9). Screening of 4,000 exconjugants carrying TnModKm-O resulted in detection of three classes of mutants (Table 1). The first class comprised two mutants deficient in acyl-HSL production (SPR144 and SPR244) (48). The second class contained four PCN-deficient mutants (SPR544, SPR644, SPR744, and SPR844), and the third class contained a single mutant which produced a significantly larger amount of green PCN crystals (SPR344). Southern blot analyses demonstrated that each isolate had a different chromosomal insertion site for TnModKm-O (data not shown). The DNA regions flanking the insertion were cloned in E. coli JM109 and sequenced with primers annealing to the ends of TnMod to determine the plasposon's point of insertion. Database searches indicated that all of the clones carried TnMod-OKm insertions in genes encoding proteins with significant similarity to proteins from Pseudomonas aeruginosa PAO1 or to proteins from different P. chlororaphis strains (Table 2).
TABLE 2.
Putative functions of gene products affected by TnMod insertions in P. chlororaphis derivatives deduced from database searches
| Strain | Organism encoding similar gene product | Accession no. | Putative protein function | % Identitya | % Similarityb |
|---|---|---|---|---|---|
| SPR144 | P. aeruginosa | AE004872 | RNA 3′-terminal phosphate cyclase | 75 | 88 |
| SPR244 | P. chlororaphis O6 | AF192795 | Two-component histidine sensor kinase GacS | 98 | 99 |
| SPR344 | P. aeruginosa | AE004913 | Glycosyl transferase | 69 | 75 |
| SPR544 | P. aeruginosa | AE004556 | Transcription regulator | 45 | 64 |
| SPR644 | P. aeruginosa | AE004709 | Heat shock protein HtpX | 90 | 94 |
| SPR744 | P. chlororaphis PCL1391 | AF195615 | Phenazine biosynthesis protein PhzF | 99 | 99 |
| SPR844 | P. chlororaphis PCL1391 | AF195615 | Phenazine biosynthesis protein PhzE | 100 | 100 |
Based on deduced amino acids matched with identical amino acids.
Based on deduced amino acids matched with similar groups of amino acids.
The deduced amino acid sequence of the gene product affected by the TnMod insertion in SPR144 most closely resembled an RNA 3′-terminal phosphate cyclase. Such enzymes catalyze the conversion of 3′-phosphate to a 2′,3′-cyclic phosphodiester at the end of RNA (44). SPR244 carried the insertion in the gacS gene encoding a two-component sensor kinase, which has been sequenced previously in a different P. chlororaphis strain. The gene flanking the TnMod insertion in SPR344 probably codes for a glycosyl transferase, and there are two classes of glycosyl transferases. The processive glycosyl transferases transfer multiple sugar residues, and the nonprocessive glycosyl transferases catalyze the transfer of a single sugar to the repeat unit of a polysaccharide. Hydrophobic cluster analysis revealed that there are two conserved regions (37). Whereas domain A is conserved in all glycosyl transferases, domain B is present only in processive enzymes. Since the QXXRW motif characteristic of domain B is not present in the SPR044 glycosyl transferase, this protein probably belongs to the class of nonprocessive enzymes (37). In the case of SPR544 the gene flanking the insertion most likely encodes a transcriptional regulator. The gene product shows 58% similarity and 42% identity to VanR of Acinetobacter sp. strain ADP1, a member of the GntR family of repressors (30). The TnMod insertion in strain SPR644 is located in htpX, which encodes a putative heat shock protein. This protein exhibits 64% similarity and 49% identity to HtpX of E. coli. Production of the 32-kDa HtpX protein is induced by a temperature upshift in E. coli. However, its role in the heat shock response is not obvious, because E. coli cells carrying an htpX disruption have no apparent phenotype at temperatures up to 47°C (22). The TnMod insertions of SPR744 and SPR844 are located in phzF and phzE, respectively. The gene products were previously identified as components of the phenazine biosynthesis pathway.
Identification of autoinducers produced by P. chlororaphis.
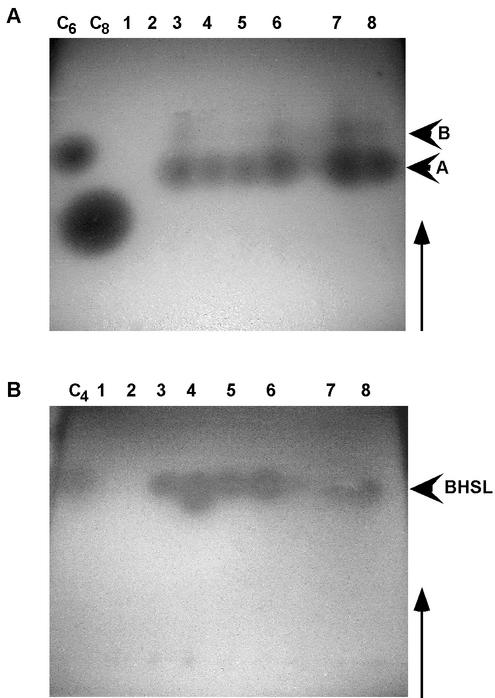
To analyze the production of autoinducers by SPR044 and its TnMod insertion derivatives, ethyl acetate extracts of the cell-free supernatants of stationary-phase cultures were separated by TLC. The plates were overlaid with A. tumefaciens A136(pCF218)(pCF372) seeded in LB agar. Two bioactive compounds that induced β-galactosidase activity were detected in extracts from SPR044 and all of the insertion mutants except SPR144 and SPR244 (Fig. 1A). The most prominent signal was the signal caused by compound A (Rf, 0.5), which migrated at a position between the positions of synthetic HHSL and OHSL. Compound B caused a relatively weak response of the test strain and migrated faster than synthetic HHSL. Synthetic BHSL was not detected, which is in accord with the low sensitivity of the detection strain for BHSL (52). The Rf values and the spot-like migration of compounds A and B detected by A136(pCF218)(pCF372), as well as the inability of these compounds to induce the quorum-sensing system of P. aureofaciens 30-84I (see below), suggest that they are probably C-3-hydroxy-substituted acyl-HSLs. Based on the relative Rf values of acyl-HSLs from P. fluorescens 2-79, compound A probably corresponds to N-(3-hydroxy-octanoyl)-homoserine lactone (3). However, compound B could not be identified, and its Rf is similar to that of an unidentified acyl-HSL produced by strain 2-79. To identify additional acyl-HSLs, TLC overlay assays were performed with P. aureofaciens 30-84I. With this analysis we identified an acyl-HSL with an Rf identical to that of synthetic BHSL in supernatants of SPR044 and all of the TnMod insertion mutants except SPR144 and SPR244 (Fig. 1B). Compounds similar to HHSL and OHSL were not detected in ethyl acetate extracts of supernatants from SPR044 cultures, even though the sensitivity of the test strains allowed detection of these compounds at concentrations as low as 0.5 and 50 μM, respectively.
FIG. 1.
Analysis of acyl-HSLs produced by P. chlororaphis SPR044 and TnMod insertion derivatives. Ethyl acetate extracts from cell supernatants and synthetic reference acyl-HSLs (lane C4, BHSL; lane C6, HHSL; lane C8, OHSL) were separated on C18 reversed-phase TLC plates in methanol-water (60:40). Bioassays were conducted with the reporter strains A. tumefaciens A136(pCF218)(pCF372) (A) and phzI-negative P. aureofaciens 30-84I (B). Lane 1, SPR144; lane 2, SPR244; lane 3, SPR344; lane 4, SPR544; lane 5, SPR644; lane 6, SPR744; lane 7, SPR844; lane 8, SPR044. The arrowheads indicate signals induced by acyl-HSLs from strain SPR044. The arrows indicate the direction of solvent migration.
Quantification of BHSL production by P. chlororaphis and its insertion derivatives.
After identification of acyl-HSLs produced by strain SPR044 and its TnMod insertion derivatives, well diffusion assays with P. aureofaciens 30-84 were performed to quantify BHSL production (34). Based on the colored surface areas induced by reference amounts of synthetic BHSL, the amounts produced by strain SPR044 and its TnMod insertion derivatives were determined (Table 3). SPR044 produced 16 μM BSHL in supernatants of cells grown to the stationary phase, whereas the two acyl-HSL-deficient mutants did not produce detectable amounts of this compound. The acyl-HSL production by SPR344 was 26% of the production by the wild type. Similarly, the PCN-negative mutants with TnMod insertions in the phz genes produced much smaller amounts. Strain SPR644 produced amounts of BHSL comparable to the amounts produced by the wild type, whereas the amount produced by SPR544 was 65% of the amount produced by the wild type. These results indicate that other acyl-HSL-regulated traits may be affected in a quantitative manner in the different TnMod insertion mutants.
TABLE 3.
BHSL production by P. chlororaphis SPR044 and its TnMod insertion derivatives
| Strain | Relative concn of BHSL (μM)a | % BHSL compared to SPR044 |
|---|---|---|
| SPR044 | 16.0 | 100 |
| SPR144 | 0 | |
| SPR244 | 0 | |
| SPR344 | 4.1 | 26 |
| SPR544 | 10.0 | 63 |
| SPR644 | 15.5 | 97 |
| SPR744 | 6.0 | 37 |
| SPR844 | 6.4 | 40 |
BHSL production was quantified in well diffusion assays by using cell-free supernatants of stationary-phase cultures. Representative results from four experiments are shown.
Analysis of the production of protease, phenazine, and antimicrobial secondary metabolites.
Since protease production and antibiotic production are often quorum sensing controlled and are considered to be important for root colonization, they were analyzed in SPR044 and its derivatives. Agar well diffusion assays (34) were used to assess the inhibition of C. violaceum as an example of gram-negative bacteria and the inhibition of B. subtilis as an example of gram-positive bacteria. In both assays, the inhibition caused by cell-free supernatant of SPR644 was similar to that of the wild type, whereas SPR144 and SPR244 did not produce antibiotics and the other derivatives produced reduced amounts (Table 4). The presence of proteases in cell-free supernatants was analyzed in agar supplemented with skim milk powder, and proteases were present in all strains except SPR144 and SPR244 (Table 4). Protease production could not be analyzed in a semiquantitative manner, because increasing the amount of supernatant did not lead to larger diameters of the lysis zones (data not shown). Production of antibiotics was clearly correlated with acyl-HSL production, showing that the biosynthesis is probably quorum sensing regulated. Phenazine production was scored based on colony pigmentation, and the pigments were identified by measuring the absorption spectra of chloroform extracts from culture supernatants (28). As expected, the two acyl-HSL-deficient mutants, as well as SPR744 and SPR844 carrying insertions in phzF and phzE, did not produce PCN and its precursor, phenazine-1-carboxylic acid (PCA).
TABLE 4.
Production of quorum-sensing-regulated metabolites by P. chlororaphis SPR044 and its TnMod derivativesa
| Strain | Protease productionb | PCA productionc | PCN productiond | Inhibition of gram-negative bacteriae | Inhibition of gram-positive bacteriaf |
|---|---|---|---|---|---|
| SPR044 | +g | ++ | + | ++ | ++ |
| SPR144 | − | − | − | − | − |
| SPR244 | − | − | − | − | − |
| SPR344 | + | + | +++ | + | + |
| SPR544 | + | ++ | − | + | + |
| SPR644 | + | ++ | − | ++ | ++ |
| SPR744 | + | − | − | + | + |
| SPR844 | + | − | − | + | + |
Representative data from four experiments are shown.
Protease activity was tested in well diffusion assays by using cell-free supernatants of stationary-phase cultures and agar supplemented with skim milk powder.
PCA production was measured by spectral analysis (200 to 500 nm) of chloroform extracts of cell-free supernatants.
PCN production was analyzed by visual estimation of the amount of green PCN crystals per colony.
A semiquantitative analysis was performed by using agar well diffusion assays with cell-free supernatants of stationary-phase cultures and cultures of C. violaceum.
A semiquantitative analysis was performed by using agar well diffusion assays with cell-free supernatants of stationary-phase cultures and cultures of B. subtilis.
+, present; −, absent. The number of plus signs indicates the production or inhibition compared to the production or inhibition by the wild type.
Influence of the production of acyl-HSLs and PCN on rhizosphere colonization by P. chlororaphis.
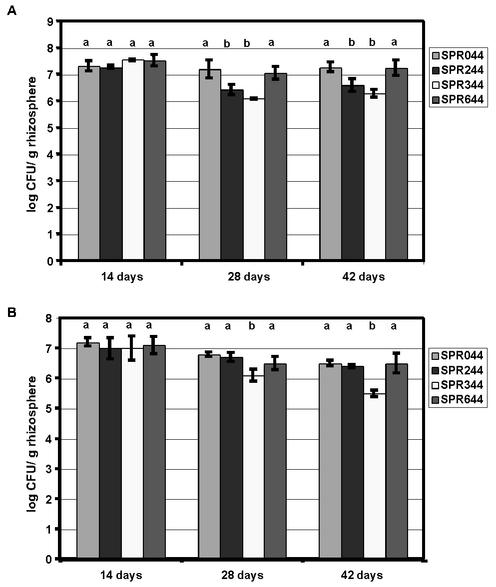
Many quorum-sensing-regulated metabolic activities are important for rhizosphere colonization. We chose one mutant from each class of plasposon derivatives to assess the effects of acyl-HSL and phenazine production on colonization. SPR244 carrying the TnMod insertion in the gene encoding the sensor GacS was chosen as an acyl-HSL-negative mutant. SPR644 was chosen for the analysis of the effects of PCN production on colonization because it produced PCA. The amounts of acyl-HSLs and antimicrobial agents produced were similar to the amounts produced by the wild type, whereas the other PCN-deficient mutants had pleiotropic defects. SPR344 was used to assess the effects of PCN overproduction. The growth rates of the TnMod insertion derivatives in LB medium were identical to the growth rate of the wild type (data not shown). This was a prerequisite for further studies since a slightly reduced growth rate was shown previously to negatively affect rhizosphere growth (43). Strains were inoculated into sterile soil microcosms, and colonization of the rhizosphere of A. thaliana was monitored. After 14 days the size of the population of SPR044 was approximately 107.3 CFU/g of root and did not differ significantly from the sizes of the populations of the variants (Fig. 2A). After 28 days, however, the size of the population of SPR044 (107.3 CFU/g of root) was similar to that of PCN-negative strain SPR644 but significantly larger than that of SPR244 or SPR344. After 42 days the sizes of the populations of the wild type and SPR644 were still similar, whereas the sizes of the populations of SPR244 and SPR344 were significantly smaller. The difference between the sizes of the populations of strains SPR344 and SPR044 increased with each harvest cycle, until after 42 days the size of the population of SPR344 was 1 order of magnitude smaller than the size of the population of the wild type. In contrast, after 42 days the size of the population of SPR244 was only fourfold smaller than the size of the population of SPR044. To determine the cultivation success, the cell numbers were analyzed by a culture-independent method in parallel as described previously (40). No divergence between the results obtained by the culture-dependent method and the results obtained by the culture-independent method was observed (data not shown) (38). This indicates that rhizosphere colonization is strongly affected by increased PCN production, whereas defects in acyl-HSL production do not have similarly pronounced effects.
FIG. 2.
Rhizosphere colonization by SPR044 and its insertion derivatives (A) and by SPR044 in cocultures with TnMod insertion derivatives (B). P. chlororaphis SPR044, its quorum-sensing-negative derivative SPR244, the PCN overproducer SPR344, or the PCN-negative mutant SPR644 was inoculated to an optical density at 600 nm of 0.01 together with 50 A. thaliana seeds into sterile soil microcosms. For coculture experiments, one mutant belonging to each class was inoculated together with the wild type and 50 A. thaliana seeds. Population sizes were determined after 14, 28, and 42 days. The mean values for six independent experiments were calculated. Means labeled with a different letter are significantly different (P = 0.05) from the mean value for SPR044 at that time point, as determined by the Kruskal-Wallis test.
Influence of acyl-HSL and PCN production on rhizosphere colonization in coinoculation experiments.
When SPR044 was coinoculated with the variants, comparable population sizes for this strain were obtained for all three combinations, and the sizes ranged from 107.3 CFU/g of root after 14 days to 106.4 CFU/g of root after 42 days (Fig. 2B). The significantly reduced growth compared to the growth observed after solitary colonization as described above may have been due to competition between the coinoculated strains for limiting nutrients. Compared to solitary growth, the difference in population sizes between SPR044 and SPR344 was even more pronounced under these conditions. The size of the population of SPR344 was almost 2 orders of magnitude smaller than the size of the population of the wild type after 42 days. The sizes of the populations of SPR244 and SPR644, however, were similar to the size of the wild-type population even after 42 days. Since in solitary growth experiments the size of the population of SPR244 was significantly less than the size of the population of the wild type, it was quite surprising that the population sizes were similar in coinoculation experiments. These results suggest that the absence of GacS per se is not detrimental to the fitness of rhizosphere-grown cells.
Acyl-HSL-deficient mutants have a significantly reduced lag phase.
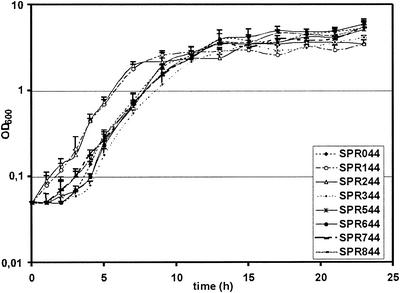
To gain further insight into the physiological basis for this phenomenon and into the relative fitness of SPR044, growth was analyzed in M9 minimal medium supplemented with 0.5% glucose. Bacterial multiplication was strongly reduced under these conditions compared to growth in LB medium, and this permitted a more detailed analysis. The mean generation time was determined in the logarithmic growth phase, and the analysis included a minimum of five independent experiments. The growth rates of the variants were similar to the growth rate of SPR044, and a typical sigmoidal growth curve was observed in all cases (Fig. 3). In the case of SPR144 and SPR244, however, the lag phase was significantly reduced to less than 1 h instead of the 3 h observed for the wild type and the other derivatives. This effect was independent of the medium used for growth of the overnight cultures (LB medium or M9 minimal medium), and it was still observed when conditioned medium was used (that is, in the presence of wild-type acyl-HSLs) (data not shown).
FIG. 3.
Growth of P. chlororaphis SPR044 and its TnMod insertion derivatives in liquid culture. Cells were grown in M9 minimal medium supplemented with 0.5% glucose under aerobic conditions at 26°C. OD600, optical density at 600 nm.
Complementation of the TnMod insertion in the gacS gene.
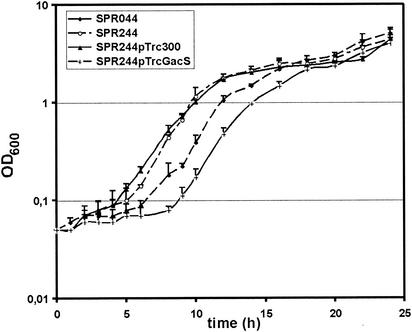
To analyze this effect in more detail, complementation of SPR244 by expression of gacS in trans was assessed. gacS was PCR amplified from SPR044 genomic DNA, and the PCR product was cloned into pTrc300 under control of the trc promoter. Transformation of SPR244 with pTrcGacS restored acyl-HSL production, as well as PCN production, even in the absence of isopropyl-β-d-thiogalactopyranoside (IPTG), which was probably due to leakiness of the promoter (data not shown). To further slow bacterial growth down and to avoid possible effects of catabolite repression, the growth of SPR044 and the growth of SPR244, as well as the growth of SPR244 carrying pTrc300 or pTrcGacS, were analyzed in M9 minimal medium supplemented with 0.5% myo-inosit (Fig. 4). Under these conditions, SPR244 had a lag phase of 3 h, compared to a lag phase of 6 h in case of the wild type, which is in accord with the results described above. Expression of gacS in trans increased the lag phase of SPR244(pTrcGacS) to 8 h, which is even longer than that of the wild type. The prolonged lag phase of SPR244(pTrcGacS) probably was a consequence of overexpression of gacS. However, the observed reduction in the lag phase (i.e., the fast restart of growth) of SPR244 in fresh medium indicated that only minor adaptations of the physiology of this organism are necessary. One possible explanation is that the cells are still in the exponential phase or do not fully enter the stationary phase.
FIG. 4.
Expression of gacS in trans compensates for the reduced lag phase of strain SPR244. P. chlororaphis SPR044 and SPR244, as well as SPR244 carrying pTrc300 or pTrcGacS, were grown in liquid M9 minimal medium supplemented with 0.5% myo-inosit under aerobic conditions at 26°C. OD600, optical density at 600 nm.
Synthesis of σs protein in SPR044 and its derivatives.
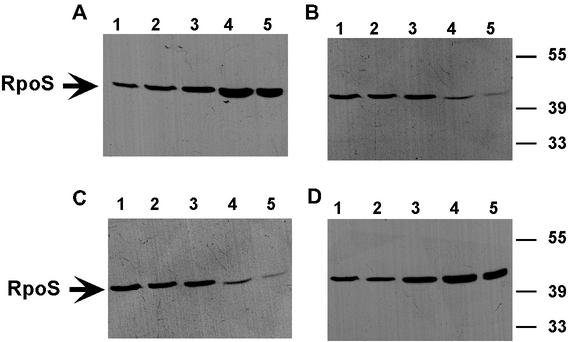
To test this hypothesis, the cellular level of the stationary-phase-specific sigma factor σs was analyzed. It is well established that σs (RpoS) is unstable in the exponential phase (half-life, 14 min) but is stabilized at the onset of the stationary phase in E. coli (19, 24). To determine the level of cell-bound σs in P. chlororaphis, samples were taken periodically from cells grown in minimal medium as described above, and cell lysates were analyzed with RpoS-specific antiserum. This analysis revealed that there were significant differences in the cellular σs levels in SPR044 and SPR244 carrying pTrc300 or pTrcGacS (Fig. 5). The RpoS-specific signal strongly increased after 18 h of growth in SPR044 and in SPR244(pTrcGacS) cultures. In SPR244 and SPR244(pTrc300) cultures, however, the RpoS-specific signal remained constant or even decreased after 24 h. This supports the assumption that the GacS-negative strains do not fully enter the stationary phase under these conditions, which may permit a faster restart of growth after inoculation into fresh media.
FIG. 5.
Levels of RpoS during growth of strain SPR044 and derivatives of this strain. Cells were grown in M9 minimal medium supplemented with 0.5% myo-inosit under aerobic conditions at 26°C, harvested after 6 h (lane 1), 10 h (lane 2), 14 h (lane 3), 18 h (lane 4), or 24 h (lane 5), and lysed in sample buffer. Equal amounts of proteins in cell lysates were subjected to SDS-PAGE and Western blot analysis with RpoS-specific antiserum. (A) SPR044; (B) SPR244; (C) SPR244(pTrc300); (D) SPR244(pTrcGacS). Molecular masses of reference proteins are given on the right.
DISCUSSION
The aim of this study was to analyze the importance of acyl-HSL and PCN production for rhizosphere colonization by P. chlororaphis SPR044. As a first step, acyl-HSLs produced by strain SPR044 and variants of this strain defective in acyl-HSL and/or PCN biosynthesis were characterized, and their regulatory functions for production of antimicrobial agents and extracellular enzymes were assessed (29). Analysis of cell-free culture supernatants revealed that SPR044 produces three acyl-HSLs, but HHSL, the main autoinducer described for P. chlororaphis PCL1391 (6), was not detected. In contrast, BHSL was identified as a major acyl-HSL in strain SPR044, but it was also present in strain PCL1391 in addition to HHSL. Although a detailed chemical analysis was beyond the scope of this work, the identities of acyl-HSLs from strain SPR044 could be partially determined by using previously described criteria (3, 41). Based on the spot-like migration during TLC (3-oxo derivatives give comet-like signals), the failure to induce the quorum-sensing system of P. aureofaciens 30-84I, and the deviation of the Rf values from the Rf values of nonsubstituted HHSL and OHSL, compounds A and B are probably C-3-hydroxy acyl-HSLs. The Rf of 0.5 observed in the case of compound A suggests that it is probably 3-hydroxy-OHSL (3). In contrast, fractionation of compound B did not allow assignment, and more extensive analytical chemistry is required to identify the structure of this compound.
Quantitative analysis of acyl-HSL production by the different TnMod insertion derivatives revealed a correlation between the amounts of autoinducer and the amounts of antimicrobial metabolites, phenazines, and extracellular proteases, showing that the regulation is similar to that of strain PCL1391 (6). Strain SPR344, isolated because of the increased amounts of green PCN crystals per colony, deviated from this scheme. The production of acyl-HSLs and antimicrobial agents was not greater, suggesting that acyl-HSL-controlled processes are not generally up-regulated. Possibly, overproduction of PCN results in down-regulation of different biochemical pathways at the expense of other metabolites. A similar effect was described in the case of a PCN-overproducing phzM mutant of P. aeruginosa, which synthesizes reduced amounts of other phenazines (28). Based on sequence analysis of the gene affected by the insertion, however, an alternative explanation is likely. The gene product exhibits 55% similarity to the protein encoded by lpsE of Sinorhizobium meliloti and 55% similarity to an RbfU-related protein from E. coli. These proteins are glycosyl transferases involved in lipopolysaccharide (LPS) core biosynthesis. Overproduction of green PCN crystals could be a consequence of the altered LPS, favoring the efflux of PCN and/or the formation of green crystals. In support of this hypothesis, LPS was isolated and, indeed, the LPS from SPR344 proved to be different from the LPS from the wild type (data not shown).
One mutant belonging to each phenotypic class (acyl-HSL negative, PCN negative, PCN overproducer) was chosen for studies of A. thaliana rhizosphere colonization. The goal of our work was to analyze subtle effects caused by alterations of such colonization factors. We therefore chose a gnotobiotic system due to its relative simplicity and reproducibility and to the fact that the results obtained with such systems mimic those obtained with natural soil (43). The absence of PCN production by strain SPR644 did not negatively affect its ability to colonize. In addition, coculture with other efficient colonizers was not affected (38). This is in contrast to previous results showing that a complete loss of phenazine production reduced the ability of P. aureofaciens to survive in soil (29). However, SPR644 lost only the ability to produce PCN, and it was still able to synthesize PCA. Conversion to the last product of the pathway is obviously not required for efficient colonization. The antifungal activity, which was reduced in a PCN-negative PCL391 derivative, was not examined in the present study (4).
The growth of SPR344 was not reduced in liquid culture under different conditions tested in this study, showing that this bacterium does not have a major fitness defect. However, its ability to survive in the A. thaliana rhizosphere was markedly reduced, which may have been due to increased PCN accumulation in the area around the cell. Diffusion of phenazines across the membrane results in acceptance of single electrons and disruption of the respiratory chain. The increased extracellular accumulation of PCN could also result in overproduction of O2− and H2O2 to levels which may exceed the capacities of the cellular superoxide dismutase (16, 17). The altered LPS composition of strain SPR344 is probably not the main reason for its reduced growth, since colonization of the higher parts of tomato roots by an O-antigenic mutant of P. fluorescens was not affected (10). In any case, this strain was unable to compete with other bacteria upon prolonged cultivation. Chin-A-Woeng et al. showed that engineering the phenazine pathway towards PCN production extends the biocontrol ability of Pseudomonas spp., which do not naturally synthesize it (5). Our results indicate that great care should be taken, because massive overproduction of PCN may negatively affect bacterial fitness.
Rhizosphere colonization by GacS-defective strain SPR244 was significantly reduced compared to rhizosphere colonization by the wild type when the two strains were inoculated separately. In contrast, after coinoculation, the population sizes were similar. To explain this observation, a detailed analysis of the growth in combination with an immunological analysis of the stationary-phase sigma factor RpoS was performed. The reduction in the lag phase observed for the quorum-sensing-negative strains was shown to be a consequence of delayed and incomplete entry into stationary-phase physiology. Our results are consistent with those of Latifi et al. (25) and You et al. (51), who showed that addition of acyl-HSLs to exponentially growing cultures leads to repression of cell growth and production of stationary-phase-specific RpoS regardless of the cell density. We thus suggest the following explanation for the finding that the size of the population of SPR244 was significantly smaller than that of the wild type in solitary growth experiments but was similar during cocultivation experiments. Due to an inability to produce quorum-sensing-regulated extracellular enzymes, the SPR244 population is dependent on readily available carbon sources. In solitary growth experiments, this leads to nutrient depletion and a decrease in the growth rate after 14 days. During cocultivation, however, extracellular enzymes from the wild type could provide access to easily degradable carbon sources, and this may relieve nutrient limitation for SPR244. Diffusion of acyl-HSLs is not likely to complement the gacS defect, because the GacA-GacS regulon affects many acyl-HSL-independent pathways (18, 33). In addition, experiments with conditioned medium also showed that there was a reduction in the lag phase and postponed entry into the stationary phase for the quorum-sensing-negative mutants (data not shown). Significantly, the postponed entry into the stationary phase may allow relatively fast growth of SPR244, whereas it may cause entry into the stationary phase by the wild type. This hypothesis is supported by the results of Bull et al., who inoculated P. fluorescens wild-type strain CHAO (99%) with the CHA89 gacA mutant (1%) and found that the proportion of gacA mutants increased to 4% before entry into the stationary phase (1). This indicates that in rich media the gacA mutants have a competitive advantage, which may be related to delayed entry into the stationary phase. Similar observations were made by Sánchez-Contreras et al., who showed that phase variation and phenotypic selection during alfalfa root colonization by P. fluorescens are often correlated with point mutations in gacA (36). In addition, Duffy and Défago observed accumulation of spontaneous gacA or gacS mutants during biocontrol inoculum production (12). The mutants may have an advantage, because they continue to grow exponentially, whereas the wild type enters the stationary phase due to the high concentration of acyl-HSLs.
Taken together, our data support the notion that the presence of the GacA-GacS system may be disadvantageous to rhizosphere bacteria under some conditions. This may be due to the fact that in addition to the regulation of colonization, this system promotes entry into the growth-retarding stationary phase. A balance of GacA-GacS-positive and -negative strains may therefore be optimal for the survival and adaptability of a community. From the applied point of view it, would be interesting to assess whether it is advantageous to use acyl-HSL-negative strains as inoculants. The insensitivity of a GacA-GacS-defective inoculum to a high concentration of acyl-HSLs may have a positive effect on the population of the inoculated strain, resulting in enhanced delivery of catabolic plasmids to an indigenous population and in stimulation of bioremediation.
Acknowledgments
We thank Clay Fuqua and Leland S. Pierson for AHL detection strains, K. Tanaka for the gift of RpoS-specific antiserum, and August Böck for continued support and discussions. Natalie Domke is thanked for construction of plasmid pTrc300, and the support of different members of the MECBAD consortium (BIO4-CT98-0099) is gratefully acknowledged.
This work was supported by the BMBF (Biomonitor/Molekulare Mikrobenökologie grant 0311940).
REFERENCES
- 1.Bull, C. T., B. Duffy, C. Voisard, G. Défago, C. Keel, and D. Haas. 2001. Characterization of spontanous gacS and gacA regulatory mutants of Pseudomonas fluorescens biocontrol strain CHAO. Antonie Leeuvenhoek 79:327-336. [DOI] [PubMed] [Google Scholar]
- 2.Bull, C. T., D. M. Weller, and L. S. Thomashow. 1991. Relationship between root colonization and suppression of Graeumannomyces graminis var. tricitii by Pseudomonas fluorescens strain 79-2. Phytopathology 81:954-959. [Google Scholar]
- 3.Cha, C., P. Gao, Y.-C. Chen, P. D. Shaw, and S. K. Farrand. 1998. Production of acyl-homoserine lactone quorum-sensing signals by Gram-negative plant associated bacteria. Mol. Plant-Microbe Interact. 11:1119-1129. [DOI] [PubMed] [Google Scholar]
- 4.Chin-A-Woeng, T. F. C., G. V. Bloemberg, I. H. M. Mulders, L. C. Dekkers, and B. J. J. Lugtenberg. 2000. Root colonization by phenazine-1-carboxamide-producing bacterium Pseudomonas chlororaphis PCL1391 is essential for biocontrol of tomato foot and root rot. Mol. Plant-Microbe Interact. 13:1340-1345. [DOI] [PubMed] [Google Scholar]
- 5.Chin-A-Woeng, T. F. C., J. E. Thomas-Oates, B. J. J. Lugtenberg, and G. V. Bloemberg. 2001. Introduction of the phzH gene of Pseudomonas chlororaphis PCL1391 extends the range of biocontrol ability of phenazine-1-carboxylic acid-producing Pseudomonas spp. strains. Mol. Plant-Microbe Interact. 14:1006-1015. [DOI] [PubMed] [Google Scholar]
- 6.Chin-A-Woeng, T. F. C., D. van den Broek, G. de Voer, K. M. G. M. van der Drift, S. Tuinman, J. E. Thomas-Oates, B. J. J. Lugtenberg, and G. V. Bloemberg. 2001. Phenazine-1-carboxamide production in the biocontrol strain Pseudomonas chlororaphis PCL1391 is regulated by multiple factors secreted into the growth medium. Mol. Plant-Microbe Interact. 14:969-979. [DOI] [PubMed] [Google Scholar]
- 7.Ching-A-Woeng, T. F. C., G. V. Bloemberg, A. J. van der Bij, K. M. G. M van der Drift, J. Schripsema, B. Kroon, R. J. Scheffer, C. Keel, P. A. H. M. Bakker, H.-V. Tichy, F. J. de Bruijn, J. E. Thomas-Oates, and B. J. J. Lugtenberg. 1998. Biocontrol by phenazine-1-carboxamide-producing Pseudomonas chlororaphis PCL1391 of tomato root rot caused by Fusarium oxysporium f. sp. radicis-lycopersici. Mol. Plant-Microbe Interact. 11:1069-1077. [Google Scholar]
- 8.Corbell, N., and J. E. Loper. 1995. A global regulator for secondary metabolite production in Pseudomonas fluorescens Pf-5. J. Bacteriol. 177:6230-6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 64:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Weger, L. A., P. A. H. M. Bakker, B. Schippers, M. C. M. van Loosdrecht, and B. J. J. Lugtenberg. 1989. Pseudomonas ssp. with mutational changes in the O-antigenic side chain of their lipopolysaccharide are affected in their ability to colonize potato roots, p. 197-202. In B. J. J. Lugtenberg (ed.), Signal molecules in plant microbe interactions, vol. H36. Springer Verlag, Berlin, Germany.
- 11.de Weger, L. A., C. I. M. van der Vlugt, A. H. M. Wijfjes, P. A. H. M. Bakker, B. Schippers, and B. J. J. Lugtenberg. 1987. Flagella of plant growth-stimulating Pseudomonas fluorescens are required for colonization of potato roots. J. Bacteriol. 169:2769-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duffy, B. K., and G. Défago. 2000. Controlling instability in gacS-gacA regulatory genes during inoculum production of Pseudomonas fluorescens biocontrol strains. Appl. Environ. Microbiol. 66:3142-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727-751. [DOI] [PubMed] [Google Scholar]
- 14.Fuqua, C., and S. C. Winans. 1996. Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J. Bacteriol. 178:435-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray, K. M. 1997. Intercellular communication and group behaviour in bacteria. Trends Microbiol. 5:184-188. [DOI] [PubMed] [Google Scholar]
- 16.Hassan, H. M., and I. Fridovich. 1980. Mechanism of the antibiotic action of pyocyanine. J. Bacteriol. 141:156-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hassett, D. J., H. P. Schweizer, and D. E. Ohman. 1995. Pseudomonas aeruginosa sodA and sodB mutants defective in manganese- and iron-cofactored superoxide dismutase activity demonstrate the importance of the iron-cofactored form in aerobic metabolism. J. Bacteriol. 177:6330-6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heeb, S., C. Blumer, and D. Haas. 2002. Regulatory RNA as mediator in GacS/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHAO. J. Bacteriol. 184:1046-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hengge-Aronis, R. 1993. Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell 72:165-168. [DOI] [PubMed] [Google Scholar]
- 20.Ka, J. O., W. E. Holben, and J. M. Tiedje. 1994. Analysis of competition in soil among 2,4-dichlorophenoxyacetic acid-degrading bacteria. Appl. Environ. Microbiol. 60:1121-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kessler, B., V. de Lorenzo, and K. N. Timmis. 1992. A general system to integrate lacZ fusions into the chromosomes of gram negative eubacteria: regulation of the Pm promotor of the TOL plasmid studied with all controlling elements in monocopy. Mol. Gen. Genet. 233:293-301. [DOI] [PubMed] [Google Scholar]
- 22.Kornitzer, D., D. Teff, S. Altuvia, and A. B. Oppenheim. 1991. Isolation, characterization and sequence of an Escherichia coli heat shock gene, htpX. J. Bacteriol. 173:2944-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 24.Lange, R., and R. Hengge-Aronis. 1994. The cellular concentration of the σs-subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 8:1600-1612. [DOI] [PubMed] [Google Scholar]
- 25.Latifi, A., M. Foglino, K. Tanaka, P. Williams, and A. Lazdunski. 1996. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhlR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol. Microbiol. 21:1137-1146. [DOI] [PubMed] [Google Scholar]
- 26.Laville, J., C. Voisard, C. Keel, M. Maurhofer, G. Défago, and D. Haas. 1992. Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc. Natl. Acad. Sci. USA 89:1562-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lugtenberg, B. J. J., L. Dekkers, and G. V. Bloemberg. 2001. Molecular determinants of rhizosphere colonization by Pseudomonas. Annu. Rev. Phytopathol. 39:461-490. [DOI] [PubMed] [Google Scholar]
- 28.Mavrodi, D. V., R. F. Bonsall, S. M. Delaney, M. J. Soule, G. Philipps, and L. S. Thomashow. 2002. Functional analysis of genes for biosynthesis of phycocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 183:6454-6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mazzola, M., R. J. Cook, L. S. Thomashow, D. M. Weller, and L. S. Pierson III. 1992. Contribution of phenazine antibiotic biosynthesis to the ecological competence of fluorescent pseudomonads in soil habitats. Appl. Environ. Microbiol. 58:2616-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moawski, B., A. Segura, and N. Ornston. 2000. Repression of Acinetobacter vanillate demethylase synthesis by VanR, a member of the GntR family of transcriptional regulators. FEMS Microbiol. Lett. 187:65-68. [DOI] [PubMed] [Google Scholar]
- 31.Natsch, A., C. Keel, H. A. Pfirter, D. Haas, and G. Défago. 1994. Contribution of the global regulator gene gacA to persistence and dissemination of Pseudomonas fluorescens biocontrol strain CHAO introduced into soil microcosms. Appl. Environ. Microbiol. 60:2553-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newman, E. I. 1978. Root microorganisms: their significance in the ecosystem. Biol. Rev. 53:511-554. [Google Scholar]
- 33.Pierson, L. S., III, D. W. Wood, E. A. Pierson, and S. T. Chancey. 1998. N-Acyl-homoserine lactone mediated gene regulation in biological control by fluorescent pseudomonads: current knowledge, and future work. Eur. J. Plant Pathol. 104:1-9. [Google Scholar]
- 34.Ravn, L., A. B. Christensen, S. Molin, M. Givskov, and L. Gram. 2001. Methods for detecting acylated homoserine lactones produced by Gram-negative bacteria and their application in studies of AHL-production kinetics. J. Microbiol. Methods 44:239-251. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 36.Sánchez-Contreras, M., M. Martín, M. Villacieros, F. O'Gara, I. Bonilla, and R. Rivilla. 2002. Phenotypic selection and phase variation occur during alfalfa root colonization by Pseudomonas fluorescens F113. J. Bacteriol. 184:1587-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saxena, I. M., R. M. Brown, M. Fevre, A. G. Roberto, and B. Henrissat. 1995. Multidomain architecture of β-glycosyltransferases: implications for mechanism of action. J. Bacteriol. 177:1419-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt-Eisenlohr, H., and C. Baron. 2003. The competitiveness of Pseudomonas chlororaphis carrying pJP4 is reduced in the Arabidopsis thaliana rhizosphere. Appl. Environ. Microbiol. 69:1827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt-Eisenlohr, H., N. Domke, and C. Baron. 1999. TraC of IncN plasmid pKM101 associates with membranes and extracellular high molecular weight structures in Escherichia coli. J. Bacteriol. 181:5563-5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmidt-Eisenlohr, H., M. Rittig, and C. Baron. 2001. Biomonitoring of pJP4-carrying Pseudomonas chlororaphis with Trb protein specific antisera. Environ. Microbiol. 3:720-730. [DOI] [PubMed] [Google Scholar]
- 41.Shaw, P. D., P. Gao, S. L. Daly, C. Cha, J. Cronan, Jr., K. L. Rinehart, and S. K. Farrand. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. USA 94:6036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simons, M., H. P. Permentier, L. A. de Weger, C. A. Wijffelman, and B. J. J. Lugtenberg. 1997. Amino acid synthesis is necessary for tomato root colonization by Pseudomonas fluorescens strain WCS365. Mol. Plant-Microbe Interact. 10:102-106. [Google Scholar]
- 43.Simons, M., A. van der Bij, I. Brand, L. A. de Weger, C. A. Wijffelman, and B. J. J. Lugtenberg. 1996. Gnotobiotic systems for studying rhizosphere colonization by plant growth-promoting Pseudomonas bacteria. Mol. Plant-Microbe Interact. 9:600-607. [DOI] [PubMed] [Google Scholar]
- 44.Stover, C. K., X.-Q. T. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowali, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Wetsbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Lerbig, R. M. Lim, K. A. Smith, D. H. Spencer, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomona aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 45.Swift, S., J. P. Throup, P. Williams, G. P. C. Salmond, and G. S. A. B. Stewart. 1996. Quorum sensing: a population density component in the determination of bacterial phenotype. Trends Biochem. Sci. 21:214-219. [PubMed] [Google Scholar]
- 46.Timms-Wilson, T. M., R. J. Ellis, A. Renwick, D. J. Rhodes, D. V. Mavrodi, D. M. Weller, L. S. Thomashow, and M. J. Bailey. 2000. Chromosomal insertion of phenazine-1-carboxylic acid biosynthetic pathway enhances efficacy of damping-off disease control by Pseudomonas fluorescens. Mol. Plant-Microbe Interact. 13:1293-1300. [DOI] [PubMed] [Google Scholar]
- 47.Weller, D. M. 1988. Biological control of soilborne plant pathogens in the rhizosphere with bacteria. Annu. Rev. Phytopathol. 26:379-407. [Google Scholar]
- 48.Wood, D. W., F. Gong, M. M. Daykin, P. Williams, and L. S. Pierson. 1997. N-Acyl-homoserine lactone-mediated regulation of phenazine gene expression by Pseudomonas aureofaciens 30-84 in the wheat rhizosphere. J. Bacteriol. 179:7663-7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wood, D. W., and L. S. Pierson. 1996. The phzI gene of Pseudomonas aureofaciens 30-84 is responsible for the production of a diffusible signal required for phenazine antibiotic production. Gene 168:49-53. [DOI] [PubMed] [Google Scholar]
- 50.Yanisch-Perron, C., J. Viera, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp18 and pUC18 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 51.You, Z., J. Fukushima, K. Tanaka, S. Kawamoto, and K. Okuda. 1998. Induction of stationary growth phase in Pseudomonas aeruginosa by N-acylhomoserine lactone. FEMS Microbiol. Lett. 164:99-106. [DOI] [PubMed] [Google Scholar]
- 52.Zhu, J., J. W. Bearber, M. I. Moré, C. Fuqua, A. Eberhard, and S. C. Winans. 1998. Analogs of the autoinducer 3-oxooctanoyl-homoserine lactone strongly inhibit activity of the TraR protein of Agrobacterium tumefaciens. J. Bacteriol. 180:5398-5405. [DOI] [PMC free article] [PubMed] [Google Scholar]