Abstract
The effect of the large catabolic IncP plasmid pJP4 on the competitiveness of Pseudomonas chlororaphis SPR044 and on its derivatives SPR244 (GacS deficient), SPR344 (phenazine-1-carboxamide overproducer), and SPR644 (phenazine-1-carboxamide deficient) in the Arabidopsis thaliana rhizosphere was assessed. Solitary rhizosphere colonization by the wild type, SPR244, and SPR644 was not affected by the plasmid. The size of the population of SPR344 carrying pJP4, however, was significantly reduced compared to the size of the population of the plasmid-free derivative. The abiotic stress caused by phenazine-1-carboxamide overproduction probably resulted in a selective disadvantage for cells carrying pJP4. Next, the effect of biotic stress caused by coinoculation of other bacteria was analyzed. Cells carrying pJP4 had a selective disadvantage compared to plasmid-free cells in the presence of the efficient colonizer Pseudomonas fluorescens WCS417r. This effect was not observed after coinoculation with a variety of other bacteria, and it was independent of quorum sensing and phenazine-1-carboxamide production. Thus, the presence of large catabolic plasmids imposes a detectable metabolic burden in the presence of biotic stress. Plasmid transfer in the A. thaliana rhizosphere from P. chlororaphis and its derivatives to Ralstonia eutropha was determined by using culture-dependent and culture-independent techniques. With the cultivation-independent technique we detected a significantly higher portion of exconjugants, but pJP4 transfer was independent of the quorum-sensing system and of phenazine-1-carboxamide production.
Intensive industrial and agricultural exploitation and the use of pesticides and fertilizers lead to the contamination of soils with xenobiotic compounds. Diverse microbial communities present in terrestrial habitats metabolize a wide range of such chemicals (12, 13, 22). However, many synthetic compounds persist in the environment for extended periods, because the catabolic capacity is not present or the degradative population is too small or not active enough to remove them. Introduction of plasmid-borne catabolic genes into contaminated sites is a promising strategy for enhancing the breakdown of xenobiotic compounds. The introduced microorganisms or indigenous organisms could degrade a pollutant, but the latter possibility requires efficient plasmid transfer (4, 5, 7, 18, 29, 30).
A model compound for the study of degradation by plasmid-encoded catabolic functions is the herbicide 2,4-dichlorophenoxyacetic acid (2,4-D), which is a chlorinated phenol that functions as an auxin analog and is used for weed control. Most of the 2,4-D degradative pathway is encoded by plasmid pJP4 isolated from Ralstonia eutropha JMP134 (8, 9). This 80-kb broad-host-range self-transmissible plasmid belongs to the IncPβ group and determines resistance to mercury ions and phenyl mercury acetate and partial catabolism of 2,4-D, 2-methyl-4-chlorophenoxic-acid, and 3-chlorobenzoate (10). Complete degradation requires the production of the enzyme maleylacetate reductase by a host, such as R. eutropha.
Transfer of pJP4 to indigenous soil organisms has been reported previously, but the transfer rates were low, so that bioremediation strategies based on plasmid spread seemed to have limited potential (17). Introduction of nutrients or of high concentrations of 2,4-D enhanced plasmid transfer (4, 14, 18), but such procedures are hardly feasible under field conditions or would result in additional cost. The reasons for the limited spread of plasmids are largely unknown, but they may be related to slow growth or the stationary-phase physiology of the introduced bacteria. Therefore, we analyzed plasmid transfer in the rhizosphere, which is an environment with higher concentrations of nutrients than bulk soil (3).
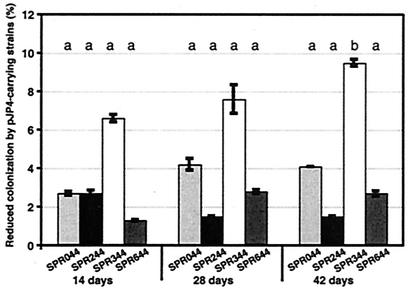
Limited work has been done to systematically assess the influence of the additional metabolic load imposed by large catabolic plasmids on the competitiveness of strains introduced into the environment. To address this question, pJP4 was introduced into SPR044 and derivatives of this strain affected in acyl-homoserine lactone (acyl-HSL) or phenazine-1-carboxamide (PCN) production by conjugative transfer from Escherichia coli (25). The presence of pJP4 was confirmed by reisolation of the plasmid and restriction analysis. The growth rates of the transformants were comparable to those of the plasmid-free strains in liquid Luria-Bertani (LB) and minimal media (data not shown). To assess the effects of pJP4 on the ability of P. chlororaphis to colonize the Arabidopsis thaliana rhizosphere, autoclaved soil microcosms containing 25 g (dry weight) of soil (Floraton 1; Florgard) were amended with 100 ml of water containing a culture in the logarithmic growth phase. Fifty A. thaliana seeds were surface sterilized by immersion in bleach and distributed randomly on the soil surface (25). Ten plants were harvested randomly after 14, 28, and 42 days. After removal of unattached soil, the root weight was determined; this was followed by transfer into flasks containing 10 ml of phosphate-buffered saline (pH 7.2) and 0.5 g of glass beads (diameter, 0.25 to 0.3 mm; B. Braun Biotech International) and vigorous shaking for 20 min. Serial dilutions were then plated on LB medium. After 14 days, the sizes of the populations of plasmid-free strain SPR044 and strain SPR244 were approximately 107.3 CFU/g of roots and the sizes of the populations of SPR344 and SPR644 were approximately 107.5 CFU/g of roots. In comparison, the sizes of the populations of the strains carrying pJP4 were reduced between 1.3 and 6.6% and thus did not differ significantly (P > 0.05) in all cases (Fig. 1). Similarly, after 28 days we observed reductions in the sizes of the populations of the plasmid-carrying cells ranging from 1.5% in case of SPR244 to 7.6% in the case of SPR344. Whereas the reduction in the size of the SPR344 population was obvious, it was not statistically significant according to the Mann-Whitney U test (SPSS10 for Macintosh). However, after prolonged incubation for 42 days, the sizes of the populations of the pJP4-carrying cells of SPR044, SPR244, and SPR644 were reduced by 4.1, 1.5, and 2.7%, respectively, compared to the sizes of the populations of the plasmid-free strains. These reductions were significantly less pronounced than the statistically significant reduction of 9.5% observed in case of SPR344, which corresponded to a difference of almost 2 orders of magnitude between the plasmid-free strain and the pJP4-carrying strain (Fig. 1). These results show that pJP4 does not have a significant effect on rhizosphere colonization by the wild type and the acyl-HSL- and PCN-deficient mutants. In contrast, a negative effect on the PCN-overproducing strain was detected. Since the changes in lipopolysaccharide composition alone were probably not responsible for the effects observed in case of SPR344 (6, 24), a likely interpretation is that the abiotic stress caused by PCN overproduction resulted in the reduced competitiveness of SPR344 carrying pJP4.
FIG. 1.
Rhizosphere colonization by pJP4-carrying SPR044 and its derivatives. The average log CFU values for six independent cultivation experiments with pJP4-carrying cells were determined at different times after sowing and were divided by the corresponding values for parallel cultures of plasmid-free cells. The reduction in colonization efficiency is the difference from a ratio of 1:1. Differences between means for the same cycle labeled with the same letter are not statistically significant.
In contrast to the results described above, reduced fitness of Pseudomonas fluorescens carrying pQBR103 was observed during initial colonization of the sugar beet phytosphere under greenhouse and field conditions but not after further development of the plants (15). Since sterile microcosms and fast-growing A. thaliana were used in the present study, it is possible that such a period of reduced fitness had already passed prior to the first sampling time. Alternatively, the difference may have been due to the plant-bacterium interaction and plasmid used, so that the experiments cannot be compared directly.
As a next step, we analyzed rhizosphere colonization by strains carrying pJP4 in the presence of other microorganisms, which more closely reflected a natural ecosystem. Strain SPR344 was not investigated further, because the presence of pJP4 per se conferred a selective disadvantage. For identification of TnMod insertion derivatives recovered from coinoculation experiments, LB agar was supplemented with kanamycin (100 μg/ml). The typical colony pigmentation was exploited for identification of Pseudomonas aureofaciens (orange) and P. chlororaphis (yellow with green PCN crystals). When strains SPR044, SPR244, and SPR644 were inoculated simultaneously with P. aureofaciens 30-84 or with a mixture of P. aureofaciens 30-84, R. eutropha JMP134, Pantoea agglomerans, and Stenotrophomonas maltophilia (Table 1), no significant difference was detected between the relative sizes of the populations of pJP4-containing and plasmid-free bacteria after 14, 28, or 42 days (data not shown). In contrast to these observations, a rapid decline in the size of the population of pJP4-carrying strain P. putida UWC3 in soil was reported (4), but this may have been due to the different experimental setup used. First, the stationary-phase cultures used as the inoculum were probably metabolically less active, and even 2,4-D-amended soil did not support bacterial growth as efficiently as the rhizosphere. Second, the experiments were performed in nonsterile soil, and biotic stress is generally higher under these conditions.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype or descriptiona | Reference |
|---|---|---|
| Strains | ||
| Pseudomonas chlororaphis SPR044 | Natural isolate | 25 |
| SPR244 | SPR044, gacS::TnMod | 24 |
| SPR344 | SPR044, TnMod insertion in gene for putative glycosyl transferase | 24 |
| SPR644 | SPR044, htpX::TnMod | 24 |
| Pseudomonas aureofaciens 30-84 | Natural isolate, phz+ HSL+ | 31 |
| Pantoea agglomerans | Natural isolate | This study |
| Stenotrophomonas maltophilia | Natural isolate | This study |
| Ralstonia eutropha JMP134 | Derivative of JMP134 cured of pJP4 | DSM5450 |
| Pseudomonas fluorescens WCS417r | Natural isolate | 19 |
| Plasmid | ||
| pJP4 | IncPβ plasmid, tfd+ 3-CBA+ Hgr | 8 |
phz+, genes coding for enzymes of phenazine biosynthesis are present; HSL+, acylated homoserine lactones are produced; tfd+genes coding for enzymes for partial degradation of 2,4-D are present; 3-CBA+, genes coding for enzymes for partial degradation of 3-chlorobenzoic acid are present; Hgr, determinants for mercury chloride resistance are present.
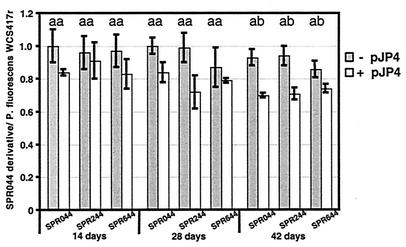
The results of coinoculation experiments with P. fluorescens WCS417r, however, were quite intriguing. This strain was described as a particularly well-adapted colonizer of the A. thaliana rhizosphere (19), and it could be discriminated from other bacteria by its ability to survive on LB agar supplemented with multiple antibiotics (13 μg of chloramphenicol per ml, 100 μg of carbenicillin per ml, 50 μg of cycloheximide per ml, and 150 μg of rifampin per ml). After 14 days of cocultivation, the ratios of plasmid-free strains SPR044, SPR244, and SPR644 to P. fluorescens WCS417r ranged from 0.96 to 1.0, showing similar initial colonization abilities (Fig. 2). The ratios of pJP4-carrying strains SPR044, SPR244, and SPR644 to P. fluorescens ranged from 0.89 to 0.93, reflecting slight reductions compared to the plasmid-free strains. Similar ratios were determined after 28 days of cocultivation. After 42 days, however, significant differences between the ratios of plasmid-carrying and plasmid-free derivatives to P. fluorescens WCS417r were observed (P < 0.05). The ratio of SPR044 to the coinoculated strain was 0.93, compared to a ratio of 0.7 in the case of pJP4-carrying SPR044. Similarly, the values for SPR244 were 0.94 and 0.71, respectively, and for SPR644 the values were 0.86 and 0.76, respectively. P. fluorescens WCS417r is known to be well adapted to the A. thaliana rhizosphere, and cocultivation may therefore impose biotic stress similar to that in a natural environment. Nevertheless, the mechanism is not immediately apparent, because all the organisms tested had comparable growth rates in liquid media and did not inhibit each other. P. fluorescens WCS417r is a biocontrol agent and has been shown to trigger induced systemic resistance against the leaf pathogen Pseudomonas syringae pv. tomato in A. thaliana (20). Chemicals released from the plant may impose stress on rhizosphere bacteria, and this may result in a competitive disadvantage for SPR044 and derivatives of this strain carrying pJP4, similar to the effect of increased PCN production on SPR344. Alternatively, biocontrol agent WCS417r and A. thaliana plants may live in a mutualistic association, and rhizosphere colonization could be stimulated by the plant. Streit and Phillips (27) showed that vitamin cofactors derived from the plant stimulated root colonization by Sinorhizobium meliloti, and a similar explanation may be applicable here. Therefore, WCS417r could have a slight competitive advantage over P. chlororaphis, which may become apparent upon nutrient limitation after prolonged rhizosphere colonization. Taken together, these studies revealed that the additional metabolic load of pJP4 per se does not necessarily affect rhizosphere colonization efficiency. In addition, neither the absence of acyl-HSL production nor the absence of PCN production resulted in a measurable difference. However, in the presence of additional abiotic or biotic stress the reduced fitness of plasmid-carrying cells compared to plasmid-free cells was detectable.
FIG. 2.
Rhizosphere colonization by pJP4-carrying SPR044 and its derivatives in competition with other bacteria. The log CFU values for SPR044 and its derivatives retrieved from the rhizosphere at different times divided by the log CFU values for the coinoculated strain are shown. The results are the averages for at least four coinoculation experiments with P. fluorescens WCS417r. Differences between means for the same cycle labeled with the same letter are not statistically significant.
Finally, it was interesting to analyze the effects of production of acyl-HSLs on plasmid transfer. Acyl-HSLs act as signal molecules for cell density in many gram-negative bacteria, a phenomenon known as quorum sensing (11). Conjugative transfer of the Ti plasmid of Agrobacterium tumefaciens is dependent on a quorum-sensing system, and plant-produced metabolites serve as additional triggers (21). Whereas the transfer machinery of the A. tumefaciens Ti plasmid and the transfer machinery of the IncP plasmids are closely related (1), virtually no information is available on the effects of quorum sensing on the transfer of broad-host-range plasmids. As shown above, plasmid pJP4 affects host physiology, but the effects of host physiology on plasmid replication and propagation are largely unknown (2). Therefore, the effect of quorum sensing on pJP4 transfer from SPR044 was analyzed next.
The donor strains P. chlororaphis SPR044 and its derivatives carrying pJP4 were coinoculated into the rhizosphere with the recipient R. eutropha, and transfer efficiencies were determined after 14, 24, and 42 days. R. eutropha colonies were discriminated from P. chlororaphis cells on the basis of their white pigmentation and their diameters, which were 0.5 to 1 mm, compared to a diameter of at least 2 mm for colonies of P. chlororaphis (which were yellow with green PCN crystals). Exconjugants (R. eutropha pJP4) were selected on M9 minimal medium supplemented with 2,4-D (30 μg/ml) as a carbon source, because the plasmid-free recipient R. eutropha and the donor P. chlororaphis(pJP4) did not grow on this medium. Since the donor grew on M9 minimal medium supplemented with 0.5% glucose or myo-inosit, we concluded that in contrast to R. eutropha, this strain probably lacks chromosomally encoded maleylacetate reductase, which is required for complete degradation of 2,4-D (14). The presence of the plasmid in exconjugants from selective agar plates was controlled by random isolation of clones, followed by restriction analysis (23) and plating on HgCl2-containing medium. In addition, the presence of the putative minor component of pJP4-determined pili (TrbF) was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis of cell lysates followed by Western blotting and detection with TrbF-specific antiserum (25). No exconjugants were detected when dilutions from control microcosms inoculated with SPR044 donors or R. eutropha only were plated on selective media. By using the plate count method, it was determined that P. chlororaphis donated pJP4 with an efficiency of 2% to R. eutropha exconjugants. After further cocultivation for 28 or 42 days, more exconjugants (up to 4% of the recipients) were detected. However, there was not a significant difference between SPR044 and its derivatives, showing that the quorum-sensing system does not affect pJP4 transfer efficiency (Table 2). The observed transfer rates were 10-fold higher than the rates observed in the case of IncPα plasmid RP4 transfer between pseudomonads in sterile loam soil (28). This was probably a consequence of the prolonged incubation time and of the nutrients released by exudation from the plant roots. In addition, the nutrients may have attracted bacterial cells, so that the donor and recipient were in close proximity on the root surface, which is a prerequisite for plasmid transfer (26).
TABLE 2.
Conjugative transfer of plasmid pJP4 between P. chlororaphis and of derivatives of this organism as donors and R. eutropha as the recipient in the A. thaliana rhizosphere
| Strain | Incubation time (days) | No. of bacteria/g of rootsa
|
Transfer efficiency (%)b | ||
|---|---|---|---|---|---|
| Donors (108 CFU) | Recipients (107 CFU) | Exconjugants (106 CFU) | |||
| SPR044 | 14 | 3.8 | 5.5 | 8.7 | 2 |
| 28 | 2.2 | 6.9 | 9.1 | 4 | |
| 42 | 1.4 | 4.5 | 5.8 | 4 | |
| SPR244 | 14 | 2.8 | 5.1 | 6.7 | 2 |
| 28 | 2.6 | 8.4 | 8.4 | 3 | |
| 42 | 2.6 | 8.8 | 9.5 | 4 | |
| SPR644 | 14 | 1.8 | 2.2 | 4.2 | 2 |
| 28 | 1.3 | 5.0 | 3.6 | 3 | |
| 42 | 1.5 | 8.6 | 3.6 | 3 | |
Average values for six replicates.
Ratio of exconjugants to donor cells.
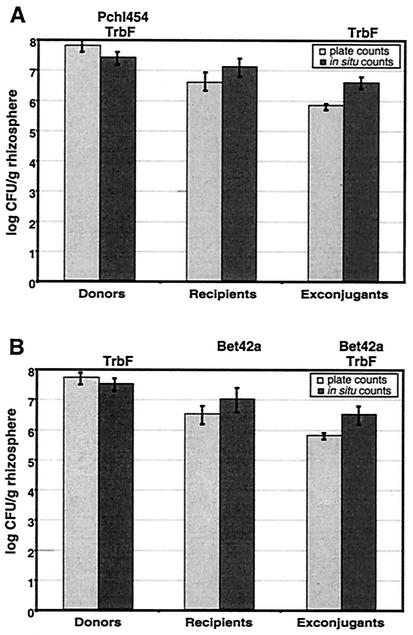
To avoid artifacts due to plate mating of the bacteria after retrieval from the rhizosphere or due to reduced viability of plasmid-carrying cells, the number of exconjugants was analyzed by a previously described culture-independent method in parallel (25). Briefly, the samples were simultaneously analyzed in situ by 4′,6′-diamidino-2-phenylindole (DAPI) and immunofluorescence staining with an antiserum specific for TrbF, the putative minor component of pJP4-encoded pili. In addition, hybridization with fluorescently labeled oligonucleotides Pchl454 (detecting P. chlororaphis) or Bet42a (16) (detecting R. eutropha) allowed differentiation of donor and exconjugant. Cells retrieved from the rhizosphere were fixed by addition of paraformaldehyde to a final concentration of 4% (vol/vol) and incubation for 30 min at room temperature. The suspension was filtered through black polycarbonate membrane filters (pore size, 0.25 μm; Millipore) to concentrate the extracted bacteria. For further analysis by immunofluorescence microscopy, the membrane filters were transferred upside down onto polylysine-coated cover slides, and this was followed by rehydration of the filters and air drying. Microscopic images were recorded by using a Spot-RT camera (Visitron) with a Zeiss Axioplan microscope and Spot (version 3.0) and IPLab (version 3.5) software. Zeiss filter sets Nr. 1 and Nr. 9 and the HQ filter set for Cy3, tetramethyl rhodamine isothiocyanate, and Alexa 546 (AHF Analysentechnik) were used for analysis of DAPI-, Oregon green-, and Cy3-stained cells, respectively. Enumeration of donors, recipients, and exconjugants was carried out visually by using screen images of the data acquired from the microscope analysis. Mean values for DAPI counts were transformed into total cell numbers by multiplication with the magnification factors. As determined by the cultivation-independent method, the number of exconjugants detected after 42 days was approximately eightfold higher than the number detected on selective agar plates (Fig. 3). This was obviously an inherent feature of the exconjugants in the gene transfer experiment, because there was no difference between the results of cultivation-dependent and in situ determinations for pJP4-carrying donors (Fig. 3A) and plasmid-free recipients (Fig. 3B). Similar results were obtained independent of the oligonucleotide probe used for hybridization. Therefore, the effect probably was not due to problems with penetration of oligonucleotide Pchl454 into the fixed donor cells, which would have resulted in overestimation of the number of recipients. As an additional control, pJP4-carrying R. eutropha cells grown in the rhizosphere alone were quantified, and there was no difference between the results of the culture-dependent method and the results of the culture-independent method (data not shown). One interpretation of the discrepancy described above is that expression of the tfd genes encoding the enzymes for degradation of 2,4-D in the recipient is delayed compared to expression of the genes in the trb operon. As a consequence, the in situ method detected the presence of pJP4-encoded Trb proteins in a higher proportion of the recipient cells, because not all of the cells were able to grow on selective minimal medium with 2,4-D. Similar conclusions could be drawn from a previous analysis of the transfer of 2,4-D degradative genes to soil populations of rhizobia. Transconjugants were selected based on the expression of plasmid-borne mercury resistance genes, and the genes required for 2,4-D degradation were not expressed (14). Thus, in situ detection of exconjugants with Trb protein-specific antisera is obviously superior to classical plate count methods for quantification of exconjugants.
FIG. 3.
Comparison of culture-dependent and culture-independent quantification of gene transfer in the A. thaliana rhizosphere. Donor strain SPR044(pJP4) and the R. eutropha recipient were coinoculated into the rhizosphere, and exconjugants were retrieved after 42 days. The numbers of donors, recipients, and exconjugants were determined by plating on rich medium and minimal medium with 2,4-D for selection of R. eutropha(pJP4). Alternatively, cells were detected in situ by a combination of fluorescent in situ hybridization with P. chlororaphis-specific oligonucleotide Pchl454 (A) or R. eutropha-specific oligonucleotide Bet42a (B) and TrbF-specific immunofluorescence staining. The data are the results of six independent experiments, and the mean deviations are indicated.
The results of this study demonstrate that quorum sensing does not affect pJP4 transfer efficiency. The independence of pJP4 transfer from quorum sensing is in stark contrast to the transfer of the Ti plasmid, in spite of the resemblance of the transfer regions. It is therefore tempting to speculate that conjugative transfer of other broad-host-range plasmids, which do not possess a plasmid-encoded quorum-sensing system, may also not be regulated by the quorum-sensing system of the host. Analysis of the regulation of plasmid transfer is of interest for other studies, because efficient plasmid transfer is required for environmental spread of degradative capabilities. Application of plasmid donor strains via the rhizosphere of plants may enhance the introduction of catabolic plasmids, thus supplementing current phytoremediation strategies for cleanup of polluted ecosystems.
Acknowledgments
We are indebted to Corné Pieterse for donation of P. fluorescens WCS417r and to August Böck for continued support and discussions. The support of different members of the MECBAD consortium (BIO4-CT98-0099) is gratefully acknowledged.
This work was supported by the BMBF (Biomonitor/Molekulare Mikrobenökologie grant 0311940).
REFERENCES
- 1.Alt-Mörbe, J., J. L. Stryker, C. Fuqua, P.-L. Li, S. K. Farrand, and S. C. Winans. 1996. The conjugal transfer system of Agrobacterium tumefaciens octopine-type Ti plasmid is closely related to the transfer system of an IncP plasmid and distantly related to Ti plasmid vir genes. J. Bacteriol. 178:4248-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Camacho, E. M., and J. Casadesus. 2002. Conjugal transfer of the virulence plasmid of Salmonella enterica is regulated by the leucine-responsive regulatory protein and DNA adenine methylation. Mol. Microbiol. 44:1589-1598. [DOI] [PubMed] [Google Scholar]
- 3.Curl, E. A., and B. Truelove. 1986. The rhizosphere. Springer Verlag, Berlin, Germany.
- 4.Dejonghe, W., J. Goris, S. el Fantroussi, M. Höfte, P. de Vos, W. Verstraete, and E. M. Top. 2000. Effect of dissemination of 2,4-dichlorophenoxyacetic acid (2,4-D) degradation plasmids on 2,4-D degradation and on bacterial community structure in two different soil horizons. Appl. Environ. Microbiol. 66:3297-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Rore, H., K. Demolder, K. de Wilde, E. Top, F. Houwen, and W. Verstraete. 1994. Transfer of the catabolic plasmid RP4::Tn4371 to indigenous soil bacteria and its effect on respiration and biphenyl breakdown. FEMS Microbiol. Ecol. 15:71-78. [Google Scholar]
- 6.de Weger, L. A., P. A. H. M. Bakker, B. Schippers, M. C. M. van Loosdrecht, and B. J. J. Lugtenberg. 1989. Pseudomonas ssp. with mutational changes in the O-antigenic side chain of their lipopolysaccharide are affected in their ability to colonize potato roots, p. 197-202. In B. J. J. Lugtenberg (ed.), Signal molecules in plant microbe interactions, vol. H36. Springer Verlag, Berlin, Germany.
- 7.DiGiovanni, G. D., J. W. Neilson, I. L. Pepper, and N. A. Sinclair. 1996. Gene transfer of Alcaligenes eutrophus JMP134 plasmid pJP4 to indigenous soil recipients. Appl. Environ. Microbiol. 62:2521-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Don, R. H., and J. M. Pemberton. 1985. Genetic and physical map of the 2,4-dichlorophenoxyacetic acid-degradative plasmid pJP4. J. Bacteriol. 161:466-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Don, R. H., and J. M. Pemberton. 1981. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J. Bacteriol. 145:681-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Don, R. H., A. J. Weightman, H.-J. Knackmuss, and K. N. Timmis. 1985. Transposon mutagenesis and cloning analysis of the pathways for degradation of 2,4-dichlorophenoxyacetic acid and 3-chlorbenzoate in Alcaligenes eutrophus JMP134(pJP4). J. Bacteriol. 161:85-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727-751. [DOI] [PubMed] [Google Scholar]
- 12.Ghosal, D., I. S. You, D. K. Chatterjee, and A. M. Chakrabarty. 1985. Microbial degradation of halogenated compounds. Science 228:135-142. [DOI] [PubMed] [Google Scholar]
- 13.Ka, J. O., W. E. Holben, and J. M. Tiedje. 1994. Analysis of competition in soil among 2,4-dichlorophenoxyacetic acid-degrading bacteria. Appl. Environ. Microbiol. 60:1121-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kinkle, B. K., M. J. Sadowsky, E. L. Schmidt, and W. C. Koskinen. 1993. Plasmids pJP4 and r68.45 can be transferred between populations of bradyrhizobia in nonsterile soil. Appl. Environ. Microbiol. 59:1762-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lilley, A. K., and M. Bailey. 1997. The acquisition of indigenous plasmids by genetically marked pseudomonad population colonizing the sugar beet phytosphere is related to local environmental conditions. Appl. Envrion. Microbiol. 63:1577-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K.-H. Schleifer. 1992. Phylogenetic oligonucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 17.Neilson, J. W., K. L. Josephson, I. L. Pepper, R. B. Arnold, G. D. D. DiGiovanni, and N. A. Sinclair. 1994. Frequency of horizontal gene transfer of a large catabolic plasmid (pJP4) in soil. Appl. Environ. Microbiol. 60:4053-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newby, D. T., K. L. Josephson, and I. L. Pepper. 2000. Detection and characterization of plasmid pJP4 transfer to indigenous soil bacteria. Appl. Environ. Microbiol. 66:290-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pieterse, C. M. J., S. C. M. van Wees, E. Hoffland, J. A. van Pelt, and L. C. van Loon. 1996. Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell 8:1225-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pieterse, C. M. J., S. C. M. van Wees, J. A. van Pelt, M. Knoester, R. Laan, H. Gerrits, P. J. Weisbeek, and L. C. van Loon. 1998. A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10:1571-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piper, K. R., S. Beck von Bodman, and S. K. Farrand. 1993. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature 362:448-450. [DOI] [PubMed] [Google Scholar]
- 22.Ramos, J. L., E. Diaz, D. Dowling, V. de Lorenzo, S. Molin, F. O'Gara, C. Ramos, and K. N. Timmis. 1994. The behavior of bacteria designed for biodegradation. Bio/Technology 12:1349-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 24.Schmidt-Eisenlohr, H., A. Gast, and C. Baron. 2003. Inactivation of gacS does not affect the competitiveness of Pseudomonas chlororaphis in the Arabidopsis thaliana rhizosphere. Appl. Environ. Microbiol. 69:1817-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt-Eisenlohr, H., M. Rittig, and C. Baron. 2001. Biomonitoring of pJP4-carrying Pseudomonas chlororaphis with Trb protein specific antisera. Environ. Microbiol. 3:720-730. [DOI] [PubMed] [Google Scholar]
- 26.Smit, E., J. D. van Elsas, J. A. van Veen, and W. M. de Vos. 1991. Detection of plasmid transfer from Pseudomonas fluorescens to indigenous bacteria in soil by using bacteriophage R2f for donor counterselection. Appl. Environ. Microbiol. 57:3482-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Streit, W. R., and D. A. Phillips. 1997. A biotin-regulated locus, bioS, in a possible survival operon of Rhizobium meliloti. Mol. Plant-Microbe Interact. 10:933-937. [DOI] [PubMed] [Google Scholar]
- 28.Trevors, J. T., and J. D. van Elsas. 1997. Quantification of gene transfer in soil and the rhizosphere, p. 500-508. In C. J. Hurst, G. R. Knudsen, M. J. McInerney, L. D. Stetzenbach, and M. V. Walter (ed.), Manual of environmental microbiology. ASM Press, Washington, D.C.
- 29.van der Bij, A. J., L. A. de Weger, W. T. Tucker, and B. J. J. Lugtenberg. 1996. Plasmid stability in Pseudomonas fluorescens in the rhizosphere. Appl. Environ. Microbiol. 62:1076-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Veen, J. A., L. S. van Overbeek, and J. D. van Elsas. 1997. Fate and activity of microorganisms introduced into soil. Microbiol. Mol. Biol. Rev. 61:121-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wood, D. W., and L. S. Pierson. 1996. The phzI gene of Pseudomonas aureofaciens 30-84 is responsible for the production of a diffusible signal required for phenazine antibiotic production. Gene 168:49-53. [DOI] [PubMed] [Google Scholar]