Abstract
Anaerobic arginine catabolism in Saccharomyces cerevisiae was genetically modified to allow assimilation of all four rather than just three of the nitrogen atoms in arginine. This was accomplished by bypassing normal formation of proline, an unusable nitrogen source in the absence of oxygen, and causing formation of glutamate instead. A pro3 ure2 strain expressing a PGK1 promoter-driven PUT2 allele encoding Δ1-pyrroline-5-carboxylate dehydrogenase lacking a mitochondrial targeting sequence produced significant cytoplasmic activity, accumulated twice as much intracellular glutamate, and produced twice as much cell mass as the parent when grown anaerobically on limiting arginine as sole nitrogen source.
Stuck wine fermentations are a recurring problem in the wine industry, and inadequate grape nitrogen is commonly believed to be a major cause. Although arginine is often the most abundant amino acid in grapes, only three of its four nitrogens are assimilated by Saccharomyces cerevisiae during vinification. The fourth is incorporated into proline, which cannot be used as a nitrogen source in the absence of oxygen. To test the possibility that yeast can use arginine more efficiently, the arginine utilization pathway was genetically modified to allow assimilation of all four arginine nitrogens under anaerobic conditions.
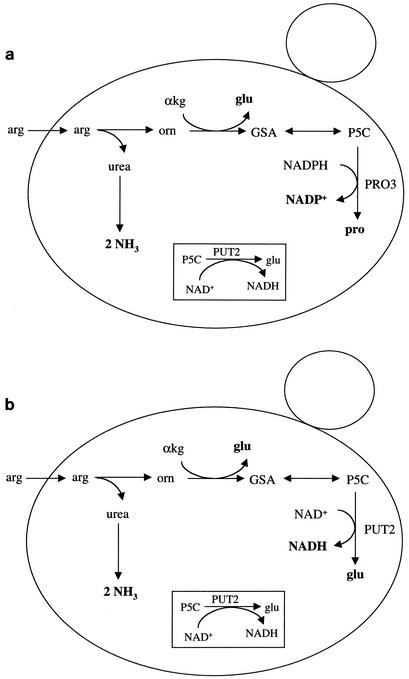
Arginine catabolism in S. cerevisiae (10) initially involves hydrolysis of arginine by arginase (CAR1) to form urea and ornithine. The urea is carboxylated by urea amidolyase (DUR1,2) to produce two molecules of ammonia via allophanate. Ornithine is transaminated by ornithine transaminase (CAR2) via α-ketoglutarate to produce glutamate-γ-semialdehyde and glutamate. Glutamate-γ-semialdehyde spontaneously forms Δ1-pyrroline-5-carboxylate (P5C), which is reduced to proline (6) via NADPH-dependent P5C reductase (PRO3). Under aerobic conditions, P5C can be imported into the mitochondria, where it is converted to glutamate by P5C dehydrogenase encoded by PUT2.
Under anaerobic conditions, the net reaction (Fig. 1a) is arginine + NADPH → 2 ammonia + glutamate + proline + NADP+. While ammonia and glutamate are readily assimilated forms of nitrogen under both anaerobic and aerobic conditions, proline cannot be used as a nitrogen source in the absence of oxygen, because the first step in its degradation is catalyzed by oxygen-dependent proline oxidase encoded by PUT1 (20). Thus, normal arginine degradation under anaerobic conditions permits assimilation of only three of its four nitrogen atoms.
FIG. 1.
Arginine uptake and utilization in wild-type S. cerevisiae (a) and in the engineered strain 3221-2c/pOM (b) under anaerobic conditions. Relevant end products are shown in boldface. The promitochondrion is boxed. arg, arginine; orn, ornithine; αkg, α-ketoglutarate; glu, glutamate; GSA, glutamate-γ-semialdehyde; P5C, Δ′pyrroline-5-carboxylate; pro, proline.
Here we describe a genetically engineered pathway for anaerobic arginine degradation that bypasses proline formation and permits assimilation of all four nitrogens.
MATERIALS AND METHODS
Yeast strain.
S. cerevisiae 3221-2c (MATa ure2Δ11::LEU2 leu2 pro3::TRP1 ura3 trp1?) was obtained as a segregant following tetrad dissection of a cross between ure2 and pro3 parental strains. The pro3 allele was from strain DT1100 (19) and lacks the entire open reading frame (ORF) beyond codon 83. The ure2 allele was from strain P40-3C (11) and contains LEU2 in place of a SacII-PvuII ORF fragment.
Enzyme assays.
Glyceraldehyde-3-phosphate dehydrogenase was assayed as described by the manufacturer (Worthington Biochemical Corporation, Freehold, N.J.) based on the method of Krebs (15). One unit of specific activity caused an initial rate of reduction of 1 μmol of NAD per min/mg of protein. The slope of A340 versus time was linear and based on readings taken every minute over 5 min. P5C dehydrogenase activity was assayed as described previously (5) with substrate prepared according to the method of Williams and Frank (21). One unit of specific P5C dehydrogenase activity was defined as 1 nmol of iodonitrotetrazolium violet formed per min/mg of protein. The slope of A492 versus time was linear and based on readings taken every 30 s over 5 min. Protein was assayed with bovine serum albumin as a standard (3).
Cell extracts.
Cytoplasmic and mitochondrial yeast extracts were prepared essentially as described previously (24) with the following modifications. Cells were grown aerobically in 500-ml flasks at 30°C and 250 rpm in 200 ml of yeast nitrogen base without amino acids (Difco, Detroit, Mich.) containing glucose (20 g/liter) and proline (1 g/liter) and were harvested in log phase (A600 = 0.7 to 1.2). The crude mitochondrial pellet was used without further purification in a density gradient. Mitochondria were sonicated in 10 1-s intervals at 0°C (setting 15 in a Fisher model 60 Sonic Dismembrator; Fisher Scientific) with a 60-s hold at 0°C between sonications.
Construction of PUT2 lacking mitochondrial targeting sequence.
A PUT2 ORF was constructed lacking the first 16 amino acids, MLSARCLKSIYFKRSF, presumed to be the mitochondrial targeting sequence, based on computer-assisted sequence analysis (17, 18), as nuclear genes encoding mitochondrial proteins do not share a consensus targeting sequence (8, 13). The 1.7-kb ORF was generated by PCR such that it lacked DNA encoding the first 16 amino acids and contained added 5′-terminal EcoRI and 3′-terminal XhoI sites and an artificial ATG start codon. In addition, an internal EcoRI site was mutated by replacing an A residue in position 1098 with a G, such that the altered codon still specified glutamate (GAA → GAG). Mutagenic PCR was performed in two separate reactions in a Robocycler 96 temperature cycler (Stratagene, La Jolla, Calif.) in 20 μl of Pfu buffer overlaid with mineral oil, containing 1 U of cloned Pfu DNA polymerase (Stratagene), 0.2 mM (each) deoxynucleoside triphosphates, 10 ng of yeast template DNA, and 0.2 μM primers. The mixture for reaction 1 contained primers PUT2L, 5′-CTCGAGTTATTCATAATTCGATGGATA-3′, and PUT2UIN, 5′-GAGTCAAAAAGTGAAGAGTTCTTATCCGA-3′, and reaction 1 generated a 0.6-kb fragment. The mixture for reaction 2 contained primers PUT2U, 5′-GAATTCAATATGTCACAACTGGGACAC-3′, and PUT2LIN, 5′-TCGGATAAGAACTCTTCACTTTTTGACAC-3′, and generated a 1.1-kb fragment. Reaction conditions were as follows: 2 min at 95°C followed by 3 cycles of 45 s at 94°C, 1 min at 45°C, and 2 min at 72°C and 27 cycles of 45 s at 94°C, 1 min at 57°C, and 3 min at 72°C, followed by a final 8-min extension at 72°C. Extension of the mutagenic PCR products was performed in the same temperature cycler in 20 μl of Pfu buffer overlaid with mineral oil, containing 1 U of cloned Pfu DNA polymerase, 0.2 mM (each) deoxynucleoside triphosphates, 5 μl of each purified mutagenic PCR product (MinElute PCR purification kit; Qiagen, Valencia, Calif.), and no primers. Reaction conditions were as follows: five cycles of 45 s at 94°C and 90 min at 72°C. The final 1.7-kb ORF was generated as described for the two mutagenic PCRs except that 10 ng of gel-purified (QIAquick gel extraction kit; Qiagen) PCR extension product served as template, the primers used were PUT2L and PUT2U, and the following reaction conditions were used: 30 cycles of 45 s at 94°C, 1 min at 57°C, and 3 min at 72°C, followed by a final 8-min extension at 72°C.
The 1.7-kb PCR product was cloned into pCR-Blunt (Invitrogen, Carlsbad, Calif.), digested with EcoRI and XhoI, and ligated into EcoRI- and XhoI-digested pJC1 (12), downstream of the PGK1 promoter and upstream of the PGK1 transcriptional terminator carried on this URA3, 2μm-based high-copy-number expression vector. The construct, hereafter referred to as pOM, was amplified in and extracted from Escherichia coli DH5α and sequenced to confirm that the added ATG codon was in frame with the truncated PUT2 ORF (data not shown). Strain 3221-2c (MATa ure2Δ11::LEU2 leu2 pro3::TRP1 ura3 trp1?) was transformed (9) with pOM and separately with pJC1, the latter serving as an empty vector control.
Cell yield on limiting arginine as sole nitrogen source.
The engineered and control strains were grown at 30°C in yeast nitrogen base without amino acids and ammonium sulfate (Difco), supplemented with glucose (20 g/liter), proline (1 g/liter), ergosterol (10 mg/liter), Tween 80 (1 mg/liter), and variable amounts of arginine as the sole catabolizable nitrogen source in an anaerobic jar (BBL GasPak Plus; Becton Dickinson, Sparks, Md.) in the presence of a methylene blue indicator strip. Medium ingredients were sterilized by filtration except for the ergosterol and Tween 80, which were prepared as a combined concentrated stock in 70% ethanol. Proline was added to satisfy an auxotrophic requirement and could not be used as a nitrogen source in the absence of oxygen. Indeed, no growth was observed when arginine was not added. Cell yield was measured as a function of added arginine after 4 days of anaerobic growth. Liquid media (1 ml/tube) were inoculated to an initial concentration of 105 cells/ml with the use of, as inoculum, cells pregrown anaerobically at 30°C in the same medium containing limiting arginine, 25 μg/ml. Final cell densities were determined by relating measured A600 values to dry weight with a plot of A600 versus dry weight of cells previously determined for strain 3221-2c.
Glutamate pool.
The engineered and control strains were grown at 30°C in about 100 ml of yeast nitrogen base without amino acids and ammonium sulfate (Difco), supplemented with glucose (20 g/liter), proline (50 mg/liter) and arginine (250 mg/liter), ergosterol (10 mg/liter), and Tween 80 (1 mg/liter) in flasks held in an anaerobic jar (BBL GasPak Plus) in the presence of a methylene blue indicator strip or aerobically in 500-ml flasks shaken at 250 rpm. Ergosterol and Tween 80 were not added to the aerobic cultures. The anaerobic and aerobic cultures were inoculated to an initial concentration of 105 cells/ml with the use of, as inoculum, cells pregrown anaerobically or aerobically, respectively, in the same medium but containing proline at 20 mg/liter. Cells were harvested in log phase (A600 = 0.05 to 0.35) by filtration through a 0.45-μm-pore-size filter and were washed with 100 ml of ice-cold water. A600 values and culture volumes were carefully measured at harvest to permit determination of cell mass as dry weight with the A600-versus-dry weight plot as noted above. Filters with cells were placed in 1.7-ml microcentrifuge tubes to which 500 μl of 5% trichloroacetic acid was added. The tubes were gently agitated to resuspend the cells and were then slightly inclined and shaken on ice at 250 rpm for 30 min. The tubes were centrifuged briefly at 12,000 × g to pellet cell debris, and 400 μl of supernatant was ultrafiltered through a 10,000-molecular-weight centrifugal ultrafilter (Ultrafree-MC; Millipore, Bedford, Mass.) at 4°C for 30 min. The filtrates were stored at −20°C until analysis. Glutamate was determined enzymatically (Boehringer Mannheim, Darmstadt, Germany) in samples preadjusted to about pH 8.5 with a known volume of 5 N NaOH.
RESULTS AND DISCUSSION
Three genetic modifications were introduced into a haploid laboratory strain of S. cerevisiae in order to permit use of all four nitrogen atoms in arginine under anaerobic conditions. The first was to mutate PRO3 in order to block proline formation from arginine. Because grape juice normally contains a significant amount of proline, proline auxotrophy was not envisioned as a potential handicap in adapting the described method for future use in commercial wine strains. The second was to introduce high-level cytoplasmic expression of NAD+-dependent P5C dehydrogenase (PUT2), which converts P5C into glutamate. Cytoplasmic Put2 activity was considered preferable to the mitochondrial matrix—its normal location (7)-as it was uncertain if nonrespiring promitochondria would contain significant Put2 activity or would take up P5C from the cytoplasm during anaerobic vinification. The third was to mutate URE2 to allow proline uptake in the presence of preferred nitrogen sources, as Ure2 is a transcriptional corepressor that plays an essential role in nitrogen catabolite repression (16, 22). Proline auxotrophs are unusual in being unable to grow in proline-containing rich media such as yeast extract-peptone-dextrose due to the presence of preferred nitrogen sources (i.e., ammonia, asparagine, and glutamine), which presumably inhibit proline uptake (4). Indeed, we found that ure2 suppressed the inability of a pro3 mutant to grow in yeast extract-peptone-dextrose, i.e., the double mutant grew (data not shown). This finding has relevance to winemaking, as even nitrogen-deficient grape juice initially contains sufficient preferred forms of nitrogen to prevent proline uptake and therefore to prevent growth of a proline auxotroph in the presence of wild-type URE2.
Anaerobic catabolism of arginine in the genetically modified strain (Fig. 1b) is expected to yield the following: arginine + NAD+ → 2 ammonia + 2 glutamate + NADH.
Engineered strain exhibits significant cytoplasmic Put2 activity.
In order to confirm that the truncated PUT2 allele was expressed and resulted in cytoplasmic activity, P5C dehydrogenase was assayed in Ura+ transformants of the constructed and control strains (Table 1). As expected, mitochondrial P5C dehydrogenase activity was found to be the same in both strains (t test, P < 0.05), as both carried a wild-type PUT2 allele and Put2 is a mitochondrial matrix enzyme (7). In contrast, significant cytoplasmic activity was found only in the engineered strain; none was detected in the control. Glyceraldehyde-3-phosphate dehydrogenase (cytoplasmic enzyme) served as a cytoplasmic control. Activities detected in the cytoplasm of the constructed and control strains were not significantly different (t test, P < 0.05). As expected, no glyceraldehyde-3-phosphate dehydrogenase activity was detected in the mitochondrial extracts.
TABLE 1.
Specific P5C dehydrogenase and glyceraldehyde-3-phosphate dehydrogenase activities of the engineered and control strainsa
| Strain | P5C dehydrogenaseb
|
Glyceraldehyde-3-phosphate dehydrogenasec
|
||
|---|---|---|---|---|
| Cytoplasm | Mitochondria | Cytoplasm | Mitochondria | |
| 3221-2c/pOM (engineered) | 1,650 ± 339 | 753 ± 40 | 775 ± 35 | NDd |
| 3221-2c/pJC1 (control) | ND | 504 ± 71 | 950 ± 71 | ND |
Data are means ± standard deviation of specific activities for two replicates.
One unit of specific P5C dehydrogenase activity is defined as 1 nmol of iodonitrotetrazolium violet formed per min/mg of protein.
One unit of specific glyceraldehyde-3-phosphate dehydrogenase activity is defined as the reduction of 1 μmol of NAD per min/mg of protein.
ND, not detected.
Cell yield on limiting arginine in engineered strain twice that of control.
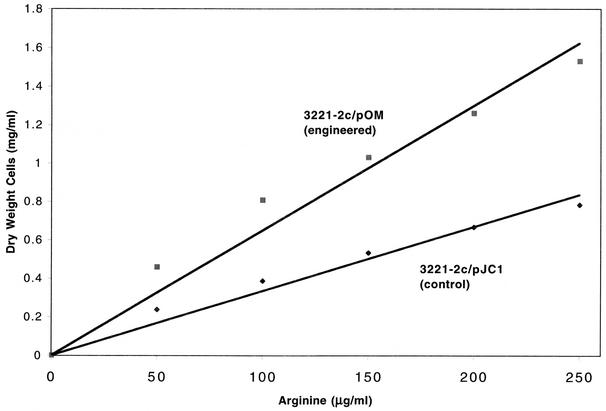
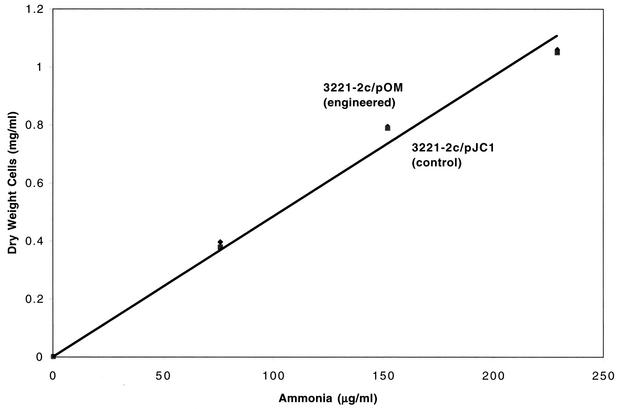
The cell yield of the engineered strain grown anaerobically on arginine as a nitrogen source was found to be twice that of the control (slopes were 0.0059 and 0.0031 for pOM and pJC1, respectively [Fig. 2]) which was 50% higher than the expected 4/3 improvement (use of four rather than three arginine nitrogens). In contrast, cell yields of the engineered and control strains grown anaerobically on ammonia (added as [NH4]2SO4) were the same (Fig. 3). The significant increase in growth yield in the engineered strain which exhibited cytoplasmic Put2 activity indicates that, under anaerobic conditions, promitochondrial Put2 is either inactive or inaccessible to cytoplasmically produced P5C.
FIG. 2.
Cell yield as a function of limiting arginine as sole nitrogen source under anaerobic conditions. Anaerobic cultures were initiated at the indicated arginine concentrations, and growth (dry weight) was determined after 4 days. Data are means of two replicates. Standard deviations were less than 10% of the means.
FIG. 3.
Cell yield as a function of limiting ammonia (added as ammonium sulfate) as sole nitrogen source under anaerobic conditions. Anaerobic cultures were initiated at the indicated ammonia concentrations, and growth (dry weight) was determined after 4 days. Data are means of two replicates. Standard deviations were less than 10% of the means.
Glutamate pool higher in engineered strain than in control.
Intracellular glutamate was measured to determine if the glutamate pool in the engineered strain was higher than that of the control when grown on arginine as the sole usable nitrogen source under anaerobic conditions. The glutamate pool was not significantly different in either strain under aerobic conditions (2.2 ± 0.4 and 3.4 ± 1.2 μg/mg [dry weight] of cells for engineered and control strains, respectively), whereas under anaerobic conditions, the pool in the engineered strain was twice that of the control, 7.0 ± 1.4 versus 3.2 ± 0.5 μg/mg (dry weight) of cells, respectively (P ≤ 0.05, t test; data are means ± standard deviations of four experiments). The finding that the pools were lower in both strains under aerobic conditions, 2 to 3 μg/mg (dry weight) of cells, suggests that demand for glutamate may be higher during the faster growth observed in the presence of oxygen (data not shown).
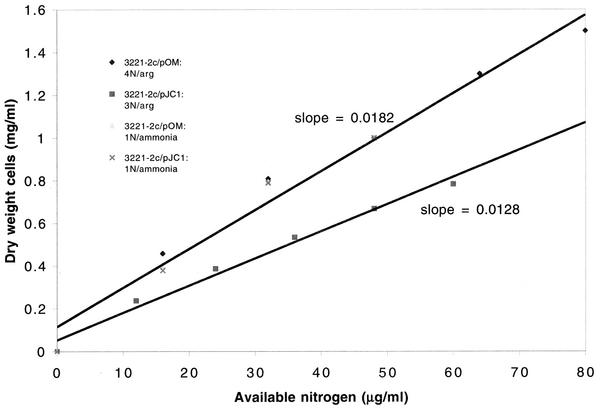
Catabolism of arginine in the control strain was expected to yield urea, glutamate, and P5C. The last compound was presumed to accumulate or be excreted, as it could not be converted into proline via PRO3 or into glutamate via PUT2. Farrant et al. (14) recently reported in vitro evidence that P5C acted as an endogenous vitamin B6 antagonist in a child with hyperprolinemia (lacking P5C dehydrogenase) due to reactivity of P5C with pyridoxal phosphate. Whether the same occurs in S. cerevisiae and might depress growth yield of a pro3 mutant has not been determined. As noted above, the cell yield of the engineered strain grown on arginine was 2-fold, rather than the expected 1.3-fold, higher than that of the control. This is consistent with two general possibilities: the engineered strain actually had a higher cell yield than expected, or the control had a lower-than-expected cell yield. As discussed below, the latter possibility seems more likely. The data reported in Fig. 2 and 3 suggest possible inhibition by P5C in the control strain when growth is replotted as a function of presumed available nitrogen, regardless of source. On such a graph (Fig. 4), the slopes of the control and engineered strains grown on ammonium sulfate (one nitrogen per ammonium sulfate) and the engineered strain grown on arginine assuming four available nitrogens were the same, 0.0182. In contrast, the slope of the control strain grown on arginine assuming three available nitrogens was 30% lower, consistent with the possibility that accumulation of arginine-derived P5C in this strain led to growth inhibition and incomplete utilization of available arginine. What distinguishes the control strain from the engineered strain when both are grown on arginine is that only the control accumulates P5C. The engineered strain converts this putative inhibitor into glutamate.
FIG. 4.
Cell yield as a function of presumed available nitrogen, based on replotting of data shown in Fig. 2 and 3.
The doubling times of the two strains were the same, 2.7 h, during aerobic growth in yeast nitrogen base without amino acids supplemented with 2% glucose and 0.1% proline and were about the same as that of a wild-type laboratory strain isogenic to S288C, grown in the same medium, but without added proline, 2.4 h (23). In these media, ammonium sulfate served as the nitrogen source. Brandriss and Magasanik (6) reported that a pro3 mutant grew six times more slowly than the wild type under different conditions (2% glucose, 0.1% ornithine as sole nitrogen source, and 0.1% proline), consistent with the possibility that accumulation of P5C or a P5C derivative is toxic. Growth on ornithine as a nitrogen source is expected to lead to P5C formation, whereas growth on ammonia is not. Although not addressed in this study, the net production of NADH in the engineered arginine catabolic pathway might adversely affect product yield, as excess NADH is normally generated during fermentation and increases formation of glycerol, succinate, and fusel oils at the expense of ethanol (1, 2). Further imbalance in the direction of excess NADH production would be tempered during vinification because arginine is never a sole source of nitrogen in grape juice.
This study demonstrates that it is possible to improve utilization of arginine as a nitrogen source in S. cerevisiae under anaerobic conditions. In order to exploit this improvement in winemaking, it will be important to demonstrate (i) that the engineered strain carrying pro3 and ure2 mutations (in addition to the PUT2 allele lacking a mitochondrial targeting sequence) exhibits a net increase in growth yield on arginine relative to an isogenic strain carrying wild-type alleles at PRO3 and URE2 and (ii) that introduction of these modifications into a wine strain yields a similar improvement without compromising overall fermentation performance or the sensory character of the resultant wine.
Acknowledgments
We thank Emile van Zyl for plasmid pJC1, Bryan L. Ford for the mutagenic PCR protocol, Mike Penner and Gary Merrill for useful discussions, Chris Mathews and Linda Wheeler for access to the sonicator, Francine Messenguy for the extraction protocol for measuring amino acid pools, Gary Merrill for critically reviewing a previous version of the manuscript, and Rino Zara for designing Fig. 1.
This work was supported by grants from USDA-CSREES (Pacific NW Center for Small Fruits Research), the Agricultural Research Foundation, and the Oregon State University Research Council.
Footnotes
Technical paper no. 11913 of the Oregon Agricultural Experiment Station.
REFERENCES
- 1.Albers, E., C. Larsson, G. Lidén, C. Niklasson, and L. Gustafsson. 1996. Influence of the nitrogen source on Saccharomyces cerevisiae anaerobic growth on product formation. Appl. Environ. Microbiol. 62:3187-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderlund, M., T. L. Nissen, J. Nielsen, J. Villadsen, J. Rydström, B. Hahn-Hägerdal, and M. C. Kielland-Brandt. 1999. Expression of the Escherichia coli pntA and pntB genes, encoding nicotinamide nucleotide transhydrogenase, in Saccharomyces cerevisiae and its effect on product formation during anaerobic glucose fermentation. Appl. Environ. Microbiol. 65:2333-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Brandriss, M. C. 1979. Isolation and preliminary characterization of Saccharomyces cerevisiae proline auxotrophs. J. Bacteriol. 138:816-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandriss, M. C. 1983. Proline utilization in Saccharomyces cerevisiae: analysis of the cloned PUT2 gene. Mol. Cell. Biol. 3:1846-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandriss, M. C., and B. Magasanik. 1980. Proline: an essential intermediate in arginine degradation in Saccharomyces cerevisiae. J. Bacteriol. 143:1403-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandriss, M. C., and K. A. Krzywicki. 1986. Amino-terminal fragments of Δ1-pyrroline-5-carboxylate dehydrogenase direct β-galactosidase to the mitochondrial matrix in Saccharomyces cerevisiae. Mol. Cell. Biol. 6:3502-3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunner, M., C. Klaus, and W. Neupert. 1994. The mitochondrial processing peptidase, p. 73-86. In G. V. Heijne (ed.), Signal peptidases. R. G. Landes Co., Austin, Tex.
- 9.Burke, D. A., D. Dawson, and T. Stearns. 2000. Methods in yeast genetics: a laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 10.Cooper, T. G. 1982. Nitrogen metabolism in Saccharomyces cerevisiae, p. 39-99. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), The molecular biology of the yeast Saccharomyces, metabolism and gene expression. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 11.Coschigano, P. W., and B. Magasanik. 1991. The URE2 gene product of Saccharomyces cerevisiae plays an important role in the cellular response to the nitrogen source and has homology to glutathione S-transferases. Mol. Cell. Biol. 11:822-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crous, J. M., I. S. Pretorius, and W. H. van Zyl. 1995. Cloning and expression of an Aspergillus kawachii endo-1,4-β-xylanase gene in Saccharomyces cerevisiae. Curr. Genet. 28:467-473. [DOI] [PubMed] [Google Scholar]
- 13.Emanuelsson, O., G. V. Heijne, and G. Schneider. 2001. Analysis and prediction of mitochondrial targeting peptides. Methods Cell Biol. 65:175-187. [DOI] [PubMed] [Google Scholar]
- 14.Farrant, R. D., V. Walker, G. A. Mills, J. M. Mellor, and G. J. Langley. 2001. Pyridoxyl phosphate de-activation by pyrroline-5-carboxylic acid. J. Biol. Chem. 276:15107-15116. [DOI] [PubMed] [Google Scholar]
- 15.Krebs, E. 1955. Glyceraldehyde-3-phosphate dehydrogenase from yeast. Methods Enzymol. 1:407-411. [DOI] [PubMed] [Google Scholar]
- 16.Kulkarni, A. A., A. T. Abul-Hamd, R. Rai, H. El Berry, and T. G. Cooper. 2001. Gln3p nuclear localization and interaction with Ure2p in Saccharomyces cerevisiae. J. Biol. Chem. 276:32136-32144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider, G., J. Schuchhardt, and P. Wrede. 1995. Peptide design in machina: development of artificial mitochondrial protein precursor cleavage sites by simulated molecular evolution. Biophys. J. 68:434-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneider, G., S. Sjöling, E. Wallin, P. Wrede, E. Glaser, and G. von Heijne. 1998. Feature-extraction from endopeptidase cleavage sites in mitochondrial targeting peptides. Proteins 30:49-60. [PubMed] [Google Scholar]
- 19.Tomenchok, D. M., and M. C. Brandriss. 1987. Gene-enzyme relationships of the proline biosynthetic pathway in Saccharomyces cerevisiae. J. Bacteriol. 169:5364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, S.-S., and M. C. Brandriss. 1987. Proline utilization in Saccharomyces cerevisiae: sequence, regulation, and mitochondrial localization of the PUT1 gene product. Mol. Cell. Biol. 7:4431-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams, I., and L. Frank. 1975. Improved chemical synthesis and enzymatic assay of Δ1-pyrolline-5-carboxylic acid. Anal. Biochem. 64:85-97. [DOI] [PubMed] [Google Scholar]
- 22.Xu, S., D. A. Falvey, and M. C. Brandriss. 1995. Roles of URE2 and GLN3 in the proline utilization pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:2321-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu, X., J. D. Wightman, B. L. Geller, D. Avram, D., and A. T. Bakalinsky. 1994. Isolation and characterization of sulfite mutants of Saccharomyces cerevisiae. Curr. Genet. 25:488-496. [DOI] [PubMed] [Google Scholar]
- 24.Yaffe, M. P. 1991. Analysis of mitochondrial function and assembly. Methods Enzymol. 194:627-643. [DOI] [PubMed] [Google Scholar]