Abstract
The goal of this study was to identify bacteria involved in soil suppressiveness against the plant-parasitic nematode Heterodera schachtii. Since H. schachtii cysts isolated from the suppressive soil can transfer this beneficial property to nonsuppressive soils, analysis of the cyst-associated microorganisms should lead to the identification of the causal organisms. Our experimental approach was to identify bacterial rRNA genes (rDNA) associated with H. schachtii cysts obtained from soil mixtures with various levels of suppressiveness. We hypothesized that we would be able to identify bacteria involved in the suppressiveness by correlating population shifts with differing levels of suppressiveness. Soil treatments containing different amounts of suppressive and fumigation-induced nonsuppressive soils exhibited various levels of suppressiveness after two nematode generations. The 10%-suppressive-soil treatment contained numbers of eggs per gram of soil similar to those of the 100%-suppressive-soil treatment, indicating that the suppressive factor(s) had been transferred. Bacterial rDNA associated with H. schachtii cysts were identified using a culture-independent method termed oligonucleotide fingerprinting of rRNA genes. Bacteria from five major taxonomic groups (Actinobacteria, Cytophaga-Flexibacter-Bacteroides, α-Proteobacteria, β-Proteobacteria, and γ-Proteobacteria) were identified. Three bacterial rDNA groups contained clones that were more prevalent in the highly suppressive soil treatments than in the less suppressive treatments, indicating a potential involvement in the H. schachtii suppressiveness. When these three groups were examined with specific PCR analyses performed on H. schachtii cysts that developed in soils treated with three biocidal compounds, only one bacterial rDNA group with moderate to high sequence identity to rDNA from several Rhizobium species and uncultured α-proteobacterial clones was consistently associated with the highly suppressive treatments. A quantitative PCR analysis confirmed the association of this Rhizobium-like rDNA group with the H. schachtii suppressiveness.
Suppressive soils have been described as soils in which a pathogen cannot establish, as soils in which a pathogen is found but does not cause disease, and as soils in which a pathogen causes disease which subsequently diminishes with continuous culture of the crop (1). In biological-control research, suppressive soils can provide a source of potentially beneficial microorganisms. In this approach, organisms are isolated from suppressive soils and then examined to assess their potential to control pests. For nematode pests, suppressiveness has been attributed to biotic and abiotic properties of soil. Although a wide range of organisms can parasitize nematodes or reduce their populations by other mechanisms, only fungi and bacteria have been shown to cause nematode suppressiveness (20).
Several studies have examined bacteria from nematode-suppressive soils. Rhizosphere bacteria isolated from Swiss chard grown in a Heterodera schachtii-suppressive soil were able to significantly reduce the amount of nematode infection (13). A larger percentage of rhizosphere bacteria isolated from nematode-antagonistic plants reduced Meloidogyne incognita and Heterodera glycines disease incidence compared to bacteria isolated from soybean (10). Specific changes in rhizosphere bacteria were associated with M. incognita suppressiveness induced by chitin amendments (6). Pseudomonas sp. strain BG33R, which was isolated from a soil suppressive against peach tree short life, reduced multiplication of Mesocriconema xenoplax and inhibited egg hatch (11, 25). Finally, changes in rhizosphere bacteria have been associated with velvetbean crop rotations, which have been shown to decrease levels of Meloidogyne arenaria (14).
The objective of this study was to identify bacteria involved in soil suppressiveness against H. schachtii. The suppressive soil is from field 9E, located at the University of California Riverside Agricultural Research Station. This soil suppressiveness is biological in nature, since fumigation with various broad-spectrum chemicals (methyl bromide, methyl iodide, and metam sodium) and exposure to aerated steam reduced suppressiveness to undetectable levels (26). The suppressive nature of the soil has been transferred via nematode cysts that developed in the 9E field soil (27). This crucial feature should greatly assist investigations directed towards the identification of the organisms involved in the suppressiveness, as the microbial communities associated with the cysts are likely to be much less complex than the microbial communities associated with the soil.
The experimental approach for this work involved identifying bacteria from nematode cysts obtained from soil mixtures with various degrees of suppressiveness. We postulated that we would be able to determine which bacteria were involved in the H. schachtii suppressiveness by identifying species whose population levels correlated positively with the suppressiveness. The experimental plan had three phases. The first phase was to identify bacteria from cysts that developed in soils possessing various degrees of suppressiveness, which were generated by mixing different amounts of suppressive and fumigation-induced nonsuppressive soil. The bacteria were identified with a culture-independent method termed oligonucleotide fingerprinting of rRNA genes (OFRG), which is an array-based method that allows extensive analysis of bacterial community composition (24). rRNA gene (rDNA) sequences that were more abundant in cysts from the highly suppressive soils than in cysts from the less suppressive soils were considered candidate sequences. The second phase of this experimental plan was to verify the results obtained from phase one. For this work, we designed selective PCR primers for each of the candidate rDNA sequences. These primers were used to determine the relative amounts of the candidate sequences in cysts from soils possessing various levels of suppressiveness, which were produced by mixing different quantities of suppressive and fumigation-induced nonsuppressive soil and by three biocidal treatments of the suppressive soil. We theorized that we would be able to identify the organisms causing the suppressiveness and remove false positives by identifying those rDNA sequences whose population levels consistently correlated with suppressiveness in these two complementary studies. The rationale for this approach was that any single greenhouse experiment might lead to the identification of organisms whose population levels correlate with suppressiveness but which exhibit this trend for reasons unrelated to the suppressiveness. In phase three, we used quantitative PCR analyses to confirm the correlations between the levels of these rDNA sequences and H. schachtii suppressiveness. A parallel study was also performed to examine the fungal rDNA associated with the H. schachtii suppressiveness (B. Yin, L. Valinsky, X. Gao, J. O. Becker, and J. Borneman, submitted for publication).
MATERIALS AND METHODS
Greenhouse trial with five 9E soil mixtures.
H. schachtii-suppressive soil was collected from the upper 10 cm of field 9E soil at the University of California Riverside Agricultural Research Station. The 9E soil was a Hanford fine sandy loam (60.9% sand, 29.6% silt, 9.5% clay, pH 8) (27). The H. schachtii suppressiveness developed at the 9E site after several years of continuous cropping to hosts of the nematode (26). After screening with a sieve with 6-mm openings, one part of the soil was fumigated with 2 ml of methyl iodide (690 kg/ha) for 4 days in 19-liter polyethylene buckets (3). At 3 days after fumigation, all fumigated and untreated soils were mixed 10:1 with silica sand. Soil mixtures were then made by amending fumigated 9E soil with 0.1%, 1.0%, and 10% (dry wt/wt) nonfumigated 9E soil. Nonfumigated and fumigated 9E soil served as the suppressive and nonsuppressive controls, respectively. Soils were placed into 6-inch-diameter pots and sown with five seeds of mustard greens of Brassica juncea (cv. Florida broadleaf; Lockhart Seeds). All pots were placed in a greenhouse under natural light at 23 ± 3°C in a randomized complete block design with five replicates. Plants were watered with tap water. After emergence, the seedlings were thinned to one per pot and fertilized with 6 g of slow-release fertilizer (Sierra 17-6-10 plus Minors; Scotts-Sierra Horticultural Products Company, Marysville, Ohio). At 4 weeks after seeding, each pot was inoculated with 10,000 second-stage juveniles of H. schachtii. Juveniles were collected and enumerated using a zinc chloride hatching protocol as previously described (17). Starting at 3 weeks after inoculation, each plant was fertilized with 50 ml of a nutrient solution (Miracle-Gro 15-30-15; Scotts Miracle-Gro Products, Marysville, Ohio) every 2 weeks. At 11 weeks after inoculation, plant tops were cut off at soil level and weighed after oven drying at 80°C for 2 days. Soil with the root system was collected and placed in a container. Roots were cut into pieces (∼0.5 cm) and mixed thoroughly with soil. Two 350-g subsamples of this soil mixture were collected using a modified Fenwick flotation can method (5) for cyst extraction. Cysts were counted and then broken in a tissue homogenizer for egg counting. A total of 200 g of the soil mixture was collected, sieved using a screen with 1.0-mm openings, and stored at −20°C for bacterial rDNA analysis.
Transfer of suppressiveness with H. schachtii cysts.
The procedures for this trial were similar to those previously described (27). Cycle 1-suppressive 9E soil was collected and screened, and portions were fumigated and mixed with sand as described above. The fumigated and nonfumigated 9E soils were placed into root observation chambers (height, width, and depth were 26 cm, 23 cm, and 3.8 cm, respectively), with five replicates for each soil treatment. One side of the root observation chamber was constructed of clear plastic, and the other was constructed of black plastic. Soils were sown with 15 seeds of B. juncea. The chambers were placed in a greenhouse under natural light at 23 ± 3°C in a randomized complete block design with the translucent side of the chambers facing downward at a 45° angle. Plants were watered with tap water. After emergence, the seedlings were thinned to five per chamber and the plants were fertilized with 6 g of slow-release fertilizer (Sierra 17-6-10 plus Minors; Scotts-Sierra Horticultural Products Company). At 4 weeks after seeding, each chamber was inoculated with 15,000 second-stage juveniles of H. schachtii. At 10 weeks after inoculation, the chambers were opened and the cysts were manually collected from the roots. Cysts from the five replicates were pooled. A portion of these cysts was stored at −20°C for bacterial rDNA analysis, and the rest were used for the next cycle of this experiment. Cycle 2-fumigated 9E soil was mixed 1:1 with silica sand containing H. schachtii cysts that were established after two nematode generations in the greenhouse. Styrofoam cups (350 ml) were filled with 360 g (wet weight) of this soil mixture, which contained approximately 70 cysts (50,000 H. schachtii eggs). Five treatments were examined. To each cup, we added either (i) three cysts from the suppressive soil treatment from cycle 1, (ii) three cysts from the nonsuppressive soil treatment from cycle 1, (iii) three cysts from the suppressive soil treatment from cycle 1 that were treated with 10% bleach for 1 min and then washed six times with sterile H2O, (iv) 5 g of suppressive 9E soil (positive control), or (v) no amendment (negative control). All cups were placed in a greenhouse under natural light at 23 ± 3°C in a randomized complete block design with five replicates. Cups were watered as needed with tap water. At 1 month later, soil in each cup was mixed thoroughly in a plastic bag and 350 g (wet weight) was placed back into the cup. Five B. juncea seeds were sown in each cup. Plants were thinned to one per cup after emergence and fertilized with 2.5 g of slow-release fertilizer (Sierra 17-6-10 plus Minors; Scotts-Sierra Horticultural Products Company). At 12 weeks after planting, plants were harvested and cysts and eggs were extracted and counted as described in the previous section.
Biocidal treatments of 9E soil.
Suppressive 9E soil was collected, screened, and mixed with sand as described above. This greenhouse trial had four treatments: (i) fludioxonil (Syngenta, Greensboro, N.C.), (ii) formaldehyde, (iii) methyl iodide, and (iv) untreated suppressive 9E soil. Each treatment had six replicates. For the fludioxonil treatment, 240 ml of a suspension containing fludioxonil (100 mg/liter) was added to 15-cm-diameter pulp pots filled with 2,000 cm3 of soil and incubated at room temperature for 7 days. For the formaldehyde treatment, 5.47 ml of formaldehyde (37%) was added to 15-cm-diameter pulp pots filled with 2,000 cm3 of soil and incubated at room temperature for 2 weeks. Methyl iodide treatment was done as described above (3). After completing the biocidal treatments, the soils were mixed separately in polyethylene bags (50 cm × 45 cm) and then repotted. Four seeds of Swiss chard (Beta vulgaris [cv. Large white ribbed; Lockard Seeds, Stockton, Calif.]) were sown in each pot. Each pot also received 6 g of slow-releasing fertilizer (Sierra 17-6-10 plus Minors; Scotts-Sierra Horticultural Products Company). The pots were arranged in a randomized complete block design in the greenhouse at 24 ± 1°C under ambient light. After emergence, the seedlings were thinned to one per pot. Each pot was infested with 5,000 second-stage juveniles of H. schachtii 45 days after sowing. At 12 weeks after infestation, the experiment was harvested as described above.
Data analysis for greenhouse trials.
The cyst and egg data were log (x + 1) transformed. The transformed cyst and egg data and the plant top weight data were subjected to analysis of variance. Fisher's least significant difference (LSD) test was used to analyze the means at P = 0.05 when the treatment F had a P value of ≤0.05.
Isolation of H. schachtii cysts from soil for DNA extraction and photography.
Soil samples were air dried for 2 days at room temperature. For each treatment, amounts of 25 g of soil from each replicate sample were pooled. Cysts were extracted from these soils with the modified version of the cyst floatation method (17). Soil samples were put in 200-ml glass beakers. A strip of water-saturated 0.35-mm-thick chromatography paper was placed in the beaker such that it covered the entire inside perimeter of the top third of the beaker. Water was added to the beaker until it covered half of the chromatography paper. The soil mixture was stirred gently with a glass rod. One drop of Tween 20 was added to the center of the water surface, which caused the floating cysts to move towards the paper. The chromatography paper with adhering cysts was carefully removed from the beaker. Cysts were manually collected under a dissecting microscope and photographed at ×40 magnification.
DNA extraction from H. schachtii cysts.
Cysts were washed with sterile water by moving a cyst-water mixture in and out of a pipette tip approximately five times with three water changes. For each treatment, DNA from 20 individual H. schachtii cysts was extracted using a proteinase K method as previously described (22), except the cysts were crushed with a pipette tip instead of glass slides.
Analysis of bacterial rDNA associated with H. schachtii cysts.
The bacteria associated with H. schachtii cysts were examined using OFRG as previously described (24). A PCR procedure was used to amplify rDNA from the cysts. For each soil treatment, the PCR templates were composed of pooled DNA isolated from 20 cysts. Amplification reactions (200 μl) were performed in 10-μl glass capillary tubes with a 1002 RapidCycler (Idaho Technologies, Idaho Falls, Idaho), and the reaction mixtures contained the following reagents: 50 mM Tris (pH 8.3), 500 μg of bovine serum albumin/ml, 2.5 mM MgCl2, 250 μM each deoxynucleoside triphosphate, 400 nM each primer (363, TCAAGAGRRTTTGATYHTGGYTCAG; 364, AACTCGBTACCTTGTTACGACTT), 20 μl of DNA isolated from the cysts, and 10 U of Taq DNA polymerase. The primers are modified versions of 27f and 1392R (12), which generate DNA fragments approximately 1,400 bp in length and selectively amplify bacterial small-subunit rDNA. The cycling parameters were as follows: 94°C for 2 min and 30 cycles of 94°C for 15 s, 48°C for 20 s, and 72°C for 45 s, followed by 72°C for 2 min. The PCR products were gel isolated and purified with a QIAquick PCR purification kit (Qiagen, Chatsworth, Calif.), ligated into pGEM-T (Promega), transformed into competent Escherichia coli JM109 (Promega), and plated on Luria-Bertani agar plates (containing 100 μg of ampicillin/ml) that were surface spread with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and IPTG (isopropyl-β-d-thiogalactopyranoside). For each treatment, 301 white colonies were randomly selected for OFRG analysis. The rDNA fragments from these clones were subsequently PCR amplified, arrayed on nylon membranes, and hybridized, using previously described protocols (23, 24), to 10 nucleotide probes. Clones that did not hybridize to the reference probe were excluded from further analysis. Hybridization signals were used to generate OFRG fingerprints as previously described (23, 24). The fingerprints were clustered using an unweighted pair group method with arithmetic mean (UPGMA), default parameters, and PAUP 4.0 beta 8 software. Clones were identified in this analysis by their association with known rDNA sequences within the tree. This was accomplished by including fingerprints from known rDNA sequences in the UPGMA analysis and by nucleotide sequence analysis of representative clones distributed throughout the tree. Bacterial groups that contained clones predominately from the highly suppressive soils were identified by a visual examination of the UPGMA tree.
Group-selective PCR.
Group-selective PCR primers were developed for the three bacterial rDNA groups which were more prevalent in the highly suppressive treatments than in the less suppressive treatments as determined by the OFRG analysis. The primers were designed using Pretty software (GCG) to locate DNA sequences that were conserved among rDNA clones (at least five per group) within each bacterial group and using BLAST (National Center for Biotechnology Information) to locate DNA sequences that had few, if any, identical matches to rDNA sequences as listed in GenBank. The primer sequences for the three groups were as follows: group 1 (G1F, CTTCGGGGGAAAGATTTAT; G1R, AGTAATGGACCAGTGAG), group 2 (G2F, TTGCGTAGGTGGTTAGTA; G2R, GCACAGTTTCCTTTCGG), and group 3 (G3F, AAGAAATCCCCTGGGTTA; G3R, GCACTTCCACATCTCTG). The sizes of the predicted PCR products were as follows: G1, 527 bp; G2, 454 bp; and G3, 579 bp. Amplification reactions (10 μl) were performed in glass capillary tubes with a 1002 RapidCycler (Idaho Technologies), and the reaction mixtures contained the following reagents: 50 mM Tris (pH 8.3), 500 μg of bovine serum albumin/ml, 2.5 mM MgCl2, 250 μM each deoxynucleoside triphosphate, 400 nM each primer, 0.2 to 1 μl of DNA isolated from the cysts, and 0.5 U of Taq DNA polymerase. For each group-selective primer pair, the template volumes for all samples were the same. The cycling parameters for the amplification reaction mixtures containing the group 1 primers were as follows: 94°C for 2 min and 40 cycles of 94°C for 15 s, 62°C for 20 s, and 72°C for 40 s, followed by 72°C for 2 min. The cycling parameters for the amplification reaction mixtures containing the group 2 and 3 primers were as follows: 94°C for 2 min and 40 cycles of 94°C for 15 s, 65°C for 20 s, and 72°C for 40 s, followed by 72°C for 2 min. To ensure that negative PCR results were not caused by inhibitory factors in the amplification reactions, templates that did not produce a PCR product were reamplified after amendment with a positive-control template; in all cases, these experiments produced an amplification product (data not shown).
Competitive PCR.
The group 1 bacterial rDNA sequences identified in this report were quantified using competitive PCR as described previously (9). Competitors were generated by low-stringency PCR. These reaction mixtures contained the same components as described in the group-selective PCR section. For each 10-μl reaction mixture, 0.2 μl of template DNA was used; the template DNA was extracted from an avocado grove soil. The PCR parameters were as follows: 94°C for 2 min and 40 cycles of 94°C for 10 s, 35°C for 2 min, and 72°C for 30 s, followed by 72°C for 2 min. The amplification products were resolved on 1% agarose gels. DNA fragments approximately 100 bp larger or smaller than the target amplicon were gel isolated and cloned into pGEM-T (Promega) to create the competitors. Quantification of the bacterial rDNA from the cysts was accomplished via competitive PCR experiments, which used three to four amplification reaction mixtures containing a constant amount of DNA isolated from the cysts with a dilution series of plasmid containing the competitor sequence. Each amplification reaction was performed three times. PCR was performed as described in the group-selective PCR section. These reaction products were resolved on 1.0% agarose gels and stained with ethidium bromide, and the amount of DNA in each band was enumerated by densitometry. Bacterial rDNA levels were quantified by determining the competitor amount at the equivalence point in a plot of the log of the competitor/cyst bacterial rDNA ratio versus the log of the level of competitor DNA. These data were subjected to analysis of variance. Fisher's LSD test was used to analyze the means at P = 0.05 when the treatment F had a P ≤ 0.05.
Sequence analysis of bacterial rDNA clones.
Nucleotide sequences of at least five rDNA clones from groups 1, 2, and 3 were determined using an ABI PRISM BigDye Terminator v3.0 cycle sequencing kit and a 3100 Genetic Analyzer (ABI) and assembled using ContigExpress (Vector NTI). Sequence identities were determined using BLAST (National Center for Biotechnology Information) and AlignX (Vector NTI).
Nucleotide sequence accession numbers.
The nucleotide sequences of the following rDNA clones identified in this work have been deposited in the GenBank database: group 1 (Rhizobium-like) rDNA, accession no. AF525837 to AF525841; group 2 rDNA, accession no. AF525832 to AF525836; and group 3 rDNA, accession no. AF525822 to AF525831.
RESULTS
Greenhouse trial with five 9E soil mixtures.
Soil treatments containing different amounts of suppressive and fumigation-induced nonsuppressive 9E soils exhibited various levels of suppressiveness after two nematode generations (Table 1). Suppressiveness was assessed by enumerating the number of eggs and cysts per gram of soil. Fewer eggs or cysts indicated greater suppressiveness. The 10%-suppressive-soil treatment contained numbers of eggs per gram of soil similar to those seen with the 100%-suppressive-soil treatment, indicating that the suppressive factor(s) had been transferred. Numbers of cysts per gram of soil were similar for the 1%-, 10%-, and 100%-suppressive-soil treatments. No significant differences in plant weights were observed, which is likely due to minimal plant stress resulting from optimal irrigation, fertilization, and temperature in this greenhouse trial.
TABLE 1.
H. schachtii population and plant yield data in relation to various proportions of suppressive 9E soila
| % of suppressive soil | No. of cysts per g of soil | No. of eggs per g of soil | Plant top dry wt (g) |
|---|---|---|---|
| 0 | 1.9c | 237.3b | 63.1a |
| 0.1 | 1.4bc | 180.2b | 64.8a |
| 1 | 1.0ab | 134.1b | 64.2a |
| 10 | 0.8a | 71.6a | 71.9a |
| 100 | 0.8a | 63.5a | 59.4a |
For each column to the right of the leftmost column, results represented by entries with identical suffixed letters were not significantly different when analyzed with the Fisher's protected LSD test at P = 0.05.
Transfer of suppressiveness with H. schachtii cysts.
Cysts that developed in the suppressive 9E soil transferred H. schachtii suppressiveness to fumigation-induced, nonsuppressive 9E soil. Nonsuppressive soil amended with cysts from the 9E soil had numbers of eggs per gram of soil (40.8) equivalent to that seen with the positive-control treatment (39.2), which was a 9E soil amendment, and significantly less than that seen with the treatment amended with cysts that developed in the fumigated 9E soil (65.7).
Biocidal treatments of 9E soil.
Three biocidal treatments produced soils with larger numbers of eggs per gram of soil than those seen with the untreated soil (Table 2). The greatest reduction in suppressiveness resulted from the methyl-iodide and formaldehyde treatments. Compared to that seen with the untreated 9E soil, the fludioxonil treatment also caused a slight reduction in suppressiveness when the number of eggs per gram of soil was used as the measure of suppressiveness.
TABLE 2.
H. schachtii population data from 9E suppressive soil treated with three biocidal compounds and cropped to B. vulgaris for two nematode generationsa
| Soil treatment | No. of cysts per g of soil | No. of eggs per g of soil |
|---|---|---|
| Fludioxonil | 1.6a | 170.4b |
| Formaldehyde | 7.7b | 625.2c |
| Methyl iodide | 11.0b | 818.6c |
| Fumigated 9E soil | ||
| Untreated 9E soil | 3.4a | 49.6a |
For each column to the right of the leftmost column, results represented by entries with identical suffixed letters were not significantly different when analyzed with the Fisher's protected LSD test at P = 0.05.
Bacterial rDNA associated with H. schachtii cysts.
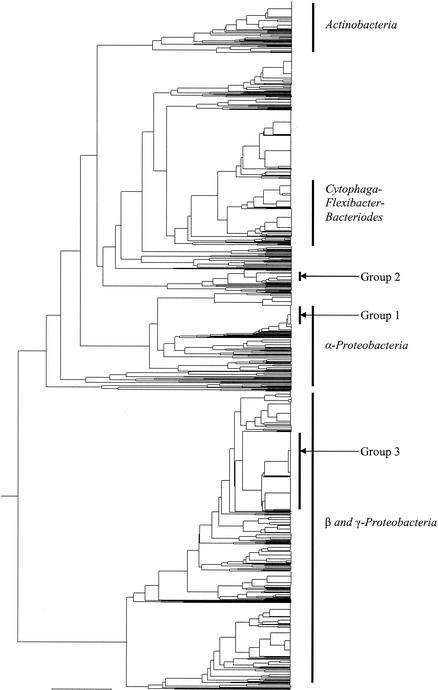
OFRG was used to identify bacterial rDNA sequences associated with the H. schachtii cysts that developed in soil mixtures containing various proportions of the suppressive 9E soil (Fig. 1 and Table 3). UPGMA analysis of the hybridization fingerprints from 1,424 clones produced a tree consisting of 376 clusters (Fig. 1); a cluster was defined as a group of clones with the same fingerprint. Five major taxonomic groups were identified: Actinobacteria, Cytophaga-Flexibacter-Bacteroides, α-Proteobacteria, β-Proteobacteria, and γ-Proteobacteria. Three assemblages of clones, designated groups 1, 2, and 3, contained clones predominantly from the highly suppressive soil treatments (Fig. 1 and Table 4), indicating a potential involvement in the H. schachtii suppressiveness.
FIG. 1.
Taxonomic depiction of bacterial rDNA associated with H. schachtii cysts that developed in soil mixtures with various proportions of suppressive 9E soil (Table 1). The UPGMA tree was constructed from rDNA hybridization fingerprints generated by OFRG. The major taxonomic groups and three treatment-specific bacterial groups (groups 1 to 3) are indicated.
TABLE 3.
Taxonomic distribution of bacterial rDNA clones from H. schachtii cysts that developed in soil mixtures composed of varying proportions of suppressive 9E soila
| % of suppressive 9E soil | No. of Actinobacteria clones | No. of Cytophaga-Flexibacter-Bacterioides clones | No. of α-Proteobacteria clones | No. of β- and γ-Proteobacteria clones | No. of clones of uncertain affiliation |
|---|---|---|---|---|---|
| 0 | 27 | 7 | 22 | 46 | 161 |
| 0.1 | 31 | 14 | 28 | 138 | 69 |
| 1 | 25 | 73 | 23 | 98 | 70 |
| 10 | 13 | 30 | 35 | 147 | 65 |
| 100 | 10 | 19 | 29 | 180 | 64 |
The data in the columns to the right of the leftmost column were obtained by adding the numbers of clones in the taxonomic groups depicted in the UPGMA tree from the OFRG analysis (Fig. 1).
TABLE 4.
Number of rDNA clones from three bacterial groups associated with H. schachtii cysts that developed in soil mixtures composed of various proportions of suppressive 9E soil
| % of suppressive soil | No. of group 1 (Rhizobium-like rDNA) clonesa | No. of group 2 clonesa | No. of group 3 clonesa |
|---|---|---|---|
| 0 | 3 | 0 | 0 |
| 0.1 | 4 | 0 | 5 |
| 1 | 1 | 0 | 7 |
| 10 | 21 | 0 | 47 |
| 100 | 10 | 26 | 108 |
A visual examination of the UPGMA tree obtained by the OFRG analysis (Fig. 1) identified three taxonomic groups that contained clones predominantly from the most suppressive soil treatments (10 and 100%). Data were obtained by adding the numbers of clones in each of the three groups.
Sequence analysis of rDNA groups associated with suppressiveness.
A nucleotide sequence analysis was performed on the three rDNA groups that contained clones predominately from the cysts isolated from the most suppressive soils (Fig. 1 and Table 4). At least five clones from each group were examined. This analysis showed that group 1 bacterial rDNA has 98.2 to 99.6% sequence identity to rDNA from several Rhizobium species and uncultured α-proteobacterial clones (accession no. AB054953, AF502218, AJ440749, U86344, and X74915). Group 2 rDNAs have 94.7 to 94.8% sequence identity to an uncultured bacterial clone, SBR2096 (accession no. AF268998). SBR2096 is a member of the candidate division TM7, which is a lineage of bacteria with no known cultured representatives (8). Group 3 rDNA has 97.9 to 98.1% sequence identity to an uncultured bacterial clone, GOUTA14 (accession no. AY050585), which is a member of the α-Proteobacteria. A pairwise sequence analysis of the clones within the same bacterial group identified some sequence variations for the Rhizobium-like rDNA (94.0 to 99.9%), group 2 rDNA (99.7 to 100%), and group 3 rDNA (97.7 to 99.9%). For the Rhizobium-like rDNA (group 1), the level of sequence variation suggests that this group contains multiple bacterial species.
Group-selective PCR analysis.
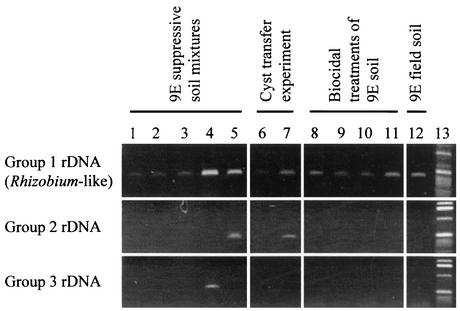
To verify the association of the three bacterial groups identified by the OFRG analysis (Table 4 and Fig. 1) with H. schachtii suppressiveness, DNA from cysts isolated from soils possessing various levels of suppressiveness were subjected to a group-selective PCR analysis (Fig. 2). The cysts used in this analysis were from the three greenhouse trials described above and the 9E field soil. This group-selective PCR analysis showed that the Rhizobium-like rDNA group (group 1) was the only group found in cysts from all the highly suppressive soils (Fig. 2, lanes 4, 5, 7, 8, 11, and 12). For the cysts that developed in the five soil mixtures, the strongest PCR products came from the 10%- and 100%-suppressive-9E-soil treatments (Rhizobium-like rDNA; lanes 4 and 5). Cysts that transferred suppressiveness to nonsuppressive soil produced a stronger PCR product (lane 7) than those that developed in the nonsuppressive soil and did not transfer suppressiveness (lane 6). For the cysts that developed in soils treated with three biocidal compounds, the strongest PCR products came from the most suppressive soil treatments (Rhizobium-like rDNA; lanes 8 and 11). Amplification products were also obtained from cysts collected from the suppressive 9E field soil (Rhizobium-like rDNA; lane 12). Since group 2 and 3 rDNA was not detected in the cysts from any of the soils from the biocidal treatment trial or the 9E field soil, no further analyses were performed on these sequences.
FIG. 2.
Amplification of bacterial rDNA associated with H. schachtii cysts by using group-selective PCR. Amplification reactions were performed using PCR primers selective for the three bacterial groups that were most abundant in cysts from the soil treatments exhibiting the most H. schachtii suppressiveness (Table 4). PCR experiments were performed on DNA from H. schachtii cysts from five 9E soil mixtures (Table 1) (lanes 1 to 5), the cyst transfer experiment (lanes 6 to 7), three biocidal treatments of 9E soil (Table 2) (lanes 8 to 11), and the 9E field soil (lane 12). Lanes: 1, 0% 9E soil, 2, 0.1% 9E soil; 3, 1% 9E soil; 4, 10% 9E soil; 5, 100% 9E soil; 6, methyl iodide-fumigated 9E soil; 7, untreated 9E soil; 8, fludioxonil-treated 9E soil; 9, formaldehyde-treated 9E soil; 10, methyl iodide-treated 9E soil; 11, untreated 9E soil; 12, 9E field soil; 13, 1-kb DNA ladder (Life Technologies). Each percentage indicates the proportion of suppressive soil.
Quantification of Rhizobium-like rDNA associated with H. schachtii cysts.
Competitive PCR experiments were used to quantify the Rhizobium-like rDNA associated with H. schachtii cysts from the soil mixture (Table 1) and biocidal treatment trials (Table 2). The relative distribution of the rDNA obtained by the competitive PCR experiments (Table 5) corroborated the distribution obtained by the OFRG analysis (Table 4). When cysts from the biocidal treatments were examined, significantly higher levels of Rhizobium-like rDNA were found in the soils exhibiting the highest levels of H. schachtii suppressiveness (Table 6).
TABLE 5.
Quantification of Rhizobium-like rDNA from H. schachtii cysts that developed in soil mixtures composed of various proportions of suppressive 9E soil
| % of suppressive soil | Rhizobium-like rDNA (group 1) (pg of amplicon/cyst)a |
|---|---|
| 0 | 12.9c |
| 0.1 | 11.5c |
| 1 | 13.6c |
| 10 | 30.7a |
| 100 | 25.6b |
The data in the column to the right were obtained by measuring the amount of Rhizobium-like rDNA associated with the cysts from soils described in Table 1. For the column to the right, results represented by entries with identical suffixed letters were not significantly different when analyzed with the Fisher's protected LSD test at P = 0.05.
TABLE 6.
Quantification of Rhizobium-like rDNA from H. schachtii cysts that developed in soils treated with three biocidal compounds
| Soil treatment | Rhizobium-like rDNA (group 1) (pg of amplicon/cyst)a |
|---|---|
| Fludioxonil | 18.5a |
| Formaldehyde | 4.3c |
| Fumigated 9E soilb | 5.6b |
| Untreated 9E soil | 18.9a |
The data in the column to the right were obtained by measuring the amount of Rhizobium-like rDNA associated with the cysts from soils described in Table 2. For the column to the right, results represented by entries with identical suffixed letters were not significantly different when analyzed with the Fisher's protected LSD test at P = 0.05.
Methyl iodide treated.
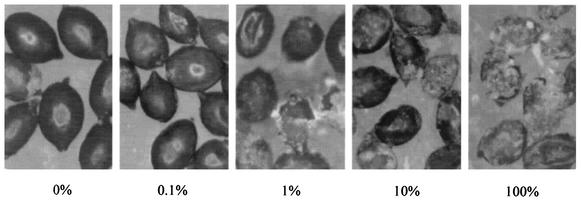
Crystalline layer associated with H. schachtii cysts.
A crystalline layer associated with H. schachtii cysts was observed (Fig. 3). The quantity of this material appeared to correlate with the level of suppressiveness.
FIG. 3.
Crystalline layer associated with H. schachtii cysts that developed in soil treatments containing various proportions of the suppressive 9E soil (×40 magnification). The values indicate the percentages of suppressive soil.
DISCUSSION
Bacteria associated with H. schachtii cysts were identified using the culture-independent OFRG method. The bacterial diversity associated with the H. schachtii cysts was considerable. From an analysis of 1,424 rDNA clones, 376 clusters were identified, each of which likely represents a different bacterial species (24). This analysis provides one of the first depictions of bacteria associated with nematode cysts. In a search of the literature, we found one other report that identified actinomycetes from H. trifolii cysts (7). The bacteria identified in our study were either firmly attached to the outside of the cysts or contained within the structure, as the cysts were washed repeatedly with water before being analyzed. Since a recent OFRG analysis of the bacterial rDNA from the suppressive 9E soil generated a distribution of clones among the major taxa (Actinobacteria, 185 clones; α-Proteobacteria, 142 clones; and β- and γ-Proteobacteria, 87 clones) (24) which was different than that found in H. schachtii cysts that developed in the same soil (Table 3), this suggests that the cyst environment selects a subset of the organisms found in soil. For example, rDNA of actinobacteria was the dominant bacterial rDNA found in the soil, while relatively few of these sequences were identified in the cysts that developed in the 100%-suppressive-9E-soil treatment. In contrast, the cysts that developed in the 100%-suppressive-9E-soil treatment were dominated by β- and γ-Proteobacteria, which comprised only the third most prevalent group in the soil.
The primary objective of this study was to identify bacteria involved in soil suppressiveness against the plant-parasitic nematode H. schachtii. The ability of the H. schachtii cysts to transfer the suppressiveness made identifying organisms involved in the suppressiveness a more feasible task, since the microbial communities associated with the cysts were likely to be less complex than the microbial communities associated with the soil. Even so, give the complexity of the bacterial communities in the cysts (Fig. 1 and Table 3), identifying one or a few organisms from such complex communities was a considerable challenge. When we analyzed the cyst-associated bacteria from five soil mixtures possessing various levels of suppressiveness, three bacterial rDNA groups were identified that contained clones predominately from the most suppressive soils (Fig. 1 and Table 4). When these three bacterial rDNA groups were analyzed by a group-selective PCR analysis, which examined cysts from the suppressive 9E field soil and the three greenhouse trials (Fig. 2), only the group 1 rDNA was consistently associated with high levels of suppressiveness. This result suggested that the group 1 bacteria are involved in the H. schachtii suppressiveness. Since groups 2 and 3 were not detected in the cysts that were directly isolated from the suppressive 9E field soil, it is likely that they are not involved in the natural field suppressiveness. However, the variations in the numbers of group 2 and 3 clones among the three greenhouse trials might also suggest that the different soil treatments produced conditions selecting for differing suppressive organisms. To confirm the role these bacteria have in the H. schachtii suppressiveness, future experimentation should include isolating them and then assessing their abilities to reproduce the suppressiveness in greenhouse and field studies. Overall, these results demonstrate the importance of developing an experimental plan that produces various levels of suppressiveness in more than one way, as the design of any single experiment might lead to the identification of organisms that appear to be associated with suppressiveness but which might be associated with an unintended factor(s).
Although numerous bacteria have been reported to affect nematodes through parasitism or by inhibiting hatching or root penetration (19, 20), only a small number of reports have suggested that Rhizobium or related bacteria were involved in these processes. A review by Siddiqui and Mahmood listed 13 studies reporting a nematode-inhibitory effect of root-nodulating bacteria and 3 studies that reported a nematode-stimulatory effect (18). Mechanisms by which these organisms might affect nematode populations include changing the host nutritional status and the production of toxic metabolites (18). Additional evidence suggesting a role for Rhizobium and related organisms in the H. schachtii suppressiveness was provided by an OFRG analysis of soil bacteria, which found a much larger number of α-proteobacteria in the suppressive 9E soil than in an adjacent nonsuppressive soil (24).
A crystalline layer associated with the exterior of H. schachtii cysts was observed after harvesting the cysts from the greenhouse trials. A similar structure was first described by Schmidt in 1871 (15). It has been suggested that this layer is derived from past cuticles (21) or from nematode exudates (15, 16) and that it might be a useful taxonomic indicator (2). Chemical analysis determined that it is composed of several compounds, including n-tetracosanoic acid, hexacosanoic acid, other aliphatic acids, and calcium salts (4). Brown et al. suggested that a symbiotic fungus that metabolizes excretory products from the nematode might produce this crystalline layer (4). However, this structure has also been reported to occur under sterile conditions (28). In this study, we observed that the amount of crystalline layer associated with the H. schachtii cysts correlated with the level of suppressiveness. Future investigations would be needed to determine whether this material is involved in the suppressiveness or whether it is produced by an organism(s) involved in the suppressiveness.
Acknowledgments
We thank John Darsow for his technical assistance and Alexandra J. Scupham, Elizabeth Bent, and Rabiu Olatinwo for their critical reviews of the manuscript.
This research was funded by in part by grants from the University of California Integrated Pest Management program and the University of California Center for Pest Management Research and Extension. L.V. was supported by Vaddia-BARD Postdoctoral Award no. FI-306-00 from BARD, The United States-Israel Binational Agricultural Research and Development Fund.
REFERENCES
- 1.Baker, K. F., and R. J. Cook. 1974. Biological control of plant pathogens. Freeman, San Francisco, Calif.
- 2.Baldwin, J. G., and M. Mundo-Ocampo. 1991. Heteroderinae, cysts and non-cyst-forming nematodes, p. 275-362. In W. R. Nickle (ed.), Manual of agricultural nematology. Marcel Dekker, New York, N.Y.
- 3.Becker, J. O., H. D. Ohr, N. M. Grech, and M. E. Sims. 1998. Evaluation of methyl iodide as a soil fumigant in container and small field plot studies. Pestic. Sci. 52:58-62. [Google Scholar]
- 4.Brown, G., R. K. Callow, C. D. Green, F. G. W. Jones, J. H. Rayner, A. M. Shephed, and T. D. Williams. 1971. The structure, composition and origin of the sub-crystalline layer in some species of the genus Heterodera. Nematologica 17:590-599. [Google Scholar]
- 5.Caswell, E. P., I. J. Thomason, and H. E. McKinney. 1985. Extraction of cysts and eggs of Heterodera schachtii from soil with an assessment of extraction efficiency. J. Nematol. 17:337-340. [PMC free article] [PubMed] [Google Scholar]
- 6.Hallmann, J., R. Rodriguez-Kabana, and J. W. Kloepper. 1999. Chitin-mediated changes in bacterial communities of the soil, rhizosphere and within roots of cotton in relation to nematode control. Soil Biol. Biochem. 31:551-560. [Google Scholar]
- 7.Hay, F. S., and R. A. Skipp. 1993. Fungi and actinomycetes associated with cysts of Heterodera trifolii Goffart (Nematoda: Tylenchida) in pasture soils in New Zealand. Nematologica 39:376-384. [Google Scholar]
- 8.Hugenholtz, P., G. W. Tyson, R. I. Webb, A. M. Wagner, and L. L. Blackall. 2001. Investigation of candidate division TM7, a recently recognized major lineage of the domain Bacteria with no known pure-culture representatives. Appl. Environ. Microbiol. 67:411-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jansson, J. K., and T. Lesser. 1996. Quantitative PCR of environmental samples, p. 2.7.4.1-2.7.4.19. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 10.Kloepper, J. W., R. Rodriguez-Kabana, J. A. McInroy, and R. W. Young. 1992. Rhizosphere bacteria antagonistic to soybean cyst (Heterodera glycines) and root-knot (Meloidogyne incognita) nematodes: identification by fatty acid analysis and frequency of biological control activity. Plant Soil 139:75-84. [Google Scholar]
- 11.Kluepfel, D. A., T. M. McInnis, and E. I. Zehr. 1993. Involvement of root-colonizing bacteria in peach orchard soils suppressive of the nematode Criconemella xenoplax. Phytopathology 83:1240-1245. [Google Scholar]
- 12.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, New York, N.Y.
- 13.Neipp, P. W., and J. O. Becker. 1999. Evaluation of biocontrol activity of rhizobacteria from Beta vulgaris against Heterodera schachtii. J. Nematol. 31:54-61. [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez-Kabana, R., J. W. Kloepper, D. G. Robertson, and L. W. Wells. 1992. Velvetbean for the management of root-knot and southern blight in peanut. Nematropica 22:75-80. [Google Scholar]
- 15.Schmidt, A. 1871. Ueber den Rüben-Nematoden (Heterodera schachtii A.S.). Z. Ver. RübenZucker-Ind. dt. Reich. 21:1-19.
- 16.Schmidt, A. 1872. Zweiter Bericht über die Rübennematoden. Z. Ver. RübenZucker-Ind. dt. Reich. 22:67-75.
- 17.Shepherd, A. M. 1970. Extraction and estimation of Heterodera, p. 23-33. In J. F. Southey (ed.), Laboratory methods for work with plant and soil nematodes. Her Majesty's Stationery Office, London, United Kingdom.
- 18.Siddiqui, Z. A., and I. Mahmood. 1995. Role of plant symbionts in nematode management: a review. Bioresour. Technol. 54:217-226. [Google Scholar]
- 19.Siddiqui, Z. A., and I. Mahmood. 1999. Role of bacteria in the management of plant parasitic nematodes: a review. Bioresour. Technol. 69:167-179. [Google Scholar]
- 20.Stirling, G. R. 1991. Biological control of plant parasitic nematodes: progress, problems and prospects. CAB International, Wallingford, United Kingdom.
- 21.Strubell, A. 1888. Untersuchungen über den Bau und die Entwicklung des Rübennematoden Heterodera schachtii Schmidt. Bibliotheca Zoolog. 2:1-52.
- 22.Subbotin, S. A., L. Waeyenberge, I. A. Molokanova, and M. Moens. 1999. Identification of Heterodera avenae group species by morphometrics and rDNA-RFLPs. Nematology 1:195-207. [Google Scholar]
- 23.Valinsky, L., G. Della Vedova, T. Jiang, and J. Borneman. 2002. Oligonucleotide fingerprinting of ribosomal RNA genes for analysis of fungal community composition. Appl. Environ. Microbiol. 68:5999-6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valinsky, L., G. Della Vedova, A. J. Scupham, S. Alvey, A. Figueroa, B. Yin, J. Hartin, M. Chrobak, D. E. Crowley, T. Jiang, and J. Borneman. 2002. Analysis of bacterial community composition by oligonucleotide fingerprinting of rRNA genes. Appl. Environ. Microbiol. 68:3243-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westcott, S. W. I., and D. A. Kluepfel. 1993. Inhibition of Criconemella xenoplax egg hatch by Pseudomonas aureofaciens. Phytopathology 83:1245-1249. [Google Scholar]
- 26.Westphal, A., and J. O. Becker. 1999. Biological suppression and natural population decline of Heterodera schachtii in a California field. Phytopathology 89:434-440. [DOI] [PubMed] [Google Scholar]
- 27.Westphal, A., and J. O. Becker. 2001. Components of soil suppressiveness against Heterodera schachtii. Soil Biol. Biochem. 33:9-16. [Google Scholar]
- 28.Zunke, U., and J. D. Eisenback. 1998. Morphology and ultrastructure, p. 31-56. In S. B. Sharma (ed.), The cyst nematodes. Chapman & Hall, London, United Kingdom.