Abstract
The potential for oxidation of ammonia in anoxic marine sediments exists through anaerobic oxidation by Nitrosomonas-like organisms, utilizing nitrogen dioxide, coupling of nitrification, manganese reduction, and anaerobic oxidation of ammonium by planctomycetes (the Anammox process). Here we describe the presence of microbial communities with the potential to carry out these processes in a natural marine sediment system (Loch Duich, Scotland). Natural microbial communities of Planctomycetales-Verrucomicrobia and β- and γ-proteobacterial ammonia-oxidizing bacteria were characterized by analysis of 16S rRNA genes amplified using group-specific primers by PCR- and reverse transcription-PCR amplification of 16S rDNA and RNA, respectively. Amplification products were analyzed by sequencing of clones and by denaturant gradient gel electrophoresis (DGGE). Amplification of primers specific for Planctomycetales-Verrucomicrobia and β-proteobacterial ammonia-oxidizing bacteria generated products at all sampling sites and depths, but no product was generated using primers specific for γ-proteobacterial ammonia-oxidizing bacteria. 16S rDNA DGGE banding patterns indicated complex communities of β-proteobacterial ammonia-oxidizing bacteria in anoxic marine sediments. Phylogenetic analysis of sequences from clones and those excised from DGGE gels suggests dominance of Nitrosospira cluster 1-like organisms and of strains belonging to a novel cluster represented in dominant bands in 16S rRNA DGGE banding patterns. Their presence indicates a group of organisms closely related to recognized β-proteobacterial ammonia-oxidizing bacteria that may be selected in anoxic environments and may be capable of anoxic ammonia oxidation. Sequence analysis of planctomycete clone libraries and sequences excised from DGGE gels also demonstrated a diverse microbial community and suggested the presence of new subdivisions, but no sequence related to recognized Anammox organisms was detected.
The concept of biogeochemical zones is a useful tool for the description and prediction of microbiological and geochemical processes in aquatic sediments (14, 15, 24). Molecular oxygen is one of the most important mediators in biogeochemical cycles since it serves as the major electron acceptor for microbial energy conservation processes. In aquatic sediments, the depth of oxygen penetration through diffusion is controlled mainly by the consumption of degradable organic matter within the sediment and in coastal ecosystems rarely exceeds more than a few millimeters (24). Consequently, microbial processes depending on the availability of free dissolved oxygen are constrained to the uppermost surface (7, 45) or, in deeper sediment layers, are coupled to irrigation and bioturbation processes of burrowing organisms (1, 64). Central to the geochemical cycling of nitrogen is nitrification, the oxidation of ammonium to nitrate via nitrite, which is thought to be controlled mainly by autotrophic bacteria (6, 19). Ammonia-oxidizing bacteria (AOB) carry out the first, often rate-limiting oxidation step but can also reduce nitrite to nitric oxide (NO) and nitrous oxide (N2O) at low oxygen tension (4, 16, 39). Classical geochemical models restrict ammonia oxidation to oxic environments or to oxic-anoxic interfaces and do not consider ammonia oxidation in completely anoxic environments (7, 19). However, recent evidence indicates that anaerobic ammonia oxidation coupled to nitrate reduction may contribute 24 to 67% of the total N2 production at two continental shelf sites at Skagerrak, Denmark (56), and Mortimer et al. (40) provided strong evidence that active nitrification processes are responsible for the formation of reoccurring nitrate peaks throughout the anoxic iron reduction zone in the marine sediments of Loch Duich, Scotland.
Physiological studies demonstrate complete anoxic conversion of ammonia by Nitrosomonas europaea and Nitrosomonas eutropha with nitrite as electron acceptor (4) and by N. eutropha with NO2 as electron acceptor (48, 49). Autotrophic AOB have also been detected in temporarily and permanently anoxic rhizosphere habitats (5). Further potential for ammonia oxidation in aquatic sediments may lie in the activity of anaerobic AOB within the Planctomycetales, utilizing nitrite as the electron acceptor, as demonstrated in wastewater treatment processes (the Anammox process [22, 54]).
The application of molecular techniques, in particular analysis of 16S rRNA genes, provides new opportunities for the assessment of ammonia-oxidizing populations in aquatic sediments. Phylogenetic analysis of 16S rRNA genes of pure and mixed cultures places AOB in three groups. Organisms involved in the Anammox process belong to the order Planctomycetales (47, 54). Although planctomycetes are present in anoxic marine sediments (31, 34), no sequence with a close phylogenetic association with those of recognized Anammox organisms has yet been reported. The second group forms a deep branch within the γ-proteobacteria of the class Proteobacteria and consists of only three recognized, closely related Nitrosococcus species (44, 62). The third group, including the majority of cultured strains, forms a monophyletic group within the β-proteobacteria and comprises two clades, Nitrosomonas and Nitrosospira. Phylogenetic analysis of 16S rDNA sequences (44, 53) indicates the existence of clusters within these two clades, and their distribution has been investigated in a number of environments (3, 26, 27, 28, 36, 37, 42, 43, 52). Pure-culture representatives have been isolated for all groups (44), with the exception of Nitrosospira cluster 1 and Nitrosomonas cluster 5, for which only clone sequences are available from marine environments (3, 37, 42).
Molecular analysis of ammonia oxidizer 16S rDNA fragments, amplified from environmental DNA, by denaturing gradient gel electrophoresis (DGGE) characterizes community structure, enables rapid analysis of clone libraries, and, through excision, reamplification, and sequencing of bands from gels, provides information on species composition. This approach has demonstrated differences in populations of Nitrosomonas-like β-proteobacterial AOB associated with polluted marine fish farm sediments (37) and marine aggregates (43) and domination by Nitrosospira-like organisms in Arctic Ocean waters (3). Analysis of 16S rDNA does not discriminate between metabolically active and quiescent cells. Analysis of environmental 16S rRNA, using reverse transcription-PCR (RT-PCR), provides greater sensitivity, because of the higher target copy number, and may indicate which members of the community are more metabolically active, if active cells contain larger numbers of ribosomes (13, 27, 58). The aim of this study was to determine the distribution and community composition of all three groups of AOB within an anoxic marine sediment system showing strong evidence of anoxic nitrification (40). As a first step in assessing their potential contribution to anoxic ammonia oxidation, 16S rRNA genes were amplified from DNA and RNA and their products analyzed by DGGE analysis and sequencing, with detailed phylogenetic analysis of almost complete 16S rRNA gene sequences from clone libraries.
MATERIALS AND METHODS
Sample location and collection.
Sediment cores were collected from Loch Duich, an organic-rich sea loch on the west coast of Scotland, at a water depth of ca. 120 m, using a stainless steel box corer with minimal disturbance of the surface. Sediment biogeochemistry at this site has been studied in detail (18, 40), and dissolved O2 penetrates only a few millimeters into sediment (P. Anschütz, personal communication). Subcores (diameter, 10 cm; length, up to 40 cm) were immediately taken from box cores and sliced at 1-cm intervals for the first 10 cm, 2-cm intervals for the subsequent 10 cm, and 4-cm intervals thereafter. Further analysis was restricted to undisturbed sediment at the center of the subcores, which were immediately frozen at −70°C until required. Analysis was carried out on samples from replicate subcores from within the same box core and on replicate samples from the same subcore. Replicate subcores, analyzed using high-resolution diffuse equilibrium in thin-film gel probes, showed distinct peaks in nitrate concentration at depths of 9 and 20 cm (0.95 and 0.6 mM NO3−, respectively) (R. C. J. Mortimer, personal communication).
DNA and RNA extraction.
The water content of sediments was reduced to ca. 5% (wt/vol) of the total water content by centrifugation at 16,000 × g for 10 min. Nucleic acids were extracted from 0.5 g of compacted sediment by disruption with a Ribolyser cell disruptor (Hybaid, Ltd., Ashford, United Kingdom) (speed 4, two 10-s bursts) in 2-ml screw cap Blue Matrix Ribolyser tubes (Hybaid) containing a mixture of ceramic and silica beads, 0.5 ml of hexadecyltrimethylammonium bromide (CTAB; Sigma, Poole, United Kingdom), extraction buffer, and 0.5 ml of phenol-chloroform-isoamyl alcohol (25:24:1) (pH 8.0). Preparation of CTAB buffer and further extraction and precipitation of nucleic acids were carried out as described by Griffiths et al. (17). Extracted nucleic acids were resuspended in 50 μl of RNase-free sterile water and divided into two aliquots for the preparation of DNA or RNA templates. DNA templates were further purified using 0.5-ml of Vivaspin concentrator columns (Vivascience, Sartorius, Germany).
PCR amplification of environmental 16S rDNA and rRNA fragments.
Extracted DNA and cDNA from RT reactions (see below) were amplified by nested PCR for DGGE analysis using 16S rDNA primer sets specific for β-proteobacterial AOB (CTO189f-GC and CTO654r [28]), the NOC1-45f and NOC2-1168r primer set specific for γ-proteobacterial AOB (57, 61), and the Planctomycetales-Verrucomicrobia assay (PV assay [8]) using the PLA40f primer in combination with the general eubacterial primer pf1053r (11). Alignments of PLA40f and pf1053r with sequences of recognized Anammox strains revealed no mismatches. However, mismatches in the NOC primer set (62), following alignments of γ-proteobacterial AOB from the GenBank database, led to modification of primer NOC1-45f to CGTYGGAATCTGGCCTCTAGA and of NOC2-1168r to AGATTAGCTCCGCATCGCTG (modifications are shown in bold). PCR products were reamplified using the general eubacterial 357f-GC and 518r primers (41). Prior to secondary amplification, PCR products were diluted 1:50 to prevent the amplification of nontarget bacteria without affecting DGGE community profiles. All specific primer sets were tested for stringency against Planctomyces maris AL (NCIMB 2232), Planctomyces brasiliensis VL32 (NCIMB 13185), Nitrosococcus oceanus (NCIMB 11848), Nitrosomonas europaea (NCIMB 11850), Nitrosospira multiformis (NCIMB 11849), and Escherichia coli (NCIMB 86).
RT-PCR was done by the method of Griffiths et al. (17) with some modifications. DNA-free RNA for RT-PCR analysis was obtained by treating 8 μl of extracted RNA with 2 U of RQ1 RNase-free DNase (Promega Corp., Madison, Wis.) for 30 min at 37°C as specified by the manufacturer. Prior to RT, the RNA secondary structure was melted by incubating the RNA samples with reverse 16S rRNA PCR primer (see above) at 0.9 μM at 70°C for 10 min. Samples of annealed primer-template were snapped on ice, and 8 μl of RT reaction mixture was added as specified by the manufacturer (SuperScript RT RNase H-Reverse Transcriptase; GibcoBRL). RT was carried out at 42°C for 50 min, and the enzyme was subsequently heat-inactivated for 15 min at 70°C. Performing PCR on RNA samples after DNase treatment confirmed the amplification of RNA templates free of DNA contamination. Following transcription of 16S rRNA templates into cDNA, PCR was carried out as described above for DNA templates. PCR-amplified fragments were resolved by electrophoresis on 1% (wt/vol) horizontal agarose gels with visualization of DNA by ethidium bromide fluorescence and with Hyperladder DNA fragment size markers (Bioline). PCR amplification was also carried out on pure cultures or clones representative of each recognized cluster of β-proteobacterial AOB (53) for use as reference sequences in CTO189f-GC plus CTO654r DGGE analysis, as follows: EnvB1-8 (Nitrosospira cluster 1), pH4.2A/27 (Nitrosospira cluster 2), pH4.2A/4 (Nitrosospira cluster 3), pH7B/C3 (Nitrosospira cluster 4), EnvA1-21 (Nitrosomonas cluster 5), EnvC1-19 (Nitrosomonas cluster 6), and N. europaea (Nitrosomonas cluster 7).
Cloning inserts of 1.1 and 1.4 kb specific for β-proteobacterial AOB and Planctomycetales-Verrucomicrobia, respectively, were created by direct PCR amplification of DNA extracts of sample “core 8/depth 20” with the βAMO161f and βAMO1301r primer set, which is selective but not completely specific for β-proteobacterial AOB (38), or by the PV assay, in conjunction with the general eubacterial primer 1492r (8, 11). PCR fragments were purified by standard agarose electrophoresis, and excised DNA bands were cleaned using QIAquick columns (Qiagen, Hilden, Germany). Primer pairs and PCR conditions for all amplifications used are summarized in Table 1. Individual reagents and their concentrations were as follows: 1× PCR buffer (Bioline), 1 U of BioTaq DNA polymerase (Bioline), 1.5 mM MgCl2, each primer at a concentration of 0.2 μM, and each deoxynucleoside triphosphate dNTP (Bioline) at a concentration of 250 μM. Amplification was performed with a total volume of 50 μl using an Omn-E thermal cycler (Hybaid), applying the thermal cycle conditions shown in Table 1.
TABLE 1.
Primers and PCR amplification conditions
| Primer set (fragment length) | Target group | PCR approach | Thermocycling programb
|
||
|---|---|---|---|---|---|
| Initial denaturation (min) | Annealing temp (°C) | Extension time(s) | |||
| βAMO161f-βAMO1301r (1.1 kb) | β-Proteobacterial ammonia oxidizers | Cloning, direct amplification | 5 | 55 | 80 |
| PLA40f-1492r (1.4 kb) | Planctomycetales-Verrucomicrobia | Cloning, direct amplification | 5 | 61 | 100 |
| CTO189f-GCa-CTO654r (465 bp) | β-Proteobacterial ammonia oxidizers | DGGE, nested amplification, first stage | 5 | 55 | 45c |
| NOC1-45f-NOC2-1168r (1.1 kb) | γ-Proteobacterial ammonia oxidizers | DGGE, nested amplification, first stage | 5 | 60 | 80 |
| PLA40f-pf1053r (1.0 kb) | Planctomycetales-Verrucomicrobia | DGGE, nested amplification, first stage | 5 | 61 | 70 |
| 357f-GCa-518r (161 bp) | Eubacteria | DGGE, nested amplification, second stage | 2 | 55 | 30c |
The general thermocycling program was used as follows: 5 min at 94°C, followed by 10 cycles of 30 s at 94°C, 30 s at the specified annealing temperature, and the specified extension time at 72°C, followed by 25 cycles of 30 s at 92°C, 30 s at the specified annealing temperature, and the specified extension time at 72°C (increasing by 1 s per cycle), followed by a 10-min final extension at 72°C. All specific amplifications were performed with a hot start by adding the Taq polymerase after the initial denaturation step; all nested second-stage nonspecific amplifications were started by placing cooled tubes containing the PCR mix into a preheated (94°C) PCR block.
Final extension time, 5 min.
DGGE analysis.
DGGE was carried out as described previously (28), using the DCode universal mutation detection system (Bio-Rad Laboratories) and 1.5-mm 8% polyacrylamide gels containing denaturant gradients of 40 to 60%, for separation of CTO189f-GC plus CTO654r PCR products and of 35 to 62% for analysis of 357f-GC plus 518r PCR products. The gels were stained for 20 min in ethidium bromide and destained twice for 10 min in 1× TAE buffer (48.22 g of Tris base, 2.05 g of anhydrous sodium acetate, and 1.86 g of disodium EDTA · 2H2O [pH 8] in 1 liter of distilled H2O) prior to UV transillumination. The gels were also silver stained for better visual resolution of complex community banding patterns.
Cloning and sequence analysis.
Three purified PCR products resulting from amplification with primers βAMOf plus βAMOr or PLA40f plus 1492r were pooled to minimize PCR drift (59), ligated into the pGEM T-vector system (Promega Ltd., Southampton, United Kingdom), and transformed into XL1-Blue MRF Kan supercompetent E. coli cells (Stratagene Inc., Maidstone, United Kingdom) as specified by the manufacturer. Transformed colonies were screened for inserts of the correct size by PCR amplification with the specific primers described above. For each PCR amplicon, 150 clones with the correct fragment size insert were subjected to nested PCR with the primer sets described above and the 357f-GC plus 518r PCR products were screened by DGGE for sequence differences within the hypervariable V3 region. For β-proteobacterial AOB, clone libraries selected for full-length sequencing of the entire insert represent all recognized clones with different DGGE migration patterns. Ten replicate clones of the recognized main β-proteobacterial sequences showing identical migration patterns on the DGGE gels were also selected for sequencing. Clones from libraries generated by the PV assay were selected at random for full-length sequencing of the entire insert. The selected clones were amplified with vector primers M13f and M13r (Promega Ltd.), and M13 PCR products were purified by standard preparative agarose gel electrophoresis as described above. To ensure complete full-length sequence read without any ambiguities of sequences of potentially closely related β-proteobacterial AOB, the M13 vector products were sequenced in both reading directions using the SP6 and T7 vector primers (Promega Ltd.), the internal 16S rDNA primer CTO654r, and 518f (reverse complement to the above described 518r). M13 vector products derived from the PV assay were sequenced using vector primers SP6 and T7 and the internal 16S rDNA primer pf1053r.
Individual DGGE bands were excised, and acrylamide slices were crushed and resuspended overnight at 4°C in 30 μl of sterile water to elute the DNA. Reamplified PCR products were again subjected to DGGE analysis to ensure purity and correct migration within the gels. Reamplified PCR products showing single bands during DGGE analysis were purified as described above and sequenced with the 518r primer. Sequencing reactions were performed using the BigDye Terminator cycle-sequencing kit (PE Biosystems, Warrington, United Kingdom), and the cycle-sequencing products were analyzed with a model ABI377 automated sequencer (PE Biosystems).
Sequence assembly and manual refinement of alignments were carried out using the Sequencer 4.1 program (Genes Codes Corp., Ann Arbor, Mich.) or the multiple-alignment algorithms as implemented in Clustalx 1.81 (EMBL, Heidelberg, Germany). For phylogenetic analysis, 1.1- and 1.4-kb clone sequences for β-proteobacterial AOB and Planctomycetales-Verrucomicrobia, respectively, were aligned with closely related 16S rDNA sequences retrieved from the GenBank database at the National Center for Biotechnology Information (35) using the Basic Local Alignment Search Tool (BLASTn [2]). Mismatches between clone sequences, sequences derived from DGGE bands, and sequences retrieved from GenBank were analyzed using the B12Seq algorithm as implemented in BLASTn, and DNA-DNA sequence similarity percentages were calculated using the sequence similarity matrix function as implemented in BioEdit (freeware by Tom Hall, Department of Microbiology, University of North Carolina). Sequence inserts that were present in a minority of aligned sequences were omitted from further analysis. Phylogenetic relationships between pairs of 16S rDNA clone sequences were calculated using maximum parsimony (Phylip 3.62) and distance analysis as implemented in PAUP 4.1 (55). Maximum-parsimony analysis was carried out using the global rearrangement criterion and randomized input of sequence order with 1,000 times bootstrap resampling. A maximum-likelihood analysis of the most parsimonious tree, constructed from a subset of sequences representing the major sequence groups, was used to estimate the proportion of variable sites needed for the analysis of LogDet/Paralinear distances (29) of variable sites (32). LogDet/Paralinear distances were calculated using the minimal evolution criterion, and data were bootstrapped 1,000 times. Phylogenetic trees were constructed from distance analysis using the neighbor-joining method (46). Additional distance matrix corrections (Kimura2; Jukes and Cantor) were used to infer established tree topologies. To exclude chimeric 16S rDNA primary structures of clone sequences prior to phylogenetic analysis, sequences were analyzed using CHIMERA CHECK of the Ribosomal Database Project (RDP [35]), and the terminal 500 to 600 nucleotide sequence positions of the 5′ and 3′ ends were used in separate phylogenetic analyses, conducted as described above.
Nucleotide sequence accession numbers.
The partial clone sequences determined in this study have been deposited in the GenBank database under accession no. AY114311 to AY114349.
RESULTS
Nucleic acid extraction and PCR amplification.
Total sediment DNA consisted of high-molecular-weight DNA fragments (10 to 20 kb). Crude nucleic acid extracts showed little contamination, and PCR amplification was generally successful without further purification of the extract. However, results were more consistent and PCR product yields were greater after one dialysis “cleanup” step, which was superior to dilution of crude nucleic acid extracts. Direct PCR amplification of purified extracts with the CTO189f plus CTO654r primers, the βAMO161f plus βAMO1301r primers, and the PV assay generated products at almost all sampling sites and depths (10 to 20 ng μl−1). All nested PCR amplifications yielded strong PCR amplification signals with product concentrations in the range of 25 to 35 ng μl−1. Aliquots of nucleic acid extracts were of sufficient quality to generate direct amplicons of high yield in RT-PCR amplifications without further purification. However, only weak products or products of spurious fragment size were obtained using βAMO1301r or 1492r as the transcription primer during RT-PCR, preventing the generation of large-fragment RT-PCR products suitable for cloning and detailed phylogenetic analysis. No direct or nested DNA PCR or RT-PCR product was obtained using the NOC1-45f plus NOC2-1168r primer set.
16S rDNA and rRNA DGGE analysis.
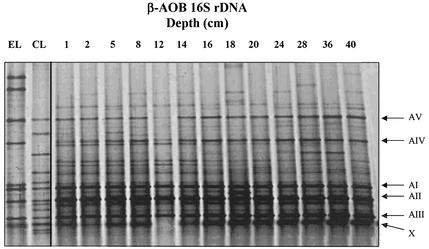
DGGE banding patterns of CTO189f-GC plus CTO654r 16S rDNA PCR products, specific for β-proteobacterial AOB, were similar for all replicates and sampling depths. Two major bands were present in all samples (Fig. 1), with the upper band migrating to a position between Nitrosomonas cluster 5 and 6 controls and the lower band migrating to a position between Nitrosospira cluster 1 and 4 controls. However, PCR products generated using CTO primers did not comigrate with any of the control sequences representing recognized β-proteobacterial AOB clusters. In addition, the presence of multiple bands, resulting from degeneracy of the CTO primer set, hampered reliable identification of closely migrating bands (28, 63). DGGE analysis was then carried out on nested 160-bp 16S rDNA PCR products, generated using primers 357f-GC plus 518r and spanning only the hypervariable V3 region. These PCR products were shorter, and their denaturing characteristics were more suitable for DGGE analysis. Banding patterns proved to be more discriminative and demonstrated diverse communities of β-proteobacterial AOB. More than 15 distinct DGGE bands were obtained per sample, five of which were dominant (Fig. 2), comprising three closely migrating bands of high intensity (AI, AII, and AIII) and two bands of lower intensity (AIV and AV). DGGE banding profiles provided no evidence of major changes in community structure with sediment depth. Dominant bands were present in all samples at all depths, and variation of the banding pattern was apparent only through the presence or absence of less prominent bands. Variation in the intensity of major DGGE bands, indicating changes in relative abundance, were less pronounced than in CTO-derived profiles, probably due to saturation of PCR templates in the nested PCR approach.
FIG. 1.
DGGE analysis of direct-amplification 465-bp β-proteobacterial AOB 16S rDNA sequences amplified from DNA extracted from Loch Duich marine sediments and representative cluster controls. Lanes: CL1, EnvB1-8 (Nitrosospira cluster 1); CL2, pH4.2A/27 (Nitrosospira cluster 2); CL3, pH4.2A/4 (Nitrosospira cluster 3); CL4, pH7B/C3 (Nitrosospira cluster 4); CL5, EnvA1-21 (Nitrosomonas cluster 5); CL6, EnvC1-19 (Nitrosomonas cluster 6); CL7, N. europaea (Nitrosomonas cluster 7). The right-hand lanes show a depth profile of Loch Duich sediment samples.
FIG. 2.
DGGE analysis of nested 160-bp β-proteobacterial ammonia oxidizer 16S rDNA sequences amplified from DNA extracted from Loch Duich marine sediments and reference sequences retrieved from Loch Duich sediments by cloning and by sequencing of excised DGGE bands. Lanes: EL, environmental library (DGGE sequences excised and reamplified from 357f-GC plus 518r 16S rDNA PCR products); CL, clone library (βAMO161f plus βAMO1301 clone sequences retrieved from Loch Duich sediments and reamplified with 357f-GC plus 518r for DGGE analysis); 1 to 40, depth profile of Loch Duich 16S rDNA 357f-GC plus 518r PCR products. Clone sequences shown in lane CL are (from top to bottom) LD1-B6, LD1-B37, LD1-A40, LD1-A3, LD1-A1, LD1-A2, LD1-B20, LD1-B10, LD1-B28, LD1-A10, LD1-A15, LD1-B9, and LD1-A20; DGGE excised sequences shown in lane EL are (from top to bottom) LD1-env-A12, LD1-env-B12, LD1-env-B11, LD1-env-B5, LD1-env-A8, LD1-env-A5, LD1-env-A1, and LD1-env-A2. Arrows (right) mark the prominent bands as referred to in the text. No sequence was generated from band X when it was excised and reamplified.
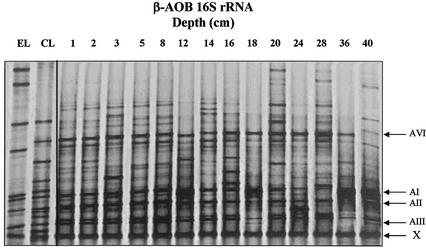
RNA-derived DGGE banding patterns, generated by RT-PCR using primers 357f-GC and 518r, showed similar diversity to those derived from DNA (Fig. 3), but not all dominant bands present in rDNA-based profiles were observed. Bands AIV and AV were absent or present only as faint bands, and, although bands AI to AIII were dominant, their intensities were more variable than in rDNA-derived profiles. A sixth band (AVI) was also recognized as dominant in almost all rRNA RT-PCR product banding profiles. Generally, differences in banding patterns between individual samples were greater than for rDNA-derived profiles.
FIG. 3.
DGGE analysis of nested 160-bp RT-PCR products derived from β-proteobacterial ammonia oxidizer 16S rRNA extracted from Loch Duich marine sediments and reference sequences retrieved from Loch Duich sediments by cloning and excising bands from DGGE bands. Lane designations are as in the legend to Fig. 2. Arrows (right) mark the prominent bands as referred to in text. No sequence was generated from band X when it was excised and reamplified.
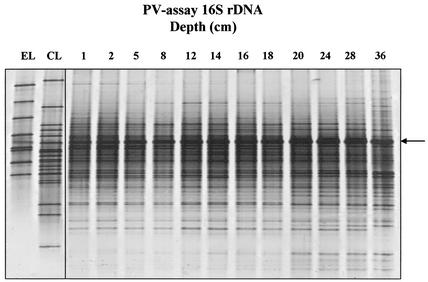
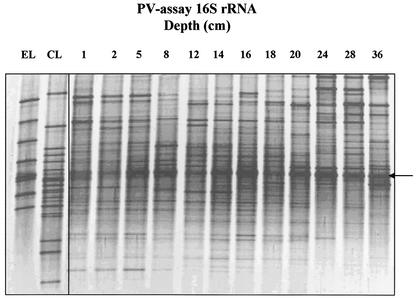
16S rDNA PCR products generated with the PV assay also demonstrated diverse microbial communities, with up to 40 individual bands per lane. The reproducibility of DGGE profiles in replicate samples was similar to that for β-proteobacterial AOB (Fig. 4), and bands were generally of similar intensity, indicating even relative abundance of different sequence types. Individual bands, however, showed trends of increasing or decreasing signal intensity with sampling depth. RNA-derived DGGE profiles were of comparable complexity (Fig. 5), but, as with RNA-derived AOB profiles, differences in banding patterns between individual samples were more distinct through the presence or absence of dominant bands in individual samples.
FIG. 4.
DGGE analysis of nested 160-bp Planctomycetales-Verrucomicrobia 16S rDNA sequences amplified from DNA extracted from Loch Duich marine sediments and reference sequences retrieved from Loch Duich sediments by cloning and sequencing of excised bands from DGGE gels. Lanes EL, environmental library (DGGE sequences excised and reamplified from 357f-GC plus 518r 16S rDNA PCR products); CL, clone library (PV clone sequences retrieved from Loch Duich sediments, reamplified with 357f-GC plus 518r for DGGE analysis); 1 to 36, depth profile 16S rDNA PCR products of Loch Duich sediment samples. Clone sequences in lane CL are (from top to bottom) LD1-PA42, LD1-PA15, LD1-PB1, LD1-PA13, LD1-PA26, LD1-PB2, LD1-PA40, LD1-PA32, LD1-PA16, LD1-PB20, LD1-PA30, LD1-PA20, LD1-PA21, LD1-PB12, LD1-PA11, LD1-PB3, LD1-PA50, LD1-PA34, and LD1-27; DGGE excised sequences in lane EL are (from top to bottom) LD1-env-P8, LD1-env-P7, LD1-env-P6, LD1-env-P1, LD1-env-P4, LD1-env-P3, LD1-env-P2, and LD1-env-P5. No sequence was generated from the band marked with an arrow when it was excised and reamplified.
FIG. 5.
DGGE analysis of nested 160-bp Planctomycetales-Verrucomicrobia 16S rDNA products amplified from RNA extracted from Loch Duich marine sediments and reference sequences retrieved from Loch Duich sediments by cloning and sequencing excised bands from DGGE gels. Lane designations are as in the legend to Fig. 4. No sequence was generated from the band marked with an arrow when it was excised and reamplified.
Sequence and phylogenetic analysis of clones and DGGE bands. (i) β-Proteobacterial AOB.
The 150 AOB 1.1-kb clones retrieved from anoxic sediment at a sampling depth of 20 cm showed 13 distinct migration patterns when subjected to DGGE analysis. The majority (85%) comigrated with clone LD1-A10, and 10 representatives of these clones, sampled randomly, showed 100% sequence identity. This indicates that in this case, migration is a reliable measure of sequence similarity.
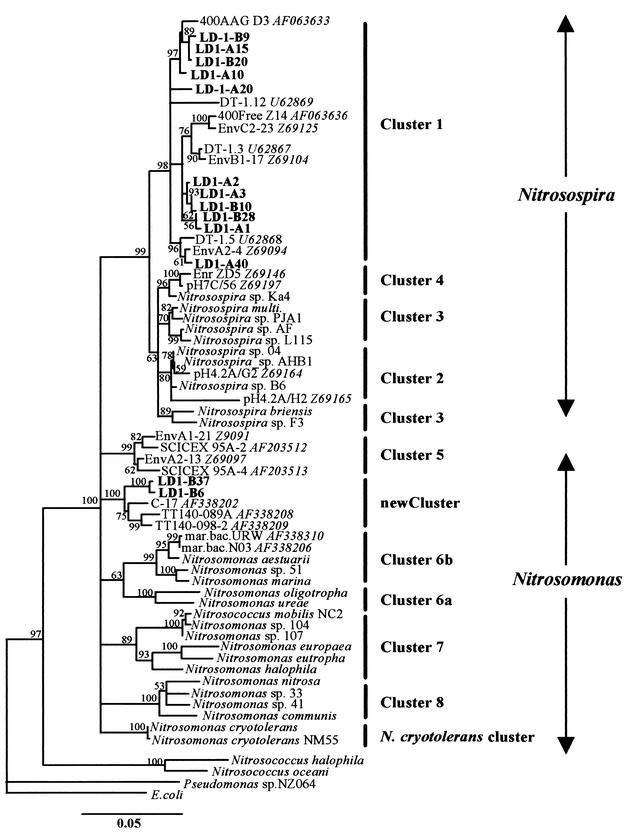
The sequence similarity of all β-proteobacterial AOB clones generated in this study was high, on average 98.6%, indicating closely related clones. Phylogenetic and 16S rDNA sequence similarity analysis showed that the clones fell within two major groups. Group 1 contained closely related clones with sequence similarities between 99.7 and 97.9%, divided into five subgroups (Fig. 6). Sequence similarities within subgroups containing two or more clones were greater than 99.4%, and differences may have resulted from sequencing or cloning errors. However, the maximum average similarity among the five subgroups was 98.1 to 98.5%, indicating independent sequences. Group 1 clones fell within Nitrosospira cluster 1, representatives of which have previously been amplified from marine environments (28, 37, 43). The phylogenetic discrimination of cluster 1 within the nitrosospiras was better supported in all tree topologies than that between all other Nitrosospira subclusters. The closest recognized AOB relative to cluster 1 clone LD1-A1 was Nitrosospira sp. strain 04 (97.5% sequence similarity).
FIG. 6.
Evolutionary distance dendrogram showing the positions of environmental 16S rDNA 1.1-kb clone sequences (labels in bold), recovered from anoxic marine sediments, in relation to representative members within the β-proteobacterial ammonia oxidizers. The tree is based on results of neighbor-joining analysis of LogDet/Paralinear distances. The multifurcation connects branches for which a relative order could not be determined unambiguously in the majority of bootstrap resamplings (1,000 replicates). Conservative bootstrap values of maximum parsimony or LogDet/Paralinear distances are indicated. The scale bar indicates an estimated 0.05 change per nucleotide position. Cluster designation are as suggested by Purkhold et al. (44) and Stephen et al. (53). GenBank accession numbers of closely related clone sequences are shown in italics.
Group 2 contained clones LD1-B6 and LD1-B37, with 99.6% sequence similarity to each other but with only 97.5% sequence similarity when compared using the V3 region only. The maximum sequence similarity to other clones generated in this study was 95.4% (clone LD1-A1). A BLASTn search revealed that the closest association was with three sequences generated from marine isolates (Table 2) (57, 60). The similarities of clones LD1-B6 and LD1-B37 to retrieved sequences were low (96.6 to 97.4%) in comparison to those clustering with Nitrosospira. The closest sequence similarity to a recognized AOB (Nitrosomonas cryotolerans NM55) was 96.3%. LogDet/Paralinear distances did not support the association of clones LD1-B6 and LD1-B37 (together with retrieved sequences) with any previously reported Nitrosospira or Nitrosomonas cluster. The closest sequence similarity to N. cryotolerans was reflected by a shared node of the new cluster in all tree topologies inferred, but bootstrap support for this association was below 50% for LogDet/Paralinear distances. The new cluster was supported in all clustering methods used, with good bootstrap support even when using only partial sequences comprising the first or last 500 bp of clone sequences. Clustering was more stable and had better bootstrap support than for other Nitrosospira clusters and was stable when phylogenetic analysis was performed with only 160-bp fragments spanning the V3 region.
TABLE 2.
Similarities of 16S rDNA sequences from excised DGGE bands representative of β-proteobacterial AOB and Planctomycetales-Verrucomicrobia 16S rDNA sequences (160 bp) to sequences retrieved from GenBank or clone sequences generated in this study
| DGGE analysis | Closest matcha | Cluster/linage association | 16S rDNA sequence similarity (%) |
|---|---|---|---|
| β-Proteobacterial AOB sequences | |||
| Env-A1 | LD1-B20 | Cluster 1 | 99.3 |
| Env-A2 | LD1-A10 | Cluster 1 | 99.3 |
| Env-A5 | LD1-A2 | Cluster 1 | 99.3 |
| Env-A8 | LD1-A1 | Cluster 1 | 99.3 |
| Env-A12 | N. europaea | Cluster 7 | 98.1 |
| Env-B5 | TT140-098-2 AF338209 | Novel cluster | 96.2b |
| Env-B11 | LD1-B6 | Novel cluster | 99.3b |
| Env-B12 | Nitrosospira sp. strain D27 | Cluster 6 | 95.2 |
| PV sequences | |||
| Env-P11 | Sva0585 AJ297463 | Verrucomicrobium | 91.9 |
| Env-P2 | PA-30 | WS3 | 98.1 |
| Env-P3 | PA-16 | WS3 | 96.2 |
| Env-P4 | PA-30 | WS3 | 91.3 |
| Env-P5 | Sludge H28 AF234749 | Planctomyces | 74.1 |
| Env-P6 | PA-21 | BRC1 | 79 |
| Env-P7 | Sva0500 AJ241008 | Verrucomicrobium | 87.3 |
| Env-P8 | PA-15 | Verrucomicrobium | 93.7 |
GenBank clone sequence accession numbers are shown in italics.
Similarities to the suggested new β-proteobacterial AOB cluster are shown in bold type. The highest similarity of DGGE sequences generated by the PV assay to the Anammox cluster was 71%.
DGGE analysis provides an indication of the relative abundance of sequences with different migration characteristics within a clone library and an estimate of the relative abundance of corresponding organisms in environmental samples. Alignment of clone sequences with those retrieved from DGGE bands and comparison of DGGE profiles of clone library fragments, excised DGGE bands, and environmental DGGE profiles enabled an assessment of environmental relevance of clone sequences. Of eight DGGE bands from environmental samples, five comigrated with clone sequence fragments and showed almost 100% sequence alignment with comigrating clone fragment sequences (Table 3; Fig. 2 and 3). All six dominant bands present in rDNA- or rRNA-derived DGGE gels were represented either as clone sequences or as sequences excised from DGGE gels. Nitrosospira cluster 1-like sequences comigrated with dominant bands AI to AIII, and sequences associated with the new cluster sequences (Table 3) comigrated with dominant bands AV and AVI. An additional sequence associated with the proposed new cluster was amplified from a dominant DGGE band (AIV) with closest sequence similarity to the marine isolate TT140-098-2 (57, 60). Clone LD1-B37 was not represented as a dominant band in rDNA-derived DGGE but comigrated with dominant band AVI in rRNA-derived gels (Fig. 3). In contrast, clone LD1-B6 and DGGE sequence env-B5 dominated in rDNA-derived but not in rRNA-derived DGGE profiles. The new cluster was therefore represented by clone sequences and sequences generated from dominant DGGE bands. Sequence fragments with closest matches to Nitrosomonas spp. were also generated from DGGE bands of weak signal intensity (env-A12 and env-B12).
TABLE 3.
Closest-match similarities of 1.1-kb 16S rDNA clone sequences of proposed novel β-proteobacterial ammonia oxidizer cluster with closest-match sequences retrieved from GenBank.
| Clone sequence or straina | Similarity (%) to novel cluster:
|
||||
|---|---|---|---|---|---|
| LD1-B37 | LD1-B6 | TT140-089A | TT140-098-2 | C-17 | |
| LD1-B6 | 99.6 | ||||
| TT140-089A AF338208 | 96.9 | 96.9 | |||
| TT140-098-2 AF338209 | 97.5 | 97.7 | 98.5 | ||
| C-17 AF338202 | 97.4 | 97.4 | 97.6 | 98.4 | |
| Nitrosomonas cryotolerans NM55 | 96.3 | 96.1 | 95.3 | 95.9 | 96.2 |
| Nitrosomonas cryotolerans | 96.2 | 96.0 | 95.4 | 96.0 | 96.3 |
| Nitrosospira sp. strain Ka4 | 96.1 | 95.9 | 95.5 | 96.1 | 96.1 |
| Nitrosospira sp. strain 40KI | 95.9 | 95.7 | 95.0 | 95.6 | 95.9 |
| Nitrosospira multiformis | 95.5 | 95.3 | 94.6 | 95.2 | 95.4 |
The proposed novel β-proteobacterial ammonia oxidizer clusters are shown in bold type. GenBank clone sequence accession numbers are in italics.
(ii) Planctomycetales-Verrucomicrobia.
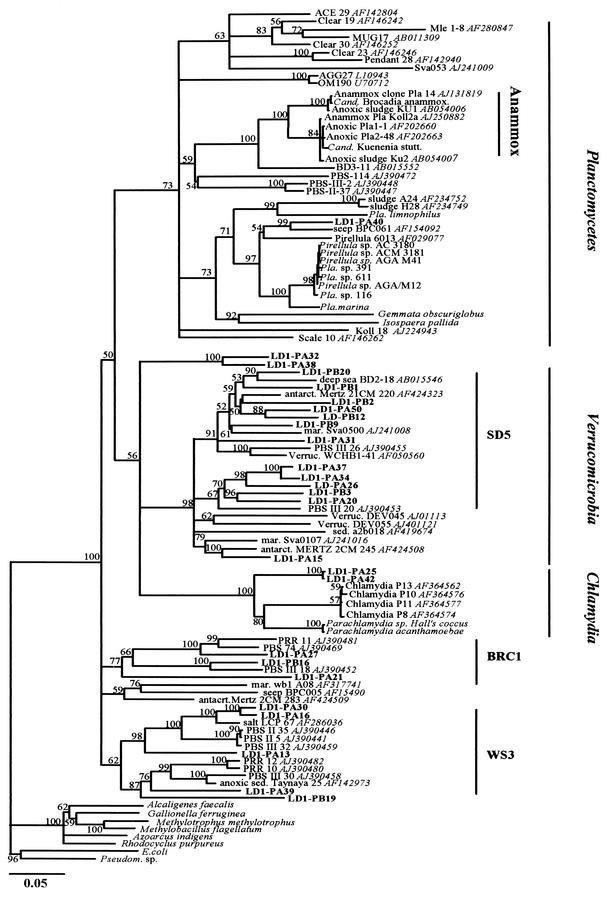
DGGE analysis of the V3 region revealed highly diverse clone sequences generated by the PV assay. For 150 clones analyzed, ca. 40 different DGGE migration patterns were observed at similar frequencies. Sequences of the 1.4-kb insert, in 26 clones selected at random, were highly dissimilar (average 78.2% sequence similarity). Clone sequences were aligned with the closest matching sequences obtained from GenBank, using BLASTn, and with recognized Anammox sequences. None of the 26 clones contained the 20-nucleotide insertion (E. coli position 157, helix 9 insertion [47]) found in Anammox organisms and closely related clone sequences.
Phylogenetic analysis of all sequences necessitated the restriction of alignments with 1.0-kb sequence fragments to the closest matches from GenBank. Phylogenetic analysis of 1.4- and 1.0-kb alignments yielded stable lineage or division association of clone sequences in all tree topologies inferred, even when using only partial sequences (the first or last 0.5 kb). The majority of PV clone sequences were associated with subdivision 5 of the Verrucomicrobia division or with bacterial candidate division WC1 and proposed bacterial candidate division BRC1 (Fig. 7). Clones LD1-PA32 and LD1-PA38 (99.3% sequence similarity to each other) may represent a new subdivision within the Verrucomicrobia since the closest match (clone wb1 A08) had only 80.2% sequence identity and phylogenetic analysis demonstrated no association with previously recognized subdivisions. Clones LD1-PA25 and LD1-PA42 may represent a previously unrecognized subdivision within Chlamydia, since both clones (99.3% sequence similarity to each other) were equally distant from recognized Chlamydia and Parachlamydia (87% sequence similarity). Of all analyzed clones, only LD1-P40 was associated with the Planctomycetales division. The closest match to this clone was marine seep clone BPC016 (89.0% sequence similarity), and the closest match to a recognized planctomycete was 87.8% to Planctomycetales sp. strain 116. Anammox-related sequences were grouped as a stable cluster in all tree topologies inferred (even if the helix 9 insertion was excluded for phylogenetic analyses), but in all tree topologies, none of the clones generated in this study was associated with the Anammox cluster (closest similarity, 77.5% for clone LD1-PA13).
FIG.7.
Evolutionary distance dendrogram showing the positions of environmental 16S rDNA 1.4- and 1.0-kb clone sequences (labels in bold) recovered from anoxic marine sediments, in relation to recognized and associated Anammox sequences (47), representative members of the order Planctomycetales, the divisions Verrucomicrobia and Chlamydia, and closely related bacterial lineages. The tree is based on results of neighbor-joining analysis of LogDet/Paralinear distances. The multifurcation connects branches for which a relative order could not be determined unambiguously in the majority of bootstrap resamplings (1,000 replicates). Conservative bootstrap values of maximum parsimony or LogDet/Paralinear distances are indicated. Subdivision (SD5) and bacterial lineage candidate designations (BRC1, WS3) are suggested by Hugenholtz et al. (20), Doika et al. (9), and Derakshani et al. (8). GenBank accession numbers of closely related clone sequences are shown in italics. The scale bar indicates an estimated 0.05 change per nucleotide position.
From eight DGGE bands derived from environmental samples, no sequence was associated with Anammox-related sequences. Five sequences were associated as the closest match with highest sequence similarity to clone sequence fragments generated in this study (Table 3). DGGE sequences were associated with the Verrucomicrobia and bacterial candidate divisions WR3 and BRC1, and one sequence was associated as closest match with a Planctomyces clone isolated from a wastewater treatment system.
DISCUSSION
Ammonia oxidation is generally ignored in traditional models of geochemical cycling in anoxic marine sediments, despite the demonstration of bacterial oxidation of ammonia under anoxic conditions in laboratory culture. The aim of this study was to determine the potential for ammonia oxidation in anoxic environments by characterization of communities of organisms capable of anoxic ammonia oxidation in marine sediments in Loch Duich. Ammonia oxidation in anoxic environments has been demonstrated in comparable marine systems (56) and in previous studies of Loch Duich sediments from the same site (40). Evidence for ammonia oxidation in anoxic sediments analyzed in this study is provided by pronounced nitrate peaks in anoxic regions (Mortimer, personal communication) and by15N-based techniques (J. Barnes, personal communication). Three bacterial groups were targeted. β-Proteobacterial ammonia oxidizers utilize NO2 in anaerobic oxidation of ammonia (50), while planctomycetes include the Anammox organisms, which carry out anaerobic ammonia oxidation in wastewater treatment systems (12, 22). There is no report of anaerobic ammonia oxidation by γ-proteobacterial ammonia oxidizers, but they are known to be present in marine environments (6, 62). Primers designed to be specific for these groups were used to amplify 16S rRNA genes from sediment samples, with subsequent analysis by cloning, sequencing, and DGGE. No γ-proteobacterial sequence was obtained. This group is represented by a small number of pure cultures isolated from marine environments, and few studies have attempted to retrieve representative sequences from natural environments (62). The inability to detect sequences from this group may be due to inadequacy of the primer set used, low population levels, or biases associated with the DNA extraction procedure. However, since γ-proteobacterial ammonia oxidizers are assumed to be ubiquitous in marine planktonic systems (62), their absence from the investigated sediments may indicate that communities of β-proteobacterial AOB are adapted to the geochemical conditions within anoxic marine sediments rather than representing relics of marine sedimentation processes. Nevertheless, this study provided no evidence for the potential role of γ-proteobacterial AOBs in ammonia oxidation in anoxic sediments. Furthermore, Nold et al. (42), analyzing amoA gene sequences, which discriminate between β- and γ-proteobacterial AOB, found no evidence for the presence of γ-proteobacterial AOB in northwest Pacific marine sediments. Sequences representative of β-proteobacterial AOB and planctomycetes were obtained from all samples and are discussed below.
β-Proteobacterial ammonia oxidizers.
DGGE analysis, sequencing, and phylogenetic analysis of cloned 16S rRNA genes demonstrated the presence of two groups of β-proteobacterial ammonia oxidizer communities, each with significant sequence diversity. Sequences belonging to the first group, Nitrosospira cluster 1, have previously been retrieved from other marine environments (3, 28, 43, 53), but no laboratory isolate representative of this group has yet been obtained. The second group contained sequences of particular interest (LD1-B6 and LD1-B37), which formed a novel cluster, not previously demonstrated, within the β-proteobacterial ammonia oxidizers together with sequences obtained from ammonia-oxidizing isolates from seawater (TT140-098-2 and TT140-098-A) and marine sediments (C17) (57, 60). Isolates TT140-098-2, TT140-098-A, and C17 have been described as rod shaped and showed immunological reactivity with both Nitrosococcus oceani and Nitrosomonas marina antisera (C17) or no pronounced reactivity with either serum (TT140-098-2 and TT140-098-A) (60). Additionally, ammonia oxidation has recently been demonstrated in an estuarine isolate with high sequence affinity to the clone sequences falling within this clade (U. Purkhold, M. Wagner, and H.-P. Koops, personal communication). The novel cluster has low sequence similarity to currently recognized AOB clusters, forms a deeply branching clade based on 1.1 kb of 16S rDNA sequence, and may represent a new species. The association of LD1-B6 and LD1-B37 with isolates for which nitrifying activity has been demonstrated supports the assumption that the organisms from which these sequences are derived are ammonia oxidizers. Additionally, amoA gene sequences from marine sediments that showed no clear association with Nitrosomonas or Nitrosospira lineages (42) may belong to representatives of this novel cluster.
Inferring the AOB tree topology, based on 1.1-kb 16S rDNA sequences (382 informative base pairs), with the Log/Det Paralinear distance function yielded the most conservative bootstrap values of all methods applied, and an unambiguous association of the novel cluster, clones of Nitrosomonas cluster 5, and N. cryotolerans (retrieved only from marine environments) with the Nitrosomonas or Nitrosospira clusters was not possible. The uncertain association of N. cryotolerans within the Nitrosomonas clade is supported by phylogenetic analysis of full-length 16S rDNA and amoA gene sequences (25, 44) and its characteristic morphological features (23). Our experience indicates that the association of the N. cryotolerans cluster with Nitrosomonas is strongly influenced by the inclusion or exclusion of the new cluster sequences within the analysis. We suggest a review of the designation of N. cryotolerans within the genus Nitrosomonas once the available cultured representatives of the novel cluster have been characterized in detail. Furthermore, our results indicate that Nitrosomonas cluster 5 may represent a new species and is not closely associated with other Nitrosomonas clusters (36). However, support for this hypothesis requires analysis of full-length gene sequences from cultured representatives of all proposed clusters and additional information from DNA-DNA hybridization and physiological studies.
Environmental relevance of clone sequences.
One source of error in 16S rRNA-based analyses of microbial communities is that of sequence errors introduced during cloning and PCR (51). To assess the significance of this problem, DGGE analysis was carried out both on PCR products of environmental DNA and on clones generated by a different primer approach. In general, there was good correspondence between profiles from the two approaches, indicating that clone libraries represented the natural population well. Almost all clone sequences generated in this study have been confirmed by the two different primer approaches and by their presence as DGGE bands in PCR amplicons from environmental samples. In this sense, we question the relevance of closely related clone sequences that were not represented by DGGE migration patterns in any of the environmental samples (LD1-A15, LD1-B9, and LD1-A20).
Planctomycete sequences.
DGGE profiles and sequence analysis of amplified planctomycete 16S rRNA genes indicated high diversity within this group, but no sequence fell within the currently recognized Anammox group and all sequences showed low similarity to Anammox organism sequences and Anammox-associated clone sequences (47) in the database. There was therefore no evidence for their role in anaerobic ammonia oxidation in these sediments. The inability to detect Anammox sequences may be due to biases associated with DNA extraction and PCR amplification or to a low relative abundance within planctomycete sequences amplified using the PV assay. Additionally, only 26 of >40 different clone types indicated in DGGE profiles and only 8 of the environmental DGGE bands were sequenced. A nested PCR approach would provide the sensitivity to detect low copy numbers of Anammox-related sequences in natural environments in future studies. The majority of studies on Anammox organisms have involved laboratory enrichments or systems related to wastewater treatment plants (12, 22, 54). The relatively few environments from which enrichment cultures have been obtained, the phylogenetic depth within these lineages, and the assumption that potential Anammox planctomycetes adapted to marine sediments may be evolutionarily distant, limit the assessment of the true extent of the phylogenetic distribution of Anammox organisms within the order Planctomycetales. This is the first study targeting planctomycete clone sequences from marine environments. The PV assay was designed by Derakshani et al. (8) on the basis of signature nucleotides described as indicative of Planctomycetales- and Verrucomicrobia-like 16S rDNA sequences for which a common ancestry had been assumed (30). Using this assay, Derakshani et al. (8) isolated three clones from anoxic soil systems that were associated with the Anammox cluster but also showed a great evolutionary distance (PBS-114, PBS-III-2, and PBS-II-37 [Fig. 7]). The majority of sequences retrieved in this study were associated with the Verrucomicrobia and the deep-branching bacterial lineages WS3 and BRC1, supporting them as candidate divisions. However, it should be emphasized that the PV assay enabled the detection of previously unrecognized sequences that had evaded detection with general bacterial primers in studies of comparable environments (31, 34). None of the clone or DGGE sequences could be associated with high sequence similarity to previously described clone sequences or sequences of cultured representatives, and almost all clone sequences retrieved as closest-match sequences from GenBank have been isolated from anoxic or marine environments, suggesting their ecological relevance in these habitats.
Presence of β-proteobacterial AOB and potential activity.
The detection of 16S rRNA gene sequences amplified from DNA provides strong evidence for the presence of β-proteobacterial AOB in anoxic marine sediments but does not necessarily demonstrate their activity. However, sediments were sampled to a depth of 40 cm, and bioturbation and mixing are believed to be minimal in these systems (18, 40). The sedimentation rate has been estimated to be 4 mm year−1, and sediment at 40 cm deep is estimated to be approximately 100 years old (18). The presence of ammonia oxidizers at this depth indicates either their survival for prolonged periods under anoxic conditions or their ability to grow during this period. Analysis of 16S rRNA gene sequences derived from 16S rRNA may provide information about which components of the population were growing or active, if ribosome levels increase with growth rate and if ribosome turnover is rapid following cessation of growth (10). There are indications that rRNA turnover in ammonia oxidizers is slow following substrate exhaustion (58), and it has been suggested that rRNA-based activity studies might not be suitable for slow-growing Anammox- and Nitrosomonas-like AOB (47, 50, 58). Nevertheless, the AOB in the anoxic Loch Duich sediments apparently maintain high 16S rRNA levels for decades, and significant differences were found between DGGE profiles of DNA- and RNA-derived PCR products. In particular, DNA- and RNA-derived PCR products of sequences associated with the new cluster differed, suggesting either differences in growth and activity of closely related AOB or alternative strategies for rRNA synthesis and turnover. This, together with the diversity of both DNA- and RNA-derived gene sequences, suggests spatial and temporal heterogeneity in growth and activity within the community, presumably resulting from heterogeneity in physicochemical characteristics within the sediments and the existence of microenvironments.
In conclusion, this study demonstrates the potential for ammonia oxidation in anoxic marine sediments. The presence of both β-proteobacterial AOB 16S rDNA and rRNA suggests that anoxic metabolism may enable their growth, activity, and survival in such environments. There was no evidence for the presence or activity of γ-proteobacterial ammonia oxidizers or Anammox organisms in these sediments, although this may indicate limitations of the methodological approaches used.
Established pathways for anaerobic oxidation by β-proteobacterial ammonia oxidizers and planctomycetes require substrates (NO2 or NO2−) derived from aerobic processes, which are unlikely to penetrate to the depths sampled in this study. Luther et al. (33) and Hulth et al. (21), however, provided indirect evidence for the potential for coupled anoxic nitrification-denitrification and manganese oxidation-reduction in marine sediments whereby Mn2+ was oxidized by nitrate and ammonia was oxidized by manganous oxides. Mortimer et al. (40) suggested that these processes occur throughout the iron reduction zone of the Loch Duich sediments and that the observed pronounced nitrate peaks may result from locally uncoupled nitrification-denitrification processes or locally increased concentrations of manganous oxides. The demonstrated presence and activity of ammonia oxidizers in these environments add further evidence for coupled anoxic nitrification-manganese reduction processes and have significant implications for descriptions and predictions of nitrogen turnover and geochemical cycling in marine sediments.
Acknowledgments
This study was supported by grant F122/BF from the Leverhulme Trust.
We thank P. Hayes and I. Davies (Marine Laboratory, Aberdeen), R. Mortimer, M. Krom, and S. Harris (University of Leeds), and J. Barnes (University of Newcastle) for helpful discussions and collection of samples.
REFERENCES
- 1.Aller, R. C. 1988. Benthic fauna and biogeochemical processes in marine sediments: the role of burrow structures, p. 301-339. In T. H. Blackburn and J. Soerensen (ed.), Nitrogen cycling in coastal marine environments. John Wiley & Sons, Chichester, United Kingdom.
- 2.Altschul, S. F., W. Gish, E. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Bano, N., and J. T. Hollibaugh. 2000. Diversity and distribution of DNA sequences with affinity to ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in the Arctic Ocean. Appl. Environ. Microbiol. 66:1960-1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bock, E., I. Schmidt, R. Stüven, and D. Zart. 1995. Nitrogen loss caused by denitrifying Nitrosomonas cells using ammonium or hydrogen as electron donors and nitrite as electron acceptor. Arch. Microbiol. 163:16-20. [Google Scholar]
- 5.Bodelier, P. L., J. A. Libochant, C. W. P. M. Blom, and H. J. Laanbroek. 1996. Dynamics of nitrification and denitrification in root-oxygenated sediments and adaptation of ammonia-oxidizing bacteria to low-oxygen or anoxic habitats. Appl. Environ. Microbiol. 62:3100-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bothe, H., G. Jost, M. Schloter, B. B. Ward, and K.-P. Witzel. 2000. Molecular analysis of ammonia oxidation and denitrification in natural environments. FEMS Microbiol. Rev. 24:673-690. [DOI] [PubMed] [Google Scholar]
- 7.Brune, A., P. Frenzel, and H. Cypionka. 2000. Life at the oxic-anoxic interface: microbial activities and adaptations. FEMS Microbiol. Rev. 24:691-710. [DOI] [PubMed] [Google Scholar]
- 8.Derakshani, M., T. Lukow, and W. Liesack. 2001. Novel bacterial lineages at the (sub) division level as detected by signature nucleotide-targeted recovery of 16S rRNA genes from bulk soil and rice roots of flooded rice microcosms. Appl. Environ. Microbiol. 67:623-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doika, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duineveld, B. M., G. A. Kowalchuk, A. Keijzer, J. D. van Elsas, and J. A. van Veen. 2001. Analysis of bacterial communities in the rhizosphere of Chrysanthemum via denaturing gradient gel electrophoresis of PCR-amplified 16S rRNA as well as DNA fragments coding for 16S rRNA. Appl. Environ. Microbiol. 67:172-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edwards, U., T. Rogall, H. Blöcker, M. Emde, and E. C. Böttger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egli, K., U. Fanger, P. J. Avarez, H. Siegrist, J. R van der Meer, and A. J. B. Zehnder. 2001. Enrichment and characterization of an Anammox bacterium from a rotating biological contactor treating ammonium-rich leachate. Arch. Microbiol. 175:198-207. [DOI] [PubMed] [Google Scholar]
- 13.Felske, A., A. Wolterink, R. van Lis, and A. D. L. Akkermans. 1998. Phylogeny of the main bacterial 16S rRNA sequences in Drentse A grassland soils (The Netherlands). Appl. Environ. Microbiol. 64:871-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fenchel, T., and R. J. Riedl. 1970. The sulfide system: a new biotic community underneath the oxidized layer of marine sand bottoms. Mar. Biol. 7:255-268. [Google Scholar]
- 15.Froelich, P. N., G. P. Klinkhammer, M. L. Blender, N. A. Luedtke, G. R. Heath, D. Cullen, and P. Dauphin. 1979. Early oxidation of organic matter in pelagic sediments of the eastern equatorial Atlantic: suboxic diagenesis. Geochim. Cosmochim. Acta 43:1075-1090. [Google Scholar]
- 16.Goreau, T. J., W. A. Kaplan, S. C. Wofsy, M. B. McElroy, F. W. Valois, and S. W. Watson. 1980. Production of NO2− and N2O by nitrifying bacteria at reduced concentrations of oxygen. Appl. Environ. Microbiol. 40:526-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffiths, R. I., A. S. Whiteley, A. G. O'Donnell, and M. J. Bailey. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayes, P. 2001. Diagenetic processes and metal mobilisation in an organic rich Scottish fjord. Ph.D. thesis. Department of Earth Sciences, The University of Leeds, Leeds, United Kingdom.
- 19.Herbert, R. A. 1999. Nitrogen cycling in coastal marine ecosystems. FEMS Microbiol. Rev. 23:563-590. [DOI] [PubMed] [Google Scholar]
- 20.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hulth, S., R. C. Aller, and F. Gilbert. 1999. Coupled anoxic nitrification/manganese reduction in marine sediments. Geochim. Cosmochim. Acta 63:49-66. [Google Scholar]
- 22.Jetten, M. S. M., M. Strous, K. T. van de Pas-Schoonen, J. Schalk, U. G. J. M. van Dongen, A. A. van de Graaf, S. Logemann, G. Muyzer, M. C. M. van Loosdrecht, and J. G. Kuenen. 1999. The anaerobic oxidation of ammonium. FEMS Microbiol. Rev. 22:421-437. [DOI] [PubMed] [Google Scholar]
- 23.Jones, R. D., R. Y. Morita, H.-P. Koops, and S. W. Watson. 1988. A new marine ammonium-oxidizing bacterium, Nitrosomonas cryotolerans sp. nov. Can. J. Microbiol. 34:1122-1128. [Google Scholar]
- 24.Jørgensen, B. B. 1983. Processes at the sediment-water interface, p. 477-506. In B. Bolin and R. B. Cook. (ed.), The major biogeochemical cycles and their interactions. Scope 21. Wiley, Chichester, United Kingdom.
- 25.Koops, H.-P., and A. Pommerening-Rösner. 2001. Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol. Ecol. 37:1-9. [Google Scholar]
- 26.Kowalchuk, G. A., P. L. E. Bodelier, G. H. J. Heilig, J. R. Stephen, and H. J. Laanbroek. 1998. Community analysis of ammonia-oxidizing bacteria, in relation to oxygen availability in soils and root-oxygenated sediments, using PCR, DGGE and oligonucleotide probe hybridization. FEMS Microbiol. Ecol. 27:339-350. [Google Scholar]
- 27.Kowalchuk, G. A., Z. S. Naoumenko, P. J. L. Derikx, A. Felske, J. R. Stephen, and I. A. Arkhipchenko. 1999. Molecular analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in compost and composted materials. Appl. Environ. Microbiol. 65:396-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kowalchuk, G. A., J. R. Stephen, W. De Boer, J. I. Prosser, T. M. Embley, and J. W. Woldendorp. 1997. Analysis of ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and PCR-amplified 16S ribosomal DNA fragments. Appl. Environ. Microbiol. 63:1489-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lake, J. A. 1994. Reconstructing evolutionary trees from DNA and protein sequences: paralinear distances. Proc. Natl. Acad. Sci. USA 91:1455-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liesack, W., and E. Stackebrandt. 1992. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J. Bacteriol. 174:5072-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Llobet-Brossa, E., R. Rosselló-Mora, and R. Amann. 1998. Microbial community composition of Wadden Sea sediments as revealed by fluorescence in situ hybridization. Appl. Environ. Microbiol. 64:2691-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lockhard, P. J., A. W. D. Larkum, M. A. Steel, P. J. Waddel, and D. Penny. 1996. Evolution of chlorophyll and bacteriochlorophyll: the problem of invariant sites in sequence analysis. Proc. Natl. Acad. Sci. USA 93:1930-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luther, G. W., III, B. Sundby, B. L. Lewis, P. J. Brendel, and N. Silverberg. 1997. Interactions of manganese with the nitrogen cycle: alternative pathways to dinitrogen. Geochim. Cosmochim. Acta 61:4043-4052. [Google Scholar]
- 34.Madrid, V. M., G. T. Taylor, M. I. Scranton, and A. Y. Chistoserdov. 2001. Phylogenetic diversity of bacterial and archaeal communities in the anoxic zone of the Cariaco Basin. Appl. Environ. Microbiol. 67:1663-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCaig, A. E., T. M. Embley, and J. I. Prosser. 1994. Molecular analysis of enrichment cultures of marine ammonia oxidizers. FEMS Microbiol. Lett. 120:363-368. [DOI] [PubMed] [Google Scholar]
- 37.McCaig, A. E., C. J. Phillips, J. R. Stephen, G. A. Kowalchuk, S. M. Harvey, R. A. Herbert, T. M. Embley, and J. I. Prosser. 1999. Nitrogen cycling and community structure of proteobacterial beta-subgroup ammonia-oxidizing bacteria within polluted marine fish farm sediments. Appl. Environ. Microbiol. 65:213-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCaig, A. E., J. I. Prosser, and T. M. Embley. 1994. Molecular analysis of enrichment cultures of marine ammonia-oxidizers. FEMS Microbiol. Lett. 120:363-368. [DOI] [PubMed] [Google Scholar]
- 39.Miller, D. J., and D. J. D. Nicholas. 1985. Characterization of a soluble cytochrome oxidase/nitrite reductase of Nitrosomonas europaea. J. Gen. Microbiol. 131:2851-2854. [Google Scholar]
- 40.Mortimer, R. J. G., M. D. Krom, S. J. Harris, P. J. Hayes, I. M. Davies, W. Davison, and H. Zhang. 2002. Evidence for complex recycling processes within sedimentary biogeochemical zones. Mar. Ecol. Prog. Ser. 236:31-35. [Google Scholar]
- 41.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nold, S. C., J. Zhou, A. H. Devol, and J. M. Tiedje. 2000. Pacific Northwest marine sediments contain ammonia-oxidizing bacteria in the β subdivision of the Proteobacteria. Appl. Environ. Microbiol. 66:4532-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phillips, C. J., Z. Smith, T. M. Embley, and J. I. Prosser. 1999. Phylogenetic differences between particle-associated and planktonic ammonia-oxidizing bacteria of the β-subdivision of the class Proteobacteria in the northwestern Mediterranean Sea. Appl. Environ. Microbiol. 65:779-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Purkhold, U., A. Pommerening-Röser, S. Juretschko, M. C. Schmid, H.-P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:6368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Risatti, J. B., W. C. Capman, and D. A. Stahl. 1994. Community structure of a microbial mat: the phylogenetic dimension. Proc. Natl. Acad. Sci. USA 91:10173-10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 47.Schmid, M., S. Schmitz-Esser, M. Jetten, and M. Wagner. 2001. 16S-23S rDNA Intergenic spacer and 23S rDNA of anaerobic ammonium-oxidizing bacteria: implications for phylogeny and in situ detection. Environ. Microbiol. 3:450-459. [DOI] [PubMed] [Google Scholar]
- 48.Schmidt, I., and E. Bock. 1997. Anaerobic ammonia oxidation with nitrogen dioxide by Nitrosomonas eutropha. Arch Microbiol. 167:106-111. [PubMed] [Google Scholar]
- 49.Schmidt, I., and E. Bock. 1998. Anaerobic ammonia oxidation by cell-free extracts of Nitrosomonas eutropha. Antonie Leeuwenhoek 73:271-278. [DOI] [PubMed] [Google Scholar]
- 50.Schmidt, I., O. Sliekers, M. Schmidt, I. Cirpus, M. Strous, E. Bock, J. G. Kuenen, and M. S. M. Jetten. 2002. Aerobic and anaerobic ammonia oxidizing bacteria—competitors or natural partners? FEMS Microbiol. Ecol. 39:175-181. [DOI] [PubMed] [Google Scholar]
- 51.Specksnijder, A. G., G. A. Kowalchuck, S. de Jong, E. Kline, J. R. Stephen, and H. J. Laanbroek. 2001. Microvariation artifacts introduced by PCR and cloning of closely related 16S rRNA gene sequences. Appl. Environ. Microbiol. 67:469-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Specksnijder, A. G., G. A. Kowalchuck, K. Roest, and H. J. Laanbroek. 1998. Recovery of a Nitrosomonas-like 16S rDNA sequence group from freshwater habitats. Syst. Appl. Microbiol. 21:321-330. [DOI] [PubMed] [Google Scholar]
- 53.Stephen, J. R., A. E. McCaig, Z. Smith, J. I. Prosser, and T. M. Embley. 1996. Molecular diversity of soil and marine 16S rRNA gene sequences related to beta-subgroup ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 62:4147-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strous, M., J. A. Fuerst, E. H. M. Kramer, S. Logemann, G. Muyzer, K. T. van de Pas-Schoonen, R. Webb, J. G. Kuenen, and M. S. M. Jetten. 1999. Missing lithotroph identified as new planctomycetes. Nature 400:446-449. [DOI] [PubMed] [Google Scholar]
- 55.Swofford, D. L. 1998. PAUP∗: phylogenetic analysis using parsimony (∗and other methods). Sinauer Associates, Sunderland, Mass.
- 56.Thamdrup, B., and T. Dalsaard. 2002. Production of N2 through anaerobic ammonium oxidation coupled to nitrate reduction in marine sediments. Appl. Environ. Microbiol. 68:1312-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Voytek, M. A. 1996. Relative abundance and species diversity of autotrophic ammonia oxidizing bacteria in aquatic systems. Ph.D. thesis. University of California, Santa Cruz.
- 58.Wagner, M., G. Rath, R. Amann, H.-P. Koops, and K.-H. Schleifer. 1995. In-situ identification of ammonia-oxidizing bacteria. Syst. Appl. Microbiol. 18:251-264. [Google Scholar]
- 59.Wagner, A., N. Blackstone, P. Cartwright, M. Dick, B. Misof, P. Snow, G. P. Wagner, J. Bartels, M. Murtha, and J. Pendelton. 1994. Surveys of gene families using polymerase chain reaction: PCR selection and PCR drift. Syst. Biol. 43:250-261. [Google Scholar]
- 60.Ward, B. B., and A. F. Carlucci. 1985. Marine ammonia- and nitrite-oxidizing bacteria: serological diversity determined by immunofluorescence in culture and in the environment. Appl. Environ. Microbiol. 50:194-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ward, B. B., D. P. Martino, M. C. Diaz, and S. B. Joye. 2000. Analysis of ammonia-oxidizing bacteria from hypersaline Mono Lake, California, on the basis of 16S rRNA sequences. Appl. Environ. Microbiol. 66:2873-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ward, B. B., and G. D. Mullan. 2002. Worldwide distribution of Nitrosococcus oceani, a marine ammonia oxidizing γ-proteobacterium, detected by PCR and sequencing of 16S rRNA and amoA genes. Appl. Environ. Microbiol. 68:4153-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Webster, G., T. M. Embley, and J. I. Prosser. 2002. Grassland management regimes reduce small-scale heterogeneity and species diversity of β-proteobacterial ammonia oxidizer populations. Appl. Environ. Microbiol. 68:20-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ziebis, W., S. Foster, M. Huettel, and B. B. Jørgensen. 1996. Complex burrows of the mud shrimp Callianassa truncata and their geochemical impact in the sea bed. Nature 382:619-622. [Google Scholar]