Abstract
Localization of soluble endoplasmic reticulum (ER) resident proteins is likely achieved by the complementary action of retrieval and retention mechanisms. Whereas the machinery involving the H/KDEL and related retrieval signals in targeting escapees back to the ER is well characterized, other mechanisms including retention are still poorly understood. We have identified a protein disulfide isomerase (Dd-PDI) lacking the HDEL retrieval signal normally found at the C terminus of ER residents in Dictyostelium discoideum. Here we demonstrate that its 57 residue C-terminal domain is necessary for intracellular retention of Dd-PDI and sufficient to localize a green fluorescent protein (GFP) chimera to the ER, especially to the nuclear envelope. Dd-PDI and GFP-PDI57 are recovered in similar cation-dependent complexes. The overexpression of GFP-PDI57 leads to disruption of endogenous PDI complexes and induces the secretion of PDI, whereas overexpression of a GFP-HDEL chimera induces the secretion of endogenous calreticulin, revealing the presence of two independent and saturable mechanisms. Finally, low-level expression of Dd-PDI but not of PDI truncated of its 57 C-terminal residues complements the otherwise lethal yeast TRG1/PDI1 null mutation, demonstrating functional disulfide isomerase activity and ER localization. Altogether, these results indicate that the PDI57 peptide contains ER localization determinants recognized by a conserved machinery present in D. discoideum and Saccharomyces cerevisiae.
INTRODUCTION
The endoplasmic reticulum (ER) is the crucial site where newly synthesized proteins enter the central vacuolar system. Secretory, lumenal, and integral membrane proteins are processed in the ER in several sequential or concomitant steps, including folding, formation and reshuffling of disulfide bonds, glycosylation and deglycosylation, and oligomerization (Hammond and Helenius, 1995; Hong, 1998). These reactions are escorted by soluble and membrane-bound resident ER proteins such as BiP (Haas and Wabl, 1983; Munro and Pelham, 1986), calreticulin, calnexin (Michalak et al., 1992; Helenius, 1994), and protein disulfide isomerase (PDI) (Noiva and Lennarz, 1992; Freedman et al., 1994), which function as chaperones and quality-control machinery (Ellgaard et al., 2000). Processed proteins are then sorted from ER residents and delivered to their final destination. Despite their high concentration, estimated at nearly 100 mg/ml (Koch, 1987), lumenal resident proteins are not secreted and are hardly detectable in the Golgi apparatus. This highly selective steady-state distribution is likely achieved by two complementary systems, one ensuring retention in the ER and one acting to retrieve escapees from the Golgi compartment back into the ER. The ER resident transmembrane type I, II, and III proteins are retrieved by discrete ER-targeting motifs, such as the cytoplasmic KKXX and RR or the lumenal HDEL (Teasdale and Jackson, 1996). Most of the ER-soluble resident proteins are characterized by a C-terminal peptide, H/KDEL, that prevents their secretion (Munro and Pelham, 1987; Pelham et al., 1988; Pelham, 1989, 1990). The retrieval mechanism of soluble H/KDEL proteins depends on the H/KDEL receptor ERD2p, itself an integral membrane protein (Lewis et al., 1990; Semenza et al., 1990). Several reports have shown that this retrieval machinery can be saturated (Dean and Pelham, 1990; Bu et al., 1997).
In comparison, it remains unclear how soluble resident proteins lacking this or a related motif are retained in the ER. It was recently shown that the KKXX retrieval motif can also serve as a retention signal (Andersson et al., 1999). Another possibility is the formation of heterooligomers with H/KDEL-containing proteins (Koivu et al., 1987; Pihlajaniemi et al., 1987; Vuori et al., 1992; Zhen et al., 1995). However, accumulating evidence indicates that ER localization of resident proteins is attributable not only to their retrieval from post-ER compartments but also to true retention mechanisms. Calreticulin and BiP lacking the KDEL sequence leak from the ER but are secreted more slowly than naturally secreted proteins. Another piece of evidence is that retrieval appears to be independent of Ca2+ (Wilson et al., 1993), whereas perturbation of intracellular Ca2+ stores by ionophores induces secretion of resident proteins (Booth and Koch, 1989). Calcium is suggested to serve as a stabilizer of a reticular matrix of resident soluble and membrane proteins. This matrix may constitute an immobile phase through which secretory proteins percolate (Hammond and Helenius, 1995).
PDI is a highly abundant ER resident enzyme representing ∼0.4% of total cellular proteins. Its basic activity is to unscramble nonnative disulfide bonds in oxidation/reduction cycles catalyzed by its thioredoxin boxes; it also functions as a chaperone (Noiva and Lennarz, 1992). In mammalian cells, PDI is present mainly as a homodimer, but it is also found as a homotetramer (Gilbert, 1998). PDI is also a component of a complex that catalyzes the transfer of triglyceride and cholesteryl ester between membranes (Wetterau et al., 1990, 1991) and works as β-subunit of the α2β2 prolyl 4-hydroxylase complex (Pihlajaniemi et al., 1987). Classic PDI is constituted of five domains, denoted a, b, b′, a′, and c. The a and a′ domains are homologous to thioredoxin, and each contains the CXXC red/ox active site motif. A recently reported nuclear magnetic resonance structure revealed that the b domain is structurally homologous to the a domain (Kemmink et al., 1997), and some degree of sequence conservation can also be discerned (Ferrari et al., 1998). The role of the acidic and Ca2+-binding c domain as a potential peptide-binding site is debated (Noiva et al., 1993; Dai and Wang, 1997; Darby et al., 1998).
PDI and related proteins form a complex superfamily that includes at least six families: PDI, ERp57, ERp72, PDIR, P5, and the newly identified PDI-D (Ferrari and Söling, 1999). Briefly, the latest classification is based on the number of active thioredoxin boxes (a and a′), of intervening inactive thioredoxin folds (b and b′), and the presence of the D domain. Although most of the PDI and other ER resident proteins have a C-terminal acidic c domain and a KDEL-related signal, members of the PDI-D family, including some proteins previously defined as P5 or PDI proper, do not follow this rule (Figure 1) (Monnat et al., 1997). Therefore, a general mechanism of ER retention for such abundant proteins must exist. We have recently cloned in Dictyostelium discoideum a PDI (Dd-PDI) member of the PDI-D family. This protein contains the two thioredoxin motifs but lacks the b, b′, and c domains as well as the H/KDEL-type retrieval sequence (Monnat et al., 1997).
Figure 1.
Sequence alignment of the C termini of Dd-PDI and other ER proteins. The recently described PDI-D domain is ∼100 residues long (Ferrari and Söling, 1999), and the highest conservation is found in the C-terminal half. Identical residues are highlighted in dark gray, homologies in light gray. The KDEL-type retrieval signal is boxed. PDI57 and PDI13 represent the fragments used to construct GFP chimeras (see text). The two asterisks indicate the positions of mutations in the Drosophila windbeutel gene. The species of origin and accession numbers for these sequences are as follows. PDI-Dα subfamily: DdPDI: D. discoideum, AF019112; alfPDI: Medicago sativa, P38661; aspPDI: Aspergilus nidulans, Q00216; SpPDI: PDI2, S. pombe, O13811; AtPDI: Arabidopsis thaliana, O22263; TabPDI: Nicotiana tabacum, P93358. PDI-Dβ subfamily: Erp28: Homo sapiens Erp28, X94910; ER29: Rattus norvegicus, P52555; Wind: Drosophila melanogaster windbeutel, O44342; ScMPD2: S. cerevisiae, D34634. Other: ScERO1: ER oxidative protein, S. cerevisiae, z50178.
Here we describe the identification of a 57-residue domain present at the C terminus of Dd-PDI that is necessary and sufficient for its ER localization. We report evidence from the secretion/retention of constructions generated by deletion as well as green fluorescent protein (GFP) fusions. Our results indicate that the HDEL retrieval system operates in parallel with the localization mechanism reported here to achieve the steady-state distribution of ER residents in D. discoideum. Investigation of the biochemical properties of lumenal complexes suggests potential retention mechanisms. Using complementation in yeast, we show that the 57-residue domain plays a ubiquitous role in ER localization conserved during evolution.
MATERIALS AND METHODS
Construction and Expression of PDI Truncations and GFP Chimeras
PDI truncated of its last 57 residues was generated by PCR with a sense primer complementary to the leader peptide sequence and an antisense primer introducing a stop codon just before the second half of the C-terminal D domain. To introduce an HDEL signal, the antisense primer contained a sequence coding for HDEL before the stop codon. The two constructs were cloned into the pDXA D. discoideum expression vector (Manstein et al., 1995) and the pDS2 yeast expression vector (Guenther et al., 1993) (see Yeast Complementation below).
GFPmut2 (Cormack et al., 1996) was fused in frame with the signal sequence of CsA (Faix et al., 1992), followed by a small polylinker, and the sequences coding for HDEL, KDEL, the last 13 (AKKLNILESFKSK) or the last 57 (IEGSYYVKVMKTIAEKSIDFVTTEIARITKLVSGSMSGKKADEFAKKLNILESFKSK) C-terminal amino acids of Dd-PDI (Monnat et al., 1997). These GFP chimeras were then introduced into the expression vector pAC6 (Fasel et al., 1992). AX2 cells were transfected by electroporation (Howard et al., 1988), and stable transformants were selected by resistance to 10 μg/ml G418 (Life Technologies, Grand Island, NY). Single transformants were further selected for their similar level of expression and cloned on DDSM 1/2 agar plated with a lawn of Klebsiella aerogenes and then grown in nutrient medium HL5c (Sussman, 1987) supplemented with 10 μg/ml G418.
Secretion/Retention Assays
Preliminary experiments showed that secretion, even of GFP-sec, was very slow compared with the rates observed in other systems. In addition, because of the quality of available antibodies, it proved difficult to optimize the immunoprecipitation of GFP-tagged proteins. Therefore, two methods were used to determine the relative secretion/retention of the studied proteins. First, a metabolic labeling pulse/chase protocol was used (Mierendorf and Dimond, 1983) to follow the fate of PDI constructs by immunoprecipitation. Briefly, 108 cells were labeled with 5 mCi of [35S]Met for 30 min in 7.5 ml of HL5c diluted 1:3 with Soerensen buffer (15 mM KH2PO4, 2 mM Na2HPO4, pH 6) and then chased in 25 ml of HL5c for the indicated times, up to 8 h. Cells were separated from the medium by centrifugation and lysed in RIPA buffer (20 mM Tris, pH 7.4, 150 mM NaCl, 1% Na deoxycholate, 1% Triton X-100, 0.1% SDS). RIPA 5× buffer was also added to the medium to an end concentration of 1×. Immunoprecipitation was carried out with a cocktail of mAbs to PDI (Monnat et al., 1997) or a polyclonal anti-GFP (MBL, Nagoya, Japan), followed by ProteinAplusG beads (Novagen, Madison, WI). The pellets were analyzed by SDS-PAGE and phosphorimager (see below).
Second, an immunoblotting method was used to quantify secretion/retention of GFP constructs. Clones expressing the different GFP constructs were grown in suspension in HL5c medium at 190 rpm at 23°C to a density of 2 × 106 cells/ml. The cells were centrifuged for 5 min at 500 × g, washed in Soerensen buffer, and resuspended in 4 ml of HL5c medium at a density of 7.5 × 105 cells/ml. The cells were then allowed to adhere on 6-cm Petri dishes for 30 min. Unattached cells were removed, and adherent cells were refed with HL5c medium supplemented with 20 μg of BSA as carrier and cultivated for 8 h. After incubation, the medium was removed and centrifuged at 1000 × g for 10 min at 4°C to eliminate a few dead cells. The proteins contained in the medium were precipitated with 10% trichloroacetic acid (TCA) for 1 h on ice and centrifuged at maximum speed in a tabletop centrifuge for 30 min. The pellet was resuspended and boiled in SDS sample buffer. The living cells were washed with ice-cold Soerensen buffer, harvested in the same buffer, centrifuged, and boiled immediately in 1 ml of SDS sample buffer. The whole medium proteins and 1% of total cell lysate were analyzed by SDS-PAGE (Laemmli, 1970), immunoblotted with the anti-GFP (Clontech, Palo Alto, CA), anti-PDI (Monnat et al., 1997), and anti-calreticulin mAbs (a generous gift of Drs. B. Knoblach and R. Mutzel, Konstanz University, Konstanz, Germany), and visualized with the use of the ECL detection system (Amersham, Arlington Heights, IL). To ensure that GFP secreted in the medium was not degraded during the time of the experiment, we incubated wild-type cells with a small amount (200 pg) of recombinant GFP and 20 μg of BSA as a carrier. After an incubation of 20 h, recombinant GFP was quantitatively recovered.
Quantitative Analysis of Immunoprecipitation and Immunoblotting
After SDS-PAGE and blotting on nitrocellulose membranes, immunoprecipitations from metabolically labeled cell extracts were quantified with the help of a phosphorimager (Fuji Film, Tokyo, Japan) and accompanying software (ImageGauge). To quantify results from anti-GFP immunoblotting, each gel was loaded with recombinant GFP standards in a range of quantities comparable to the amounts expected from the experiment performed. Therefore, experimental values were interpolated between values obtained for standards and not extrapolated. For PDI and calreticulin, dilutions of cell extracts rather than recombinant protein were used to ensure that quantitation was performed in the linear range of the method. The use of ECL-Plus substrate allowed for better consistency between different exposures on HyperFilm (Amersham). For each blot, a range of exposures were scanned and analyzed with NIH Image densitometry software. Alternatively, signals were recorded directly by a LAS-1000 Luminescent Image Analyzer (Fuji Film), allowing for quantification of signal intensities within a broad linear range. Because of a slightly variable level of GFP chimera expression, the value for secreted GFP was first calculated as the percentage of the total amount present in each strain. Then, these values were normalized to the value obtained for the GFP-sec strain representing 100% secretion. The total amounts of calreticulin and PDI were the same in every strain. Therefore, the values obtained for the secreted proteins were normalized only to the values obtained for the GFP-sec strain. The graphs present the average ± SD of at least three independent experiments.
Immunofluorescence and Microscopy
D. discoideum cells expressing the GFP constructs were allowed to adhere for 30 min to a glass coverslip. The fixation/permeabilization for immunofluorescence was performed as described previously by plunging the coverslips directly into methanol at −85°C followed by warming to −35°C (Neuhaus et al., 1998). The coverslips were then plunged rapidly into PBS at room temperature and processed for immunocytochemistry. Secondary goat anti-mouse immunoglobulin G antibodies conjugated to Cyanine 3.29-OSu (Rockland, Gilbertsville, PA) were used to detect binding of the primary mAbs directed against PDI, calreticulin, and a Golgi marker (Bertholdt et al., 1985). After mounting in Mowiol, stained cells were observed and documented with a DM/IRB Leica (Bensheim, Germany) confocal microscope with the use of a 63x Plan-Apo objective with a numerical aperture of 1.40.
Cycloheximide Treatment and Live Microscopy
Cells expressing the GFP constructs were treated for up to 4 h with 1.6 mM cycloheximide, a dose sufficient to block protein synthesis by >98% (Müller-Taubenberger et al., 1988). Live cells were observed with an Axiophot 2 (Zeiss, Thornwood, NY) microscope equipped with a water-immersion 100× objective. Frames were taken with a cooled charge-coupled device camera (PCO SensiCam, Till Photonics, Kelheim, Germany) at the given time points and saved in TIF format with the use of NIH Image 1.62 software.
Electron Microscopy
For transmission electron microscopy, cells were plated on glass coverslips and fixed with paraformaldehyde/picric acid (Humbel and Biegelmann, 1992) followed by embedding in Epon (Neuhaus et al., 1998). Thin, 100-nm sections were observed and documented in a Philips (Eindhoven, the Netherlands) 400 T transmission electron microscope.
Triton X-114 Phase Separation and Fractionation Experiments
Cells were cultivated in suspension in HL5c medium, shaken at 190 rpm at 23°C, and harvested at a density of 2 × 106 cells/ml. Cells (5 × 108) were washed in 40 ml of ice-cold buffer (50 mM Tris, pH 7.4, 250 mM sucrose, 1 mM EDTA, 1 mM DTT, containing 100, 150, or 200 mM KCl, with [TKMS] or without [TKS] 5 mM MgCl2) and broken with a ball homogenizer (European Molecular Biology Laboratory, Heidelberg, Germany) on ice in 4 ml of TKMS or TKS buffer supplemented with complete (EDTA-free) protease inhibitors cocktail (Roche, Basel, Switzerland). The cell lysate was then centrifuged for 10 min at 1000 × g at 4°C. The postnuclear supernatant was centrifuged for 45 min at 180,000 × g at 4°C in a TLA 100 rotor (Beckman, Fullerton, CA). Triton X-114 phase separation was carried out according to the method of Bordier (1981) with some modifications. The crude membrane pellet was dissolved by adding 2 ml of ice-cold TKM or TK buffer containing 2% Triton X-114 (Sigma Chemical, St. Louis, MO) and supplemented with protease inhibitors. The solubilized pellet was warmed to 30°C for 3 min and centrifuged at 10,000 × g for 2 min at room temperature. The aqueous and detergent phases were then separated, and the concentration of Triton X-114 was adjusted to 2% in the aqueous phase. A second cycle of phase separation was carried, out and fractions were pooled and adjusted to identical volumes with TKM or TK buffer. Equal volumes of each sample were loaded and analyzed by SDS-PAGE and quantitative immunoblotting.
Gel Filtration
Aliquots of 500 μl of the aqueous phase of the Triton X-114 phase separation, containing ∼5 mg of total proteins, were applied to a gel filtration column (Superdex 200 HR 10/30, Pharmacia, Piscataway, NJ) with the use of TKM or TK buffer as eluant. The 0.3-ml fractions were analyzed by SDS-PAGE and quantitative immunoblotting.
Cross-Linking Experiments on Intact Cells
GFP-PDI57 and GFP-sec strains were grown on Petri dishes in HL5c medium to subconfluence, washed, and resuspended in ice-cold Soerensen buffer at a density of 5 × 106 cells/ml. Glutaraldehyde (8% stock solution; Sigma) was added to the cells at a final concentration of 0.15%, and the mixtures were incubated on ice for the indicated times. For each time point, 200 μl of cell suspension (106 cells) were drawn and the cross-link reaction was stopped by adding Tris, pH 8.0, to a final concentration of 100 mM. Cells were centrifuged at 1500 × g for 10 min at 4°C, lysed in sample buffer, and analyzed by either uniform 10% or 4–15% gradient SDS-PAGE and quantitative immunoblotting.
Yeast Plasmids, Complementation, and Secretion Assay
First, a Dd-PDI mutant harboring a C-terminal HDEL motif was created by PCR with the Pfu polymerase and appropriate oligonucleotides and controlled by sequencing. The coding sequences of Dd-PDI and Dd-PDI-HDEL, as well as Dd-PDIΔC and Dd-PDIΔC-HDEL, were inserted into the BamHI site of the pDS2 multicopy yeast vector containing the HIS3 selection marker (Guenther et al., 1993). As positive and negative controls, yPDI, the wild-type yeast PDI, and yPDImc (with both thioredoxin CGHC boxes mutated to SGHS) coding sequences were inserted into the BamHI and SalI sites of pDS2 vector under the control of the yeast GAL1 promoter. The resulting vectors were used for complementation of a yeast TRG1/PDI1 null mutant strain, BK203-15B (Mata, ura3, leu2, his3, trp1, pdi::leu2) rescued by a centromer plasmid constitutively expressing yPDI (pWBK-PDI) (B. Kramer, unpublished data). Transformation was carried out as described (Kramer et al., 1989). Yeast strains were grown for 3–5 d at 30°C on synthetic complete (SC) minimal medium (Campbell et al., 1975) supplemented with 2% glucose (SDC). Five to 10 colonies were transferred and grown for 2–4 d at 30°C on SC minimal medium supplemented with 2% galactose (SGC). Thereafter, the colonies were transferred by stamping on SC medium supplemented with 2% galactose and 0.1% 5-FOA (SGC/FOA). From this plate, colonies were streaked and grown for 2–6 d at 30°C on rich media containing 1% yeast extract, 2% bactotryptone, and 2% galactose (YPG) or 2% glucose (YPD) or on SC medium lacking uracil and supplemented with 2% galactose or glucose (SGC/−Ura and SDC/−Ura, respectively). Extracts were prepared from cells grown in suspension culture in YPG and analyzed for their content in the different Dd-PDI constructs by immunoblotting with anti-PDI antibodies.
Retention/secretion was investigated as follows. First, yeast expressing Dd-PDI, Dd-PDI-HDEL, Dd-PDIΔC, and Dd-PDIΔC-HDEL were cultivated overnight at 30°C in YPG. Then, cells were washed twice in PBS, resuspended in fresh YPG, and incubated at 30°C with shaking. At the indicated times, 1-ml aliquots were retrieved and centrifuged at 15,000 × g for 15 min at 4°C. The medium was then concentrated by precipitation with 10% TCA for 1 h on ice, followed by centrifugation. The pellets were washed with ethanol and boiled in sample buffer. After SDS-PAGE and immunoblotting with anti-PDI antibodies, the signals were quantified as described above.
RESULTS
The C Terminus of Dd-PDI Is Conserved among a Group of ER Resident Proteins
We recently discovered in D. discoideum a divergent member of the PDI family. A growing number of PDI-related proteins share with Dd-PDI common structural peculiarities, such as the lack of both a C-terminal acidic domain and a classic KDEL-type ER retrieval motif (Monnat et al., 1997). Database searches revealed that their C-terminal domains of ∼100 residues are well conserved among a subgroup of ER proteins. Together, these observations led to the recent proposal of a novel class of PDIs, PDI-D, defined by the presence of this D domain (Ferrari and Söling, 1999). The C-terminal half of the D domains appears to be best conserved in sequence (between 29 and 38% identity, and between 37 and 51% homology) and to form an independently folding unit composed of three α-helices (Ferrari and Söling, 1999). Figure 1 shows a sequence alignment of this domain of ER residents from diverse organisms. The first 9 sequences were among the top 14 from FASTA and BLAST searches and were chosen to reflect conservation throughout evolution (missing are sequences from mouse, rice, Acetabularia, and Neurospora crassa). The bottom two sequences have a lower score but are the best two homologies in Saccharomyces cerevisiae. The residues highlighted in dark and light gray indicate identity in at least seven of the top nine sequences and homology in at least five of the top nine sequences, respectively. Some of these domains end with a canonical retrieval signal. Interestingly, the Drosophila windbeutel gene encodes an ER resident protein homologous to human Erp28 and rat Erp29 (Konsolaki and Schupbach, 1998). Two mutations resulting in severe developmental defects were identified in the 70-residue C-terminal peptide (Figure 1, asterisks). The windM46 allele contained a single base change generating a stop codon after Gln-187, amputating the whole domain homologous to PDI57. The windT6 allele had a single base change in the NIL motif, resulting in the conversion of Leu-239 to a Pro. The Leu-to-Pro change might disrupt the α-helical structure of the C-terminal peptide. Because we demonstrated previously that Dd-PDI is truly localized to the ER (Monnat et al., 1997), our working hypothesis was that the conserved D domain might play a role in retrieval or retention of ER residents independent of an H/KDEL motif.
The Last 57 Amino Acids of Dd-PDI Are Necessary for Intracellular Retention
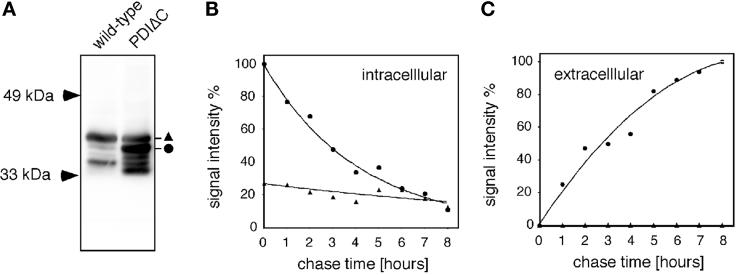
In light of the windbeutel mutations, we reasoned that the last 57 conserved residues of Dd-PDI might play a role in the retention of this PDI in the cell. Therefore, a PDI construct lacking these residues (Dd-PDIΔC) was stably expressed in wild-type cells. Quantification of expression levels showed that Dd-PDIΔC was expressed at about a threefold higher level than the endogenous PDI (Figure 2A). After a pulse/chase metabolic labeling with [35S]Met, the secretion/retention of the Dd-PDIΔC construct and the endogenous PDI were monitored by immunoprecipitation (Figure 2, B and C). Whereas Dd-PDI was relatively stable (half-life > 8 h; Figure 2B, ▴) and was undetectable in the medium (Figure 2C, ▴), the intracellular level of Dd-PDIΔC decreased much more rapidly (half-life < 3 h; Figure 2B, ●) and was clearly detected in the medium (Figure 2C, ●). Interestingly, the relatively slow secretion kinetics suggested that, even truncated of half of its D domain, Dd-PDI might still interact with other proteins in the ER.
Figure 2.
The C-terminal half of the D domain is necessary for intracellular retention of Dd-PDI. (A) A stable clonal cell line was created and shown to express about threefold more Dd-PDIΔC (●) than the endogenous Dd-PDI (▴), as revealed by anti-PDI immunoblotting. This cell line was then investigated for secretion/retention of Dd-PDI and recombinant Dd-PDIΔC by pulse/chase labeling with [35S]Met followed by anti-PDI immunoprecipitation. At the indicated times, aliquots were retrieved and cells (B, intracellular) were separated from medium (C, extracellular) by centrifugation and analyzed separately. After quantification by phosphorimager, the normalized signals corresponding to full-length Dd-PDI (▴) and Dd-PDIΔC (●) were plotted as a function of time.
The Second Half of the D Domain Confers ER Retention to GFP
Our aim was then to determine whether the C-terminal half of the D domain would be able to function in trans and retain a reporter protein inside the cell, and also to approximately map the minimal motif required. We generated stably transformed cell lines expressing GFP fused to either the last 13 (GFP-PDI13) or 57 (GFP-PDI57) C-terminal amino acids. GFP-PDI13 contains the best conserved NIL motif. As negative and positive controls for the retention assay, we used a secreted form of GFP (GFP-sec), a GFP-KDEL known to be the efficient signal in mammalian cells, and a GFP-HDEL functional in yeast and found at the C terminus of D. discoideum ER proteins such as calreticulin (Monnat et al., 1997), a classic PDI and a BiP homologue (accession numbers C91024 and C25542, respectively; J. Monnat and T. Soldati, unpublished information). To minimize the influence of the expression levels on subcellular localization, we selected clones with similar intracellular contents of GFP (Figure 3A, cells).
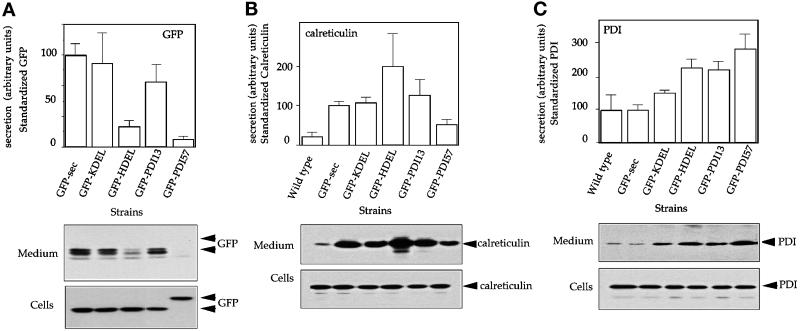
Figure 3.
Retention of different GFP chimeras and effect of their expression on the secretion of endogenous calreticulin and PDI. Stable clones transfected with GFP-sec, GFP-KDEL, GFP-HDEL, GFP-PDI13, and GFP-PDI57 constructs were selected as containing similar intracellular levels of GFP. Cells were grown for 8 h on Petri dishes in HL5c medium, and then cells and medium were separated by centrifugation. Proteins from the cell culture supernatant were TCA-precipitated, whereas cells were directly lysed in SDS sample buffer. One percent of total lysate and the whole supernatant were separated on 12% SDS-PAGE under reducing conditions and finally analyzed by quantitative immunoblotting with anti-GFP (A), anti-calreticulin (B), or anti-PDI (C) mAbs. (A) Retention analysis of GFP-sec, GFP-KDEL, GFP-HDEL, GFP-PDI13, and GFP-PDI57. The multiple bands of the GFP signal in medium likely result from posttranslational modifications during transit through the secretory pathway. The effects of the expression of GFP constructs on the secretion of endogenous calreticulin (B) or endogenous PDI (C) in the respective strains are shown. The results (average ± SD of at least three independent experiments) were quantified as described in MATERIALS AND METHODS.
To determine the level of secretion/retention of the different GFP constructs, we set up a pulse/chase experiment similar to the one used for PDI and PDIΔC. The labeling of the proteins was relatively weak, and the commercially available anti-GFP antibodies did not allow reliable detection of the GFP constructs released in the medium. Nevertheless, we could determine that the half-lives of GFP-HDEL and GFP-PDI57 in the cell were >8 h (explaining the low incorporation of radioactivity and the poor detection in the medium) and that the half-life of GFP-sec was <3 h. These intracellular values include loss by degradation and secretion and indicated that GFP-HDEL and GFP-PDI57 were very stable and that GFP-sec was likely released by the cells, although relatively slowly.
To assess retention levels, wild-type cells and cells expressing heterologous GFP fusions were plated and allowed to adhere for 30 min before fresh medium was added. After 8 h, cells and medium were separated by centrifugation. The whole supernatant and 1% of the total lysate were analyzed by SDS-PAGE and quantitative immunoblotting (see MATERIALS AND METHODS). Figure 3A presents both the quantification of the amount of GFP chimeras released in the medium and a representative immunoblot from such a retention/secretion experiment. The two main bands visible for the GFP chimeras secreted in the medium likely resulted from undefined processing, such as glycosylation, sulfation, or phosphorylation, occurring as GFP traveled through the secretory pathway. Indeed, this doublet was detectable neither in the cells nor when recombinant GFP was incubated with wild-type cells for 20 h. To facilitate direct comparison, each value was normalized to the value obtained for the strain expressing GFP-sec as representing 100% secretion. GFP-KDEL and GFP-sec were comparably secreted, indicating that the KDEL signal was not recognized efficiently by the D. discoideum retrieval system. Conversely, GFP-HDEL was almost totally retained, demonstrating the specificity of the HDEL retrieval signal. The PDI13 peptide reduced the secretion of GFP ∼25%, whereas the PDI57 peptide mediated nearly quantitative retention of GFP. Because of experimental variations, the presented results are not highly significant, but the same trend was observed in each separate experiment. Together, the results indicate that the major determinants for efficient retention of Dd-PDI are not confined to a discrete conserved motif such as NIL but that more complex features of the C-terminal domain are involved.
A First Hint for Two Independent and Saturable Retrieval/Retention Mechanisms
There is now compelling evidence for a saturable retrieval system in yeast and mammalian cells (Dean and Pelham, 1990; Lewis and Pelham, 1992; Bu et al., 1997). Therefore, we analyzed the expression of the various GFP constructs to determine if it could somehow affect the retention of the endogenous HDEL-bearing calreticulin (Figure 3B). Compared with the wild-type situation, secretion of calreticulin was slightly increased in every GFP-expressing cell line, even though the steady-state concentration of each protein in the cell appeared unchanged, ruling out an unfolded protein response (Sidrauski et al., 1998). Nevertheless, the highest secretion of calreticulin was observed in the GFP-HDEL–expressing strain, indicating that the retrieval system can be saturated specifically by the HDEL- but not by the KDEL-bearing protein. The lower level of secretion of calreticulin in the GFP-PDI57–expressing strain might indicate that the effect of GFP overexpression was somewhat “compartmentalized” and had less influence on HDEL-mediated retrieval (see below). The effect of overexpressing GFP chimeras on the retention of PDI was also examined (Figure 3C). A stronger secretion of endogenous PDI was observed as a consequence of the overexpression of GFP-PDI57, although the differences between the constructs were less manifest than for secretion of calreticulin. These results support the idea that a saturable machinery, different from the HDEL receptor retrieval system, is necessary for full intracellular retention of PDI.
Subcellular Localization of GFP Chimeras Reveals a Subdomain of the ER
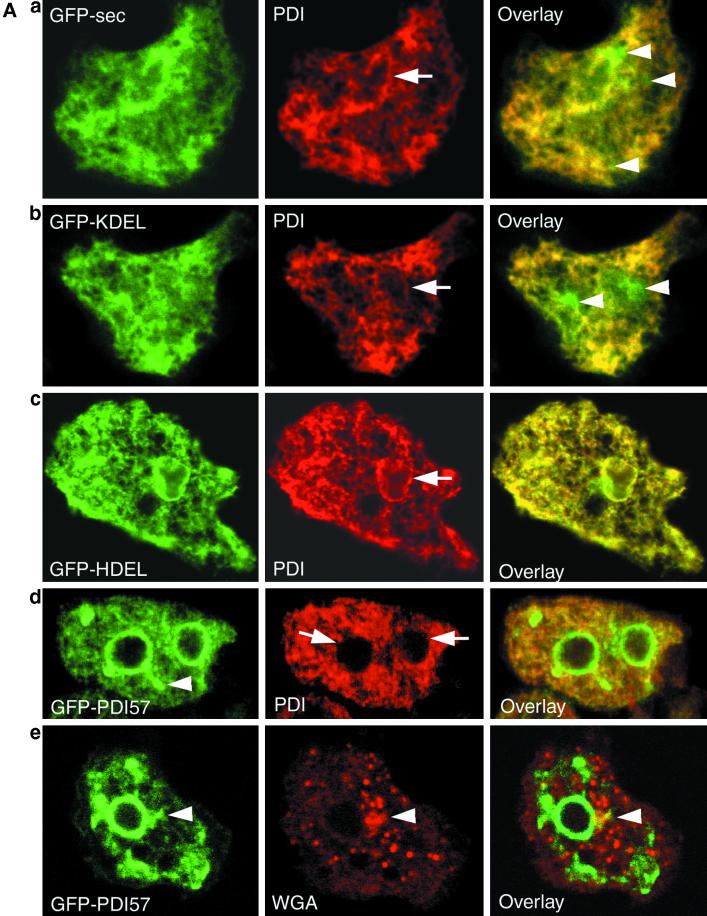
The intracellular distribution of the GFP chimeras was analyzed by fluorescence microscopy concomitantly with immunolocalization of calreticulin (our unpublished observation) and PDI (Figure 4A). GFP-sec and GFP-KDEL colocalized only partially with PDI (Figure 4A, a and b) or calreticulin, as best revealed in the overlay by the presence of an additional green “spot” typical for the Golgi morphology in D. discoideum (Bertholdt et al., 1985). These results are indicative of their transport to the Golgi compartment as well as to subsequent stages of the secretory pathway, visible as green punctations scattered in the cytoplasm (Figure 4Aa, arrowheads). In contrast, GFP-HDEL displayed a perfect colocalization with PDI (Figure 4Ac) and calreticulin, with only rare Golgi staining. GFP-PDI57 exhibited prominent staining of the nuclear envelope with an additional “finger” of fluorescence (Figure 4A, d and e, arrowheads) pointing toward or inside the Golgi region (visualized by wheat germ agglutinin staining). Notably, in cells expressing GFP-PDI57, the perinuclear staining of PDI was almost completely absent (Figure 4Ad, arrows), whereas it was visible in most cells expressing other GFP chimeras (compare Figure 4A, a–c). The phenomenon was specific, because calreticulin was still present in the nuclear envelope.
Figure 4.
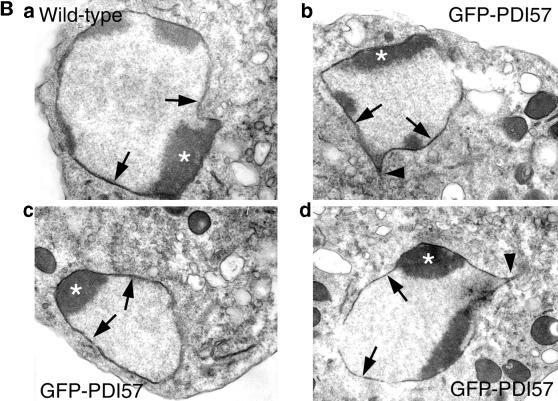
Subcellular localization of the GFP chimeras. Cells expressing different GFP constructs were immunostained with either anti-PDI antibodies or with wheat germ agglutinin as Golgi marker and analyzed by confocal microscopy. (A) The colocalization of GFP-sec (a) and GFP-KDEL (b) with PDI is only partial, as illustrated by major green “spots” on the overlays (a and b, arrowheads) indicating the transport to the Golgi compartment and more peripheral dots likely representing transport to the plasma membrane (b, arrowheads at the cell periphery). GFP-HDEL (c) colocalized perfectly with PDI. PDI is present in the nuclear envelope (arrows in a–c) and throughout the reticular network of the ER. GFP-PDI57 (d and e) localized predominantly around the nucleus, with an extension pointing toward the Golgi region (arrowheads in d and e). This “finger” shows partial colocalization of wheat germ agglutinin labeling (WGA; arrowhead in e). In the GFP-PDI57–expressing cells, the absence of perinuclear staining for PDI is clearly discernible (arrows in d). (B) Thin section electron micrographs of wild-type and GFP-PDI57–expressing cells. In both cell types, the nuclear envelope has the same appearance (arrows). Depending on the sections, most nuclei of wild-type and GFP-PDI57–expressing cells (arrowheads in b and d) are “pinched” in the direction of the microtubule-organizing center/Golgi region, likely reflecting the tight association of the centrosome with the nuclear membrane. The asterisks indicate the dark, nucleoli-like chromatin regions called nuclear caps.
To exclude the possibility that the perinuclear accumulation of GFP-PDI57 was due to tight “wrapping” of ER cisternae around the nucleus, nuclear morphology was studied by transmission electron microscopy. As shown in Figure 4B, the nuclei of cells expressing GFP-PDI57 were indistinguishable from wild-type cells and showed no sign of formation of karmellae-like structures. Note that in both cell types, most nuclei have a protrusion extending in the direction of the microtubule-organizing center/Golgi (Daunderer et al., 1999; E.M. Neuhaus and T. Soldati, unpublished observation), likely correlating with the finger seen in Figure 4A, d and e.
These observations, together with the secretion/retention data, are strong indications for the existence of a saturable mechanism for ER localization of PDI. The C terminus of GFP-PDI57 competes with PDI for common binding sites present in limiting amounts in the nuclear envelope.
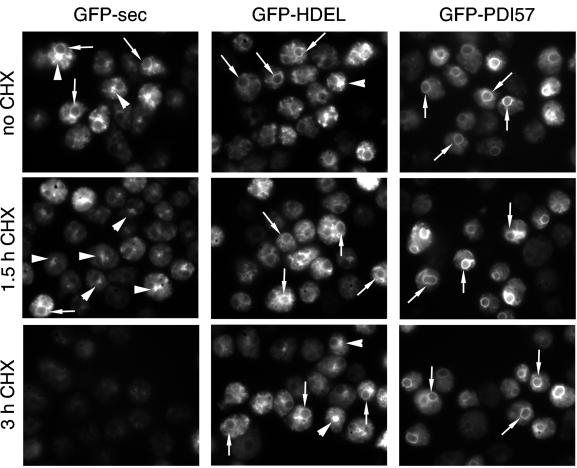
Cycloheximide Treatment and Live Microscopy
To confirm the secretion/retention data at the morphological level, the fate of the GFP constructs was analyzed after blockage of protein synthesis (Figure 5). As already shown in Figure 4A, because of relatively slow secretion kinetics, even the secreted GFP-sec was present throughout the ER, in the nuclear envelope, and in the Golgi complex. GFP-HDEL had a similar localization with rarer presence in the Golgi, whereas GFP-PDI57 was found predominantly in the nuclear envelope. After 90 min of cycloheximide treatment, the localization of GFP-HDEL and GFP-PDI57 was barely affected, and after 180 min, the fluorescence was slightly weaker for both proteins and GFP-HDEL was found more often in the Golgi. In sharp contrast, at 90 min, GFP-sec was disappearing from the ER reticulum and nuclear envelope and was found predominantly in the Golgi complex. After 180 min of protein synthesis block, GFP-sec was almost quantitatively lost from the cells.
Figure 5.
Cycloheximide treatment and live microscopy. Cells expressing GFP-sec, GFP-HDEL, and GFP-PDI57 were observed live, and the GFP fluorescence was recorded with a charge-coupled device camera. In the absence of cycloheximide (no CHX), the steady-state distribution of all three chimeras was as shown in Figure 4A. GFP-sec and GFP-HDEL were clearly found throughout the ER reticulum, in the nuclear envelope (arrows), and in the Golgi (arrowheads). In contrast, GFP-PDI57 was found predominantly in the nuclear envelope. After 1.5 and 3 h of incubation with cycloheximide (1.5 h CHX and 3 h CHX, respectively), the distribution of GFP-HDEL and GFP-PDI57 was barely affected, whereas GFP-sec first accumulated in the Golgi and then disappeared from the cells.
These data perfectly corroborated the biochemical analysis of secretion/retention and indicated that the machinery retaining GFP-PDI57 was of similar efficiency as the HDEL-mediated system.
PDI and GFP-PDI57 Are Soluble Components of the ER Lumen
Some ER residents have been reported to associate peripherally with the lumenal leaflet of the ER membrane. The mammalian lysyl hydroxylase (Myllyla et al., 1991) and the yeast ERO1 protein (Frand and Kaiser, 1998; Pollard et al., 1998) lack the H/KDEL retrieval signal and do not interact with other H/KDEL-tagged proteins that might participate indirectly in their retrieval (Vuori et al., 1992; Zhen et al., 1995). In a Triton X-114 phase separation experiment at low ionic strength, lysyl hydroxylase was recovered exclusively in the detergent phase, whereas PDI partitioned in the aqueous phase (Kellokumpu et al., 1994).
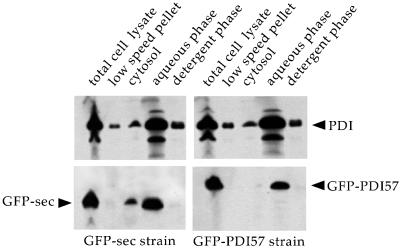
Similarly, association of Dd-PDI and GFP-PDI57 with the ER membrane might contribute to their retention. Therefore, we analyzed their partitioning by Triton X-114 phase-separation experiments. The assay was performed in isotonic conditions with 150 mM KCl and 5 mM MgCl2, because complexes containing Dd-PDI and GFP-PDI57 were detected under these conditions (see Figures 7 and 8 and text below). Dd-PDI was recovered mainly in the aqueous phase (Figure 6, upper panels). This partitioning was identical in wild-type cells and strains expressing GFP-sec or GFP-PDI57. To compare partitioning of GFP-sec and GFP-PDI57 with that of Dd-PDI, the immunoblot was subsequently probed with anti-GFP antibodies. Both GFP chimeras fractionated similarly to Dd-PDI (Figure 6, lower panels). This result supports the hypothesis that Dd-PDI retention is mediated by proteinaceous machinery rather than by direct electrostatic interactions with lipids. Additionally, the phase separation allowed quantitative and gentle recovery of potential lumenal complexes of PDI and GFP-PDI57.
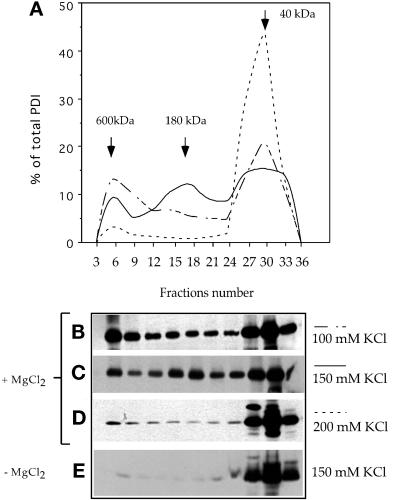
Figure 7.
PDI forms several salt-dependent complexes. A crude membrane fraction from wild-type AX2 cells was subjected to Triton X-114 phase partitioning in the presence or absence of MgCl2 and 100, 150, or 200 mM KCl. The aqueous phase was loaded onto a Superdex 200 HR 10/30 gel filtration column. The fractions were electrophoresed on 12% SDS-PAGE under reducing conditions and immunoblotted with anti-PDI antibodies. (A) Elution profiles showing progressive disruption of the high-molecular-mass complexes by increasing KCl concentrations. Representative immunoblots of elution profiles performed in 100 mM KCl (B), 150 mM KCl (C), and 200 mM KCl (D). At 150 mM salt, three forms were present at 600, 180, and 40 kDa, but at 200 mM, only the monomeric form of 40 kDa was visible. (E) Extraction and elution were performed with 150 mM KCl but in the absence of MgCl2. Under these conditions, PDI was found almost exclusively as a monomer. Molecular mass standards: thyroglobulin, 680 kDa; ferritin, 420 kDa; catalase, 205 kDa; and aldolase, 165 kDa. Densitometric quantifications were performed with NIH Image 1.61. The amount of PDI and GFP in each fraction is represented as the percentage of total eluted protein.
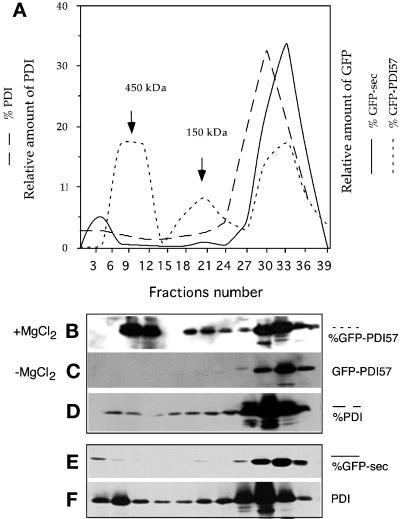
Figure 8.
Inhibition of PDI complex formation by expression of GFP-PDI57. The aqueous phase of a Triton X-114 extraction was obtained from GFP-PDI57– and GFP-sec–expressing strains in the presence of 150 mM KCl, with or without 5 mM MgCl2, and subjected to gel filtration. (A and B) The elution profile of GFP-PDI57 showed the presence of three forms: two complexes of 450 and 150 kDa and the monomeric form of ∼35 kDa. (A and C) The elution profile of GFP-PDI57 extracted without MgCl2 revealed the absence of high-molecular-mass complexes. (A and D) The presence of PDI in high-molecular-mass complexes was inhibited by expression of GFP-PDI57. (A and E) The elution profile of GFP-sec showed its exclusive presence in the 30- to 35-kDa fractions. (A and F) The elution profile of PDI in extracts of GFP-sec–expressing cells showed the presence of high-molecular-mass complexes as in the wild-type strain (see Figure 6, B and C).
Figure 6.
Subcellular fractionation and partitioning of PDI and GFP chimeras. Cells expressing GFP-sec or GFP-PDI57 were mechanically broken in TKM buffer (150 mM KCl, 5 mM MgCl2). The total cell lysate was centrifuged at 1000 × g for 10 min to obtain the low-speed pellet. The supernatant was ultracentrifuged, yielding the cytosol fraction. The crude membrane pellet was subjected to Triton X-114 phase separation (aqueous phase and detergent phase). Equal amounts of each fraction, allowing direct comparison, were loaded on 12% SDS-PAGE under reducing conditions and analyzed by immunoblotting with the use of anti-PDI (upper panels) and anti-GFP (lower panels) antibodies.
PDI Forms Several Salt-dependent Complexes
Binding of PDI to several ER resident proteins has been described previously (Koivu et al., 1987; Pihlajaniemi et al., 1987; Wetterau et al., 1990, 1991). Therefore, the presence of protein complexes containing Dd-PDI was investigated. Protein extracts from wild-type cells obtained as the aqueous phase of Triton X-114 phase separations performed in buffers with increasing concentrations of KCl, with or without 5 mM MgCl2, were applied on a gel filtration column. The column fractions were analyzed by SDS-PAGE and anti-PDI immunoblotting. To facilitate visualization and comparison of the elution profiles presented in Figure 7, B–D, the immunoblots were quantified and represented together as curves in Figure 7A. Although the major form at every salt concentration was the monomer of ∼40 kDa, two distinct complexes of 600 and 180 kDa were visible at 100 and 150 mM KCl (Figure 7C). Stepwise increases in ionic strength led to disruption of these complexes (Figure 7, B–D). Remarkably, the complexes were also completely absent from protein extracts generated with 1 mM EDTA without MgCl2 (Figure 7E). It was not possible to test whether CaCl2 would substitute for MgCl2, because the addition of CaCl2 results in enhanced proteolysis. Nevertheless, the results suggested that the interactions of Dd-PDI with other components of the ER were divalent cation–dependent.
Disruption of Endogenous PDI Complexes by Overexpression of GFP-PDI57
Similar gel filtration experiments were performed with extracts from strains expressing GFP-PDI57 and GFP-sec obtained in buffer with 150 mM KCl in the presence or absence of MgCl2. The resulting fractions were analyzed by SDS-PAGE and anti-GFP or anti-PDI immunoblotting (Figure 8, B–F). Selected quantified profiles are represented in Figure 8A for ease of comparison. The elution profiles revealed that GFP-PDI57 was present in two major forms, a complex of 450 kDa and a monomer of ∼35 kDa, with a third peak at 150 kDa (Figure 8, A and B). In these conditions, ∼40–60% of GFP-PDI57 was present in the form of high-molecular-mass complexes. As described above for endogenous PDI, the high-molecular-mass complexes were absent from extracts generated without MgCl2 (Figure 8C). The blot revealing the presence of GFP-PDI57 complexes (Figure 8B) was subsequently probed with anti-PDI antibodies, leading to the observation of an almost complete disappearance of the high-molecular-mass complexes of endogenous PDI (Figure 8D). In sharp contrast, in GFP-sec–expressing cells, GFP-sec was found mostly in its monomeric form (Figure 8E), and a significant proportion of PDI was present in high-molecular-mass complexes (Figure 8F). This observation indicated that similar or identical divalent cation–dependent interactions drive the formation of PDI and GFP-PDI57 complexes. These results strengthen the previous results showing an increased secretion of PDI in GFP-PDI57–expressing cells (Figure 3C) and the disappearance of the perinuclear localization of endogenous PDI (Figure 4A), hence supporting the hypothesis that the PDI57 peptide competes with endogenous PDI for a common binding partner.
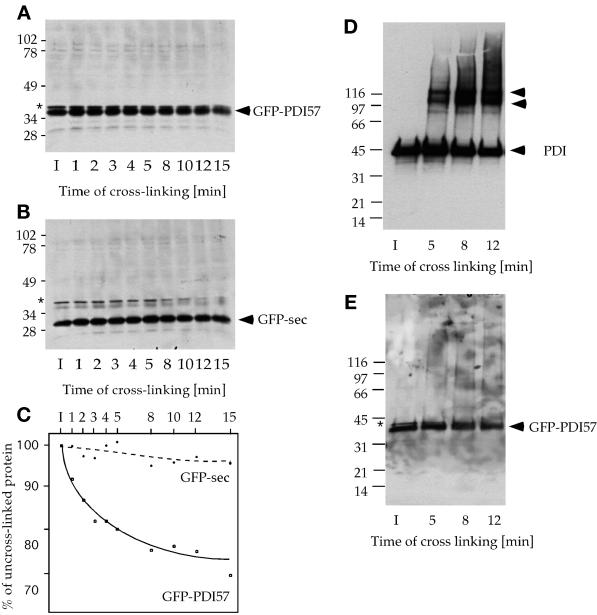
The PDI57 Domain Is Necessary to Obtain Cross-Link Products of GFP
To confirm and characterize further the high-molecular-mass complexes detected by gel filtration, glutaraldehyde cross-linking experiments were performed on intact cells expressing either GFP-sec or GFP-PDI57. The cross-linked samples were loaded on either uniform 10% (Figure 9, A and B) or 4–15% (Figure 9, D and E) gradient gels. The data presented in Figure 9, A and E, indicated that the 35-kDa band of GFP-PDI57 disappeared in a time-dependent manner. Quantification of such gels revealed that ∼30% of GFP-PDI57 protein was cross-linked after 15 min (Figure 9, A and C), whereas GFP-sec used as a negative control did not cross-link under these conditions (Figure 9, B and C). Interestingly, that 30% roughly corresponds to the percentage of GFP-PDI57 present in high-molecular-mass complexes (40–60%; Figure 8) and might correspond to the fraction of GFP-PDI57 present in the nuclear envelope. Unfortunately, likely because of their large size, no clear cross-link product of higher molecular mass was observed on either uniform 10% or 4–15% gradient gels. Alternatively, the complexes might be too heterogeneous to be resolved in discrete bands; rather, they appeared as smears (faintly visible in Figure 9E). To determine which cross-link products may be formed by endogenous PDI, the membrane used to visualize GFP-PDI57 (Figure 9D) was subsequently probed with anti-PDI antibodies. After cross-linking, a small portion of PDI was shifted into two main products of 100 and 120 kDa that did not contain GFP-PDI57. We conclude that the high-molecular-mass complexes of GFP-PDI57 detected by gel filtration were mediated by the PDI57 domain but were not attributable to interactions with endogenous PDI.
Figure 9.
In vivo cross-linking experiments in cells expressing GFP-sec or GFP-PDI57. Cross-links were performed with 106 intact cells with the use of 0.15% glutaraldehyde for 1–15 min as indicated (I, input). Samples were separated either on uniform 10% (A and B) or 4–15% gradient (D and E) SDS-PAGE under reducing conditions and immunoblotted with anti-GFP or anti-PDI antibodies. The 35-kDa GFP-PDI57 band decreased as a function of time (A), whereas GFP-sec was stable in these conditions (B). The band visible in A and B (asterisks) is a minor cross-reaction product of the anti-GFP antibodies. The resulting cross-link products were not resolved on the gels (see text). (C) Densitometric quantification of data in A and B. Samples from similar cross-linking experiments were separated on 4–15% gradient gels. Immunoblotting against PDI revealed the generation of two major complexes of ∼100 and 120 kDa (D), which did not contain detectable GFP-PDI57 (E). In addition, GFP-PDI57 was incorporated into heterogeneous cross-link products visible only as a weak smear (E).
The C-terminal Half of the D Domain Is Necessary for Dd-PDI to Complement a Yeast PDI Null Mutant
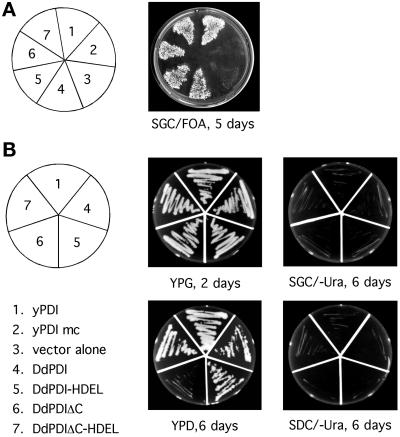
The TRG1/PDI1 gene encodes an ER resident protein structurally related to PDI and essential for growth. A deletion of the C-terminal 38 residues, including the retention signal HDEL, has also been shown to be lethal if the mutant TRG1 gene was expressed at low level under the control of the constitutive ADC1 promoter. However, the same mutant gene fused to the stronger GAL1 promoter sustained growth on galactose (Guenther et al., 1991). These results indicated that, at a low expression level, the trg1 protein must be localized to the ER to ensure cell viability. Hence, complementation of the TRG1 deficiency is a stringent test for both the disulfide isomerase activity of a protein and its concentration in the ER. For this purpose, we introduced both a full-length Dd-PDI and the Dd-PDIΔC (see Figure 2 and text) construct into the high-copy-number vector pDS2 (Guenther et al., 1993) downstream of the strong galactose-inducible GAL1 promoter. To ensure that both proteins are enzymatically active and able to complement the TRG1 deficiency when retained in the ER by the classic Erd2p-mediated pathway, an HDEL signal was introduced at the C terminus of both the Dd-PDI and Dd-PDIΔC coding sequences, resulting in the Dd-PDI-HDEL and Dd-PDIΔC-HDEL constructs. As positive and negative controls, we used wild-type yeast PDI (yPDI) and an inactive mutant form (yPDImc, with both thioredoxin CGHC boxes mutated to SGHS), respectively. The complementation experiments were performed in BK203-15B cells carrying a lethal deletion of the TRG1 gene (B. Kramer, unpublished data) and that are rescued by the pWBK-PDI centromer plasmid (Kramer et al., 1989). This centromer vector carries a URA3 gene, which allows positive selection for growth in the absence of uracil and negative selection in the presence of 5-FOA. As shown in Figure 10A, irrespective of the presence of the HDEL signal, all four Dd-PDI constructs supported growth as efficiently as yPDI on a plate containing 5-FOA and supplemented with galactose. As expected, the inactive mutant yPDImc was unable to grow under these conditions. This indicated that yeast can grow perfectly in the absence of the yPDI copy of the centromer plasmid as long as Dd-PDI is expressed at high levels. To test whether Dd-PDI would also complement at lower expression levels, we streaked colonies from the 5-FOA plate onto other plates with rich medium in the presence of galactose (YPG) or glucose (YPD). On YPG plates, all five constructs complemented equally well, and growth was vigorous, as shown after 2 d (Figure 10B). On YPD plates (shown after 6 d), the growth rates of the different cell lines were affected to different extents. Cells complemented with yPDI had only slightly reduced growth on YPD compared with growth on YPG. Cells with Dd-PDIΔC-HDEL grew almost as fast, whereas the growth of cells expressing the full-length Dd-PDI constructs (with or without HDEL) was slowed about threefold. In sharp contrast, Dd-PDIΔC exhibited virtually no complementation in this interval. None of the strains picked from the 5-FOA plate grew on plates lacking uracil (−Ura), demonstrating that growth on YPG and YPD plates was not due to the presence of the centromer plasmid pWBK-PDI (Figure 10B).
Figure 10.
Yeast complementation experiments. The strain BK203-15B carries a lethal deletion of the TRG1 gene but is rescued by the centromer plasmid pWBK-PDI, which contains a copy of the wild-type TRG1 gene and a URA3 selection marker. BK203-15B was transformed with different pDS2 constructs (1 = yPDI, 2 = yPDImc, 3 = vector alone, 4 = Dd-PDI, 5 = Dd-PDI-HDEL, 6 = Dd-PDIΔC, and 7 = Dd-PDIΔC-HDEL) under the control of the yeast galactose-inducible GAL1-promoter. Clones from each strain were then transferred onto a plate containing galactose and 5-FOA (SGC/FOA) to counterselect for the presence of the centromer plasmid (A). Of the seven strains, only the five with functional PDI complemented the TRG1 deletion. Finally, these five strains were streaked as indicated in the diagram on rich medium with galactose or glucose (B). All five strains grew at indistinguishable speed on galactose (YPG, 2 d). In contrast, on glucose (YPD, 6 d), only yPDI grew rapidly, followed in order by Dd-PDIΔC-HDEL and then by both Dd-PDI and Dd-PDI-HDEL. Dd-PDIΔC exhibited virtually no complementation in this period. None of the strains picked from the 5-FOA plate grew on plates lacking uracil (−Ura), demonstrating that growth on YPG and YPD plates was not due to the presence of the centromer plasmid. yPDI, wild-type yeast PDI; yPDImc, yeast PDI with both thioredoxin CGHC boxes mutated to SGHS.
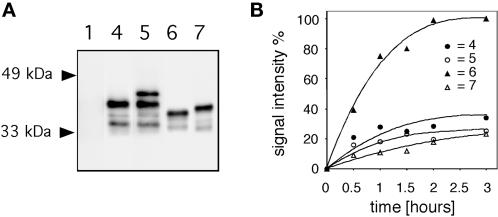
Extracts from cells grown in YPG were analyzed by immunoblotting with anti-PDI antibodies, revealing the intracellular steady-state levels of the four constructs based on Dd-PDI (Figure 11A). Note the absence of cross-reacting protein in wild-type yeast (lane 1). The major band in Dd-PDI– (lane 4) and Dd-PDI-HDEL– (lane 5) expressing cells had similar migration and were of the same size as endogenous PDI in D. discoideum. There was an additional product of higher apparent molecular mass in Dd-PDI-HDEL–expressing cells (lane 5). Dd-PDIΔC (lane 6) was of the same size as when expressed in D. discoideum, whereas the major signal in Dd-PDIΔC-HDEL–expressing cells (lane 7) was of slower mobility. The nature of the slower-migrating product of Dd-PDI-HDEL is not clear yet but could result from a modification (e.g., glycosylation) acquired during cycling between the ER and the Golgi. The same may apply to Dd-PDIΔC-HDEL. On average, the four constructs were expressed at similar levels, Dd-PDI was slightly less abundant than the two forms of Dd-PDI-HDEL together, and Dd-PDIΔC and Dd-PDIΔC-HDEL were almost identically expressed.
Figure 11.
Retention/secretion of different Dd-PDI constructs in yeast. (A) Extracts from cells grown in YPG were analyzed by immunoblotting with anti-PDI antibodies. Note the absence of signal in lane 1. The major band was in Dd-PDI– (lane 4) and Dd-PDI-HDEL– (lane 5) expressing cells, but there was an additional, minor, slowly migrating band in Dd-PDI-HDEL–expressing cells (lane 5). Dd-PDIΔC (lane 6) was of the expected size, but Dd-PDIΔC-HDEL (lane 7) showed a major signal at a slightly slower mobility. (B) Yeast cell lines expressing the different Dd-PDI constructs (nomenclature identical to that in Figure 10) were washed extensively and further cultivated in fresh YPG. At the indicated times, aliquots were taken and the medium was analyzed for the content of released PDI. Dd-PDIΔC was secreted at a more than threefold higher rate than all of the other constructs. Dd-PDI was retained as efficiently as the HDEL-tagged forms.
The secretion/retention of these constructs was then investigated by anti-PDI immunoblotting simply by monitoring their appearance in the medium as a function of time. As can be seen in Figure 11B, the rate of release of Dd-PDIΔC was significantly higher than the rates for the three other constructs. Therefore, it can be concluded that even at the high expression levels resulting from galactose induction, Dd-PDI was as efficiently retained in the cell as its HDEL-tagged counterpart.
DISCUSSION
The steady-state distribution of specific proteins, and thereby the identity of the endomembrane compartments forming the biosynthetic and endocytic systems, is achieved and maintained by the concerted and balanced action of three mechanisms: sorting of exported protein, retention of residents, and retrieval of escapees. Among these mechanisms, the best understood are the retrieval of soluble ER resident proteins bearing the H/KDEL-type signal (Pelham, 1990) and the retrograde transport of transmembrane proteins bearing either cytosolic KKXX or RR motifs or a lumenal HDEL signal (Teasdale and Jackson, 1996). On the other hand, mounting evidence indicates that the retrieval motifs cannot be the sole determinants of the steady-state distribution of ER residents. First, the KKXX motif has now been shown to function as a retention signal (Andersson et al., 1999). Second, some transmembrane proteins, e.g., ERGIC53, which carries an ER retention motif, are not localized to the ER (Schindler et al., 1993). Third, the acidic C-terminal domains of calreticulin and BiP were shown to participate somehow in their retention, possibly by a (Ca2+-dependent) interaction with the reticular matrix (Munro and Pelham, 1987; Zagouras and Rose, 1989). A fourth line of evidence favoring a model of ER retention comes from the continuous discovery of soluble ER resident proteins, among them Dd-PDI, without any identifiable retrieval signal. Until now, the primary sequences of such proteins have not revealed a consensus motif responsible for their retention. Only one unique motif, VEKPFAIAKE, has been identified in s-cyclophilin and is involved in its localization to a subcompartment of the ER (Arber et al., 1992).
The goal of the present work was to determine whether the C-terminal half of the D domain contains a functional ER localization motif. We tested whether it was both necessary for the intracellular retention of Dd-PDI and sufficient for the retention of a reporter protein, GFP. Using deletion analysis, we first showed that the domain is indeed crucial to the retention of PDI in the cell. Next, we speculated that the NIL motif present in the 13-amino acid C-terminal peptide could serve as an ER-localization motif. This tripeptide is found in every member of the recently recognized PDI-D subfamily, which includes both enzymes active as disulfide isomerase (PDI-Dα) and other less well-characterized ER proteins (PDI-Dβ). Although the peptide was not sufficient to mediate complete retention of GFP-PDI13, it consistently reduced its rate of secretion. Remarkably, the Drosophila windbeutel is an ER resident protein homologous to human Erp28 and rat Erp29 (Konsolaki and Schupbach, 1998), and mutations that compromise the integrity of the 70-residue C-terminal peptide result in severe developmental defects (Figure 1). Hence, although the NIL motif was insufficient to mediate complete ER retention in D. discoideum, the presence of the domain equivalent to PDI57 and the integrity of the NIL motif both appear crucial for the correct function of a Drosophila ER protein.
In contrast to the shorter peptide, the last 57 amino acids of Dd-PDI were able to mediate full retention in D. discoideum. Several facts support the conclusion that we are dealing with a true retention mechanism rather than a retrieval process. First, confocal analysis showed that GFP-PDI57 was concentrated mainly in an ER subdomain, the nuclear envelope. Second, after a complete block of protein synthesis, GFP-PDI57 was stably retained in the cell for hours at least as efficiently as GFP-HDEL, whereas GFP-sec was secreted. Hints of the mechanism responsible for this retention are revealed by the gel filtration experiments, which indicated that PDI and GFP-PDI57 form salt-dependent complexes. These complexes also depend on the presence of divalent cations, in agreement with previous reports demonstrating the role of Ca2+ in preventing secretion of reticuloplasmins (Booth and Koch, 1989). In contrast, retrieval processes seem independent of Ca2+ and thus distinguishable from true retention (Wilson et al., 1993). It was also shown that interactions between calreticulin and PDI are Zn2+-dependent (Baksh et al., 1995).
It has been suggested that the ER resident proteins, present at millimolar concentration, form a low-affinity, high-capacity reticular matrix that can accept large amounts of newly synthesized proteins and would be difficult to saturate (Sonnichsen et al., 1994). Here we obtained several indications that PDI retention is mediated by a saturable mechanism. First, in wild-type and every other GFP-expressing strain, endogenous PDI was localized in the perinuclear region and throughout the reticular ER network. On the contrary, in the GFP-PDI57 strain, endogenous PDI was absent around the nucleus, replaced by GFP-PDI57, correlating with an increased secretion of PDI in these cells. We estimated that the expression levels of the GFP chimeras were between 0.1 and 0.6% of total proteins, levels comparable to the cellular contents of PDI and calreticulin. GFP-PDI57 concentrated in the nuclear envelope may compete efficiently with endogenous PDI in the formation of retention complexes. As a control, the HDEL motif was found to efficiently retrieve proteins in D. discoideum and the mechanism was saturable, as shown previously in yeast and mammalian cells (Dean and Pelham, 1990; Lewis and Pelham, 1992; Bu et al., 1997). The saturation of the retention system was more difficult to observe than for HDEL retrieval, possibly because only a small amount of total PDI is localized in the perinuclear region and is involved in the competition with GFP-PDI57. Alternatively, secretion of PDI chased out of the nuclear envelope by GFP-PDI57, or secretion of PDIΔC, might be delayed because of the barrier formed by the reticular matrix. The broader distribution of PDI compared with GFP-PDI57 may be caused by interactions of the a and b domains with other ER residents and newly synthesized proteins, whereas the C-terminal domain would be responsible solely for its retention in the nuclear envelope.
Saturation of the retention system was also supported by gel filtration experiments. The high-molecular-mass complexes containing PDI (∼40–60% of total PDI) and present in wild-type and GFP-sec strains were almost completely absent from GFP-PDI57 strains, indicative of competition between endogenous PDI and the 57-amino acid C-terminal peptide for the formation of a complex. Altogether, the results argue against a low-specificity/low-affinity interaction of the C terminus of PDI with ER proteins, as is generally proposed for the reticular matrix. Alternatively, ER residents might be retained by electrostatic binding to the ER membrane in regions unable to form vesicles (Bannykh et al., 1996). However, our fractionation experiments revealed that PDI and GFP-PDI57 do not interact strongly with membranes.
To isolate one or several binding partners of PDI, we overexpressed GST-PDI57 in Escherichia coli and performed affinity chromatography with protein extracts from D. discoideum. Although some proteins were retained specifically by the C terminus of PDI, the results were too variable to allow reliable recognition of stoichiometric components. Therefore, to identify labile interactions, we performed cross-linking experiments in intact cells. Surprisingly, despite significant disappearance of the chimera, no distinct cross-link products of GFP-PDI57 were observed, even on 4–15% gradient gels. It is not clear yet whether the cross-linked complexes are too large (consistent with the complexes detected by gel filtration) or too heterogeneous to be resolved on the gels. By the same method, we determined that cross-linking of PDI generated two distinct products of 100 and 120 kDa containing no detectable GFP-PDI57. Because stable PDI complexes were almost absent from the gel filtration analysis of GFP-PDI57–expressing strains, these complexes may represent labile interactions of PDI with its substrates or with other multimeric enzymatic complexes. These results demonstrate that the PDI57 peptide is necessary and sufficient to induce the formation of ER lumenal complexes of GFP, but these complexes are not due to a direct interaction with PDI.
Because the D domain appears to be present throughout evolution, and because the windbeutel mutations in Drosophila (see above) were particularly suggestive, we speculated that the ER retention function might be conserved too. As a preliminary experiment, we expressed GFP-sec and GFP-PDI57 in human embryonic kidney cells and analyzed their localization by immunofluorescence (our unpublished results). Unlike in D. discoideum, the fluorescence of GFP-sec was barely detectable in the cell, likely because of the different folding kinetics of the GFPmut2 mutant (Cormack et al., 1996) at 37°C. Nevertheless, at steady state, GFP-PDI57 was detected in a punctate distribution, partially but not completely overlapping with structures labeled by antibodies against intermediate compartment markers such as β′-COP and p53.
Using another approach, we exploited the wealth of knowledge about retrieval/retention mechanisms in S. cerevisiae and made use of its powerful genetics. The yeast TRG1 gene was shown to be essential for cell viability. A mutant trg1 protein lacking its last C-terminal 38 residues, including the HDEL retrieval signal, and expressed at a low level was unable to complement the TRG1 null mutant, indicating that efficient ER retention is necessary for cell viability. This TRG1 mutant was able to promote yeast growth only when strongly overexpressed (Guenther et al., 1993). Similarly, it has been shown that mammalian Erp61 bearing a QDEL retrieval signal did not promote cell viability (Guenther et al., 1993). Hence, we complemented this yeast null mutant and demonstrated that Dd-PDI and Dd-PDIΔC were perfectly able to perform the disulfide isomerase activity of the wild-type yeast PDI. We also showed that, at an expression level similar to that of Dd-PDI-HDEL, Dd-PDI but not Dd-PDIΔC was sufficient to allow for growth on glucose. These results demonstrate that Dd-PDI is retained in the yeast ER equally well as when fused to the efficient yeast retrieval signal HDEL. This is not the case for Dd-PDIΔC or for other PDI proteins such as P5 and Erp72 (B. Kramer, unpublished data).
Altogether, our results demonstrate that the C-terminal end of Dd-PDI contains determinants required for ER retention. In addition, they suggest the presence of a previously unrecognized retention machinery present in D. discoideum and yeast, and they further emphasize that correct location of a given protein along the secretory pathway can be necessary for cell viability.
ACKNOWLEDGMENTS
A special thank you goes to Dr. H.-D. Söling (Max-Planck-Institute for Biophysical Chemistry, Göttingen) for his support and discussions during the course of this project, and to Dr. I. Haas (Department of Biochemistry, University of Heidelberg) for critical reading of the manuscript. We also thank the members of the laboratory for their support and suggestions, and U. Buhre and J. Schleich for their technical assistance. J.M. and E.M.N. were the recipients of a postdoctoral and a doctoral fellowship, respectively, from the Max-Planck-Gesellschaft, and M.S.P. was supported by a grant from the Deutsche Forschungsgemeinschaft (SPP 312) to T.S.
REFERENCES
- Andersson H, Kappeler F, Hauri HP. Protein targeting to endoplasmic reticulum by dilysine signals involves retention in addition to retrieval. J Biol Chem. 1999;274:15080–15084. doi: 10.1074/jbc.274.21.15080. [DOI] [PubMed] [Google Scholar]
- Arber S, Krause KH, Caroni P. s-Cyclophilin is retained intracellularly via a unique COOH-terminal sequence and colocalizes with the calcium storage protein calreticulin. J Cell Biol. 1992;116:113–125. doi: 10.1083/jcb.116.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baksh S, Burns K, Andrin C, Michalak M. Interaction of calreticulin with protein disulfide isomerase. J Biol Chem. 1995;270:31338–31344. doi: 10.1074/jbc.270.52.31338. [DOI] [PubMed] [Google Scholar]
- Bannykh SI, Rowe T, Balch WE. The organization of endoplasmic reticulum export complexes. J Cell Biol. 1996;135:19–35. doi: 10.1083/jcb.135.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertholdt G, Stadler J, Bozzaro S, Fichtner B, Gerisch G. Carbohydrate and other epitopes of the contact site A glycoprotein of Dictyostelium discoideum as characterized by monoclonal antibodies. Cell Differ. 1985;16:187–202. doi: 10.1016/0045-6039(85)90516-0. [DOI] [PubMed] [Google Scholar]
- Booth C, Koch GL. Perturbation of cellular calcium induces secretion of lumenal ER proteins. Cell. 1989;59:729–737. doi: 10.1016/0092-8674(89)90019-6. [DOI] [PubMed] [Google Scholar]
- Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- Bu G, Rennke S, Geuze HJ. ERD2 proteins mediate ER retention of the HNEL signal of LRP's receptor-associated protein (RAP) J Cell Sci. 1997;110:65–73. doi: 10.1242/jcs.110.1.65. [DOI] [PubMed] [Google Scholar]
- Campbell DA, Fogel S, Lusnak K. Mitotic chromosome loss in a disomic haploid of Saccharomyces cerevisiae. Genetics. 1975;79:383–396. doi: 10.1093/genetics/79.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- Dai Y, Wang C. A mutant truncated protein disulfide isomerase with no chaperone activity. J Biol Chem. 1997;272:27572–27576. doi: 10.1074/jbc.272.44.27572. [DOI] [PubMed] [Google Scholar]
- Darby NJ, Penka E, Vincentelli R. The multi-domain structure of protein disulfide isomerase is essential for high catalytic efficiency. J Mol Biol. 1998;276:239–247. doi: 10.1006/jmbi.1997.1504. [DOI] [PubMed] [Google Scholar]
- Daunderer C, Schliwa M, Gräf R. Dictyostelium discoideum: a promising centrosome model system. Biol Cell. 1999;91:313–320. doi: 10.1016/s0248-4900(99)80092-6. [DOI] [PubMed] [Google Scholar]
- Dean N, Pelham HR. Recycling of proteins from the Golgi compartment to the ER in yeast. J Cell Biol. 1990;111:369–377. doi: 10.1083/jcb.111.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L, Molinari M, Helenius A. Setting the standards: quality control in the secretory pathway. Science. 2000;286:1882–1888. doi: 10.1126/science.286.5446.1882. [DOI] [PubMed] [Google Scholar]
- Faix J, Gerisch G, Noegel AA. Overexpression of the csA cell adhesion molecule under its own cAMP-regulated promoter impairs morphogenesis in Dictyostelium. J Cell Sci. 1992;102:203–214. doi: 10.1242/jcs.102.2.203. [DOI] [PubMed] [Google Scholar]
- Fasel N, Begdadirais C, Bernard M, Bron C, Corradin G, Reymond CD. Dictyostelium discoideum as an expression host for the circumsporozoite protein of Plasmodium falciparum. Gene. 1992;111:157–163. doi: 10.1016/0378-1119(92)90683-g. [DOI] [PubMed] [Google Scholar]
- Ferrari DM, Söling H-D. The protein disulphide isomerase family: unraveling a string of folds. Biochem J. 1999;339:1–10. [PMC free article] [PubMed] [Google Scholar]
- Ferrari DM, Van Nguyen P, Kratzin HD, Söling HD. ERp28, a human endoplasmic-reticulum-lumenal protein, is a member of the protein disulfide isomerase family but lacks a CXXC thioredoxin-box motif. Eur J Biochem. 1998;255:570–579. doi: 10.1046/j.1432-1327.1998.2550570.x. [DOI] [PubMed] [Google Scholar]
- Frand AR, Kaiser CA. The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol Cell. 1998;1:161–170. doi: 10.1016/s1097-2765(00)80017-9. [DOI] [PubMed] [Google Scholar]
- Freedman RB, Hirst TR, Tuite MF. Protein disulphide isomerase: building bridges in protein folding. Trends Biochem Sci. 1994;19:331–336. doi: 10.1016/0968-0004(94)90072-8. [DOI] [PubMed] [Google Scholar]
- Gilbert HF. Protein disulfide-isomerase. Methods Enzymol. 1998;290:26–50. doi: 10.1016/s0076-6879(98)90005-2. [DOI] [PubMed] [Google Scholar]
- Guenther R, Brauer C, Janetzky B, Foerster HH, Ehbrecht IM, Lehle L, Kuentzel H. The Saccharomyces cerevisiae Trg1 gene is essential for growth and encodes a lumenal endoplasmic reticulum glycoprotein involved in the maturation of vacuolar carboxypeptidase. J Biol Chem. 1991;266:24557–24563. [PubMed] [Google Scholar]
- Guenther R, Srinivasan M, Haugejorden S, Green M, Ehbrecht IM, Kuentzel H. Functional replacement of the Saccharomyces cerevisiae trg1-pdi1 protein by members of the mammalian protein disulfide isomerase family. J Biol Chem. 1993;268:7728–7732. [PubMed] [Google Scholar]
- Haas IG, Wabl M. Immunoglobulin heavy chain binding protein. Nature. 1983;306:387–389. doi: 10.1038/306387a0. [DOI] [PubMed] [Google Scholar]
- Hammond C, Helenius A. Quality control in the secretory pathway. Curr Opin Cell Biol. 1995;7:523–529. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- Helenius A. How N-linked oligosaccharides affect glycoprotein folding in the endoplasmic reticulum. Mol Biol Cell. 1994;5:253–265. doi: 10.1091/mbc.5.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W. Protein transport from the endoplasmic reticulum to the Golgi apparatus. J Cell Sci. 1998;111:2831–2839. doi: 10.1242/jcs.111.19.2831. [DOI] [PubMed] [Google Scholar]
- Howard PK, Ahern KG, Firtel RA. Establishment of a transient expression system for Dictyostelium discoideum. Nucleic Acids Res. 1988;16:2613–2623. doi: 10.1093/nar/16.6.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbel BM, Biegelmann E. A preparation protocol for postembedding immunoelectron microscopy of Dictyostelium discoideum cells with monoclonal antibodies. Scanning Microsc. 1992;6:817–825. [Google Scholar]
- Kellokumpu S, Sormunen R, Heikkinen J, Myllyla R. Lysyl hydroxylase, a collagen processing enzyme, exemplifies a novel class of luminally oriented peripheral membrane proteins in the endoplasmic reticulum. J Biol Chem. 1994;269:30524–30529. [PubMed] [Google Scholar]
- Kemmink J, Darby NJ, Dijkstra K, Nilges M, Creighton E. The folding catalyst protein disulfide isomerase is constructed of active and inactive thioredoxin modules. Curr Biol. 1997;7:239–245. doi: 10.1016/s0960-9822(06)00119-9. [DOI] [PubMed] [Google Scholar]
- Koch GL. Reticuloplasmins: a novel group of proteins in the endoplasmic reticulum. J Cell Sci. 1987;87:491–492. doi: 10.1242/jcs.87.4.491. [DOI] [PubMed] [Google Scholar]
- Koivu J, Myllyla R, Helaakoski T, Pihlajaniemi T, Tasanen K, Kivirikko KI. A single polypeptide acts both as the beta subunit of prolyl 4-hydroxylase and as a protein disulfide-isomerase. J Biol Chem. 1987;262:6447–6449. [PubMed] [Google Scholar]
- Konsolaki M, Schupbach T. Windbeutel, a gene required for dorsoventral patterning in Drosophila, encodes a protein that has homologies to vertebrate proteins of the endoplasmic reticulum. Genes Dev. 1998;12:120–131. doi: 10.1101/gad.12.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer B, Kramer W, Williamson MS, Fogel S. Heteroduplex DNA correction in Saccharomyces cerevisiae is mismatch specific and requires functional PMS genes. Mol Cell Biol. 1989;9:4432–4440. doi: 10.1128/mcb.9.10.4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lewis MJ, Pelham HR. Sequence of a second human KDEL receptor. J Mol Biol. 1992;226:913–916. doi: 10.1016/0022-2836(92)91039-r. [DOI] [PubMed] [Google Scholar]
- Lewis MJ, Sweet DJ, Pelham HR. The ERD2 gene determines the specificity of the luminal ER protein retention system. Cell. 1990;61:1359–1363. doi: 10.1016/0092-8674(90)90699-f. [DOI] [PubMed] [Google Scholar]
- Manstein DJ, Schuster HP, Morandini P, Hunt DM. Cloning vectors for the production of proteins in Dictyostelium discoideum. Gene. 1995;162:129–134. doi: 10.1016/0378-1119(95)00351-6. [DOI] [PubMed] [Google Scholar]
- Michalak M, Milner RE, Burns K, Opas M. Calreticulin. Biochem J. 1992;285:681–692. doi: 10.1042/bj2850681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierendorf RCJ, Dimond RL. Functional heterogeneity of monoclonal antibodies obtained using different screening assays. Anal Biochem. 1983;135:221–229. doi: 10.1016/0003-2697(83)90754-6. [DOI] [PubMed] [Google Scholar]
- Monnat J, Hacker U, Geissler H, Rauchenberger R, Neuhaus EM, Maniak M, Soldati T. Dictyostelium discoideum protein disulfide isomerase, an endoplasmic reticulum resident enzyme lacking a KDEL-type retrieval signal. FEBS Lett. 1997;418:357–362. doi: 10.1016/s0014-5793(97)01415-4. [DOI] [PubMed] [Google Scholar]
- Müller-Taubenberger A, Hagmann J, Noegel A, Gerisch G. Ubiquitin gene expression in Dictyostelium is induced by heat and cold shock, cadmium, and inhibitors of protein synthesis. J Cell Sci. 1988;90:51–58. doi: 10.1242/jcs.90.1.51. [DOI] [PubMed] [Google Scholar]
- Munro S, Pelham HR. An Hsp70-like protein in the ER: identity with the 78 kd glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell. 1986;46:291–300. doi: 10.1016/0092-8674(86)90746-4. [DOI] [PubMed] [Google Scholar]
- Munro S, Pelham HRB. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1987;48:899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- Myllyla R, Pihlajaniemi T, Pajunen L, Turpeenniemi-Hujanen T, Kivirikko KI. Molecular cloning of chick lysyl hydroxylase: little homology in primary structure to the two types of subunit of prolyl 4-hydroxylase. J Biol Chem. 1991;266:2805–2810. [PubMed] [Google Scholar]
- Neuhaus EM, Horstmann H, Almers W, Maniak M, Soldati T. Ethane-freezing/methanol-fixation of cell monolayers: a procedure for improved preservation of structure and antigenicity for light and electron microscopies. J Struct Biol. 1998;121:326–342. doi: 10.1006/jsbi.1998.3971. [DOI] [PubMed] [Google Scholar]
- Noiva R, Freedman RB, Lennarz WJ. Peptide binding to protein disulfide isomerase occurs at a site distinct from the active sites. J Biol Chem. 1993;268:19210–19217. [PubMed] [Google Scholar]
- Noiva R, Lennarz WJ. Protein disulfide isomerase: a multifunctional protein resident in the lumen of the endoplasmic reticulum. J Biol Chem. 1992;267:3553–3556. [PubMed] [Google Scholar]
- Pelham HR. Control of protein exit from the endoplasmic reticulum. Annu Rev Cell Biol. 1989;5:1–23. doi: 10.1146/annurev.cb.05.110189.000245. [DOI] [PubMed] [Google Scholar]
- Pelham HR. The retention signal for soluble proteins of the endoplasmic reticulum. Trends Biochem Sci. 1990;15:483–486. doi: 10.1016/0968-0004(90)90303-s. [DOI] [PubMed] [Google Scholar]
- Pelham HRB, Hardwick KG, Lewis MJ. Sorting of soluble ER proteins in yeast. EMBO J. 1988;7:1757–1762. doi: 10.1002/j.1460-2075.1988.tb03005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pihlajaniemi T, Helaakoski T, Tasanen K, Myllyla R, Huhtala M-L, Koivu J, Kivirikko KI. Molecular cloning of the β-subunit of human prolyl 4-hydroxylase: this subunit and protein disulphide isomerase are products of the same gene. EMBO J. 1987;6:643–649. doi: 10.1002/j.1460-2075.1987.tb04803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard MG, Travers KJ, Weissman JS. Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol Cell. 1998;1:171–182. doi: 10.1016/s1097-2765(00)80018-0. [DOI] [PubMed] [Google Scholar]
- Schindler R, Itin C, Zerial M, Lottspeich F, Hauri HP. ERGIC-53, a membrane protein of the ER-Golgi intermediate compartment, carries an ER retention motif. Eur J Cell Biol. 1993;61:1–9. [PubMed] [Google Scholar]
- Semenza JC, Hardwick KG, Dean N, Pelham HR. ERD2, a yeast gene required for the receptor-mediated retrieval of luminal ER proteins from the secretory pathway. Cell. 1990;61:1349–1357. doi: 10.1016/0092-8674(90)90698-e. [DOI] [PubMed] [Google Scholar]
- Sidrauski C, Chapman R, Walter P. The unfolded protein response: an intracellular signaling pathway with many surprising features. Trends Cell Biol. 1998;8:245–249. doi: 10.1016/s0962-8924(98)01267-7. [DOI] [PubMed] [Google Scholar]
- Sonnichsen B, Fullekrug J, Van Nguyen P, Diekmann W, Robinson DG, Mieskes G. Retention and retrieval: both mechanisms cooperate to maintain calreticulin in the endoplasmic reticulum. J Cell Sci. 1994;107:2705–2717. doi: 10.1242/jcs.107.10.2705. [DOI] [PubMed] [Google Scholar]
- Sussman M. Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. Methods Cell Biol. 1987;28:9–29. doi: 10.1016/s0091-679x(08)61635-0. [DOI] [PubMed] [Google Scholar]
- Teasdale RD, Jackson MR. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the Golgi apparatus. Annu Rev Cell Dev Biol. 1996;12:27–54. doi: 10.1146/annurev.cellbio.12.1.27. [DOI] [PubMed] [Google Scholar]
- Vuori K, Pihlajaniemi T, Myllyla R, Kivirikko KI. Site-directed mutagenesis of human protein disulphide isomerase: effect on the assembly, activity and endoplasmic reticulum retention of human prolyl 4-hydroxylase in Spodoptera frugiperda insect cells. EMBO J. 1992;11:4213–4217. doi: 10.1002/j.1460-2075.1992.tb05515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetterau JR, Combs KA, McLean LR, Spinner SN, Aggerbeck LP. Protein disulfide isomerase appears necessary to maintain the catalytically active structure of the microsomal triglyceride transfer protein. Biochemistry. 1991;30:9728–9735. doi: 10.1021/bi00104a023. [DOI] [PubMed] [Google Scholar]
- Wetterau JR, Combs KA, Spinner SN, Joiner BJ. Protein disulfide isomerase is a component of the microsomal triglyceride transfer protein complex. J Biol Chem. 1990;265:9801–9807. [PubMed] [Google Scholar]
- Wilson DW, Lewis MJ, Pelham HR. pH-dependent binding of KDEL to its receptor in vitro. J Biol Chem. 1993;268:7465–7468. [PubMed] [Google Scholar]
- Zagouras P, Rose JK. Carboxy-terminal SEKDEL sequences retard but do not retain two secretory proteins in the endoplasmic reticulum. J Cell Biol. 1989;109:2633–2640. doi: 10.1083/jcb.109.6.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen L, Rusiniak ME, Swank RT. The beta-glucuronidase propeptide contains a serpin-related octamer necessary for complex formation with egasyn esterase and for retention within the endoplasmic reticulum. J Biol Chem. 1995;270:11912–11920. doi: 10.1074/jbc.270.20.11912. [DOI] [PubMed] [Google Scholar]