Abstract
Our goal was to develop a field soil biodegradation assay using 13C-labeled compounds and identify the active microorganisms by analyzing 16S rRNA genes in soil-derived 13C-labeled DNA. Our biodegradation approach sought to minimize microbiological artifacts caused by physical and/or nutritional disturbance of soil associated with sampling and laboratory incubation. The new field-based assay involved the release of 13C-labeled compounds (glucose, phenol, caffeine, and naphthalene) to soil plots, installation of open-bottom glass chambers that covered the soil, and analysis of samples of headspace gases for 13CO2 respiration by gas chromatography/mass spectrometry (GC/MS). We verified that the GC/MS procedure was capable of assessing respiration of the four substrates added (50 ppm) to 5 g of soil in sealed laboratory incubations. Next, we determined background levels of 13CO2 emitted from naturally occurring soil organic matter to chambers inserted into our field soil test plots. We found that the conservative tracer, SF6, that was injected into the headspace rapidly diffused out of the soil chamber and thus would be of little value for computing the efficiency of retaining respired 13CO2. Field respiration assays using all four compounds were completed. Background respiration from soil organic matter interfered with the documentation of in situ respiration of the slowly metabolized (caffeine) and sparingly soluble (naphthalene) compounds. Nonetheless, transient peaks of 13CO2 released in excess of background were found in glucose- and phenol-treated soil within 8 h. Cesium-chloride separation of 13C-labeled soil DNA was followed by PCR amplification and sequencing of 16S rRNA genes from microbial populations involved with 13C-substrate metabolism. A total of 29 full sequences revealed that active populations included relatives of Arthrobacter, Pseudomonas, Acinetobacter, Massilia, Flavobacterium, and Pedobacter spp. for glucose; Pseudomonas, Pantoea, Acinetobacter, Enterobacter, Stenotrophomonas, and Alcaligenes spp. for phenol; Pseudomonas, Acinetobacter, and Variovorax spp. for naphthalene; and Acinetobacter, Enterobacter, Stenotrophomonas, and Pantoea spp. for caffeine.
Achieving a mechanistic understanding of microorganisms where they dwell, in terrestrial and aquatic field habitats, is one of the major goals of microbial ecology; such understanding is facilitated by an ability to directly measure microbial metabolic processes and to identify microorganisms responsible for particular field biogeochemical reactions (4, 24, 35, 41, 48). But a variety of methodological obstacles have traditionally prevented investigators from simultaneously documenting identity and activity in real-world habitats such as soil. The most notable obstacles are the size discrepancy between humans and microorganisms, incomplete understanding of microhabitat physicochemical characteristics, a large reservoir of inactive, but potentially responsive cells in environmental samples, and the related propensity for microbial communities to change physiologically and in their population structure after removal from their in situ context (36). Increasingly, however, the obstacles are falling away. Recent success in linking identity with activity (3, 9, 17, 41, 50) can largely be attributed to the development of sophisticated multidisciplinary techniques and their application to field study sites that are often relatively simple in biogeochemical terms. One approach that attempts to identify active microorganisms relies upon laboratory-incubated model systems containing radioisotopically labeled substrates in sealed experimental vessels (6, 33, 53). While the environmental realism of laboratory-based model systems is somewhat controversial (7, 10, 11, 35, 43, 44, 51), the advantage of successfully merging microbial metabolic-activity data with microautoradiographic visualization of active cells (13, 28, 42) is clear.
When microbial processes are examined directly in field study sites and samples derived therefrom, the relevance of the data to in situ processes cannot be questioned. In some instances, stable isotopically labeled biomarkers are fortuitously present in the habitat of interest and these allow both biogeochemical processes and the active microorganisms to be documented (3, 17, 41). However, under routine circumstances, detection and quantification of metabolic processes in soil, water, and sediments becomes problematic because field sites are open systems where mass-balance approaches for measuring biogeochemical change is a challenge. In field-based investigations, the prospects for radiolabeling of active microbial populations are dim due to the unlikelihood of quantitative isotope retrieval from the field and to safety and environmental regulations. If experiments that release substrates to field sites are implemented, then nonradioactive surrogate compounds are likely to be employed (with or without conservative tracers that assist in mass-balance accounting [16, 49]).
Radajewski et al. (45) introduced stable isotope probing of community-extracted DNA as a laboratory-based means of identifying microbial populations involved in 13C-substrate metabolism. In the present investigation, we combined the realism of field-released 13C-substrates with extraction and sequencing of soil DNA. Our goals were to develop a 13C-based field respiration assay for testing biodegradability, apply the assay to a range of organic compounds representing diverse molecular structures, examine the contrast between the results of the field release assays and those of parallel assays conducted in sealed chambers, and link the 13C field release assay to DNA extraction analyses of the active microbial populations.
MATERIALS AND METHODS
Field study site.
The study site was located at the Cornell University Agricultural Experimental Station, Ithaca, N.Y. The predominant soil series was Collamer Silt loam. The plots were free of vegetation, were level, and were kept in the shade of a table (0.8 m high) so that direct sunlight did not impinge on the jars.
Field treatments.
The open-bottom soil cover treatments were prepared by inserting truncated 250-ml glass canning jars 3 to 5 cm into the soil, enclosing 28 cm2 of the surface. Other treatments involved removal of soil adjacent to the soil cover tests and placement of intact soil peds (∼30 g), soil cores (removed from soil after insertion of a 3-cm section of a 60-ml plastic syringe), or hand-crushed soil (5 g) into the same type of jars but with the bottom intact. The jars, with and without severed bottoms, were sealed with metal screw-cap canning-jar lids fitted with Teflon-coated butyl rubber septa. Within 5 min of the deployment of containers and soils in the four treatments (soil cover or jars containing ped, core, or crushed soil), aqueous solutions (∼0.3 ml) of 13C-labeled substrates (glucose, phenol, caffeine [∼450 μg total], or naphthalene [10 μg total]) were added dropwise to the soil to uniformly dispense the substrate over a circle of soil ∼4 cm in diameter. Screw-cap rings and lids were installed, and headspace sampling (250-μl gas-tight syringe) was begun and continued for 24 h. Three to five replicates of each treatment were prepared. After each sampling, the syringes were shuttled to the laboratory for gas chromatography/mass spectrometry (GC/MS) analysis.
Laboratory treatments.
Collamer Silt loam (5 g) was manually crushed and added to 38-ml serum bottles, which were closed with Teflon-faced septa and aluminum crimp seals. The soil was amended with 0.2- to 0.5-ml aqueous solutions of [13C]glucose, [13C]phenol, [13C]naphthalene, or [13C]caffeine to reach initial concentrations of 50, 50, 2, and 50 ppm, respectively. Control treatments included soil only, water only, and the same substrates but without 13C enrichment.
Chemicals.
The 13C-labeled substrates were glucose (13C6; 99% purity; Isotec Inc., Miamisburg, Ohio), phenol (13C6; 99% purity; Isotec Inc.), naphthalene (13C6; 99% purity; Cambridge Isotope Laboratories, Inc., Cambridge, Mass,), and caffeine (trimethyl-13C3; 99% purity; Cambridge Isotope Laboratories, Inc.). For preparation of the aqueous naphthalene 1 day prior to an experiment, 1 to 3 ml of water was added to a glass Teflon-sealed screw-cap vial. After autoclaving, approximately five crystals of [13C]naphthalene were transferred to the warm water. After a day at room temperature, high-performance liquid chromatography analysis verified that this produced a 20- to 24-ppm solution (∼75% saturation). This was dispensed with a glass pipette. In the tracer test, SF6 (Matheson Gas Products, Montgomeryville, Pa.) was injected with a 3-ml syringe and mixed by withdrawing and refilling four times.
GC/MS assays.
A Hewlett-Packard 5971A GC/MS equipped with a Hewlett-Packard Pora Plot Q column (25 m by 0.32 mm; 10-μm film thickness; He as the carrier gas) in the splitless mode was utilized to separate gaseous components. The detector was operated at 1.10−5 torr and 70 eV (56). The GC oven program was isothermal at 60°C. CO2 eluted at 2.5 min. Using the single ion monitoring mode, the detector was able to simultaneously quantify 12CO2 (m/z = 44) from 13CO2 (m/z = 45). SF6 eluted at 1.3 min. CO2 quantification was based on standards (Scott Specialty Gases Inc., Plumsteadville, Pa.) and serial dilutions prepared therefrom.
DNA extraction and CsCl fractionation of [13C]DNA.
After headspace sampling had been completed, the soil that received substrate was removed with a sterile spatula. Approximately 10 g from each jar was transferred to a sterile 35-ml centrifuge tube containing 10 ml of phosphate buffer (1 mM), which was then placed on dry ice, transported to the lab on dry ice, and stored at −80°C. After thawing, 0.25 g of sodium dodecyl sulfate was added to each tube and vortexed (1 min). The tubes were incubated for 30 min at 70°C, with rigorous mixing and vortexing every 5 min. Soil and cell debris were pelleted (10 min; 11,951 × g; 4°C). After transfer to a clean centrifuge tube, the supernatant was extracted twice with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) and extracted again with an equal volume of chloroform-isoamyl alcohol (24:1). The DNA was precipitated (two times the volume) in ethanol and washed twice with 70% ethanol (20 ml), dried at room temperature in a hood, and then dissolved in 1 ml of Tris-EDTA (TE) buffer.
As a positive control for [13C]DNA and [12C]DNA, Pseudomonas putida strain G7 was grown in two mineral salts media, one with 100% [13C]glucose and the other with nonenriched (i.e., 12C) glucose, and DNA was extracted as described above. One milliliter of the DNA solution was diluted to 4.5 ml with TE buffer, and 4.5 g of CsCl was added and shaken gently until dissolved. Ethidium bromide (100 μl; 10 mg/ml) was added to each ultracentrifuge tube, which was then sealed. Tubes were centrifuged at 140,000 × g (Vti 81 rotor; 41,900 rpm) for 66 h at 20°C. Resultant bands were clearly separated (9 to 12 mm). The centrifuge tubes were pierced, using standard methods (46), with an 18-gauge needle in two locations: 2 mm below where we could see the 12C band and 2 mm below the band depth that matched that of another tube containing a 13C standard. In processing DNA from the [13C]naphthalene-treated soil, we compensated for partial 12C labeling of DNA by sampling approximately 3 mm above the 13C band. In processing DNA from the [13C]caffeine-treated soil, no such compensation was made because methyl groups were the presumed substrate. Approximately 0.5 to 0.7 ml of CsCl solution containing the DNA was withdrawn and transferred to another centrifuge tube. Ethidium bromide was extracted from the DNA by the addition of 10 to 20 volumes (e.g., 6 to 12 ml to 0.6 ml) of TE-saturated 1-butanol by gentle mixing. The organic layer was discarded and the extraction was repeated five to six times, and then the volume of DNA was brought to 3 ml in TE. DNA precipitation occurred overnight at −20°C by addition of 300 μl of 3 M sodium acetate (pH 4.6) and twice the volume of ethanol. After pelleting at 13,000 to 15,000 × g for 30 min, the DNA was washed twice with 70% ethanol, centrifuged at the same speed for 10 min, resuspended in 50 to 100 μl of TE, and stored at −20°C.
PCR cloning, restriction digestion, and sequencing.
PCR amplification of 16S rRNA genes (rDNA) in the [13C]DNA fraction utilized universal eubacterial primers (27f and 1492r) by methods described previously (2). The product was ligated into the vector pCR2.1 (TA cloning; Invitrogen, Carlsbad, Calif.) by following the manufacturer's recommended protocol. Following transformation of plasmids into host cells and blue/white screening, colonies with inserts were verified by PCR with primers 27f and 1492r. The amplicons were digested with HaeIII and HhaI. Restriction fragment length polymorphism (RFLP) patterns were analyzed on 3% MetaPhore agarose gels (BioWhittaker; Molecular Applications, Rockland, Maine) with a 100-bp/1-kb ladder (Promega) as a marker. Clones containing unique RFLP patterns were selected for sequencing, grown overnight in 5 ml of Luria-Bertani broth with appropriate antibiotics (kanamycin and ampicillin), and pelleted, and plasmids were purified (QiaPrep spin miniprep kit; Qiagen, Santa Clarita, Calif.). PCR primers 27f, 533f, and 1492r (16 μl; 1 pM/μl) were used by the Cornell DNA sequencing facility (Ithaca, N.Y.). Raw sequence data from both strands were assembled into full-length sequences by using the SeqMan II program (DNASTAR, Inc.). After assembly, the consensus sequence was verified manually by referring to the corresponding ABI chromatograms of the sequencing reactions. The computational tools of the Ribosomal Database-II project (http://www.cme.msu.edu/RDP/html/analyses.html) were used to check chimeras and to calculate the similarity values for individual rDNA sequences by using the SEQUENCE_MATCH program. A BLAST search (http://www.ncbi.nlm.nih.gov.BLAST) was also used to identify the additional related sequences. The closest relatives identified from both searches were included in further phylogenetic analysis. Sequences were then imported into the ARB rRNA software (Technical University of Munich, Munich, Germany; http://www.arb-home.de) for phylogenetic tree construction.
Statistics.
Data produced from replicated treatments were averaged. The differences between treatments at discrete sampling times were assessed with Student's t test.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported here have been submitted to GenBank under accession no. AF534190 to AF534218.
RESULTS
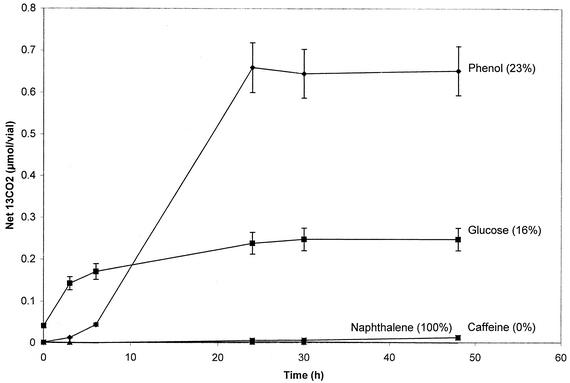
Microcosms demonstrated the utility of GC/MS in documenting net 13CO2 release from 13C-labeled glucose, phenol, naphthalene, and caffeine (Fig. 1). Although 12CO2 and 13CO2 coelute in the same peak from the GC column, single ion monitoring of 13CO2 (m/z = 45) and 12CO2 (m/z = 44) allowed both forms of CO2 to be quantified. 13CO2 that evolved from native soil organic matter in the water-only treatment was subtracted from 13CO2 produced by the 13C-substrate-amended soil. The soil microbial community readily mineralized glucose, phenol, and naphthalene but not caffeine within 24 h. The low water solubility of naphthalene limited its addition to soil; 13CO2 evolution was correspondingly diminished. Perhaps due to contrasts in the solubility, sorption, and biochemical characteristics of the substrates, the extent of metabolism for the three mineralized compounds was greater for naphthalene than for phenol and greater for phenol than for glucose. The assay (Fig. 1) was repeated three times with consistent results.
FIG. 1.
Evolution of 13CO2 from 13C-labeled substrates added to soil (naphthalene, 2 ppm; other substrates, 50 ppm) in sealed chambers incubated in the laboratory. Data show net production of 13CO2 evolved from 5 g of soil. Net production was computed by subtracting 13CO2 in the water-only control from total 13CO2. The percentage in parentheses shows the proportion of the total of each added substrate recovered as 13CO2. Averages of three replicate treatments are shown; error bars indicate standard deviations.
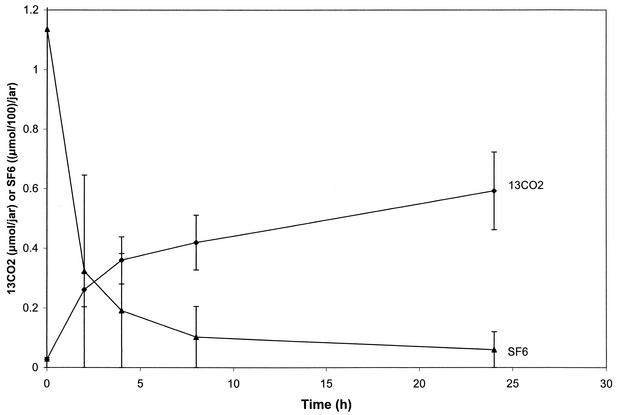
We examined the characteristics of the in situ, open-bottom soil cover assay system in the absence of added 13C-labeled substrates by monitoring accumulation of 13CO2 (Fig. 2). The field chambers captured CO2 originating from an unknown volume (perhaps 600 cm3) beneath. 13CO2 concentrations in the field chambers installed over unamended soil rose slightly for the first 10 h and remained constant (Fig. 2). Equilibration of headspace 13CO2 concentrations reflected the open nature of the assay chambers. A dynamic equilibrium was reached between soil-generated CO2 diffusing up into the chambers and its loss through the unsealed porous matrix below. The high initial concentration of SF6 rapidly diffused from the headspace through the soil (Fig. 2).
FIG. 2.
Behavior of gases in soil cover field chamber. SF6 was injected at the beginning of the experiment. 13CO2 was derived from native soil organic matter. Averages of three replicate treatments are shown; error bars indicate standard deviations.
When 13C-substrates are added to soil, quantification of their biodegradation depends on background control treatments receiving water and/or the non-13C-enriched substrates (12C-substrates). We sought another measure for background 13CO2, i.e., interpolation of 13CO2 production based on the fixed ratio of 13C to 12C in naturally occurring C pools. The natural abundance of 13C in nature is 1.11% (18). We verified this abundance by analyzing 53 headspace samples derived from chambers placed on the soil surface in which the mass of injected soil-derived CO2 ranged from 1.28 to 42 nmol (data not shown). A linear relationship (r = 0.997; slope = 0.0117) was found between 13CO2 (m/z = 45) and 12CO2 (m/z = 44). Because the amount of background 13CO2 derived from soil organic matter was constantly 1.17% of the 12CO2 peak area, we had a means for inferring background 13CO2 production from measurements of 12CO2 produced from soils amended with substrates uniformly labeled with 13C. For substrates containing both 12C and 13C atoms, the same inference (though conservative) applies (see below).
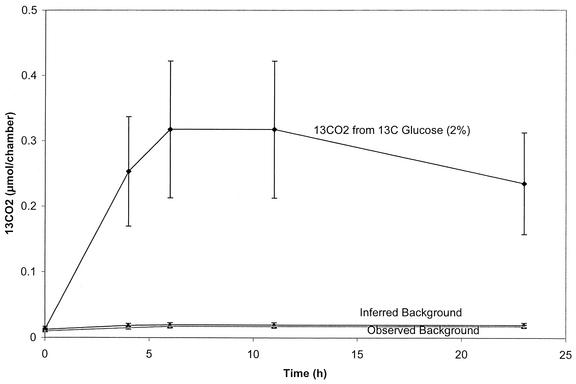
To evaluate the above-mentioned extrapolation approach for predicting background levels of 13CO2, we initiated both laboratory microcosm and field assays. In laboratory incubations analogous to those that produced the results shown in Fig. 1, treatments included unamended soil and soil amended with water only or [12C]phenol or [13C]phenol. After a 14-day incubation (data not shown), the total 13CO2 evolved in the [13C]phenol treatment was 1.2 ± 0.2 μmol (equivalent to 35% of added substrate). The amounts of total 13CO2 evolved in all three of the control treatments (0.2 ± 0.06 μmol) were statistically indistinguishable from one another and from the background value interpolated from the concentration of 12CO2 produced in the [13C]phenol treatment. If the 13C-phenol substrate had had a mixture of 12C and 13C atoms, then the total pool of measured 12CO2 would have been slightly larger, the corresponding background values for inferred 13CO2 would also have been higher, and the estimate of net 13CO2 from 13C-substrate would be conservative. A field-based experiment using the open-bottom soil cover assay also compared inferred (1.17% of 12CO2) and directly measured amounts of 13CO2 (Fig. 3). A burst of [13C]glucose respiration was found in these open chambers after 6 h, corresponding to 2% of the total added. But most importantly, the background level of 13CO2 measured beneath chambers lacking [13C]glucose matched the 13CO2 concentration inferred from the 12CO2 concentration measured in the treatment receiving [13C]glucose.
FIG. 3.
Field respiration experiment verifies that interpolated (inferred) 13CO2 matches that of measured controls. In the field release open-bottom soil cover assay, treatments were with water only and 400 μg of [13C]glucose. The percentage in parentheses shows the proportion of the total substrate recovered as 13CO2. The inferred background was 1.17% of the 12CO2 in the 13C-glucose treatment. Each treatment was performed in triplicate; error bars indicate standard deviations. (Note: the ambient temperature during this assay was ∼10°C cooler than for the ones shown in Fig. 4.)
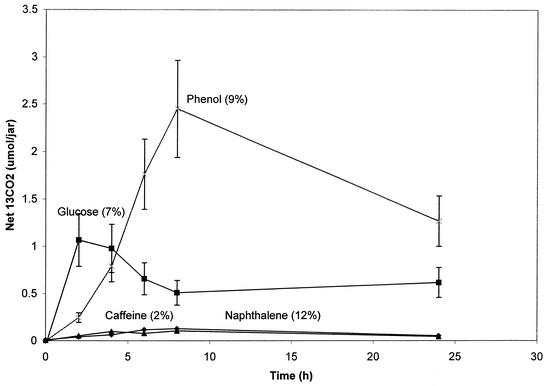
The net biodegradation of four 13C-substrates (glucose, phenol, naphthalene, and caffeine) was assayed (24 h) by coupling the open-bottom soil cover assay with the inferred background procedure (Fig. 4). Maximum concentrations of 13CO2 occurred early in the experiments and then diminished because of the constant flux of CO2 through the headspace. Glucose was rapidly metabolized, reaching a maximum of nearly 7% of the total 13C added within 2 h. Phenol was also rapidly metabolized, reaching a maximum of 9% after 8 h. 13CO2 production in excess of the background level in the naphthalene-treated soil (Fig. 4) appeared to reach 12% of the total added naphthalene; however, because of the low solubility of naphthalene, high variability in the replicates, and the relatively high proportion of background 13CO2, net production was not significant (t test; P < 0.05). In the caffeine-treated soil, the net amount of 13CO2 produced was also not different from the background level.
FIG. 4.
Results of field release open-bottom soil cover assay. All four 13C-labeled substrates were added to field soil and covered with a chamber. GC/MS analysis monitored both 12CO2 and 13CO2 concentrations. Net 13CO2 reflects total 13CO2 minus inferred background 13CO2. The percentage in parentheses shows the proportion of the total of each added substrate recovered as 13CO2. Three to five replicate treatments were prepared; error bars indicate standard deviations.
Simultaneous to the assays that produced the data shown in Fig. 4, we prepared three additional 13C-amended soil treatments in sealed glass chambers, which were all incubated adjacent to the open-bottom soil cover assay in the field: (i) an intact soil ped (∼30 g); (ii) a soil core; and (iii) ∼5 g of crushed soil. Table 1 provides a summary of the results of all 16 field respiration treatments (four substrates and four soil treatments). Among the four treatments with glucose listed in Table 1, that with crushed soil within the sealed chamber had the highest metabolic activity: the total 13CO2, net 13CO2, proportion of substrate respired (68%), and rates were consistently at least three times greater than those for the other glucose treatments. Predictably, the values for the respiration of glucose in the open-bottom soil cover assay, especially the total recovered as 13CO2, were the lowest among the four treatments. Nonetheless, the initial rate of respiration in the soil cover assay nearly matched that of the sealed chambers containing intact ped and core.
TABLE 1.
Summary of 13CO2 field respiration assays conducted in the agricultural field test plots
| Substratea | Treatmentb | Total evolved 13CO2 (μm) | 13CO2 blankc (μm) | Net 13CO2 (μm) | % of added 13C respired | Initial rate (μm/h) |
|---|---|---|---|---|---|---|
| Glucose | Soil cover | 3.4 ± 0.3 | 2.3 ± 0.37 | 1.1 | 6.8 | 0.25 |
| Sealed chamber | ||||||
| Intact ped | 7.4 ± 10 | 4.5 ± 6.3 | 2.9d | 20d | 0.28d | |
| Core | 4.8 ± 1.9 | 1.7 ± 0.39 | 3.1 | 22 | 0.27 | |
| Crushed soil | 16 ± 3.7 | 5.7 ± 1.2 | 9.8 | 68 | 1.2 | |
| Phenol | Soil cover | 8.1 ± 0.8 | 5.7 ± 1.0 | 2.5 | 9.1 | 0.2 |
| Sealed chamber | ||||||
| Intact ped | 7.2 ± 4.1 | 1.8 ± 0.63 | 5.4 | 20 | 0.2 | |
| Core | 5.5 ± 3.7 | 1.2 ± 1.1 | 4.2 | 16 | 0.1 | |
| Crushed soil | 13.5 ± 1.2 | 3.5 ± 0.56 | 9.5 | 37 | 0.5 | |
| Naphthalene | Soil cover | 2.6 ± 0.45 | 2.4 ± 0.43 | 0.11d | ||
| Sealed chamber | ||||||
| Intact ped | 0.87 ± 0.73 | 0.87 ± 0.69 | 0d | |||
| Core | 0.81 ± 0.34 | 0.79 ± 0.34 | 0.02d | |||
| Crushed soil | 2.0 ± 0.26 | 1.9 ± 0.25 | 0.1d | |||
| Caffeine | Soil cover | 3.2 ± 1.6 | 3.0 ± 1.5 | 0.13d | ||
| Sealed chamber | ||||||
| Intact ped | 1.4 ± 0.84 | 1.4 ± 0.81 | 0.03d | |||
| Core | 1.1 ± 0.45 | 1.0 ± 0.42 | 0.09d | |||
| Crushed soil | 2.5 ± 0.51 | 2.4 ± 0.49 | 0.08d |
Aqueous substrates (glucose, phenol, and caffeine) were added as 450 μg in 0.1 ml. Naphthalene was added as 0.5 ml of a 20-ppm aqueous solution.
Four treatments were prepared. “Soil cover” designates field release with open-bottom chamber cover installed to trap 13CO2. The three sealed chamber treatments were incubated in the field alongside the soil cover experiment. All treatments were prepared in triplicate.
Blank was interpolated from 12CO2 evolved simultaneously with 13CO2.
Net was not significantly different from zero as judged by Student's t test comparing final total and background 13CO2.
The four phenol-treated respiration assays (Table 1) exhibited trends much like those for the glucose treatments: crushed soil within chambers yielded high respiratory activity. The activity of the remaining enclosed soil treatments was lower by a factor of about 2, and respiration in the open-bottom soil cover chamber was lowest (especially percentage of recovery) but the rate was comparable to those for the noncrushed enclosed treatments. The four soil treatments with naphthalene and caffeine yielded data, but the relatively high background production of 13CO2 prevented the net respiration from being determined for these two substrates.
After monitoring of the 13CO2 production in the open-bottom soil cover field chambers (Fig. 4), the surface soil from both 13C and 12C treatments was collected and DNA was fractionated. Although only the [12C]DNA band was visible, we removed fluid from the ultracentrifuge tubes in the location where [13C]DNA was expected. In all paired treatments of 12C- and 13C-amended soils, an amplicon of 16S rDNA (∼1,500 bp) was obtained only from the location of [13C]DNA in 13C-substrate-treated soil. Because no amplicons resulted from our attempts to amplify the 16S rDNA from the location of [13C]DNA in 12C-substrate treatments, we concluded that amplicons from the 13C-substrate-treated soils represented microbial populations that directly or indirectly metabolized and grew on the 13C-substrates in situ.
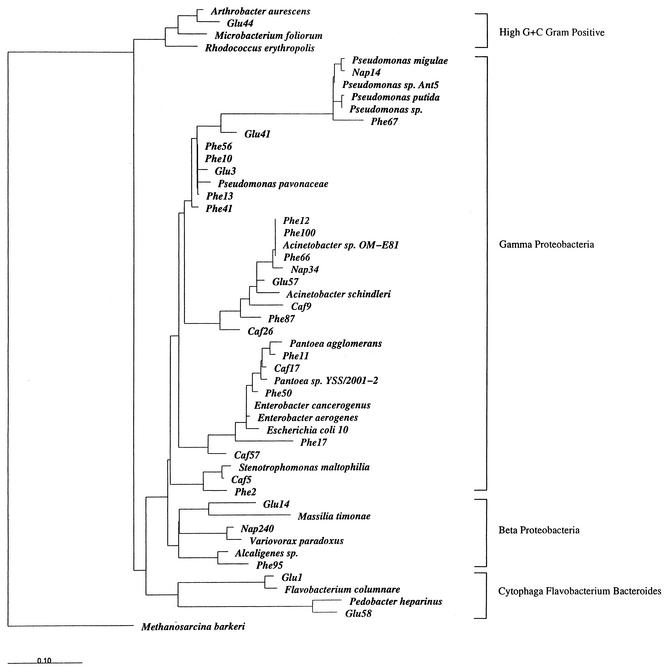
Following cloning of the mixture of 16S rDNA amplicons, ∼100 white colonies were screened from each soil extract by their RFLP patterns and 29 unique clones were selected for sequencing. A phylogenetic tree was constructed from the full sequences and 23 reference strains. Sequences derived from the seven glucose-metabolizing populations were related to Arthrobacter spp. (high G+C gram-positive cluster); to Pseudomonas, Acinetobacter, and Massilia spp. (γ and β proteobacteria); and to Flavobacterium and Pedobacter spp. (Cytophaga/Flavobacterium/Bacteroides group). The 14 active phenol-degrading populations were related to members of the γ and β proteobacteria: Pseudomonas, Alcaligenes, Acinetobacter, Pantoea, Enterobacter, and Stenotrophomonas. The five caffeine-degrading populations were restricted exclusively to the γ proteobacteria: Acinetobacter, Pantoea, Enterobacter, and Stenotrophomonas. The three naphthalene-degrading populations (γ and β proteobacteria) were related to Pseudomonas, Acinetobacter, and Variovorax.
DISCUSSION
Biodegradation of organic chemical pollutants is one of the many important processes effected in field sites by microorganisms. Knowledge of how naturally occurring microorganisms respond to anthropogenic compounds is ecologically significant and can make bioremediation technologies more robust (37, 39, 40). Thus, testing the biodegradability of organic pollutant compounds has industrial, regulatory, biogeochemical, and biotechnological implications (20, 21, 31, 32). From an industrial point of view, biodegradability testing is a means of screening commercially used compounds for potential threats of environmental toxicity and persistence (15, 23, 54). Such biodegradability and ecological fate estimates are often required for regulatory approval of chemicals intended for release to aquatic and terrestrial habitats (26, 27).
Approaches to assess biodegradation have traditionally relied upon laboratory-based measures of metabolic potential (10, 14, 34). As mentioned in the introduction, a long-standing criticism of this approach is that resultant data may be misleading because of artifacts imposed on microbial reactions by unrealistic laboratory conditions (34, 55). Other approaches to measure biodegradation have ranged from empirical observation of diminished effectiveness of intentionally released chemicals (e.g., herbicides [38]) to rare field release of 14C-labeled substrates (8, 30) to the use of conservative tracers during injection and retrieval of substrates in subsurface environments (16, 22, 25, 47). Issues surrounding the effectiveness and interpretations of such assays have been previously addressed (34, 39).
To our knowledge, this report is the first to implement methodologies that simultaneously assess the activity and identity of soil microorganisms metabolizing 13C-labeled substrates in situ. The metabolic activity assay is based on the detection of substrate-derived 13CO2 in excess of background 13CO2 respired from soil organic matter. The limits of sensitivity reflect the interactions among total 13C-substrate added, the size of native soil microbial populations adapted for substrate metabolism, the ambient rate of 13CO2 produced from soil organic matter, and the volume of soil contributing to headspace 13CO2. We had hoped that SF6 might act as a conservative tracer that would allow calculation of 13CO2 recovery; however, this proved futile because a single dose of SF6 in the headspace does not mimic a constant source of CO2 beneath. Preclusion of a mass-balance budget for the added substrate has obvious drawbacks, especially for compounds that are only slowly metabolized. Despite such drawbacks, the data presented here provide a proof of principle that the open-bottom soil cover approach is able to document in situ substrate mineralization. Predictably, when the physical soil structure was destroyed, respiration rates were very high (Table 1); this disturbance artifact has been extensively documented previously (5, 12, 52). Surprisingly, the initial rates in the open-bottom soil cover assay nearly matched the rates detected in sealed adjacent vessels in which the soil structure had been kept intact (Table 1). This lack of departure between true in situ and adjacent ex situ assays may have important implications. It may be that disturbance artifacts implicit in the carrying out of laboratory-based soil biodegradation assays can be minimized by maintaining soil structure and carefully mimicking genuine field conditions. This possibility needs to be verified and could add needed robustness to biodegradation testing techniques.
The [13C]DNA assay used here, aimed at identification of active populations, was pioneered by Radajewski et al. (45) and is analogous to a lipid biomarker assay developed by Boschker et al. (4). The principle (extraction, separation, and PCR amplification of 16S [13C]rDNA sequences in soil) is elegant. In the original application, [13C]methanol was added in two doses at a relatively high concentration (50 μl to 10 g of soil). Furthermore, in order to be certain that a strong [13C]DNA band appeared in the CsCl gradient, the laboratory incubation was for 44 days. It is likely that the incubation conditions chosen by Radajewski et al. (45) caused substantial alteration in the active populations (enrichment); furthermore, by the end of the assay the 13C label may have migrated beyond 1° degraders to other members of the microbial community. Our field strategy avoided the long incubation. This was enforced by the fact that the burst of 13CO2 from added substrate was not likely to be detectable beyond the first day of incubation. Despite the relatively brief exposure period to 13C-substrates used here (1 day), we cannot be certain that the 16S rDNA sequences that we recovered represented the organisms active in the first step of community metabolism.
The data in Fig. 4 and 5 show that glucose and phenol were rapidly converted to 13CO2 in soil and that the 13C fraction of soil DNA identified active populations. Surprisingly, even when no 13CO2 was detected in excess of the background produced from soil organic matter (naphthalene and caffeine treatments), the [13C]DNA assay revealed active populations. There are two ways to explain this apparent anomaly. The first involves questioning the validity of our strategy for identifying the active cells. We only analyzed amplicons from the location of the [13C]DNA band in 13C-treated soil when the corresponding CsCl location from 12C-treated soil failed to contain amplifiable 16S rDNA. The logic here seems sound and directs us to the next explanation: we speculate that the microbial populations able to grow on naphthalene and caffeine were present, active, and doubled at least twice within 24 h. Two doublings are necessary to obtain a pool of DNA fully labeled with13C. If the populations were small enough, then they may have produced PCR-detectable 16S rDNA but failed to produce 13CO2 in excess of that emitted from organic matter in soil beneath the chamber.
FIG. 5.
Dendrogram showing phylogenetic relationships among 29 16S rDNA sequences obtained from extracted 13C-labeled soil DNA and 23 relevant reference strains. The sequences from treatments receiving [13C]glucose, [13C]phenol, [13C]caffeine, and [13C]naphthalene are indicated by the prefixes “Glu,” “Phe,” “Caf,” and “Nap,” respectively.
One issue that arose in the implementation and interpretation of these experiments is the volume of soil, and hence the size of natural populations that actually contacted and metabolized aqueous-phase substrates added to the soil matrix. The four soil treatments examined for each substrate (Table 1) were prepared from different masses of soil (from 5 g of crushed soil enclosed in a jar to ∼700 g of soil beneath the open-bottom treatment), but in no case did the soil become saturated. Net respiration appeared to be independent of soil mass (Table 1). Our interpretation is that in all treatments the soil matrix provided sufficient volume to contain the aqueous substrates; thus, the effective population sizes exposed to the 13C-substrates were equivalent. The portion of soil adjacent to that which was in contact with the aqueous-phase 13C-substrates apparently only influenced the biodegradation assay by contributing background CO2.
Many previous investigations have reported soil enrichment and/or plating and isolation experiments that identify bacterial cultures capable of growing on a variety of organic compounds on defined media in the laboratory (19). Also, previous studies have surveyed substrate-responsive populations in laboratory-incubated environmental samples (29). However, to our knowledge, no prior study has produced a list of bacterial genera active in situ in field soil on particular carbon substrates. While PCR primer and cell lysis biases must be acknowledged as a potential influence in the information produced (Fig. 5), it is still noteworthy that this approach delivered sequence data on members of 11 genera with overlapping niches (6 genera active on glucose, 6 on phenol, 4 on caffeine, and 3 on naphthalene). Perusal of Bergey's Manual (19) and the GenBank database indicates that 10 of the genera retrieved from field soil have routinely been isolated as chemoorganotrophs from soil, water, and/or sewage habitats. The 11th, Massilia, has representatives from human and environmental samples. Thus, there is remarkable agreement between the results reported here and those from prior culture-based investigations. The results of this investigation, which introduced C substrates to soil, confirm the notion that the genera whose sequences were found are ecological opportunists (r selected [1]). The amendment-based approach utilized in this study would not likely be able to identify less responsive, slow-growing (K-selected [1]) members of the soil microbial community.
Acknowledgments
This work was supported by the National Science Foundation (MCB #0084175 awarded to E.L.M.), the Federation of European Chemical Industries Long-Range Research Initiative (CEFIC-LRI), and AstraZeneca Global SHE. P. Padmanabhan was supported by an Overseas Associateship from the Department of Biotechnology, Ministry of Science and Technology, India.
REFERENCES
- 1.Atlas, R. M., and R. Bartha. 1998. Microbial ecology, 4th ed. Addison Wesley Longman, Inc., Menlo Park, Calif.
- 2.Bakermans, C., and E. L. Madsen. 2003.. Diversity of 16S rRNA and naphthalene dioxygenase genes from coal tar waste-contaminated aquifer waters. Microb. Ecol. 44:95-106. [DOI] [PubMed] [Google Scholar]
- 3.Boetius, A., K. Ravenschlag, C. J. Schubert, D. Rickert, F. Widdel, A. Gieseke, R. Amann, B. B. Jorgensen, U. Witte, and O. Pfannkuche. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623-626. [DOI] [PubMed] [Google Scholar]
- 4.Boschker, H. T. S., S. C. Nold, P. Wellsbury, D. Bos, W. D. Graaf, R. Pel, R. J. Parkes, and T. E. Cappenberg,. 1998. Direct linking of microbial populations to specific biogeochemical processes by 13C-labelling of biomarkers. Nature 392:801-805. [Google Scholar]
- 5.Brady, N. C., and R. R. Weil. 1990. The nature and properties of soil, 12th ed. Prentice Hall, Upper Saddle River, N.J.
- 6.Brock, T. D., and M. L. Brock. 1966. Autoradiography as a tool in microbial ecology. Nature 209:734-736. [DOI] [PubMed] [Google Scholar]
- 7.Brockman, F. J., S. W. Li, J. K. Fredrickson, D. B. Ringelberg, T. L. Kieft, C. M. Spadoni, J. P. McKinley, and D. C. White. 1998. Post-sampling changes in microbial community structure and activity in a subsurface paleosol. Microb. Ecol. 36:152-164. [DOI] [PubMed] [Google Scholar]
- 8.Burauel, P., and F. Fuhr. 2000. Formation and long-term fate of non-extractable residues in outdoor lysimeter studies. Environ. Pollut. 108:45-52. [DOI] [PubMed] [Google Scholar]
- 9.Capone, D. A., J. P. Zehr, H. W. Paerl, B. Bergman, and E. J. Carpenter. 1997. Trichodesmium, a globally significant marine cyanobacterium. Science 276:1221-1229. [Google Scholar]
- 10.Chapelle, F. H., and D. R. Lovley. 1990. Rates of microbial metabolism in deep coastal plain aquifers. Appl. Environ. Microbiol. 56:1865-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunbar, J., S. White, and L. Forney. 1997. Genetic diversity through the looking glass: effect of enrichment bias. Appl. Environ. Microbiol. 63:1326-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elmholt, S. 1992. Effect of propiconazole on substrate amended soil respiration following laboratory and field application. Pestic. Sci. 34:139-146. [Google Scholar]
- 13.Fliermans, C. B., and E. L. Schmidt. 1975. Autoradiography and immunofluorescence combined for autoecological study of single cell activity with Nitrobacter as a model system. Appl. Microbiol. 30:676-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Focht, D. D. 1994. Microbiological procedures for biodegradation research, p. 407-426. In R. W. Weaver et al. (ed.), Methods of soil analysis, part 2. Microbiological and biochemical properties. SSSA book series, no. 5. SSSA, Madison, Wis.
- 15.Gouin, T., D. Mackay, E. Webster, and F. Wania. 2000. Screening chemicals for persistence in the environment. Environ. Sci. Technol. 34:881-884. [Google Scholar]
- 16.Hageman, K. J., J. D. Istok, J. A. Field, T. E. Buscheck, and L. Semprini. 2001. In situ anaerobic transformation of trifluoroethene in trichloroethene-contaminated groundwater. Environ. Sci. Technol. 35:1729-1735. [DOI] [PubMed] [Google Scholar]
- 17.Hinrichs, H.-U., J. M. Hayes, S. P. Sylva, P. G. Brewer, and E. F. DeLong. 1999. Methane-consuming archaebacteria in marine sediments. Nature 398:802-805. [DOI] [PubMed] [Google Scholar]
- 18.Hoefs, J. 1987. Stable isotope geochemistry, 3rd ed. Springer-Verlag, New York, N.Y.
- 19.Holt, J. G., N. R. Krieg, P. H. A. Sneath, J. T. Staley, and S. T. Williams (ed.). 1994. Bergey's manual of determinative bacteriology, 9th ed. Williams & Wilkins, Baltimore, Md.
- 20.Howard, P. H., R. S. Boethling, W. F. Jarvis, W. M. Meyland, and E. M. Michalenko. 1991. Handbook of environmental degradation rates. Lewis, Chelsea, Mich.
- 21.Hutzinger, O. (ed.). 2001. The handbook of environmental chemistry: biodegradation and persistence, vol. 2. Reactions and processes. Springer Verlag, New York, N.Y.
- 22.Istok, J. D., M. D. Humphrey, M. H. Schroth, M. R. Hyman, and K. T. O'Reilly. 1997. Single well “push-pull” test for in situ determination of microbial activities. Ground Water 35:619-631. [Google Scholar]
- 23.Jansson, B., R. L. Lipnick, D. Mackay, and M. Petreas. 2001. Identification of persistent, bioaccumulative and toxic substances, chapter 1. In R. L. Lipnick, D. L. G. Muir, and K. C. Jones (ed.), Persistent, bioaccumulative and toxic chemicals. I. Assessment and emerging new chemicals. American Chemical Society Symposium Series 773. Oxford University Press, New York, N.Y.
- 24.Karl, D. M. 1986. Determination of in situ microbial biomass, viability, metabolism, and growth, p. 81-176. In J. S. Poindexter and E. R. Leadbetter (ed.), Bacteria in nature. Plenum Press, New York, N.Y.
- 25.Kim, H., H. F. Hemond, L. R. Krumholz, and B. A. Cohen. 1995. In situ biodegradation of toluene in a contaminated stream. 1. Field studies. Environ. Sci. Technol. 29:108-116. [DOI] [PubMed] [Google Scholar]
- 26.Klecka, G., R. Boethling, J. Franklin, L. Grady, D. Graham, P. H. Howard, K. Kannan, R. Larson, D. Mackay, D. Muir, and D. van de Meent. 2000. Evaluation of persistence and long-range transport of organic chemicals in the environment. SETAC Press, Pensacola, Fla.
- 27.Larson, R. J., and C. E. Cowan. 1995. Quantitative application of biodegradation data to environmental risk and exposure assessments. Environ. Toxicol. Chem. 14:1433-1442. [Google Scholar]
- 28.Lee, N., P. H. Nielsen, K. H. Andreasen, S. Juretschko, J. L. Nielsen, K.-H. Schleifer, and M. Wagner. 1999. Combination of fluorescent in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 65:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lipson, D. A., C. W. Schadt, and S. K. Schmidt. 2002. Changes in soil microbial community structure and function in an alpine dry meadow following spring snow melt. Microb. Ecol. 43:307-314. [DOI] [PubMed] [Google Scholar]
- 30.MacIntyre, W. C., C. P. Antworth, T. B. Stauffer, and J. M. Boggs. 1993. Degradation kinetics of aromatic organic solutes introduced into a heterogeneous aquifer. Water Resour. Res. 29:4045-4051. [Google Scholar]
- 31.Mackay, D., W.-Y. Shiu, and K.-C. Ma. 1997. Illustrated handbook of physical-chemical properties of environmental fate for organic chemicals, vol. II. CRC Press, Boca Raton, Fla.
- 32.Mackay, D., S. Paterson, A. Di Guardo, and C. E. Cowan. 1996. Evaluating the environmental fate of types of chemicals using the EQC model. Environ. Toxicol. Chem. 15:1627-1637. [Google Scholar]
- 33.Madan, A. P., and S. A. Nierzwicki-Bauer. 1993. In situ detection of transcripts for ribulose-1,5-bisphosphate carboxylase in cyanobacterial heterocysts. J. Bacteriol. 175:7301-7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madsen, E. L. 1991. Determining in situ biodegradation: facts and challenges. Environ. Sci. Technol. 25:1662-1673. [Google Scholar]
- 35.Madsen, E. L. 1996. A critical analysis of methods for determining the composition and biogeochemical activities of soil microbial communities in situ, p. 287-370. In G. Stotzky and J.-M. Bollag (ed.), Soil biochemistry, vol. 9. Marcel Dekker, New York, N.Y.
- 36.Madsen, E. L. 1998. Epistemology of environmental microbiology. Environ. Sci. Technol. 32:429-539. [Google Scholar]
- 37.Madsen, E. L. 2002. Methods for determining biodegradability, p. 903-915. In C. J. Hurst et al. (ed.), Manual of environmental microbiology, 2nd ed. ASM Press, Washington, D.C.
- 38.Moorman, T. B., M. W. Broder, and W. C. Koskinen. 1992. Kinetics of EPTC biodegradation and the effects of the inhibitor dietholate in solids. Soil Biol. Biochem. 24:121-127. [Google Scholar]
- 39.National Research Council. 2000. Natural attenuation for groundwater remediation. National Academy Press, Washington, D.C.
- 40.National Research Council. 1993. In situ bioremediation: when does it work? National Academy Press, Washington, D.C.
- 41.Orphan, V. J., C. H. House, K.-U. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2001. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293:484-487. [DOI] [PubMed] [Google Scholar]
- 42.Ouverney, C. C., and J. A. Fuhrman. 1999. Combined microautoradiography-16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl. Environ. Microbiol. 65:1746-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parkin, T. B., H. F. Kaspar, A. J. Sexstone, and J. M. Tiedje. 1984. A gas-flow soil core method to measure field denitrification rates. Soil Biol. Biochem. 16:323-330. [Google Scholar]
- 44.Phelps, T. J., E. M. Murphy, S. M. Pfiffner, and D. C. White. 1994. Comparison between geochemical and biological estimates of subsurface microbial activities. Microb. Ecol. 28:335-349. [DOI] [PubMed] [Google Scholar]
- 45.Radajewski, S., R. Ineson, N. R. Parekh, and J. C. Murrell. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646-649. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Schroth, M. H., J. D. Istok, G. T. Conner, M. R. Hyman, R. Haggerty, and K. T. O'Reilly. 1998. Spatial variability in in situ aerobic respiration and denitrification rates in a petroleum-contaminated aquifer. Ground Water 36:925-937. [Google Scholar]
- 48.Staley, J. T., and A. Konopka. 1985. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 39:321-346. [DOI] [PubMed] [Google Scholar]
- 49.Thierrin, J., G. B. Davis, and C. Barber. 1995. A ground-water tracer test with deuterated compounds for monitoring in situ biodegradation and retardation of aromatic hydrocarbons. Ground Water 33:469-475. [Google Scholar]
- 50.Vaulot, D., D. Marie, R. J. Olson, and S. W. Chisholm. 1995. Growth of Prochlorococcus, a photosynthetic prokaryote, in the Equatorial Pacific Ocean. Science 268:1480-1482. [DOI] [PubMed] [Google Scholar]
- 51.Venrick, E. L., J. R. Beers, and J. F. Heinbokel. 1977. Possible consequences of containing microplankton for physiological rate measurements. J. Exp. Mar. Biol. Ecol. 26:55-76. [Google Scholar]
- 52.Visser, S., J. Fujikawa, C. L. Griffiths, and D. Parkinson. 1984. Effect of topsoil storage on microbial activity, primary production and decomposition potential. Plant Soil 82:41-50. [Google Scholar]
- 53.Ward, B. B. 1984. Combined autoradiography and immunofluorescence for estimation of single cell activity by ammonium-oxidizing bacteria. Limnol. Oceanogr. 29:402-410. [Google Scholar]
- 54.Webster, E., D. Mackay, and F. Wania. 1998. Evaluating environmental persistence. Environ. Toxicol. Chem. 17:2148-2158. [Google Scholar]
- 55.Wesnigk, J. B., M. Kekin, W. Jonas, K. Figge, and G. Rheinheimer. 2001. Predictability of biodegradation on the environment: limits of prediction from experimental data, p. 253-290. In B. Beek (ed.), The handbook of environmental chemistry: biodegradation and persistence, vol. 2K. Reactions and processes. Springer Verlag, New York, N.Y.
- 56.Wilson, M. S., and E. L. Madsen. 1996. Field extraction of a unique intermediary metabolite indicative of real time in situ pollutant biodegradation. Environ. Sci. Technol. 30:2099-2103. [Google Scholar]