Abstract
In higher eukaryotes, the origin recognition complex (ORC) lacks sequence-specific DNA binding, and it remains unclear what other factors specify an origin of DNA replication. The Epstein–Barr virus origin of plasmid replication (OriP) recruits ORC, but the precise mechanism of ORC recruitment and origin activation is not clear. We now show that ORC is recruited selectively to the dyad symmetry (DS) region of OriP as a consequence of direct interactions with telomere repeat factor 2 (TRF2) and ORC1. TRF-binding sites within DS stimulate replication initiation and facilitate ORC recruitment in vitro and in vivo. TRF2, but not TRF1 or hRap1, recruits ORC from nuclear extracts. The amino-terminal domain of TRF2 associated with a specific region of ORC1 and was necessary for stimulation of DNA replication. These results support a model in which TRF2 stimulates OriP replication activity by direct binding with ORC subunits.
Keywords: ORC, TRF2, origin, replication, EBV
Introduction
Origins of eukaryotic DNA replication are regulated spatially and temporally to complete chromosome duplication once in a single cell cycle (Raghuraman et al, 2001; Weinreich et al, 2004; Blow & Dutta, 2005). These origins are recognized by the origin recognition complex (ORC), which orchestrates the formation of a multi-protein pre-replication complex (Bell, 2002; Bell & Dutta, 2002; DePamphilis, 2003; Mendez & Stillman, 2003; Diffley, 2004; Prasanth et al, 2004). The mechanism of origin-site-selection by ORC is not completely understood. In Saccharomyces cerevisiae, ORC can bind in an ATP-dependent manner to a specific 11 bp sequence found in many autonomous replicating sequences (Bell & Stillman, 1992). In Schizosaccharomyces pombe, the ORC binds to AT-rich sequences in replication origins through an AT-hook domain unique to the S. pombe ORC4 subunit (Chuang & Kelly, 1999). However, in higher eukaryotes, ORC lacks sequence-specific DNA-binding activity, and the factors that specify ORC binding at chromosomal sites and stimulate DNA replication initiation remain elusive (Stillman, 2001; Wyrick et al, 2001; Gilbert, 2004; Cvetic & Walter, 2005). Studies in Drosophila have shown that ORC can be directed to a replication origin by sequence-specific DNA-binding proteins, such as Dm-myb (Beall et al, 2002), and to heterochromatin by interactions with heterochromatin protein 1 (HP1; Pak et al, 1997). In more complex vertebrates, ORC binding and origin selection seem to be regulated by poorly understood epigenetic factors (McNairn & Gilbert, 2003).
The Epstein–Barr virus (EBV) origin of plasmid (OriP) replication contains an ∼120 bp element, referred to as the dyad symmetry (DS), which has been shown to recruit ORC in vivo (Chaudhuri et al, 2001; Dhar et al, 2001; Schepers et al, 2001; Ritzi et al, 2003; Wang & Sugden, 2005). The DS binds the ORC and minichromosome maintenance (MCM) proteins, and is controlled by cell-cycle licensing factors that include geminin and cdt1 (Chaudhuri et al, 2001; Dhar et al, 2001; Schepers et al, 2001). OriP-dependent DNA replication also requires EBV nuclear antigen 1 (EBNA1), the EBV-encoded origin-binding protein. EBNA1 binds to several regions of the EBV genome, but initiates replication only at DS, in which the binding sites are spaced precisely 21 bp from centre to centre (Bashaw & Yates, 2001). EBNA1 can co-immunoprecipitate with the ORC subunits in nuclear extracts, but no direct physical interaction between EBNA1 and the ORC subunits has been described (Schepers et al, 2001; Ritzi et al, 2003). In addition to EBNA1-binding sites, the OriP minimal replicator contains three telomere repeat factor (TRF)-binding sites, which flank the EBNA1 21-bp spaced sites (Niller et al, 1995; Deng et al, 2002, 2003). Mutation of TRF-binding sites reduces OriP replication activity, but the precise function of TRFs at OriP remains unclear (Vogel et al, 1998; Deng et al, 2002, 2003; Julien et al, 2004). It is also not known how ORC is specifically recruited to the DS region of OriP. Here, we show that TRF2 has a direct role in the recruitment of ORC to the DS region of OriP.
Results and Discussion
To investigate the DNA sequence requirements for ORC recruitment, we compared the ability of three different EBV genome fragments containing EBNA1-binding sites to recruit ORC using a DNA affinity purification assay in vitro (Fig 1). EBNA1 binds to at least three different high-affinity sites in the EBV genome (Fig 1A). The family of repeats (FR) and DS comprise the maintenance and replicator elements of OriP, whereas the Qp-binding sites function in transcription regulation (Leight & Sugden, 2000). Raji cell nuclear extracts were incubated with magnetic beads coupled to DNA fragments containing the EBNA1-binding sites from DS, FR, Qp or control vector sequence BKS, and analysed by western blot for the recruitment of ORC2 (Fig 1B). We found that ORC2 bound predominantly to the DS DNA and only very weakly to FR; ORC2 did not bind to Qp or BKS DNA. The same binding pattern was observed with TRF2. In contrast, EBNA1 bound most abundantly to FR, followed by DS and Qp, with no detectable binding to BKS. These results indicate that ORC binding correlates with replication function, and that EBNA1 binding by itself is not sufficient for ORC recruitment to FR or Qp.
Figure 1.
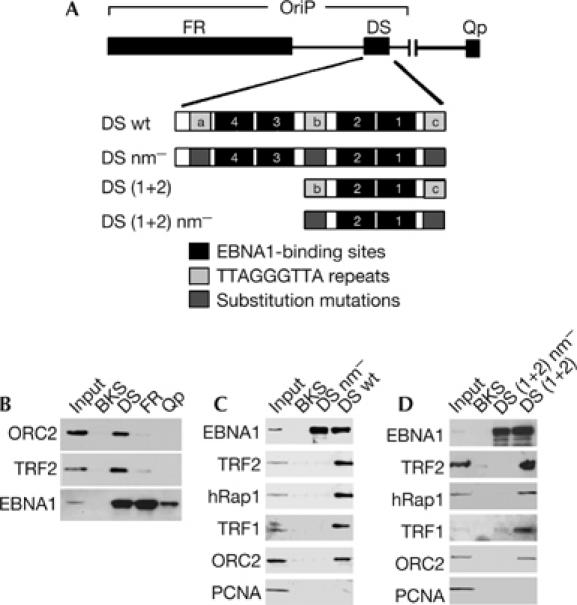
Telomere repeats recruit ORC in vitro. (A) Schematic of EBV DNA fragments and telomere repeat substitution mutations used for DNA affinity. (B) DNA affinity assay with FR, DS or Qp template DNA and Raji nuclear extracts. Bound proteins were monitored by western blotting with antibodies to ORC2, TRF2 or EBNA1. (C) DNA affinity with control BKS, DS nm− or DS wt DNA and Raji nuclear extracts was analysed with antibodies to EBNA1, TRF2, hRap1, TRF1, ORC2 and PCNA. (D) DNA affinity as in (C) with templates for control BKS, DS (1+2) nm− or DS (1+2). EBV, Epstein–Barr virus; EBNA1, EBV nuclear antigen 1; DS, dyad symmetry; FR, family of repeats; ORC, origin recognition complex; OriP, origin of plasmid replication; PCNA, proliferating-cell nuclear antigen; TRF, telomere repeat factor; wt, wild type.
To explore the possibility that the telomere repeats contribute to ORC recruitment at DS, we compared wild-type DS (DS wt), DS with substitution mutations in all three telomere repeats (also referred to as nonamer repeats; DS nm−) or control vector sequence (BKS; Fig 1C). Bound proteins were analysed by western blotting with antibodies to EBNA1, TRF2, hRap1, TRF1, ORC2 or proliferating-cell nuclear antigen (PCNA). We found that ORC2 bound to DS wt but not to DS nm− or BKS, indicating that telomere repeats were required for ORC recruitment in vitro. As expected, EBNA1 bound to DS wt and DS nm− but not to control BKS DNA. Telomere repeat-binding factors TRF2, hRap1 and TRF1 bound to DS wt but did not bind to DS nm− or BKS DNA, which lack telomere repeats. As a control for specificity, PCNA did not bind to any of these DNA fragments tested. Essentially identical results were observed when DNA affinity was measured using a minimal replicator sequence containing only two EBNA1-binding sites surrounded by two TRF sites (Fig 1D). These results indicate that ORC binds to a minimal replicator element and is dependent on telomere repeats for stable recruitment in vitro.
Previous studies by others and ourselves have shown that telomere repeats contribute to the replication activity of DS in the context of other OriP sequence, including FR. We now demonstrate that the telomere repeats stimulate replication activity of the OriP minimal replicator by ∼2.2-fold (Fig 2A). We next assessed, by chromatin immunoprecipitation (ChIP) assay, whether substitution mutations in the telomere repeats abrogated protein assembly at DS in vivo (Fig 2B). We found that mutation of the telomere repeats reduced ORC2 binding by ∼2.0-fold and MCM3 binding by ∼3.2-fold. This was considered significant, because TRF2 binding was reduced by only ∼2.3-fold in the absence of telomere repeat-binding sites. We also observed a slight decrease in EBNA1 binding (∼1.3-fold), suggesting that TRF2 sites may stabilize EBNA1 binding to DS in vivo. No significant binding of EBNA1, TRF2, ORC2 or MCM3 was observed at the control ampicillin gene (AMP) located on the same plasmid DNA (supplementary Fig 1A online).
Figure 2.
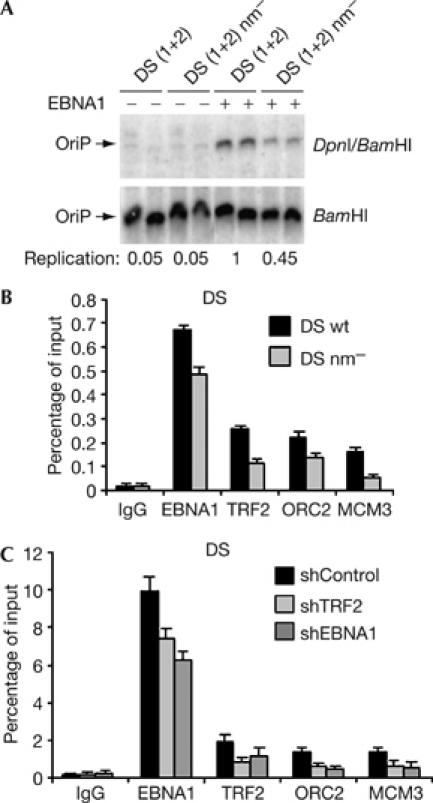
Telomere repeats function in OriP replication and ORC recruitment in vivo. (A) DNA replication assay for DS (1+2) or DS (1+2) nm− in the absence (−) or presence (+) of EBNA1 transfected into HeLa cells. (B) ChIP assay of EBNA1, TRF2, ORC2, MCM3 or control IgG at DS wt or DS nm− in transfected D98/HR1 cells with primers specific for the plasmid-based DS region. (C) ChIP assay of EBNA1, TRF2, ORC2, MCM3 or control IgG binding to DS in EBV-positive ZKO-293 cells transfected with shTRF2, shEBNA1 or control plasmids. ChIP, chromatin immunoprecipitation; DS, dyad symmetry; EBNA1, Epstein–Barr virus nuclear antigen 1; MCM, minichromosome maintenance; ORC2, origin recognition complex 2; OriP, origin of plasmid replication; TRF2, telomere repeat factor 2; wt, wild type.
ORC and MCM recruitment to OriP was next examined in cells in which TRF2 and EBNA1 were depleted by small hairpin (sh) RNA (Fig 2C). We have previously shown that shRNA depletion of TRF2 inhibits OriP-dependent DNA replication (Deng et al, 2003). ChIP assays showed that shRNA depletion of TRF2 caused ∼2.4-fold reduction in TRF2 binding, and a similar fold loss of ORC2 and MCM3 from DS (Fig 2C). TRF2 depletion caused a small loss of EBNA1 binding (∼1.2-fold), supporting the idea that TRF2 cooperatively binds with EBNA1 at DS. Similar results were observed for shRNA depletion of EBNA1. No specific binding of EBNA1, TRF2, ORC2 or MCM3 was observed at the control OriLyt region of the EBV genome (supplementary Fig 1C online). These results indicate that TRF2 depletion leads to a specific loss of ORC2 and MCM3 binding to the DS region of the EBV episome in latently infected cells.
To determine whether one of the three well-characterized telomere repeat proteins is capable of binding to ORC, we expressed and purified TRF2, hRap1 and TRF1 as glutathione S-transferase (GST) fusion proteins from Escherichia coli (Fig 3A) and assayed for their ability to bind to ORC2 from Raji nuclear extracts (Fig 3B). We found that GST–TRF2 bound to ORC2 significantly better than GST–TRF1, GST–hRap1 or GST alone (Fig 3B). Ethidium bromide (EtBr), which disrupts nonspecific DNA bridging, did not inhibit the association between ORC2 and GST–TRF2 (Fig 3B). We next determined whether TRF2 and ORC2 form a stable complex in vivo, using immunoprecipitation (Fig 3C). We also tested whether EBNA1 was a stable component of this complex, as we found evidence for cooperative binding of TRF2 and EBNA1 in vivo (Fig 2) and an ability of these two proteins to interact in vitro (supplementary Figs 2,3 online). We found that antibodies to EBNA1 co-immunoprecipitated with ORC2 and TRF2, but antibodies to TRF2 co-immunoprecipitated with only ORC2, suggesting that TRF2 associates with ORC in complexes both with and without EBNA1 (Fig 3C). To further test this possibility, we compared the co-immunoprecipitation of TRF2 with ORC2 in EBNA1-positive Raji cells and EBNA1-negative HeLa cells (Fig 3D). ORC2 was detected in the TRF2 immunoprecipitates from both cell types, indicating that a stable complex exists between ORC2 and TRF2, which is independent of EBNA1 protein or OriP DNA. GST–TRF2 was also capable of binding to a partially purified FLAG–ORC2 complex (Fig 3E), suggesting that TRF2 binds to ORC subunits directly or through proteins that are tightly associated with ORC2 in human cells. These findings suggest that TRF2 is the telomere protein involved in ORC recruitment to DS.
Figure 3.
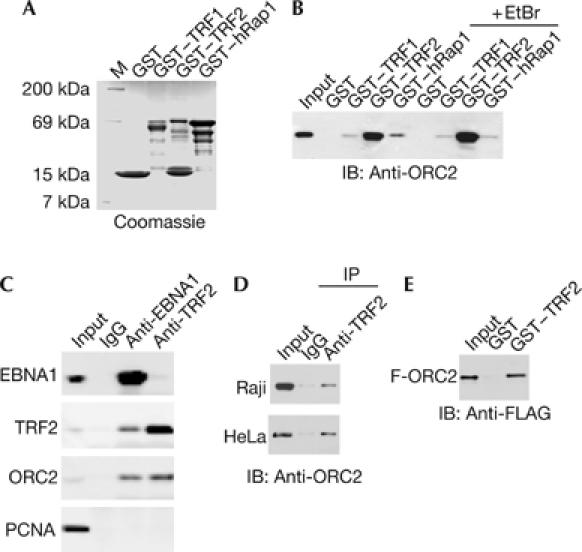
ORC interacts with TRF2. (A) Coomassie stain of purified GST, GST–hRap1, GST–TRF2 and GST–TRF1. (B) Western blot with ORC2 antibodies of GST pull-down assays using Raji nuclear extract and various GST proteins shown in (A). Ethidium bromide (EtBr; 100 μg/ml) was included in the binding reactions, as indicated. (C) Co-immunoprecipitation with antibodies to EBNA1, TRF2 or control IgG. Immunoprecipitates were assayed by western blot with antibodies to EBNA1, TRF2, ORC2 or control PCNA. (D) Co-immunoprecipitation assay with anti-TRF2 and western blot with anti-ORC2 in Raji (top) or HeLa (bottom) nuclear extracts. (E) FLAG–ORC2 was purified from stable cell lines and assayed for direct interaction with purified GST or GST–TRF2 by western blot with anti-FLAG. EBNA1, Epstein–Barr virus nuclear antigen 1; GST, glutathione S-transferase; IB, immunoblotting; IP, immunoprecipitation; ORC, origin recognition complex; PCNA, proliferating-cell nuclear antigen; TRF, telomere repeat factor.
To identify a subdomain of TRF2 responsible for ORC recruitment, we fused TRF2 subfragments to GST and assayed their ability to bind to ORC2 from HeLa nuclear extracts (Fig 4). All of the GST–TRF2 truncation mutants were expressed and purified to similar levels (Fig 4B). GST–TRF2 (2–454) lacking the myb DNA-binding domain or GST–TRF2 (2–90) lacking the DNA-binding and homodimerization domain bound to ORC2 similarly to full-length GST–TRF2 (Fig 4A). Mutants lacking the amino (N)-terminal basic domain, including TRF2 (45–500), TRF2 (45–454), TRF2 (245–454) and TRF2 (245–500), failed to bind to ORC2 (Fig 4A). These findings indicate that the N-terminal region (amino acids (aa) 2–90) of TRF2 is necessary and sufficient for recruiting ORC2 from HeLa nuclear extracts (summarized in Fig 4C).
Figure 4.
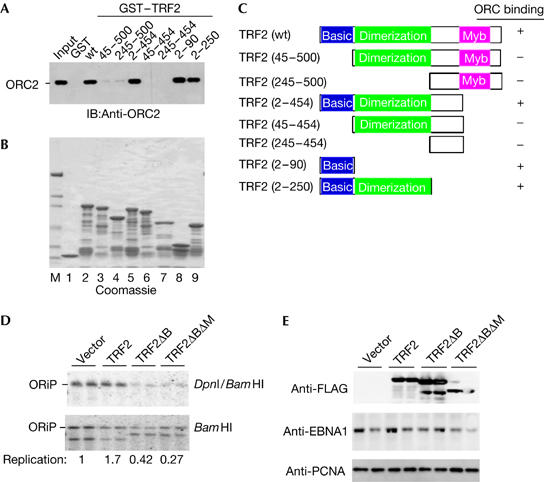
The basic region of TRF2 interacts with ORC and is required for OriP replication function. (A) Truncation mutants of TRF2 were fused to GST and assayed for ORC2 recruitment with Raji nuclear extracts. Bound ORC2 was identified by western blot with anti-ORC2. (B) Coomassie stain of GST–TRF2 truncation mutants, as indicated in (A). (C) Summary of ORC2 binding to GST–TRF2 truncation mutants. (D) DNA replication assay of OriP in ZKO-293 cells co-transfected with FLAG-tagged TRF2, TRF2ΔB, TRF2ΔBΔM or vector control. (E) Protein expression was confirmed by western blotting of whole-cell extracts derived from transfected ZKO-293 cells used in replication assay of (D) and probed with anti-FLAG, anti-EBNA1 or anti-PCNA loading control. EBNA1, Epstein–Barr virus nuclear antigen 1; GST, glutathione S-transferase; IB, immunoblotting; ORC, origin recognition complex; OriP, origin of plasmid replication; PCNA, proliferating-cell nuclear antigen; TRF2, telomere repeat factor 2.
To determine whether ORC recruitment by TRF2 is important for OriP replication, we analysed the effect of TRF2 deletion mutants on OriP DNA replication. The deletion mutant TRF2ΔBΔM, which lacks the N-terminal basic (B) and myb (M) DNA-binding domain, has been shown to function as a dominant-negative inhibitor of native TRF2 (van Steensel et al, 1998) and inhibit OriP DNA replication (Deng et al, 2003). We now show that transient overexpression of TRF2ΔB, which lacks the basic N-terminal domain and is incapable of binding to ORC, inhibits OriP DNA replication similar to TRF2ΔBΔM (Fig 4D,E). This suggests that the ORC recruiting function of TRF2 N terminus is essential for OriP replication.
We next set to determine whether TRF2 basic region can bind directly to any subunit of ORC (Fig 5). Previous studies have shown that ORC1 physically interacts with a basic motif in the AIF-C1 repressor/origin-binding protein at the rat aldolase B gene (Saitoh et al, 2002). AIF-C1 was found to interact with a small region (aa 210–239) between the conserved BAH and ATPase domain of ORC1 (Fig 5A). As the TRF2 N-terminal domain basic region shares some common features with AIF-C1, we tested whether a similar region of ORC1 also bound to TRF2. We first expressed and purified GST, GST–ORC1(1–200), GST–ORC1(201–511) and GST–ORC1(512–862) from E. coli (Fig 5B), and assayed these purified proteins for their ability to bind to native TRF2 from Raji nuclear extracts (Fig 5C). We found that GST–ORC1(201–511) bound to TRF2 strongly, whereas GST, GST–ORC1(1–200), and GST–ORC1(512–862) had little detectable binding. As a negative control, PCNA was detected in the input but did not bind to any of the GST–ORC1 fusion peptides (Fig 5C, lower panel). To determine whether the interaction between TRF2 and ORC1 was direct, we purified a hexahistidine (H6)-tagged ORC1 (201–511) and assayed it for specific binding to purified GST or GST–TRF2 (13–47; supplementary Fig 4A online). We found that GST–TRF2 (13–47) bound to H6–ORC (201–511) efficiently, whereas GST had no detectable binding (supplementary Fig 4B online). These data indicate that purified fragments from TRF2 N terminus (aa 13–47) can bind to a purified fragment of ORC1 (aa 201–511), and suggest that the TRF2 recruitment of ORC is mediated, in part, by this direct protein interaction. The functional significance of this interaction was investigated by overexpressing the ORC1 fragment (aa 201–511) and assaying its effect on OriP DNA replication (Fig 5D). We found that ORC1 (aa 201–511) inhibited OriP replication by ∼3-fold. Overexpression of ORC1 (aa 201–511) had no observable effect on cell cycle progression, and therefore was unlikely to inhibit OriP by indirectly blocking cellular DNA synthesis (data not shown). This result was similar to that observed by overexpression of TRF2ΔB (Fig 4D), and suggests that disruption of ORC recruitment to DS by overexpression of truncated forms of TRF2 or ORC1 can block OriP replication.
Figure 5.
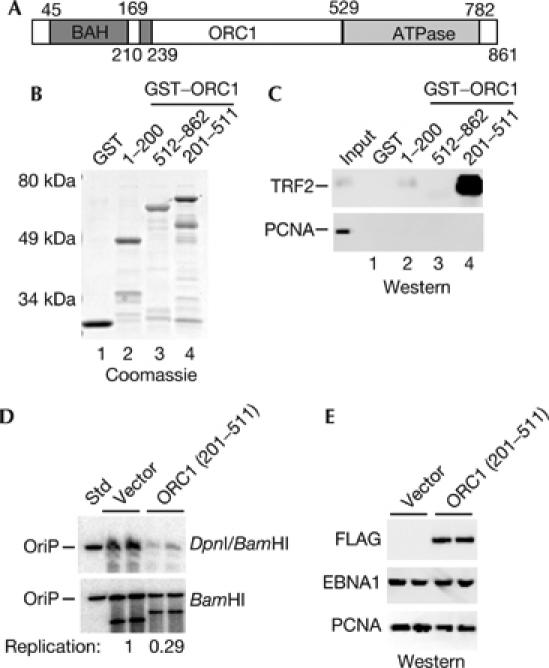
TRF2 basic region binds directly to a subdomain of ORC1. (A) Diagram of the known functional domains of ORC1, including the BAH (aa 1–201), the ATPase (529–782) and the AIF-C1 interacting domain (210–239). (B) Coomassie stain of GST, and GST–ORC1 amino acids (1–200), (512–862) and (201–511). (C) HeLa cell nuclear extracts were incubated with GST or GST–ORC1 fragments as indicated, and analysed by western blot with an antibody specific for TRF2. Input represents 5% of the HeLa nuclear extract used for GST interaction assays. (D) DNA replication assay for OriP in ZKO-293 cells co-transfected with CMV–FLAG vector control or FLAG–ORC1 (aa 201–511). (E) Western blot with antibodies to FLAG–ORC1 (aa 201–511), EBNA1 or control PCNA corresponding to replication assay shown in (D). EBNA1, Epstein–Barr virus nuclear antigen 1; GST, glutathione S-transferase; ORC, origin recognition complex; OriP, origin of plasmid replication; PCNA, proliferating-cell nuclear antigen; TRF2, telomere repeat factor 2.
ORC recruitment is an essential and early event in the establishment of a eukaryotic replication origin (Bell & Dutta, 2002). Stable recruitment of ORC to a specific DNA sequence may be sufficient to establish a replication origin, as fusion of ORC1, ORC2 or CDC6 to the GAL4 DNA-binding domain confers origin functions to a plasmid containing five GAL4-binding sites (Takeda et al, 2005). Our findings are consistent with this model, as origin activity correlates well with ORC recruitment to DS. However, the mechanism of ORC recruitment to DS may be complex. It is well established that replication activity of DS is dependent on EBNA1 (Leight & Sugden, 2000), and that EBNA1 can form a stable immunoprecipitable complex with ORC subunits (Schepers et al, 2001; Ritzi et al, 2003). However, EBNA1-binding sites at FR and Qp do not recruit ORC and do not initiate DNA replication. It is also known that the spacing of EBNA1 sites at DS is crucial for replication activity and that EBNA1 induces DNA bending at DS (Yates et al, 2000; Bashaw & Yates, 2001). Our data indicate that TRF2 enhances EBNA1 binding to DS in vivo (Fig 2B–E) and in vitro (Deng et al, 2002, 2003). We have shown here that TRF2 can bind to ORC1 directly. However, we also found that EBNA1 can bind to TRF2 directly, and that the RGG-like domain of EBNA1 (aa 330–380), which is essential for DNA replication function, mediates an interaction with the TRF2 dimerization domain (aa 45–250; supplementary Figs 2,3 online; Sears et al, 2004). Thus, it is likely that EBNA1 and TRF2 function cooperatively to provide a highly stable interaction surface for ORC recruitment to DS and that no single factor is sufficient for stable origin formation.
TRF2 has a central role in telomere length maintenance and end protection (de Lange, 2004) but has no known function in DNA replication or origin formation. ORC has a fundamental role in DNA replication and origin formation, and may also be important in chromatin structure (Bell, 2002). How the interaction between TRF2 and ORC that we have described in this work affects TRF2 function at telomeres remains to be determined. We found that the TRF2 basic N-terminal domain can bind directly to ORC1. Previous studies have shown that overexpression of TRF2 lacking this domain (TRF2ΔB) causes a loss of telomere repeat length and generates telomere circles (Wang et al, 2004). At DS, TRF2ΔB overexpression inhibits DNA replication. It will be important to determine whether TRF2 can recruit ORC to telomeres, and whether the telomere defect caused by TRF2ΔB overexpression is a consequence of a failure to interact with ORC. It is also possible that TRF2 interaction with ORC is important for chromatin structure at OriP, as well as at cellular telomeres.
Methods
D98/HR1 and ZKO-293 (gift from H.-J. Delecluse) are EBV genome-positive adherent cell lines. Raji is an EBV-positive Burkitt's lymphoma suspension culture cell line. HeLa N2 and 293 HEK cells are EBV-negative adherent cell lines. Plasmids containing DS (1+2) and DS (1+2) nm− were generated by cloning a 72 bp oligonucleotide DS (1+2) or DS (1+2) nm− fragment into the BamHI/HindIII site of pBluescript-KS vector. pCMV-FLAG-TRF2 (aa 1–500) and pCMV-FLAG-TRF2ΔB (aa 45–500) were cloned into HindIII–BamHI sites of p3xFLAG-CMV (Sigma–Aldrich, MO, USA). PCMV-FLAG-ORC2 was generated by PCR amplification of ORC2 complementary DNA and cloning into the BglII–Asp718 site of p3xFLAG-CMV. GST–hRap1, GST–TRF1, GST–TRF2 and ORC1 truncation mutants were generated by PCR amplification and subcloning in the pGEX-2T vector (Amersham Biosciences, Piscataway, NJ, USA). ORC1 subdomains containing amino-acid residues 1–200, 201–511 and 512–862 were subcloned as BamHI–HindIII fragments into pRSETA in-frame with hexahistidine N-terminal tag (Invitrogen, CA, USA). Further details with regard to plasmids are available on request. DNA affinity pull-down assays have been described previously (Deng et al, 2002; Atanasiu et al, 2005). DNA replication assay, ChIP assays, GST interaction assays and transfection protocols have been described previously (Deng et al, 2002). Detailed methods for the above assays and additional experimental details are included in the Supplementary materials available online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
supplementary Figures and Materials
Acknowledgments
We acknowledge the technical support of Wistar Cancer Center Core Facilities, and financial support from the Leukemia Research Foundation (C.A.), the Leukemia-Lymphoma Society (Z.D.), the NIH Training Program in Genetics (5-T32-GM08216-18; J.N.), the Pennsylvania Department of Health, the DOD (DAMD17-03-1-0313) and the NIH (CA93606) to P.M.L.
References
- Atanasiu C, Lezina L, Lieberman PM (2005) DNA affinity purification of Epstein–Barr virus OriP-binding proteins. Methods Mol Biol 292: 267–276 [DOI] [PubMed] [Google Scholar]
- Bashaw JM, Yates JL (2001) Replication from oriP of Epstein–Barr virus requires exact spacing of two bound dimers of EBNA1 which bend DNA. J Virol 75: 10603–10611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall EL, Manak JR, Zhou S, Bell M, Lipsick JS, Botchan MR (2002) Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature 420: 833–837 [DOI] [PubMed] [Google Scholar]
- Bell SP (2002) The origin recognition complex: from simple origins to complex functions. Genes Dev 16: 659–672 [DOI] [PubMed] [Google Scholar]
- Bell SP, Dutta A (2002) DNA replication in eukaryotic cells. Annu Rev Biochem 71: 333–374 [DOI] [PubMed] [Google Scholar]
- Bell SP, Stillman B (1992) ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357: 128–134 [DOI] [PubMed] [Google Scholar]
- Blow JJ, Dutta A (2005) Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol 6: 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri B, Xu H, Todorov I, Dutta A, Yates JL (2001) Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein–Barr virus. Proc Natl Acad Sci USA 98: 10085–10089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang RY, Kelly TJ (1999) The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proc Natl Acad Sci USA 96: 2656–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetic C, Walter JC (2005) Eukaryotic origins of DNA replication: could you please be more specific? Semin Cell Dev Biol 16: 343–353 [DOI] [PubMed] [Google Scholar]
- de Lange T (2004) T-loops and the origin of telomeres. Nat Rev Mol Cell Biol 5: 323–329 [DOI] [PubMed] [Google Scholar]
- Deng Z, Lezina L, Chen CJ, Shtivelband S, So W, Lieberman PM (2002) Telomeric proteins regulate episomal maintenance of Epstein–Barr virus origin of plasmid replication. Mol Cell 9: 493–503 [DOI] [PubMed] [Google Scholar]
- Deng Z, Atanasiu C, Burg JS, Broccoli D, Lieberman PM (2003) Telomere repeat binding factors TRF1, TRF2, and hRAP1 modulate replication of Epstein–Barr virus OriP. J Virol 77: 11992–12001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis ML (2003) The ‘ORC cycle': a novel pathway for regulating eukaryotic DNA replication. Gene 310: 1–15 [DOI] [PubMed] [Google Scholar]
- Dhar SK, Yoshida K, Machida Y, Khaira P, Chaudhuri B, Wohlschlegel JA, Leffak M, Yates J, Dutta A (2001) Replication from oriP of Epstein–Barr virus requires human ORC and is inhibited by geminin. Cell 106: 287–296 [DOI] [PubMed] [Google Scholar]
- Diffley JF (2004) Regulation of early events in chromosome replication. Curr Biol 14: R778–R786 [DOI] [PubMed] [Google Scholar]
- Gilbert DM (2004) In search of the holy replicator. Nat Rev Mol Cell Biol 5: 848–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien MD, Polonskaya Z, Hearing J (2004) Protein and sequence requirements for the recruitment of the human origin recognition complex to the latent cycle origin of DNA replication of Epstein–Barr virus oriP. Virology 326: 317–328 [DOI] [PubMed] [Google Scholar]
- Leight ER, Sugden B (2000) EBNA-1: a protein pivotal to latent infection by Epstein–Barr virus. Rev Med Virol 10: 83–100 [DOI] [PubMed] [Google Scholar]
- McNairn AJ, Gilbert DM (2003) Epigenomic replication: linking epigenetics to DNA replication. Bioessays 25: 647–656 [DOI] [PubMed] [Google Scholar]
- Mendez J, Stillman B (2003) Perpetuating the double helix: molecular machines at eukaryotic DNA replication origins. Bioessays 25: 1158–1167 [DOI] [PubMed] [Google Scholar]
- Niller HH, Glaser G, Knuchel R, Wolf H (1995) Nucleoprotein complexes and DNA 5-ends at OriP of Epstein–Barr virus. J Biol Chem 270: 12864–12868 [DOI] [PubMed] [Google Scholar]
- Pak DT, Pflumm M, Chesnokov I, Huang DW, Kellum R, Marr J, Romanowski P, Botchan MR (1997) Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell 91: 311–323 [DOI] [PubMed] [Google Scholar]
- Prasanth SG, Mendez J, Prasanth KV, Stillman B (2004) Dynamics of pre-replication complex proteins during the cell division cycle. Philos Trans R Soc Lond B Biol Sci 359: 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL (2001) Replication dynamics of the yeast genome. Science 294: 115–121 [DOI] [PubMed] [Google Scholar]
- Ritzi M, Tillack K, Gerhardt J, Ott E, Humme S, Kremmer E, Hammerschmidt W, Schepers A (2003) Complex protein–DNA dynamics at the latent origin of DNA replication of Epstein–Barr virus. J Cell Sci 116: 3971–3984 [DOI] [PubMed] [Google Scholar]
- Saitoh Y, Miyagi S, Ariga H, Tsutsumi K (2002) Functional domains involved in the interaction between Orc1 and transcriptional repressor AlF-C that bind to an origin/promoter of the rat aldolase B gene. Nucleic Acids Res 30: 5205–5212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers A, Ritzi M, Bousset K, Kremmer E, Yates JL, Harwood J, Diffley JF, Hammerschmidt W (2001) Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein–Barr virus. EMBO J 20: 4588–4602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears J, Ujihara M, Wong S, Ott C, Middeldorp J, Aiyar A (2004) The amino terminus of Epstein–Barr virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J Virol 78: 11487–11505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B (2001) DNA replication. Genomic views of genome duplication. Science 294: 2301–2304 [DOI] [PubMed] [Google Scholar]
- Takeda DY, Shibata Y, Parvin JD, Dutta A (2005) Recruitment of ORC or CDC6 to DNA is sufficient to create an artificial origin of replication in mammalian cells. Genes Dev 19: 2827–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T (1998) TRF2 protects human telomeres from end-to-end fusions. Cell 92: 401–413 [DOI] [PubMed] [Google Scholar]
- Vogel M, Wittmann K, Endl E, Glaser G, Knuchel R, Wolf H, Niller HH (1998) Plasmid maintenance assay based on green fluorescent protein and FACS of mammalian cells. BioTechniques 24: 540–544 [PubMed] [Google Scholar]
- Wang J, Sugden B (2005) Origins of bidirectional replication of Epstein–Barr virus: models for understanding mammalian origins of DNA synthesis. J Cell Biochem 94: 247–256 [DOI] [PubMed] [Google Scholar]
- Wang RC, Smogorzewska A, de Lange T (2004) Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119: 355–368 [DOI] [PubMed] [Google Scholar]
- Weinreich M, Palacios DeBeer MA, Fox CA (2004) The activities of eukaryotic replication origins in chromatin. Biochim Biophys Acta 1677: 142–157 [DOI] [PubMed] [Google Scholar]
- Wyrick JJ, Aparicio JG, Chen T, Barnett JD, Jennings EG, Young RA, Bell SP, Aparicio OM (2001) Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science 294: 2357–2360 [DOI] [PubMed] [Google Scholar]
- Yates JL, Camiolo SM, Bashaw JM (2000) The minimal replicator of Epstein–Barr virus oriP. J Virol 74: 4512–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary Figures and Materials


