Abstract
In order to evaluate the in situ degradative capabilities of microorganisms in an underground reactor facility housing two flowthrough columns filled with aquifer soil, we examined the distribution and phylogeny of gene transcripts encoding enzymes capable of catalyzing the cleavage of the chlorinated aromatic ring during transformation of the main pollutant, chlorobenzene. Initial biostimulation of the autochthonous bacteria in the originally anaerobic reactor columns was achieved by injecting nitrate and oxygen in the form of H2O2. Two broad-range primer pairs were used for reverse transcriptase PCR (RT-PCR) of partial subunit genes of chlorocatechol 1,2-dioxygenase and catechol 2,3-dioxygenase from RNA directly extracted from different groundwater and aquifer samples. Samples retrieved from the lowermost sections of the reactor columns, which were operated in upflow mode, were positive for the presence of chlorocatechol 1,2-dioxygenase and catechol 2,3-dioxygenase mRNA. On the other hand, chlorocatechol 1,2-dioxygenase RT-PCR products were detected in a larger part of each reactor column, up to a zone 5.5 m above the bottom. Phylogenetic analyses of these chlorocatechol 1,2-dioxygenase sequences clearly separated them into two main clusters, one of which was closely affiliated with the broad-spectrum chlorocatechol 1,2-dioxygenase from Pseudomonas chlororaphis RW71. Analysis of sequences obtained from RT-PCR products amplified with catechol 2,3-dioxygenase primers revealed that their closest relative was the chlorocatechol 2,3-dioxygenase gene cbzE from Pseudomonas putida GJ31 (A. E. Mars, J. Kingma, S. R. Kaschabek, W. Reineke, and D. B. Janssen, J. Bacteriol. 181:1309-1318, 1999), with sequence similarities between 97.8 and 99.0%.
In situ investigation of the microbial communities from environmental samples taking part in biodegradation processes has proved to be a difficult challenge for microbiologists. This is because the traditional approach, based on cultivation of bacteria from environmental samples, usually catches only a minor fraction of the total bacterial diversity and often produces biased results (7). In contrast, the ideal assay should ascertain the identities and functions of the naturally occurring bacteria that are immediately fixed in the field. DNA- and RNA-based approaches have helped workers overcome many former limitations. One approach for studying specific microbial activities is to investigate the expression of genes by analyzing mRNA transcripts. The usefulness of transcription analysis is based on the short half-life of mRNA, which is sometimes on the order of minutes (2, 5). Thus, regulation at the transcription level almost immediately affects the rate of protein synthesis, and detection of gene expression can be used for investigation of real and specific microbial processes (i.e., phenomena occurring under in situ conditions). Thus, this method is particularly useful for tracking genes encoding enzymes involved in the degradation of xenobiotic compounds.
The aims of this study were to analyze the biodegradative capabilities of microorganisms residing in a reactor column filled with aquifer material located at the on-site underground reactor facility used in the SAFIRA project (Sanierungs-Forschung In Regional kontaminierten Aquiferen). The main pollutant at this site is chlorobenzene. During a stimulation experiment, in which hydrogen peroxide was added as an oxygen-releasing compound, directly extracted RNA was used for reverse transcriptase PCR (RT-PCR) to analyze the expression of genes encoding enzymes involved in the catabolism of chlorobenzene. Cleavage of the aromatic ring, which is catalyzed by dioxygenases, is a key step in the catabolic pathways for halogenated aromatic compounds under aerobic conditions. Two types of enzymes, referred to as intradioldiol dioxygenases for the ortho-cleavage pathway or modified ortho-cleavage pathway and extradiol dioxygenases for the meta-cleavage pathway, are known to be used by bacteria. Therefore, chlorocatechol dioxygenase genes are good targets for monitoring bacteria involved in the ring cleavage of chlorocatechols. Although degradation of chlorinated compounds generally occurs via the modified ortho-cleavage pathway (26, 27), we also used broad-range primers that were initially designed for amplification of different catechol 2,3-dioxygenase gene fragments in order to detect bacteria using the recently described meta-cleavage pathway for the conversion of chlorinated substrates (19, 20). To evaluate the specificity of our approach and to examine the phylogeny, selected RT-PCR products were cloned and screened by restriction fragment length polymorphism (RFLP) analysis, and selected clones were then sequenced.
MATERIALS AND METHODS
Site description and sample collection.
The physicochemical characteristics of the Bitterfeld aquifer and test site have been described in detail elsewhere (21, 30). The reactor columns of shaft 5 (columns 1a and 1b) are 12 m tall, and each has a diameter of 600 mm (30). Native aquifer sediment was used as the reactor column fill material, and both reactor columns were operated under conditions very similar to the in situ conditions. Both reactor columns were operated in flowthrough mode from bottom to top with a flow rate of 4.7 l h−1, resulting in a retention time of approximately 10 days for water according to a conductivity tracer test (30). The reactor columns were put into operation in June 1999, and after approximately 250 days a chemical steady state for chlorobenzene was reached. The potential for stimulating microbial chlorobenzene degradation by adding substrates was investigated during different experimental phases. Concentrated hydrogen peroxide and a nitrate solution were stored in tanks (capacity, 60 liters) made of stainless steel and were added to the inflowing groundwater by using a flexible-tube pump and feed lines made of stainless steel at a rate of 1.04 liters day−1. Starting on operation day 363, reactor column 1b was amended with 2.94 mM hydrogen peroxide and 2 mM nitrate, and then on operation day 719 the hydrogen peroxide concentration was reduced to 0.88 mM. Reactor column 1a served as an anaerobic control reactor column and was operated without hydrogen peroxide but with nitrate (2 mM). On operation days 733 and 775, groundwater samples for RNA extraction were taken from different reactor column zones (Table 1). Between 100 and 300 ml of each water sample was immediately filtered through 0.22-μm-pore-size filters (Durapore; Millipore, Bedford, Mass.). After processing, the filters were frozen and stored at −80°C until extraction. Aquifer samples (for sediment composition, see reference 30) were taken between operation days 803 and 895 from different reactor column zones with a sample lance, immediately transferred to a box filled with dry ice, and stored at the lab at −80°C until they were processed (Table 1 and see below).
TABLE 1.
Detection by RT-PCR of chlorocatechol dioxygenase genes from groundwater and aquifer samples retrieved from different sectors of in situ reactor columns 1a (RA4) and 1b (Z, RB3, RB4, RB5, RB7, RB9)
| Sample type | Operation day | Gene | RT-PCR products detected at the following sampling stationsa:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Z | RB3 | RB4 | RB5 | RB7 | RB9 | RA4 | |||
| Ground water | 733 | Chlorocatechol 1,2-dioxygenase | + | + | + | NDb | + | ND | ND |
| Chlorocatechol 2,3-dioxygenase | + | − | − | ND | − | ND | ND | ||
| 775 | Chlorocatechol 1,2-dioxygenase | + | + | + | − | − | ND | ND | |
| Chlorocatechol 2,3-dioxygenase | +c | − | − | − | − | ND | ND | ||
| Aquifer | 803 | Chlorocatechol 1,2-dioxygenase | +c | ND | − | − | − | − | |
| Chlorocatechol 2,3-dioxygenase | +c | ND | − | − | − | − | |||
| 845 | Chlorocatechol 1,2-dioxygenase | +c | ND | ND | + | + | ND | ||
| Chlorocatechol 2,3-dioxygenase | + | ND | ND | − | − | ND | |||
| 859 | Chlorocatechol 1,2-dioxygenase | + | + | + | + | + | ND | ||
| Chlorocatechol 2,3-dioxygenase | + | +c | − | − | − | ND | |||
| 895 | Chlorocatechol 1,2-dioxygenase | + | + | + | + | − | − | ||
| Chlorocatechol 2,3-dioxygenase | + | − | − | − | − | − | |||
The sampling stations were the following distances from the reactor bottom: Z, 0 m; RB3, 0.1 m; RB4, 0.35 m; RB5, 0.8 m; RB7, 2.5 m; RB9, 5.5 m; and RA4, 0.35 m.
ND, not determined.
The RT-PCR products were cloned and screened for variety by RFLP analysis, and selected clones were sequenced.
Measurement of chlorobenzene concentrations.
Chlorobenzene was analyzed by automated headspace gas chromatography with a Varian 3800 gas chromatograph (Varian, Palo Alto, Calif.) equipped with a CP SIL 5 CB capillary column (film thickness, 0.12 μm; inside diameter, 0.25 mm; length, 25 m) and a flame ionization detector. The chromatographic conditions were as follows: injector temperature, 250°C (split 1:2); detector temperature, 300°C; and an oven temperature program consisting of 35°C for 3 min, followed by increases at a rate of 10°C min−1 to 65°C and then at a rate of 30°C min−1 to 260°C. Liquid test samples (diluted 1:10 or 1:20 in 1.6 mM H2SO4; final volume, 10 ml) were prepared in 20-ml glass vials. The samples were incubated for 20 min at 35°C in an agitator (rotation regime, 250 rpm for 5 s and no rotation for 2 s) prior to analysis, and 1 ml of each sample's headspace was injected. For calibration, diluted standards of chlorobenzene prepared from stock solutions were treated in the same way as the samples. The stock solutions were prepared in pure methanol.
RNA extraction and RT-PCR.
One- to six-gram portions of reactor column sediments were used for RNA extraction, and samples were prepared by using an RNA extraction method described by Borneman and Triplett (6) and a FastPrep RNA kit (Qbiogene Inc., Carlsbad, Calif.). To extract the water samples, the filters were cut into small pieces by using an aseptic technique. The pieces were placed in a capped tube with extraction buffer and beads as described above for sediment samples. After extraction, 50-μl portions were treated with DNase I and RNase inhibitor (Ambion, Austin, Tex.) according to the manufacturer's specifications to obtain DNA-free RNA for subsequent RT-PCR.
The following primers for selective amplification of chlorocatechol 1,2-dioxygenase genes targeting a variety of members of the Proteobacteria were used as described by Leander et al. (18): forward primer CCDb (5′GTITGGCA[CT]TCIACICCIGA[CT]GG3′) and reverse primer CCDe (5′CCICC[CT]TCGAAGTAGTA[CT]TGIGT3′). The second PCR primer pair used was originally designed for specific amplification of a wide variety of catechol 2,3-dioxygenase genes and was described by Sei et al. (25); the forward primer was C230f (5′AAGAGGCATGGGGGCGCACCGGTTCGATCA3′), and the reverse primer was C230r (5′CCAGCAAACACCTCGTTGCGGTTGCC3′). The expected sizes of the amplification products were 270 to 279 bp for chlorocatechol 1,2-dioxygenase genes and around 380 bp for catechol 2,3-dioxygenase genes (18, 25). Reverse transcription and subsequent PCR amplification were performed by using a one-step reaction scheme carried out sequentially in the same tube (Qiagen OneStep RT-PCR kit; Qiagen Inc., Valencia, Calif.).
RT amplification was carried out by using the protocol recommended by the manufacturer with 50-μl reaction mixtures containing each deoxynucleoside triphosphate at a concentration of 400 μM, each primer at a concentration of 0.6 μM, and about 0.02 to 2 μg of template RNA with a PTC200 thermal cycler (MJ Research, Inc., Waltham, Mass.). The thermal cycle parameters were as follows: 30 min at 50°C for the reverse transcription reaction, followed by 15 min at 95°C to inactivate the RT and to activate the HotStartTaq DNA polymerase. For PCR amplification of partial chlorocatechol 1,2-dioxygenase genes, 40 cycles of 45 s at 95°C, 45 s at 50°C, and 1 min 30 s at 72°C were used, while for the catechol 2,3-dioxygenase genes 40 cycles of 45 s at 95°C, 45 s at 55°C, and 2 min at 72°C were used. For both reactions, the cycles were followed by 5 min of incubation at 72°C. To verify that the RT-PCR products did not originate from contaminating DNA, for each sample a negative control was subjected to PCR without RT. Products were electrophoresed on 0.8% agarose gels and visualized with ethidium bromide.
Cloning, variant screening, and DNA sequence analysis.
Selected RT-PCR products in the expected size range were cut out of the gels and purified (QIAquick gel extraction kit; Qiagen Inc.). Purified DNA fragments were TA cloned into pCR2.1 (Invitrogen Corp., Carlsbad, Calif.) by using the protocols provided by the manufacturer. Plasmid DNA was isolated (Qiagen plasmid kit; Qiagen Inc.), and clones were screened for the presence of inserts by PCR by using vector-specific primers. Portions (5 μl) of the ribosomal DNA amplicons derived from the clones were digested with restriction endonuclease HaeIII and/or AluI for 3 h. The reaction was stopped by incubating the samples at 65°C for 20 min, and the products in 10-μl portions of the restriction digests were separated by using Metaphor agarose (FMC Corp., Rockland, Maine).) For each clone library 20 clones were screened for variation by RFLP analysis. Clone patterns that occurred more than once and most representatives with unique RFLP types were selected and sequenced from both sites. Sequencing was carried out with an ABI PRISM 310 genetic analyzer (Applied Biosystems, Foster City, Calif.) by using dye terminators. The closest relatives of nucleic acid sequences and deduced amino acid sequences were obtained by using the sequence similarity search tools BLASTN and BLASTP (Basic Local Alignment Search Tool) (3) of the National Center for Biotechnology Information (NCBI). For phylogenetic analysis, deduced amino acids were aligned by using Clustal W as provided by the OMIGA 1.1 software (Oxford Molecular Group, Oxford, United Kingdom), followed by visual inspection of the alignment. Neighbor-joining trees applying gamma distribution as the distance method were computed with the MEGA 2.1 software package (17).
Nucleotide sequence accession numbers.
The sequences obtained have been deposited in the NCBI databases under accession numbers AF527737 to AF527753.
RESULTS AND DISCUSSION
Reactor column operation and chlorobenzene concentrations.
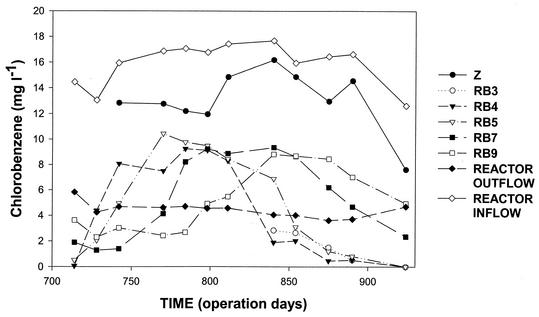
Stimulation of the autochthonous bacteria in reactor column 1b was achieved by injecting nitrate and oxygen in the form of H2O2. Previously (starting on operation day 363), reactor column 1b was operated with 2 mM nitrate and 2.94 mM H2O2; during this period chlorobenzene was completely degraded in the lower reactor column section (C. Vogt, A. Alfreider, H. Lorbeer, D. Hoffmann, L. Wünsche, and W. Babel, submitted for publication). Starting on operation day 719, the hydrogen peroxide concentration in reactor column 1b was lowered to 0.88 mM, whereas the nitrate concentration remained constant (2 mM). This modification in the oxygen supply was accompanied by a subsequent increase in the chlorobenzene concentrations in the lower part of the reactor column, and the maximum chlorobenzene concentrations, which were slightly less than 10 mg liter−1 around operation day 800, were followed by decreases in the chlorobenzene concentrations (Fig. 1). On operation day 924, chlorobenzene was no longer detectable at sampling stations RB3, RB4, and RB5. The chlorobenzene concentrations in the upper part of the reactor column did not exhibit this pattern, presumably because biodegradation processes were masked by chlorobenzene sorption and desorption by lignite, which accounted for an important fraction of the aquifer sediments (30). At sampling port Z, which was located directly at the entrance of the reactor column, where the groundwater was already enriched with H2O2 and nitrate, about 20% of the chlorobenzene was degraded during the entire investigation period. Reactor column 1a served as an anaerobic control reactor column and was operated without hydrogen peroxide but with nitrate (2 mM). No biodegradation of chlorobenzene was observed in reactor column 1a (data not shown).
FIG. 1.
Time course of chlorobenzene concentrations in groundwater samples from different zones of reactor column 1b. The locations of the sampling ports (Z, RB3 to RB9) are described in Table 1.
Detection of RT-PCR products in the reactor column samples.
The results of RT-PCR amplification of extracted RNA that produced fragments of the expected size from different groundwater samples (operation days 733 and 775) and aquifer samples (operation days 803 to 895) from the reactor columns are summarized in Table 1. In groundwater samples from station Z, located directly at the inlet of the reactor column, transcripts for both types of genes, coding for ortho- and meta-cleavage dioxygenases, were amplified. Groundwater retrieved from the lower section of reactor column 1b tested positive for the presence of chlorocatechol 1,2-dioxygenase mRNA, but catechol 2,3-dioxygenase mRNA was never detected in the pore water of the reactor column. On the other hand, aquifer samples retrieved from the same sampling port, RB3, produced RT-PCR products with both primer pairs. During the study period, the presence of catechol 2,3-dioxygenase genes was limited to this reactor column section; the only exception was operation day 859, when catechol 2,3-dioxygenase genes were also detected in a reactor column zone 0.35 m above the reactor column bottom. Chlorocatechol 1,2-dioxygenase genes were also found in sediments from other reactor column parts on operation days 845 and 859, as well as in sediments obtained from sampling port RB9, which was situated 5.5 m above the reactor column bottom. As anticipated, samples obtained from anaerobic reactor column 1a (sampling port RA4) never yielded positive RT-PCR products (Table 1).
Several previously described broad-range primer pairs for amplification of partial chlorocatechol 1,2-dioxygenases genes (described in references 8, 15, and 18) were tested with different reactor column samples (data not shown). The best primer pair in terms of signal intensity and specificity, which was originally designed to amplify fragments of bacterial genes that code for chlorocatechol 1,2-dioxygenases from different members of the Proteobacteria (18), was used for further investigation. Recently, some gram-positive bacteria, including Arthrobacter and Rhodococcus species with a chlorocatechol-dependent catechol-degrading ortho-cleavage pathway, have been isolated from various environments. Phylogenetic analysis of the chlorocatechol 1,2-dioxygenase sequence from the gram-positive bacterium Rhodococcus opacus 1CP indicated that the gene which encodes this enzyme is phylogenetically distinct from those in Proteobacteria, presumably due to functionally convergent evolution (10, 24). It should therefore be mentioned that the primer pair used in this study is not suitable for amplifying chlorocatechol 1,2-dioxygenase genes of gram-positive bacteria, which probably represented a minority of the bacteria inhabiting the reactor column (1).
The second primer pair used in the present study was initially designed for amplification of specific fragments of a wide variety of catechol 2,3-dioxygenase genes (25). This pair of primers does not contain dITP residues or multiple bases to compensate for the possible presence of different bases and mismatches with certain genes at positions where there is sequence divergence. However, the optimal primability and stability of the primers were tested with PCR simulation software, and the results were verified by successful specific amplification of partial catechol 2,3-dioxygenase genes from miscellaneous bacterial strains (25).
Sequence analysis of ortho- and meta-cleavage pathway genes.
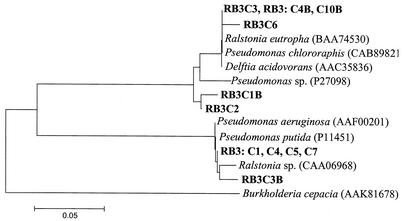
Chlorocatechol 1,2-dioxygenase sequences obtained from two clone libraries from sampling port RB3 were clearly separated into two main clusters, and there was a high degree of homology within (but not between) the clusters (Fig. 2). On operation day 803, four identical chlorocatechol 1,2-dioxygenase-encoding sequences (RB3C1, RB3C4, RB3C5, and RB3C7) derived from sampling station RB3 showed 100% similarity with chlorocatechol dioxygenase genes analyzed from Pseudomonas putida (11) and Pseudomonas aeruginosa (M. E. Corbella, A. Garrido-Pertierra, and A. Puyet, unpublished data.). Clone RB3C3 from operation day 845 was also found to cluster with the sequences from Pseudomonas sp. (sequence identity at 87 of 88 deduced amino acids) and from Ralstonia sp. strain JS705 (sequence identity at 86 of 88 deduced amino acids) isolated from a chlorobenzene-contaminated aquifer (28). The second clearly separated cluster contained three identical sequences (RB3C3, RB3C4B, and RB3C10B) and clone RB3C6. These sequences grouped together with sequences encoding amino acid sequences of Delftia acidovorans (12), Ralstonia eutropha (22), and Pseudomonas chlororaphis (23) obtained from public databases. The sequences of this cluster were also closely affiliated the sequences of partial chlorocatechol 1,2-dioxygenase genes from clones RB3C1B and RB3C2, for which the deduced amino acid sequences exhibited three and two mismatches, respectively (Fig. 2).
FIG. 2.
Neighbor-joining tree based on deduced amino acid sequences of chlorocatechol 1,2-dioxygenase genes derived from the clone libraries obtained in this study (boldface type) and amino acid sequences from the NCBI database. The scale bar represents an estimated change of 5%. The clone designations indicate the sampling stations. For example, clone RB3C3 was obtained from sampling port 3 of reactor 1b. C1, C2, C3, etc. indicate the consecutive numbers of the clones.
An initial BLASTN analysis of all six cloned nucleic acid sequences obtained from three clone libraries with the RT-PCR products amplified by using catechol 2,3-dioxygenase primers indicated that the closest listed relative is the chlorocatechol 2,3-dioxygenase gene from P. putida GJ31 (19), which exhibited sequence similarities of 97.8 to 99.0% (data not shown). Phylogenetic analysis of the deduced amino acids confirmed the results of the nucleic acid data analysis and revealed one distinct cluster containing the sequences obtained in this study and chlorocatechol 2,3-dioxygenase from P. putida GJ31. All other affiliated amino acid sequences retrieved from public databases showed sequence similarities of 82% or less (data not shown).
Mars et al. (19) investigated the catabolic pathway of P. putida strain GJ31 grown on chlorobenzene; they discovered that enzymes of the modified ortho-cleavage pathway were not expressed, but there was high meta-cleavage pathway catechol 2,3-dioxygenase activity. The corresponding enzyme, designated CbzE, efficiently cleaves 3-chlorocatechol in the meta position to 2-hydroxymuconate, which is further metabolized (13). Until recently, it was postulated that the modified ortho-cleavage pathway is the exclusive route for degradation of chlorobenzene. When chlorocatechols are converted by meta-cleavage diooxygenases, these enzymes are usually rapidly inactivated (4, 14, 29). The degradation of 3-chlorocatechol via the meta-cleavage pathway is important if bacteria are confronted with additional aromatic substances. Methylated aromatic compounds (e.g., toluene) are known to be generally mineralized via the meta-cleavage pathway, because metabolism via the ortho-cleavage pathway would produce the dead end product methyllactone (19). As shown by Mars et al. (19), P. putida strain GJ31 is able to grow on a mixture of chlorobenzene and toluene, degrading both compounds via meta-cleavage pathways and thus avoiding any of the incompatibilities mentioned above. In the groundwater of the SAFIRA pilot plant, chlorobenzene is the main pollutant (30), and methylated aromatic compounds have not been detected. Therefore, activation of the meta-cleavage pathway in the lower reactor column section does not result from a necessity to convert chloroaromatic compounds and methylaromatic compounds at the same time. Based on the phylogenetic affiliation with other catechol 2,3-dioxygenase amino acid sequences, we cannot derive additional information concerning the biochemical or functional properties of the enzyme involved. This is because the closest relatives of this enzyme are phylogenetically clearly separated extradiol dioxygenases belonging to so-called subgroup C, which have no distinct biochemical preferences in terms of particular oxygen or catechol concentrations (9, 16, 20). Our findings show that the meta-cleavage pathway was induced only near the bottom of the reactor column, a zone in which chlorobenzene was completely degraded. Chlorocatechol 1,2-dioxygenase was observed in all reactor column sections, including higher zones where chlorobenzene was not degraded at all (or was degraded only at very low rates), probably because of oxygen limitations. This conclusion is supported by the observed changes in the abundance of potential chlorobenzene degraders, which decreased by 2 to 3 orders of magnitude from sampling stations RB3 to RB7 (as measured by the most-probable-number method [Vogt et al., submitted]).
Acknowledgments
We thank Anett Heidtmann and Renate Boetz for their technical assistance in the laboratory and the staff of the Industrial and Mining Landscapes Department for their collaboration.
This work was funded by the Federal Ministry of Education and Research (BMBF) as part of the SAFIRA groundwater remediation project.
REFERENCES
- 1.Alfreider, A., C. Vogt, and W. Babel. 2002. Microbial diversity in an in situ reactor system treating monochlorobenzene contaminated groundwater as revealed by 16S ribosomal DNA analysis. Syst. Appl. Microbiol. 25:232-240. [DOI] [PubMed] [Google Scholar]
- 2.Alifano, P., C. B. Bruni, and M. S. Carlomagno. 1994. Control of mRNA processing and decay in prokaryotes. Genetica 94:157-172. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Bartels, I., H.-J. Knackmuss, and W. Reineke. 1984. Suicide inactivation of catechol 2,3-dioxygenase from Pseudomonas putida mt-2 by halocatechols. Appl. Environ. Microbiol. 47:500-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belasco, J. G., and C. F. Higgins. 1988. Mechanisms of mRNA decay in bacteria. A perspective. Gene 72:15-23. [DOI] [PubMed] [Google Scholar]
- 6.Borneman, J., and E. W. Triplett. 1997. Rapid and direct method for extraction of RNA from soil. Soil Biol. Biochem. 29:1621-1624. [Google Scholar]
- 7.Brock, T. D. 1987. The study of microorganisms in situ: progress and problems. Symp. Soc. Gen. Microbiol. 41:1-17. [Google Scholar]
- 8.Cavalca, L., A. Hartmann, N. Rouard, and G. Soulas. 1999. Diversity of tfdC genes: distribution and polymorphism among 2,4-dichlorophenoxyacetic acid degrading soil bacteria. FEMS Microb. Ecol. 29:45-58. [Google Scholar]
- 9.Eltis, L. D., and J. T. Bolin. 1996. Evolutionary relationships among extradiol dioxygenases. J. Bacteriol. 178:5930-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eulberg, D., E. M. Kourbatova, L. A. Golovleva, and M. Schlömann. 1998. Evolutionary relationship between chlorocatechol catabolic enzymes from Rhodococcus opacus 1CP and their counterparts in proteobacteria: sequence divergence and functional convergence. J. Bacteriol. 180:1082-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frantz, B., and A. M Chakrabarty. 1987. Organization and nucleotide sequence determination of a gene cluster involved in 3-chlorocatechol degradation. Proc. Natl. Acad. Sci. USA 84:4460-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffmann, D., S. Kleinsteuber, R. H. Mueller, and W. Babel. 2001. Development and application of PCR primers for the detection of the tfd genes in Delftia acidovorans P4a involved in the degradation of 2,4-D. Acta Biotechnol. 21:321-331. [Google Scholar]
- 13.Kaschabek, S. R., T. Kasberg, D. Müller, A. E. Mars, D. B. Janssen, and W. Reineke. 1998. Degradation of chloroaromatics: purification and characterization of a novel type of chlorocatechol 2,3-dioxygenase of Pseudomonas putida GJ31. J. Bacteriol. 180:296-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klecka, G. M., and D. T. Gibson. 1981. Inhibition of catechol 2,3-dioxygenase from Pseudomonas putida by 3-chlorocatechol. Appl. Environ. Microbiol. 41:1159-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleinsteuber, S., D. Hoffmann, R. H. Müller, and W. Babel. 1998. Detection of chlorocatechol 1,2-dioxygenase genes in proteobacteria by PCR and gene probes. Acta Biotechnol. 3:231-240. [Google Scholar]
- 16.Kukor, J. J., and R. H. Olsen. 1996. Catechol 2,3-dioxygenases functional in oxygen-limited (hypoxic) environments. Appl. Environ. Microbiol. 62:1728-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 18.Leander, M., T. Vallaeys, and R. R. Fulthorpe. 1998. Amplification of putative chlorocatechol dioxygenase gene fragments from alpha- and beta-proteobacteria. Can. J. Microbiol. 44:482-486. [DOI] [PubMed] [Google Scholar]
- 19.Mars, A. E., J. Kingma, S. R. Kaschabek, W. Reineke, and D. B. Janssen. 1999. Conversion of 3-chlorocatechol by various catechol 2,3-dioxygenases and sequence analysis of the chlorocatechol dioxygenase region of Pseudomonas putida GJ31. J. Bacteriol. 181:1309-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mars, A. E., T. Kasberg, S. R. Kaschabek, M. H. van Agteren, D. B. Janssen, and W. Reineke. 1997. Microbial degradation of chloroaromatics: use of the meta-cleavage pathway for mineralization of chlorobenzene. J. Bacteriol. 179:4530-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merkel, P., G. Teutsch, H. Weiss, and H. H. Rijnaart. 2000. Innovative reactive barrier technologies for regional contaminated groundwater, p. 532-540. In Proceedings of the Seventh International FZK/TNO Conference on Contaminated Soil (ConSoil 2000), vol. 1. Thomas Telford Publishing, London, United Kingdom.
- 22.Ogawa, N., and K. Miyashita. 1999. The chlorocatechol-catabolic transposon Tn5707 of Alcaligenes eutrophus NH9, carrying a gene cluster highly homologous to that in the 1,2,4-trichlorobenzene-degrading bacterium Pseudomonas sp. strain P51, confers the ability to grow on 3-chlorobenzoate. Appl. Environ. Microbiol. 65:724-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Potrawfke, T., J. Armengaud, and R. M. Wittich. 2001. Chlorocatechols substituted at positions 4 and 5 are substrates of the broad-spectrum chlorocatechol 1,2-dioxygenase of Pseudomonas chlororaphis RW71. J. Bacteriol. 182:997-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlömann, M. 1994. Evolution of chlorocatechol catabolic pathways. Conclusions to be drawn from comparisons of lactone hydrolases. Biodegradation 5:301-321. [DOI] [PubMed] [Google Scholar]
- 25.Sei, K., K. Asano, N. Tateishi, K. Mori, M. Ike, and M. Fujita. 1999. Design of PCR primers and gene probes for the general detection of bacterial populations capable of degrading aromatic compounds via catechol cleavage pathways. Biosci. Bioeng. 88:542-550. [DOI] [PubMed] [Google Scholar]
- 26.van der Meer, J. R. 1994. Genetic adaptation of bacteria to chlorinated aromatic compounds. FEMS Microbiol Rev. 15:239-249. [DOI] [PubMed] [Google Scholar]
- 27.van der Meer, J. R. 1997. Evolution of novel metabolic pathways for the degradation of chloroaromatic compounds. Antonie Leeuwenhoek 71:159-178. [DOI] [PubMed] [Google Scholar]
- 28.van der Meer, J. R., C. Werlen, S. F. Nishino, and J. C. Spain. 1998. Evolution of a pathway for chlorobenzene metabolism leads to natural attenuation in contaminated groundwater. Appl. Environ. Microbiol. 64:4185-4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Hylckama Vlieg, J. E., G. J. Poelarends, A. E. Mars, and D. B. Janssen. 2000. Detoxification of reactive intermediates during microbial metabolism of halogenated compounds. Curr. Opin. Microbiol. 3:257-262. [DOI] [PubMed] [Google Scholar]
- 30.Vogt, C., A. Alfreider, H. Lorbeer, J. Ahlheim, B. Feist, O. Böhme, H. Weiss, W. Babel, and L. Wünsche. 2002. Two pilot plant reactors designed for the in situ bioremediation of chlorobenzene-contaminated ground water: hydrogeological and chemical characteristics and bacterial consortia. Water Air Soil Pollut. Focus 2:161-170. [Google Scholar]