Abstract
The chaperone–usher pathway directs the formation of adhesive surface fibres in numerous pathogenic Gram-negative bacteria. The fibres or pili consist exclusively of protein subunits that, before assembly, form transient complexes with a chaperone in the periplasm. In these chaperone:subunit complexes, the chaperone donates one β-strand to complete the imperfect immunoglobulin-like fold of the subunit. During pilus assembly, the chaperone is replaced by a polypeptide extension of another subunit in a process termed ‘donor strand exchange' (DSE). Here we show that DSE occurs in a concerted reaction in which a chaperone-bound acceptor subunit is attacked by another chaperone-bound donor subunit. We provide evidence that efficient DSE requires interactions between the reacting subunits in addition to those involving the attacking donor strand. Our results indicate that the pilus assembly platforms in the outer membrane, referred to as ushers, catalyse fibre formation by increasing the effective concentrations of donor and acceptor subunits.
Keywords: chaperone–usher pathway, FimA, FimC, type 1 pili
Introduction
A broad variety of Gram-negative pathogens use the chaperone–usher pathway to assemble adhesive surface fibres, which are composed of up to several thousand protein subunits (Vetsch & Glockshuber, 2005). Such adhesive pili target pathogenic bacteria to the site of infection and thus have a crucial role during pathogenesis. Assembly of these adhesive fibres in vivo requires a soluble periplasmic chaperone and a molecular assembly platform (usher) that is embedded in the outer membrane. The periplasmic chaperones bind to non-native subunits, catalyse subunit folding (Vetsch et al, 2004) and remain bound to the native subunits to direct them to the usher (Dodson et al, 1993). The usher then mediates incorporation of pilus subunits into the growing fibre and translocation of folded subunits to the cell surface (Norgren et al, 1987). X-ray structures of chaperone:subunit complexes have shown that the subunits of adhesive pili share an immunoglobulin-like fold that lacks the carboxy-terminal β-strand, which creates a deep hydrophobic groove on the surface of the subunits. In chaperone:subunit complexes, this groove is occupied by a β-strand segment of the chaperone that complements the fold of the subunit. During incorporation of the subunit into the pilus, the chaperone is replaced by an amino-terminal, previously disordered extension of the incoming subunit in a reaction termed ‘donor strand exchange' (DSE; Choudhury et al, 1999; Sauer et al, 1999, 2002; Zavialov et al, 2003).
Although the incorporation of subunits into the growing fibre is the central process of pilus assembly, the mechanism of DSE has remained unknown. Two alternative models of DSE have been proposed (Zavialov et al, 2003): (i) a sequential mechanism in which release of the chaperone that caps the acceptor subunit precedes association with another subunit; and (ii) a concerted mechanism in which the chaperone bound to the acceptor subunit is released concurrently with the insertion of the donor strand from the attacking subunit into the hydrophobic groove of the acceptor subunit.
Here we have used the type 1 pilus system from uropathogenic Escherichia coli to investigate the mechanism of DSE. We found that the main subunit FimA is in a highly dynamic binding equilibrium with the chaperone FimC. Monomeric FimA quickly binds to the chaperone, but it is virtually unable to accept the donor strand of another subunit. Our results indicate that the acceptor subunit remains bound to the chaperone while interactions with the donor subunit are established, and that the donor strand of the attacking subunit replaces the donor strand of the chaperone in a concerted reaction.
Results
The FimC:FimA binding equilibrium is highly dynamic
To get an insight into the kinetic stability of pilus chaperone:subunit complexes, we focused on the interaction between the chaperone FimC and the main structural subunit FimA from type 1 pili. We performed a competition experiment in which we mixed the purified FimCHis:FimA complex (FimCHis is a functional His-tagged variant of FimC; supplementary Fig 1 online) with a ninefold molar excess of FimC. Analysis of the reaction products by fast ion-exchange chromatography showed that relaxation towards the new equilibrium was completed after 3 min (Fig 1A,B). Once the new equilibrium was attained, ∼90% and ∼10% of FimA were found to be in complex with FimC and FimCHis, respectively, demonstrating that the affinity of FimA towards FimC and FimCHis is the same (Fig 1B). The fact that the sum of the concentrations of FimCHis:FimA and FimC:FimA was constant throughout the individual reactions shows that virtually no FimC:FimA:FimA complex is formed on the timescale relevant for chaperone:subunit dissociation. To test whether the transfer of FimA from FimCHis to FimC occurred through direct attack of FimCHis:FimA by FimC or through sequential dissociation of the subunit from the chaperone and subsequent rebinding, we performed the reaction with two different initial protein concentrations, but with the same molar ratio of the reactants (Fig 1B). The two reactions yielded virtually identical time constants (τ), proving that τ depends only on the dissociation rate (koff) and the ratio between FimC and FimCHis. Consequently, chaperone exchange occurred through dissociation of the FimCHis:FimA complex, followed by rebinding of FimA to FimC (Fig 1C). The half-life of the complex (ln 2/koff) was 35 s at 25°C and less than 5 s at 37°C (supplementary Fig 2 online). Therefore, capping of the hydrophobic groove of FimA is much more dynamic than anticipated (Sauer et al, 2002).
Figure 1.
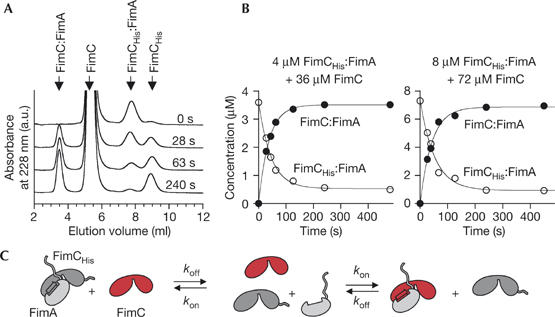
Dissociation dynamics of the FimCHis:FimA complex at 25°C. For determination of the dissociation rate (koff), the FimCHis:FimA complex was mixed with a ninefold excess of FimC. Relaxation towards equilibrium was measured by ion-exchange chromatography. (A) Representative elution profiles. (B) Kinetics of chaperone exchange at initial FimCHis:FimA concentrations of 4 and 8 μM yielded time constants of 43±2 s and 49±6 s, respectively. (C) Reaction scheme for the replacement of FimCHis (grey) by FimC (red) in a chaperone:subunit complex.
We found that freshly purified FimA loses its ability to bind to FimCHis during storage (supplementary Fig 3 online). As this loss of binding competence of isolated FimA strongly impedes titration experiments with FimCHis, we deduced the affinity of the FimCHis:FimA complex in a sample containing equal amounts of both proteins. Freshly purified FimCHis:FimA complex (4 μM, 25°C) was subjected to fast ion-exchange chromatography. The peak corresponding to the complex accounted for 90% of the total absorbance, whereas the peak representing monomeric FimCHis contributed to 6% of the total absorbance (supplementary Fig 4A online, profile at top). These numbers translate into a dissociation constant (KD) of less than 40 nM. This value represents an upper limit for the KD, as it cannot be ruled out that a small fraction of FimCHis:FimA dissociated during chromatography. Together with the measured koff, the estimated KD defines a lower limit for the association rate (kon in Fig 1C) of 5 × 105 M−1 s−1.
Donor strand exchange occurs in a concerted reaction
To analyse the mechanism of DSE, oligomerization of FimA was followed by ion-exchange chromatography. We first incubated the FimCHis:FimA complex alone. The elution profiles (supplementary Fig 4A online) show that the concentration of FimCHis:FimA decreased very slowly on the timescale of hours, while monomeric FimCHis appeared with the same rate (Fig 2A). Under these conditions, not only FimCHis:FimA:FimA complexes are formed, but also, as the reaction progresses, larger complexes with three or more subunits bound to FimCHis. To reduce the complexity of the reaction, we repeated the experiment in the presence of a 9- or 18-fold molar excess of FimAA (Fig 2B,C). FimAA is an artificial FimA variant that is elongated at its C terminus by a short linker and a further FimA donor strand that stabilizes the fold of the subunit through intramolecular donor strand complementation. Therefore, FimAA can neither bind to the chaperone nor self-polymerize, but it should retain the ability to interact with FimA (cf. Fig 2G). Indeed, we found that addition of a ninefold excess of FimAA to FimCHis:FimA led to markedly faster release of FimCHis from the FimCHis:FimA complex (compare Fig 2A and B), suggesting that incubation of FimCHis:FimA with an excess of FimAA primarily yields monomeric FimCHis and FimAA:FimA heterodimers.
Figure 2.
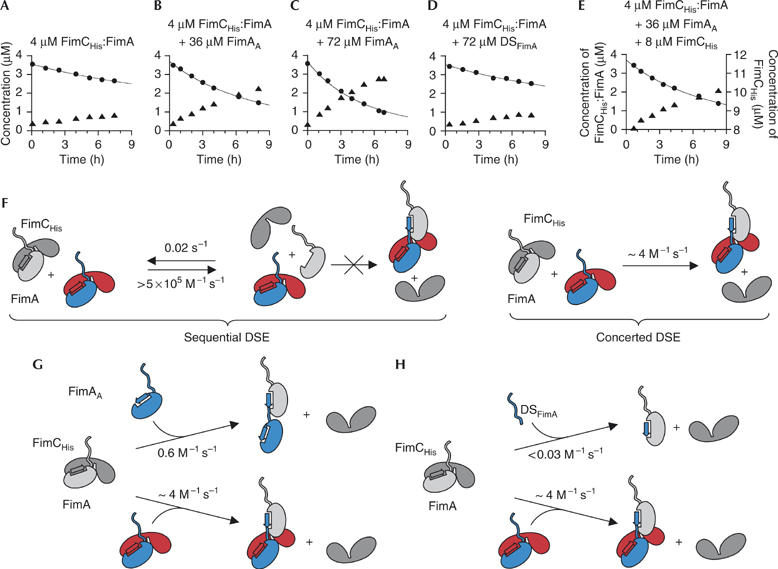
Kinetic analysis of donor strand exchange at 25°C. (A–E) The FimCHis:FimA complex was incubated either alone or with FimAA, a mixture of FimAA and FimCHis or the donor strand of FimA (DSFimA). After different reaction times, the concentrations of FimCHis:FimA and free FimCHis were measured by ion-exchange chromatography (filled circles and triangles, respectively). The solid lines represent fits to a second-order reaction (complex alone) or parallel second-order and pseudo-first-order reactions (all other reactions). (F–H) DSE reaction schemes. Components with the attacking donor strand are coloured and the FimCHis:FimA complex under attack is depicted in grey. In (F), the two alternative mechanisms of DSE are shown for the reaction in which FimCHis:FimA alone undergoes DSE. When FimAA or DSFimA is also present, parallel reactions occur (G,H). DSE, donor strand exchange.
To distinguish between a sequential and a concerted mechanism of DSE (Fig 2F), we repeated the above reaction in the presence of an excess of free FimCHis (4 μM FimCHis:FimA, 36 μM FimAA, 8 μM FimCHis). The KD of the FimCHis:FimA complex (<40 nM) predicts that the concentration of monomeric FimA is decreased by more than 19-fold in the presence of an excess of FimCHis. Consequently, if monomeric FimA was a reaction intermediate in DSE, the excess of FimCHis should strongly decrease the rate of DSE. However, we observed that the DSE reaction proceeded with exactly the same rate as in the absence of excess FimCHis (Fig 2E). This provides strong evidence that DSE occurs through a concerted mechanism, in which only chaperone-bound FimA can accept the attacking donor strand of another subunit (Fig 2F, right). The data also show that monomeric FimA, although existing at equilibrium (Fig 1), is inactive as an acceptor subunit. Consequently, the kinetics of all DSE experiments were evaluated on the basis of a second-order mechanism.
The rate constant of DSE obtained is ∼4 M−1 s−1 for the reaction in which only the FimCHis:FimA complex was present (Fig 2F). In contrast, DSE leading to formation of FimAA:FimA complex occurs with a rate constant of only 0.6 M−1 s−1 (Fig 2G; see also the Methods section). The fact that DSE is about sevenfold faster when the donor subunit is bound to FimC indicates that FimC bound to the attacking subunit facilitates DSE, possibly by positioning the attacking donor strand in close proximity to the hydrophobic groove of the acceptor subunit. To test the hypothesis that additional interactions to those formed by the attacking donor strand are required for efficient DSE, we examined how fast a synthetic peptide corresponding to the isolated donor strand of FimA (DSFimA) replaces the chaperone from the FimCHis:FimA complex. Fig 2D shows that an 18-fold molar excess of the peptide increased the consumption of the FimCHis:FimA only marginally. DSFimA is therefore at least 20-fold less efficient than FimAA in replacing the chaperone from FimA (Fig 2G,H). Head-to-tail interactions between the two reacting pilus subunits are consequently important for efficient DSE, potentially because these interactions can be established before the actual DSE.
Role of the N-terminal usher domain during DSE
To define the role of the periplasmic, N-terminal domain of the usher FimD (FimDN) that specifically recognizes FimC:subunit complexes (Nishiyama et al, 2003), we again used the reaction between 4 μM FimCHis:FimA and 36 μM FimAA, which primarily yields the FimA:FimAA complex. When this experiment was performed in the presence of FimDN, formation of the FimA:FimAA complex was strongly suppressed (Fig 3A). This demonstrates that FimDN cannot be the site of the usher that binds to the chaperone:subunit complex to be attacked. Instead, FimDN probably represents the binding site for incoming chaperone:subunit complexes. This role is in good agreement with the full exposure of FimDN to the periplasm (Nishiyama et al, 2003), the predicted flexible orientation of the domain relative to the membrane-embedded part of the usher (Nishiyama et al, 2005) and the observation that the tail of FimDN seems to tether the donor strand of the chaperone to the interactive groove of the subunit (Fig 3B).
Figure 3.
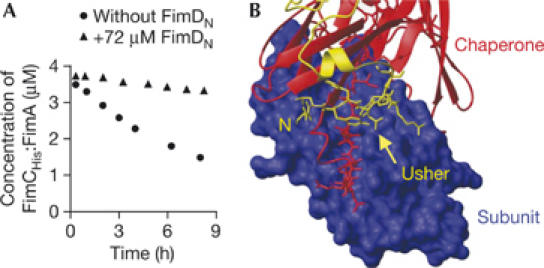
Role of the amino-terminal usher domain (FimDN) during donor strand exchange. (A) A mixture of 4 μM FimCHis:FimA and 36 μM FimAA was incubated at 25°C in the absence or presence of 72 μM FimDN. The concentration of FimCHis:FimA during the reaction was measured by ion-exchange chromatography. (B) Close-up of the X-ray structure of the ternary complex FimDN:FimC:FimHp, highlighting the binding interface between the subunit FimHP (blue surface representation), FimC (red; ribbon diagram, except for the donor strand, for which the side chains are depicted) and FimDN (yellow; ribbon diagram, except for the N-terminal nine residues, for which the side chains are shown). FimHP, the pilin domain of the subunit FimH, has the same fold as FimA. N, amino terminus of FimDN.
Discussion
The key observation in our DSE experiments is the finding that an excess of the free chaperone does not influence the kinetics of DSE. This has two important implications. First, it shows that the formation of FimA:FimA complexes is virtually irreversible, further corroborating measurements indicating that the thermodynamic stability of subunit:subunit complexes is much higher than that of chaperone:subunit complexes (Vetsch et al, 2004; Zavialov et al, 2005). Second, it argues strongly for a concerted mechanism of DSE. Moreover, our data show that efficient DSE requires interactions between the two reacting chaperone:subunit complexes in addition to the contacts involving the donor strand of the incoming subunit. The fact that the FimCHis:FimA complex attacks another FimCHis:FimA complex about sevenfold more efficiently than FimAA indicates that the chaperone bound to the donor subunit increases the reactivity of the attacking subunit during DSE, potentially by interacting with the acceptor subunit. This interaction might be similar to the contacts observed in the X-ray structure of the ternary Caf1M:Caf1:Caf1 complex from the F1 antigen system of Yersinia pestis (Zavialov et al, 2003), in which the chaperone Caf1M bound to the donor subunit also interacts with the acceptor subunit (560 Å2 interface). The same X-ray structure shows a defined head-to-tail interface between the two subunits. Such interactions between donor and acceptor subunit seem to be crucial for DSE as well, because the isolated donor strand peptide (DSFimA) replaced FimCHis from the FimCHis:FimA complex at least 20-fold slower than FimAA.
Co-purification experiments with ushers indicated that the last incorporated subunit in the pilus remains capped by the chaperone (Saulino et al, 2000; Ng et al, 2004). It is thus conceivable that DSE occurs in a concerted manner also in the presence of the usher in vivo. Pilus assembly in vivo occurs within minutes (Dodd & Eisenstein, 1984), whereas DSE proceeds on a timescale of hours in vitro (Fig 2). It is therefore likely that the usher catalyses DSE in vivo. The simplest mechanism by which the usher could act as a catalyst is that it brings an attacking chaperone:subunit complex in close proximity to the chaperone-capped, last incorporated subunit of the growing pilus, which would increase the effective concentrations of donor and acceptor subunits. This model (Fig 4) predicts that the chaperone that capped the donor subunit during a DSE cycle becomes the chaperone that caps the acceptor subunit at the growing pilus end in the next catalytic cycle.
Figure 4.
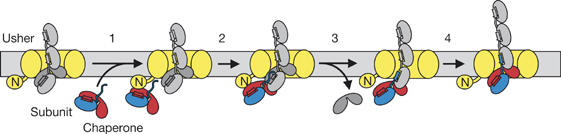
Model of donor strand exchange and pilus elongation in vivo. An incoming chaperone:subunit complex binds to the amino-terminal domain (N) of the usher (step 1). The donor subunit (blue) then attacks (step 2) the acceptor subunit (grey). The transient complex in which the acceptor subunit is in contact with both the donor strand of the chaperone and the donor strand of the attacking subunit might represent a transition state or a short-lived reaction intermediate. After release of the chaperone from the acceptor subunit (step 3), the pilus translocates by the length of one subunit towards the cell surface (step 4).
It is likely that the ability of the usher to induce high local concentrations of chaperone:subunit complexes is a key component of its catalytic power. The usher could further increase the efficiency of DSE by stabilizing interactions between the attacking and the attacked chaperone:subunit complex or by destabilizing the interactions between the acceptor subunit and its capping chaperone. The mechanism of fibre formation described here represents a valuable working model for addressing these intriguing questions on usher-catalysed pilus assembly by use of an in vitro reconstituted assembly system.
Methods
Protein expression and purification. The methods used are described at EMBO reports online (http://www.emboreports.org).
Kinetic and thermodynamic stability of the FimCHis:FimA complex. To measure the dissociation rate of FimCHis:FimA, the complex was mixed with a ninefold molar excess of FimC and incubated in 20 mM Tris–HCl, pH 8.0, at 25°C. After different reaction times, samples were withdrawn, rapidly cooled (<4°C) and immediately applied to a Resource S 1 ml column in 20 mM MOPS–NaOH, pH 6.7, at 4°C. FimC:FimA and FimCHis:FimA eluted at 30 mM (2.0 min) and 110 mM NaCl (4.8 min), respectively. Protein concentrations were calculated using the corresponding extinction coefficients at 228 nm (FimC, 176,000 M−1 cm−1; FimCHis, 179,000 M−1 cm−1; FimC:FimA, 244,000 M−1 cm−1; FimCHis:FimA, 247,000 M−1 cm−1) and the known total concentrations of FimA, FimC and FimCHis. The observed kinetics were described by single exponential functions. The dissociation rate constant koff is a function of the measured time constant (τ) and the total concentrations of FimC and FimCHis according to
To estimate the association rate between FimCHis and FimA, the freshly purified complex was incubated (3 min) in 20 mM Tris–HCl, pH 8.0, at 25°C and applied to a Resource S 1 ml column in 20 mM MOPS–NaOH, pH 6.7, at 4°C. Whereas both FimCHis:FimA and FimCHis bind to the column under these conditions, monomeric FimA (extinction coefficient at 228 nm: 67,000 M−1 cm−1) does not bind and is therefore removed in seconds. The bound components were eluted with a linear NaCl gradient (FimCHis:FimA at 110 mM (2.9 min), FimCHis at 140 mM (3.4 min)) and their concentrations were calculated as described above.
Kinetics of donor strand exchange. Freshly purified FimCHis:FimA (4 μM) was incubated in 20 mM Tris–HCl, pH 8.0, at 25°C either alone or in the presence of FimAA (36 or 72 μM), DSFimA (72 μM, identical to the first 19 residues of FimA, >95% pure, purchased from AnaSpec, San Jose, CA, USA), FimCHis (8 μM) or FimDN (72 μM). After various times, samples were withdrawn and analysed as described above. The initial phase of the reaction involving exclusively FimCHis:FimA was described as a second-order reaction generating predominantly FimCHis:FimA:FimA. The reactions in which FimAA was also present were fitted according to the following equation, which takes into account that FimCHis:FimA reacts with itself (second-order reaction) and also with excess FimAA (pseudo-first-order reaction):
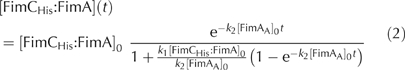 |
[FimCHis:FimA]0 and [FimAA]0 are the initial concentrations of FimCHis:FimA and FimAA, respectively; k1 and k2 are the association rates between two FimCHis:FimA complexes and between FimAA and FimCHis:FimA, respectively.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Figures
Supplementary methods
Acknowledgments
We thank A. Fritz for excellent technical assistance, R. Brunisholz (Functional Genomics Center Zurich) for Edman sequencing and U. Grauschopf and E. Weber-Ban for valuable advice and critical reading of the manuscript. This work was supported by the Swiss National Science Foundation (SNSF, 3100A0-100787) and the NCCR Structural Biology Program of the SNSF.
References
- Choudhury D, Thompson A, Stojanoff V, Langermann S, Pinkner J, Hultgren SJ, Knight SD (1999) X-ray structure of the FimC–FimH chaperone–adhesin complex from uropathogenic Escherichia coli. Science 285: 1061–1066 [DOI] [PubMed] [Google Scholar]
- Dodd DC, Eisenstein BI (1984) Kinetic analysis of the synthesis and assembly of type 1 fimbriae of Escherichia coli. J Bacteriol 160: 227–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson KW, Jacob-Dubuisson F, Striker RT, Hultgren SJ (1993) Outer-membrane PapC molecular usher discriminately recognizes periplasmic chaperone–pilus subunit complexes. Proc Natl Acad Sci USA 90: 3670–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng TW, Akman L, Osisami M, Thanassi DG (2004) The usher N terminus is the initial targeting site for chaperone–subunit complexes and participates in subsequent pilus biogenesis events. J Bacteriol 186: 5321–5331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama M, Vetsch M, Puorger C, Jelesarov I, Glockshuber R (2003) Identification and characterization of the chaperone–subunit complex-binding domain from the type 1 pilus assembly platform FimD. J Mol Biol 330: 513–525 [DOI] [PubMed] [Google Scholar]
- Nishiyama M et al. (2005) Structural basis of chaperone–subunit complex recognition by the type 1 pilus assembly platform FimD. EMBO J 24: 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norgren M, Baga M, Tennent JM, Normark S (1987) Nucleotide sequence, regulation and functional analysis of the papC gene required for cell surface localization of Pap pili of uropathogenic Escherichia coli. Mol Microbiol 1: 169–178 [DOI] [PubMed] [Google Scholar]
- Sauer FG, Futterer K, Pinkner JS, Dodson KW, Hultgren SJ, Waksman G (1999) Structural basis of chaperone function and pilus biogenesis. Science 285: 1058–1061 [DOI] [PubMed] [Google Scholar]
- Sauer FG, Pinkner JS, Waksman G, Hultgren SJ (2002) Chaperone priming of pilus subunits facilitates a topological transition that drives fiber formation. Cell 111: 543–551 [DOI] [PubMed] [Google Scholar]
- Saulino ET, Bullitt E, Hultgren SJ (2000) Snapshots of usher-mediated protein secretion and ordered pilus assembly. Proc Natl Acad Sci USA 97: 9240–9245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetsch M, Glockshuber R (2005) Formation of adhesive pili by the chaperone–usher pathway. In Protein Folding Handbook, Buchner J, Kiefhaber T (eds) Vol 2, pp 965–986. Weinheim, Germany: Wiley-VCH [Google Scholar]
- Vetsch M, Puorger C, Spirig T, Grauschopf U, Weber-Ban EU, Glockshuber R (2004) Pilus chaperones represent a new type of protein-folding catalyst. Nature 431: 329–333 [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Berglund J, Pudney AF, Fooks LJ, Ibrahim TM, MacIntyre S, Knight SD (2003) Structure and biogenesis of the capsular F1 antigen from Yersinia pestis: preserved folding energy drives fiber formation. Cell 113: 587–596 [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Tischenko VM, Fooks LJ, Brandsdal BO, Aqvist J, Zav'yalov VP, Macintyre S, Knight SD (2005) Resolving the energy paradox of chaperone/usher-mediated fibre assembly. Biochem J 389: 685–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary methods


