Abstract
On starvation, Dictyostelium cells form a terminally differentiated structure, known as the fruiting body, which comprises stalk and spore cells. Their precursors–prestalk and prespore cells–are spatially separated and accessible in a migratory structure known as the slug. This simplicity and manipulability has made Dictyostelium attractive to both experimental and theoretical developmental biologists. However, this outward simplicity conceals a surprising degree of developmental sophistication. Multiple prestalk subtypes are formed and undertake a co-ordinated series of morphogenetic cell movements to generate the fruiting body. This review describes recent advances in understanding the signalling pathways that generate prestalk-cell heterogeneity, focusing on the roles of the prestalk-cell inducer differentiation-inducing factor-1 (DIF-1), the tip inducer cAMP and the transcription factors that mediate their actions; these include signal transducer and activator of transcription (STAT) proteins, basic leucine zipper (bZIP) proteins and a Myb protein of a class previously described only in plants.
Keywords: bZIP, Dictyostelium, Myb, prestalk, STAT
Introduction
As with vertebrate embryos, morphogenetic cell movement is a major shaping force in the primary stages of Dictyostelium pattern formation. However, during culmination, formation and extension of the stalk occurs in a position-dependent manner that resembles pattern formation in non-regulative animal embryos and higher plants. This mechanistic diversity accords well with the wide array of signalling proteins, some typical of animals, others typical of plants, that direct differentiation in Dictyostelium.
Dictyostelium live as isolated amoebae in soil and leaf litter. When their bacterial food source is exhausted, as many as 100,000 cells aggregate in response to pulsatile cAMP signalling. Cells within the aggregate differentiate into prestalk or prespore cells and form a slug-shaped structure. After a variable period of migration, the slug cells undertake culmination, which is a final round of differentiation and morphogenetic cell movement that results in formation of the fruiting body. This contains two basic cell types: spore and stalk cells. Spore cells are dormant, environmentally resistant cells that are located at the top of the stalk. Stalk cells are highly vacuolated dead cells that form the stalk itself, and also the conical basal disc into which the foot of the stalk is embedded.
The prestalk and prespore cells can be conveniently studied at the migratory slug stage. Most of the prestalk cells are located in the front one-fifth of the slug, whereas all of the prespore cells are located in the rear four-fifths (Fig 1). One apparent difference between the prespore and prestalk populations is in their degree of heterogeneity: there are several prestalk-cell subtypes, whereas, apart from an anteroposterior gradient in their utilization efficiency of certain prespore–promoter constructs (Haberstroh & Firtel, 1990), the prespore cells seem to be a homogeneous population.
Figure 1.
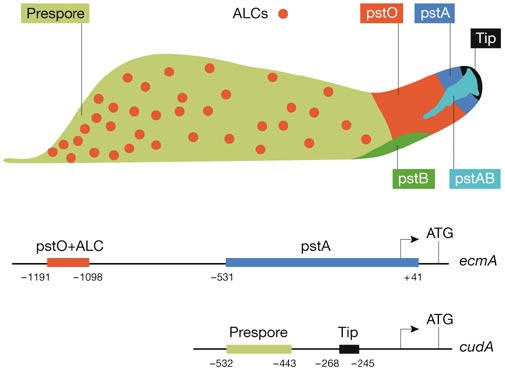
Distribution of the prestalk-cell types, and organization of the ecmA and cudA promoters.This figure depicts a Dictyostelium slug and the various cell types in a highly schematic form. The cells in the prestalk region are represented as blocks of colour, as are the prespore cells. The anterior-like cells (ALCs) are shown individually, but the drawing is not to scale. Below the slug are representations of two of the three promoter fragments that have been used–as part of reporter constructs–to deduce this organization of cell types. The other paradigmatic prestalk promoter, the ecmB promoter, is not shown, but directs strong expression in pstAB cells and weaker expression in pstB cells.
The prestalk-cell subtypes
The prestalk zone stains strongly with vital dyes, such as neutral red, and the cells can be seen to move down through the prespore mass to form the stalk at culmination. Anterior-like cells (ALCs; Fig 1) are scattered in the rear prespore region of the slug and also stain with vital dyes. They too terminally differentiate into stalk cells, but, during culmination, move to form cup-shaped structures that cradle the spore head. Studies using promoter fusions have extended the early vital-dye studies by revealing the existence of multiple prestalk-cell subtypes that differ in their patterns of gene expression.
Initially, the promoters of two genes encoding closely related extracellular-matrix proteins, ecmA and ecmB, were used to identify and study these cell types. Cap-site proximal ecmA promoter sequences (Early et al, 1993) direct reporter expression in the most anterior prestalk cells, the pstA cells (Fig 1). A more distal promoter region directs expression in the cells that make up the rear of the prestalk region, the pstO cells, and also in the ALCs. A reporter fusion containing the promoter of the ecmB gene (Ceccarelli et al, 1991) is expressed in the pstB and pstAB cells (Fig 1). These two cell types move at culmination to form, respectively, the inner and outer parts of the basal disc (Jermyn & Williams, 1996; Dormann et al, 1996). These subtypes have now been confirmed by in situ hybridization by using probes from many different genes (Maeda et al, 2003; Yamada et al, 2005). Indeed, these studies suggest that the prestalk subtypes are further divisible, because some probes hybridize to regions that do not fully overlap with the previously defined subregions.
A more complete catalogue of the cell-type-specific protein sequences, deduced from further in situ studies, will help to define the biological significance of the subtypes. However, there are clues from their behaviour during slug formation, migration and culmination. One feature that clearly distinguishes the subtypes is their different patterns of movement. In addition to the differences between pstB and pstAB cells noted above, the pstO cells orbit around the long axis of the slug (Abe et al, 1994; Siegert & Weijer, 1992). These differences in movement pattern might partly explain the need for multiple prestalk-cell subtypes, each of which presumably expresses the gene products needed to generate its particular pattern of cell movement.
DIF-1 signalling and pstO cell differentiation
Differentiation-inducing factor-1 (DIF-1) is a chlorinated hexaphenone (Fig 2A) that induces stalk-cell differentiation in a monolayer-assay system (reviewed by Kay et al, 1999). DIF-1 is produced by the prespore cells (Kay & Thompson, 2001). Its concentration in the slug is regulated by a negative-feedback loop, whereby it rapidly induces the production of DIF-1 dechlorinase (DIFase), which catalyses DIF-1 inactivation (Fig 2B; Kay et al, 1993). DIF-1 acts antagonistically to cAMP by repressing prespore differentiation and directing a proportion of the cell population to differentiate as prestalk cells (Kay & Jermyn, 1983). As an early part of the latter process, DIF-1 induces transcription of the ecmA gene (Williams et al, 1987).
Figure 2.
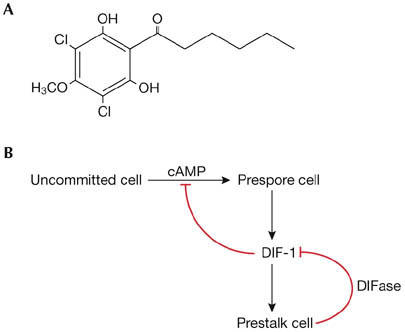
DIF-1 and the DIF-1-control circuit.(A) DIF-1 structure. (B) Cell-type-specific metabolism of DIF-1 and the DIF-1 cAMP antagonists.
The final step in DIF-1 biosynthesis is the addition of a methyl group. A null mutant for the cognate methyl transferase, the dmtA− mutant, produces no detectable DIF-1 (Thompson & Kay, 2000). These cells are defective in gene expression directed by the complete ecmA promoter, but only in pstO cells, as expression in pstA cells occurs normally (Thompson & Kay, 2000). Furthermore, dmtA− slugs transformed with a pstO-specific reporter lacZ construct, which contains a distal segment of the ecmA promoter (Fig 1), display no expression. However, expression of a pstO-specific marker is induced when the mutant slugs are exposed to DIF-1. These findings suggest that pstO differentiation is DIF-1-dependent, whereas pstA differentiation is not. This implies a separate signalling molecule that acts to induce pstA differentiation; pharmacological evidence suggests that this factor is, similarly to DIF-1, a polyketide (Serafimidis & Kay, 2005).
The situation becomes more complex when other pstO-specific markers and prespore markers are examined (Maeda et al, 2003). These results suggest that DIF-1 is essential for fully repressing prespore differentiation and for inducing complete pstO differentiation, but is dispensable for the expression of some pstO-specific genes. The identification and analysis of transcription factors that mediate the effects of DIF-1 signalling also indicate an obligate link between DIF-1 signalling and pstO-specific expression of the ecmA gene.
Transcriptional control of prestalk differentiation
Basic leucine zipper proteins. DimA is a member of the basic leucine zipper (bZIP) family of transcription factors, and the dimA-null mutant is defective in DIF-1-induced stalk-cell differentiation (Thompson et al, 2004). As assayed using the complete ecmA promoter, null mutants for dimA display a foreshortened prestalk zone, which is a phenotype that again suggests a selective effect of DIF-1 on pstO differentiation. DimB is a closely related bZIP protein that heterodimerizes with DimA (Huang et al, 2006). DimB binds to two widely separated parts of the ecmA promoter in vitro (Fig 3). In response to DIF-1, it accumulates rapidly in the nucleus and becomes associated with the ecmA promoter, as shown by chromatin immunoprecipitation (ChIP) studies (Zhukovskaya et al, 2006).
Figure 3.
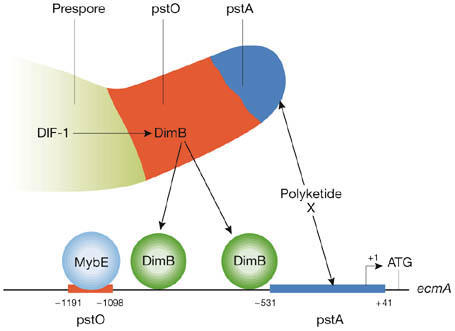
The ecmA promoter and its regulation.This is a representation of the promoter of the ecmA gene, showing the approximate positions of the characterized pstO and pstA regions. There is a second, less well-defined, pstO-specific region in which the known DimB-binding sites lie (Early et al, 1993; Zhukovskaya et al, 2006). DimB was identified in two laboratories (Huang et al, 2006; Zhukovskaya et al, 2006). Both studies showed that the ecmA gene is not DIF-1-inducible in dimB-null strains, but different results were obtained during normal development. This inconsistency presumably arose because the two studies used different Dictyostelium strains that are known to display patterning differences.
DimA fused to green fluorescent protein (GFP) also translocates to the nucleus in response to DIF-1, and the ecmA gene is not DIF-inducible in the dimA-null and dimB-null strains. These observations suggest that DimA and DimB are DIF-1-regulated transcription factors that induce pstO-specific differentiation. Although there is no direct evidence that DimA binds to the ecmA promoter, its heterodimerization with DimB suggests that it also forms part of the transcription complex. The two bZIP proteins also seem likely to function in concert with a novel Myb domain protein, MybE.
MybE. MybE was isolated by affinity chromatography by using a small DNA fragment from within the cap-site distal region of the ecmA promoter that directs DIF-1-inducible prestalk-specific gene expression (Fukuzawa et al, 2006). Most Myb transcription factors have either two or three copies of a DNA-binding domain that contains three symmetrically spaced tryptophan residues. MybE belongs to the SHAQKY family–a large group of green plant proteins that contain a single Myb domain, in which the third conserved tryptophan residue is replaced by a sequence including the consensus motif SHAQKY (Jin & Martin, 1999). MybE is the first SHAQKY family member to be described and characterized outside plants.
The ecmA gene is not DIF-1-inducible in the mybE-null strain, and, at the slug stage, ecmA expression is tightly restricted to the pstA cells–the pstO cells and the ALCs show negligible expression. Again, these data support a specific role for DIF-1 in regulating pstO, but not pstA, cell differentiation. mybE is expressed during growth and throughout development, and a MybE:GFP fusion protein seems to be constitutively nuclear (Y. Yamada and J.G.W., unpublished data). Thus, one possibility, depicted in Fig 3, is that MybE is constitutively bound to the ecmA promoter where it co-operates with DimB in some way to direct DIF-1-inducible transcription.
Signal transducer and activator of transcription c (STATc). The signal transducer and activator of transcription (STAT) proteins are important mediators of phosphotyrosine-regulated signalling in animal cells. Thus far, Dictyostelium is the only non-metazoan in which STATs have been identified and functionally characterized (reviewed in Williams, 2003). STATc is tyrosine phosphorylated and accumulates in the nucleus when cells are treated with DIF-1 (Fukuzawa et al, 2001). Nuclear accumulation occurs as a result of the repression of STATc nuclear export (Fukuzawa et al, 2003). This repression is triggered by the DIF-1-induced phosphorylation of STATc through an as-yet-unidentified tyrosine kinase. As might be expected from its activation by DIF-1, STATc is selectively enriched in the nuclei of pstO cells at the slug stage. Moreover, the STATc-null strain has a defect in pstO differentiation, such that a pstA-specific reporter construct, derived from the ecmA promoter, is ectopically expressed in pstO cells (Fukuzawa et al, 2001).
The above observations suggest that gene expression in the pstO region is controlled by an interplay between MybE, DimA, DimB and STATc. This does not exclude the involvement of other transcription factors and will almost certainly not apply in precisely this form to every prestalk-specific gene. It does, however, provide a framework within which to begin to understand one form of prestalk differentiation. Another prestalk-cell subtype, the differentiation of which is beginning to be understood at a molecular level, is the tip cell (Fig 1).
cAMP signalling, STATa, CudA and tip formation
The slug tip controls behaviour in much the same way as the organizer region of a vertebrate embryo–when grafted to the rear of a recipient slug, it recruits a mass of prespore cells and forms a secondary slug (Raper, 1940). The tip also regulates the choice between continued slug migration and fruiting-body formation (Smith & Williams, 1980) and is the region that directs slug phototaxis (reviewed by Fisher, 1997).
Initially, the tip was defined only by functional assays, using techniques such as grafting. However analysis of CudA, which is a protein that regulates the slug/fruiting-body switch, showed it to be localized in the prespore zone and in a cone of cells at the extreme anterior of the slug (Fukuzawa et al, 1997). Two pieces of genetic evidence suggest that the latter population is made up of tip cells. First, the cudA-null mutant remains as a slug under conditions in which the wild-type slugs culminate; such a mutant is termed a ‘slugger', and, as already mentioned, this switch is controlled by the tip cells. Second, and more tellingly, the slugger phenotype is reversed when CudA is expressed under the control of the ecmA promoter (Fukuzawa et al, 1997). The cone of CudA expression overlaps with the anterior-most pstA cells, suggesting sequential differentiation of a subset of the latter into tip cells.
The only homologues of CudA occur in the related amoebozoan Entamoeba histolytica. The function of CudA is unknown, but it is likely to be a transcription factor because it is located in the nucleus, and the cotC prespore-specific gene shows greatly reduced expression in a cudA-null strain (Fukuzawa et al, 1997). The cudA promoter contains separate regions that direct its expression in prespore and tip cells (Fukuzawa & Williams, 2000). The tip-specific region contains an essential dyad that is a binding site for STATa, which negatively regulates the expression of ecmB (Mohanty et al, 1999), and also regulates several other prestalk-specific and stalk-specific genes (Shimada et al, 2004a, 2004b). STATa is enriched in nuclei located in the slug tip (Araki et al, 1998). In a STATa-null mutant, the expression of CudA in the prespore region is normal, but there is no CudA expression in the tip (Fukuzawa & Williams, 2000). These facts suggest that STATa is the direct activator of CudA expression.
The STATa protein is tyrosine phosphorylated and translocates to the nucleus when cells developing in suspension are treated with cAMP (Araki et al, 1998). The slug tip is believed to control the behaviour of the slug by acting as a source of extracellular cAMP signalling. If cAMP is diffused from a needle into the rear of a slug expressing STATa:GFP, the fusion protein accumulates rapidly in prespore nuclei surrounding the needle (Dormann et al, 2001). A similar phenomenon occurs when the ACA adenylyl cyclase is overexpressed from a semi-constitutive promoter (Verkerke-van Wijk et al, 2001). Moreover, in such slugs, cudA is ectopically expressed. During normal development, expression of the ACA gene becomes restricted to cells at the front of the slug. These observations suggest the pathway shown in Fig 4, whereby localized expression of ACA leads to localized cAMP accumulation, which activates STATa to direct transcription of cudA.
Figure 4.

Regulation of tip-specific gene expression.This figure depicts a proposed pathway for tip-cell-specific gene expression of cudA. The tip-specific element contains an imperfect dyad, AGATAATCT, which is essential for expression and binds specifically to STATa (Fukuzawa & Williams, 2000). The proposed pathway postulates an elevated cAMP concentration in the tip region, caused by the tissue-specific expression of the ACA adenylyl cyclase. This leads to localized activation of STATa and expression of cudA.
Conclusions and perspectives
The development of Dictyostelium presents a microcosm of pattern-formation processes in higher organisms. Although the extracellular signalling molecules that direct the process are different, some of the intracellular signalling components are conserved. One remaining challenge is to link the transcription factors that direct pstO and tip-specific gene expression to upstream-signalling components. Another is to start defining the transcriptional processes that direct prespore pstA, pstB and pstAB differentiation.

Jeffrey G. Williams
References
- Abe T, Early A, Siegert F, Weijer C, Williams J (1994) Patterns of cell movement within the Dictyostelium slug revealed by cell type-specific, surface labeling of living cells. Cell 77: 687–699 [DOI] [PubMed] [Google Scholar]
- Araki T, Gamper M, Early A, Fukuzawa M, Abe T, Kawata T, Kim E, Firtel RA, Williams JG (1998) Developmentally and spatially regulated activation of a Dictyostelium STAT protein by a serpentine receptor. EMBO J 17: 4018–4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli A, Mahbubani H, Williams JG (1991) Positively and negatively acting signals regulating stalk cell and anterior-like cell differentiation in Dictyostelium. Cell 65: 983–989 [DOI] [PubMed] [Google Scholar]
- Dormann D, Siegert F, Weijer CJ (1996) Analysis of cell movement during the culmination phase of Dictyostelium development. Development 122: 761–769 [DOI] [PubMed] [Google Scholar]
- Dormann D, Abe T, Weijer CJ, Williams J (2001) Inducible nuclear translocation of a STAT protein in Dictyostelium prespore cells: implications for morphogenesis and cell-type regulation. Development 128: 1081–1088 [DOI] [PubMed] [Google Scholar]
- Early AE, Gaskell MJ, Traynor D, Williams JG (1993) Two distinct populations of prestalk cells within the tip of the migratory Dictyostelium slug with differing fates at culmination. Development 118: 353–362 [DOI] [PubMed] [Google Scholar]
- Fisher PR (1997) Genetics of phototaxis in a model eukaryote, Dictyostelium discoideum. BioEssays 19: 397–407 [DOI] [PubMed] [Google Scholar]
- Fukuzawa M, Williams JG (2000) Analysis of the promoter of the cudA gene reveals novel mechanisms of Dictyostelium cell type differentiation. Development 127: 2705–2713 [DOI] [PubMed] [Google Scholar]
- Fukuzawa M, Hopper N, Williams J (1997) cudA: a Dictyostelium gene with pleiotropic effects on cellular differentiation and slug behaviour. Development 124: 2719–2728 [DOI] [PubMed] [Google Scholar]
- Fukuzawa M, Araki T, Adrian I, Williams JG (2001) Tyrosine phosphorylation-independent nuclear translocation of a Dictyostelium STAT in response to DIF signaling. Mol Cell 7: 779–788 [DOI] [PubMed] [Google Scholar]
- Fukuzawa M, Abe T, Williams JG (2003) The Dictyostelium prestalk cell inducer DIF regulates nuclear accumulation of a STAT protein by controlling its rate of export from the nucleus. Development 130: 797–804 [DOI] [PubMed] [Google Scholar]
- Fukuzawa M, Zhukovskaya NV, Yamada Y, Araki T, Williams JG (2006) Regulation of Dictyostelium prestalk-specific gene expression by a SHAQKY family MYB transcription factor. Development 133: 1715–1724 [DOI] [PubMed] [Google Scholar]
- Haberstroh L, Firtel RA (1990) A spatial gradient of expression of a cAMP-regulated prespore cell-type-specific gene in Dictyostelium. Genes Dev 4: 596–612 [DOI] [PubMed] [Google Scholar]
- Huang E, Blagg SL, Keller T, Katoh M, Shaulsky G, Thompson CR (2006) bZIP transcription factor interactions regulate DIF responses in Dictyostelium. Development 133: 449–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jermyn KA, Williams JG (1996) The initiation of basal disc formation in Dictyostelium discoideum is an early event in culmination. Development 122: 753–760 [DOI] [PubMed] [Google Scholar]
- Jin H, Martin C (1999) Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol 41: 577–585 [DOI] [PubMed] [Google Scholar]
- Kay RR, Jermyn KA (1983) A possible morphogen controlling differentiation in Dictyostelium. Nature 303: 242–244 [DOI] [PubMed] [Google Scholar]
- Kay RR, Thompson CRL (2001) Cross-induction of cell types in Dictyostelium: evidence that DIF-1 is made by prespore cells. Development 128: 4959–4966 [DOI] [PubMed] [Google Scholar]
- Kay RR, Large S, Traynor D, Nayler O (1993) A localized differentiation-inducing-factor sink in the front of the Dictyostelium slug. Proc Natl Acad Sci USA 90: 487–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay RR, Flatman P, Thompson CRL (1999) DIF signalling and cell fate. Sem Cell Dev Biol 10: 577–585 [DOI] [PubMed] [Google Scholar]
- Maeda M, Sakamoto H, Iranfar N, Fuller D, Maruo T, Ogihara S, Morio T, Urushihara H, Tanaka Y, Loomis WF (2003) Changing patterns of gene expression in Dictyostelium prestalk cell subtypes recognized by in situ hybridization with genes from microarray analyses. Eukaryot Cell 2: 627–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty S, Jermyn KA, Early A, Kawata T, Aubry L, Ceccarelli A, Schaap P, Williams JG, Firtel RA (1999) Evidence that the Dictyostelium Dd-STATa protein is a repressor that regulates commitment to stalk cell differentiation and is also required for efficient chemotaxis. Development 126: 3391–3405 [DOI] [PubMed] [Google Scholar]
- Raper KB (1940) Pseudoplasmodium formation and organization in Dictyostelium discoideum. J Elisha Mitchell Sci Soc 56: 241–282 [Google Scholar]
- Serafimidis I, Kay RR (2005) New prestalk and prespore inducing signals in Dictyostelium. Dev Biol 282: 432–441 [DOI] [PubMed] [Google Scholar]
- Shimada N, Maeda M, Urushihara H, Kawata T (2004a) Identification of new modes of Dd-STATa regulation of gene expression in Dictyostelium by in situ hybridisation. Int J Dev Biol 48: 679–682 [DOI] [PubMed] [Google Scholar]
- Shimada N, Nishio K, Maeda M, Urushihara H, Kawata T (2004b) Extracellular matrix family proteins that are potential targets of Dd-STATa in Dictyostelium discoideum. J Plant Res 117: 345–353 [DOI] [PubMed] [Google Scholar]
- Siegert F, Weijer CJ (1992) Three-dimensional scroll waves organize Dictyostelium slugs. Proc Natl Acad Sci USA 89: 6433–6437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Williams K (1980) Evidence for tip control of the “slug/fruit” switch in slugs of Dictyostelium discoideum. J Embryol Exp Morphol 57: 233–240 [PubMed] [Google Scholar]
- Thompson CRL, Kay RR (2000) The role of DIF-1 signaling in Dictyostelium development. Mol Cell 6: 1509–1514 [DOI] [PubMed] [Google Scholar]
- Thompson CRL, Fu Q, Buhay C, Kay RR, Shaulsky G (2004) A bZIP/bRLZ transcription factor required for DIF signaling in Dictyostelium. Development 131: 513–523 [DOI] [PubMed] [Google Scholar]
- Verkerke-van Wijk I, Fukuzawa M, Devreotes PN, Schaap P (2001) Adenylyl cyclase A expression is tip-specific in Dictyostelium slugs and directs StatA nuclear translocation and CudA gene expression. Dev Biol 234: 151–160 [DOI] [PubMed] [Google Scholar]
- Williams JG (2003) The STAT proteins of Dictyostelium. In Signal Transducers and Activators of Transcription (STATs). Activation and Biology, Sehgal PB, Levy DE, Hirano T (eds), pp 105–121. Boston, MA, USA: Kluwer Academic [Google Scholar]
- Williams JG, Ceccarelli A, McRobbie S, Mahbubani H, Kay RR, Farly A, Berks M, Jermyn KA (1987) Direct induction of Dictyostelium prestalk gene expression by DIF provides evidence that DIF is a morphogen. Cell 49: 185–192 [DOI] [PubMed] [Google Scholar]
- Yamada Y, Sakamoto H, Ogihara S, Maeda M (2005) Novel patterns of the gene expression regulation in the prestalk region along the antero-posterior axis during multicellular development of Dictyostelium. Gene Expr Patt 6: 63–68 [DOI] [PubMed] [Google Scholar]
- Zhukovskaya NV, Fukuzawa M, Yamada Y, Araki T, Williams JG (2006) The Dictyostelium bZIP transcription factor DimB regulates prestalk-specific gene expression. Development 133: 439–448 [DOI] [PubMed] [Google Scholar]


