Abstract
TraM is essential for F plasmid-mediated bacterial conjugation, where it binds to the plasmid DNA near the origin of transfer, and recognizes a component of the transmembrane DNA transfer complex, TraD. Here we report the 1.40 Å crystal structure of the TraM core tetramer (TraM58–127). TraM58–127 is a compact eight-helical bundle, in which the N-terminal helices from each protomer interact to form a central, parallel four-stranded coiled-coil, whereas each C-terminal helix packs in an antiparallel arrangement around the outside of the structure. Four protonated glutamic acid residues (Glu88) are packed in a hydrogen-bonded arrangement within the central four-helix bundle. Mutational and biophysical analyses indicate that this protonated state is in equilibrium with a deprotonated tetrameric form characterized by a lower helical content at physiological pH and temperature. Comparison of TraM to its Glu88 mutants predicted to stabilize the helical structure suggests that the protonated state is the active form for binding TraD in conjugation.
Introduction
Bacterial conjugation diversifies the bacterial genome and accounts for rapid acquisition of antibiotic resistance in many medically important human pathogens. Bacterial conjugation is defined as the unidirectional transfer of single-stranded DNA from a donor to a recipient cell in response to an uncharacterized mating signal generated by intimate cell-to-cell contact (Willetts and Wilkins, 1984; Lanka and Wilkins, 1995). TraM is a highly conserved cytoplasmic protein that is required for conjugative transfer of a variety of F-like plasmids, but is not essential for either the nicking reaction that prepares DNA for transfer or formation of the transmembrane DNA transfer complex (Kingsman and Willetts, 1978; Everett and Willetts, 1980; Howard et al, 1995; Nelson et al, 1995; Fekete and Frost, 2000; Karl et al, 2001). Instead, TraM has been thought to regulate F conjugation by performing a crucial signaling function (Willetts and Wilkins, 1984).
Part of the function of TraM is to bind near the origin of transfer (oriT). The sequence, arrangement, and number of TraM binding sites are not conserved in F-like plasmids. For example, R1 TraM binds two large sites near oriT (Schwab et al, 1991), whereas R100 TraM binds four sites in the vicinity of its oriT (Abo and Ohtsubo, 1993). F plasmid TraM cooperatively binds to three sites (sbmA, sbmB, sbmC) within oriT in the F plasmid (Di Laurenzio et al, 1992; Fekete and Frost, 2002). No conserved sequence motif has been identified in these three sites so far. TraM represses its own gene promoter by binding to sbmA and sbmB, but sbmC, which is the site of the lowest affinity to TraM and is separated by an approximately 90-bp AT-rich region upstream of the DNA transfer initiation site (the nic site), is the most important site of the three for F conjugation (Fu et al, 1991). DNA transfer requires unwinding of this AT-rich region upstream of the nic site to activate the helicase function of TraI (Matson et al, 2001; Csitkovits et al, 2004).
TraM also contacts TraD, an inner membrane coupling protein that interacts with type IV secretion systems that specifically function in conjugative DNA transfer (also called the ‘type IVA subfamily' (Disque-Kochem and Dreiseikelmann, 1997; Christie et al, 2005). TraM–TraD interactions are thought to recruit the plasmid to the secretion pore, and indeed, the R1 TraD cytoplasmic domain can bind TraM–DNA complexes in vitro (Beranek et al, 2004). The structure of the cytoplasmic domain of one member of the TraD family, TrwB, has been determined, revealing a hexameric ring with structural similarity to the nucleotide binding domains of DNA-dependent ring helicases (Gomis-Ruth et al, 2001). TrwB is thought to use energy from ATP hydrolysis to transport single-stranded DNA through its central channel (Tato et al, 2005). Whereas part of the TraD cytoplasmic domain bears strong sequence conservation to TrwB, the C-terminal 140 amino acids of TraD, which define the specificity of TraM–coupling protein interactions and contain the very C-terminal 38 amino acids that bind TraM, are not present in TrwB (Sastre et al, 1998; Beranek et al, 2004).
Limited trypsin digestion cleaves TraM into two functional fragments. The N-terminal fragment (residues 2–55) adopts a helical, likely dimeric structure (Verdino et al, 1999; Stockner et al, 2001; Miller and Schildbach, 2003). Mutations have been uncovered in this domain that lead to a loss of DNA binding and concomitant transcriptional repression, but do not affect its oligomeric state, suggesting that this domain directly contacts DNA within oriT (Lu et al, 2004). The C-terminal fragment (residues 58–127) adopts a tetrameric, helical structure (Miller and Schildbach, 2003). Mutations within this domain generally disrupt tetramerization and lead to a loss of DNA binding activity in the context of the full-length protein (Lu et al, 2004). A single mutation (K99E) has been uncovered in the C-terminal domain, which affects the ability of TraM to stimulate conjugation and interact with TraD, yet does not affect its oligomeric state or its ability to repress transcription (Lu and Frost, 2005). These data suggest that the C-terminal domain directly contacts TraD via a surface involving Lys99. The ability of TraM to simultaneously bind multiple sites in oriT and the TraD hexamer led us to propose that TraM might change conformation in response to the mating signal transferred from TraD, to induce melting of the AT-rich region upstream of the nic site to trigger conjugative DNA transfer (Lu and Frost, 2005).
It has not yet been possible to obtain a detailed structure of Escherichia coli F plasmid TraM or its homologues, either alone or in complex with DNA. An NMR structure of the N-terminal DNA binding domain (residues 2–56) of F-like R1 plasmid TraM has been obtained as a monomer at pH 4.0; however, the functional state of this fragment in F-like TraM appears to be dimeric and requires further tetramerization involving the C-terminal domain of TraM in order to bind DNA (Stockner et al, 2001; Miller and Schildbach, 2003; Lu et al, 2004). We have therefore cloned, expressed, and purified the trypsin-resistant tetramerization domain of TraM, which interacts with TraD, and determined its X-ray crystal structure to 1.4 Å resolution. Based on this structure, we used biochemical, biophysical, and genetic assays to show that protonation of four glutamate residues buried within the central four helices regulates structural flexibility of TraM. We suggest that the protonated state of TraM is conjugation-proficient, and that de-protonation of TraM may provide a mechanism to downregulate conjugation in response to environmental stress.
Results
Overall structure of the TraM core tetramer (TraM58–127)
To determine the structure of the tetramer core domain, we cloned, produced, and crystallized the region of TraM corresponding to the trypsin-resistant C-terminal domain (residues 58–127, and referred to here as TraM58–127; Figure 1A). The crystal structure was determined by MAD phasing and refined to a resolution of 1.40 Å (see Materials and methods). The crystal structure of TraM58–127 is a symmetrical eight-stranded helical bundle composed of four intertwined protomers that each donates a pair of α-helices (Figure 1B and C). The N-terminal helix of each protomer (residues 63–89) interacts at the center of the structure to form a parallel, four-stranded coiled coil, wrapped in a left-handed superhelix. The C-terminal helix of each protomer (residues 102–120) is packed in a right-handed arrangement around the outside of the structure, antiparallel to the N-terminal helices. Overall, the structure resembles a cylinder, ∼36 Å in diameter and ∼42 Å in height, with the N- and C-termini at one end and the loops linking the two helices of each protomer at the opposite end of the cylinder.
Figure 1.
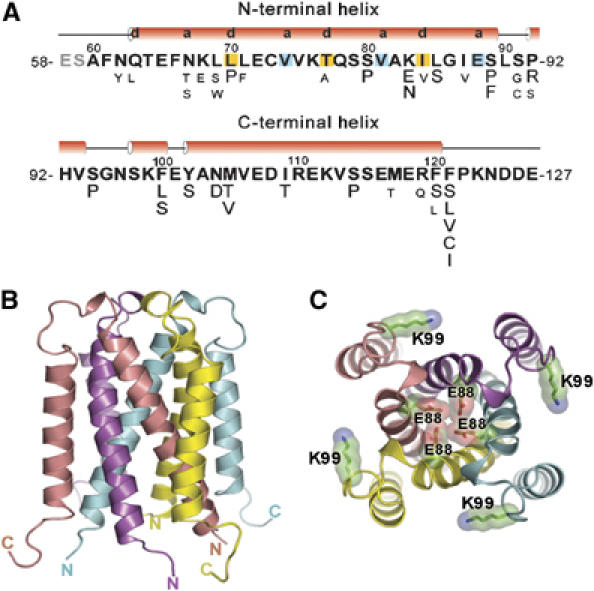
Primary sequence and overall three-dimensional structure of the TraM58–127 homotetramer. (A) Amino-acid sequence of TraM58–127. Secondary structural elements were obtained from the X-ray structure. The a and d positions of the heptad repeats in the N-terminal helices forming the central coiled coils are highlighted. Previously selected missense mutations affecting TraM autoregulation and F conjugation in TraM58–127 are shown under corresponding residues (Lu et al, 2004). Strong mutations, which complemented pOX38-MK3 (a traM-deficient F plasmid derivative) for conjugation to a frequency lower than 10−5 transconjugants per donor, are shown in larger font size. (B) Side view of TraM58–127. The N- and C-termini of the four protomers are labeled. (C) Front view from the loop end of TraM58–127. The side chains of the four protonated glutamic acid residues (E88) buried in the central coiled coils and the four lysine residues (K99) involved in interactions with TraD (Lu and Frost, 2005) are highlighted. Oxygen atoms are colored in red and nitrogen atoms in blue.
Mechanism of TraM tetramerization
The N-terminal helix contains an unusual distribution of residues at the a and d positions of the helical heptad repeat, which constitute the core of the four-stranded coiled-coil (for a review of coiled-coil structure, see Lupas, 1996). The first two turns of the N-terminal helix are not packed against its symmetry-related partners, probably because of the presence of hydrophilic residues (Gln63 and Asn67) at the first d and a positions at the N-terminal end, and proper coiled-coil packing is only established at Leu70, at the following d position (Figure 2A). Three out of four of the following a position residues are β-branched, a feature that stabilizes the packing of dimeric and trimeric coiled-coils, but not tetrameric coiled-coils, where non-β-branched residues at this position are favored (Harbury et al, 1993). This suboptimal packing creates a space at the coiled-coil interface between the a position Val74 and Thr77 at the following d position. This space is occupied by a layer of water molecules, inaccessible to the bulk solvent. This network of waters is anchored to the protein through hydrogen bonding interactions to the backbone carbonyl of Val74 and the side-chain hydroxyl of Thr77 in each of the four protomers (Figure 2C). Coiled-coil packing is terminated at the C-terminal end by the a position Glu88. Surprisingly, this residue is tightly packed against its symmetry-related partners such that the oxygens of the side-chain carboxylic acid groups are within hydrogen bonding distance (2.70 Å) of their neighbors (Figure 2B and D). This packing arrangement could only be accommodated with the protonation of Glu88, implying that the quaternary structure of TraM dramatically elevates the pKa of this residue.
Figure 2.
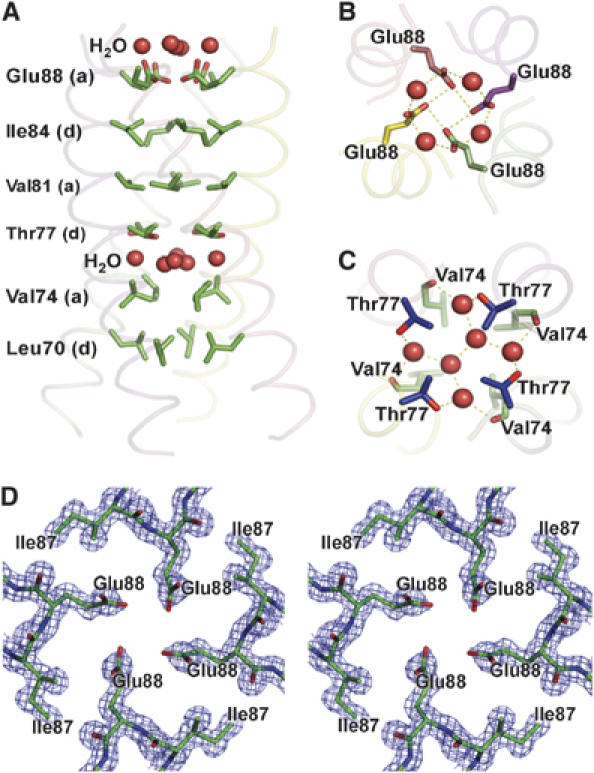
Structure arrangement of the central four-helix bundle of TraM58–127. (A) Layers of a and d residues of the heptad repeats and water molecules. (B) Hydrogen bonding of the protonated Glu88 residues. (C) Hydrogen bonding of the central water molecules with the side-chain oxygen of Thr77 and the main-chain oxygen of Val74. (D) A stereo view of the 2Fo−Fc electron density map contoured at 1.5σ, showing residues Ile87 and Glu88 arranged around the four-fold axis of the tetramer.
The presence of buried water molecules and glutamic acid residues at the coiled-coil interface suggests that this structure is not stable without supporting interactions from the peripheral C-terminal helices. We previously generated an extensive set of mutants in the TraM C-terminal domain that disrupt its function and tetrameric structure (Lu et al, 2004; Figure 1A). This mutational analysis suggests that packing of the C-terminal helices against the N-terminal helices provides a dominant force to stabilize the TraM tetramer. Proximal to the loop at one end of the structure, residues Phe100, Tyr102, Met105, and Ile109 of the C-terminal helix pack against Leu85 of the N-terminal helix from the same protomer and Ile87 from the N-terminal helix of another protomer. Missense mutations at any of these sites affect TraM autoregulation and F conjugation. At the opposite end of the structure, proximal to the chain termini, Phe120 and Phe121 of the C-terminal helix pack against Phe61 and Phe66 of an N-terminal helix from a different protomer. A number of distinct substitutions at both Phe120 and Phe121 are highly defective for autoregulation and F conjugation. Interestingly, Phe121 seems particularly critical as substitution of this residue with other hydrophobic residues (Leu or Val) leads to a loss of function (Figure 1A). Deletion of the N-terminal four residues (58–61) of TraM58–127 resulted in a fragment that was no longer trypsin resistant; however, deletion of just the N-terminal two residues (58–59) of TraM58–127 yielded a fragment that maintained trypsin resistance (data not shown), suggesting that Phe61 is required for structural integrity of TraM. A salt bridge on the surface of the structure between Lys83 of an N-terminal helix and Asp108 of a C-terminal helix from a different protomer may also be critical for the structural stability of the tetramer as mutation of Lys83 to Glu or Asn strongly affects TraM autoregulation and F conjugation (Figure 1A).
Although most of the mutations at the C-terminal region of TraM can be explained in terms of destabilization of the tetramer structure, one conjugation-defective mutant cannot be explained. Mutation of Lys99 to Glu has been shown to decrease conjugation and TraD interactions; however, DNA binding, TraM autoregulation, and tetramerization are not affected (Lu and Frost, 2005). Lys99 therefore likely defines part of the TraD contact surface. This residue is found in the loop at the end of the helical bundle, exposed on the outside of the tetramer and making no electrostatic interactions with the rest of the structure (Figure 1C). Its substitution by glutamic acid is therefore unlikely to result in a perturbation of the tetramer structure. We conclude that the end of the cylindrical helical bundle that is distal to the chain termini is likely a key point of contact to the conjugative pore; however, additional regions may also be important, as TraM-K99E still shows significant interactions with TraD (Lu and Frost, 2005).
Deprotonation of Glu88 relaxes TraM structure
We have been unable to discover any other four-stranded coiled-coils in the structure database that contain four glutamic or aspartic acid residues packed together at the coiled-coil interface. To assess the role of Glu88 in TraM folding, we monitored the structural stability of TraM58–127 by CD spectroscopy, and compared the results to those obtained from a TraM58–127 variant bearing a Glu–Gln or a Glu–Leu substitution at position 88. Gln88 should allow a similar hydrogen bonding geometry at position 88 as in the wild-type structure but without the potential charge repulsion effect of Glu88, whereas Leu is predicted to further stabilize the coiled-coil (TraM58–127-E88L; Harbury et al, 1993). The crystal structure of TraM58–127-E88L was solved to 1.8 Å resolution and revealed the overall structure of both the monomer (asymmetric unit) and the crystallographic/biological tetramer to be almost identical to that of the wild-type structure with an RMSD of 0.5 Å for the 68 Cα main-chain atoms of the monomer (Supplementary Figure 1A and B). Small deviations of the main-chain occur at the N- and C-termini and in the loop. The substituted leucine at position 88 adopts the same rotomer as the wild-type glutamic acid, and packs well against its symmetry-related partners around the four-fold axis of the tetramer (Figure 3A).
Figure 3.
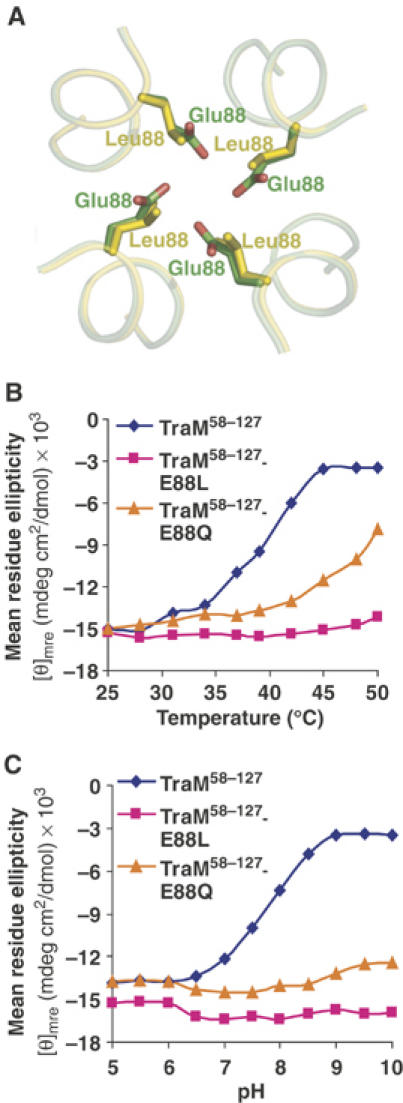
Structure comparison and circular dichroism analysis of TraM58–127, TraM58–127-E88L, and TraM58–127-E88Q. (A) Structural alignment of the side chains of Glu88 in TraM58–127 and Leu88 in TraM58–127-E88L. (B) Mean residue ellipticity at 220 nm of TraM58–127, TraM58–127-E88L, and TraM58–127-E88Q at different pH values and at a fixed temperature (37°C). (C) Mean residue ellipticity at 220 nm of TraM58–127, TraM58–127-E88L, and TraM58–127-E88Q at different pH values and at a fixed temperature (37°C).
We found that the wild-type TraM58–127 helical structure is highly sensitive to increases in temperature, exhibiting complete unfolding by 45°C at pH 7.5 (Figure 3B), consistent with previous studies that showed that TraM58–127 undergoes unfolding at relatively low concentrations of chemical denaturant (Miller and Schildbach, 2003). In matched experiments, TraM58–127-E88Q displayed greater thermal stability but did lose significant helical structure at the highest temperatures, whereas TraM58–127-E88L showed no detectable thermal unfolding up to 50°C. TraM58–127 structure is also highly sensitive to elevations in pH (Figure 3C). Whereas the mean residue ellipticity is very similar between TraM58–127, TraM58–127-E88Q, and TraM58–127-E88L below pH 6, as the pH is raised, the structure of the wild-type protein is destabilized with an apparent transition midpoint of pH 7.7 at 37°C. In contrast, TraM58–127-E88Q and TraM58–127-E88L are stable over the entire range of pH under the same conditions. This indicates that the pKa of Glu88 is elevated by the folding of TraM from ∼4 (the pKa of free glutamic acid) to ∼7.7. Under physiological conditions, charge repulsions associated with the deprotonation of Glu88 lead to a destabilization of TraM helical structure.
We next used size-exclusion chromatography and multi-angle laser light scattering (MALLS) to determine the effects of Glu88 protonation on TraM size and oligomerization. As pH is increased from 7.5 to 9.0, TraM58–127 was eluted earlier from the column, suggesting a larger hydrodynamic radius (RH; Figure 4A). Consistent with this, in-line dynamic light scattering data over the entire peak were also consistent with an increase in the size of the complex, although noise in these data precluded a precise determination of RH (not shown). Interestingly, in-line MALLS demonstrated that the increase in hydrodynamic radius is not owing to an increase in the oligomeric state. Under all pH conditions, TraM58−127 remained tetrameric with a molar mass of ∼32 kDa; however, at pH 9.0, a small amount of a late-eluting complex was detected with MALLS, possibly owing to pH-induced denaturation. A full CD scan of TraM58–127 at the same temperature (25°C) as used in the MALLS experiments revealed only a small decrease in α-helicity as the pH is increased from 7.5 to 9.0 (data not shown). Taken together, these results indicate that TraM becomes less helical when Glu88 is deprotonated, yet maintains its overall tetrameric oligomeric state.
Figure 4.
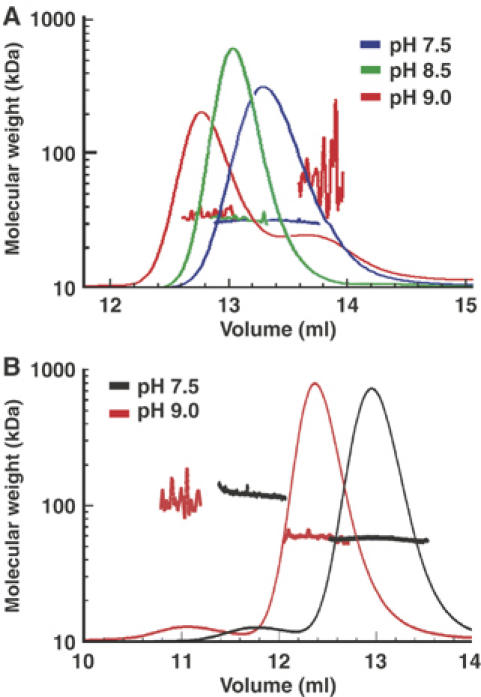
(A) Size-exclusion chromatography and molecular weight determination of TraM58–127 at pH 7.5 (blue), pH 8.5 (green), and pH 9.0 (red) by MALLS. Colored lines superimposed on the different peaks indicate the molecular weight of the proteins as shown on the Y-axis. (B) Size-exclusion chromatography and molecular weight determination of TraM at pH 7.5 (black) and pH 9.0 (red). Colored lines superimposed on the different peaks indicate the molecular weight of the proteins as shown on the Y-axis.
We also used MALLS to probe structural changes in full-length TraM as a function of pH. The increased size of the full-length TraM tetramer provided better quality dynamic scattering data that enabled an estimation of the apparent hydrodynamic radius. The major elution peak at either pH 7.5 or 9.0 contained complexes ∼58 kDa in size, consistent with a tetrameric structure, whereas the hydrodynamic radius of TraM was 3.6 (±0.1) nm at pH 7.5 and 4.4 (±0.5) nm at pH 9.0, indicating an increase in the size of the complex with increasing pH (Figure 4B). In addition, small amounts of a higher molecular weight species (∼100–150 kDa) were also detected at earlier elution times in the full-length protein, as previously reported (Verdino et al, 1999; Lu et al, 2004), but these species were not detected in TraM58−127 samples. This suggests that the N-terminal domain is likely responsible for the stabilization of a small proportion of higher oligomeric states, consistent with previous studies that have shown that the N-terminal DNA binding domain dimerizes and stabilizes the structure of full-length TraM (Miller and Schildbach, 2003; Lu et al, 2004). Overall, this work suggests that the protonation state of Glu88 governs a dynamic equilibrium between a compact form of the TraM tetramer, likely represented by the pH 6.5 crystal structure, and more extended or ‘relaxed' deprotonated forms under physiological conditions.
Role of Glu88 protonation in F conjugation
To understand the role of Glu88-dependent structural flexibility in TraM for F conjugation, we mutated Glu88 of TraM in pRFM200, a plasmid that expresses the wild-type TraM constitutively at a low but functional level (Lu et al, 2003, 2004). Two pRFM200 mutant derivatives (E88L and E88Q) were introduced into XK1200 cells containing pOX38-MK3 (a traM-deficient F plasmid) to compare the abilities of the TraM mutants to facilitate F conjugation.
Wild-type, E88L, and E88Q TraM-transformed cells were all equally capable of donating F plasmids under physiological conditions (pH 7.5 and 37°C; Figure 5A and B). Cells containing pRFM200-Mdel (a traM-deficient pRFM200 derivative) instead of pRFM200 had no detectable donor ability (<10−7 transconjugants per donor) in the same assay. Our biophysical studies have demonstrated that the TraM protonated state is destabilized by elevated temperature and pH, whereas the E88L and E88Q variants are much more stable. This suggested that deprotonation-induced structural changes might serve as a pH/temperature sensor, to reduce conjugation under non-optimal growth conditions. To test this hypothesis, we assessed the abilities of TraM and its position 88 mutants to effect conjugative transfer over a range of temperature (Figure 5A) and pH (Figure 5B). The results clearly demonstrate that as either temperature or pH is raised above physiological values, the conjugation efficiency of wild-type TraM cells drops off dramatically compared to either E88L or E88Q cells. The effect appears to be more dramatic for temperature than for pH, suggesting that the cells may be able to buffer their internal pH against that of the external media in this assay. We also observed a pronounced loss of conjugation activity at temperatures below 37°C for all the TraM variants, which could be explained by loss of mating pair formation at low temperature (Walmsley, 1976). Levels of intracellular TraM or its mutants remained comparable in donor cells incubated at different temperatures for 45 min during the mating assays (Figure 6A), indicating that high temperatures did not affect TraM stability. Overall, our data strongly indicate that the compact, protonated form of the TraM tetramer is the active conformational state for F conjugation.
Figure 5.
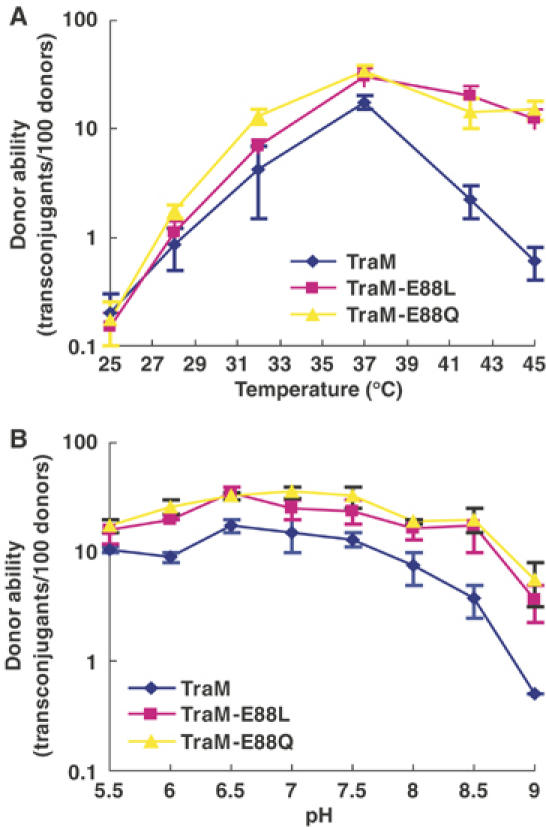
In vivo activity of TraM and its mutants for autoregulation and F conjugation. (A) Donor ability of cell containing pOX38-MK3 and pRFM200 (or one of its mutant derivatives) as a function of temperature. (B) Donor ability of cell containing pOX38-MK3 and pRFM200 (or one of its mutant derivatives) as a function of pH.
Figure 6.
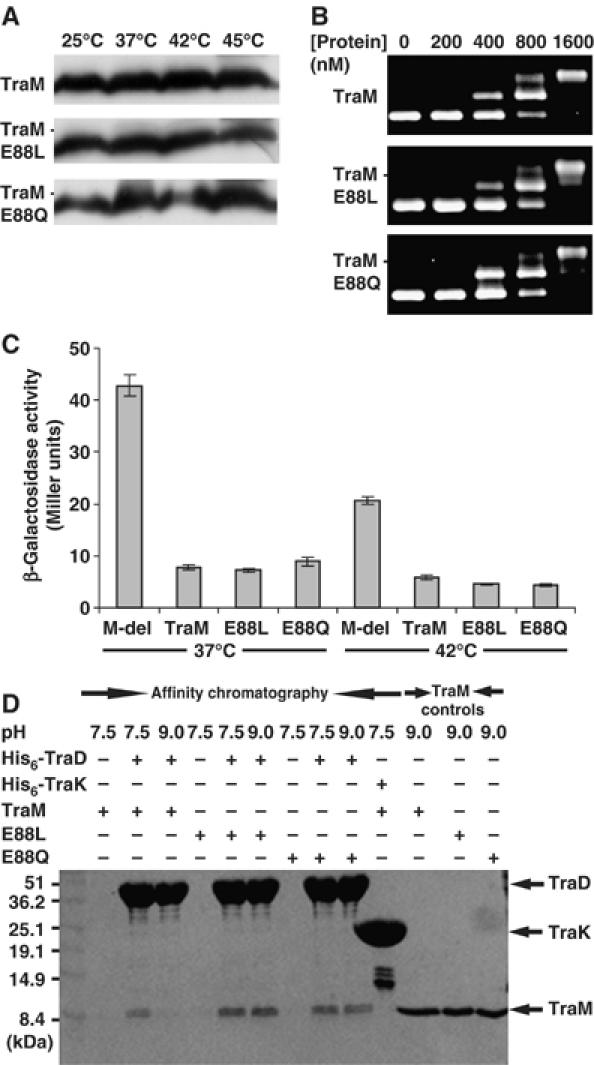
(A) TraM and its mutants detected by immunoblot with the anti-TraM antiserum. 0.1 OD600 of cells incubated at different temperatures (indicated above the figure) for 45 min were loaded in each lane. (B) Binding of TraM or its mutants to sbmABC (containing all three TraM binding sites) as determined using EMSA. TraM concentration is indicated above each lane. (C) β-Galactosidase activity of cells containing the traM promoter-lacZ reporter plasmid pACPM24fs::lacZ and a second plasmid expressing either wild type, E88L, or E88Q TraM or no protein (M-del) at 37 and 42°C. (D) Interactions between His6-TraDcyt and TraM58–127 (or its E88L or E88Q mutants) as determined by affinity chromatography at pH 7.5 and 9.0. TraM, E88L, and E88Q represent TraM58–127, TraM58–127-E88L, and TraM58–127-E88Q, respectively. Proteins were visualized by Coomassie blue staining. Protein molecular weight markers are indicated to the right of the figure. His6-TraK is used as a negative control. A 5 μg portion of the indicated TraM58−127 variants was loaded in the lanes on the left of the gel.
Both DNA binding and interaction with TraD are required for TraM to function during F conjugation (Lu et al, 2004; Lu and Frost, 2005). TraM and its E88L and E88Q mutants shifted sbmABC DNA (containing all three TraM binding sites) into two different species at similar protein concentrations as monitored by electrophoretic mobility shift assays (EMSA) (Figure 6B), indicating that all three proteins bind to cognate DNA efficiently and cooperatively. TraM binding to sbmA and B represses the expression of the traM promoter. To compare the ability of TraM and its variants to regulate transcription of the traM promoter, we utilized a traM promoter-lacZ fusion plasmid, pACPM24fs::lacZ, and monitored β-galactosidase activity in cells expressing different TraM variants. At either 37 or 42°C, cells expressing wild-type TraM, or the E88L or E88Q variants, demonstrated similar levels of β-galactosidase activity in a quantitative solution assay, whereas cells lacking TraM (Mdel) had an ∼5-fold higher level of β-galactosidase activity at the corresponding temperatures (Figure 6C). Taken together, these data indicate that the structural instability of the wild-type protein does not significantly affect its ability to bind its promoter DNA, or to inhibit transcription. Cells grown at 42°C in the absence of TraM had lower levels of β-galactosidase activity compared to cells grown at 37°C, suggesting that high temperatures may affect β-galactosidase expression.
Affinity chromatography was used to determine whether Glu88 deprotonation could affect TraM–TraD interactions. The same amount of Ni-NTA agarose bound with His-tagged TraD cytoplasmic domain (His6-TraDcyt) was mixed with an equivalent amount of purified TraM or its mutants at pH 7.5 or 9.0. After extensive washing, the bound proteins were eluted from the Ni-NTA agarose (see Materials and methods). More wild-type TraM was co-eluted with His6-TraDcyt at pH 7.5 than at pH 9.0 (Figure 6D). In contrast, comparable amounts of TraM-E88L (or TraM-E88Q) were eluted at pH 7.5 and 9.0, suggesting that Glu88 deprotonation reduced the ability of TraM to interact with TraD at the elevated pH. In control experiments, no detectable TraM was co-eluted with His6-tagged TraK (a component of F conjugation machinery), indicating the specificity of TraM–TraD interaction. Therefore, our results suggest that the compact, Glu88-protonated, form of TraM interacts with TraD to activate F conjugation.
Discussion
The protonated form of the TraM C-terminal domain bears an intriguing resemblance to the core helical bundle structures of class I viral envelop proteins, such as influenza hemagglutinin and HIV gp41 (Wilson et al, 1984; Bullough et al, 1994; Chan et al, 1997; Weissenhorn et al, 1997). While these proteins are trimeric, TraM has a similar helical bundle topology, where the N-terminal helix of each protomer occupies the center of the structure, and the C-terminal helix wraps around the outside to complete the bundle. Particularly interesting is the fact that many of these envelop proteins also undergo a pH-dependent conformational reorganization of the helical bundle upon exposure to the low-pH environment of the endosome (for a review, see Eckert and Kim, 2001).
A key question remains as to the nature of the equilibrium between the deprotonated and protonated forms of TraM under physiological conditions. Our biophysical data suggest that while increasing pH leads to a progressive loss of helical structure and increasing RH, the protein remains a tetramer. Only at very high pH (9.0) do we see the beginning of unfolding to aggregation-prone monomeric forms. We have not been able to obtain a crystal structure of a high-pH (deprotonated) form of TraM, and we believe it is unlikely that there exists a single, discrete deprotonated form of the tetramer, unlike the viral envelop proteins, which exist in either of two distinct, pH-dependent conformations. Our data are consistent with a model where the protonated form visualized in our crystal structure represents the active conformation. The observation that the two mutants that stabilize the low-pH form of TraM, E88L and E88Q, not only show no detectable functional defects, but actually demonstrate a somewhat higher competence for conjugation than the wild type provides strong support for this model.
The conformational plasticity associated with Glu88 appears to affect interactions with TraD to a greater extent than DNA binding or transcriptional repression (Figure 6). This is consistent with a model where the tetramerization domain directly contacts TraD but not DNA. TraD likely binds a surface on the TraM tetramer that involves Lys99, at the same end of the tetramer as Glu88 (Lu and Frost, 2005). The charge repulsion that would be associated with Glu88 deprotonation could alter the structure and/or stability of this region of the tetramer, reducing the affinity for TraD. However, this raises the question as to why Glu88, and its associated conformational plasticity, has been highly conserved in TraM. As conjugation is a non-essential, yet metabolically costly process, it is downregulated in natural F-like plasmids via a fertility inhibition system, FinOP, or by H-NS silencing of the transfer region (Arthur et al, 2003; Will et al, 2004). During periods of derepression, the temperature and pH sensitivity of TraM suggests a mechanism for the F conjugation system to sense environmental stress and quickly downregulate conjugation in a manner independent of gene regulation.
Materials and methods
Growth media and bacterial strains
Cells were grown in LB (Luria-Bertani) broth or on LB solid medium containing appropriate antibiotics. X-gal (5-bromo-4-chloro-3-indolyl-β-D-galactoside) was used at a final concentration of 100 μg/ml. Isopropylthio-β-D-galactoside (IPTG) was used at a final concentration of 1 mM. The following E.coli strains were used: XK1200 (Moore et al, 1987), ED24 (Willetts and Finnegan, 1970), DH5α (Hanahan, 1983), and BL21-DE3 (Stratagene).
Plasmids and plasmid construction
Plasmids pRFM200, pRFM200-Mdel, pACPM24fs::lacZ (Lu et al, 2003), pOX38-MK3 (Penfold et al, 1996), and pT7-7 (Tabor and Richardson, 1985) were described previously. Plasmids pRFM200-E88L and pRFM200-E88Q are pRFM200 derivatives generated by site-directed mutagenesis of traM as described below. Plasmid pJLTraM58–127, which encodes a TraM tryptic fragment (TraM58–127, residues 58–127 of TraM), was constructed by ligating the 2.5-kb EcoRI–HindIII fragment of pT7-7 to the 0.5-kb EcoRI–BamHI fragment of DNA amplified from pRFM200 using JLU251 (5′-TAG AAT TCA CCA TCA CCA TCA CCA Taa aGA GTC TGC ATT TAA TCA GAC-3′) and JLU4 (5′-CGA TAA GCT TGG GCT GCA GG-3′) as primers. Because TraM58–127 is trypsin-resistant (Miller and Schildbach, 2003; J Lu and JNM Glover, unpublished results), an arginine codon for a trypsin cleavage site (lowercase letter) was included in JLU251 between the 6-His tag (underlined) and the TraM58–127 coding regions for cleaving off the His-tag during purification of TraM58–127. Plasmid pJLTraM60–127 or pJLTraM62–127, which encodes a TraM58–127 equivalent with a deletion of two or four N-terminal residues, was constructed similarly except by using JLU252 (5′-TAG AAT TCA CCA TCA CCA TCA CCA Tcg tGC ATT TAA TCA GAC TGA GTT TAA T-3′) or JLU253 (5′-TAG AAT TCA CCA TCA CCA TCA CCA Tcg tea TCA GAC TGA GTT TAA TAA ATT GC-3′) instead of JLU251 as the forward PCR primer, respectively. Plasmid pJLTraM58–127-E88L and pJLTraM58–127-E88Q were also constructed in a similar way as pJLTraM58–127 except using pRFM200-E88L and pRFM200-E88Q as the PCR templates, respectively.
Site-directed mutagenesis
Site-directed mutagenesis by overlap extension (Ho et al, 1989) was used to construct different traM mutants. The primer pairs used for introducing point mutations into traM are 5′-GCG AAA ATT TTG GGT ATT TTG TCT CTC AGT CCT CAT G-3′ and 5′-CAT GAG GAC TGA GAG ACA AAA TAC CCA AAA TTT TCG C-3′ for E88L, and 5′-GCG AAA ATT TTG GGT ATT CAG TCT CTC AGT CCT CAT G-3′ and 5′-CAT GAG GAC TGA GAG ACT GAA TAC CCA AAA TTT TCG C-3′ for E88Q. Primers JLU3 (5′-CTA TAG GGA GAC CGG AAT TCG-3′) and JLU4 (5′-CGA TAA GCT TGG GCT GCA GG-3′) were used with the above primers to amplify the mutated DNA fragments from pRFM200. The EcoRI–BamHI fragments of the mutagenesis PCR products were cloned back to replace traM in pRFM200, resulting in different pRFM200 derivatives named after their mutations.
Protein expression and purification
To overexpress His-tagged TraM58–127, BL21-DE3 cells containing pJLTraM58–127 were grown in 1 l of LB broth containing ampicillin at 37°C with vigorous shaking. After 3 h, IPTG was added and the culture was grown for another 5 h at 30°C before harvesting by centrifugation. The cell pellet was suspended in 100 ml of lysis buffer (50 mM Tris–HCl, 10 mM imidazole, 250 mM NaCl, pH 7.5) with one tablet of Complete, Mini protease inhibitor mixture, EDTA-free (Roche Applied Science). The suspension was lysed by sonication on ice for 3 min (30 s with a 30-s break, repeated six times) at maximum output. After centrifugation at 27 000 g for 60 min, the supernatant of the lysate was loaded on a column with 3 ml Ni-NTA agarose (Qiagen) pre-equilibrated with 30 ml of lysis buffer. After washing with 30 ml of buffer (50 mM Tris–HCl, 20 mM imidazole, 250 mM NaCl, pH 7.5), the protein bound to the Ni-NTA agarose was eluted with 12 ml of elution buffer (50 mM Tris–HCl, 250 mM imidazole, 250 mM NaCl, pH 7.5) in 2 ml fractions. SDS–polyacrylamide gel electrophoresis was used to separate 5 μl samples from each fraction, and the protein was visualized by Coomassie blue staining (Sambrook et al, 1989). The fractions containing His6-TraM58–127 were pooled and mixed with 100 μg of trypsin at room temperature for 16 h. The trypsin-digested mixture was further separated by size-exclusion chromatography (Hiload® 26/60 Superdex 75 prep grade column, Amersham Biosciences), and proteins were eluted with buffer (50 mM Tris–HCl, 150 mM NaCl, 1 mM DTT, pH 7.5). The peak containing TraM58–127 was concentrated and buffer-exchanged to 0.5 M ammonium acetate and 2 mM DTT by using an Amicon® ultracentrifuge filter (Millipore). Protein concentration was determined using BCA protein assays (Pierce) following the manufacturer's instructions. TraM60–127, TraM58–127-E88L, or TraM58–127-E88Q was purified similarly, except using BL21-DE3 cells containing pJLTraM60–127, pJLTraM58−127-E88L, or pJLTraM58−127-E88Q for overexpression, respectively. Selenomethionyl TraM58−127 was produced as previously described (Doublie, 1997) and purified in the same manner as the native protein. Full-length TraM, TraM-E88L, and TraM-E88Q were overexpressed and purified as previously described (Lu et al, 2004). The molecular weight of each protein was confirmed by mass spectroscopy at the Institute for Biomolecular Design, University of Alberta. Cloning and purification of His-tagged TraD cytoplasmic domain (residues 137–717; J Lu and LS Frost, unpublished data) and TraK (J Manchak and LS Frost, unpublished data) will be described elsewhere.
Crystallization and data collection
TraM58–127, selenomethionyl TraM58–127, or TraM58–127-E88L was concentrated to 25 mg/ml in 0.5 M ammonium acetate and 2 mM DTT. Crystals were grown at room temperature (22–25°C) using the hanging drop vapor diffusion technique. A measure of 1 μl of 25 mg/ml protein in 0.5 M ammonium acetate and 2 mM DTT was mixed with 1 μl of well solution (100 mM MES, pH 6.6, 30% PEG 400, 4 mM DTT) to produce tetragonal crystals within 3–7 days. All three proteins crystallized in space group I4 with similar unit cells.
Data were collected from exposure of single crystals at beamline 8.3.1 of the Advanced Light Source, Lawrence Berkeley National Laboratory. A three-wavelength MAD data set was collected in inverse-beam mode to 1.42 Å resolution for selenomethionyl TraM58−127, and a native data set to 1.40 Å for TraM58−127 and to 1.80 Å for TraM58−127-E88L. Data for TraM58−127 were processed and scaled using the HKL package (Table I; Otwinowski and Minor, 1997). Data for the TraM58−127-E88L structure were processed and scaled using ELVES (Holton and Alber, 2004).
Table 1.
Data collection, phasing, and refinement statistics
| Native TraM58–127 | SeMet-TraM58–127 | Native TraM58–127-E88L | ||
|---|---|---|---|---|
| Data collection | ||||
| Space group | I4 | I4 | I4 | |
| Cell dimensions | ||||
| a, b, c (Å) | 51.73, 51.73, 49.59 | 51.68, 51.68, 49.73 | 50.94, 50.94, 46.55 | |
| α, β, γ (deg) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | |
| Inflection | Remote | |||
| Wavelength | 1.116975 | 0.979617 | 1.019440 | 1.115869 |
| Resolution (Å) | 1.40 | 1.42 | 1.34 | 1.80 |
| Rsym or Rmerge | 0.035 (0.373) | 0.051 (0.448) | 0.055 (0.567) | 0.047 (0.362) |
| I/σI | 27.6 (1.8) | 23.8 (3.4) | 23.7 (2.4) | 25.1 (2.4) |
| Completeness (%) | 99.1 (86.6) | 98.1 (96.6) | 97.8 (96.1) | 98.6 (84.1) |
| Redundancy | 3.6 (2.8) | 4.1 (3.9) | 4.1 (3.9) | 3.6 (2.2) |
| Refinement | ||||
| Resolution (Å) | 36.6–1.40 | 36.0–1.80 | ||
| No. of reflections | 12 201 | 5288 | ||
| Rwork/Rfree | 0.193/0.224 | 0.182/0.244 | ||
| No. of atoms | ||||
| Protein | 551 | 543 | ||
| Ligand/ion | 0 | 0 | ||
| Water | 55 | 42 | ||
| B-factors | ||||
| Protein | 16.6 | 23.6 | ||
| Ligand/ion | ||||
| Water | 24.6 | 27.2 | ||
| RMS deviations | ||||
| Bond lengths (Å) | 0.010 | 0.012 | ||
| Bond angles (deg) | 1.233 | 1.193 | ||
| Highest resolution shell for the native TraM58–127 crystal is 1.42–1.40, for the selenomethionine derivative is 1.44–1.42 (inflection) and 1.36–1.34 (remote), and for the native TraM58–127-E88L crystal is 1.83–1.80. | ||||
Structure determination
The program SOLVE (Terwilliger and Berendzen, 1999) located both expected selenium atoms in the asymmetric unit using data from only two wavelengths (inflection and remote). Data collected at the peak wavelength were not included owing to an unresolved scaling problem. The best solution from SOLVE had a Z-score of 5.7 and a mean figure of merit of 0.64 for all reflections to 1.5 Å resolution. Maximum likelihood density modification in RESOLVE (Terwilliger, 2000) was used to improve initial phases yielding an overall figure of merit of phasing of 0.76. Automatic model building in RESOLVE (Terwilliger, 2002) placed 57 of the 68 ordered residues in the monomer. The RESOLVE-built model was used as the basis for further automated model building in ARP/wARP (Morris et al, 2003), which built 63 residues in two chains. Iterative cycles of refinement in REFMAC (Murshudov et al, 1997) against the low-energy remote data to 2.0 Å resolution and manual model building using XFIT (McRee, 1999) were used to complete and refine the model to an R-factor of 0.24 and free R-factor of 0.28. At this point, refinement was continued against the native data to 1.40 Å resolution using REFMAC with TLS refinement. The final model of the crystallographic asymmetric unit contains one 68-residue protomer and 55 water molecules; the biologically relevant tetramer is generated from this model by rotation about the crystallographic four-fold axis. The final R-factor and free R-factors are 0.193 and 0.224, respectively. A total of 95% of residues fall within the most favored regions of the Ramachandran plot, with the remaining three residues in the additionally allowed regions. Refinement statistics are summarized in Table I. The TraM58−127-E88L structure was solved by molecular replacement (Vagin and Teplyakov, 1997) using the wild-type structure as the search model. Differences in density clearly revealed the leucine substitution. Like the wild-type structure, the biologically relevant tetramer is formed around the crystallographic four-fold axis. The structure was refined to final R-factor and free R-factors of 0.182 and 0.244, respectively. The coordinates and structure factors have been submitted to the RCSB (PDB ID 2G7O and 2G9E for the wild type and the E88L mutant, respectively).
Circular dichroism spectroscopy
Spectra were recorded in a Jasco J720 spectropolarimeter in thermostated, jacketed cells of 0.05 cm pathlength. Concentrations of solutions were calculated based on protein concentrations measured using BCA assays and data were converted to mean residue ellipticity using a mean residue weight of 113.9 for TraM58–127, 113.7 for TraM58–127-E88L, and 113.9 for TraM58–127-E88Q, calculated from the known sequence of TraM (Frost et al, 1994). Effects of temperature and pH on the secondary structure of the proteins were assessed by monitoring the ellipticity at 220 nm. For each temperature or pH data point, the sample was first equilibrated for 5 min before a 5-min data collection.
Size-exclusion chromatography/multi-angle laser light scattering
Samples in 50 mM Tris–HCl and 150 mM NaCl at different pH values were applied to a SuperoseTM 12 HR 10/30 column eluted with the same buffer. The effluent from this column was run directly through in-line DAWN EOSTM MALLS and Optilab rEXTM differential refractive index detectors (Wyatt Technologies, Santa Barbara, CA). ASTRA v. 4.90 software was used to process these data. Average values of the RH for complexes were calculated from data collected over the elution peak (from 1/2 maximum peak height to the peak maximum) and the average and standard deviation were calculated. To systematically remove outlier data that were artificially high owing to aggregation, we removed the data points that deviated more than 2 standard deviations from the mean, and re-calculated the mean and standard deviation.
β-Galactosidase assays
β-Galactosidase activity was assayed in cells containing the traM promoter-lacZ reporter plasmid pACPM24fs::lacZ and plasmid pRFM200, pRFM200-E88L, or pRFM200-E88Q, which expresses wild type, E88L, or E88Q TraM, respectively, for determining the autoregulation function of TraM or its mutants. A fresh, single colony was inoculated into LB broth containing appropriate antibiotics and grown at 37 or 42°C with shaking for 16 h. A 200 μl sample was used for determining β-galactosidase activity as described by Miller (1972) and reported as Miller units.
Donor ability assays
E. coli XK1200 and ED24 cells were grown in LB broth to a mid-exponential phase at 37°C and used as donors and recipients for the mating assay, respectively. The donor cells were incubated in 1 ml of LB at different pH values or temperatures for 15 min before recipient cells were added for mating for 30 min at corresponding pH or temperatures. The mating experiments were performed as previously described (Lu et al, 2002). Donor ability was calculated as the number of transconjugants divided by the number of donors. Each data point was obtained from four replicates.
Analysis of TraM–TraD interactions using affinity chromatography
All the following procedures were performed at room temperature (22–25°C). Purified His6-TraDcyt or His-tagged TraK (200 μg) in 100 μl of binding buffer (50 mM Tris–HCl (pH 7.5), 10 mM imidazole, 250 mM NaCl) was mixed with 20 μl of 50% Ni-NTA agarose resin (Qiagen) gently for 2 h. The resin was pelleted by centrifugation at 15 000 g for 10 s and was washed two times with pH 7.5 or 9.0 buffer (50 mM Tris–HCl, 10 mM imidazole, 50 mM NaCl). The resin washed with pH 7.5 or 9.0 buffer was suspended in 0.3 ml of pH 7.5 or 9.0 buffer, respectively. BSA (150 μg) and 150 μg of purified TraM58–127 (or one of its mutant proteins) were added to the suspension and mixed gently for 4 h. The resin was pelleted by centrifugation at 15 000 g for 10 s and washed with 100 μl of same pH buffers. The washed resin was eluted with 25 μl of elution buffer (50 mM Tris–HCl (pH 7.8), 500 mM imidazole, 250 mM NaCl). A 5 μl volume was run on a 15% SDS–polyacrylamide gel and proteins were detected by Coomassie blue staining.
Other assays
Immunoblot analysis of TraM and EMSA were performed as previously described (Lu et al, 2004).
Supplementary Material
Supplementary Figure 1A and B
Acknowledgments
We thank other members of the Glover lab for their help during the progress of this project. We also thank Joanne Lemieux and the staff at Advanced Light Source (ALS) Beamline 8.3.1 for help during X-ray diffraction data collection. This work was supported by Canadian Institutes for Health Research (CIHR). The Alberta Synchrotron Institute (ASI) synchrotron access program is supported by grants from the Alberta Science and Research Authority (ASRA) and the Alberta Heritage Foundation for Medical Research (AHFMR). JL is supported by a postdoctoral fellowship from AHFMR.
References
- Abo T, Ohtsubo E (1993) Repression of the traM gene of plasmid R100 by its own product and integration host factor at one of the two promoters. J Bacteriol 175: 4466–4474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur DC, Ghetu AF, Gubbins MJ, Edwards RA, Frost LS, Glover JNM (2003) FinO is an RNA chaperone that facilitates sense–antisense RNA interactions. EMBO J 22: 6346–6355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beranek A, Zettl M, Lorenzoni K, Schauer A, Manhart M, Koraimann G (2004) Thirty-eight C-terminal amino acids of the coupling protein TraD of the F-like conjugative resistance plasmid R1 are required and sufficient to confer binding to the substrate selector protein TraM. J Bacteriol 186: 6999–7006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullough PA, Hughson FM, Skehel JJ, Wiley DC (1994) Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371: 37–43 [DOI] [PubMed] [Google Scholar]
- Chan DC, Fass D, Berger JM, Kim PS (1997) Core structure of gp41 from the HIV envelope glycoprotein. Cell 89: 263–273 [DOI] [PubMed] [Google Scholar]
- Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E (2005) Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol 59: 451–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csitkovits VC, Dermic D, Zechner EL (2004) Concomitant reconstitution of TraI-catalyzed DNA transesterase and DNA helicase activity in vitro. J Biol Chem 279: 45477–45484 [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L, Frost LS, Paranchych W (1992) The TraM protein of the conjugative plasmid F binds to the origin of transfer of the F and ColE1 plasmids. Mol Microbiol 6: 2951–2959 [DOI] [PubMed] [Google Scholar]
- Disque-Kochem C, Dreiseikelmann B (1997) The cytoplasmic DNA-binding protein TraM binds to the inner membrane protein TraD in vitro. J Bacteriol 179: 6133–6137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublie S (1997) Preparation of selenomethionyl proteins for phase determination. Methods Enzymol 276: 523–530 [PubMed] [Google Scholar]
- Eckert DM, Kim PS (2001) Mechanisms of viral membrane fusion and its inhibition. Annu Rev Biochem 70: 777–810 [DOI] [PubMed] [Google Scholar]
- Everett R, Willetts N (1980) Characterisation of an in vivo system for nicking at the origin of conjugal DNA transfer of the sex factor F. J Mol Biol 136: 129–150 [DOI] [PubMed] [Google Scholar]
- Fekete RA, Frost LS (2000) Mobilization of chimeric oriT plasmids by F and R100-1: role of relaxosome formation in defining plasmid specificity. J Bacteriol 182: 4022–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete RA, Frost LS (2002) Characterizing the DNA contacts and cooperative binding of F plasmid TraM to its cognate sites at oriT. J Biol Chem 277: 16705–16711 [DOI] [PubMed] [Google Scholar]
- Frost LS, Ippen-Ihler K, Skurray RA (1994) Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol Rev 58: 162–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YH, Tsai MM, Luo YN, Deonier RC (1991) Deletion analysis of the F plasmid oriT locus. J Bacteriol 173: 1012–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis-Ruth FX, Moncalian G, Perez-Luque R, Gonzalez A, Cabezon E, de la Cruz F, Coll M (2001) The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409: 637–641 [DOI] [PubMed] [Google Scholar]
- Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166: 557–580 [DOI] [PubMed] [Google Scholar]
- Harbury PB, Zhang T, Kim PS, Alber T (1993) A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262: 1401–1407 [DOI] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77: 51–59 [DOI] [PubMed] [Google Scholar]
- Holton J, Alber T (2004) Automated protein crystal structure determination using ELVES. Proc Natl Acad Sci USA 101: 1537–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard MT, Nelson WC, Matson SW (1995) Stepwise assembly of a relaxosome at the F plasmid origin of transfer. J Biol Chem 270: 28381–28386 [PubMed] [Google Scholar]
- Karl W, Bamberger M, Zechner EL (2001) Transfer protein TraY of plasmid R1 stimulates TraI-catalyzed oriT cleavage in vivo. J Bacteriol 183: 909–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsman A, Willetts N (1978) The requirements for conjugal DNA synthesis in the donor strain during Flac transfer. J Mol Biol 122: 287–300 [DOI] [PubMed] [Google Scholar]
- Lanka E, Wilkins BM (1995) DNA processing reactions in bacterial conjugation. Annu Rev Biochem 64: 141–169 [DOI] [PubMed] [Google Scholar]
- Lu J, Fekete RA, Frost LS (2003) A rapid screen for functional mutants of TraM, an autoregulatory protein required for F conjugation. Mol Genet Genomics 269: 227–233 [DOI] [PubMed] [Google Scholar]
- Lu J, Frost LS (2005) Mutations in the C-terminal region of TraM provide evidence for in vivo TraM-TraD interactions during F-plasmid conjugation. J Bacteriol 187: 4767–4773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Manchak J, Klimke W, Davidson C, Firth N, Skurray RA, Frost LS (2002) Analysis and characterization of the IncFV plasmid pED208 transfer region. Plasmid 48: 24–37 [DOI] [PubMed] [Google Scholar]
- Lu J, Zhao W, Frost LS (2004) Mutational analysis of TraM correlates oligomerization and DNA binding with autoregulation and conjugative DNA transfer. J Biol Chem 279: 55324–55333 [DOI] [PubMed] [Google Scholar]
- Lupas A (1996) Coiled coils: new structures and new functions. Trends Biochem Sci 21: 375–382 [PubMed] [Google Scholar]
- Matson SW, Sampson JK, Byrd DR (2001) F plasmid conjugative DNA transfer: the TraI helicase activity is essential for DNA strand transfer. J Biol Chem 276: 2372–2379 [DOI] [PubMed] [Google Scholar]
- McRee DE (1999) XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J Struct Biol 125: 156–165 [DOI] [PubMed] [Google Scholar]
- Miller DL, Schildbach JF (2003) Evidence for a monomeric intermediate in the reversible unfolding of F factor TraM. J Biol Chem 278: 10400–10407 [DOI] [PubMed] [Google Scholar]
- Miller JH (1972) Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory [Google Scholar]
- Moore D, Wu JH, Kathir P, Hamilton CM, Ippen-Ihler K (1987) Analysis of transfer genes and gene products within the traB-traC region of the Escherichia coli fertility factor, F. J Bacteriol 169: 3994–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RJ, Perrakis A, Lamzin VS (2003) ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol 374: 229–244 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Nelson WC, Howard MT, Sherman JA, Matson SW (1995) The traY gene product and integration host factor stimulate Escherichia coli DNA helicase I-catalyzed nicking at the F plasmid oriT. J Biol Chem 270: 28374–28380 [PubMed] [Google Scholar]
- Otwinowski L, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Penfold SS, Simon J, Frost LS (1996) Regulation of the expression of the traM gene of the F sex factor of Escherichia coli. Mol Microbiol 20: 549–558 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Press [Google Scholar]
- Sastre JI, Cabezon E, de la Cruz F (1998) The carboxyl terminus of protein TraD adds specificity and efficiency to F-plasmid conjugative transfer. J Bacteriol 180: 6039–6042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab M, Gruber H, Hogenauer G (1991) The TraM protein of plasmid R1 is a DNA-binding protein. Mol Microbiol 5: 439–446 [DOI] [PubMed] [Google Scholar]
- Stockner T, Plugariu C, Koraimann G, Hogenauer G, Bermel W, Prytulla S, Sterk H (2001) Solution structure of the DNA-binding domain of TraM. Biochemistry 40: 3370–3377 [DOI] [PubMed] [Google Scholar]
- Tabor S, Richardson CC (1985) A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA 82: 1074–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tato I, Zunzunegui S, de la Cruz F, Cabezon E (2005) TrwB, the coupling protein involved in DNA transport during bacterial conjugation, is a DNA-dependent ATPase. Proc Natl Acad Sci USA 102: 8156–8161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC (2000) Maximum-likelihood density modification. Acta Crystallogr D 56 (Part 8): 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC (2002) Automated structure solution, density modification and model building. Acta Crystallogr D 58: 1937–1940 [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J (1999) Automated MAD and MIR structure solution. Acta Crystallogr D 55 (Part 4): 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A (1997) MOLREP: an automated program for molecular replacement. J Appl Crystallogr 30: 1022–1025 [Google Scholar]
- Verdino P, Keller W, Strohmaier H, Bischof K, Lindner H, Koraimann G (1999) The essential transfer protein TraM binds to DNA as a tetramer. J Biol Chem 274: 37421–37428 [DOI] [PubMed] [Google Scholar]
- Walmsley RH (1976) Temperature dependence of mating-pair formation in Escherichia coli. J Bacteriol 126: 222–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC (1997) Atomic structure of the ectodomain from HIV-1 gp41. Nature 387: 426–430 [DOI] [PubMed] [Google Scholar]
- Will WR, Lu J, Frost LS (2004) The role of H-NS in silencing the F transfer region during entry into stationary phase. Mol Microbiol 54: 769–782 [DOI] [PubMed] [Google Scholar]
- Willetts N, Wilkins B (1984) Processing of plasmid DNA during bacterial conjugation. Microbiol Rev 48: 24–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willetts NS, Finnegan DJ (1970) Characteristics of E. coli K12 strains carrying both an F prime and an R factor. Genet Res 16: 113–122 [DOI] [PubMed] [Google Scholar]
- Wilson IA, Niman HL, Houghten RA, Cherenson AR, Connolly ML, Lerner RA (1984) The structure of an antigenic determinant in a protein. Cell 37: 767–778 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1A and B


