Abstract
Regulation of Wnt transcriptional targets is thought to occur by a transcriptional switch. In the absence of Wnt signaling, sequence-specific DNA-binding proteins of the TCF family repress Wnt target genes. Upon Wnt stimulation, stabilized β-catenin binds to TCFs, converting them into transcriptional activators. C-terminal-binding protein (CtBP) is a transcriptional corepressor that has been reported to inhibit Wnt signaling by binding to TCFs or by preventing β-catenin from binding to TCF. Here, we show that CtBP is also required for the activation of some Wnt targets in Drosophila. CtBP is recruited to Wnt-regulated enhancers in a Wnt-dependent manner, where it augments Armadillo (the fly β-catenin) transcriptional activation. We also found that CtBP is required for repression of a subset of Wnt targets in the absence of Wnt stimulation, but in a manner distinct from previously reported mechanisms. CtBP binds to Wnt-regulated enhancers in a TCF-independent manner and represses target genes in parallel with TCF. Our data indicate dual roles for CtBP as a gene-specific activator and repressor of Wnt target gene transcription.
Keywords: Armadillo, CtBP, TCF, Wingless, Wnt
Introduction
The Wnt/β-catenin pathway is a signaling cascade that is highly conserved among metazoans (Cadigan and Nusse, 1997; Primus and Freeman, 2004). This pathway is used throughout animal development to control a variety of cell fate decisions (Cadigan and Nusse, 1997; Logan and Nusse, 2004). Mutations causing constitutive activation of Wnt/β-catenin signaling have been identified in many human cancers (Polakis, 2000). Several studies suggest that Wnt/β-catenin signaling promotes oncogenesis by maintaining a proliferative, stem cell fate (Willert et al, 2003; Pinto and Clevers, 2005). In addition, perturbation of this pathway has been linked to abnormal bone density and vascular defects of the eye in humans (Logan and Nusse, 2004).
Wnt/β-catenin signaling is regulated by the stability and cellular location of a pool of β-catenin that is distinct from the β-catenin associated with adherens complexes. In the absence of Wnt stimulation, this pool of β-catenin is small and largely cytosolic. This is due to constitutive phosphorylation by a so-called degradation complex, which contains Axin, the adenomatous polyposis coli (APC) protein, glycogen synthase kinase 3 (GSK3) and casein kinase 1 (Ding and Dale, 2002). Phosphorylated β-catenin is then targeted to the ubiquitin/proteosome degradation pathway (Daniels et al, 2001). Upon Wnt stimulation, the degradation complex is inactivated, causing the accumulation of hypophosphorylated β-catenin. This stabilized β-catenin then translocates into the nucleus where it complexes with transcription factors, most notably members of the TCF family of DNA-binding proteins (Roose and Clevers, 1999).
In the absence of Wnt signaling, TCFs are thought to function as repressors of Wnt target gene expression, as has been suggested for other transcription factors mediating signaling (Barolo and Posakony, 2002). It has been shown that TCFs can form a repressive complex by interacting with transcriptional co-repressors of the Groucho/TLE (Gro) family (Cavallo et al, 1998; Roose et al, 1998). β-catenin and Gro bind competitively to TCF through overlapping binding sites, suggesting that β-catenin displaces Gro once it enters the nucleus, relieving transcriptional repression (Daniels and Weis, 2005).
In addition to relieving TCF repression, β-catenin is thought to activate directly Wnt target gene expression by recruiting additional proteins to TCF-bound chromatin. In Drosophila, Legless (Lgs) acts as an adaptor between Armadillo (Arm), the fly β-catenin, and Pygopus (Pygo), which promotes transcriptional activation (Kramps et al, 2002; Thompson, 2004; Hoffmans et al, 2005). Lgs binds to Arm repeats in the N-terminal half of Arm (Hoffmans and Basler, 2004), consistent with the finding that the N-terminal half of β-catenin has potent transcriptional activation activity (Hsu et al, 1998). In addition, both β-catenin and Arm have been shown to possess a distinct transcription activation domain at their C-terminus (van de Wetering et al, 1997; Hsu et al, 1998; Cox et al, 1999). This portion of Arm/β-catenin has been shown to bind to CBP/p300 (Hecht et al, 2000; Takemaru and Moon, 2000) as well as with the TRRAP/TIP60 and mixed-lineage leukemia (MLL1/MLL2) SET1-type chromatin-modifying complexes (Sierra et al, 2006). This region of Arm/β-catenin can also bind to the chromatin remodeler Brg-1 (Barker et al, 2001) and the zinc-finger protein Teashirt (Gallet et al, 1999). These interactions are thought to contribute to the ability of TCF/β-catenin to activate Wnt target genes, supporting the model that β-catenin/Arm converts TCFs from repressors to transcriptional activators (van Es et al, 2003).
In this report, we focus on the role of C-terminal-binding protein (CtBP) in the regulation of Wnt target genes in Drosophila. CtBPs are well-characterized transcriptional corepressors that have sequence homology to D2-hydroxyacid dehydrogenases and are known to bind to several DNA-binding proteins (Chinnadurai, 2002). Mammals have two CtBP genes, while flies have only one, which is alternatively spliced to form a shorter and longer isoform, both containing the dehydrogenase domain but differing in their C-termini (Nibu et al, 1998; Poortinga et al, 1998).
CtBPs can directly interact with some vertebrate TCFs, and overexpression of CtBP inhibits TCF-mediated transcriptional activation of reporter genes (Brannon et al, 1999; Valenta et al, 2003). These observations support a model where TCF bound by CtBP and Gro silences Wnt target genes in unstimulated cells. However, another report found no interaction between CtBP and TCF, and provided evidence that CtBP antagonizes Wnt signaling by binding to APC and diverting β-catenin/Arm away from TCF (Hamada and Bienz, 2004). This is further supported by data that a APC/CtBP complex is recruited to a Wnt transcriptional target, where it somehow dislodges β-catenin from TCF to suppress Wnt signaling (Sierra et al, 2006).
Both models for CtBP function are based largely on studies with reporter genes containing concatermerized TCF binding sites, which may not reflect the regulation of endogenous Wnt targets. In this report, we find that CtBP is required for basal repression of naked cuticle (nkd), a direct transcriptional target of TCF/Arm. Our loss of function data suggests that CtBP acts in parallel to TCF and Gro to repress basal nkd expression. Our data cannot be explained by increased access of Arm to TCF, ruling out the mechanism proposed involving APC/CtBP interaction with Arm (Hamada and Bienz, 2004; Sierra et al, 2006). In unstimulated cells, CtBP binds the nkd Wnt-regulated enhancer (WRE), but this binding is TCF-independent. Our data are consistent with CtBP acting on elements distinct from the TCF binding sites within the nkd control region to repress nkd expression in the absence of Wnt stimulation.
In addition to its already postulated role in Wnt target gene repression, we demonstrate that CtBP is required for the activation of some Wnt targets, both in wing imaginal discs and cultured cells. CtBP is recruited to WREs by Wnt stimulation, and can be recruited to a reporter gene by the full-length Arm or the N-terminal half of Arm. These data argue strongly for a previously unsuspected role for CtBP as a gene-specific Wnt transcriptional activator.
Results
CtBP both represses and activates Wingless signaling in Drosophila
Expression of wingless (wg), a fly Wnt, via the GMR promoter in the developing eye results in a severe reduction in adult eye size (Figure 1A; Cadigan et al, 2002). This GMR/wg background was used to screen for genes that could suppress the small eye phenotype when overexpressed. Random genes were placed under the control of Gal4-dependent promoters using a bidirectional EP element known as P[GSV] (Toba et al, 1999). Two GSV transposon insertions (P[GSV]A396 and P[GSV]A132) located in the first intron of CtBP strongly suppress the GMR/wg phenotype (Figure 1A–C). Both inserts drive the expression of CtBP in a Gal4-dependent manner, as judged by immunostaining (see Supplementary data, Supplementary Figure S1 and data not shown). CtBP expression also strongly suppressed the effects of an activated form of Arm (Arm*; Figure 1D–F), which cannot be phosphorylated by the Axin/APC/GSK3/CKI degradation complex (Freeman and Bienz, 2001). The effect of CtBP was specific for Wnt signaling, as P[GSV]A396 and P[GSV]A132 did not suppress the small eye phenotype generated by GMR-hid, a potent activator of apoptosis (data not shown). These data suggest that overexpression of CtBP can block the Wg pathway downstream of Arm stabilization.
Figure 1.
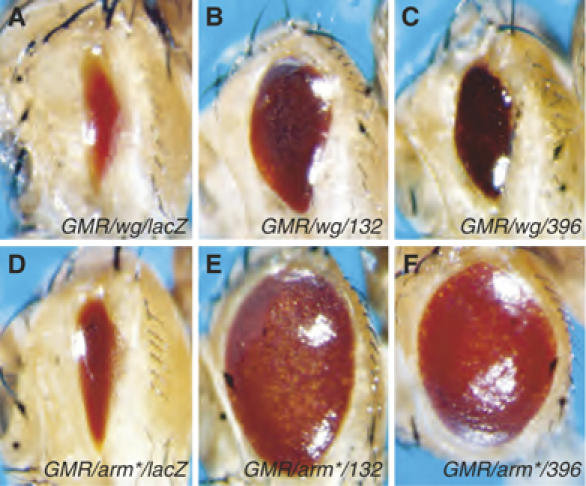
Two GSV insertions in the CtBP locus suppress Wg and Arm-dependent signaling in the eye. Micrographs of adult fly heads containing P[GMR-Gal4] and P[UAS-wg] (A–C) or P[GMR-Arm*] (D–F) and the following transposons: (A, D) P[UAS-lacZ], (B, E) P[GSV]A132 or (C, F) P[GSV]A396. Expression of wg via the GMR promoter produces an eye that is severely reduced in size (A), and this phenotype is suppressed by A132 or A396 (B, C). Expression of an activated form of Arm (Arm*) also causes eye size reduction (D) that is dramatically suppressed by A132 or A396 (E, F). Each transgene is present in one copy/fly and flies were reared at 25°C.
To examine the effect of CtBP overexpression on endogenous Wg signaling, we turned to the wing imaginal disc. In this larval tissue, a stripe of Wg expression at the dorsal/ventral (D/V) boundary of the wing blade primordia (the wing pouch) regulates target genes such as senseless (sens) and Distal-less (Dll) (Cadigan, 2002; Parker et al, 2002). Endogenous CtBP is predominately localized to the nucleus and its expression/localization does not appear to be regulated by Wg signaling (see Supplementary data and Supplementary Figure S1).
During late third instar larvae, Wg signaling activates Sens expression in two stripes adjacent to the Wg D/V stripe (Nolo et al, 2000; Parker et al, 2002; see Figure 2A and B). Expression of CtBP in the posterior compartment of the wing pouch using Engrailed (En)-Gal4 (Figure 2I) had no effect on Wg expression (Figure 2E), but caused a severe reduction in Sens levels (Figure 2F). In contrast to Sens, Dll and a Dll-lacZ reporter are expressed in a broader domain surrounding the Wg D/V stripe (Figure 2C and D). CtBP overexpression (Figure 2K) caused a significant expansion of the Dll-lacZ expression domain (Figure 2H) and a more subtle expansion of endogenous Dll protein (Figure 2G). Since there was no obvious expansion of the Wg stripe at the D/V boundary (Figure 2E), the effect on Dll expression is unlikely to be due to ectopic Wg expression.
Figure 2.
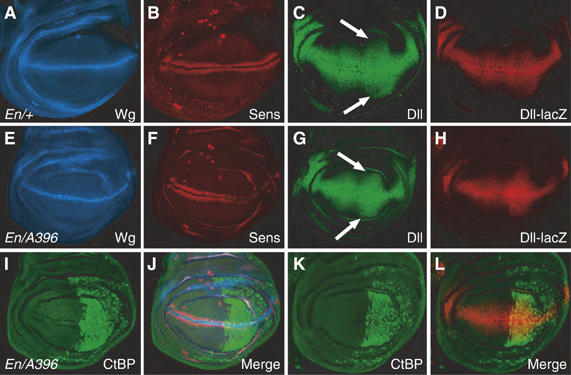
CtBP overexpression can activate and repress Wg targets in the wing imaginal discs. Confocal images of wing imaginal discs from late third instar larva. (A, B) P[En-Gal4]/+ disc immunostained for Wg (blue) and Sen (red) displaying the wild-type expression pattern. (C, D) P[En-Gal4]/P[Dll-lacZ] discs stained for Dll (green) and lacZ (red) showing the normal broad expression surrounding the D/V boundary. (E, F, I, J) P[En-Gal4]/P[GSV]A396 disc, where CtBP (green) is overexpressed in the posterior compartment. Wg expression (blue) is unaffected, while Sens (red) is sharply reduced in the CtBP-expressing domain. (G) P[En-Gal4]/P[GSV]A396 disc stained for Dll (green), displaying a subtle but reproducible expansion of Dll expression in the posterior compartment (compare arrows in panel G with those in panel C). (H, K, L) P[En-Gal4] P[GSV]A396 P[Dll-lacZ] disc stained for CtBP (green) and lacZ (red), exhibiting a significantly wider Dll-lacZ expression domain in the posterior compartment.
The experiments described in Figures 1 and 2 are complicated by the fact that they are based on overexpression of CtBP. In addition, it is also possible that the bidirectional P[GSV] EP element is driving expression of other genes in addition to CtBP. To determine whether endogenous CtBP is regulating Wg targets, mitotic clones of a strong allele of CtBP (CtBP87De-10; Poortinga et al, 1998) were examined for Sens and Dll expression. These clones had no detectable signal with anti-CtBP antisera (see Supplementary data and Supplementary Figure S1) and did not disrupt Wg expression (data not shown). For Sens, the effect of removing CtBP was different depending on the developmental stage. The double row of Sens on either side of the Wg D/V stripe is initiated during mid-third instar. It starts in the middle of the pouch, at the anterior/posterior boundary, and expands both anteriorly and posteriorly over the next 18 h. By 6 h prior to pupariation, the Sens double row spans the entire wing pouch (DS Parker and KM Cadigan, unpublished). Thus, the Sens pattern is a sensitive indicator of the developmental stage of the wing imaginal discs.
The effect of loss of CtBP on Sens expression is stage-specific. At 12–15 h before pupariation, Sens expression is absent in CtBP clones (Figure 3A–C), even though the Sens double row is present on either side of the clones. However, in later discs, Sens is expressed normally inside CtBP clones, with no obvious expansion of the normal Sens domain (Figure 3D–F). These data suggest a lag in Wg activation of Sens in cells lacking CtBP. There was also a consistent (90%) reduction of Dll levels in CtBP clones (Figure 3G–I), which is more pronounced in cells further away from the Wg stripe (see arrowheads in Figure 3G–I). The CtBP clonal analysis suggests a positive role for CtBP in Wg signaling.
Figure 3.
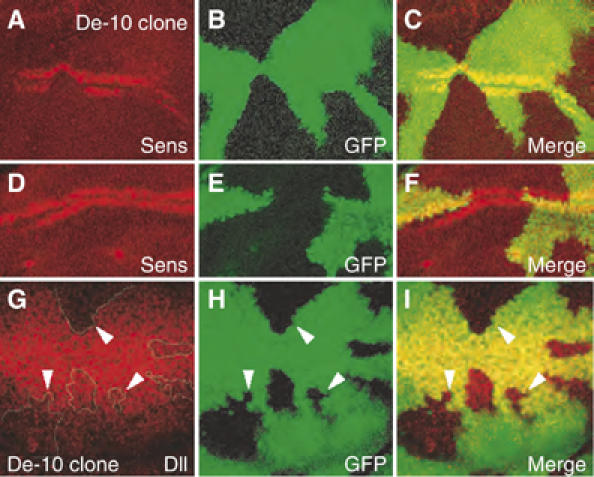
Loss of endogenous CtBP results in a reduction in activation of Wg targets in the wing imaginal discs. CtBP activity was removed by creating mitotic clones of CtBP87De-10 (De-10), a strong CtBP allele. Clones are marked by the absence of GFP (green). Clones displayed a highly penetrant (100%, n=8) loss of Sens expression at ∼12–15 h before pupariation (A–C), which was not observed in clones from discs that were a few hours prior to pupariation (D–F). A reduction in Dll expression (90% penetrance, n=30) was observed in late third instar discs (G–I), which was more pronounced in clones further from the D/V boundary (white arrowheads).
CtBP is a gene-specific repressor and activator of Wg targets in cultured cells
To examine the mechanism by which CtBP regulates Wg target gene expression, we utilized a cell culture model that appears to faithfully recapitulate Wg signaling. Kc167 (Kc) cells are derived from embryonic hemocytes (Goto et al, 2001) and are responsive to Wg signaling, as judged by reporter genes (Lum et al, 2003; DasGupta et al, 2005). To identify endogenous targets of the pathway, we performed microarray analysis with control cells and cells stimulated by Wg-conditioned media (Wg-CM). Several activated targets were identified (T Blauwkamp and K Cadigan, unpublished) and two of them, nkd and CG6234, are described here in detail. nkd is a Wg antagonist that is activated by Wg signaling in flies (Zeng et al, 2000) and CG6234 is predicted to encode a membrane protein of unknown function (http://flybase.bio.indiana.edu). Wg-CM induced the transcript levels of CG6234 and nkd seven- to 15- or 15- to 30-fold, respectively (Figure 4A, B, E and F). RNA interference (RNAi)-mediated knockdown of TCF or the coactivators pygopus (pygo) and arm blocked Wg activation of these genes (Figure 4A and B), indicating that nkd and CG6234 are activated through the canonical β-catenin/Arm pathway.
Figure 4.
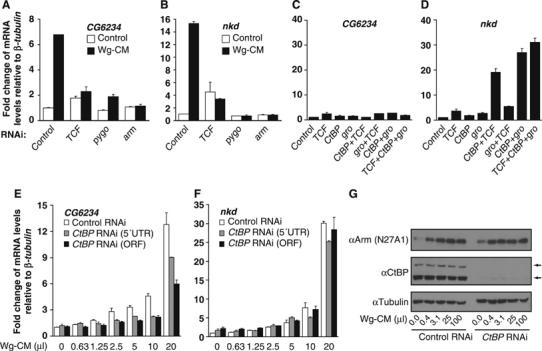
CtBP represses as well as activates endogenous Wg targets in Kc cells. (A, B) Kc cells were treated with control dsRNA or sequences corresponding to TCF, pygo and arm for 4 days before the addition of control media or Wg-CM for 4 h. Transcript levels of CG6234 (A) and nkd (B) were measured by quantitative RT–PCR as described in Materials and methods. Results were normalized to β-tubulin56D expression. The RNAi efficiencies for each gene were monitored by Western analysis to ensure that the corresponding protein levels were significantly reduced (data not shown). The induction of CG6234 and nkd expression by Wg-CM was severely reduced by TCF, Pygo or Arm depletion. (C, D) Synergistic derepression of nkd but not CG6234 by CtBP RNAi combined with the RNAi of TCF or gro. Kc cells were treated with the indicated dsRNAs for 4 days before harvesting and expression analysis as described above. (E, F) Cells were incubated with control dsRNA or duplexes specific for the 5′UTR or ORF of CtBP for 4 days before stimulation with increasing amounts of Wg-CM for 4 h before analysis of CG6234 (E) and nkd (F) expression as described above. Each bar represents the mean of duplicate cultures and duplicate transcript determinations, with the lines indicating the standard error. All experiments have been performed at least three separate times and representative experiment is shown. (G) Western analysis with anti-Arm and anti-CtBP antibody in control or CtBP RNAi-treated cells with increasing amounts of Wg-CM. CtBP RNAi severely affected CtBP expression (the two arrows indicate the short and long CtBP isoforms) but had no effect on Arm stabilization by Wg-CM. αTubulin levels are used as a loading control. The gels shown are representative of three separate experiments.
The standard model predicts that in the absence of Wnt stimulation, transcriptional targets of the pathway are repressed by TCFs bound by corepressors such as Gro (Cavallo et al, 1998; Roose et al, 1998) and CtBP (Brannon et al, 1999; Valenta et al, 2003). To test whether repression was occurring in Kc cells, we inhibited CtBP, TCF and gro using RNAi. Depletion of any of these genes individually had a minimal effect on CG6234 basal expression (Figure 4C). nkd was slightly more sensitive, with CtBP or gro inhibition causing a two- to three-fold increase in expression, and TCF a three- to five-fold increase (Figure 4B and D). When multiple genes were knocked down, a more dramatic difference between the two Wg targets was observed. CG6234 depression was never more than three-fold (Figure 4C), while that of nkd could exceed 30-fold (Figure 4D). Interestingly, gro/TCF RNAi treatment only slightly increased nkd expression compared to TCF alone, suggesting that these proteins act together to repress nkd. Synergistic derepression was only observed when CtBP depletion was combined with either gro or TCF (Figure 4D). This suggests that CtBP is acting in parallel to TCF/Gro to repress the basal level of nkd expression, not through TCF as proposed previously.
In addition to its role in repression in the absence of Wg signaling, CtBP is also required for maximal activation of CG6234 expression by Wg (Figure 4E). In several experiments, CtBP depletion caused a two- to three-fold reduction in the activation of CG6234 by intermediate levels of Wg-CM, and usually less than two-fold at higher Wg-CM concentrations (Figure 4E and data not shown). Inhibition of CtBP did not affect the ability of Wg to stabilize Arm (Figure 4G), and had no effect on the activation of nkd expression by Wg (Figure 4F). As is the case in the wing imaginal disc, CtBP is required for optimal activation of Wg targets, although the data with nkd indicates that this effect is gene-specific.
CtBP binding to WREs in endogenous Wg transcriptional targets
To determine whether CtBP directly regulates Wg transcriptional regulation of endogenous targets, we characterized the WREs in CG6234 and nkd. We identified clusters of putative TCF binding sites in the intergenic and intronic regions of nkd and CG6234 using an online tool called Target Explorer (http://trantor.bioc.columbia.edu/Target_Explorer; Sosinsky et al, 2003). The matrix that defined the search criteria is based on the DNA-binding data from the sloppy pair1, even-skipped, Ultrabithorax and decapentapleigic Wg-dependent enhancers (Riese et al, 1997; Lee and Frasch, 2000; Yang et al, 2000; Knirr and Frasch, 2001). For CG6234, two regions containing TCF sites were identified (C#1 & C#2), located approximately 2.8 and 1.8 kb upstream from the transcription initiation site respectively (Figure 5A). A 1.8 kb fragment containing several TCF sites was fused upstream of the hsp70 core promoter driving luciferase (pCG6234). This construct was activated 10- to 20-fold by cotransfection with Arm*. The WRE was further localized to a 1.15 kb fragment containing the C#1 sites and three other potential TCF sites (pCG6234A). Mutation of all these sites (pCG6234Amut) abolishes the reporter's responsiveness to Arm* (Figure 5A). Similar data were obtained for a 420 bp fragment containing TCF cluster N#5 in the first intron of nkd (Figure 5D and J Li and K Cadigan, in preparation). Thus, both Wg targets contain functional TCF sites that respond to Wg signaling, suggesting that they are direct targets of the pathway.
Figure 5.
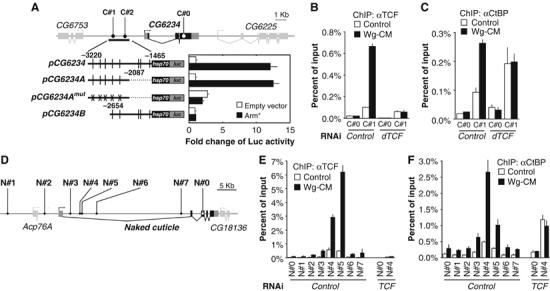
CtBP binding to WREs is activated by Wg signaling, but is TCF-independent in the absence of Wg. (A) Schematic diagram of the CG6234 locus showing the location of the predicted TCF sites (C#1 and 2) and a coding region control site (C#0) used for ChIP analysis. pCG6234 is a 1.7 kb fragment containing the predicted TCF sites (vertical lines) fused to a hsp70 core promoter and luciferase. This reporter construct, and one containing the 5′ 1133 bp of the fragment (pCG6234A) are activated by co-transfection of Arm*. This activation is abolished by mutations of all the potential TCF sites (pCG6234Amut). Each bar is the mean of duplicate transfections where luciferase activity was determined in duplicate, with the standard error indicated by the lines. The data shown is a representative example from three separate experiments. (B) TCF binding to C#1 and the C#0 control, as assayed by ChIP in cells treated with control or TCF dsRNA and stimulated with control or Wg-CM for 4 h. Wg stimulation increases TCF binding, and little signal is observed in TCF-depleted cells. (C) Cells treated as in (B) analyzed for CtBP binding. Wg stimulation increased the CtBP ChIP signal on the WRE, and this increase is abolished in TCF-depleted cells. However, CtBP binding in the absence of Wg is TCF-independent. (D) nkd locus with the predicted TCF site clusters (N#1–7) and a coding region control site (N#0) for ChIP analysis. (E, F) ChIP analysis shows overlapping binding of TCF (E) and CtBP (F) to the nkd control region. Binding of both proteins is increased by a 4 h treatment of Wg-CM, and CtBP Wg-dependent recruitment is blocked in TCF-depleted cells. As with CG6234, CtBP binding in the absence of Wg is TCF-independent. For both loci, in the absence of Wg signaling, there is a reproducible increase in the CtBP ChIP signal in TCF-depleted cell extracts. It should also be noted that relative strength of the ChIP signal for N#4 and N#5 sites in nkd varies from experiment to experiment for both TCF and CtBP. The proximity of these primers (352 bp) is less than the resolution of ChIP (based on the size of the sonicated fragments). However, these primer sets always give higher signals than primers corresponding to other regions of the nkd locus. The bars for each ChIP signal are the mean of duplicate precipitations, with duplicate Q-PCR reactions. Standard errors are indicated by the lines and each experiment has been carried out at least three times with similar results.
Further support for direct regulation of CG6234 and nkd by TCF comes from chromatin immunoprecipitation (ChIP) studies using antibodies against endogenous TCF. Strong TCF binding was observed on the C#1 site upstream of the CG6234 transcriptional unit, compared to the CG6234 ORF (C#0; Figure 5B). Preferential binding to two TCF binding site clusters in the nkd intron (N#4 and N#5) is also observed (Figure 5D and E), consistent with these sequences containing a WRE. TCF binding to the WREs in both genes is greatly enhanced after a 4 h treatment of Wg-CM. TCF expression is not activated by Wg signaling (data not shown) and the mechanism of this Wg-dependent increase of TCF binding to the WREs is under investigation. The TCF ChIP signal is dramatically reduced by RNAi depletion of TCF, indicating that it is specific for TCF (Figure 5B and E).
Following identification of bona fide WREs in CG6234 and nkd, ChIP analysis using antibodies against endogenous CtBP was performed to determine whether CtBP occupies this region of the chromatin. Preferential CtBP binding was found for both the C#1 site in CG6234 (Figure 5C) and N#4 and N#5 in nkd (Figure 5F). Stimulation of the cells with Wg-CM for 4 h reveals a marked increase in CtBP binding to these sites (Figure 5C and F). These data indicate that CtBP is physically present on these WREs both in the absence and presence of Wg signaling, consistent with CtBP playing a direct role in both repression and activation of Wg targets.
The pattern of TCF and CtBP binding to the CG6234 and nkd regulatory regions is consistent with TCF recruitment of CtBP to DNA (Figure 5B, C, E and F). To test this, CtBP binding to chromatin was determined in cells that were depleted for TCF via RNAi. Two results were observed. First, CtBP still binds to the WREs of both genes after TCF depletion (Figure 5C and F), under conditions where TCF binding was greatly reduced (Figure 5B and E). In fact, a 1.5- to 2.0-fold increase in the CtBP ChIP signal was consistently observed in TCF-depleted versus control cells, the cause of which is not clear. The data strongly argue that, in the absense of Wg, CtBP recruitment to the WREs is TCF-independent. The second result is that the Wg-dependent increase in CtBP binding to the WREs was abolished by TCF depletion (Figure 5C and F). This suggests that the Wg-dependent increase in CtBP recruitment to the WREs requires TCF.
CtBP mediates transcriptional activation through the N-terminal half of Arm
To further explore the activating role of CtBP in Wg signaling, we examined a simple reporter gene system where the requirement for TCF is bypassed by fusing Arm to the Gal4 DNA-binding domain (Gal4DBD). As shown for Gal4-β-catenin (Hsu et al, 1998), Gal4Arm can activate a UASluc reporter (Figure 6B, C, E and F). Arm contains at least two transcriptional activation domains, one in the N-terminal half (Gal4ArmN) and another in the C-terminal portion of Arm (Gal4ArmC; see Figure 6A–C). Coexpression of either the short or long isoform of CtBP with Gal4Arm or Gal4ArmN consistently enhanced (five- to 12-fold) their ability to activate UASluc (Figure 6B), although the effects were greatest when Gal4Arm or Gal4ArmN activation of UASluc was kept to a moderate level (five- to 10-fold over Gal4DBD; data not shown). However, CtBP expression had no effect on the ability of Gal4ArmC to activate the reporter (Figure 6B), regardless of the level of Gal4ArmC expressed (data not shown). Conversely, CtBP depletion via RNAi reduced the activity of Gal4Arm and Gal4ArmN four- to eight-fold, but not Gal4ArmC (Figure 6C). The RNAi effect on endogenous CtBP is specific, as judged by the ability of a CtBP transgene not targeted by the dsRNA (corresponding to the 5′UTR of the endogenous transcripts) to rescue the CtBP RNAi defect of Gal4Arm transcriptional activation (Figure 6E). These data indicate that CtBP acts through the N-terminus of Arm to activate transcription of UASluc.
Figure 6.
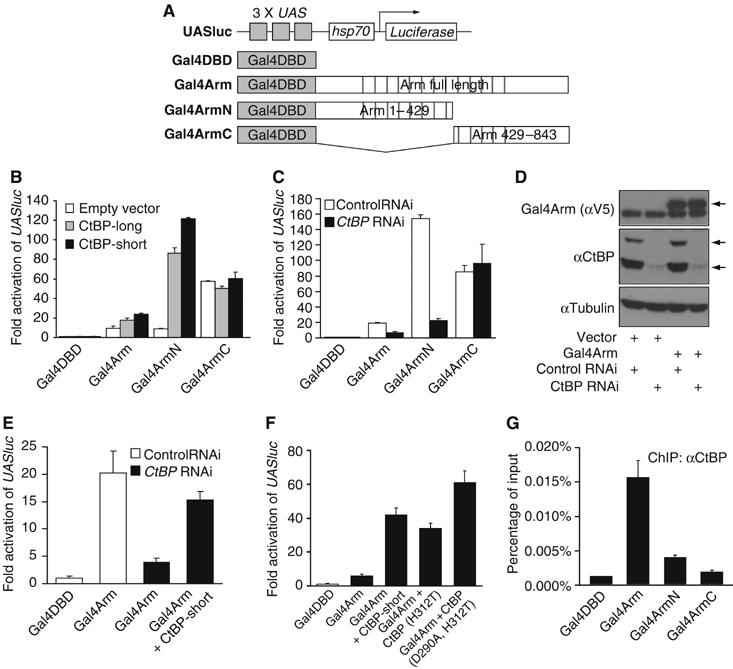
CtBP is required for Arm-dependent transcription activity. (A) Schematic diagram of the UAS reporter and the Gal4 expression vectors used in the following experiments. (B) Overexpression of both short and long forms of CtBP (500 ng/well) enhances the transcription activities of Gal4Arm (50 ng/well) and Gal4ArmN but not Gal4ArmC (each 20 ng/well) on the UASluc reporter in Kc cells. Cells were transfected and luciferase and β-galactosidase activities assayed as described in Materials and methods. (C) CtBP RNAi using a dsRNA corresponding to the 5′UTR diminishes the transcription activities of Gal4Arm and Gal4ArmN but not Gal4ArmC (all 100 ng/well) on UASluc reporter. (D) Western blot analysis shows the expression levels of Gal4Arm (V5 tagged) and short and long isoforms of CtBP in cells with RNAi and transfection as indicated. CtBP RNAi severely reduced the amount of endogenous CtBP, but had no effect on transfected Gal4Arm (the band below Gal4Arm is non-specific). (E) The effect of CtBP RNAi (5′UTR) can be rescued by a CtBP transgene containing a heterologous 5′UTR. (F) Mutations of conserved catalytic residues (D290A and H312T) in CtBP had no effect on enhancement on UASluc reporter activity with Gal4Arm (5 ng/well). (G) CtBP ChIP in cells co-transfected with UASluc (1 ng/well) and Gal4DBD, Gal4Arm, Gal4ArmN or Gal4ArmC constructs (500 ng/well each) shows enhanced occupancy of CtBP on the UAS sites of UASluc in cells containing Gal4Arm and Gal4ArmN compared with Gal4DBD. Gal4ArmC did not show a significant difference with Gal4DBD in several experiments. Each bar is the mean of duplicate transfections and duplicate luciferase or Q-PCR reactions, with the lines indicating the standard errors. Each experiment was performed at least three times and similar results were obtained in each experiment.
CtBPs exhibit sequence homology to D2-hydroxyacid dehydrogeneases (Schaeper et al, 1995) and biochemical studies have shown that human CtBP1 (hCtBP1) is a functional dehydrogenase (Kumar et al, 2002). Mutations in the catalytic site of hCtBP1 blocked its ability to interact with known binding partners such as E1A to repress transcription (Kumar et al, 2002). Because the dehydrogenase domain is highly conserved between hCtBP1 and fly CtBP (72% identity, 84% similarity), we took advantage of the crystal structure of hCtBP1 (Kumar et al, 2002) to examine whether dehydrogenase activity is required for the ability of fly CtBP to augment Gal4Arm transcriptional activation. The conserved residues aspartate 290 and histidine 312, crucial for substrate binding and catalysis (Kumar et al, 2002), were converted to alanine and threonine, respectively. Two mutant proteins (CtBP-H312T and CtBP-D290A, H312T) were able to activate Gal4Arm activity as effectively as wild-type CtBP (Figure 6F). Both mutant proteins were expressed at similar levels as the wild type (data not shown). These data suggest that dehydrogenase activity is not required for CtBP's ability to enhance Arm transcriptional activation.
We have tested whether Arm and CtBP interact when coexpressed in Kc cells, but no association was detected (data not shown). This raises the possibility that CtBP's effect on Arm activity is indirect. To address this, ChIP was performed on cells transfected with different Gal4Arm fusions and UASluc. The ability of anti-CtBP antisera to pull down UASluc was enhanced almost 10-fold by Gal4Arm compared to Gal4DBD (Figure 6G). In several experiments, transfection of Gal4ArmN gave a reproducible three-fold increase in CtBP ChIP signal compared to Gal4DBD, while Gal4ArmC was not consistently higher than the negative control (Figure 6G and data not shown). These data are consistent with the functional interaction between CtBP and the N-terminus of Arm, and indicate a physical association of CtBP and Arm on WREs.
Discussion
CtBP represses Wnt target genes independently of TCF and Arm
CtBP has previously been identified as a repressor of Wnt signaling, as measured by TCF reporter genes in cultured cells (Valenta et al, 2003; Hamada and Bienz, 2004). Consistent with this, we identified CtBP in an overexpression screen via its ability to suppress Wg and Arm action in the developing eye (Figure 1). In wing imaginal discs, CtBP overexpression also inhibited the Wg target Sens (Figure 2F). Consistent with this overexpression data, the reduction of CtBP in cultured cells via RNAi is also consistent with a role for CtBP in repressing some Wnt targets (Figure 4D).
Our working model for CtBP repression of Wnt target gene expression is summarized in Figure 7A. CtBP is bound to the same area of the nkd and CG6234 loci as TCF, but this binding is TCF-independent (Figure 5C and F). Consistent with this, knock down of CtBP and TCF or gro synergistically derepressed nkd expression (Figure 4D). No synergism was seen with TCF/gro double depletions (Figure 4D). The RNAi and ChIP data together favor a model where CtBP acts in parallel with TCF/Gro to repress nkd expression in the absence of Wg stimulation. Because CtBP has no detectable ability to bind nucleic acids (Chinnadurai, 2002), we assume that unknown DNA-binding protein(s) recruit CtBP to the WRE (Figure 7A).
Figure 7.
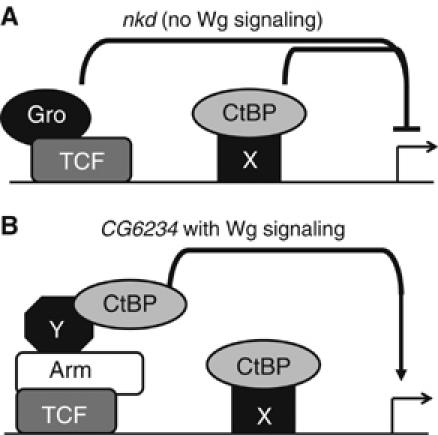
Model depicting CtBP functions in the absence or presence of Wg signaling. (A) In the absence of Wg signaling, CtBP (presumably recruited to the WRE by an unknown protein) acts in parallel to TCF/Gro to repress nkd gene expression. (B) Upon Wg stimulation, CtBP is recruited by Arm and other factors to the TCF binding sites, where it contributes to activation of targets such as CG6234.
The existing models for CtBP antagonism of Wnt signaling cannot explain our data. TCF-independent recruitment of CtBP to WREs is not consistent with work suggesting direct binding of CtBP to TCF (Brannon et al, 1999; Valenta et al, 2003). The alternative mechanism, where a CtBP/APC complex diverts Arm/β-catenin away from TCF (Hamada and Bienz, 2004; Sierra et al, 2006), also is inconsistent with our results. In this model, the activation of nkd expression after CtBP RNAi treatment would be dependent on TCF and arm. Because the derepression of nkd occurred when both CtBP and TCF were depleted (Figure 3D) and was not affected when arm was also inhibited (data not shown), we do not favor this model to explain the effects of CtBP depletion on nkd expression. These distinct mechanisms for CtBP repression are not mutually exclusive and may all occur in some contexts.
There is a qualitative difference in the amount of derepression found between the two Wg targets studied in Kc cells. Depletion of CtBP and TCF/gro causes a large (20- to 30-fold) increase in nkd basal expression (Figure 3D), but has a much more modest (<3-fold) effect on CG6234 (Figure 3C). These differences may reflect a fundamental difference in the way TCF/Gro and CtBP act on various Wnt targets in unstimulated cells, but it is equally likely that the surrounding cis-elements in these targets have a strong influence on the degree of derepression that can be observed.
CtBP plays a direct role in transcriptional activation of Wnt targets
In addition to defining a novel mechanism for CtBP repression of Wg targets, we provide strong evidence for CtBP playing a role in Wg-mediated transcriptional activation. In the wing imaginal discs, loss of CtBP resulted in a lag in Wg-dependent activation of Sens (Figure 3A and D) and a reduction in Dll expression (Figure 3G). In cultured Kc cells, CtBP depletion caused a two- to three-fold reduction in the ability of Wg to activate CG6234 expression (Figure 4E). The ability of Gal4-Arm chimeras to activate a Gal4 reporter gene is also highly dependent on CtBP levels (Figure 6B and C). In all these contexts, CtBP is not absolutely required for Wg signaling, but is necessary for maximal activation of Wg/Arm transcriptional activation.
The positive effect of CtBP on Wg signaling is direct, as judged by ChIP. Assuming that ChIP is measuring the degree of occupancy of CtBP on the chromatin, and not simply antigen accessibility, Wg stimulation promotes the association of CtBP with the CG6234 WRE (Figure 5C). This increase in CtBP binding is not observed in TCF-depleted cells (Figure 5C). Gal4-Arm recruits endogenous CtBP to a UASluc reporter (Figure 6G). Taken together, these data support a model where TCF/Arm recruits CtBP to Wg targets. We have been unable to detect binding between Arm and CtBP by co-immunoprecipitation (data not shown), suggesting that another factor(s) may act as an adaptor between CtBP and the Arm bound to TCF (Figure 7B).
As is the case for β-catenin (Hsu et al, 1998), Arm has transcriptional activation activity in both the N- and C-terminal portions of the protein (Figure 6A–C). CtBP overexpression or RNAi depletion greatly effects the activity of the N-terminal half of Arm but has no effect on the C-terminal portion (Figure 6B and C). Consistent with this, the N-terminal portion can recruit CtBP to a reporter gene, but not the C-terminus (Figure 6G). Other factors that have been linked to the N-terminal portion of Arm include Lgs and Pygo (Kramps et al, 2002; Hoffmans and Basler, 2004) and the ATPases Pontin and Reptin (Bauer et al, 2000). It may be that CtBP acts in concert with one or more of these factors.
CtBPs have strong sequence similarity with D2-hydroxyacid dehydrogenases (Schaeper et al, 1995). hCtBP1 is a functional dehydrogenase and point mutations blocking CtBP1 dehydrogenase activity inhibit its ability to interact with binding partners and act as a transcriptional corepressor (Kumar et al, 2002). However, another group found that similar mutations had no effect on the ability of CtBP to repress transcription (Grooteclaes et al, 2003). In our report, mutation of two residues (D290A and H312T) predicted to be essential for catalytic activity had no effect on the ability of fly CtBP to potentiate Gal4-Arm transcriptional activation (Figure 6F). Further complicating the issue is data from experiments expressing the fly CtBP fused to Gal4DBD in mammalian cells (Phippen et al, 2000). In some cells, Gal4-CtBP activated a UAS reporter, while the same reporter was repressed in other cell lines. Interestingly, conversion of CtBP's catalytic histidine to glutamine abolished transcriptional activation, but not repression (Phippen et al, 2000). The heterologous nature of these experiments and the differences in the assays employed may explain the discrepancy between these studies, and further experiments will be needed on endogenous targets to determine how much dehydrogenase activity of CtBP contributes to repression and activation of Wnt targets.
Although CtBP is required for maximal activation of CG6234 expression and a Gal4-Arm-dependent reporter gene, Wg activation of nkd did not appear to require CtBP (Figure 4F). The basis for this gene-specific requirement for CtBP is not clear. CtBP is recruited to the nkd WRE in a Wg-dependent manner (Figure 5F), similar to what was observed for CG6234 (Figure 5C). It may be that CtBP is required for nkd activation, but this is masked by its role in repressing nkd expression. This hypothesis could be tested if we are able to separate CtBP's activator and repressor activities.
The requirement for CtBP in Wnt transcriptional activation may have been previously overlooked due to its well-characterized role as a co-repressor. For example, mouse embryos that lack CtBP2 have axial truncations and reduced Brachyury (T) expression that is reminiscent of Wnt3a mutants (Hildebrand and Soriano, 2002). These results suggest that the activating role for CtBP in Wnt signaling that we have identified is evolutionarily conserved.
Materials and methods
Drosophila genetics
A bidirectional EP element, P[GSV] (Toba et al, 1999), was mobilized using P[delta 2–3] as the source of P element transposase (Robertson et al, 1988). Insertions were screened for the ability to suppress P[GMR-Gal4] P[UAS-wg] and P[GMR-Gal4] P[GMR-arm*] as described previously (Parker et al, 2002). The insertions were mapped using inverse PCR as described on the Berkeley Drosophila Genome Project website (http://www.fruitfly.org/about/methods/inverse.pcr.html). CtBP87De-10, EnGal4, Dll-lacZ, UASlacZ, ywP[hsFLP]1 and P[FRT]82B P[Ubi-GFP] P[lacW]RpL141/TM6 were obtained from the Bloomington Stock Center.
Experiments with GMR-Gal4 were carried out at 25°C, while those with En-Gal4 were performed at 18°C. Clones of CtBP87De-10 were generated by mitotic recombination using hsFLP and a P[FRT]82B P[Ubi-GFP] P[lacW]RpL141chromosome carrying a Minute mutation via a 1 h 37°C heat shock at 48–72 h after egg laying.
Antibodies
Rabbit polyclonal anti-CtBP antisera was generated against bacterially produced full-length CtBP (accession no. AB011840) GST fusion protein. Antisera were affinity purified using GST-CtBP coupled to a AminoLink Plus Column (Pierce). Rabbit polyclonal anti-TCF antisera against the N-terminus of TCF were generated as described previously (Chan and Struhl, 2002). Guinea-pig anti-Sens was generated as described (Nolo et al, 2000). N2 7A1 (anti-Arm) was from Developmental Studies Hybridoma Bank at the University of Iowa. Anti-V5 epitope antibody was purchased from Invitrogen. For Western blot analysis, anti-CtBP (1:1000) and anti-Arm (1:1000) were followed by HRP-anti-rabbit IgG or HRP-anti-mouse (Amersham Bioscience), respectively. Signal was detected using the ECL kit (Amersham Bioscience). Immunostaining of wing imaginal discs were performed as described previously (Cadigan et al, 1998), using rabbit anti-CtBP (1:500), guinea-pig anti-Sens (1:500) and rabbit anti-Dll antisera (1:100) (Panganiban et al, 1995). Cy3- and Alexa 488-conjugated secondary antibodies were from Jackson Immunochemicals and Molecular Probes, respectively. Samples were examined using an Axiophot (Zeiss) coupled to a LSM 510 confocal apparatus (Zeiss).
Drosophila cell culture
Kc167 (Kc) cells were routinely cultured in the Schneider's Drosophila media (Invitrogen) containing 5% FBS at room temperature. RNAi-mediated gene knockdowns were performed essentially as described (Clemens et al, 2000). Briefly, cells were resuspended at 2 × 106/ml in Drosophila SFM (Invitrogen), seeded at 106/well and 9 μg of dsRNA added. After a 1 h incubation, 1 ml of media containing 7.5% FBS was added. Cells were harvested on the fourth day. Primers for dsRNA synthesis are available in the Supplementary data (Table I).
Table 1.
Primer sequences for dsRNA templates, RT–PCR and ChIP analysis
| dsRNA | Control | gaattaatacgactcactatagggagaatgattg aacaagatggattgcacgca |
| gaattaatacgactcactatagggagaaatatca cgggtagccaacgctatgtcct | ||
| CtBP (ORF) | gaattaatacgactcactatagggagaatgcaca aagcacctccgaaatacacga | |
| gaattaatacgactcactatagggagagcaccag gtcgcatctgtttaattgtgaat | ||
| CtBP (5′UTR) | gaattaatacgactcactatagggagaattctcg atttcaatatgaagcgccaa | |
| gaattaatacgactcactatagggagagctgttt ttcaatctgtctgctgctgtcct | ||
| groucho | gaattaatacgactcactatagggagaccattag ccctgactcgaaggtgtgctt | |
| gaattaatacgactcactatagggagagttttac tgccgatgctgctgctgttgt | ||
| armadillo | gaattaatacgactcactatagggagaatgagtt acatgccagcccagaatcgaa | |
| gaattaatacgactcactatagggagacgatggt gtgataagttgtgcagtgttccta | ||
| pygopus | ttaatacgactcactatagggagaccgctacaac cgaatttcttgc | |
| ttaatacgactcactatagggagagtgattcata tgcggcggtagtc | ||
| TCF | gaattaatacgactcactatagggagagaagatg actacgatgatgataaactaggcgga | |
| gaattaatacgactcactatagggagaaataggg tttcgggatgtgtttggcatt | ||
| RT–PCR | β-tubulin56D | agacctactgcatcgacaac |
| gacaagatggttcaggtcac | ||
| CG6234 | gctgctctgcgtgatcgtcttc | |
| tctggtgttggtgaactctcctcc | ||
| naked cuticle | taaaattctcggcggctacaa | |
| cgcacctggtggtacatcag | ||
| ChIP | C#0 | accttctggctttggagcag |
| tgggctcctcataaactggc | ||
| C#1 | tgcataatgcatacgatcgga | |
| tgttcggcggaaaagctaaa | ||
| N#0 | ccagcatcgctatcgacca | |
| gcgtccttctccttttcgct | ||
| N#1 | ttgatcccgattccccattc | |
| ttggtgttccatcacagccac | ||
| N#2 | tcaatttgcagctctggcact | |
| agtataatggaatttaataggcgcgat | ||
| N#3 | cctttgaattccccctgcat | |
| gccaggccaacactttgaac | ||
| N#4 | tcaatcagacgtcagaggtaccg | |
| ctgatggaagaaccgtgttgg | ||
| N#5 | aattttcccagaccgctttcc | |
| cgaaaaagccgccaaacatat | ||
| N#6 | tcagcatcggctacagcga | |
| aaactcttatcaaacaggagccca | ||
| N#7 | ttctaccggcaccattcacg | |
| ttcccctcgaaataattgctactg | ||
| UASluc | tgctagctcgaggccttgag | |
| gctgcgcttgtttatttgctt |
Transient transfections were carried out with Fugene 6 (Roche Applied Science) according to the manufacturer's instruction. If a transfection was combined with RNAi, cells were washed with media twice on the second day after RNAi treatment, and transfection was performed according to the same procedure (see below). pAcCtBP-long and pAcCtBP-short expression vectors were generated by subcloning the KpnI/XbaI long and short CtBP fragments with double FLAG tags at the N-terminus (gifts from Dr D Arnosti) into a pAc5.1 expression vector (Invitrogen). Mutant CtBP (CtBP-H312T and CtBP-D290A, H312T) constructs were made by PCR-based introduction of the mutation into the pAcCtBP-short vector. The Arm* expression vector was constructed by first cloning a full-length fragment of Arm into the pAc5.1 vector, followed by introducing mutations that substitute Thr52 and Ser56 to Ala via PCR. Reporter constructs of CG6234 (pCG6234, pCG6234A&B and pCG6234Amut) were made by incorporating KpnI/XmaI PCR fragments as indicated in Figure 4A into a pGL3-Basic vector (Promega). The constructing of UASluc reporter vector, Gal4DBD and Gal4Arm fusion expression vectors are described elsewhere (J Li and K Cadigan, in preparation).
For the UASluc luciferase reporter assays, a mixture of DNA containing 100 ng UASluc and the following expression vectors, 1 ng pAclacZ (Invitrogen), 20–100 ng of Gal4Arm fusions and 500–625 ng of CtBP vectors, were co-transfected. Gal4DBD or pAc5.1 vectors were used to normalize the DNA content or as controls. Cells were harvested 2 days later. Luciferase and β-galactosidase activities were assayed using the Tropix Luc-Screen and Galacto-Star kits (Applied Biosystems) and quantitated with a Chameleon plate luminometer (Hidex Personal Life Science). Transfection efficiency was normalized using the pAclacZ β-galactosidase activities. For the CG6234 reporters, 100 ng of the reporter construct, 1 ng pAclacZ and 500 ng Arm* vectors were co-transfected into 106 cells and the Luciferase activity was measured 2 days later as for UASluc.
Wg--CM was collected using stable pTubwg S2 cells, kindly provided by Dr R Nusse from Stanford University, and was typically concentrated for ∼50-fold using a Centricon tube (Millipore) and stored at −80°C. Kc cells were treated with Wg-CM for 4 h prior to harvesting.
Real-time quantitative PCR (Q-PCR)
All real-time Q-PCR analyses were carried out with iQ SYBR Green Supermix (BioRad) on a iCycler iQ real-time PCR detection system (BioRad). For RT–PCR, total RNA from 1 to 5 × 106 Kc cells was extracted with Trizol Reagent (Invitrogen), and reverse transcribed with Stratascript reverse transcriptase (Stratagene) followed by Q-PCR analysis. Sequences of the primer pairs used are listed in Table I.
ChIP
ChIP analysis was performed using a ChIP assay kit (Upstate Biotechnology) essentially as described by the manufacturer, except that we have included a initial protein–protein crosslinking step by incubating cells with a 10 mmol/l dimethyl 3,3′-dithio-bis(propionimidate) dihydrochloride (Sigma-Aldrich) solution for 30 min on ice as described (Fujita et al, 2003). A total of 2–3 × 106 cells and 5–10 μl of antisera were typically used per ChIP analysis. All resulting precipitated DNA samples were quantified with Q-PCR. Data are expressed as the percent of input DNA.
Supplementary Material
Supplementary Figure S1
Supplementary Figure Legend
Acknowledgments
We thank RJ Wessells, J Krupp and R Bodmer for help in generating the P[GSV] insertions. We also thank D Arnosti, G Struhl and H Bellen for providing the bacterial expression vectors used for the production of anti-CtBP, TCF and Sens antisera, and D Arnosti for CtBP cDNAs. Special thanks to R Nusse for the pTubwg S2 cells and the Bloomington Stock Center and Hybridoma Bank for reagents. We also thank Y Liu, D Parker and JH Chang and Y Ni for careful reading of the manuscript. This work was supported by NIH Grants RO1 GM59846 and CA95869 to KMC.
References
- Barker N, Hurlstone A, Musisi H, Miles A, Bienz M, Clevers H (2001) The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. EMBO J 20: 4935–4943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barolo S, Posakony JW (2002) Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev 16: 1167–1181 [DOI] [PubMed] [Google Scholar]
- Bauer A, Huber O, Usseglio F, Rothbacher U, Aragnol D, Kemler R, Pradel J (2000) Pontin 52 and reptin 52 function as antagonistic regulators of beta-catenin signalling activity. EMBO J 19: 6121–6130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannon M, Brown JD, Bates R, Kimelman D, Moon RT (1999) XCtBP is a XTcf-3 co-repressor with roles throughout Xenopus development. Development 126: 3159–3170 [DOI] [PubMed] [Google Scholar]
- Cadigan KM (2002) Regulating morphogen gradients in the Drosophila wing. Semin Cell Dev Biol 13: 83–90 [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Fish MP, Rulifson EJ, Nusse R (1998) Wingless repression of Drosophila frizzled 2 expression shapes the Wingless morphogen gradient in the wing. Cell 93: 767–777 [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Jou AD, Nusse R (2002) Wingless blocks bristle formation and morphogenetic furrow progression in the eye through repression of Daughterless. Development 129: 3393–3402 [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Nusse R (1997) Wnt signaling: a common theme in animal development. Genes Dev 11: 3286–3305 [DOI] [PubMed] [Google Scholar]
- Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, Peifer M, Bejsovec A (1998) Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature 395: 604–608 [DOI] [PubMed] [Google Scholar]
- Chan SK, Struhl G (2002) Evidence that armadillo transduces wingless by mediating nuclear export or cytosolic activation of pangolin. Cell 111: 265–280 [DOI] [PubMed] [Google Scholar]
- Chinnadurai G (2002) CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol Cell 9: 213–224 [DOI] [PubMed] [Google Scholar]
- Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, Hemmings BA, Dixon JE (2000) Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci USA 97: 6499–6503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RT, Pai LM, Kirkpatrick C, Stein J, Peifer M (1999) Roles of the C terminus of Armadillo in Wingless signaling in Drosophila. Genetics 153: 319–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels DL, Eklof Spink K, Weis WI (2001) Beta-catenin: molecular plasticity and drug design. Trends Biochem Sci 26: 672–678 [DOI] [PubMed] [Google Scholar]
- Daniels DL, Weis WI (2005) Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat Struct Mol Biol 12: 364–371 [DOI] [PubMed] [Google Scholar]
- DasGupta R, Kaykas A, Moon RT, Perrimon N (2005) Functional genomic analysis of the Wnt-wingless signaling pathway. Science 308: 826–833 [DOI] [PubMed] [Google Scholar]
- Ding Y, Dale T (2002) Wnt signal transduction: kinase cogs in a nano-machine? Trends Biochem Sci 27: 327–329 [DOI] [PubMed] [Google Scholar]
- Freeman M, Bienz M (2001) EGF receptor/Rolled MAP kinase signalling protects cells against activated Armadillo in the Drosophila eye. EMBO Rep 2: 157–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Jaye DL, Kajita M, Geigerman C, Moreno CS, Wade PA (2003) MTA3, a Mi-2/NuRD complex subunit, regulates an invasive growth pathway in breast cancer. Cell 113: 207–219 [DOI] [PubMed] [Google Scholar]
- Gallet A, Angelats C, Erkner A, Charroux B, Fasano L, Kerridge S (1999) The C-terminal domain of armadillo binds to hypophosphorylated teashirt to modulate wingless signalling in Drosophila. EMBO J 18: 2208–2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto A, Aoki M, Ichihara S, Kitagawa Y (2001) Alpha-, beta- or gamma-chain-specific RNA interference of laminin assembly in Drosophila Kc167 cells. Biochem J 360: 167–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grooteclaes M, Deveraux Q, Hildebrand J, Zhang Q, Goodman RH, Frisch SM (2003) C-terminal-binding protein corepresses epithelial and proapoptotic gene expression programs. Proc Natl Acad Sci USA 100: 4568–4573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada F, Bienz M (2004) The APC tumor suppressor binds to C-terminal binding protein to divert nuclear beta-catenin from TCF. Dev Cell 7: 677–685 [DOI] [PubMed] [Google Scholar]
- Hecht A, Vleminckx K, Stemmler MP, van Roy F, Kemler R (2000) The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J 19: 1839–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand JD, Soriano P (2002) Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol Cell Biol 22: 5296–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmans R, Basler K (2004) Identification and in vivo role of the Armadillo–Legless interaction. Development 131: 4393–4400 [DOI] [PubMed] [Google Scholar]
- Hoffmans R, Stadeli R, Basler K (2005) Pygopus and legless provide essential transcriptional coactivator functions to armadillo/beta-catenin. Curr Biol 15: 1207–1211 [DOI] [PubMed] [Google Scholar]
- Hsu SC, Galceran J, Grosschedl R (1998) Modulation of transcriptional regulation by LEF-1 in response to Wnt-1 signaling and association with beta-catenin. Mol Cell Biol 18: 4807–4818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knirr S, Frasch M (2001) Molecular integration of inductive and mesoderm-intrinsic inputs governs even-skipped enhancer activity in a subset of pericardial and dorsal muscle progenitors. Dev Biol 238: 13–26 [DOI] [PubMed] [Google Scholar]
- Kramps T, Peter O, Brunner E, Nellen D, Froesch B, Chatterjee S, Murone M, Zullig S, Basler K (2002) Wnt/wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear beta-catenin-TCF complex. Cell 109: 47–60 [DOI] [PubMed] [Google Scholar]
- Kumar V, Carlson JE, Ohgi KA, Edwards TA, Rose DW, Escalante CR, Rosenfeld MG, Aggarwal AK (2002) Transcription corepressor CtBP is an NAD(+)-regulated dehydrogenase. Mol Cell 10: 857–869 [DOI] [PubMed] [Google Scholar]
- Lee HH, Frasch M (2000) Wingless effects mesoderm patterning and ectoderm segmentation events via induction of its downstream target sloppy paired. Development 127: 5497–5508 [DOI] [PubMed] [Google Scholar]
- Logan CY, Nusse R (2004) The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol 20: 781–810 [DOI] [PubMed] [Google Scholar]
- Lum L, Yao S, Mozer B, Rovescalli A, Von Kessler D, Nirenberg M, Beachy PA (2003) Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science 299: 2039–2045 [DOI] [PubMed] [Google Scholar]
- Nibu Y, Zhang H, Levine M (1998) Interaction of short-range repressors with Drosophila CtBP in the embryo. Science 280: 101–104 [DOI] [PubMed] [Google Scholar]
- Nolo R, Abbott LA, Bellen HJ (2000) Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell 102: 349–362 [DOI] [PubMed] [Google Scholar]
- Panganiban G, Sebring A, Nagy L, Carroll S (1995) The development of crustacean limbs and the evolution of arthropods. Science 270: 1363–1366 [DOI] [PubMed] [Google Scholar]
- Parker DS, Jemison J, Cadigan KM (2002) Pygopus, a nuclear PHD-finger protein required for Wingless signaling in Drosophila. Development 129: 2565–2576 [DOI] [PubMed] [Google Scholar]
- Phippen TM, Sweigart AL, Moniwa M, Krumm A, Davie JR, Parkhurst SM (2000) Drosophila C-terminal binding protein functions as a context-dependent transcriptional co-factor and interferes with both mad and groucho transcriptional repression. J Biol Chem 275: 37628–37637 [DOI] [PubMed] [Google Scholar]
- Pinto D, Clevers H (2005) Wnt, stem cells and cancer in the intestine. Biol Cell 97: 185–196 [DOI] [PubMed] [Google Scholar]
- Polakis P (2000) Wnt signaling and cancer. Genes Dev 14: 1837–1851 [PubMed] [Google Scholar]
- Poortinga G, Watanabe M, Parkhurst SM (1998) Drosophila CtBP: a hairy-interacting protein required for embryonic segmentation and hairy-mediated transcriptional repression. EMBO J 17: 2067–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primus A, Freeman G (2004) The cnidarian and the canon: the role of Wnt/beta-catenin signaling in the evolution of metazoan embryos. Bioessays 26: 474–478 [DOI] [PubMed] [Google Scholar]
- Riese J, Yu X, Munnerlyn A, Eresh S, Hsu SC, Grosschedl R, Bienz M (1997) LEF-1, a nuclear factor coordinating signaling inputs from wingless and decapentaplegic. Cell 88: 777–787 [DOI] [PubMed] [Google Scholar]
- Robertson HM, Preston CR, Phillis RW, Johnson-Schlitz DM, Benz WK, Engels WR (1988) A stable genomic source of P element transposase in Drosophila melanogaster. Genetics 118: 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roose J, Clevers H (1999) TCF transcription factors: molecular switches in carcinogenesis. Biochim Biophys Acta 1424: M23–M37 [DOI] [PubMed] [Google Scholar]
- Roose J, Molenaar M, Peterson J, Hurenkamp J, Brantjes H, Moerer P, van de Wetering M, Destree O, Clevers H (1998) The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature 395: 608–612 [DOI] [PubMed] [Google Scholar]
- Schaeper U, Boyd JM, Verma S, Uhlmann E, Subramanian T, Chinnadurai G (1995) Molecular cloning and characterization of a cellular phosphoprotein that interacts with a conserved C-terminal domain of adenovirus E1A involved in negative modulation of oncogenic transformation. Proc Natl Acad Sci USA 92: 10467–10471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra J, Yoshida T, Joazeiro CA, Jones KA (2006) The APC tumor suppressor counteracts b-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev 20: 586–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosinsky A, Bonin CP, Mann RS, Honig B (2003) Target explorer: an automated tool for the identification of new target genes for a specified set of transcription factors. Nucleic Acids Res 31: 3589–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemaru KI, Moon RT (2000) The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression. J Cell Biol 149: 249–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BJ (2004) A complex of Armadillo, Legless, and Pygopus coactivates dTCF to activate wingless target genes. Curr Biol 14: 458–466 [DOI] [PubMed] [Google Scholar]
- Toba G, Ohsako T, Miyata N, Ohtsuka T, Seong KH, Aigaki T (1999) The gene search system. A method for efficient detection and rapid molecular identification of genes in Drosophila melanogaster. Genetics 151: 725–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenta T, Lukas J, Korinek V (2003) HMG box transcription factor TCF-4's interaction with CtBP1 controls the expression of the Wnt target Axin2/Conductin in human embryonic kidney cells. Nucleic Acids Res 31: 2369–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, Peifer M, Mortin M, Clevers H (1997) Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell 88: 789–799 [DOI] [PubMed] [Google Scholar]
- van Es JH, Barker N, Clevers H (2003) You Wnt some, you lose some: oncogenes in the Wnt signaling pathway. Curr Opin Genet Dev 13: 28–33 [DOI] [PubMed] [Google Scholar]
- Willert K, Brown JD, Danenberg E, Duncan AW, Weissman IL, Reya T, Yates JR III, Nusse R (2003) Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature 423: 448–452 [DOI] [PubMed] [Google Scholar]
- Yang X, van Beest M, Clevers H, Jones T, Hursh DA, Mortin MA (2000) Decapentaplegic is a direct target of dTcf repression in the Drosophila visceral mesoderm. Development 127: 3695–3702 [DOI] [PubMed] [Google Scholar]
- Zeng W, Wharton KA Jr, Mack JA, Wang K, Gadbaw M, Suyama K, Klein PS, Scott MP (2000) Naked cuticle encodes an inducible antagonist of Wnt signalling. Nature 403: 789–795 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure Legend


