Abstract
Brochothrix campestris ATCC 43754 produces a heat-stable, two-component, nonlantibiotic, class IIb bacteriocin, brochocin C (BrcC), that is active against a broad range of gram-positive bacteria, including spores of Clostridium botulinum. An improved purification method was developed for BrcC, in which n-butanol and chloroform extraction are used. Mass spectral characterization of the two components, brochocin A (BrcA) and brochocin B (BrcB), showed that both components are excreted into the medium by B. campestris as mature peptides consisting of 59 and 43 amino acids, respectively. Separate expression clones of BrcA and BrcB were constructed previously in Carnobacterium piscicola LV17C, but the products were not chemically characterized. Purification by the new protocol showed that BrcA is expressed as the mature 59-amino-acid peptide but that BrcB is produced by C. piscicola as a fragment, BrcB(10-43), which is cleaved at an internal Gly-Gly site. This fragment is not antimicrobial by itself, but in combination with BrcA it displays the full activity of the BrcC complex. Circular dichroism measurements revealed a high β-sheet content in the secondary structure of both BrcA and BrcB(10-43), as well as in a 1:1 BrcA-BrcB(10-43) mixture. Separate expression clones of brcA and brcB were also constructed in Escherichia coli, but these clones only produced multiple fragments of the desired peptides with little or no activity.
Bacteriocins are peptides secreted by lactic acid bacteria that often display antimicrobial activity against other related bacteria (10). Bacteriocins commonly occur either as unmodified peptides with a signal peptide or a double glycine type leader peptide (except for possible disulfide bridges) (13, 22) or as lantibiotics with extensive posttranslational modification (5, 11, 24). Because of the increasing demand for more natural and microbiologically safe food products, there is a need for new preservation techniques. Bacteriocins have considerable potential for food preservation, as well as for human therapy as potential supplements or replacements for currently used antibiotics. A number of both modified and unmodified bacteriocins have been reported to occur as two-peptide systems in which both components are necessary for full antibacterial activity (4). Recently, our attention has been focused on brochocin C (BrcC) from Brochothrix campestris ATCC 43754, a heat-stable two-peptide unmodified bacteriocin system originally discovered by Siragusa and Cutter (19) and characterized by McCormick et al. (12). BrcC has a broad activity spectrum comparable to that of nisin (3). It is active against a broad range of gram-positive bacteria and spores of Clostridium and Bacillus species (8). Its antibacterial activity depends on the complementary action of its two constituents, brochocin A (BrcA) and brochocin B (BrcB), which have no detectable activities on their own (12). BrcA and BrcB are class IIb ribosomally synthesized peptides processed from longer precursors, and in mature form they are predicted to consist of 59 and 43 amino acids, respectively, based on genetic analysis (Fig. 1). The entire sequence has been published previously (12).
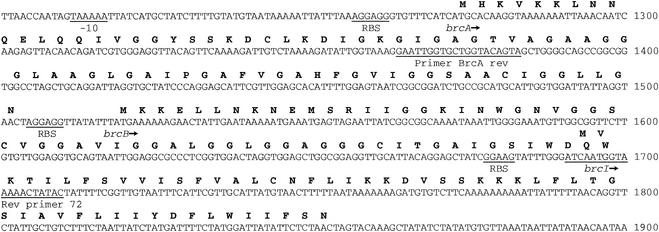
FIG. 1.
Single-strand DNA and amino acid sequences for nucleotides 1201 to 1900 of the EcoRI fragment of pAP7.4 containing the structural (brcA and brcB) and immunity (brcI) genes of brochocin C.
Since knowledge of the three-dimensional structure of bacteriocins can provide a basis for detailed studies of structure-activity relationships (29) and mechanisms of action, workers in our laboratories have employed nuclear magnetic resonance (NMR) spectroscopy to determine structures of single-peptide type IIa bacteriocins, such as leucocin A (2) and carnobacteriocin B2 (26). In the present study, as a prelude to NMR examination of two-peptide bacteriocin interactions, we characterized the peptides produced by separate Carnobacterium piscicola LV17C brcA and brcB clones and Escherichia coli brcA and brcB clones. The results show that the C. piscicola clones produce mature BrcA and a fully functional truncated fragment of BrcB missing the first nine amino acids, whereas the E. coli clones generate multiple fragments of BrcA or BrcB that are weakly active or inactive. Conformational circular dichroism studies were conducted with both the isolated peptides and a mixture of the two purified peptides. In addition, an effective and easy purification method applicable to both BrcA and BrcB was developed. The results presented in this paper provide insight into understanding how BrcC can be a potential meat preservative.
MATERIALS AND METHODS
Bacterial strains, plasmids, culture media, and growth conditions.
The plasmids and producer strains utilized in this study are listed in Table 1. Most bacteria were maintained as frozen stock cultures at −70°C in Bacto APT broth (All Purpose Tween; Difco Laboratories Inc., Detroit, Mich.) supplemented with 20% (vol/vol) glycerol; the exceptions were strains of E. coli, which were stored in Luria-Bertani (LB) broth under the same conditions. Prior to experimental use, E. coli strains were subcultured twice in LB broth and grown overnight with shaking at 37°C. All other strains were subcultured twice and grown overnight at 25°C in APT broth (first day) and in modified semidefined Casamino Acids medium (CAA medium) (second day) (6). Transformants of E. coli were selected on LB agar (Difco) with a selective concentration of ampicillin (150 μg/ml). For growth of Carnobacterium transformants on APT agar, erythromycin (5 μg/ml) was used. Solid and soft agar media were prepared by addition of granulated agar (1.5 and 0.75% [wt/vol], respectively) to the broth media.
TABLE 1.
Plasmids and producer strains utilized in this study
| Plasmid or producer strain | Relevant propertiesa | Reference or source |
|---|---|---|
| Plasmids | ||
| pT712 | Ampr, 2.8 kb, T7 polymerase expression vector | 21 |
| pJKM46 | pRW19e containing 309-bp HindIII-KpnI, dvn::brcB brcI fragment, 4.0 kb, Emr | 12 |
| pJKM61 | 292-bp Xbal-SacI fragment from pJKM53-1 and 426-bp SacI-KpnI fragment from pJKM46 cloned in XbaI and KpnI sites of pUC118, dvn::brcA dvn::brcB brcI, Ampr, 3.9 kb | 12 |
| pJKM56 | 292-bp XbaI-SacI fragment from pJKM53-1 and 444-bp SacI-KpnI background chromosomal fragment cloned in XbaI and KpnI sites of pMG36e, dvn::brcA, Emr, 4.3 kb | 12 |
| pMG36e | 3.6 kb, Emr | 23 |
| pSG1 | 292-bp XbaI-SacI fragment from pJKM61 cloned in XbaI and SalI sites of pT712, dvn::brcA, Ampr, 3.1 kb | This study |
| pSG15 | PCR fragment from pJKM46 cloned in XbaI and EcoRI sites of pT712, dvn::brcB, Ampr | This study |
| Strains | ||
| E. coli BL21 (DE3) | F−omp T rB mB int, bacteriophage DE3 lysogen carrying the T7 RNA polymerase gene controlled by the lacUV5 promoter | 20 |
| C. piscicola UAL26 | Plasmid free, produces uncharacterized bacteriocin, brochocin C sensitive | Laboratory collection |
| C. divergens LV13 | Wild-type divergicin A producer, brochocin C sensitive | 28 |
| C. piscicola LV17C | Plasmid free, brochocin C sensitive | 1 |
Ampr, ampicillin resistant; Emr, erythromycin resistant; dvn::brcA, brochocin C peptide A gene fused to DNA encoding divergicin A signal peptide; dvn::brcB, brochocin C peptide B gene fused to DNA encoding divergicin A signal peptide; brcI, brochocin C immunity gene.
DNA isolation and manipulation.
Plasmids from carnobacteria and E. coli were isolated as previously described (18, 28) and were purified by cesium chloride-ethidium bromide density gradient centrifugation (18). Restriction enzyme digestions were performed according to the manufacturer's instructions (Boehringer Mannheim, Dorval, Quebec, Canada; New England Biolabs, Mississauga, Ontario, Canada; Promega, Burlington, Ontario, Canada). Ligation and E. coli transformation were done by using standard methods (18). Agarose gel electrophoresis was conducted with Tris-borate-EDTA buffer.
Construction of brcA and brcB expression clones.
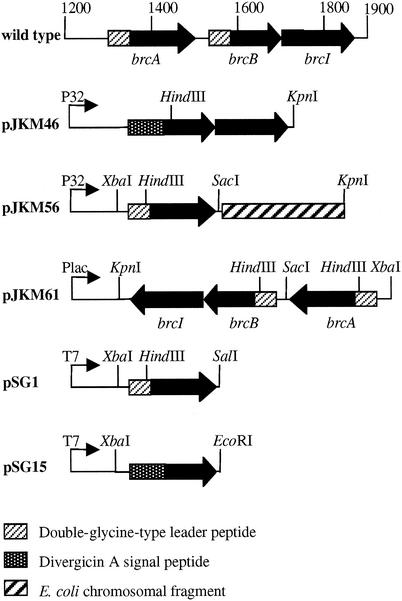
brcA and brcB expression clones were constructed in E. coli BL21(DE3). To construct plasmid pSG1, which carries brcA (Fig. 2), the dvn::brcA gene (292-bp fragment) from pJKM61 was excised with restriction enzymes XbaI and SalI and cloned into the XbaI and SalI sites of pT712. The brcB gene insert cloned in pSG15 was prepared by PCR. To construct pSG15, which carries brcB, primer 71 and reverse primer 72 were first used to amplify the dvn::brcB gene fusion from pJKM46. Primer 71 (5′-ATATTCTAGATTGGAGGTTGGTATATATG-3′) is based on the 5′ end of the nucleotide sequence encoding the divergicin A signal peptide (28) and contains an XbaI restriction site (underlined). Reverse primer 72 (5′-ATATGAATTCGTATAGTTTTTACCATTGAT-3′) is based on the 3′ end of brcB and contains an EcoRI restriction site (underlined). When the XbaI and EcoRI sites were used, the resulting PCR fragment was ligated into plasmid pT712 to obtain pSG15 (Fig. 2). Taqplus precision polymerase (Stratagene, Aurora, Ontario, Canada) was used to perform DNA amplification in PCR experiments (the denaturation, annealing, and extension temperatures were 94, 50, and 74°C, respectively). DNA sequencing confirmed the absence of mutations in the two constructs, pSG1 and pSG15, containing the genes encoding BrcA and BrcB, respectively.
FIG. 2.
Genetic organization of different expression clones of BrcA and BrcB in carnobacteria (pJKM46, pJKM56, and pJKM61) and in E. coli (pSG1 and pSG15).
DNA sequencing.
The nucleotide sequence of plasmid DNA was determined by the Taq DyeDeoxy cycle sequencing method (Department of Biochemistry, University of Alberta, Edmonton, Alberta, Canada) with an Applied Biosystems model 373A sequencer. Primer BrcA rev (5′-TACTGTACCAGCACCAATTC-3′) and forward and reverse primers 71 and 72 were used to determine the DNA sequences of pSG1 and pSG15, respectively.
T7 RNA polymerase-directed expression of BrcA and BrcB in E. coli.
Cultures of E. coli BL21(DE3) containing pSG1 and pSG15 were grown for 2 h in LB broth containing 150 μg of ampicillin per ml with shaking at 37°C to an optical density at 600 nm of 0.3. They were subsequently induced for expression of the T7 RNA polymerase gene by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) (final concentration, 0.4 mM). After 1 h of incubation at 37°C, rifampin was added to a final concentration of 200 μg/ml. After incubation for an additional 1 h, cells were harvested by centrifugation (8,000 × g, 15 min, 4°C) by using a Sorvall RC5B centrifuge with a model SLA-1500 rotor. They were washed and concentrated 100-fold in deionized water or ethanol and then lysed with a French press at 4°C. n-Butanol extraction (700 ml of n-butanol per liter of culture) of the supernatant, the French press-treated cells, and the cell debris was followed by high-performance liquid chromatography (HPLC). Both peptides were tested for activity on APT agar plates against C. piscicola LV17C containing pMG36e after each purification step.
HPLC.
The peptides produced by pSG1 and pSG15 were purified by HPLC by using a reversed-phase diphenyl column (VYDAC 219TP54; 300 Å; 5 μm; 4.6 by 250 mm) and an analytical Beckman System Gold apparatus equipped with 32karat software. All compounds were detected at 218 nm and eluted at a rate of 1 ml/min by using the following conditions: 90% H2O containing 0.075% trifluoroacetic acid (TFA) and 10% methanol containing 0.075% TFA for 2 min; increased to 70% methanol containing 0.075% TFA in 2 min; increased to 100% methanol containing 0.075% TFA in 8 min; 100% methanol containing 0.075% TFA for 13 min; decreased to 10% methanol containing 0.075% TFA in 2 min; and 10% methanol containing 0.075% TFA for 10 min. All fractions were tested for antimicrobial activity and analyzed by mass spectrometry.
Purification of BrcA and BrcB in carnobacteria.
Cultures of C. piscicola LV17C containing pJKM56 (BrcA) and pJKM46 (BrcB) were grown in a sterile modified semidefined CAA medium (no Tween 80, 5 μg of erythromycin per ml) at 25°C at a constant pH of 6.7 controlled by addition of filter-sterilized (pore size, 0.22 μm) 2 M NaOH with a Chemcadet controller (Cole-Parmer, Chicago, Ill.). After 21 h of incubation, cells were removed from the culture broth by centrifugation (8,000 × g, 20 min, 4°C). The supernatant was extracted twice for 20 min with n-butanol (500 ml/liter of culture each time), and the emulsion was broken by centrifugation (6,000 × g, 10 min, 4°C, polypropylene centrifuge bottles). The n-butanol was evaporated under reduced pressure at a low temperature (<30°C). The residue was suspended in water (10 ml/liter of culture) and extracted three times for 20 min with chloroform (150 ml/liter of culture each time). The organic layer containing the desired bacteriocin was separated by centrifugation (2,000 × g, 4 min, 4°C, Teflon centrifuge bottles) and evaporated under reduced pressure. The residue was suspended in a 1:1 methanol-chloroform mixture and loaded onto a Sephadex LH-20 (Amersham Pharmacia Biotech, Quebec, Canada) column (2.5 by 115 cm) and eluted at a rate of 0.5 ml/min (4 min/tube) with a 1:1 methanol-chloroform mixture. All fractions collected were tested for antimicrobial activity by the spot-on-lawn assay; the preparations were overlaid with LV17C(pMG36e). All pure active fractions were combined and analyzed by mass spectrometry, amino acid sequencing, and circular dichroism.
Bacteriocin detection and activity assay.
After each purification step, the number of arbitrary activity units of bacteriocin per milliliter of concentrated solution was determined by determining the reciprocal of the highest dilution that produced a zone of growth inhibition showing extracellular complementation of BrcA and BrcB peptides. Fresh overnight APT broth cultures of strains containing pJKM46 and pJKM56 were spotted onto different APT agar plates supplemented with erythromycin (5 μg/ml) and incubated for 18 h at 25°C. Beside the resulting colonies, the bacteriocins to be tested were spotted (10 μl), and an overlay of soft APT agar (6 ml) containing erythromycin (5 μg/ml) and an overnight culture of the indicator strain LV17C(pMG36e) (60 μl) was poured onto the plates. The plates were incubated again at 25°C for 18 h and examined for clear zones of inhibition.
Activity and stability of BrcA and BrcB.
Samples of both peptides from carnobacteria were tested for stability under different storage conditions (temperatures of −20, 4, and 25°C; under argon or exposed to air) after each step of the purification procedure. Residual activity was measured by the spot-on-lawn assay with LV17C(pMG36e) every 24 h for 4 to 6 days depending on the samples. It was established that the best storage conditions in all cases were under argon at −20°C. The degree of purity of a sample was also found to have an effect on its stability. The more impure the sample, the more stable it was. Pure samples of BrcA and BrcB exposed to air were found to lose all bactericidal activity in less than 48 h, whereas pure peptides kept under argon stayed active when they were stored at −20°C.
Mass spectrometry.
Mass spectrometry analyses were performed with a linear matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometer (Applied Biosystems Voyager Elite). All spectra were recorded in positive ion mode with an acceleration voltage of 20 kV in the presence of a nitrogen laser (λ, 337 nm) used for desorption and ionization of the samples. Samples were prepared by using α-cyano-4-hydroxycinnamic acid (Aldrich) as a matrix and were fixed to a gold target before analysis. Bovine insulin (MH+ = 1,046.542; Sigma) was used for calibration of the instrument, which was performed before each experiment.
N-terminal amino acid sequencing.
The amino acid sequence of partially purified BrcA was determined by the Alberta Peptide Institute (University of Alberta, Edmonton, Alberta, Canada) by automated Edman degradation with a gas phase protein sequencer (Applied Biosystems model 470A) equipped with an on-line model 120A phenylthiohydantoin amino acid analyzer.
Circular dichroism.
Circular dichroism measurement for purified samples of BrcA and BrcB(10-43) in methanol was performed (cell length, 0.02 cm) in the absence and in the presence of 50% trifluoroethanol at wavelengths between 250 and 188 nm at 25°C by using a JACSO J720 spectropolarimeter equipped with JASCO J700 software (performed by R. Luty, Department of Biochemistry, University of Alberta, Edmonton, Alberta, Canada).
Nucleotide sequence accession number.
The nucleotide sequence determined in this study has been deposited in the GenBank database under accession number AF075600 (12).
RESULTS
Purification of BrcA and BrcB in carnobacteria.
The purification procedure previously described for BrcC by McCormick et al. (12) did not allow successful isolation of both components from 3-liter fermentation cultures of C. piscicola containing pJKM56 or pJKM46 in a modified semidefined CAA medium. A new purification method consisting of n-butanol and chloroform extraction and size exclusion chromatography was developed. It is known that Tween 80, a detergent usually used to prevent the bacteriocin produced from sticking to the bacterial cells and the glassware, stops the adsorption of nisin and enterocin on polypropylene surfaces (9). As Tween 80 interferes with purification of the desired peptides and hampers mass spectral analyses, it was eliminated from the original CAA medium (6). Due to the highly hydrophobic character of both of the peptides studied, Sephadex LH-20, which can be used with organic solvents, was chosen as the solid support for size exclusion chromatography. The antimicrobial activity results observed at each stage of the purification procedure are summarized in Table 2. When this new purification procedure was used, about 2 mg of pure peptide could be recovered per liter of fermentation culture. The final specific activities of the purified BrcA and BrcB fractions were 706,000 and 800 arbitrary activity units/mg, respectively. As expected, the BrcA peptide displayed antimicrobial activity against indicator strain LV17C(pMG36e) only in the presence of BrcB or the BrcB-producing organism, whereas the BrcB peptide required BrcA or its producing organism for activity.
TABLE 2.
Purification of BrcA (pJKM56) and BrcB (pJKM46)
| Plasmid | Purification stage | Vol (ml) | Activity (AU/ml)a | Total activity (AU) | Protein concn (mg/ml) | Sp act (AU/mg) | Recovery (%) |
|---|---|---|---|---|---|---|---|
| pJKM56 | Culture supernatant | 1,000 | 3,200 | 3.2 × 106 | 100 | ||
| n-Butanol extraction (leave in H2O) | 10 | 204,800 | 2.0 × 106 | 63 | |||
| CHCl3 extraction | 1 | 819,200 | 8.2 × 105 | 26 | |||
| Sephadex LH-20 (leave in methanol) | 1.5 | 409,600 | 6.1 × 105 | 0.58 | 7.06 × 105 | 19 | |
| pJKM46 | Culture supernatant | 3,000 | 12,800 | 3.8 × 107 | 100 | ||
| n-Butanol extraction (leave in H2O) | 40 | 409,600 | 1.6 × 107 | 42 | |||
| CHCl3 extraction | 4 | 51,200 | 2.0 × 105 | 0.53 | |||
| Sephadex LH-20 (leave in methanol) | 3.22 | 1,600 | 5.2 × 103 | 2.00 | 8.00 × 102 | 0.01 |
Au, arbitrary activity units.
Mass spectrometry and amino acid sequence.
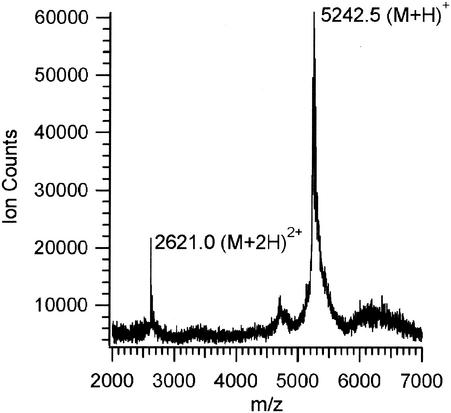
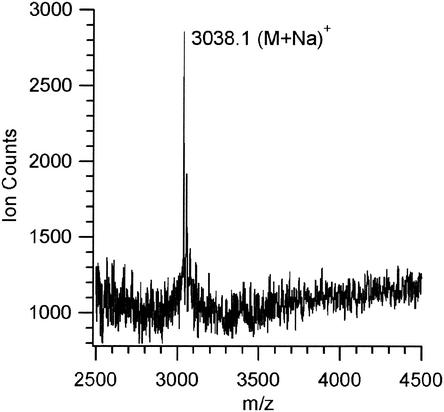
Both components of BrcC produced by B. campestris could be detected by MALDI-TOF mass spectrometry after the chloroform extraction step. Methods to detect bacteriocins by MALDI-TOF mass spectrometry in highly contaminated culture supernatants have been reported recently (16). However, BrcB is difficult to detect by mass spectrometry until a reasonable state of purity is attained. The molecular masses of BrcA and BrcB were 5,244 and 3,944 Da, respectively, values which are in good agreement with the masses predicted from the genetic sequence (12), which are 5,245 and 3,945 Da, respectively. Analogous purification of BrcA from expression clone pJKM56 in C. piscicola LV17C resulted in a pure peptide whose mass (5,245.5 Da) indicated that the desired mature peptide was isolated (Fig. 3). However, the molecular mass observed for the active peptide isolated from the BrcB expression clone pJKM46 in C. piscicola LV17C (3,038.1 Da) indicated that the expected mature peptide had been cut after the two glycines found at positions 8 and 9 and that the isolated compound was a fragment consisting of amino acids 10 to 43, which was designated BrcB(10-43) (Fig. 4). Cleavage at a double-glycine position due to protease activity concomitant with externalization by a dedicated ABC transporter is well known (13, 22). The BrcB(10-43) fragment displays the requirement expected from complementation by BrcA for antimicrobial activity. The amino acid sequence analysis of BrcA after chloroform extraction identified the first 31 residues of the desired peptide excluding the starting Y as follows: NH2-SSKDCLKDIGKGIGAGTVAGAAGGGLAAGL. No N-terminal amino acid sequence of BrcB(10-43) could be obtained due to the hydrophobicity of this peptide and solubility problems encountered when we tried to perform sequencing analyses.
FIG. 3.
Mass spectrum obtained for pure BrcA after purification of a 3-liter fermentation culture of pJKM56.
FIG. 4.
Mass spectrum obtained for pure BrcB(10-43) after purification of a 3-liter fermentation culture of pJKM46.
Expression of BrcA and BrcB in E. coli.
Although expression clones pSG1 and pSG15 in E. coli were shown to contain the correct sequence, initial extraction of peptide fractions gave sample mixtures that showed slight activity by themselves without a requirement for the complementary peptide BrcA or BrcB component. This activity might have been due to the large quantity of the fragments. MALDI-TOF mass spectrometry of both the n-butanol and chloroform extracts, as well as of fractions of these extracts that were separated by HPLC, revealed no signals for the parent BrcA or BrcB peptide. Typical mass results obtained for pSG1 and pSG15 from HPLC purification for the fractions exhibiting weak activity against the indicator strain (with no requirement for the complementary peptide) are shown in Table 3. Depending on the fermentation and extraction procedures used, peptide fragments that varied in size and distribution were obtained. These data indicate that in each case, the desired BrcA and BrcB peptides were degraded into small fragments when they were produced in E. coli.
TABLE 3.
MALDI-TOF mass spectrometry of pSG1 and pSG15a
| Plasmid | Retention time on HPLC (min) | Mol wt | Mol wt of possible cut peptide (amino acids) |
|---|---|---|---|
| pSG1 | 18.2-19.0 | 1,288.95 | |
| 1,302.87 | |||
| 2,758.56 | |||
| 3,038.77 | 3,036.58 (9-44) | ||
| 3,037.66 (23-58) | |||
| 3,039.63 (18-53) | |||
| 3,039.67 (6-41) | |||
| pSG15 | 13.4-16.0 | 1,568.91 | |
| 1,669.97 | |||
| 1,754.01 | |||
| 2,124.23 | |||
| 2,223.37 | |||
| 2,397.52 | 2,398.85 (1-28) | ||
| 2,398.88 (12-40) | |||
| 2,407.47 | |||
| 2,533.55 | 2,531.99 (4-34) |
Data were obtained after HPLC of the n-butanol extracts of French press-treated cells containing pSG1 and pSG15.
Conformational studies by circular dichroism.
To investigate the structure of the two purified peptides, BrcA and BrcB(10-43), individually and as a mixture, circular dichroism measurements were obtained in methanol with and without trifluoroethanol. It was observed that both peptides were well structured in methanol and that the presence of trifluoroethanol did not induce any changes in the degree of secondary structure. A high content of β-sheets was found in both BrcA and BrcB(10-43) and in a 1:1 mixture of the two peptides.
DISCUSSION
With the ultimate goals of determining the structure and understanding the mechanism of action of the two-peptide bacteriocin BrcC, we developed a facile new purification procedure for BrcA and BrcB. These two components are inactive by themselves, but they combine to form the bactericidal BrcC complex. The purification procedure described previously for the BrcC complex (12) was unsuccessful for the individual BrcA and BrcB(10-43) peptides due to loss at the Sephadex G-50 column chromatography step, probably due to very strong binding to the matrix. The individual components are quite intractable and bind either irreversibly or with degradative activity loss to a variety of solids, including silica gel (both normal and C18, C8, and C4 reverse phase), polystyrene, cellulose, and various HPLC column supports. Purification of either peptide can be readily achieved by using three steps: n-butanol extraction, chloroform extraction, and Sephadex LH-20 size exclusion chromatography. Acetonitrile precipitation, which is useful for BrcC purification, did not prove to be necessary for obtaining pure compounds. There was good recovery of activity after n-butanol extraction [63 and 42% for BrcA and BrcB(10-43), respectively]. Despite the loss of some peptide during chloroform extraction, this step is a key step for purification of both of these very hydrophobic compounds because it allows removal of most hydrophilic contaminants. As both pure peptides precipitate in an aqueous solution (>8% H2O) (data not shown), Sephadex LH-20, which permits the use of organic solvents, was chosen as the solid support for size exclusion chromatography. The procedure can also be used effectively up through the chloroform extraction step for the BrcC complex produced by B. campestris.
Mass spectral analysis of the two components of BrcC from B. campestris demonstrated that the mature peptides which are expected on the basis of genetic analysis (12) are in fact produced. In contrast, the expression clones in C. piscicola LV17C when the divergicin A leader (10) was used produced mature BrcA and a fragment of BrcB resulting from cleavage at a Gly-Gly site that removed the first nine amino acids after the leader sequence. Presumably, the bacteriocin production machinery present in C. piscicola (14) recognizes the expressed precursor bearing the leader as a prebacteriocin to be processed at the common Gly-Gly site. This cleavage is likely accomplished by an ABC transporter having cysteine proteinase activity (7, 25). It seems unlikely that the peptide is first exported by the sec pathway (mediated by the leader [11, 28]) and then processed by the bacteriocin transporter system, but this possibility cannot be rigorously excluded at present. Interestingly, the BrcB(10-43) fragment has all of the properties expected for the complete BrcB peptide. Most importantly, it is inactive by itself but complements BrcA to give a potent antimicrobial complex whose activity level and spectrum cannot be distinguished from those of native BrcC. The list of two-peptide bacteriocins, both unmodified and lantibiotic, is growing rapidly (4), and understanding the mode of interaction requires a detailed understanding of the three-dimensional structures of the components. Although BrcC is stable at pH values ranging from 2 to 9 at 100°C (12, 19), the BrcA and BrcB components are much less robust. The purity of a sample has a considerable effect on the stabilities of these components; although very crude extracts may be more prone to degradation, in somewhat purer samples other contaminating peptide impurities seem to protect BrcA and BrcB from decomposition. Based on this observation and on the data obtained for activity and stability of BrcA and BrcB, the best storage conditions for pure BrcA and BrcB(10-43) were determined to be a temperature of −20°C or less under an inert argon atmosphere.
Although universal labeling of bacteriocins with 13C and 15N for NMR studies can be achieved in gram-positive organisms by using complex labeled media laboriously derived from blue-green algae (cyanobacteria) (Anabaena sp.) (17), expression in E. coli permits facile isotopic labeling with defined media and simple commercial precursors. Hence, we constructed brcA and brcB expression clones in E. coli. Construction of plasmid pSG1 for expression in E. coli was achieved by cloning the dvn::brcA gene in conjunction with the gene coding for BrcA into the XbaI and SalI sites of pT712. Plasmid pT712 was chosen because of its strong T7 promoter that promotes overexpression of the desired peptides. Attempts to ligate the dvn::brcB gene and the gene coding for BrcB into pT712 digested with only SacI were unsuccessful as the genes were inserted in all cases in the wrong direction (data not shown). To construct plasmid pSG15, the dvn::brcB gene and the gene coding for BrcB were cloned into the XbaI and EcoRI sites of pT712. DNA sequencing confirmed that the correct constructs were made. Unfortunately, only multiple fragments of both peptides were recovered after purification by HPLC. The exact composition varied unpredictably depending on the fermentation conditions and the isolation procedures. Clearly, use of the sec leader is insufficient to obtain relatively large bacteriocins (i.e., 59 amino acids) intact in E. coli, and approaches in which large fusion proteins (e.g., maltose binding protein fusion) are used are likely to be successful (15).
Thus far, circular dichroism studies (27) and NMR solution structures (2, 26) of type II nonlantibiotic bacteriocins having 48 amino acids or fewer indicate that these compounds usually have random coil conformations in pure water and assume defined three-dimensional structures only upon addition of lipid micelles or trifluoroethanol. Such experiments are not feasible with BrcA and BrcB(10-43) because of their very low water solubility. However, the circular dichroism spectra of each of these pure peptides, as well as of a 1:1 mixture, show that they have defined conformations in methanol that are not detectably altered upon addition of trifluororethanol. Both peptides appear to have a significant β-sheet structure, which may in part account for their self-association and intractable properties. Studies of production of universally 13C- and 15N-labeled brochocins and of their NMR solution three-dimensional structures are in progress, with the goal of understanding the complexation of these compounds and their modes of antimicrobial action.
Acknowledgments
This work was supported by CanBiocin Inc., the Natural Sciences and Engineering Research Council of Canada, the Alberta Heritage Foundation for Medical Research (scholarship to S.G.), the Canada Research Chair in Bioorganic and Medicinal Chemistry, the Canada Foundation for Innovation, and the Killam Trusts (Izaak Walton Killam Memorial scholarship to S.G.).
REFERENCES
- 1.Ahn, C., and M. E. Stiles. 1990. Plasmid-associated bacteriocin production by a strain of Carnobacterium piscicola from meat. Appl. Environ. Microbiol. 56:2503-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fregeau Gallagher, N. L., M. Sailer, W. P. Niemczura, T. T. Nakashima, M. E. Stiles, and J. C. Vederas. 1997. Three dimensional structure of leucocin A in trifluoroethanol and dodecylphosphocholine micelles: spatial location of residues critical for biological activity in type IIa bacteriocins from lactic acid bacteria. Biochemistry 36:15062-15072. [DOI] [PubMed] [Google Scholar]
- 3.Gao, Y., M. J. van Belkum, and M. E. Stiles. 1999. The outer membrane of gram-negative bacteria inhibits antibacterial activity of brochocin-C. Appl. Environ. Microbiol. 65:4329-4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garneau, S., N. I. Martin, and J. C. Vederas. 2002. Two-peptide bacteriocins produced by lactic acid bacteria. Biochimie 84:577-592. [DOI] [PubMed] [Google Scholar]
- 5.Guder, A., I. Wiedemann, and H. G. Sahl. 2000. Posttranslationally modified bacteriocins—the lantibiotics. Biopolymers 55:62-73. [DOI] [PubMed] [Google Scholar]
- 6.Hastings, J. W., M. Sailer, K. Johnson, K. L. Roy, J. C. Vederas, and M. E. Stiles. 1991. Characterization of leucocin A-UAL and cloning of the bacteriocin gene from Leuconostoc gelidum. J. Bacteriol. 173:7491-7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Havarstein, L. S., D. B. Diep, and I. F. Nes. 1995. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol. Microbiol. 16:229-240. [DOI] [PubMed] [Google Scholar]
- 8.Hurst, A. 1972. Interactions of food starter cultures and food-borne pathogens: the antogonism between Streptococcus lactis and sporeforming microbes. J. Milk Food Technol. 35:418-423. [Google Scholar]
- 9.Joosten, H. M. L. J., and M. Nuñez. 1995. Adsorption of nisin and enterocin 4 to polypropylene and glass surfaces and its prevention by Tween 80. Lett. Appl. Microbiol. 21:389-392. [Google Scholar]
- 10.Klaenhammer. T. R. 1993. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol. Rev. 12:39-86. [DOI] [PubMed] [Google Scholar]
- 11.McAuliffe, O., R. P. Ross, and C. Hill. 2001. Lantibiotics: structure, biosynthesis and mode of action. FEMS Microbiol. Rev. 25:285-308. [DOI] [PubMed] [Google Scholar]
- 12.McCormick, J. K., A. Poon, M. Sailer, Y. Gao, K. L. Roy, L. M. McMullen, J. C. Vederas, M. E. Stiles, and M. J. van Belkum. 1998. Genetic characterization and heterologous expression of brochocin-C, an antibotulinal, two-peptide bacteriocin produced by Brochothrix campestris ATCC 43754. Appl. Environ. Microbiol. 64:4757-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nes, I. F., and H. Holo. 2000. Class II antimicrobial peptides from lactic acid bacteria. Biopolymers (Peptide Sci.) 55:50-61. [DOI] [PubMed] [Google Scholar]
- 14.Quadri, L. E. N., M. Kleerebezem, O. P. Kuipers, W. M. de Vos, K. L. Roy, J. C. Vederas, and M. E. Stiles. 1997. Genes from Carnobacterium pisicola LV17B involved in bacteriocin production and immunity: evidence for global transcriptional regulation mediated by different exported peptides. J. Bacteriol. 179:6163-6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quadri, L. E. N., L. Z. Yan, M. E. Stiles, and J. C. Vederas. 1997. Effect of amino acid substitutions on the activity of carnobacteriocin B2: overproduction of the antimicrobial peptide, its engineered variants, and its precursor in Escherichia coli. J. Biol. Chem. 272:3384-3388. [DOI] [PubMed] [Google Scholar]
- 16.Rose, N. L., P. Sporns, and L. M. McMullen. 1999. Detection of bacteriocins by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Appl. Environ. Microbiol. 65:2238-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sailer, M., G. L. Helms, T. Henkel, W. P. Niemczura, M. E. Stiles, and J. C. Vederas. 1993. 15N- and 13C-labeled media from Anabaena sp. for universal isotopic labeling of bacteriocins: NMR resonance assignments of leucocin A from Leuconostoc gelidum and nisin A from Lactococcus lactis. Biochemistry 32:310-318. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 19.Siragusa, G. R., and C. N. Cutter. 1993. Brochocin-C, a new bacteriocin produced by Brochothrix campestris. Appl. Environ. Microbiol. 59:2326-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Studier, R. W., and B. Moffat. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 21.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Belkum, M. J., and M. E. Stiles. 2000. Nonlantibiotic antimicrobial peptides from lactic acid bacteria. Nat. Prod. Rep. 17:323-335. [DOI] [PubMed] [Google Scholar]
- 23.van de Guchte, M., J. M. B. M. van der Vossen, J. Kok, and G. Venema. 1989. Construction of a lactococcal expression vector: expression of hen egg white lysozyme in Lactococcus lactis subsp. lactis. Appl. Environ. Microbiol. 55:224-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Kraaij, C., W. M. De Vos, R. J. Siezen, and O. P. Kuipers. 1999. Lantibiotics: biosynthesis, mode of action and applications. Nat. Prod. Rep. 16:575-587. [DOI] [PubMed] [Google Scholar]
- 25.Venema, K., J. Kok, J. D. Marugg, M. Y. Toonen, A. M. Ledeboer, G. Venema, and M. L. Chikindas. 1995. Functional analysis of the pediocin operon of Pediococcus acidilactici PAC1.0: PedB is the immunity protein and PedD is the precursor processing enzyme. Mol. Microbiol. 17:515-522. [DOI] [PubMed] [Google Scholar]
- 26.Wang, Y., M. E. Henz, N. L. Fregeau Gallagher, S. Chai, L. Z. Yan, M. E. Stiles, D. S. Wishart, and J. C. Vederas. 1999. Solution structure of carnobacteriocin B2 and implications for structure-activity relationships among type IIa bacteriocins from lactic acid bacteria. Biochemistry 38:15438-15447. [DOI] [PubMed] [Google Scholar]
- 27.Watson, R. M., R. W. Woody, R. V. Lewis, D. S. Bohle, A. H. Andreotti, B. Ray, and K. W. Miller. 2001. Conformational changes in pediocin AcH upon vesicle binding and approximation of the membrane-bound structure in detergent micelles. Biochemistry 40:14037-14046. [DOI] [PubMed] [Google Scholar]
- 28.Worobo, R. W., M. J. van Belkum, M. Sailer, K. L. Roy, J. C. Vederas, and M. E. Stiles. 1995. A signal peptide secretion-dependent bacteriocin from Carnobacterium divergens. J. Bacteriol. 177:3143-3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan, L. Z., A. C. Gibbs, M. E. Stiles, D. S. Wishart, and J. C. Vederas. 2000. Analogs of bacteriocins: antimicrobial specificity and interactions of leucocin A with its enantiomer, carnobacteriocin B2 and truncated derivatives. J. Med. Chem. 43:4579-4581. [DOI] [PubMed] [Google Scholar]