Abstract
We have determined distinct roles for different proteasome complexes in adenovirus (Ad) E1A-dependent transcription. We show that the 19S ATPase, S8, as a component of 19S ATPase proteins independent of 20S (APIS), binds specifically to the E1A transactivation domain, conserved region 3 (CR3). Recruitment of APIS to CR3 enhances the ability of E1A to stimulate transcription from viral early gene promoters during Ad infection of human cells. The ability of CR3 to stimulate transcription in yeast is similarly dependent on the functional integrity of yeast APIS components, Sug1 and Sug2. The 20S proteasome is also recruited to CR3 independently of APIS and the 26S proteasome. Chromatin immunoprecipitation reveals that E1A, S8 and the 20S proteasome are recruited to both Ad early region gene promoters and early region gene sequences during Ad infection, suggesting their requirement in both transcriptional initiation and elongation. We also demonstrate that E1A CR3 transactivation and degradation sequences functionally overlap and that proteasome inhibitors repress E1A transcription. Taken together, these data demonstrate distinct roles for APIS and the 20S proteasome in E1A-dependent transactivation.
Keywords: proteasome, transcription, virus
Introduction
Adenovirus (Ad) E1A protein expression is essential for both viral replication and cellular transformation (Gallimore and Turnell, 2001). Ad E1A proteins are expressed predominantly, from two splice variant transcripts, 12S and 13S, which, in Ad2/5, give rise to proteins of 243 and 289 amino acids respectively. Ad-regulated expression of viral early genes, and some cellular genes, is dependent upon conserved region 3 (CR3), a region unique to 13S E1A (Jones, 1995). Studies investigating CR3 have not only been important in elucidating E1A function, but have also served as a model system for understanding the basis of transactivation per se.
Comparison of E1A sequences from different Ad subtypes suggests that CR3 extends from residue 144 to 191 in Ad2/5 (Avvakumov et al, 2002). CR3 contains a C4 zinc-finger, a single Zn2+ ion coordinated by amino acids C154, C157, C171 and C174; mutation of any of the critical C residues results in loss of transactivation function (Culp et al, 1988; Webster et al, 1991). Residues H160 and S172 within the finger domain, residues V147, P150, Y175 and R177, which lie proximal to the finger domain, and a contiguous stretch of amino acids, encompassing residues V183–S188 within the C-terminal region of CR3, are also critical for transactivation (Webster and Ricciardi, 1991). The transactivating properties of CR3 reside in its ability to recruit the necessary components of the cellular transcription machinery specifically to transcription factor-bound promoters. Indeed, the zinc-finger binds TBP, as part of a functional holo-TFIID complex (Lee et al, 1991; Boyer and Berk, 1993; Geisberg et al, 1994). The C-terminal region of CR3 (i.e. V183–S188) also recruits, independently of TBP, multiple TAF components of TFIID (Chiang and Roeder, 1995; Geisberg et al, 1995; Mazzarelli et al, 1997). The C-terminal region of CR3 is also required for promoter targeting of competent E1A-containing transactivation complexes; CR3 binds to promoter-bound transcription factors through their DNA-binding domains (DBD) (Hurst and Jones, 1987; Lee et al, 1987; Lillie and Green, 1989; Liu and Green, 1990, 1994). Phosphorylation of residues S185 and S188 within the C-terminal domain modulates E1A-dependent transactivation (Whalen et al, 1997). The zinc-finger domain of CR3 also binds hSur2 (MED23), a component of Mediator (Boyer et al, 1999). Thus, CR3 is unable to function efficiently as a transactivator of Ad early genes in sur2−/− embryonic stem cells following viral infection or DNA transfection (Stevens et al, 2002). Indeed, the ability of mouse Ad1 to stimulate early gene transcription, and replicate, in sur2−/− mouse embryo fibroblasts is reduced significantly, relative to its capacity to perform these functions in sur2+/+ mouse embryo fibroblasts (Fang et al, 2004).
The 26S proteasome is the major non-lysosomal proteolytic machinery in eukaryotes, serving to degrade polyubiquitylated proteins in an ATP-dependent manner (Ciechanover, 1998). The 26S proteasome is assembled from two large macromolecular complexes, the 20S proteolytic particle and the 19S regulatory complex (RC). The 20S proteasome degrades non-ubiquitylated proteins in an energy-independent manner and possesses trypsin-, chymotrypsin- and postglutamyl hydrolase-like activities. The 19S RC can be functionally subdivided into base and lid components. The base complex consists of six homologous AAA ATPases (S4, S6, S6′, S7, S8 and S10b) and three non-ATPase (S1, S2 and S5a) components, whereas the lid of the RC is made of an additional eight non-ATPase subunits. The 19S ATPases regulate the assembly of the 26S proteasome, unfold substrate proteins targeted for degradation and gate protein translocation into the central chamber of the 20S proteasome (Ciechanover, 1998).
The realisation that the Saccharomyces cerevisiae protein responsible for rescue of a class of Gal4 activation domain mutants, Sug1, was a component of the 26S proteasome was crucial in establishing a potential role for the proteasome in transcription (Rubin et al, 1996). Significantly, Sug1 had previously been reported to recruit transcription factors such as Gal4 to TBP (Swaffield et al, 1995). The belief that Sug1 functions specifically in a number of diverse transcription programmes was strengthened by the observation that the mammalian orthologue of Sug1, S8, interacted with the thyroid receptor, the retinoic acid receptor (RAR) α, the oestrogen receptor (ER) and other nuclear receptors in a ligand- and AF-2 activation domain-dependent manner (Lee et al, 1995; vom Baur et al, 1996). 26S proteasome-mediated degradation is essential for both RARγ- and ER-dependent transactivation (Lonard et al, 2000; Gianni et al, 2002).
Given the emerging role of the proteasome in transcriptional regulation, and the specific ability of 13S E1A to function as a transactivator, we undertook a comprehensive study to determine whether there was a role for the proteasome in CR3-dependent transcriptional activation. Data presented here offer new insights into E1A-dependent transactivation and clearly establish roles for the 19S ATPase, S8, as a component of 19S ATPase proteins independent of 20S (APIS), and the 20S proteasome in Ad E1A-CR3-regulated transcriptional initiation and elongation in vivo. We also show for the first time that E1A transactivation capacity is dependent upon its inherent stability. CR3 transactivation and degradation sequences functionally overlap, suggesting that the 20S proteasome also regulates the termination of E1A transcription programmes. Taken together, these data define key and novel roles for proteasomal complexes in transcription programmes regulated by E1A, and offer insights into proteasome function, per se, in transcriptional regulation.
Results
The 19S ATPase S8 binds to the CR3 transactivation domain of E1A
We have previously shown that S8 binds to the N-terminal region of E1A (Turnell et al, 2000). In order to determine whether S8 can also bind to E1A through CR3, we utilised an N-terminal double point E1A mutant, L1920A, which in the context of 12S E1A does not bind S8 (Rasti et al, 2005). Using GST-w.t.-12S E1A and GST-L1920A-12S E1A fusion proteins we initially confirmed, using pull-downs from A549 cell lysates, that the N-terminal E1A binding proteins S8, TBP and Ran all bind w.t. 12S E1A but do not bind L1920A-12S E1A (Figure 1A, compare lanes 2 and 3). Interestingly, however, S8 and TBP retained significant binding capacity for L1920A-13S, whereas Ran did not (Figure 1A, compare lanes 4 and 5), suggesting that S8 and TBP bind CR3 but Ran does not. The ability of pRB to bind in equal measure to each of the GST-E1A fusion proteins validates the structural integrity of the GST proteins used (Figure 1A, upper panel).
Figure 1.
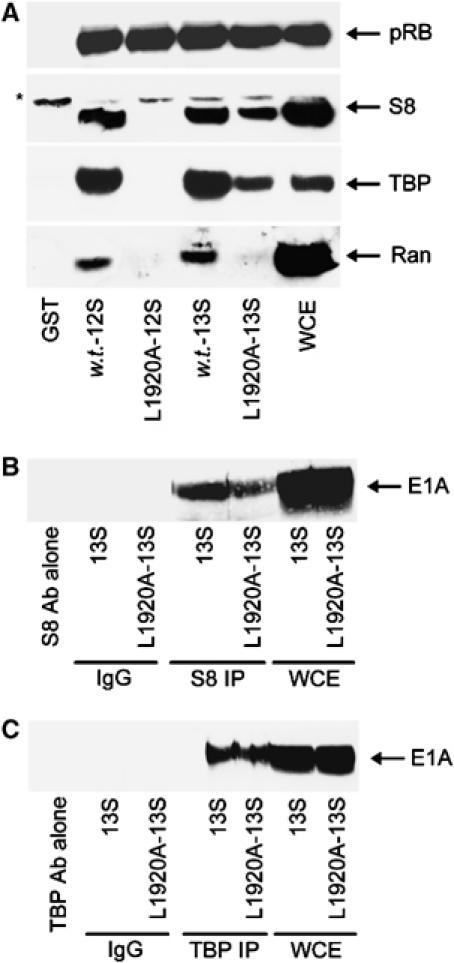
S8 binds to CR3 of Ad5 13S E1A. (A) GST pull-downs using w.t. 12S and 13S E1A, and 12S L1920A and 13S L1920A E1A mutants, were performed to assess the in vitro binding capacity of these E1A proteins for pRB, S8, TBP and Ran from A549 cellular lysates. Following pull-down, binding was assessed by Western blotting. (B, C) S8 and TBP retain the ability to bind L1920A 13S E1A in vivo. S8 and TBP complexes were immunoprecipitated from 13S E1A-, and 13S E1A L1920A-, expressing A549 cells, and subjected to Western blotting for E1A. *Nonspecific band recognised by the anti-S8 antibody. WCE, whole-cell extract.
In order to establish whether S8 binds to CR3 in vivo, we generated a number of A549-derived, clonal cell lines that constitutively express either w.t. 13S E1A or L1920A-13S E1A. Significantly, immunoprecipitation studies revealed that relative to w.t. 13S E1A, L1920A-13S E1A retained appreciable binding capacity for S8 (Figure 1B) and TBP (Figure 1C), indicating that these proteins bind CR3 specifically, in vivo. Using cell lysis conditions that dissociate the 19S proteasome from the 20S proteasome (Grand et al, 1999), we next determined, by GST pull-down, the binding site within CR3 for S8. These experiments revealed that the S8 binding site extends from amino acid 169 to 188 (Figure 2A, upper panel). S8 retained appreciable binding capacity for the zinc-finger mutant C157S (Figure 2A, upper panel), and had w.t. CR3 binding capacity for the H160Y and Y175F zinc-finger mutants (Figure 2A, middle panel). S8 also bound to CR3 species from all six human Ad subgroups (Figure 2A, lower panel), suggesting that this is a conserved function in E1A. TBP had a similar propensity to bind these same CR3 mutant proteins (Supplementary Figure S1A).
Figure 2.
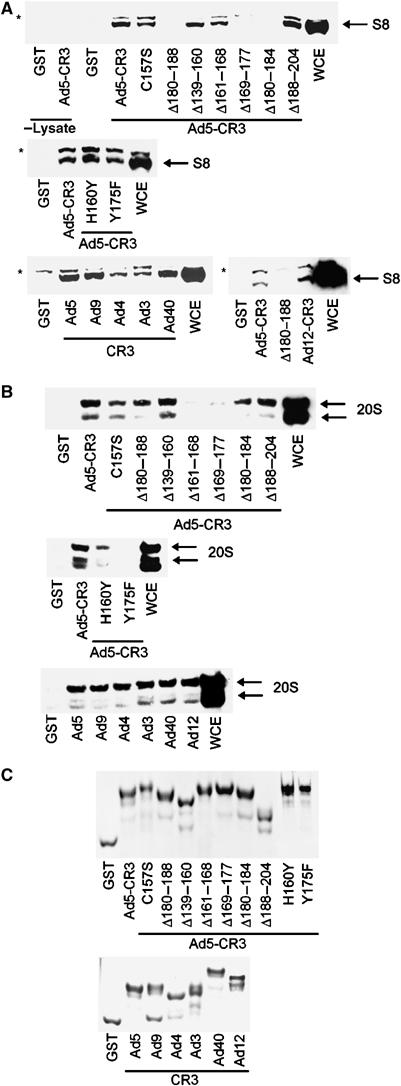
Binding of S8 and 20S proteasomal components to Ad5 CR3 E1A mutants, and to CR3s from different Ad subgroups. Following GST pull-down, binding capacity was assessed by Western blotting for S8 (A) and 20S proteasomes (B). The 20S proteasome antibody recognises α1–α3 and α5–α7 subunits. In lanes 1 and 2 of the upper panel of (A), GST and GST-Ad5 CR3 were incubated in lysis buffer alone. (C) Coomassie-stained gel showing the purity of the E1A proteins used. *Nonspecific band recognised by the anti-S8 antibody. WCE, whole-cell extract. The corresponding regions of CR3 used during this study were as follows: Ad5, residues 139–204; Ad3, residues 132–209; Ad4, residues 128–206; Ad9, residues 120–198; Ad12, residues 124–210; Ad40, residues 117–193.
As S8 and TBP physically interact (Swaffield et al, 1995), we next examined by selective siRNA treatment whether S8 and TBP bind independently to CR3. Significantly, TBP retained full binding capacity for CR3 following the knock-down of S8, and S8 retained full binding capacity for CR3 following the knock-down of TBP (Supplementary Figure S2A and B). As S8 is one of six, homologous ATPases that comprise the base of the 19S RC, we also examined the capacity of CR3 and TBP to bind S4, S6, S6′, S7, S8 and S10b in vitro. Interestingly, TBP bound to all the 19S ATPases (Supplementary Figure S2C), although it bound to S8 with the highest capacity. CR3, on the other hand, bound S8 strongly, but had little or no capacity to bind other 19S ATPases (Supplementary Figure S2C). CR3, thus binds to S8 independently of TBP, and selectively binds S8 in preference to other 19S ATPases.
The 20S proteasome binds to the CR3 transactivation domain independently of S8
Given the ability of CR3 to bind S8, we wished to determine whether 20S proteasomes similarly bound to CR3. Using the protocol outlined above, we assayed the relative abilities of the CR3-E1A mutants to bind 20S proteasomes from A549 cell lysates. Under these conditions, the 20S proteasome bound to CR3 (Figure 2B, upper panel). 20S binding to CR3 differed appreciably from that of S8. The binding site within CR3 for 20S extends from amino acid 161 to 177 (Figure 2B, upper panel). In contrast to S8, the 20S proteasome had little or no affinity for the zinc-finger mutants, H160Y and Y175F (Figure 2A and B, compare middle panels). The 20S proteasome did however bind to CR3 species from all Ad subgroups, suggesting that this is a conserved function of E1A (Figure 2A and B, compare lower panels). Taken together, these data suggest that S8 function and 20S proteasome function, in the context of E1A transcription, are separable. MED23 had a similar, but not identical, propensity as the 20S proteasome to bind these same CR3 mutants (Supplementary Figure S1B).
CR3 binds preferentially to ‘free' 20S proteasomes in vivo
Given that the 20S core particle can exist in vivo as a distinct entity, we wished to determine which 20S species were targeted by CR3. To do this, we fractionated 26S proteasomes from 20S proteasomes derived from A549 and 13S-L1920A A549 cell lysates by fast protein liquid chromatography (FPLC) (Figure 3). 26S proteasomes and 20S proteasomes from A549 and 13S-L1920A A549 cells eluted with identical profiles (cf. Figure 3A and B), suggesting that E1A binding does not affect 26S proteasome assembly. Immunoprecipitation studies confirmed this observation (Supplementary Figure S3A). Significantly, immunoprecipitation of 20S proteasomes from fractionated 13S-L1920A A549 cells revealed that E1A bound preferentially to ‘free' 20S proteasomes, and that only a minor proportion of 20S bound E1A as a component of the 26S proteasome (Figure 3C).
Figure 3.
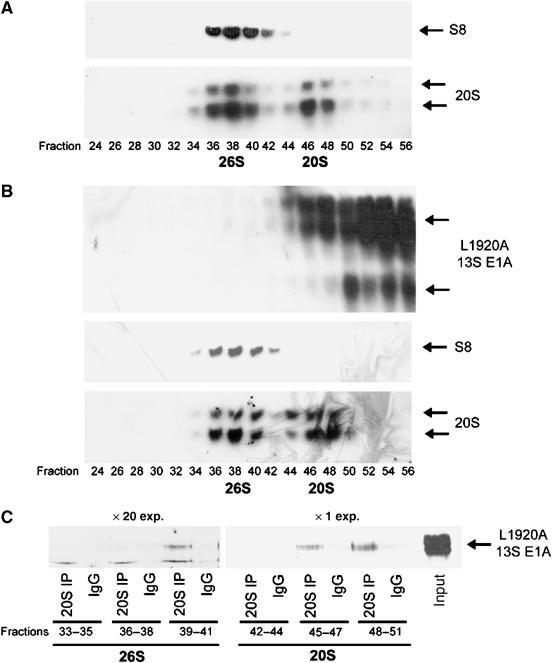
Binding of 20S proteasomes to 13S L1920A in vivo. A549 (A) and 13S-L1920A A549 (B) cellular lysates were fractionated upon a Superose 6 HR 10/30 column by FPLC (Materials and methods). E1A, and 26S, and 20S, proteasome-containing fractions were identified by Western blotting. Fractions containing 26S proteasomes or 20S proteasomes were pooled, and immunoprecipitation was performed either with an antibody that immunoprecipitates 20S proteasomal components or a normal IgG for control immunoprecipitations. Immunoprecipitates were separated upon a urea gel, and E1A identified by Western blotting (C). The blots indicate that L1920A 13S E1A binds significantly better to bulk 20S proteasomes (× 1 exposure) than 20S proteasomes associated with 26S proteasomes (× 20 exposure).
CR3 binds preferentially to APIS-associated S8 in vivo
The 19S RC exists predominantly in vivo associated with one or both ends of the 20S proteasome as functional 26S complexes. The base component of the 19S RC, of which S8 is a part, has also recently been shown to exist as a distinct functional species, APIS, that is not associated with 20S proteasomes (Gonzalez et al, 2002; Figure 4A). Using fractionated eluates from 13S-L1920A A549 cells (Figure 4A), we endeavoured to resolve whether CR3 targets S8 associated with 26S proteasomes or S8 associated with APIS. Intriguingly, immunoprecipitation studies revealed that CR3 bound preferentially to S8 associated with APIS (Figure 4B, upper panel); only a minor proportion of S8 bound to CR3 was through the 26S proteasome (Figure 4B, upper panel, compare lane 2 with lanes 6 and 8). Significantly, a ‘minor fraction' of 20S proteasomes co-eluting with APIS were found not to be associated with E1A; CR3 targeted ‘free' 20S proteasomes (Figure 4B, lower panel). To confirm that APIS components associate with E1A in vivo, we immunoprecipitated 19S base components S8, S2, S6, S6′ and S10b from 13S-L1920A A549 cells and subsequently Western blotted these samples for E1A. Immunoprecipitation revealed that 19S base components are found associated with E1A in 13S-L1920A A549 cells (Figure 4C, upper panel). Reciprocal immunoprecipitation similarly revealed the association of 19S base components S2 and S10b with E1A (Figure 4C, lower panel). Consistent with these findings, E1A expression did not affect association of S8 with other 19S base components (Supplementary Figure S3B), indicating that E1A targets the whole APIS complex. The 19S base non-ATPase component S2 co-fractionated with APIS (Figure 4A) and also bound E1A (Figure 4C), establishing S2 as an integral component of APIS. Taken in their entirety, these studies indicate that CR3 targets 20S proteasomes and APIS, independently in vivo, to perform distinct functions.
Figure 4.
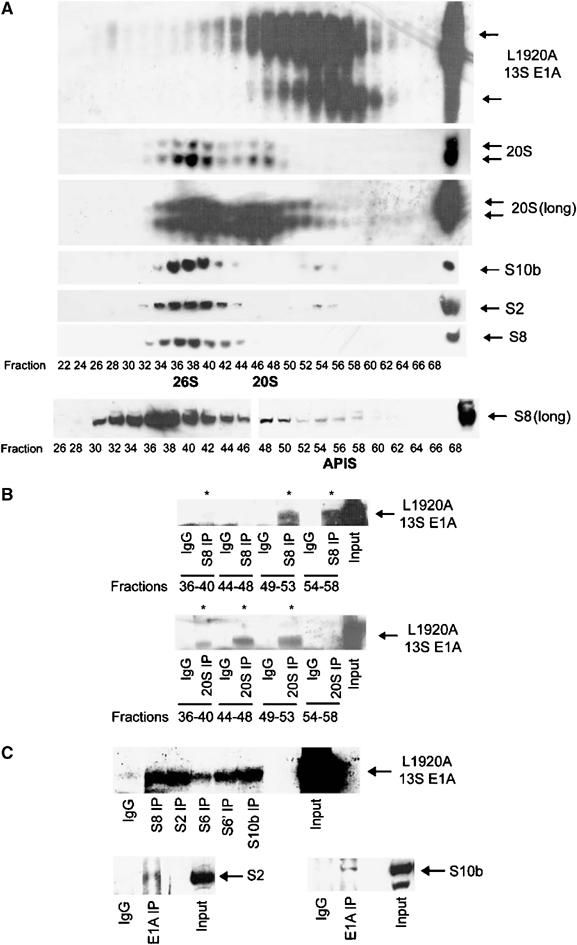
Binding of S8 as a component of APIS to 13S L1920A in vivo. (A) 13S-L1920A A549 cellular lysates were fractionated by FPLC and E1A-, 26S-, 20S- and APIS-containing fractions identified by Western blotting. Long exposures (long) were performed to identify all fractions containing 26S, 20S and APIS. Fractions comprising 26S proteasomes, or 20S proteasomes or APIS were pooled and immunoprecipitation was performed either with an antibody that immunoprecipitates S8 (B, upper panel), an antibody that immunoprecipitates 20S proteasomal components (B, lower panel) or a normal IgG for control immunoprecipitations (B). Immunoprecipitates were separated upon a urea gel, and E1A identified by Western blotting (B). The blots indicate that L1920A 13S E1A binds significantly better to S8 through APIS than through 26S proteasomes (B, upper panel), and that L1920A 13S E1A binds significantly better to bulk 20S proteasomes rather than 20S proteasomes associated with 26S (B, lower panel). L1920A 13S E1A also associates with other 19S base (APIS) components in vivo (C). S8, S2, S6, S6′ and S10b were immunoprecipitated from 13S-L1920A A549 cellular lysates and immunoprecipitated E1A* identified by Western blotting.
S8 functions to enhance CR3-dependent transactivation in vitro
To determine whether CR3 utilises S8 for its transactivation function, we initially investigated the effects of S8 overexpression upon the ability of a Gal4DBD-CR3 construct to transactivate a Gal4-responsive luciferase reporter gene in HCT116 cells. Interestingly, low-level expression of exogenous FLAG-tagged human S8 potentiated CR3 transactivation (Figure 5A, compare lanes 1 and 3 with lanes 4 and 5), whereas high-level expression of FLAG-tagged human S8 repressed CR3 transactivation (Figure 5A, compare lanes 1 and 3 with lanes 6–8). As low levels of exogenous S8 enhanced CR3 transactivation, we next determined by RNA interference (RNAi) the requirement for endogenous S8 in CR3-dependent transactivation. Treatment of HCT116 cells with non-silencing siRNAs allowed efficient transactivation by exogenously expressed Gal4DBD-CR3 (Figure 5B, compare lanes 1 and 2). Interestingly, however, knock-down of S8 expression by RNAi dramatically reduced the ability of DBD-CR3 to transactivate the Gal4-responsive reporter gene (Figure 5B, compare lanes 2 and 6). Knock-down of TBP expression by RNAi similarly reduced the ability of DBD-CR3 to transactivate the Gal4-responsive reporter gene (Figure 5B, compare lanes 2 and 4).
Figure 5.
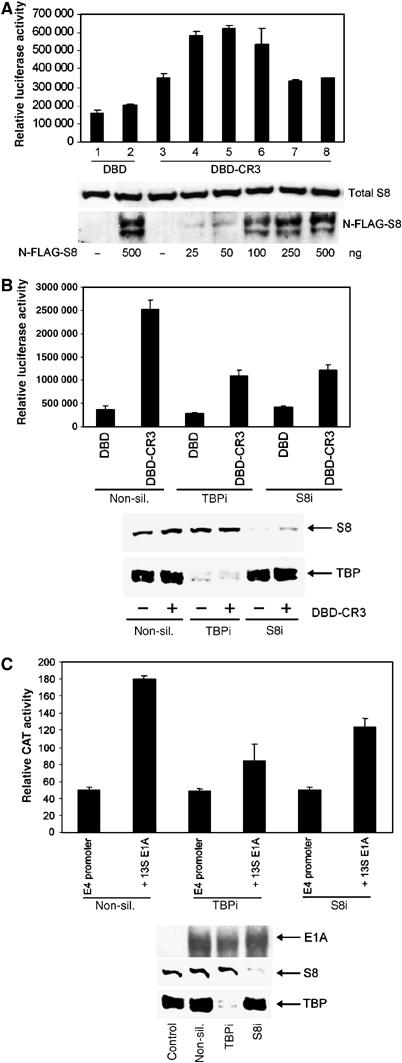
Requirement for S8 and TBP in 13S E1A transactivation function. (A) Effect of S8 expression upon E1A transactivation function. HCT116 cells were transfected with a constant amount of pcDNA3-Gal4DBD-CR3, and a Gal4-responsive luciferase reporter, with increasing amounts of pcDNA3-N-FLAG-S8. At 24 h post-transfection, cell lysates were prepared and luciferase activities measured. Total S8 and N-FLAG S8 were quantified by Western blotting. (B) Effect of TBP and S8 knock-down upon E1A transactivation. Following knock-down, HCT116 cells were transfected with a constant amount of pcDNA3-Gal4DBD and a Gal4-responsive luciferase reporter or pcDNA3-Gal4DBD-CR3 and a Gal4-responsive luciferase reporter. At 24 h post-transfection, cell lysates were prepared and luciferase activities measured. S8 and TBP levels were determined by Western blotting. (C) Following knock-down, HCT116 cells were transfected with an Ad5 E4 promoter-tethered CAT reporter and pcDNA3 Ad5 13S E1A. At 24 h post-transfection, cell lysates were prepared and CAT activities measured. In all instances, transfected DNA levels were equalised using pcDNA3 alone. Results are the mean±s.d. from three independent experiments. E1A, S8 and TBP levels were determined by Western blotting. The ability of pLE2 to transactivate the Ad5 E4 promoter was also inhibited significantly following knock-down of TBP or S8 (data not shown).
Given these findings, we next investigated whether knock-down of S8 (or TBP) expression by RNAi affected 13S E1A-dependent transactivation of the Ad5 E4 promoter. Significantly, knock-down of either S8 or TBP expression by RNAi attenuated 13S E1A-dependent transactivation of the E4 promoter (Figure 5C). Importantly, exogenous expression of FLAG-tagged human S8, or the ablation of endogenous S8 expression by RNAi, did not affect the transactivation capacity of Gal4DBD-CBP (Supplementary Figure S4A and B), suggesting that not all transactivators utilise S8 for transcription. Taken together, these experiments demonstrate that S8 function is integral to CR3 transactivation function in mammalian cells.
Functional 19S ATPases are required for CR3 transactivation in yeast
Recent studies have utilised 19S conditional mutants of S. cerevisiae to demonstrate a functional requirement for Sug1 and Sug2, as components of APIS, in transcription initiation and transcription elongation (Gonzalez et al, 2002; Ezhkova and Tansey, 2004; Lee et al, 2005). Using these same yeast mutants, we investigated whether the ability of CR3 to stimulate transcription in S. cerevisiae (Shuen et al, 2002) was dependent on the function of these 19S base proteins. Following transformation of w.t. yeast and sug1-25 and sug2-1 strains with Gal4DBD-CR3 and a Gal4-responsive reporter possessing the GAL1 regulatory region upstream of the bacterial LacZ reporter gene, and their subsequent growth selection, we assayed CR3 transactivation capacity in these cells. In w.t. cells, CR3 efficiently stimulated transcription, whereas CR3 was unable to stimulate transcription in sug1-25 and sug2-1 mutant cells (Figure 6A), despite the Gal4-CR3 protein being expressed to similar levels (Figure 6B). These data unequivocally establish a requirement for Sug1 and Sug2 in CR3-dependent transcription in yeast, and are consistent with a role for orthologous proteins in CR3-dependent transcription in human cells.
Figure 6.
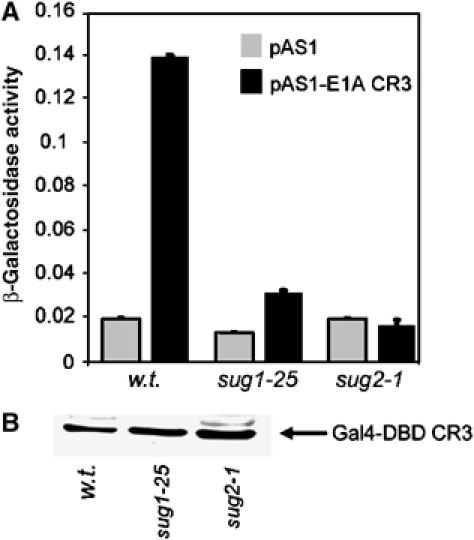
A requirement for Sug1 and Sug2 in CR3 transactivation function in yeast. W.t., sug1-25 and sug2-1 yeast strains were transformed with a Gal4-reporter construct and Gal4DBD or Gal4DBD-CR3. Cell lysates were subsequently prepared and β-galactosidase assays (A) performed to determine the transactivation capacity of CR3 in mutant sug1-25, mutant sug2-1 and the congenic w.t. strain. Western blotting analyses were also performed (B) with an anti-GAL4DBD antibody to determine the levels of E1A protein expression. Results are the mean±s.d. from three independent experiments.
S8 function is required for efficient CR3 transactivation during Ad infection
As S8 interacts directly with CR3 in vitro and in vivo, and regulates CR3 transactivation function, we wished to determine whether S8 modulated the ability of CR3 to transactivate viral early promoters during Ad infection. To do this, we initially treated A549 cells with either non-silencing siRNAs or with siRNAs directed against S8. At 24 h post-transfection, we infected cells with w.t. Ad5 at a multiplicity of infection (m.o.i.) of one plaque forming unit (PFU)/cell. At the appropriate time post-infection, we quantified Ad early gene expression. Significantly, the mRNA levels of 13S E1A, the 2.2 kb E1B transcript, E3A and E4 orf6/7 were all reduced significantly in Ad-infected cells where S8 expression was reduced by RNAi, relative to their levels in Ad-infected control cells, as measured by quantitative real-time polymerase chain reaction (PCR) (Figure 7A). Consistent with these findings, Ad early protein expression was also reduced appreciably following S8 knock-down, relative to cells expressing normal levels of S8 (Figure 7B, compare lanes 1–6 with lanes 7–12). These data provide substantive evidence to suggest that S8 functions in vivo during Ad infection to regulate CR3 transactivation function.
Figure 7.
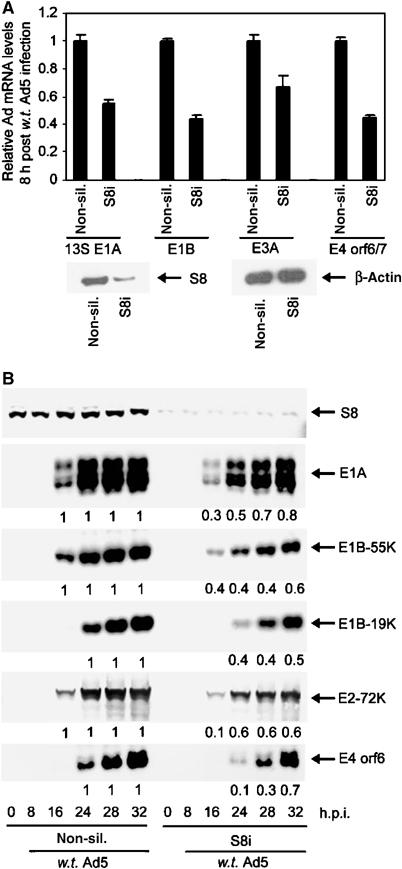
The effect of S8 knock-down upon Ad early region gene, and gene product, expression during Ad infection. Following knock-down, A549 cells were infected with w.t. Ad5 at 1 PFU/cell. At the indicated time post-infection, total RNA was prepared and the absolute levels of 13SE1A, the 2.2 kb E1B transcript, E3A and E4 orf6/7 mRNAs were determined (A) by quantitative real-time PCR. In each instance, Ad early mRNA levels in non-silencing controls were ascribed a value of 100% expression. mRNA levels in knock-down cells were calculated on this basis. Values are the mean±s.d. from three independent experiments. At the appropriate time post-infection, cell lysates were prepared and the relative levels of S8, E1A, E1B-55K, E1B-19K, E2-72K and E4ORF6 proteins were determined (B) by Western blotting.
Recruitment of S8 and 20S proteasomes to Ad early promoters during Ad infection
We next investigated by chromatin immunoprecipitation (ChIP) whether E1A, S8 or the 20S proteasome could associate with Ad early promoters during viral infection. A549 cells were thus either mock-infected or infected with w.t. Ad5 at an m.o.i. of 1 PFU/cell. At 24 h post-infection, protein–DNA complexes were recovered by ChIP and the specific recruitment of E1A, S8 and the 20S proteasome to early region promoter elements was determined by PCR (see Supplementary Figure S5 for schematic representation of promoter regions amplified). Interestingly, these analyses showed that in addition to E1A, S8, α4 and α6 subunits of the proteasome were all found associated with Ad early promoters during viral infection (Figure 8, panels 1–5, respectively; compare lanes 1–5 with lanes 6–10). The apparent difference between α4 and α6 reflects the differential abilities of the respective antibodies to immunoprecipitate the 20S proteasome (data not shown). Consistent with earlier reports that E1A does not form specific complexes with E2l promoter, we found only modest amounts of E1A and S8 associated with the E2l promoter, compared to the association of E1A and S8 with the E2e promoter (Figure 8, panel 6, compare lanes 2–4 with lanes 7–9). These studies suggest that S8, in the context of APIS, and the 20S proteasome function, in concert with E1A, to initiate transcription.
Figure 8.
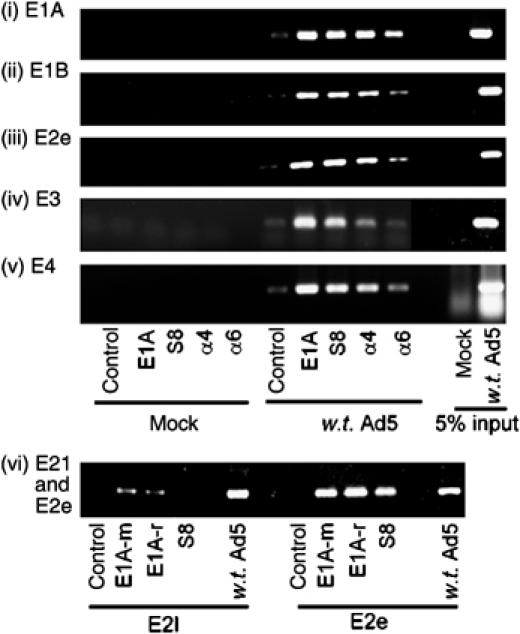
E1A, S8 and 20S proteasomal components are recruited to Ad early promoters in vivo during a w.t. Ad5 infection. Following infection, E1A, S8 and 20S protein–DNA complexes were isolated by ChIP. Typically, 10 μg of each antibody was used for ChIP. A normal rabbit IgG was used as a control antibody. E1A, E1B, E2e, E3, E4 and E2l promoter elements were detected by PCR. E1A-m, mouse monoclonal; E1A-r, rabbit polyclonal. Similar results were obtained from three independent experiments.
S8 recruitment to early region promoters during infection is dependent upon CR3
As S8 associates with Ad early promoters during infection, we next determined whether S8 recruitment was dependent upon E1A. Thus, following infection of A549 cells with either w.t. Ad5, or Ad5 dl1113 (Δ169–177; Figure 9A, upper panel) or Ad5 dl1114 (Δ180–184); Figure 9A, lower panel) mutants that do not bind S8, we determined by ChIP that association of S8 with the E3 promoter (Figure 9A, upper and middle panels, compare lane 8 with lane 12) and other Ad early promoters (data not shown) was dependent upon the expression of a 13S E1A species that bound S8. Western blotting revealed that w.t. E1A and E1A mutant proteins were expressed to similar levels following infection (Figure 9A, lower panel). These data suggest that S8 recruitment to Ad early promoters by E1A is mediated exclusively through CR3 and is not through the alternative S8 binding site located in the N-terminal region (Turnell et al, 2000).
Figure 9.
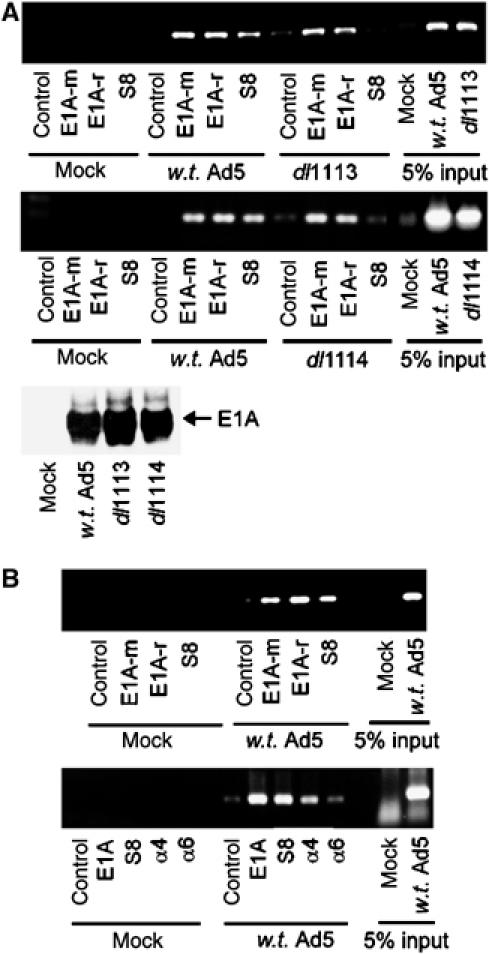
(A) Recruitment of S8 to Ad early region promoters is dependent upon binding to CR3. A549 cells were infected with either w.t. Ad5, or Ad5 mutants, dl1113 (panel 1) or dl1114 (panel 2), at an m.o.i. of 1 PFU/cell. At the appropriate time post-infection, E1A and S8 protein–DNA complexes were isolated by ChIP and E3 promoter elements detected by PCR. E1A expression was determined by Western blotting in the lower panel. (B) A role for E1A and S8 in transcriptional elongation. Following infection with w.t. Ad5, E1A, S8 and 20S protein–DNA complexes were isolated by ChIP and E1B gene elements detected by PCR. Upper panel, nucleotides 1751–2030 were amplified by PCR; lower panel, nucleotides 2591–2930 were amplified by PCR; typically 10 μg of each antibody was used for ChIP. A normal rabbit IgG was used as a control antibody. E1A-m, mouse monoclonal; E1A-r, rabbit polyclonal. Similar results were obtained from three independent experiments.
As studies in yeast have suggested that S8 and 20S proteasomes can be found associated with actively transcribing gene units (Ferdous et al, 2001; Gonzalez et al, 2002), we investigated whether E1A and proteasomal subunits could also associate with actively transcribed Ad gene regions during infection. Significantly, E1A, S8 and 20S proteasomes could be found associated with E1B gene elements across the length of the E1B gene (Figure 9B, upper and lower panels), indicating that E1A, 19S and the 20S proteasome are also required for transcriptional elongation.
Relationship between 13S E1A stability and transactivation potential
As the transactivation potential of a number of transcription factors is controlled specifically by proteasome-mediated degradation (Salghetti et al, 2000), we also addressed whether transactivation-defective 13S mutant proteins had altered half-lives relative to w.t. 13S E1A. To do this, we used A549 cells that stably expressed either w.t. 13S E1A or transactivation-defective 13S E1A mutants H160Y and Y175F. We specifically chose to investigate these two transactivation-defective mutants as they have little or no affinity for the 20S proteasome (Figure 2B). Significantly, 13S E1A mutants H160Y and Y175F had considerably longer half-lives than w.t. 13S (Figure 10A); w.t. 13S E1A had a half-life of approximately 1 h, whereas H160Y had a half-life of approximately 2 h, and Y175F had a half-life of approximately 3 h. We also determined that transactivation-defective 13S E1A mutants (Δ161–168; Δ169–177), which similarly do not bind the 20S proteasome, also had extended half-lives relative to w.t. 13S E1A (data not shown). Taken together, these experiments reveal that there is a close relationship between 13S E1A stability, 20S proteasome binding and transactivation potential. Consistent with these findings, w.t. 13S E1A protein levels were more sensitive to proteasomal inhibition than the transactivation-defective 13S E1A mutants H160Y and Y175F (Figure 10B). Indeed, selective proteasomal inhibition repressed CR3-dependent transactivation (Supplementary Figure S6), suggesting that proteasome function is important in the transactivation process.
Figure 10.
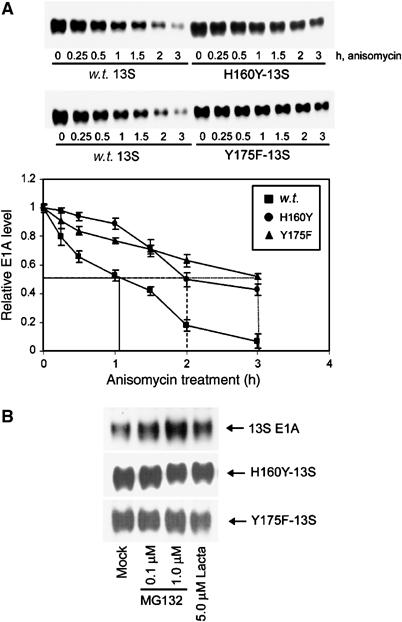
The protein half-lives of transactivation-defective 13S E1A mutants are extended relative to w.t. 13S E1A (A). Stably expressing 13S E1A-A549 cells, 13S H160Y E1A-A549 cells and 13S Y175F E1A-A549 cells were treated with the protein synthesis inhibitor anisomycin (100 μM) and harvested at the indicated times. E1A levels were determined by Western blotting and quantified using a Bio-Rad GS-800 densitometer. Results shown are the mean±s.d. from four independent experiments. The half-lives of 13S H160Y E1A and 13S Y175F E1A were similarly enhanced relative to w.t. 13S E1A when assessed by Western blotting (with E1A mAb, M2) of anisomycin-treated A549 cells transiently transfected with pLE2-expressing w.t. or mutant 13S E1A proteins (data not shown). Sensitivity of w.t. 13S E1A, H160Y-13S and Y175F-13S protein levels to treatment with proteasome inhibitors (B). Stably expressing 13S E1A-A549 cells, 13S H160Y E1A-A549 cells and 13S Y175F E1A-A549 cells were treated with the indicated amounts of proteasome inhibitors for 8 h. E1A levels were determined by Western blotting. Proteasome inhibitors also enhanced w.t. 13S E1A protein levels in A549 cells when transiently expressed using pLE2, but were unable to enhance H160Y E1A and 13S Y175F E1A protein levels when similarly expressed transiently using pLE2 (data not shown).
Discussion
Binding of 19S and 20S to the CR3 transactivation domain of E1A
We present considerable evidence to demonstrate that the 19S ATPase, S8, and the 20S proteasome bind to separate sites within CR3 (cf. Figure 2A and B). In Ad5, the binding site for S8 extends from residue 169 to 188, whereas the 20S proteasome binding site extends from residue 161 to 177. Indeed, specific point mutants H160Y and Y175F bind S8 as w.t. CR3, yet have little or no capacity to bind the 20S proteasome (cf. Figure 2A and B). Significantly, CR3 from all Ad subgroups bind both S8 and the 20S proteasome, suggesting a conserved Ad function. The ability of CR3 to target these proteins separately presumably reflects the capacity of 19S and 20S to perform distinct functions during E1A-mediated transcription.
We also obtained evidence in vivo to indicate that CR3 targets S8 and the 20S proteasome separately. Following the resolution of 26S proteasomes, 20S proteasomes and APIS from 13S-L1920A A549 cells by FPLC, we determined by immunoprecipitation that CR3 bound S8 as a component of APIS, independent of 26S proteasomes (Figure 4B), and similarly bound ‘free' 20S proteasomes, independently of 26S proteasomes (Figures 3C and 4B). These studies also revealed that other 19S ATPase components of APIS were associated with CR3 of E1A in vivo, indicating that E1A CR3 targets the entire APIS complex through binding to S8 (Supplementary Figure S2C). Previous studies have suggested that APIS minimally comprises the six 19S base-component ATPases (Gonzalez et al, 2002). Our immunoprecipitation studies reveal that the non-ATPase 19S subunit, S2, is also a component of APIS (Figure 4C).
Our in vitro binding studies suggested that the binding sites for S8 and TBP were very similar (cf. Supplementary Figure S2A and B). However, although TBP and Sug1 (S8) can functionally interact (Swaffield et al, 1995), we determined by RNAi that TBP and S8 bind CR3 independently (Supplementary Figure S2), suggesting distinct functions for these proteins in the transactivation process. We also determined that the transactivation domain of E1A bound preferentially to S8 (Supplementary Figure S2C), whereas TBP was able to bind to all six 19S ATPases, although it bound most strongly to S8 (Supplementary Figure S2C). Interestingly, the Gal4 activation domain has previously been shown to bind both Sug1 and Sug2 preferentially in yeast (Gonzalez et al, 2002). Significantly, however, both CR3 (Figure 4C) and Gal4 (Gonzalez et al, 2002) associate with all 19S ATPases in vivo. It thus appears that different transactivators target different 19S ATPases for the specific recruitment of APIS. It is perhaps not surprising, therefore, that TBP binds to all six ATPases (Supplementary Figure S2C), facilitating the specific recruitment of APIS–transactivator complexes to the basal transcriptional machinery.
Role for APIS in 13S E1A-mediated transcription initiation and elongation
Data presented within this report indicate that the proteasomal ATPase S8, as a component of APIS, is required for CR3 13S E1A transactivation function (Figures 4, 5, 6 and 7). In this regard, CR3 can utilise low levels of exogenous S8 for transactivation (Figure 5). Consistent with our findings that overexpression of exogenous S8 represses CR3 activity (Figure 5A), high levels of exogenous S8 have been postulated previously to interfere with endogenous S8 function in proteasomal-mediated protein degradation, and hence activation of the nuclear receptor RARγ2 (Gianni et al, 2002). Using RNAi, we also established a role for endogenous S8 in CR3-dependent transactivation in reporter assays (Figure 5B and C) and during w.t. Ad5 infection. Indeed, the ability of w.t. Ad5 to promote early gene mRNA expression, and consequently early gene protein expression, is reduced significantly in cells where S8 expression has been reduced by RNAi (Figure 7). The fact that expression of E1A itself is reduced in these circumstances is a significant finding and demonstrates an important feedback loop in the in vivo requirement for E1A and S8 in the transcriptional activation of the E1A promoter (Figure 7). The observation that E1A still retains transactivation potential following S8 knock-down in these instances might reflect residual S8 activity, the ability of E1A to form partially competent transactivation complexes in the absence of S8, the ability of E1A to bind APIS independently of S8 or may merely reflect 13S E1A-independent activation of these promoters. In addition to these findings, we also determined a role for 19S ATPases in CR3 transactivation function in yeast, such that CR3 could not function as a transactivator where Sug1 or Sug2, and hence APIS were functionally inactivated through mutation in their ATPase domains (Figure 6). These data are consistent with previous studies indicating a requirement for APIS in Gal4-dependent transcription in yeast (Gonzalez et al, 2002; Ezhkova and Tansey, 2004).
Recruitment of APIS and 20S proteasomes to promoters and gene elements in vivo
We also demonstrate in this report that 13S E1A, S8, and 20S proteasomes are recruited to both Ad early region promoters and Ad early region gene units during the early phase of an Ad infection (Figures 8 and 9). Roles for E1A, the 20S proteasome and APIS in transcription initiation are well documented (Jones, 1995; Morris et al, 2003; Ezhkova and Tansey, 2004). This is the first report, however, demonstrating a potential role for 13S E1A in transcriptional elongation. Pertinent to these new findings, Sug1, as a component of APIS in yeast, has recently been reported to associate not only with the activated GAL1-10 promoter but also with GAL1 gene units that are being actively transcribed (Gonzalez et al, 2002) and participate in transcriptional elongation (Ferdous et al, 2001). Indeed Sug1, through its recruitment to ubiquitylated histone H2B, can also regulate transcription initiation through the modification of histone H3 methylation (Ezhkova and Tansey, 2004). Given the specific requirement for Sug1 in RNA polymerase II-dependent transcriptional activation and elongation in eukaryotes, the ability of 13S E1A to utilise S8, as a component of APIS, for initiation and elongation of Ad early region genes suggests an evolutionarily conserved mechanism for transcription of cellular and viral genes. Moreover, recent evidence suggests that Sug1 can specifically recruit the SAGA acetyltransferase complex to transcriptional activators and enhance its recruitment to promoter elements to affect global histone H3 acetylation status (Lee et al, 2005). Interestingly, we have previously shown that CR3 recruits the SAGA complex, through binding SAGA component gcn5 acetyltransferase (Shuen et al, 2002). CR3, in this circumstance, will also bind the human gcn5 orthologue, P/CAF (Shuen et al, 2002). It will be of considerable importance in the future to establish if P/CAF and APIS interact to mediate Ad transcription function in human cells.
Role for proteasome-mediated degradation in CR3 transactivation function
The transactivation domains from a number of short-lived transcription factors functionally overlap with sequences that ultimately coordinate their destruction by the proteasome (Salghetti et al, 2000). The activation domain potency of a transcription factor is thus inversely related to the half-life of the protein; mutations that inhibit proteasome-mediated degradation inhibit transactivation function (Molinari et al, 1999). In this report, we present evidence to demonstrate that 20S proteasomes bind 13S E1A (Figures 2B, 3 and 4) and are found associated with both enhancer/promoter (Figure 8) and actively transcribed Ad early region genes (Figure 9B). Consistent with the possibility that 20S proteasome-mediated degradation might regulate 13S E1A transactivation capacity, previous reports have indicated that the 13S E1A protein is a short-lived protein, and significantly more labile than the 12S E1A protein (Spindler and Berk, 1984); 13S E1A possesses a PEST motif within CR3 that is lacking in 12S E1A (Turnell et al, 2000). Given these findings, we investigated the possibility that the transactivation function of 13S E1A is regulated by proteasome-mediated degradation. Significantly, proteasome inhibitors repressed CR3 transactivation function (Supplementary Figure S6), indicating that the proteasome directly controls 13S E1A transactivation potential. The ability of E1A to stimulate transcription is related directly to the level of E1A expression (Supplementary Figure S6). This reflects the ability of proteasome–E1A complexes to regulate the E1A promoter, and is consistent with the effects of S8 knock-down upon the level of expression of E1A following w.t. Ad5 infection (Figure 7). In consideration of whether proteasome-mediated degradation of 13S E1A controlled its inherent transactivation capacity, we established that transactivation-defective mutants H160Y and Y175F had little or no capacity to bind the 20S proteasome (Figure 2B) and had significantly longer protein half-lives than w.t. 13S E1A protein (Figure 10A). Thus, the transactivation potency of 13S E1A is related to its inherent stability. Moreover, we also determined that in contrast to w.t. 13S E1A, H160Y 13S and Y175F 13S protein levels are resistant to the use of selective proteasome inhibitors (Figure 10B), consistent with the low affinity of these proteins for 20S proteasomes.
Taken together, these data indicate that 13S E1A has evolved such that its transactivation domain functionally overlaps sequences that target the protein for degradation. Proteasome-mediated degradation of 13S E1A is required for efficient 13S E1A-mediated transcription function. We suggest that during viral infection or cellular transformation, 13S E1A utilises S8, as a component of APIS, and 20S proteasomes independently and at multiple stages in the transcription process to regulate Ad transcription programmes. In this regard, we hypothesise that E1A would recruit APIS to enhancer/promoter elements and gene elements independent of 20S proteasomes, to activate transcription in a non-proteolytic manner. As such, APIS recruitment might facilitate the recruitment of coactivators such as P/CAF and methyltransferases to the promoter to modify promoter acetylation and methylation, and hence transcriptional initiation. E1A might also recruit APIS to gene elements to promote transcriptional elongation. On the other hand, E1A might recruit 20S proteasomes, independently of APIS, to regulate transcription through proteolysis. In this regard, E1A might recruit 20S proteasomes to promoter elements to facilitate degradation of chromatin-bound transcription factors, and/or E1A, in order to promote multiple rounds of transcription initiation. E1A might similarly recruit 20S proteasomes to gene elements to establish limits for transcription through targeted degradation of regulatory proteins. The efficacy of 13S E1A-dependent transactivation is thus, in turn, dependent upon the combined activities of APIS and 20S proteasomes. The ability of 13S E1A to recruit these complexes separately allows for a temporally and spatially coordinated programme of proteasome-mediated activation and inactivation. During an Ad infection, the magnitude of the proteasome-dependent 13S E1A transactivation signal is modulated, in part, by a feedback loop involving 13S E1A itself, which regulates its own promoter. Whether APIS and 20S proteasomes can functionally cooperate in the transcription process by transiently associating on chromatin awaits further investigation. We propose a model whereby CR3 of 13S E1A binds to the basal transcriptional machinery (e.g. TBP), TBP-associated factors, transcription factors (e.g. ATF-2), transcriptional Mediator (e.g. MED23), transcriptional coactivators (e.g. P/CAF), APIS, the 20S proteasome, and potentially other transcriptional regulators to temporally coordinate E1A-stimulated gene expression.
Materials and methods
Cells and viruses
Human, small-cell lung carcinoma A549 cells and colon carcinoma HCT116 cells were grown and maintained in HEPES-buffered Dulbecco's modified Eagle's medium containing 2 mM glutamine and 8% fetal calf serum. A549-derived clones that stably express similar levels of w.t. 13S Ad5 E1A, or 13S Ad5 E1A mutant proteins were isolated after transfection and selection with G418 (800 μg/ml). w.t. Ad5 and Ad5 mutants Ad5 dl1113 and Ad5 dl1114 (Jelsma et al, 1988) were grown in Ad5 293 cells and the virus titre determined by plaque assay using Ad5 911 cells.
Ad E1A mutants
Ad 13S E1A and Ad CR3 mutants were generated by PCR in accordance with the manufacturer's instructions (Stratagene). Mutants were validated by direct sequencing of both strands of AdE1A cDNA using an ABI Prism 3100 Genetic Analyzer.
Immunoprecipitation and GST pull-downs
For immunoprecipitation, cells were lysed in buffer containing 50 mM Tris–HCl (pH 7.4), 0.825 M NaCl and 1% NP-40, sonicated and cleared by centrifugation. For GST pull-downs, 10 μg of the appropriate GST-fusion protein was mixed with either A549 cellular lysate or with the relevant in vitro-translated protein (Promega), labelled with L-α-[35S]methionine (Amersham). Immunoprecipitation and GST pull-downs were performed as described (Rasti et al, 2005).
PAGE and Western blot analysis
Samples, solubilised in 9 M urea, 50 mM Tris–HCl (pH 7.4) and 0.15 M β-mercaptoethanol, were sonicated and cleared by centrifugation. Protein concentrations were determined by Bradford assay (Bio-Rad). Samples were separated either on 12% polyacrylamide gels, run in the presence of 0.1 M Tris, 0.1 M Bicine and 0.1% SDS, or in the presence of 7 M urea, 93 mM Tris, 15 mM glycine but in the absence of SDS. For Western blotting, proteins were electroblotted onto nitrocellulose filters (Gelman Sciences), hybridised with the appropriate antibodies and visualised by enhanced chemiluminescence (Amersham Pharmacia).
Fractionation of 26S proteasomes, 20S proteasomes and APIS by FPLC
Cells were washed in ice-cold saline and solubilised by homogenisation in 20 mM Tris (pH 7.2), 5 mM ATP, 20% (v/v) glycerol and 0.2% (v/v) NP-40. Lysates were clarified by centrifugation at 40 000 r.p.m. for 30 min. Protein lysate (2 mg) was fractionated on a Superose 6 HR 10/30 column connected to an Amersham Pharmacia FPLC system. The column was eluted with 20 mM Tris (pH 7.2), 5 mM ATP, 0.1 M NaCl and 10% (v/v) glycerol at 0.25 ml/min and 0.25 ml fractions collected.
Reporter assays
DNA-LipofectAMINE complexes were incubated with cells for 6 h according to the manufacturer's instructions (Invitrogen). At the appropriate time post-transfection, cells were lysed and luciferase or CAT activities determined according to the manufacturer's instructions (Promega).
RNA interference
Purified, annealed double-stranded 21-mer RNA oligonucleotides were from Ambion. The targeted gene sequences were S8 (nucleotides 485–507): 5′-aagaagtgatcgagctgcctgtt; TBP (nucleotides 606–628): 5′-gaggataagagagccacgaactt. A non-silencing siRNA 5′-aattctccgaacgtgtcacgttt, with no known homology to any human gene, was used as a negative control. A549 cells were transfected with siRNAs by electroporation (960 μF and 220 V in 4 mm cuvettes).
Chromatin immunoprecipitation
After mock or Ad infection, cells were treated with 1% paraformaldehyde (v/v) for 10 min at 37°C to crosslink DNA–protein complexes. ChIP was then performed according to the manufacturer's instructions (Upstate Biotechnology). The oligonucleotides used for amplification were as follows: E1A promoter: forward tgacgtagtagtgtggcggaagtgt and reverse tataaatacactacacgtcagctga; E1B promoter: forward gtgtctagagaatgcaatagtagtacggat and reverse taaccaagattagcccacggcgcattatat; E2e promoter: forward tcatgtagtttattcgggttgagtagtctt and reverse atactgcgcgctgactcttaaggactagtt; E3 promoter: forward tggtgtaccaggaaagtcccgctcccacca and reverse caagcgaggagctcaccgactcgtcgttga; E4 promoter: forward gcagagcgagtatatataggactaaaaaat and reverse taccttattttggattgaagccaatatgat; E2l promoter: forward gactgtcccagtattcctcctcgtccgtgggt and reverse ccacattcttggccaattgcaagccatcaa; E1B gene 1750: forward tgcgtaacttgctggaacagagctctaaca and reverse tcttcgctccatttatcctttataaaactc; and E1B gene 2591: forward tgtaaatatcaggaattgttgctacatttc and reverse tttcttaattgaagccctgcttttggggcg. Oligonucleotides were from Alta Bioscience (The University of Birmingham).
Ad5 CR3 transactivation studies in yeast
Yeast strains, Sc507 (MATa, can1-100, ade2-1, his3-11, -15, leu2-3, -112, trp1-1, ura3-1) and congenic strains Sc660 (sug1-25) and Sc671 (sug2-1) were from Dr Tom Kodadek (UT-Southwestern Medical Center, Dallas, TX). Ad5 CR3 was cloned into the yeast expression plasmid, pAS1, such that CR3 was expressed as a Gal4DBD fusion protein (Shuen et al, 2002). The Gal4-responsive reporter construct used in these studies contains the GAL1 regulatory region upstream of the bacterial LacZ reporter gene, an ampicillin resistance gene for Escherichia coli selection, a LEU2 gene for yeast auxotrophic selection and a 2-μm yeast origin of replication. For experimental purposes, yeast strains were transformed with the Gal4-reporter construct and pAS1 or pAS1-CR3 using a modified lithium acetate procedure and plated on synthetic complete media lacking appropriate nutrients. Yeast transformants were then grown in selective media at 30°C until they reached an OD600 of 1.0. Cellular lysates were subsequently prepared for either β-galactosidase assays (Promega) or Western blotting analysis.
Quantitative real-time PCR
RNA extraction and PCR were essentially as described previously (Zhang et al, 2004). Primers used for amplification were as follows: 13S E1A F, 5′-atg ttt gtc tac agt cct gtg tct gaa; 13S E1A R, 5′-gat agc agg cgc cat ttt agg; E1B F, 5′-cgc gct gag ttt ggc tct ag; E1B R, 5′-tca aac gag ttg gtg ctc atg; E3A F, 5′-gcc gcc aca agt gct ttg; E3A R, 5′-ctc gga gag gtt ctc tcg tag act; E4 orf6/7 F, 5′-ctg ctg ccc gaa tgt aac act; E4 orf6/7 R, 5′-tcc acc ttg cgg ttg ctt aa. FAM-labelled Ad early probes were as follows: 13S E1A, 5′-cca gaa ccg gag cct gca aga cct ac; E1B, 5′-cgg cgg ctg ctc aat ctg tat ctt ca; E3A, 5′-ccg cga ctc cgt ttc aac cca g; E4 orf6/7, 5′-tga caa tgc aca agc gtg gac ttc tcc. All of the reactions were multiplexed with a preoptimised control VIC-labelled probe for 18S ribosomal RNA (PE Biosystems). Primers and probes were from Eurogentec.
Antibodies
The E1A monoclonal antibodies (mAb) M2, M73 and M58, E1B-55K mAb, 2A6, and the pRB mAb, IF8, were all obtained as a supernatant fluid from hybridoma cell lines. The E1A polyclonal rabbit antibody (pAb) was from Eurogentec, the E1B-19K rat mAb was from Oncogene Science, the E2-72K pAb was from Peter van der Vliet (University Medical Centre, Utrecht, The Netherlands), the E4orf6 mAb was from Thomas Dobner (University of Regensberg, Germany), the S8 pAb was from Wenlan Wang (AI duPont Hospital for Children, DE, USA) and the TBP pAb was from Nouria Hernandez (Cold Spring Harbor, NY, USA). The 20S mAb and α4 and α6 antibodies were from Biomol. The Ran mAb was from Transduction Laboratories. The N-FLAG mAb was from Sigma. The Gal4 mAb was from Santa Cruz.
Plasmids
Ad5 12S E1A, 13S E1A and Ad E1A mutants were cloned into pcDNA3.1 (Invitrogen) for both in vitro transcription/translation and mammalian expression, and pGEX 4T-1 (Amersham Pharmacia) for bacterial expression. Genomic w.t. and mutant Ad5 E1A clones, under the control of the E1A enhancer/promoter region, in expression vector pLE2, were also used for transient mammalian expression (Jelsma et al, 1988). Human proteasomal ATPase cDNAs in pBS were from Carlos Gorbea (University of Ohio at Salt Lake City, USA). S8 was also cloned into pcDNA3-N-FLAG. Gal4DBD, Gal4DBD-Ad5 CR3 and the Gal4-luciferase reporter were cloned into pcDNA3. The Ad5 E4 CAT reporter gene has been described (Dery et al, 1987). Sequences were verified by sequencing using an ABI Prism 3100 Genetic Analyzer.
Supplementary Material
Supplementary Information
Acknowledgments
This work was supported by CR UK programme grant C2/A1113 and CIHR grant MOP-74647. MR was sponsored by a studentship from the Iranian government. AFY is a scholar of the CIHR/UWO Strategic Training initiative in Cancer Research and Technology transfer.
References
- Avvakumov N, Wheeler R, D'Halluin JC, Mymryk JS (2002) Comparative sequence analysis of the largest E1A proteins of human and simian adenoviruses. J Virol 76: 7968–7975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer TG, Berk AJ (1993) Functional interaction of adenovirus E1A with holo-TFIID. Genes Dev 7: 1810–1823 [DOI] [PubMed] [Google Scholar]
- Boyer TG, Martin ME, Lees E, Ricciardi RP, Berk AJ (1999) Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature 399: 276–279 [DOI] [PubMed] [Google Scholar]
- Chiang CM, Roeder RG (1995) Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science 267: 531–536 [DOI] [PubMed] [Google Scholar]
- Ciechanover A (1998) The ubiquitin–proteasome pathway: on protein death and cell life. EMBO J 17: 7151–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culp JS, Webster LC, Friedman DJ, Smith CL, Huang WJ, Wu FY, Rosenberg M, Ricciardi RP (1988) The 289-amino acid E1A protein of adenovirus binds zinc in a region that is important for trans-activation. Proc Natl Acad Sci USA 85: 6450–6454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dery CV, Herrmann CH, Mathews MB (1987) Response of individual adenovirus promoters to the products of the E1A gene. Oncogene 2: 15–23 [PubMed] [Google Scholar]
- Ezhkova E, Tansey WP (2004) Proteasomal ATPases link ubiquitylation of histone H2B to methylation of histone H3. Mol Cell 13: 435–442 [DOI] [PubMed] [Google Scholar]
- Fang L, Stevens JL, Berk AJ, Spindler KR (2004) Requirement of Sur2 for efficient replication of mouse adenovirus type 1. J Virol 78: 12888–12900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdous A, Gonzalez F, Sun L, Kodadek T, Johnston SA (2001) The 19S regulatory particle of the proteasome is required for efficient transcription elongation by RNA polymerase II. Mol Cell 7: 981–991 [DOI] [PubMed] [Google Scholar]
- Gallimore PH, Turnell AS (2001) Adenovirus E1A: remodelling the host cell, a life or death experience. Oncogene 20: 7824–7835 [DOI] [PubMed] [Google Scholar]
- Geisberg JV, Chen JL, Ricciardi RP (1995) Subregions of the adenovirus E1A transactivation domain target multiple components of the TFIID complex. Mol Cell Biol 15: 6283–6290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisberg JV, Lee WS, Berk AJ, Ricciardi RP (1994) The zinc finger region of the adenovirus E1A transactivating domain complexes with the TATA box binding protein. Proc Natl Acad Sci USA 91: 2488–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianni M, Bauer A, Garattini E, Chambon P, Rochette-Egly C (2002) Phosphorylation by p38MAPK and recruitment of SUG-1 are required for RA-induced RAR gamma degradation and transactivation. EMBO J 21: 3760–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F, Delahodde A, Kodadek T, Johnston SA (2002) Recruitment of a 19S proteasome subcomplex to an activated promoter. Science 296: 548–550 [DOI] [PubMed] [Google Scholar]
- Grand RJ, Turnell AS, Mason GG, Wang W, Milner AE, Mymryk JS, Rookes SM, Rivett AJ, Gallimore PH (1999) Adenovirus early region 1A protein binds to mammalian SUG1 – a regulatory component of the proteasome. Oncogene 18: 449–458 [DOI] [PubMed] [Google Scholar]
- Hurst HC, Jones NC (1987) Identification of factors that interact with the E1A-inducible adenovirus E3 promoter. Genes Dev 1: 1132–1146 [DOI] [PubMed] [Google Scholar]
- Jelsma TN, Howe JA, Evelegh CM, Cunniff NF, Skiadopoulos MH, Floroff MR, Denma JE, Bayley ST (1988) Use of deletion and point mutants spanning the coding region of the adenovirus 5 E1A gene to define a domain that is essential for transcriptional activation. Virology 163: 494–502 [DOI] [PubMed] [Google Scholar]
- Jones N (1995) Transcriptional modulation by the adenovirus E1A gene. In The Molecular Repertoir of Adenoviruses III, Deerfler W, Bohm P (eds) pp 59–90. Berlin: Springer [DOI] [PubMed] [Google Scholar]
- Lee D, Ezhkova E, Li B, Pattenden SG, Tansey WP, Workman JL (2005) The proteasome regulatory particle alters the SAGA coactivator to enhance its interactions with transcriptional activators. Cell 123: 423–436 [DOI] [PubMed] [Google Scholar]
- Lee JW, Ryan F, Swaffield JC, Johnston SA, Moore DD (1995) Interaction of thyroid-hormone receptor with a conserved transcriptional mediator. Nature 374: 91–94 [DOI] [PubMed] [Google Scholar]
- Lee KA, Hai TY, SivaRaman L, Thimmappaya B, Hurst HC, Jones NC, Green MR (1987) A cellular protein, activating transcription factor, activates transcription of multiple E1A-inducible adenovirus early promoters. Proc Natl Acad Sci USA 84: 8355–8359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WS, Kao CC, Bryant GO, Liu X, Berk AJ (1991) Adenovirus E1A activation domain binds the basic repeat in the TATA box transcription factor. Cell 67: 65–76 [DOI] [PubMed] [Google Scholar]
- Lillie JW, Green MR (1989) Transcription activation by the adenovirus E1a protein. Nature 338: 39–44 [DOI] [PubMed] [Google Scholar]
- Liu F, Green MR (1990) A specific member of the ATF transcription factor family can mediate transcription activation by the adenovirus E1a protein. Cell 61: 1217–1224 [DOI] [PubMed] [Google Scholar]
- Liu F, Green MR (1994) Promoter targeting by adenovirus E1a through interaction with different cellular DNA-binding domains. Nature 368: 520–525 [DOI] [PubMed] [Google Scholar]
- Lonard DM, Nawaz Z, Smith CL, O'Malley BW (2000) The 26S proteasome is required for estrogen receptor-alpha and coactivator turnover and for efficient estrogen receptor-alpha transactivation. Mol Cell 5: 939–948 [DOI] [PubMed] [Google Scholar]
- Mazzarelli JM, Mengus G, Davidson I, Ricciardi RP (1997) The transactivation domain of adenovirus E1A interacts with the C terminus of human TAF(II)135. J Virol 71: 7978–7983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari E, Gilman M, Natesan S (1999) Proteasome-mediated degradation of transcriptional activators correlates with activation domain potency in vivo. EMBO J 18: 6439–6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris MC, Kaiser P, Rudyak S, Baskerville C, Watson MH, Reed SI (2003) Cks1-dependent proteasome recruitment and activation of CDC20 transcription in budding yeast. Nature 423: 1009–1013 [DOI] [PubMed] [Google Scholar]
- Rasti M, Grand RJA, Mymryk JS, Gallimore PH, Turnell AS (2005) Recruitment of CBP/p300, TBP and S8 to distinct regions at the N-terminus of adenovirus E1A. J Virol 79: 5594–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin DM, Coux O, Wefes I, Hengartner C, Young RA, Goldberg AL, Finley D (1996) Identification of the gal4 suppressor Sug1 as a subunit of the yeast 26S proteasome. Nature 379: 655–657 [DOI] [PubMed] [Google Scholar]
- Salghetti SE, Muratani M, Wijnen H, Futcher B, Tansey WP (2000) Functional overlap of sequences that activate transcription and signal ubiquitin-mediated proteolysis. Proc Natl Acad Sci USA 97: 3118–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuen M, Avvakumov N, Walfish PG, Brandl CJ, Mymryk JS (2002) The adenovirus E1A protein targets the SAGA but not the ADA transcriptional regulatory complex through multiple independent domains. J Biol Chem 277: 30844–30851 [DOI] [PubMed] [Google Scholar]
- Spindler KR, Berk AJ (1984) Rapid intracellular turnover of adenovirus 5 early region 1A proteins. J Virol 52: 706–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens JL, Cantin GT, Wang G, Shevchenko A, Shevchenko A, Berk AJ (2002) Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science 296: 755–758 [DOI] [PubMed] [Google Scholar]
- Swaffield JC, Melcher K, Johnston SA (1995) A highly conserved ATPase protein as a mediator between acidic activation domains and the TATA-binding protein. Nature 374: 88–91 [DOI] [PubMed] [Google Scholar]
- Turnell AS, Grand RJ, Gorbea C, Zhang X, Wang W, Mymryk JS, Gallimore PH (2000) Regulation of the 26S proteasome by adenovirus E1A. EMBO J 19: 4759–4773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- vom Baur E, Zechel C, Heery D, Heine MJ, Garnier JM, Vivat V, Le Douarin B, Gronemeyer H, Chambon P, Losson R (1996) Differential ligand-dependent interactions between the AF-2 activating domain of nuclear receptors and the putative transcriptional intermediary factors mSUG1 and TIF1. EMBO J 15: 110–124 [PMC free article] [PubMed] [Google Scholar]
- Webster LC, Ricciardi RP (1991) trans-dominant mutants of E1A provide genetic evidence that the zinc finger of the trans-activating domain binds a transcription factor. Mol Cell Biol 11: 4287–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster LC, Zhang K, Chance B, Ayene I, Culp JS, Huang WJ, Wu FY, Ricciardi RP (1991) Conversion of the E1A Cys4 zinc finger to a nonfunctional His2, Cys2 zinc finger by a single point mutation. Proc Natl Acad Sci USA 88: 9989–9993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen SG, Marcellus RC, Whalen A, Ahn NG, Ricciardi RP, Branton PE (1997) Phosphorylation within the transactivation domain of adenovirus E1A protein by mitogen-activated protein kinase regulates expression of early region 4. J Virol 71: 3545–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Turnell AS, Gorbea C, Mymryk JS, Gallimore PH, Grand RJ (2004) The targeting of the proteasomal regulatory subunit S2 by adenovirus E1A causes inhibition of proteasomal activity and increased p53 expression. J Biol Chem 279: 25122–25133 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


