Abstract
Y-family DNA polymerases have spacious active sites that can accommodate a wide variety of geometric distortions. As a consequence, they are considerably more error-prone than high-fidelity replicases. It is hardly surprising, therefore, that the in vivo activity of these polymerases is tightly regulated, so as to minimize their inadvertent access to primer-termini. We report here that one such mechanism employed by human cells relies on a specific and direct interaction between DNA polymerases ι and η with ubiquitin (Ub). Indeed, we show that both polymerases interact noncovalently with free polyUb chains, as well as mono-ubiquitinated proliferating cell nuclear antigen (Ub-PCNA). Mutants of polι (P692R) and polη (H654A) were isolated that are defective in their interactions with polyUb and Ub-PCNA, whilst retaining their ability to interact with unmodified PCNA. Interestingly, the polymerase mutants exhibit significantly lower levels of replication foci in response to DNA damage, thereby highlighting the biological importance of the polymerase–Ub interaction in regulating the access of the TLS polymerases to stalled replication forks in vivo.
Keywords: 26S proteasome, Rad30, translesion replication, Y-family DNA polymerases
Introduction
Ultraviolet (UV) light causes a variety of DNA lesions that block the normal replication machinery. In human cells, nucleotide excision repair (NER) removes most UV-induced DNA damage before replication takes place (Hoeijmakers, 2001; Hasty et al, 2003). However, NER rarely operates at 100% efficiency and the remaining lesions can impede the cell's replicase during genome duplication. It is now evident that under these conditions, many organisms employ specialized DNA polymerases that can accommodate a wide variety of DNA lesions, and/or geometric distortions within their active site, thus enabling bypass of the replicase-blocking lesion (reviewed in Woodgate, 1999; Kunkel et al, 2003; Friedberg et al, 2005; Lehmann, 2005; Yang, 2005). Many of the lesion-bypassing polymerases belong to the ‘Y-family' of DNA polymerases (Ohmori et al, 2001). Structural studies with Dpo4, a Y-family polymerase from Sulfolobus solfataricus (Ling et al, 2001, 2003, 2004), have revealed that the ‘trade-off' of being able to accommodate an assortment of DNA lesions, is that many Y-family polymerases are also able to accept a wide variety of noncanonical undamaged base pairs within their active sites. As a consequence, Y-family DNA polymerases often exhibit low-fidelity when replicating undamaged DNA (reviewed in Kunkel, 2004). Thus, the challenge to any cell possessing a Y-family polymerase is to minimize its use and only utilize it when absolutely necessary. Such an approach presumably helps restrict spontaneous mutagenesis to an evolutionarily acceptable level (Friedberg et al, 2002).
It appears that eukaryotes largely employ posttranslational modification to regulate the activity of error-prone translesion polymerases (Plosky and Woodgate, 2004; Friedberg et al, 2005). For example, in Saccharomyces cerevisiae, ubiquitination can direct repair pathways into either error-free or error-prone pathways. Many of the proteins in the Rad6 ‘postreplication repair' pathway are involved in the conjugation of ubiquitin (Ub) to specific target proteins. Rad6 is an E2-Ub conjugating enzyme and in conjunction with Rad18, an E3-Ub ligase, is capable of covalently linking Ub to target proteins (Bailly et al, 1994, 1997). At the present time, it is believed that the major cellular target of the Rad6/18 complex is the proliferating cell nuclear antigen (PCNA), the sliding-clamp that normally tethers the cell's main replicase to DNA (Hoege et al, 2002). Since RAD6 and RAD18 mutants are defective in polη/polζ-dependent damage-induced mutagenesis (Stelter and Ulrich, 2003), this indicates that monoubiquitination of PCNA directs the cell towards a translesion DNA synthesis (TLS) pathway involving S. cerevisiae polη and polζ. Interestingly, sumoylation of PCNA at the evolutionarily conserved K164 residue that is ubiquitinated adds an additional level of regulation that has yet to be observed in other eukaryotes (Hoege et al, 2002; Matunis, 2002; Stelter and Ulrich, 2003; Papouli et al, 2005; Pfander et al, 2005). Furthermore, the monoUb moiety that is covalently linked to PCNA can be extended into K63-linked polyubiquitin (polyUb) chains through the concerted action of Ubc13/Mms2 (Ub E2 variant, UEV) and Rad5 (E3 ligase), leading to an error-free damage-avoidance pathway (Hoege et al, 2002; Tsui et al, 2005).
Recent data suggest that ubiquitination of PCNA also regulates TLS in mammalian cells. For example, it is known that human polη interacts with the E3 ligase, Rad18 (Watanabe et al, 2004), as well as monoUb-PCNA (Kannouche et al, 2004). Such interactions are believed to help target polη to damage–induced replication foci, where a switch is proposed to occur between the cell's replicases (pols δ/ɛ) and polη, so as to facilitate TLS (Kannouche et al, 2004; Solomon et al, 2004; Watanabe et al, 2004; Garg and Burgers, 2005). The formation of K63-linked polyUb chains also appears integral in directing mammalian cells down error-free repair pathways, since cells overexpressing antisense to hMMS2 exhibit elevated levels of UV-induced mutagenesis (Li et al, 2002).
Given the extraordinary lengths that cells go through to keep their Y-family polymerases in check, it seems reasonable to speculate that human polι (the most error-prone of the Y-family polymerases), would also be tightly regulated. We hypothesized that one mechanism might be via distinct protein–protein interactions with polι's regulatory partners. To identify such partners, we used full-length human polι as the ‘bait' in the yeast ‘two-hybrid assay'. Remarkably, 25–30% of the polι-interacting clones encoded some form of Ub. Further investigation revealed that in addition to polι, polη also interacts with Ub. We subsequently isolated missense mutants of both polι and polη that are selectively defective in their ability to interact with Ub. Interestingly, both mutations lie within the respective Ub-binding motifs (UBMs) of pols ι and η that were very recently identified by Bienko et al (2005). Furthermore, both polymerase mutants exhibit lower levels of damage-induced replication foci than the wild-type polymerase, indicating that an ability to bind Ub is a prerequisite for targeting DNA polymerases ι and η to sites of cellular damage.
Results
Specific interactions between polι, polη and Ub
We used human polι as a bait to screen a human testis cDNA library in the yeast two-hybrid system. Approximately 25–30% of the clones that were found to interact with polι encoded the UbA52 or UbB ubiquitin precursor proteins (Figure 1A and B). UbA52 encodes Ub fused to the completely unrelated L40 subunit of the 60S ribosome, whereas UbB encodes three copies of Ub fused in a linear fashion. In both cases, the precursor proteins are postranslationally processed in eukaryotic cells to produce a free Ub moiety.
Figure 1.
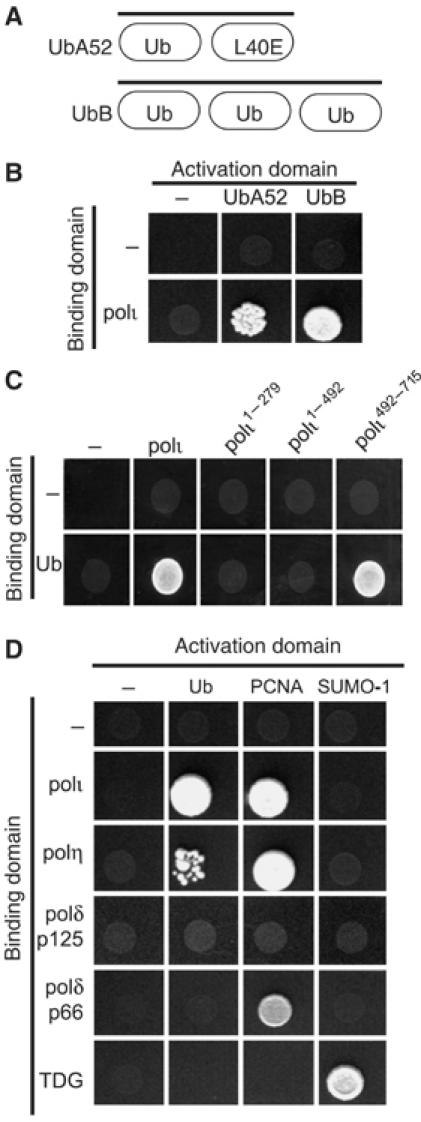
Yeast two-hybrid interactions between TLS polymerases and Ub. (A) Diagram of the gene structure of UbA52 and UbB. (B) Yeast two-hybrid assay showing the interaction (by activation of the ADE2 and HIS3 reporter genes) between full-length polι and UbA52 and UbB. In these, and all subsequent two-hybrid assays, 10 μl of an overnight culture was spotted on CSM -Leu, -Trp, -His, -Ade plates and incubated at 30°C for 5 days. (C) Deletion mapping of polι reveals that the interaction with recombinant Ub is localized to the C-terminal 224 amino acids of polι. (D) In addition to polι, polη also binds to Ub. However, neither the p125 and p66 subunits of polδ bind Ub. PCNA was used as a positive control for polι, polη, and p66 of polδ. The interaction between polι and polη and Ub is specific, as neither polymerase binds to the Ub-like protein, SUMO-1. TDG was used as a positive control for SUMO-1.
To confirm that the observed interaction was between polι and Ub, rather than the L40 ribosome subunit of UbA52, protein interactions were retested using a recombinant gene encoding a single Ub protein. Furthermore, by using a series of polι deletion mutants, we mapped the region of the Ub-interaction to the C-terminal 224 amino acids of polι (Figure 1C).
To determine if the interaction between polι and Ub was unique, or could be extended to other polymerases, or to the Ub-related SUMO-1 protein, we performed two-hybrid analysis with human polη and polδ (p125 and p66 subunits) cloned into the GAL4 binding-domain and Ub or SUMO-1 cloned into the activation domain vectors. As a positive control for these assays, we used PCNA cloned into the activation domain, since interactions between polι and polη and PCNA have previously been reported (Haracska et al, 2001; Vidal et al, 2004). Like polι, polη appears to interact with Ub (Figure 1D). Neither the p125 or p66 subunit of polδ interacts with Ub, but as expected, the p66 subunit interacts with PCNA (Figure 1D; Hughes et al, 1999). The interaction of polι and polη with Ub is therefore specific and at least in the two-hybrid system, does not extend to SUMO-1. We believe that SUMO-1 is active under our assay conditions, as we were able to detect an interaction between SUMO-1 and thymine DNA glycosylase (TDG) (Baba et al, 2005; Figure 1D).
Noncovalent interactions between full-length polι and polη and K48- and K63-linked polyUb chains
Having established that the interactions between polι and polη and Ub are specific, we wanted to determine if the interaction is through a direct covalent linkage between the pols and Ub, or is, perhaps, via a noncovalent interaction. To do so, we utilized a mutant of Ub in which the C-terminal glycines (residues 75 and 76) were changed to serine. This double mutation prevents the E3 ligase from covalently linking Ub to a target protein and from forming polyUb chains (Pickart et al, 1994). As seen in Figure 2A, neither polι nor polη interact with the G75S/G76S Ub mutant in the two-hybrid assay, suggesting that either the pols need to be physically conjugated to Ub, or that they interact noncovalently with a conjugated form of Ub, that is, polyUb chains or a ubiquitinated protein. To address the latter possibility, we mutated either K48 or K63 of Ub to Arginine. The K48R-Ub mutant can form K63-linked polyUb chains, but is unable to make K48-linked polyUb chains. Likewise, the K63R-Ub mutant forms K48-linked chains, but not K63-linked chains. Both polι and polη retain the ability to bind either Ub mutant in the two-hybrid assay, suggesting pols ι and η have no particular preference for either type of polyUb chain.
Figure 2.
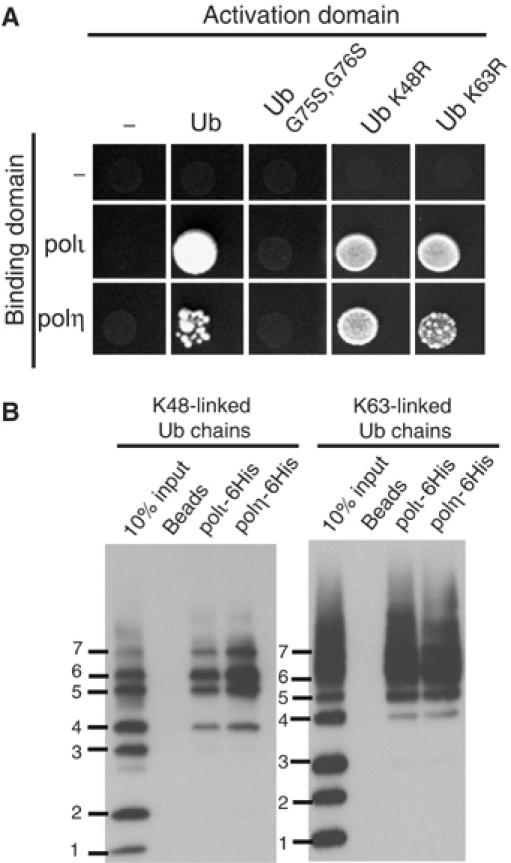
The interaction between pols ι and η and Ub is noncovalent, but requires Ub conjugation. (A) While polι and polη interact with wild-type Ub, neither interacts with the G75S/G76S Ub mutant that cannot be conjugated to a target protein or form polyUb chains. Additionally, polι and polη are able to interact with either K48R- or K63R-Ub. (B) In vitro pull down assays with purified C-terminal His-tagged polι and polη demonstrate that both polymerases interact with pure K48- and K63-linked polyUb chains ranging from four to seven Ub moieties. Bound proteins were separated on 10–20% Tris–glycine–SDS–polyacrylamide gels and transferred onto PVDF membrane and Ub chains visualized by Western blot.
To assay the pol-Ub interaction in vitro, we used commercially available mixtures of K48- and K63-linked Ub ranging in length from monoUb to chains containing 7-Ub moieties in pull-down experiments with either 6-His-tagged polι and/or 6-His-tagged polη. As seen in Figure 2B, in the absence of any Ub-conjugating proteins, polι and polη preferentially interact with both K48- and K63-linked polyUb 4–7 chains, and no binding was observed with mono-, di-, or tri-Ub. Each pull-down assay was repeated several times, and for polι, Ub binding was also tested using a full-length GST-tagged protein to confirm the observed interactions (data not shown).
Since the pull-down assays lack Ub ligase or ATP required for conjugation, they demonstrate that the interaction between polι and polη and polyUb is noncovalent in nature.
Identification of a region in polι that binds Ub
In addition to binding to Ub, polι also binds to polη (Kannouche et al, 2002). During our attempts to delineate the interface between the polymerases, we serendipitously identified a critical residue required for Polι's interaction with Ub. We originally set out to make mutations in polι, which we hoped would physically disrupt the polι–polη interface (A Vidal, unpublished observations), but much to our surprise, a P692R substitution in polι did not affect its ability to interact with wild-type polη or PCNA, but was, instead, selectively defective in its ability to interact with Ub (Figure 3A). Furthermore, while peptides encompassing P692 were able to pull-down both K48- and K63-linked polyUb chains, a peptide with the P692R substitution was unable to pull-down either form of Ub chain (Figure 3B). We therefore conclude that P692 is a key residue required for polι's ability to interact with Ub.
Figure 3.
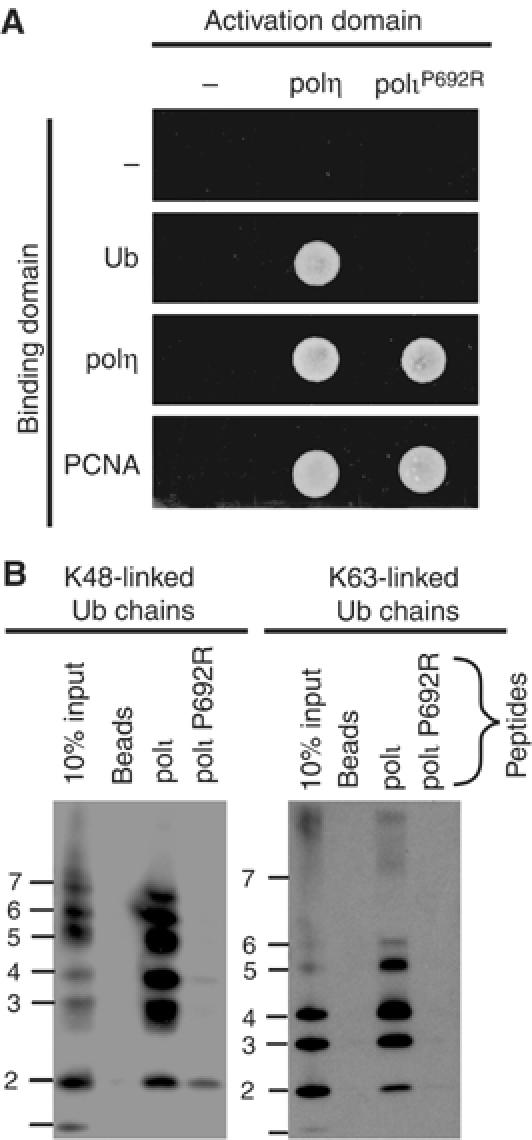
Identification of a mutant polι that cannot bind Ub. (A) polι P692R has lost the ability to bind Ub, but can still bind polη and PCNA in the yeast two-hybrid assay. (B) Pull-down assay using biotinylated peptides corresponding to the C-terminus of wild-type polι or a P692R variant and either K48- or K63-linked polyUb chains were analyzed as described in Figure 2B. Only the peptide corresponding to the wild-type polι sequence is able to bind either form of polyUb chain.
Effect of polι Ub-binding mutant and proteasome inhibitor on foci formation
As noted above, the P692R polι mutant is fully functional in its ability to interact with polη and PCNA, and in addition exhibits normal replication characteristics in vitro (Supplementary Figure S1A). We were therefore interested in determining the cellular effects that the P692R polι mutant might exhibit. The function of polι is currently unknown, but like polη (Kannouche et al, 2001), it forms foci after DNA damage that are believed to represent sites of stalled replication complexes (Kannouche et al, 2002). To determine what effect the P692R mutation might have on damage-induced foci formation, wild-type EGFP-polι and EGFP-polι P692R were transiently transfected into MRC5 cells (Figure 4A). As previously reported (Kannouche et al, 2002), EGFP-polι localizes to foci in 18% of unirradiated cells and 65% of cells treated with 7 J/m2 UV (Figure 4A). In striking contrast, the P692R mutant forms foci in only 0.5% of unirradiated cells and 5% of irradiated cells (Figure 4A). We therefore conclude that polι's ability to bind Ub is a prerequisite for its accumulation into replication foci.
Figure 4.
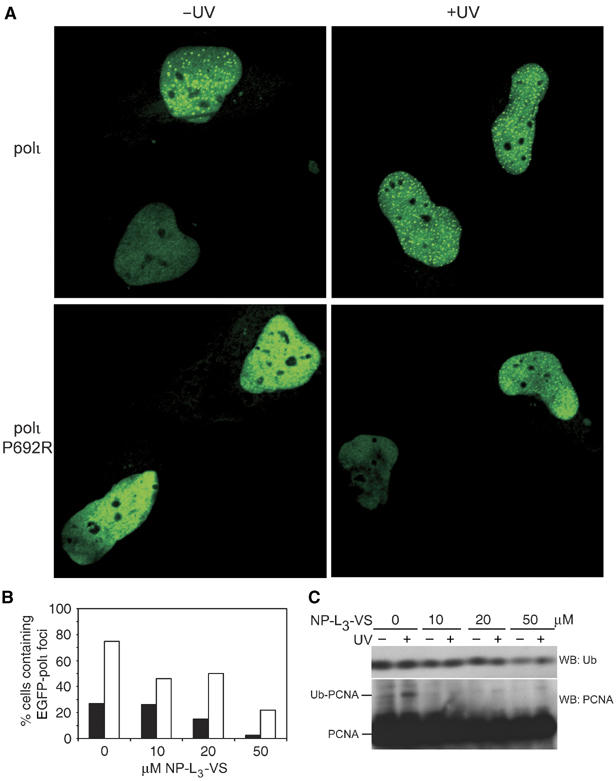
Binding to Ub is required for polι to relocalize into replication foci. (A) Wild-type pEGFP polι (upper panels) or pEGFP polι P692R (lower panels) were transfected into MRC5 cells and either mock treated (left hand panels) or irradiated with 7 J/m2 UV (right-hand panels). Nuclei from two adjacent cells are shown in each panel for comparison. (B) Histogram showing the percentage of nuclei with foci after exposure to various concentrations of the 26S proteasome inhibitor, NP-L3-VS before (closed bars) and after UV-irradiation (open bars). Numbers are based upon 200 nuclei analyzed per data point. (C) Western blot of Ub or PCNA in whole-cell extracts (50 μg) separated by SDS–PAGE on a 15% gel, after treatment with NP-L3-VS with and without UV-irradiation (7 J/m2). Note that the levels of Ub and Ub-PCNA decrease as the dose of NP-L3-VS is increased.
To further investigate the role of ubiquitination in polι-dependent foci formation, we assayed foci formation in the presence of various concentrations of the 26S proteasome inhibitor, NP-L3-VS (Figure 4B). It is thought that inhibition of the 26S proteasome stabilizes ubiquitinated proteins and thereby effectively functioning as a Ub sink, eventually depleting the pool of free Ub available for subsequent de novo Ub conjugation (Mimnaugh et al, 1997). Indeed, at the 50 μM dose of NP-L3-VS, a Western blot of whole-cell extracts reveals that levels of both free Ub and mono-ubiquitinated PCNA have decreased (Figure 4C). When cells were treated with increasing concentrations of NP-L3-VS prior to UV-irradiation, the number of nuclei exhibiting EGFP-polι foci steadily decreased (Figure 4B). Our data therefore indicate that polι foci formation depends upon de novo ubiquitination, rather than by binding to proteins ubiquitinated prior to UV-irradiation. The fact that we also saw a reduction in the number of nuclei with foci in a nonirradiated cell, also argues that at least one factor (most likely PCNA, see Discussion) must be ubiquitinated in S phase, even in the absence of exogenous DNA damage (Hoege et al, 2002; Watanabe et al, 2004).
Identification of a region in polη involved in Ub binding
Having identified a key residue required for the polι–Ub interaction, we were keen to do the same for polη. Indeed, by employing the yeast two-hybrid assay and various deletion constructs of polη, we mapped the Ub-binding site to a region spanning polη residues 587–641 (Supplementary Figure S2). This region contains a conserved C2H2 or so-called ‘Zinc-finger' motif (Vaisman et al, 2004). Since Zn-finger motifs are known to promote protein–protein interactions in a variety of proteins (Krishna et al, 2003; McCarty et al, 2003), we changed several residues of the polη Zn-finger motif to alanine and subsequently assayed the ability of the mutant protein to interact with Ub. Interestingly, a single H654A substitution in one of the conserved histidine residue of the C2H2 motif abolishes the interaction between polη and Ub (Figure 5A). The inability of the polη H654A mutant to interact with Ub cannot be due to the fact that the protein is simply unfolded or inactive, as the mutant retains its ability to bind both polι and PCNA (Figure 5A), and also exhibits catalytic efficiencies similar to the wild-type polη in vitro (Supplementary Figure S1B).
Figure 5.
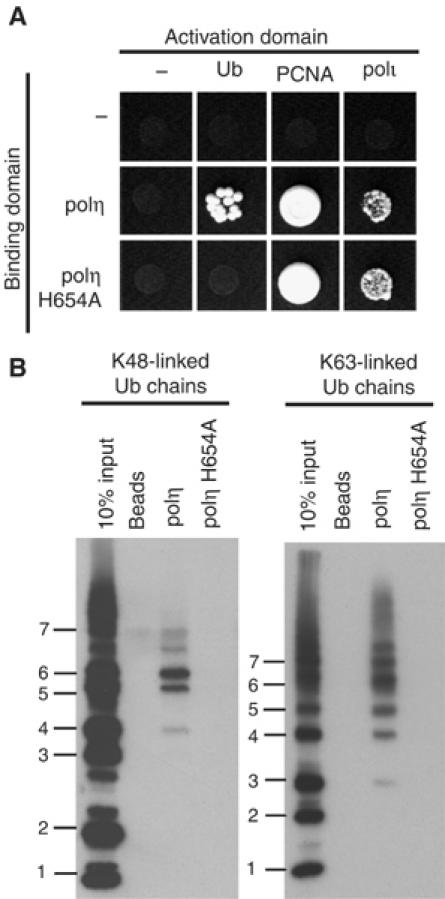
A mutant in polη's Zn-finger motif is defective in binding Ub. (A) Yeast two-hybrid assays showing that the polη H654A mutant is unable to interact with Ub, but nevertheless retains its ability to interact with full-length polι and PCNA. (B) Full-length C-terminal His-tagged polη or polη-H654A were used in pull-down assays with K48- or K63-linked Ub chains demonstrating that the H654A mutant is defective at binding either form of polyUb. Bound proteins were analyzed as described in Figure 2B.
To confirm that the region is required for the interaction between polη and Ub, we used full-length C-terminal His-tagged polη or polη-H654A and assayed the ability of each to bind Ub. As shown in Figure 5B, the wild-type protein clearly binds K48- and K63-linked polyUb chains. In contrast, the H654A polη mutant protein is unable to bind either form of polyUb.
Like the polι P692R mutant (Figure 4A), the H654A polη mutant is also defective at forming damage-induced foci. While wild-type polη forms foci in 38% of untreated cells and in 93% of UV-irradiated cells, the number of cells expressing EGFP-polη H654A foci drops dramatically in the untreated cells (∼4% cells with foci) and is reduced ∼2-fold (to 47%) of UV-irradiated cells (data not shown). The fact that 47% of the cells expressing EGFP-polη-H654A still form foci despite having a reduced ability to interact with Ub, can potentially be explained by the observation that the mutant nevertheless retains its ability to interact with both polι and PCNA (Figure 5A) and such interactions may be sufficient to help guide the mutant polη protein into damage-induced replication foci in vivo.
Ub binding enhances polymerase interactions with Ub-PCNA
It is entirely conceivable that polι and polη could bind to any ubiquitinated protein after DNA damage in order to localize at stalled replication forks. At the present time, however, Ub-PCNA appears to be the most likely candidate protein (Kannouche et al, 2004; Watanabe et al, 2004). We were therefore interested in assaying the ability of both wild-type polι and polη and their respective Ub-binding mutants to interact with Ub-PCNA in vitro. To do so, we first generated His-tagged Ub-PCNA in vitro following the protocol recently described for the ubiquitination of S. cerivisiae PCNA by Garg and Burgers (2005) (Supplementary Figure S3A and Supplementary Materials and Methods). As expected from earlier studies (Hoege et al, 2002), the His-Ub is conjugated to PCNA via K164, since a K164R PCNA mutant could not be ubiquitinated under the same assay conditions in vitro (Supplementary Figure S3B). Under our experimental conditions, only ∼30% of the PCNA is ubiquitinated in vitro (Supplementary Figure S3A). We believe that this is likely due to the inefficient loading of human PCNA onto the circular DNA template by S. cerevisiae RFC, but may also reflect differences between the yeast and human systems. However, after further purification of the His-Ub-PCNA over a nickel column, fractions were obtained that were significantly enriched for ubiquitinated PCNA (Supplementary Figure S3).
Using an N-terminal GST-fusion of full-length wild-type polι, or the polι P692R mutant, we then performed pull-down assays with PCNA or His-Ub-PCNA. Both polι and polι P692R are able to bind to unmodified PCNA (Figure 6B, left panel). However, the amount of Ub-PCNA bound by polι P692R is significantly reduced compared to wild-type polι. There is some residual binding, which is perhaps mediated through the combination of the PIP box and another potential UBM in polι (see Discussion).
Figure 6.
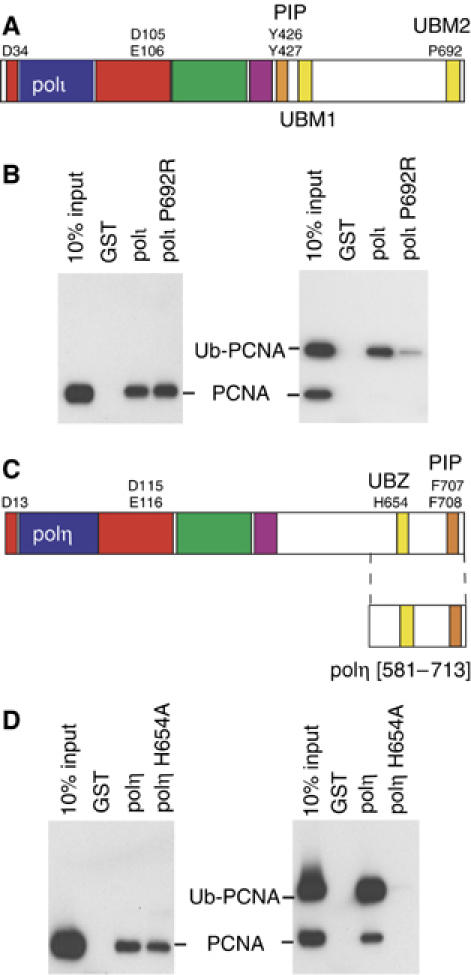
Protein–protein interactions regulating TLS. (A) Schematic representation of the domain/motif structure of polι. The polymerase domain representing conserved finger (blue) palm (red), thumb (green) and little finger (purple) spans the N-terminal portion of each protein. The PCNA binding site (PIP-Box) of each protein is colored orange and the UBMs are in yellow. UBM1 spans residues 496–524 and UBM2 spans amino acids 681–709 (Bienko et al, 2005). Residues identified in the active site of the polymerase, PIP-box or Ub interactions are shown by the single letter code. (B) Full-length wild-type GST-polι or polι-P692R mutant were used in pull-down assays with either PCNA or ubiquitinated-PCNA, separated on 10–20% Tris–glycine–SDS gels, transferred onto PVDF membrane, and PCNA was detected by Western Blot. Left: Both wild-type, and P692R polι GST-fusion proteins are able to bind to PCNA. Right: However, polι-P692R has a reduced ability to pull down Ub-PCNA. (C) Schematic representation of the domain/motif structure of polη (same color scheme as (A)), with amino acids 631–659 comprising the UBZ domain (Bienko et al, 2005). (D) Pull-down assays as described above were carried out with GST fused to the C-terminal 133 amino acids of polη or polη-H654A. Left: GST-polη[581–713] contains the PIP box and both wild-type polη, or the H654A polη mutant can bind PCNA, in accord with data shown in Figure 4B. Right: However, while the wild-type polη protein is able to pull-down Ub-PCNA, the H654A polη-Ub mutant is unable to bind ubiquitinated PCNA.
In the case of polη, the C-terminal amino acids of polη contain both the C2H2 Zn-finger motif and the PIP box (Figure 6C). It is not surprising, therefore, that a GST-fusion corresponding to the C-terminal 130 amino acids of polη (polη-[581–713]) can pull-down both unmodified PCNA (Figure 6D, left panel) and Ub-PCNA (Figure 6D, right panel). Interestingly, the amount of Ub-PCNA pulled down in these assays appears to be significantly greater than unmodified PCNA (cf. left and right panels Figure 6D with wild-type polη), suggesting that polη has a greater affinity for Ub-PCNA than it does unmodified PCNA. In contrast, while the polη H654A mutant retains a PIP box, it is nevertheless unable to pull-down ubiquitinated PCNA in the in vitro assay, thereby indicating that both the Ub-binding and PCNA-binding activities are required for an efficient interaction between polη and Ub-PCNA. Indeed, differences in the relative affinity of polη for PCNA versus Ub-PCNA may help explain, in part, why Kannouche et al (2004) were able to detect an interaction between polη and Ub-PCNA in vivo, yet were unable to detect any interaction between polη and unmodified PCNA, that we observe in vitro.
Discussion
Y-family polymerases are found in all kingdoms of life, suggesting that they play valuable cellular roles that are of great evolutionary importance. The central role that Y-family polymerases play in maintaining genomic integrity is highlighted by human polη, which appears to have evolved to protect humans from the deleterious effects of unrepaired sunlight-induced DNA lesions. Indeed, human polη replicates past cis-syn cyclobutane pyrimidine dimers with the same, or perhaps even higher efficiency and accuracy than it does undamaged DNA (McCulloch et al, 2004) and humans lacking polη display the variant form of Xeroderma Pigmentosum, a sunlight-sensitive, cancer-prone syndrome (Johnson et al, 1999; Masutani et al, 1999). Owing to its spacious active site, polη replicates undamaged DNA with low-fidelity, and it is this property that is believed to underlie its mutation-generating role in the diversification of somatic immunoglobulin genes (Rogozin et al, 2001; Zeng et al, 2001). Thus, polη plays both mutation reducing and mutation enhancing roles in human cells. It is likely that similar functions will ultimately be discovered for polι, polκ and Rev1, the three remaining human Y-family polymerases. To fulfill such diverse roles, the activity of the enzymes must presumably be exquisitely regulated, so that each enzyme only gains proper access to its appropriate substrate. How such regulation occurs in vivo is only now being fully elucidated.
Elegant genetic studies with S. cerevisiae have shown that ubiquitination of PCNA is a key step in determining whether cells utilize the inherently error-prone process of TLS or an error-free damage avoidance (DA) pathway to circumvent lesions in DNA. It is believed that Rad6/18 dependent monoubiquitination of PCNA helps promote TLS, while extension of the Ub chains by MMS2 and Ubc13 through a K63-linkage, helps promote a DA pathway (Hoege et al, 2002).
A similar picture is emerging in mammalian cells, where monoubiquitination of PCNA is believed to help target polη to sites of DNA damage (Kannouche et al, 2004; Watanabe et al, 2004). The role that polyUb chains play in regulating the activity of the TLS enzymes is, however, much less clear. Ub can be conjugated through any number of its Lysine residues to form polyUb chains (Pickart and Fushman, 2004). The best studied is the K48-linkage, which targets the substrate for proteasomal degradation (Chau et al, 1989). As with S. cerevisiae, polyUb linkage through K63 is believed to help promote the switch between TLS and DA pathways (Li et al, 2002). The mechanistic basis for the switch is, however, largely unknown.
In the present manuscript, we have shown that human Y-family polymerases ι and η bind Ub in a direct and noncovalent manner. We have also identified residues in each polymerase that are required for the polymerase–Ub interaction. While both polι and polη Ub-binding mutants exhibit wild-type biochemical activities, they are defective in their ability to relocalize into replication foci (Figure 4). Furthermore, our studies with a 26S proteasomal inhibitor indicate that such relocalization also depends upon de novo ubiquitination in vivo.
Since the interaction between polymerases ι and η with Ub is noncovalent, both polymerases could, in principal, bind to any number of repair/replication/transcription proteins that are specifically ubiquitinated, especially after DNA damage. However, within the context of TLS, the most likely target for ubiquitination is PCNA. Using His-Ub-PCNA that was generated in vitro, we are able to demonstrate that the Ub binding properties of both polι and polη are necessary to facilitate binding to ubiquitinated PCNA (Figure 6).
Taken together, our data support the notion that the ability of DNA polymerases ι and η to bind Ub, and specifically Ub-PCNA, is central for their ability to support TLS in vivo. Such conclusions are in perfect agreement with the recently reported studies of Bienko et al (2005), who have used a bioinformatics approach to identify several novel UBMs in all four human Y-family polymerases (polη, polι, polκ and Rev1). Interestingly, polι apparently has two UBMs and P692, which we identified via a genetic approach as being required for a polι-Ub interaction, lies in the very center of the UBM2 motif identified by Bienko et al.
Bienko et al (2005) also identified the C2H2 motif in polη as a novel UBM, that they termed a ‘UBZ' motif. Similar to our observations with the C2H2-H654A polη mutant, Bienko et al observed reduced UV-induced foci formation with a D652A-C2H2 polη mutant, as well as an inability of a polη UBZ mutant (C638A) to rescue the UV-sensitivity of XP-V cells. The latter observation therefore strongly supports the hypothesis that polη's ability to promote TLS in vivo is tightly linked to its ability to bind Ub.
Bienko et al (2005) also reported that both pols ι and η are themselves ubiquitinated in vivo and that ubiquitination decreases in response to UV-damage. If a similar situation occurs in S. cerevisiae, then such observations might explain why we failed to observe an interaction between human pols ι and η and the G75A–G76A Ub mutant in the two-hybrid assay (Figure 2A). Thus, it would appear that both pols are likely to be regulated by covalent and noncovalent interactions with Ub. The same is possibly true for polκ and Rev1 that also contain UBMs (Bienko et al, 2005).
How then, do such modifications affect TLS? Clearly, at one level, it helps promote re-localization, or at least accumulation of the polymerases into replication foci (Figure 4; Bienko et al, 2005), which are thought to contain several stalled replication forks (Kannouche et al, 2002). Recent biochemical data also suggest that unlike polδ, the enzymatic properties of two S. cerevisiae TLS enzymes, polη and Rev1, are stimulated in the presence of fully ubiquitinated PCNA (Garg and Burgers, 2005). Presumably, the same stimulatory effects of Ub-PCNA on the activity of human TLS enzymes will eventually be documented. While polδ may be able to utilize Ub-PCNA, it appears much more likely that ubiquitination increases the affinity of polη and polι for PCNA, thereby allowing the TLS polymerases to more effectively compete with polδ for access to the primer terminus. But what about the switch back to the main replicase, polδ after TLS? Very recently, Chui et al have shown that PCNA may become polyubiquitinated in response to DNA damage (Bradley Wouters, pers comm) and it is possible that DNA polymerases ι and η's affinity for polyUb chains may help dissociate the TLS polymerases from the 3′ primer terminus and facilitate the switch back to polδ after TLS has occurred.
Materials and methods
Two-hybrid screen
A screen for polι-interacting proteins was performed using the yeast two-hybrid Matchmaker III system (Clontech, Mountain View, CA). The entire human POLI gene was cloned as an ∼2.5 kb NcoI–BamHI fragment from p6-1 (McDonald et al, 1999) into the similarly digested GAL4 DNA-binding domain plasmid pGBKT7 (Clontech). This recombinant plasmid, called pAR110, expresses the GAL4 DNA-binding domain fused in-frame to the N-terminus of the entire polι protein. pAR110 was then transformed in the yeast strain AH109, and mated with Y187 containing a human testis cDNA-library cloned into the GAL4 activating domain vector, pACT2 (Clontech; K1604-A), as suggested by the manufacturer. Fusion proteins that potentially interact with polι were identified under medium stringency conditions by spreading the mating mixture on CSM-Trp-Leu-His+2.5 mM 3-amino-1,2,4-triazol plates (Qbiogene, Irvine, CA). Under these conditions, more than 400 independent clones were obtained and these were subsequently re-streaked on CSM-Trp-Leu-His-Ade+X-α-Gal plates (Qbiogene) to confirm the activation of all reporter genes. A number of the pACT2-library clones were randomly selected and the 5′-end of the insert determined by standard DNA sequencing protocols (Lark Technologies, Houston, TX).
Isolation of polι P692R mutants
The polι P692R mutant was generated by PCR amplification of wild-type POLI using primers AVR49 and AVR51 (Supplementary Materials and Methods), which introduces the P692R substitution and a silent SacI restriction enzyme site for subsequent analysis. The amplicon was cloned as an NcoI–SacI fragment into a pACT2-iota construct to generate pACT2-polι [P692R], (pAVR76).
Peptides
Two 46 amino-acid peptides, corresponding to polι residues 669–715 and a P692R substituted variant (underlined) (PDSVDEKITFPSDIDPQVFYELPEAVQKELLAEWKRTGSDFHIGHK) were each synthesized with an N-terminal biotin and purified to greater than 95% homogeneity by Genscript Corporation (Piscataway, NJ).
Ub and Ub-PCNA pull-down assay
Biotinylated-peptide, His-tag, and GST-tag pull-down assays of polyUb chains or Ub-PCNA were carried out in 400 μl of binding buffer (1 × PBS, 2.5 mg/ml BSA, 0.2% Triton X-100, protease inhibitors (Roche)). For Ub pull-downs, 5 μg Ub, K48- and K63-linked polyUb chains (Boston Biochem, Boston, MA) were added to each mixture, and for PCNA pull-downs, 100 ng PCNA or Ub-PCNA was added to the binding mixture. For peptides, 2 μg of each peptide were bound to 0.25 mg of streptavidin paramagnetic particles (Promega, Madison, WI). For Ni2+ pull-downs, 5 μg of 6-His-tagged protein were mixed with 15 μl of MagneHis (Promega) slurry added to binding buffer and 20 mM imidazole. For GST-pull-downs, 5 μg of each GST-tagged protein was mixed with 20 μl of glutathione sepharose slurry (Amersham Biosciences). Mixtures were incubated overnight while rotating at 4°C. The beads were washed three times with PBS containing 0.2% Triton X-100 followed by boiling in 1 × sample buffer and separation on a 10–20% Tris–glycine gel. Gels were transferred onto PVDF and Ub was detected with a polyclonal Ub antibody (Dako, Carpinteria, CA). PCNA was detected with the PC10 monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA).
Transfection and Immunofluorescence
MRC5 fibroblasts cells were plated onto Number 1 coverslips and transfected with pEGFP-polι using Fugene™6, according to the manufacturer's protocol (Roche). At 24 h after transfection, cells were irradiated at 7 J/m2 and incubated for a further 4 h. Cells were fixed using 2–3% formaldehyde in PBS (Polysciences, Inc., Warrington, PA) and mounted onto slides using Mowiol mounting medium. Fluorescence images of cell nuclei were acquired on a Zeiss LSM 510 inverted confocal microscope (Carl Zeiss) using LSM 5 software. An Argon laser was used for excitation (488 nm) and detection (above 505 nm) of EGFP. A minimum of 200 nuclei was analyzed per experiment, for each cell line and treatment. For experiments using the 26S proteasome inhibitor, NP-L3-VS (Calbiochem, La Jolla, CA), aliquots of a 10 mM stock solution (suspended in DMSO) were added to cell media to a final concentration of 10, 20 and 50 μM respectively, 1 h prior to UV-irradiation. Control cells were treated with DMSO using the same volume as that used for NP-L3-VS. At the time of irradiation, the media with NP-L3-VS was removed but saved. Cells were washed with Dulbecco's PBS warmed to 37°C. After UV (7 J/m2), media containing NP-L3-VS was returned to their respective cultures and cells were incubated an additional 4 h until cell fixation.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Materials and Methods
Acknowledgments
This work was supported by funds from the NICHD/NIH Intramural Research Program. We thank Alan Tomkinson and Sangeetha Vijayakumar for the gift of purified human PCNA; Michael Matunis for kindly providing the SUMO-1 cDNA; Kathleen Downey for the cDNA to POLD1; Vladimir Podust for cDNA to POLD3; Mike O'Donnell for the yeast RFC expression plasmid; Alan Lehmann for pEGFP-polη H654A; and Ivan Dikic, Alan Lehman, Peter Burgers and Bradley Wouters for kindly sharing data prior to publication. Microscopy Imaging was performed at the Microscopy & Imaging Core Facility (NICHD/NIH) with the assistance of Dr Vincent Schram. We dedicate this manuscript to the memory of the late Cecile Pickart. Cecile participated in many stimulating discussions during the evolution of the project and without her advice and continual encouragement many of the experiments described herein would not have been attempted.
References
- Baba D, Maita N, Jee JG, Uchimura Y, Saitoh H, Sugasawa K, Hanaoka F, Tochio H, Hiroaki H, Shirakawa M (2005) Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature 435: 979–982 [DOI] [PubMed] [Google Scholar]
- Bailly V, Lamb J, Sung P, Prakash S, Prakash L (1994) Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev 8: 811–820 [DOI] [PubMed] [Google Scholar]
- Bailly V, Lauder S, Prakash S, Prakash L (1997) Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J Biol Chem 272: 23360–23365 [DOI] [PubMed] [Google Scholar]
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, Hofmann K, Dikic I (2005) Novel ubiquitin-binding domains in Y-family polymerases regulate translesion DNA synthesis. Science 310: 1821–1824 [DOI] [PubMed] [Google Scholar]
- Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A (1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243: 1576–1583 [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Lehmann AR, Fuchs RP (2005) Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol Cell 18: 499–505 [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Wagner R, Radman M (2002) Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science 296: 1627–1630 [DOI] [PubMed] [Google Scholar]
- Garg P, Burgers PM (2005) Ubiquitinated PCNA activates translesion DNA polymerases η and REV1. PNAS 102: 18361–18366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Johnson RE, Unk I, Phillips B, Hurwitz J, Prakash L, Prakash S (2001) Physical and functional interactions of human DNA polymerase η with PCNA. Mol Cell Biol 21: 7199–7206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasty P, Campisi J, Hoeijmakers J, van Steeg H, Vijg J (2003) Aging and genome maintenance: lessons from the mouse? Science 299: 1355–1359 [DOI] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141 [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH (2001) Genome maintenance mechanisms for preventing cancer. Nature 411: 366–374 [DOI] [PubMed] [Google Scholar]
- Hughes P, Tratner I, Ducoux M, Piard K, Baldacci G (1999) Isolation and identification of the third subunit of mammalian DNA polymerase δ by PCNA-affinity chromatography of mouse FM3A cell extracts. Nucleic Acids Res 27: 2108–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Kondratick CM, Prakash S, Prakash L (1999) hRAD30 mutations in the variant form of Xeroderma Pigmentosum. Science 285: 263–265 [DOI] [PubMed] [Google Scholar]
- Kannouche P, Broughton BC, Volker M, Hanaoka F, Mullenders LH, Lehmann AR (2001) Domain structure, localization, and function of DNA polymerase η, defective in xeroderma pigmentosum variant cells. Genes Dev 15: 158–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche P, Fernandez de Henestrosa AR, Coull B, Vidal A, Gray C, Zicha D, Woodgate R, Lehmann AR (2002) Localisation of DNA polymerases η and ι to the replication machinery is tightly co-ordinated in human cells. EMBO J 21: 6246–6256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche PL, Wing J, Lehmann AR (2004) Interaction of human DNA polymerase η with monoubiquitinated PCNA; a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell 14: 491–500 [DOI] [PubMed] [Google Scholar]
- Krishna SS, Majumdar I, Grishin NV (2003) Structural classification of zinc fingers: survey and summary. Nucleic Acids Res 31: 532–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA (2004) DNA replication fidelity. J Biol Chem 279: 16895–16898 [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Pavlov YI, Bebenek K (2003) Functions of human DNA polymerases η, κ and ι suggested by their properties, including fidelity with undamaged DNA templates. DNA Repair 2: 135–149 [DOI] [PubMed] [Google Scholar]
- Lehmann AR (2005) Replication of damaged DNA by translesion synthesis in human cells. FEBS Lett 579: 873–876 [DOI] [PubMed] [Google Scholar]
- Li Z, Xiao W, McCormick JJ, Maher VM (2002) Identification of a protein essential for a major pathway used by human cells to avoid UV-induced DNA damage. Proc Natl Acad Sci USA 99: 4459–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H, Boudsocq F, Plosky BS, Woodgate R, Yang W (2003) Replication of a cis-syn thymine dimer at atomic resolution. Nature 424: 1083–1087 [DOI] [PubMed] [Google Scholar]
- Ling H, Boudsocq F, Woodgate R, Yang W (2001) Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell 107: 91–102 [DOI] [PubMed] [Google Scholar]
- Ling H, Boudsocq F, Woodgate R, Yang W (2004) Snapshots of replication through an abasic lesion; structural basis for base substitutions and frameshifts. Mol Cell 13: 751–762 [DOI] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F (1999) The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature 399: 700–704 [DOI] [PubMed] [Google Scholar]
- Matunis MJ (2002) On the road to repair: PCNA encounters SUMO and ubiquitin modifications. Mol Cell 10: 441–442 [DOI] [PubMed] [Google Scholar]
- McCarty AS, Kleiger G, Eisenberg D, Smale ST (2003) Selective dimerization of a C2H2 zinc finger subfamily. Mol Cell 11: 459–470 [DOI] [PubMed] [Google Scholar]
- McCulloch SD, Kokoska RJ, Chilkova O, Welch CM, Johansson E, Burgers PM, Kunkel TA (2004) Enzymatic switching for efficient and accurate translesion DNA replication. Nucleic Acids Res 32: 4665–4675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JP, Rapic-Otrin V, Epstein JA, Broughton BC, Wang X, Lehmann AR, Wolgemuth DJ, Woodgate R (1999) Novel human and mouse homologs of Saccharomyces cerevisiae DNA polymerase η. Genomics 60: 20–30 [DOI] [PubMed] [Google Scholar]
- Mimnaugh EG, Chen HY, Davie JR, Celis JE, Neckers L (1997) Rapid deubiquitination of nucleosomal histones in human tumor cells caused by proteasome inhibitors and stress response inducers: effects on replication, transcription, translation, and the cellular stress response. Biochemistry 36: 14418–14429 [DOI] [PubMed] [Google Scholar]
- Ohmori H, Friedberg EC, Fuchs RPP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, Prakash L, Prakash S, Todo T, Walker GC, Wang Z, Woodgate R (2001) The Y-family of DNA polymerases. Mol Cell 8: 7–8 [DOI] [PubMed] [Google Scholar]
- Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD (2005) Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p the processing of DNA lesions during replication. Mol Cell 19: 123–133 [DOI] [PubMed] [Google Scholar]
- Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S (2005) SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436: 428–433 [DOI] [PubMed] [Google Scholar]
- Pickart CM, Fushman D (2004) Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol 8: 610–616 [DOI] [PubMed] [Google Scholar]
- Pickart CM, Kasperek EM, Beal R, Kim A (1994) Substrate properties of site-specific mutant ubiquitin protein (G76A) reveal unexpected mechanistic features of ubiquitin-activating enzyme (E1). J Biol Chem 269: 7115–7123 [PubMed] [Google Scholar]
- Plosky BS, Woodgate R (2004) Switching from high-fidelity replicases to low-fidelity lesion-bypass polymerases. Curr Opin Genet Dev 14: 113–119 [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Pavlov YI, Bebenek K, Matsuda T, Kunkel TA (2001) Somatic mutation hotspots correlate with DNA polymerase η error spectrum. Nat Immunol 2: 530–536 [DOI] [PubMed] [Google Scholar]
- Solomon DA, Cardoso MC, Knudsen ES (2004) Dynamic targeting of the replication machinery to sites of DNA damage. J Cell Biol 166: 455–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter P, Ulrich HD (2003) Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425: 188–191 [DOI] [PubMed] [Google Scholar]
- Tsui C, Raguraj A, Pickart CM (2005) Ubiquitin binding site of the ubiquitin E2 variant (UEV) protein Mms2 is required for DNA damage tolerance in the yeast RAD6 pathway. J Biol Chem 280: 19829–19835 [DOI] [PubMed] [Google Scholar]
- Vaisman A, Lehmann AR, Woodgate R (2004) DNA polymerases η and ι. In DNA Repair and Replication, Yang W (ed), Vol. 69, pp. 205–228. San Diego: Elsevier/Academic Press [Google Scholar]
- Vidal AE, Kannouche P, Podust VN, Yang W, Lehmann AR, Woodgate R (2004) Proliferating cell nuclear antigen-dependent coordination of the biological functions of human DNA polymerase ι. J Biol Chem 279: 48360–48368 [DOI] [PubMed] [Google Scholar]
- Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M (2004) Rad18 guides polη to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J 23: 3886–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodgate R (1999) A plethora of lesion-replicating DNA polymerases. Genes Dev 13: 2191–2195 [DOI] [PubMed] [Google Scholar]
- Yang W (2005) Portraits of a Y-family DNA polymerase. FEBS Lett 579: 868–872 [DOI] [PubMed] [Google Scholar]
- Zeng X, Winter DB, Kasmer C, Kraemer KH, Lehmann AR, Gearhart PJ (2001) DNA polymerase η is an A–T mutator in somatic hypermutation of immunoglobulin variable genes. Nat Immunol 2: 537–541 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Materials and Methods


