Abstract
The structure of most of the peripheral stalk, or stator, of the F-ATPase from bovine mitochondria, determined at 2.8 Å resolution, contains residues 79–183, 3–123 and 5–70 of subunits b, d and F6, respectively. It consists of a continuous curved α-helix about 160 Å long in the single b-subunit, augmented by the predominantly α-helical d- and F6-subunits. The structure occupies most of the peripheral stalk in a low-resolution structure of the F-ATPase. The long helix in subunit b extends from near to the top of the F1 domain to the surface of the membrane domain, and it probably continues unbroken across the membrane. Its uppermost region interacts with the oligomycin sensitivity conferral protein, bound to the N-terminal region of one α-subunit in the F1 domain. Various features suggest that the peripheral stalk is probably rigid rather than resembling a flexible rope. It remains unclear whether the transient storage of energy required by the rotary mechanism takes place in the central stalk or in the peripheral stalk or in both domains.
Keywords: ATP synthase, mitochondria, stator, structure, function
Introduction
The F-ATPases in mitochondria, similar to their counterparts in chloroplasts and eubacteria, are multisubunit membrane-bound complexes that couple the transmembrane proton motive force to the synthesis of ATP from ADP and orthophosphate (Boyer, 1998; Walker, 1998). They consist of two main structural domains, a globular catalytic domain known as F1 and a membrane domain known as Fo, linked together by a central stalk and a peripheral stalk (Walker and Kane Dickson, 2006). In mitochondria, the F1 domain (an assembly of subunits α, β, γ, δ and ɛ in the ratio 3:3:1:1:1) extends into the matrix of the organelle. The γ-, δ- and ɛ-subunits form the central stalk in which an elongated α-helical coiled-coil in the γ-subunit penetrates into the α3β3 subcomplex along its axis of pseudo three-fold symmetry (Abrahams et al, 1994; Gibbons et al, 2000). Its lower region, including its foot, is exposed between the α3β3 subcomplex and Fo, and the foot makes an extensive area of contact with a ring of c-subunits buried in Fo (Stock et al, 1999). Synthesis of ATP in the catalytic β-subunits at interfaces with non-catalytic α-subunits depends upon the rotation of the central stalk (Noji et al, 1997) and the c-ring as an ensemble (Sambongi et al, 1999; Stock et al, 1999). The rotation is driven by the passage of protons from the inter-membrane space into the matrix via channels at the interface between the external surface of the c-ring and a hydrophobic membrane subunit known as a (or ATPase-6). The peripheral stalk links subunit a to the external surface of the F1 domain, and it is thought to act as a stator to counter the tendency of the α3β3 subcomplex to follow the rotation of the central stalk (Collinson et al, 1996; Walker and Kane Dickson, 2006). In bovine mitochondria, the peripheral stalk consists of single copies of subunits OSCP (oligomycin sensitivity conferral protein), b, d and F6 (coupling factor 6; Collinson et al, 1994, 1996). It has been visualised by electron microscopy of the intact enzyme in negative stain (Karrasch and Walker, 1999), as has the peripheral stalk in the bacterial and chloroplast F-ATPases (Böttcher et al, 1998; Wilkens and Capaldi, 1998). The subunit compositions of the bacterial and chloroplast F-ATPases differ substantially from those of the mitochondrial enzyme, being composed of one copy of subunit δ (homologous to the OSCP) and either two copies of subunit b in some eubacteria or of single copies of the homologous subunits b and b′ in photosynthetic bacteria and chloroplasts (where subunits b and b′ are usually known as subunits I and II, respectively).
The overall structure of the bovine F-ATPase has been determined to 32 Å resolution by electron cryomicroscopy of single particles (Rubinstein et al, 2003). By docking high-resolution structures of the F1 domain (Abrahams et al, 1994; Gibbons et al, 2000) and of the F1 domain with the attached c-ring (Stock et al, 1999) into this overall structure, an accurate outline structure of the peripheral stalk has been obtained. As expected, it extends from the top of the α3β3 subcomplex, along its external surface. Then, it reaches down about 45 Å to the membrane surface into the membrane domain, which it penetrates and traverses alongside the c-ring:subunit a subcomplex. A number of detailed features have been located within this outline structure. The structure of the N-terminal domain of the OSCP (residues 1–120), a bundle of six α-helices, has been placed on top of the F1-domain using biochemical data that showed that this domain interacts with the N-terminal regions of α-subunits (Hundal et al, 1983; Walker et al, 1985). These N-terminal regions of α-subunits are exposed on the top of the bovine α3β3 subcomplex, and also in the enzyme from Escherichia coli (Dunn et al, 1980). Molecular details of the interaction of the domain with the N-terminal regions of α-subunits have been established in both the bovine mitochondrial and bacterial enzymes (Carbajo et al, 2005; Wilkens et al, 2005). In Saccharomyces cerevisiae, the C-terminus of the OSCP lies about 90 Å along the surface of the α3β3 subcomplex (Rubinstein and Walker, 2002), and that of F6 (subunit h) is in the vicinity of the surface of the membrane (Rubinstein et al, 2005). The solution structure of bovine F6 has also been established (Carbajo et al, 2004). The isolated protein has a highly mobile and flexible structure consisting of two α-helices crossing at about 120° in their centres where they pack together via a loose hydrophobic core. The helices are connected by an unstructured linker. It is unlikely that this structure represents the arrangement of the α-helices in the assembled enzyme (Carbajo et al, 2004). It is much more probable that the protein has an extended structure where the helices lie in a more linear arrangement along the axis of the peripheral stalk (Rubinstein et al, 2005).
Extensive studies of the arrangement of subunits in the bovine peripheral stalk have been carried out by in vitro reconstitution of recombinant subunits and their individual domains (Collinson et al, 1994), and by deletion analysis (JA Silvester and JE Walker, unpublished results). In these studies, a stable subcomplex of the peripheral stalk has been formed that consists of single copies of the OSCP, b′ (residues 79–214; it lacks the N-terminal membrane spanning domain) d and F6. This subcomplex forms a 1:1 complex with F1-ATPase. Subunits b′ and d form a stable dimer, and the C-terminal regions of the OSCP and b′ interact (Collinson et al, 1994). Most importantly, a subcomplex consisting of residues 79–184 of subunit b, residues 1–124 of subunit d and the whole of F6 (76 residues plus an artificial initiator methionine residue) crystallised spontaneously (Silvester et al, 2006). The quality of these crystals has been improved, and the structure of this subcomplex, which represents 54% of the membrane extrinsic region of the peripheral stalk, has been determined by X-ray crystallography. Together with the structure of the N-terminal domain of the OSCP, representing a further 21% of the peripheral stalk, this structure has been incorporated into the overall structure of the bovine F-ATPase. As discussed below, it provides new insights into the properties and function of the peripheral stalk, and it provides new opportunities to study those properties.
Results and discussion
Characterisation of the overexpressed peripheral stalk subcomplex
The molecular masses of the proteins, both in the peripheral stalk subcomplex and in the subcomplex in which methionine residues had been substituted biosynthetically with selenomethionine, corresponded to the masses calculated from their sequences, assuming that the translational initiator methionine had been removed from the b and d fragments, and that it had been retained by F6 (see Supplementary Table I). There was no evidence in the mass spectra of oxidised products in either preparation, nor of partial retention of methionine in the selenomethionine-substituted proteins.
Structure determination
The structure of the b79–184d1–124F6 subcomplex was solved by multiple wavelength anomalous dispersion based on data to 3.2 Å resolution from crystals of the selenomethionine-substituted subcomplex. A preliminary model of the subcomplex consisting of 225 amino acids (out of a total of 305) was built in the experimentally phased electron density map. This model gave a clear molecular replacement solution for the native crystals (which contained two copies in the asymmetric unit compared to the four copies in the selenomethionine crystals). Further model building and refinement against the 2.8 Å resolution native data resulted in a final model consisting of residues 79–183 of subunit b, 3–123 of subunit d and 5–70 of F6. The two non-symmetry-related subcomplexes are very similar in conformation (r.m.s. deviation in main-chain atoms 0.54 Å) and they will not be distinguished in the following discussion. The final native structure was subjected to limited refinement against the selenomethionine data, to allow a comparison of the molecular conformations of the four copies in the selenomethionine crystals and the two copies in the native crystals. Data processing, phasing and refinement statistics are summarised in Table I, and a view of the electron density map is shown in Supplementary Figure 1.
Table 1.
Data collection and refinement and phasing statistics for the peripheral stalk subcomplex
| SeMet |
Native | ||
|---|---|---|---|
| Peak | Remote | ||
| Space group | P21 | P21 | |
| Unit cell: a, b, c (Å) | 98.4, 81.1, 129.5 | 50.5, 79.3, 115.7 | |
| α, β, γ (deg) | 90.0, 104.7, 90.0 | 90.0, 93.1, 90.0 | |
| Resolution range (Å) | 34–3.2 | 65–2.8 | |
| Wavelength (Å) | 0.9793 | 0.9393 | 1.771 |
| Unique reflections | 32 967 | 32 631 | 21 257 |
| Multiplicity | 7.4 (7.3) | 7.4 (7.4) | 11.0 (9.6) |
| Completeness | 97.7 (91.7) | 99.9 (100.0) | 94.1 (77.4) |
| Rmerge (%)a | 18.9 (176.4) | 99.9 (100) | 10.3 (94.2) |
| Mean 〈I/σ(I)〉 | 11.2 (1.3) | 14.2 (2.1) | 19.7 (2.4) |
| Wilson B factor (Å2) | 72.0 | ||
| Refinement | |||
| R factor (%) | 23.0 | ||
| Free R factor (%) | 29.3 | ||
| R.m.s.d. values | |||
| Bonds (Å) | 0.01 | ||
| Angles (deg) | 1.5 | ||
| Phasing | |||
| Cullis R factor (anom/iso)b | 0.774 | 0.854/0.557 | |
| Phasing power (anom/iso)b | 1.21 | 1.01/1.42 | |
| Mean figure of merit (acentric/centric) | 0.428/0.385 | ||
| Statistics for the highest resolution bin (2.95–2.80 Å) are shown in parentheses. Data beyond 2.94 Å were measured only in the corners of the detector, hence the low completeness for the highest resolution bin. | |||
a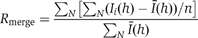
| |||
| bValues quoted for the Cullis R factor and phasing power are for acentric reflections only. | |||
Structure of the subcomplex of the peripheral stalk
The structure of the subcomplex contains extensive regions of α-helix (Figure 1). As summarised in Supplementary Table II, the presence and extent of these α-helical regions had been predicted rather accurately. The whole of the fragment of the b-subunit in the subcomplex forms a continuous curved α-helix about 160 Å long. Its N-terminal amino acid, residue 79 of the intact protein, is predicted from the hydropathic profile of the sequence of the subunit to be at the interface between the aqueous phase of the mitochondrial matrix and the inner surface of the inner mitochondrial membrane (Walker et al, 1987), and so in the intact F-ATPase, the helix in subunit b will extend in an N- to C-terminal direction upwards from the membrane towards the F1-domain.
Figure 1.
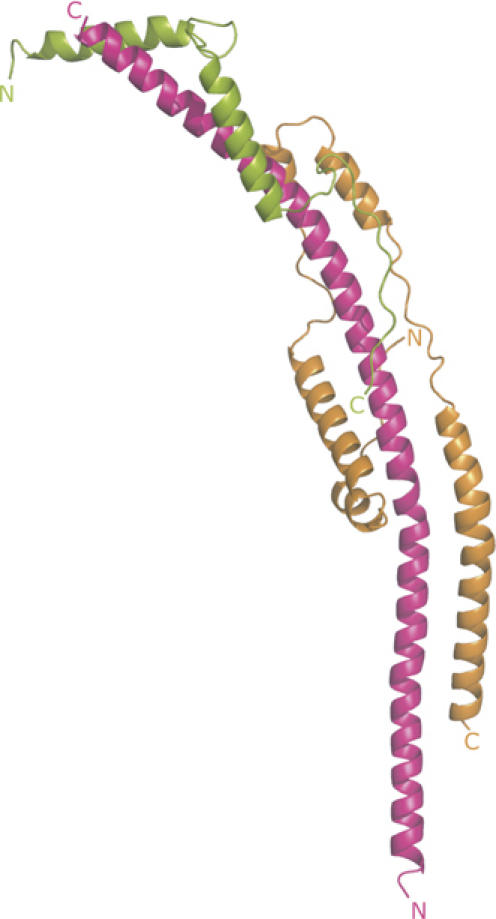
The structure of a subcomplex of the peripheral stalk of the F-ATPase from bovine mitochondria. The structure consists of residues 79–183 of subunit b, residues 3–123 of subunit d and residues 5–70 of F6, shown in magenta, orange and green, respectively. Their N- and C-terminals are indicated.
Residues 3–123 of subunit d are folded into five α-helical segments (residues 11–19, 24–44, 52–59, 64–78 and 85–122; see Supplementary Table II) linked by regions of extended structure. The fold of the protein is reminiscent of the shape of a paper clip. Its long axis is aligned with the long axis of the α-helix in subunit b, so that helix 2 lies parallel to residues 121–142 of subunit b, and helices 4 and 5 are antiparallel to residues 147–162 and 99–129, respectively. Thus, subunit d interacts with subunit b via a parallel and two antiparallel coiled-coil interactions in the vicinity of residues 99–162 of subunit b. It also interacts with the α-helix of subunit b via its extended regions (see Figure 1). The total buried surface area in the interface between subunits b and d is 2 × 831.9 Å2.
Subunit F6 is folded into two α-helices (residues 8–24 and 34–51) linked by an extended region. Its N-terminal region (residues 5–7) and especially its C-terminal region (residues 52–70) also have extended conformations. The secondary structure of the F6-subunit in the subcomplex is very similar to that determined by solution studies of the isolated subunit (Carbajo et al, 2004), but unlike this structure, where the α-helices (residues 7–22 and 34–51) cross over at an angle of about 122° near their centres and interact via hydrophobic patches (residues 27–32 and 34–51), F6 in the subcomplex has an elongated conformation. Instead of interacting with each other, the hydrophobic patches (residues F11, I15, Y18, and L39, L46, Y50) make the following interactions with subunit b: Y18-V177, L46-V161 and Y50-Y157. Residues F11 and I15 are exposed in the subcomplex, but in the intact peripheral stalk they may also form interactions with subunit b. In addition, the extended regions form specific interactions with the α-helix of the b-subunit, and the C-terminal helix of F6 makes a parallel interaction with helix 4 of subunit d. The buried surface areas in b–F6 and the d–F6 interfaces are 2 × 473.8 and 2 × 120.0 Å2, respectively.
The observed conformation of F6 is similar to the elongated arrangement predicted for this subunit in the intact peripheral stalk, where the N-terminus of the protein was proposed to be distal from the membrane with the C-terminus extending down towards the membrane surface (Rubinstein et al, 2005). From the model of the subcomplex, the C-terminus of bovine F6 would lie about 70 Å from the membrane surface, whereas the subunit equivalent to F6 in the enzyme from S. cerevisiae (subunit h) is 16 amino acids longer. Some of the additional residues are probably in the region linking the two main α-helical regions, and others contribute to a longer C-terminal region following helix 2. Therefore, subunit h can extend down almost to the membrane surface, as demonstrated by electron microscopy (Rubinstein et al, 2005).
Structure of the peripheral stalk in the F-ATPase complex
The peripheral stalk region of the 32 Å resolution structure obtained by cryo-electron microscopy of single particles of the intact enzyme complex (Rubinstein et al, 2003) was defined by docking into it the structure of the F1c10 subcomplex of the F-ATPase from S. cerevisiae (Figure 2). The resulting unoccupied region of the three-dimensional mesh, representing the surface of the complex, is the peripheral stalk region, the membrane region of subunit b, subunit a and other minor membrane subunits (e, f, g and A6L). The structure of the peripheral stalk subcomplex was docked by eye into this residual region of the mesh. This docking procedure was constrained severely by the length, curvature and shape of the subcomplex, and also by the requirement to position residue 79 of subunit b close to the boundary with the inner surface of the inner mitochondrial membrane. In this region, the structure of the peripheral stalk determined by electron cryomicroscopy narrows markedly. The structure of the N-terminal domain of the OSCP was positioned in the region on top of the α3β3 subcomplex, in close apposition with residue 184 at the C-terminal end of the fragment of subunit b in the peripheral stalk subcomplex (Figure 2).
Figure 2.
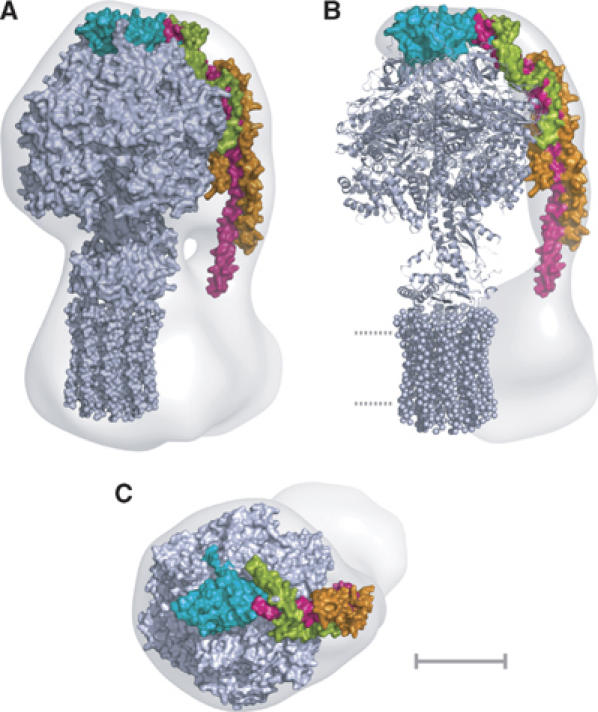
The composite structure of the ATP synthase from mitochondria. Detailed structures of the F1c10 subcomplex (grey), the N-terminal domain of the OSCP (cyan) and the peripheral stalk subcomplex (magenta, orange and green) were introduced by eye into an electron density map determined by averaging single particles of the intact bovine complex observed by electron cryo-microscopy. (A) side view; (B) residual density corresponding to the peripheral stalk and the second domain of Fo (Rubinstein et al, 2003). Dotted lines represent the lipid bilayer; (C) view looking down onto the ‘crown' of the F1 catalytic domain. The scale bar is 50 Å.
With the exception of a few residues, which protrude from the surface of the mesh facing towards the F1 domain (see Figure 3), the structure fits the mesh remarkably well. In addition, three local regions of the mesh are not occupied by the structure (Figure 3A). The first region (denoted by I in Figure 3), which lies on the outer surface of the curved part of the structure between subunits b and F6, is likely to be the site occupied by the C-terminal domain of the OSCP (residues 121–190). It is predicted to form a mixed α–β structure (Walker and Kane Dickson, 2006). Its size is compatible with the size of the unoccupied space, and its location is consistent with that of the C-terminus of the OSCP in the F-ATPase from S. cerevisiae estimated by electron microscopy (Rubinstein and Walker, 2002). The second unoccupied space in the mesh (denoted II in Figure 3A) lies in a region facing the surface of the F1 domain, where subunits b, d and F6 all come together. This space is likely to contain part of residues 185–214 of subunit b. This region of subunit b (residues 185–214) is predicted to form a turn from residues 185–187 followed by a second α-helix from residues 188–204 probably antiparallel to the long α-helix (see Figure 3B). The third space (III in Figure 3A) is likely to be the site occupied by part of residues 124–160 of subunit d in the peripheral stalk in the intact F-ATPase. This region is predicted to be folded into an α-helix from residues 132–138 followed by an extended region (residues 139–160), and because the constriction of the mesh towards the membrane surface seems to be capable of accommodating only the single α-helix of the b-subunit, the chain of the d-subunit must turn back on itself in the deleted region (Figure 3B). These additional modelled regions account reasonably well for the unoccupied regions of the mesh. This proposal is supported by a study of the interactions between the subunits using peptide arrays, which suggests an interaction between subunit d at a site in the vicinity of residues 100–160 and the C-terminal region of subunit b (V Kane Dickson and JE Walker, unpublished work).
Figure 3.
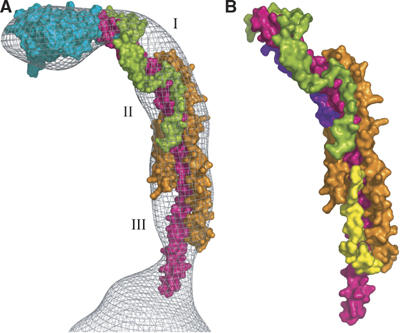
Locations of regions of the peripheral stalk subunits that were deleted in the subcomplex. Subunits b, d, F6 and OSCP are shown in magenta, orange, green, and cyan, respectively. In (A), I, II and III indicate regions that probably accommodate residues 121–190, 185–214 and 125–160 of subunits OSCP, b and d, respectively. In (B), residues 188–214 of subunit b have been modelled as an α-helix shown in purple lying antiparallel to the central α-helix. Residues 125–160 of subunit d are modelled in yellow as an extended region from residues 125–131 followed by a short α-helix (residues 132–138) approximately orthogonal to the plane of the paper and an extended region parallel to the central α-helix in the b-subunit.
At present, there is only limited information about the structure of the enzyme in the region below the point where the peripheral stalk enters the membrane. The long α-helix in subunit b is predicted to continue unbroken across the membrane (see Supplementary Table II). A turn and a second transmembrane α-helix are predicted for residues 48–54 and 33–47, respectively, and the N-terminal region (residues 1–30, where residues 19–26 are probably α-helical) is exposed in the mitochondrial matrix (Walker and Kane Dickson, 2006). Each of bovine subunits e, f, g and A6L probably contains a single membrane spanning α-helix, but the locations of their N- and C-termini in either the inter-membrane space or the matrix are not known with certainty. In S. cerevisiae, the N-terminus of subunit f is in the mitochondrial matrix and its C-terminus is in the intermembrane space (Roudeau et al, 1999). On the basis of crosslinking experiments, the a subunit in the E. coli F-ATPase has been proposed to form five transmembrane α-helices (Fillingame et al, 2003; Zhang and Vik, 2003). Helix 4 in the E. coli model contains R210 that is involved in proton translocation across the membrane (Fillingame and Dmitriev, 2002), and so these helices must be part of an interface with the ring of c-subunits, which contain an acidic amino acid side chain (D61) that also forms part of the proton translocation pathway. Currently, there is no other information about how the α-helical regions in the membrane domains of the various subunits interact with each other.
Comparison of peripheral stalks in F-ATPases
The similarities in sequences and subunit compositions of the F-ATPases in eubacteria, mitochondria and chloroplasts indicate that the structures of their F1 and Fo domains are closely related, and it is likely that the peripheral stalks will be similar also. The similarity between the structures and function of the mitochondrial OSCP and the bacterial and chloroplast δ-subunits is clear in their sequences, in the structures of their N-terminal domains and in their mode of binding to α-subunits. Similarities in predicted secondary structures of their C-terminal domains indicate that their C-terminal regions are likely to have related structures also, and this similarity is likely to extend to the mode of interaction of these C-terminal domains with other subunits in their peripheral stalks. The subcomplex of b′ (the extrinsic domain of mitochondrial subunit b), subunit d and F6 is replaced in eubacteria and chloroplasts by the extrinsic domains of either a homodimer of b-subunits or heterodimer subunits (known as b and b′) of unknown structures. Three domains have been defined in the extrinsic region of subunit b in E. coli. They are the ‘δ-subunit interaction domain' at the C-terminal end, preceded by the ‘dimerisation' and ‘tether' domains (Dunn et al, 2000). The dimerisation domain is capable of forming an α-helical coiled-coil. It has been proposed to make a right-handed coiled-coil (Dunn et al, 2000), but the crystal structure is a monomeric α-helix (Del Rizzo et al, 2002). It is unlikely that a homodimer of bacterial b-subunits can adopt a structure that is similar to the region of the mitochondrial peripheral stalk that contains the interacting b- and d-subunits, which contain both parallel and antiparallel coiled-coils. Thus, although there are clear structural similarities between F-ATPases from various sources, from the available evidence it appears that the structure of a large part of the peripheral stalk has not been conserved.
The N-terminal regions of the bacterial and chloroplast b-subunits traverse the membrane once via α-helical regions (Dmitriev et al, 1999). These two parallel α-helices interact with the a-subunit, and the two antiparallel α-helices in the mitochondrial b-subunits are likely to do so also, as crosslinking experiments suggest (Velours et al, 2000).
Properties and role of the peripheral stalk
The unit cells of the crystals of the native and selenomethionine-substituted subcomplexes of the peripheral stalk complex contain six non-symmetry-related complexes that are subject to different crystal packing interactions. For example, in some copies (Supplementary Figure 2), there are additional crystal contacts at the ends of the α-helix in subunit b. Nonetheless, their structures are very similar with r.m.s.d. values between the main-chain carbon atoms of 0.5–1.4 Å. These close structural similarities between the six complexes suggest that the peripheral stalk in the mitochondrial F-ATPase is a rather inflexible structure consisting of a single unbroken curved α-helix in the b-subunit, which is supported and augmented throughout most of its length by close interactions with both α-helical and extended regions in subunits d and F6. These interactions bury 60% of the surface area of subunit b in the subcomplex, and in the intact peripheral stalk, the buried area of the membrane extrinsic region of subunit b will be significantly higher. The proposed interactions of the upper extremity of this α-helix with the C-terminal region of the b-subunit and with the C-terminal domain of the OSCP will add to its inflexibility. Only the lower part of the long α-helix in the structure, from residues 79 to 99 immediately above the region where it enters the membrane, seems to lack support from other subunits. However, it is possible that support is provided for this exposed region by the N-terminal region of subunit b from residues 1–30 or by regions of subunits e, f and g that are exposed in the mitochondrial matrix. At the top extremity of the peripheral stalk, a groove between helices 1 and 5 in the N-terminal domain of the OSCP interacts with a region that includes most or all of residues 1–12 in the N-terminal region of a single α-subunit of the α3β3 subcomplex of the F-ATPase (Carbajo et al, 2005). In the enzyme from E. coli, the equivalent interaction has been estimated to be sufficiently strong to resist the torque of 50 kJ/mol generated by the rotary action of the enzyme (Weber et al, 2002), which might imply that there is no requirement for specific interactions between the surface of the peripheral stalk and the external surface of the F1 domain. The surfaces of both the bovine F1-ATPase and the peripheral stalk structure are both predominantly hydrophilic, and there is no evidence for complementary hydrophobic or charged regions that could form specific interactions between the two surfaces.
This emerging picture of a rather inflexible mitochondrial peripheral stalk connecting the top of the F1 domain and the membrane domain by the shortest route is entirely compatible with its role as a stator, preventing the α3β3 subcomplex from following the rotation of the central stalk. The apparent rigidity of the peripheral stalk subcomplex suggests that it may also be important for maintaining the integrity of the interface between the a-subunit and the rotating ring of c-subunits by acting as clamp keeping the a-subunit and c-ring in contact, thereby maintaining the integrity of the proton channel in the interface between them. A similar role has been proposed for the b-dimer in the bacterial enzyme (Ono et al, 2004).
This model of the mitochondrial stalk is radically different from extant models of the peripheral stalk in bacterial and chloroplast F-ATPases where it is considered to be a flexible structure, resembling a rope. These properties are inferred from the ability to shorten the extrinsic region of the b-subunit in the E. coli enzyme by making deletions up to 11 residues (Sorgen et al, 1998), to lengthen it by making insertions up to 14 residues (Sorgen et al, 1999) and to co-assemble one shortened b-subunit with a wild-type subunit (Grabar and Cain, 2003) without affecting the activity of the enzyme. However, the effects of these changes on the structure and physical properties of the bacterial peripheral stalk are not known. Moreover, mutation of residues in the dimerisation domain of the E. coli subunit b affected both dimerisation and enzyme function negatively (Cipriano et al, 2006). The possible importance of subunit b as a flexible elastic link for delivering torque more smoothly during ATP synthesis or hydrolysis, and so improve the efficiency of the enzyme, has been proposed on theoretical grounds (Oster and Wang, 2000), and the possible role of an elastic peripheral stalk for transient storage of energy between steps in the rotary motion of the enzyme has been discussed (Cherepanov et al, 1999), as has a similar role for the central stalk (Ma et al, 2002) and even the c ring itself (Meier et al, 2005). The structure of the mitochondrial peripheral stalk and the availability of various subcomplexes of it purified from recombinant sources should allow their bending, stretching and compression modes of elastic deformation to be measured directly in biophysical experiments. This should reveal the site or sites of transient storage of torsional energy during rotational catalysis.
Materials and methods
Expression, purification, characterisation and crystallisation of the subcomplex of the peripheral stalk
The recombinant expression, purification and crystallisation of the bovine peripheral stalk subcomplex are described elsewhere (Silvester et al, 2006). The subcomplex in which methionine residues were replaced by selenomethionine was produced by growing the cells of E. coli C41 (DE3) containing the expression plasmid in defined medium lacking methionine, but containing selenomethionine (van den Ent and Löwe, 2000). The level of expression was ca. 10 mg/l. It was purified as described for the native complex except that the concentration of dithiothreitol in buffers was 5 mM.
The purified subcomplex was analysed by SDS–PAGE, and proteins were stained with Coomassie brilliant blue R-250. Protein bands were digested with trypsin and analysed by peptide mass-mapping in a Micromass TofSpec 2E mass spectrometer (Carroll et al, 2003). The subunits in the purified complex were separated by reverse-phase chromatography on a column of Aquapore RP-300 (1.0 × 100 mm) in 0.1% trifluoroacetic with a gradient of acetonitrile. The eluate was introduced directly into a Quattro Ultima (Micromass) triple quadrupole instrument. A solution (10 μl) of the selenomethionine-labelled subcomplex (16–18 mg/ml) was diluted 150-fold in 50% acetonitrile containing 1% formic acid, and injected directly into a Q-TOF (Micromass) quadrupole-time of flight instrument via a nanospray interface. Positive ion spectra of both the native and selenomethionine-substituted subcomplexes were recorded over the mass to charge range 600–1800.
A native data set was collected from a crystal grown in the standard buffer plus 17.0% polyethylene glycol (PEG) 5000 monomethylether (MME) (w/v) and 1.0% γ-picoline (v/v). These crystals have two copies of the subcomplex in the asymmetric unit. Derivative data were collected from a crystal of the selenomethionine-substituted subcomplex grown under the same conditions, with 14.5% PEG 5000 MME except that the additive used was 0.9% thioxane and 5 mM dithiothreitol was present in the buffer. The cell dimensions of the Se-Met crystal were different from those of the native crystal (Table I), and they correspond approximately to a doubling of the a-axis of the unit cell. The Se-Met crystal contains four copies of the subcomplex in the asymmetric unit, and it also exhibited weaker and more anisotropic diffraction.
Data collection and structure determination
All X-ray diffraction data were collected from flash-frozen cryo-protected crystals at the European Synchrotron Radiation Facility, Grenoble, France. The native data were collected at a wavelength of 1.771 Å on beamline ID23eh1 using a MarMosaic 225 CCD detector from a crystal that had been exposed to xenon at a pressure of greater than 15 bar in an attempt to obtain a derivative. Subsequently, no xenon substitution was detected. The Se-Met data were collected at two wavelengths (peak and remote) on beamline ID29 using an ADSC Q210 CCD detector. Diffraction images were integrated with MOSFLM (Leslie, 1992) and further data reduction carried out with SCALA (Evans, 1997) and TRUNCATE (French and Wilson, 1978).
The selenium sites were located using SHELXD (Schneider and Sheldrick, 2002) and refined with SHARP (Bricogne et al, 2003), and the resulting electron density was improved with SOLOMON (Abrahams and Leslie, 1996) as implemented in SHARP. The effective resolution of the phasing was 3.6 Å. Averaging the four independent copies of the subcomplex did not improve the quality of the electron density significantly, because of deviations from exact non-crystallographic symmetry. The known positions of the selenium sites and the helix-build utility of ARP/wARP (Perrakis et al, 1999; Morris et al, 2004) facilitated building a model of one of the four copies of the subcomplex using O (Jones et al, 1991). The resulting model, consisting of 225 amino acids, was used to perform molecular replacement with the native data set, using the program Phaser (McCoy et al, 2005). The higher resolution of the native data allowed the model to be extended, and alternate rounds of rebuilding and refinement including non-crystallographic symmetry restraints were carried out with REFMAC5 (Murshudov et al, 1997). The stereochemistry of the refined model was analysed with the program PROCHECK (Laskowski et al, 1993). There are no residues in disallowed regions of the Ramachandran plot, and 90.3% lie in the most favoured regions.
In order to be able to compare the structures of the Se-Met and native subcomplexes, the final native model was placed in the Se-Met unit cell, using the program AMoRe (Navaza, 1994). A phased translation function (based on the Se-Met experimental phases) was required to position the model correctly, presumably because of the pseudo-translational symmetry of the Se-Met crystals. Limited rigid-body refinement of this model was carried out by dividing the complex into five rigid groups. The final Rfree value for this model was 37.0%. The refined coordinates were used to compare the NCS copies to those of the native form.
Images of three-dimensional structures and electron density maps were generated with the program PyMOL (DeLano, 2002).
Deposition of coordinates
The atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB code 2CLY).
Interpretation of a low-resolution model of the F-ATPase
The coordinates of the structure of the subcomplex were fitted by eye with PyMOL into the electron density map of the bovine F-ATPase obtained by averaging single particles observed by electron cryo-microscopy (Rubinstein et al, 2003).
Supplementary Material
Supplementary Information
References
- Abrahams JP, Leslie AGW (1996) Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr D 52: 30–42 [DOI] [PubMed] [Google Scholar]
- Abrahams JP, Leslie AGW, Lutter R, Walker JE (1994) Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370: 621–628 [DOI] [PubMed] [Google Scholar]
- Böttcher B, Schwarz L, Gra̋ber P (1998) Direct indication for the existence of a double stalk in CFoF1. J Mol Biol 281: 757–762 [DOI] [PubMed] [Google Scholar]
- Boyer PD (1998) ATP synthase—past and future. Biochim Biophys Acta 1365: 3–9 [DOI] [PubMed] [Google Scholar]
- Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W (2003) Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr D 59: 2023–2030 [DOI] [PubMed] [Google Scholar]
- Carbajo RJ, Kellas FA, Runswick MJ, Montgomery MG, Walker JE, Neuhaus D (2005) Structure of the F1-binding domain of the stator of bovine F1Fo-ATPase and how it binds an alpha-subunit. J Mol Biol 351: 824–838 [DOI] [PubMed] [Google Scholar]
- Carbajo RJ, Silvester JA, Runswick MJ, Walker JE, Neuhaus D (2004) Solution structure of subunit F6 from the peripheral stalk region of ATP synthase from bovine heart mitochondria. J Mol Biol 342: 593–603 [DOI] [PubMed] [Google Scholar]
- Carroll J, Fearnley IM, Shannon RJ, Hirst J, Walker JE (2003) Analysis of the subunit composition of complex I from bovine heart mitochondria. Mol Cell Proteomics 2: 117–126 [DOI] [PubMed] [Google Scholar]
- Cherepanov DA, Mulkidjanian AY, Junge W (1999) Transient accumulation of elastic energy in proton translocating ATP synthase. FEBS Lett 449: 1–6 [DOI] [PubMed] [Google Scholar]
- Cipriano DJ, Wood KS, Bi Y, Dunn SD (2006) Mutations in the dimerization domain of the b subunit from the Escherichia coli ATP synthase: deletions disrupt function but not enzyme assembly. J Biol Chem 281: 4126–4131 [DOI] [PubMed] [Google Scholar]
- Collinson IR, Skehel JM, Fearnley IM, Runswick MJ, Walker JE (1996) The F1Fo-ATPase complex from bovine heart mitochondria: the molar ratio of the subunits in the stalk region linking the F1 and Fo domains. Biochemistry 35: 12640–12646 [DOI] [PubMed] [Google Scholar]
- Collinson IR, van Raaij MJ, Runswick MJ, Fearnley IM, Skehel JM, Orriss GL, Miroux B, Walker JE (1994) ATP synthase from bovine heart mitochondria. In vitro assembly of a stalk complex in the presence of F1-ATPase and in its absence. J Mol Biol 242: 408–421 [DOI] [PubMed] [Google Scholar]
- Del Rizzo PA, Bi Y, Dunn SD, Shilton BH (2002) The ‘second stalk' of Escherichia coli ATP synthase: structure of the isolated dimerization domain. Biochemistry 41: 6875–6884 [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific [Google Scholar]
- Dmitriev OY, Jones PC, Fillingame RH (1999) Structure of the subunit c oligomer in the F1Fo ATP synthase: model derived from solution structure of the monomer and cross-linking in the native enzyme. Proc Natl Acad Sci USA 96: 7785–7790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn SD, Heppel LA, Fullmer CS (1980) The NH2-terminal portion of the alpha subunit of Escherichia coli F1 ATPase is required for binding the delta subunit. J Biol Chem 255: 6891–6896 [PubMed] [Google Scholar]
- Dunn SD, McLachlin DT, Revington M (2000) The second stalk of Escherichia coli ATP synthase. Biochim Biophys Acta 1458: 356–363 [DOI] [PubMed] [Google Scholar]
- Evans PR (1997) Scaling of MAD data. Joint CCP4 ESF-EAMCB Newsl Protein Crystallogr 33: 22–24 [Google Scholar]
- Fillingame RH, Angevine CM, Dmitriev OY (2003) Mechanics of coupling proton movements to c-ring rotation in ATP synthase. FEBS Lett 555: 29–34 [DOI] [PubMed] [Google Scholar]
- Fillingame RH, Dmitriev OY (2002) Structural model of the transmembrane Fo rotary sector of H+-transporting ATP synthase derived by solution nmr and intersubunit cross-linking in situ. Biochim Biophys Acta 1565: 232–245 [DOI] [PubMed] [Google Scholar]
- French S, Wilson KS (1978) On the treatment of negative intensity observations. Acta Crystallogr A 34: 517–525 [Google Scholar]
- Gibbons C, Montgomery MG, Leslie AGW, Walker JE (2000) The structure of the central stalk in bovine F1-ATPase at 2.4 Å resolution. Nat Struct Biol 7: 1055–1061 [DOI] [PubMed] [Google Scholar]
- Grabar TB, Cain BD (2003) Integration of b subunits of unequal lengths into F1Fo-ATP synthase. J Biol Chem 278: 34751–34756 [DOI] [PubMed] [Google Scholar]
- Hundal T, Norling B, Ernster L (1983) Lack of ability of trypsin-treated mitochondrial F1-ATPase to bind the oligomycin-sensitivity conferring protein (OSCP). FEBS Lett 162: 5–10 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Karrasch S, Walker JE (1999) Novel features in the structure of bovine ATP synthase. J Mol Biol 290: 379–384 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Moss DS, Thornton JM (1993) Main-chain bond lengths and bond angles in protein structures. J Mol Biol 231: 1049–1067 [DOI] [PubMed] [Google Scholar]
- Leslie AGW (1992) Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 ESF-EAMCB Newsl Protein Crystallogr 26: 22–33 [Google Scholar]
- Ma J, Flynn TC, Cui Q, Leslie AGW, Walker JE, Karplus M (2002) A dynamic analysis of the rotation mechanism for conformational change in F1-ATPase. Structure 10: 921–931 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ (2005) Likelihood-enhanced fast translation functions. Acta Crystallogr D 61: 458–464 [DOI] [PubMed] [Google Scholar]
- Meier T, Polzer P, Diederichs K, Welte W, Dimroth P (2005) Structure of the rotor ring of F-type Na+-ATPase from Ilyobacter tartaricus. Science 308: 659–662 [DOI] [PubMed] [Google Scholar]
- Morris RJ, Zwart PH, Cohen S, Fernandez FJ, Kakaris M, Kirillova O, Vonrhein C, Perrakis A, Lamzin VS (2004) Breaking good resolutions with ARP/wARP. J Synchrotron Radiat 11: 56–59 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Navaza J (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr A 50: 157–163 [Google Scholar]
- Noji H, Yasuda M, Yoshida M, Kinosita K Jr (1997) Direct observation of the rotation of F1-ATPase. Nature 386: 299–302 [DOI] [PubMed] [Google Scholar]
- Ono S, Sone N, Yoshida M, Suzuki T (2004) ATP synthase that lacks Fo a-subunit: isolation, properties, and indication of Fo b2-subunits as an anchor rail of a rotating c-ring. J Biol Chem 279: 33409–33412 [DOI] [PubMed] [Google Scholar]
- Oster G, Wang H (2000) Reverse engineering a protein: the mechanochemistry of ATP synthase. Biochim Biophys Acta 1458: 482–510 [DOI] [PubMed] [Google Scholar]
- Perrakis A, Morris R, Lamzin VS (1999) Automated protein model building combined with iterative structure refinement. Nat Struct Biol 6: 458–463 [DOI] [PubMed] [Google Scholar]
- Roudeau S, Spannagel C, Vaillier J, Arselin G, Graves PV, Velours J (1999) Subunit f of the yeast mitochondrial ATP synthase: topological and functional studies. J Bioenerget Biomembr 31: 85–94 [DOI] [PubMed] [Google Scholar]
- Rubinstein JL, Kane Dickson V, Runswick MJ, Walker JE (2005) ATP synthase from Saccharomyces cerevisiae: location of subunit h in the peripheral stalk region. J Mol Biol 345: 513–520 [DOI] [PubMed] [Google Scholar]
- Rubinstein JL, Walker JE (2002) ATP synthase from Saccharomyces cerevisiae: location of the OSCP subunit in the peripheral stalk region. J Mol Biol 321: 613–619 [DOI] [PubMed] [Google Scholar]
- Rubinstein JL, Walker JE, Henderson R (2003) Structure of the mitochondrial ATP synthase by electron cryomicroscopy. EMBO J 22: 6182–6192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambongi Y, Iko Y, Tanabe M, Omote H, Iwamoto-Kihara A, Ueda I, Yanagida T, Wada Y, Futai M (1999) Mechanical rotation of the c subunit oligomer in ATP synthase (FoF1): direct observation. Science 286: 1722–1724 [DOI] [PubMed] [Google Scholar]
- Schneider TR, Sheldrick GM (2002) Substructure solution with SHELXD. Acta Crystallogr D 58: 1772–1779 [DOI] [PubMed] [Google Scholar]
- Silvester JA, Kane Dickson V, Runswick MJ, Leslie AGW, Walker JE (2006) The expression, purification, crystallization and preliminary X-ray analysis of a subcomplex of the peripheral stalk of ATP synthase from bovine mitochondria. Acta Crystallogr F 62: (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorgen PL, Bubb MR, Cain BD (1999) Lengthening the second stalk of F1Fo ATP synthase in Escherichia coli. J Biol Chem 274: 36261–36266 [DOI] [PubMed] [Google Scholar]
- Sorgen PL, Caviston TL, Perry RC, Cain BD (1998) Deletions in the second stalk of F1Fo-ATP synthase in Escherichia coli. J Biol Chem 273: 27873–27878 [DOI] [PubMed] [Google Scholar]
- Stock D, Leslie AGW, Walker JE (1999) Molecular architecture of the rotary motor in ATP synthase. Science 286: 1700–1705 [DOI] [PubMed] [Google Scholar]
- van den Ent F, Löwe J (2000) Crystal structure of the cell division protein FtsA from Thermotoga maritima. EMBO J 19: 5300–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velours J, Paumard P, Soubannier V, Spannagel C, Vaillier J, Arselin G, Graves PV (2000) Organisation of the yeast ATP synthase Fo: a study based on cysteine mutants, thiol modification and cross-linking reagents. Biochim Biophys Acta 1458: 443–456 [DOI] [PubMed] [Google Scholar]
- Walker JE (1998) ATP synthesis by rotary catalysis. Angew Chem Int Ed 3: 2308–2319 [DOI] [PubMed] [Google Scholar]
- Walker JE, Fearnley IM, Gay NJ, Gibson BW, Northrop FD, Powell SJ, Runswick MJ, Saraste M, Tybulewicz VLJ (1985) Primary structure and subunit stoichiometry of F1-ATPase from bovine mitochondria. J Mol Biol 184: 677–701 [DOI] [PubMed] [Google Scholar]
- Walker JE, Kane Dickson V (2006) The peripheral stalk of the mitochondrial ATP synthase. Biochim Biophys Acta (in press) [DOI] [PubMed] [Google Scholar]
- Walker JE, Runswick MJ, Poulter L (1987) ATP synthase from bovine mitochondria: characterization and sequence analysis of two membrane associated subunits and their corresponding cDNAs. J Mol Biol 197: 89–100 [DOI] [PubMed] [Google Scholar]
- Weber J, Wilke-Mounts S, Senior AE (2002) Quantitative determination of binding affinity of delta-subunit in Escherichia coli F1-ATPase: effects of mutation, Mg2+, and pH on Kd. J Biol Chem 277: 18390–18396 [DOI] [PubMed] [Google Scholar]
- Wilkens S, Borchardt D, Weber J, Senior AE (2005) Structural characterization of the interaction of the delta and alpha subunits of the Escherichia coli F1Fo-ATP synthase by nmr spectroscopy. Biochemistry 44: 11786–11794 [DOI] [PubMed] [Google Scholar]
- Wilkens S, Capaldi RA (1998) Electron microscopic evidence of two stalks linking the F1 and Fo parts of the Escherichia coli ATP synthase. Biochim Biophys Acta 1365: 93–97 [DOI] [PubMed] [Google Scholar]
- Zhang D, Vik SB (2003) Helix packing in subunit a of the Escherichia coli ATP synthase as determined by chemical labelling and proteolysis of the cysteine-substituted protein. Biochemistry 42: 331–337 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


