Abstract
Thermodynamic parameters are reported for duplex formation of 48 self-complementary RNA duplexes containing Watson–Crick terminal base pairs (GC, AU and UA) with all 16 possible 3′ double-nucleotide overhangs; mimicking the structures of short interfering RNAs (siRNA) and microRNAs (miRNA). Based on nearest-neighbor analysis, the addition of a second dangling nucleotide to a single 3′ dangling nucleotide increases stability of duplex formation up to 0.8 kcal/mol in a sequence dependent manner. Results from this study in conjunction with data from a previous study [A. S. O'Toole, S. Miller and M. J. Serra (2005) RNA, 11, 512.] allows for the development of a refined nearest-neighbor model to predict the influence of 3′ double-nucleotide overhangs on the stability of duplex formation. The model improves the prediction of free energy and melting temperature when tested against five oligomers with various core duplex sequences. Phylogenetic analysis of naturally occurring miRNAs was performed to support our results. Selection of the effector miR strand of the mature miRNA duplex appears to be dependent upon the identity of the 3′ double-nucleotide overhang. Thermodynamic parameters for 3′ single terminal overhangs adjacent to a UA pair are also presented.
INTRODUCTION
Single and double-nucleotide overhangs on the 3′ end of RNA duplexes have previously been shown to contribute to the stability of a duplex in a sequence dependent manner (1–5). The increase in duplex stability by 3′ dangling nucleotides is attributed to stacking interactions that dangling bases form with neighboring closing base pairs in the duplex as a result of A-form helical geometry. Base identity of 3′ double-nucleotide overhangs is critical in determining thermodynamic stability of duplexes.
Understanding the thermostability of RNA duplexes with 3′ double-nucleotide overhangs is essential for understanding some of the major biological roles of this structure. The 3′ double-nucleotide overhang in the sequences used in this study mimic the structure of short interfering (siRNAs) and micro RNAs (miRNAs) (6–10). siRNAs are formed when the ribonuclease III enzyme, dicer, processes long non-coding double-stranded RNA (dsRNA) in the cell into ∼21 nt oligonucleotides containing a 19mer duplex with double-nucleotide overhangs on the 3′ ends (11–15). One strand of the siRNA duplex, called the ‘guide’ strand is then incorporated into the RNA Induced Silencing Complex (RISC) and target mRNA complementary in sequence to the siRNA, while the other strand, the ‘passenger’ strand is degraded. miRNAs are endogenous to the cell and occur when a large RNA hairpin referred to as a pri-miRNA is processed into a smaller RNA hairpin pre-miRNA. This structure is further processed by the dicer enzyme into a mature miRNA sequence containing a miR/miR* duplex ∼19 bp with 2 nt overhangs on the 3′ ends. The guide, or miR, strand of the mature miRNAs are also loaded into the RNAi effector complex RISC to target complementary mRNAs, while the miR* strand is degraded. Once incorporated into RISC, siRNAs and miRNAs pair with mRNA of complementary sequence and either induce cleavage of the target mRNA or translational repression of the mRNA message (16). Many groups have shown that both siRNAs and mature miRNAs are incorporated into RISC in an asymmetric manner which is dependent on the stability of the base pairing at the 3′ ends of each strand in the duplex (17,18). Until now there has not been an accurate method for calculating the contribution of 3′ double-nucleotide overhangs on thermodynamic stability of duplex formation and therefore no reliable way of predicting which strand of the siRNA or the miRNA will be loaded into RISC as the effector strand in RNAi.
We have shown previously for duplexes containing a CG closing base pair, a double-nucleotide overhang on the 3′ end of the duplex where the addition of a second dangling nucleotide to a single purine dangling nucleotide can enhance stabilization of the duplex. However, if the first dangling nucleotide is a pyrimidine then the addition of a second dangling nucleotide will not provide any additional stabilization of the duplex O'Toole et al. (5). Here we have included in our study all possible 3′ double-nucleotide dangling ends using Watson–Crick bases on core duplexes containing the remaining three possible orientations of Watson–Crick terminal base pairs: GC, AU and UA. The thermodynamic parameters obtained from this study along with those from our previous work have allowed us to develop a model for improved prediction of stability of a duplex with a 3′ double-nucleotide dangling end.
MATERIALS AND METHODS
RNA synthesis and purification
All oligomers were synthesized on CPG solid supports (Applied Biosystems 392 DNA/RNA Synthesizer) with phosphoramidites with the 2′ hydroxyl protected as the tert-butyl dimethylsilyl ether from Glen Reseach (Sterling VA). Oligomers underwent ammonia and fluoride deprotection, and the crude sample was purified using preparative TLC (n-propanol:ammonium hydroxide:water, 55:35:10) and Sep-Pak C18 (Waters) chromatography as previously described (19). Sample purity was determined through analytical TLC or HPLC (C-18), and was >95%.
Melting curve and data analysis
Optical melting experiments were performed using a Beckman DU 640 Spectrophotometer and High Performance Temperature Controller at 280 nm. Absorbance changes for oligomers in 1 M NaCl melt buffer (1 M NaCl, 0.01 M cacodylic acid, 0.001 M ethylenediamine tetraacetic acid, pH 7.0) were recorded as function of temperature from 90 to 5°C at a rate of 1°C/min as described previously (19). The experiment was repeated at 10 varying sample concentrations to give at least a 50-fold concentration range (10 μM–1 mM) for each sample. Absorbance versus temperature profiles were fit to a two-state model with sloping base lines using a non-linear least squares program (20). Thermodynamics parameters for the oligomers were determined from both the average of the individual melt curves and plots of the reciprocal melting temperature () versus ln(Ct) for self-complementary sequences or () versus ln(Ct/4) for non-self-complementary sequences. Parameters derived from the two methods agreed within 10%, consistent with the two-state model (21).
Phylogenetic analysis
A total of 1290 experimentally validated miRNA sequences from miRBase release 8.0 (http://microrna.sanger.ac.uk/sequences/; February 2006) were ‘conceptually diced’ using an algorithm which ‘dices’ the pre-miRNA sequences based on a 19 nt region of base pairing with a 3′ double-nucleotide overhangs. These mature miRNA sequences were then analyzed for the frequency of Watson–Crick closing base pairs with 3′ double-nucleotide overhangs. Of the 1290 sequences 1009 fit our criteria. The occurrence of each of the possible combinations of Watson–Crick closing base pairs with double-nucleotide overhang on both the miR and miR* strands of the mature miRNA sequences was determined.
RESULTS AND DISCUSSION
For duplexes containing terminal CG base pairs, stability of duplex formation has been shown to be influenced by the second nucleotide of a 3′ double-nucleotide overhang (5). The additional stability of the duplex varies based on the identity of the nucleotides in the overhang; in the order pur–pyr > pur–pur > pyr–pyr = pyr–pur = 0. To explore the generality of the trend observed previously, we examined the influence of 3′ double-nucleotide overhangs on duplexes containing all possible Watson–Crick closing base pairs.
Thermodynamic data
The measured thermodynamic parameters for all 48 of the 3′ double-nucleotide terminal overhangs are presented in Table 1. Thermodynamic parameters were determined using both melt curve analysis and the TM dependence models (20). Data from both models agreed within 10% for all sequences, consistent with the two-state model with the exception of (CCGGCG)2. The average deviations in thermodynamic parameter values are ±5.6, 6.3 and 2.0% for ΔHo, ΔSo and , respectively.
Table 1.
Thermodynamic parameters for duplex formation in 1 M NaCla
| Average of curve fits | versus log Ct plots | |||||||
|---|---|---|---|---|---|---|---|---|
| Sequences | −ΔH° (kcal/mol) | −ΔS° (eu) | (kcal/mol) | (°C) | −ΔH° (kcal/mol) | −ΔS° (eu) | (kcal/mol) | TMb (°C) |
| (CCGGAA)2 | 48.6 ± 4.3 | 134.2 ± 13.7 | 6.9 ± 0.2 | 45.1 | 52.6 ± 5.0 | 147.2 ± 15.9 | 7.0 ± 0.2 | 44.7 |
| (CCGGAG)2 | 49.3 ± 6.3 | 135.1 ± 19.8 | 7.4 ± 0.2 | 48.1 | 47.1 ± 3.7 | 128.6 ± 11.6 | 7.3 ± 0.1 | 47.8 |
| (CCGGAC)2 | 50.0 ± 4.8 | 136.1 ± 14.9 | 7.8 ± 0.3 | 50.7 | 50.3 ± 5.6 | 137.2 ± 18.5 | 7.8 ± 0.3 | 50.4 |
| (CCGGAU)2 | 50.9 ± 5.3 | 139.4 ± 17.0 | 7.7 ± 0.2 | 49.9 | 48.5 ± 2.0 | 131.7 ± 6.3 | 7.6 ± 0.1 | 50.0 |
| (CCGGAp)2c | 46.8 | 128.5 | 6.9 | 45.6 | ||||
| (CCGGGA)2 | 47.8 ± 3.9 | 129.9 ± 12.3 | 7.5 ± 0.2 | 49.5 | 52.8 ± 5.0 | 145.7 ± 15.7 | 7.6 ± 0.2 | 48.9 |
| (CCGGGG)2 | 55.9 ± 1.8 | 156.5 ± 5.0 | 7.3 ± 0.3 | 46.5 | 48.6 ± 11.3 | 133.5 ± 35.1 | 7.2 ± 1.0 | 47.0 |
| (CCGGGC)2 | 53.4 ± 2.8 | 144.4 ± 8.7 | 8.6 ± 0.1 | 54.8 | 54.5 ± 1.4 | 147.9 ± 4.3 | 8.6 ± 0.1 | 54.6 |
| (CCGGGU)2 | 48.1 ± 3.5 | 129.3 ± 10.9 | 8.0 ± 0.2 | 52.5 | 47.6 ± 7.4 | 128.0 ± 23.2 | 8.0 ± 0.4 | 52.6 |
| (CCGGGp)2d | 47.9 | 132 | 7.0 | 45.5 | ||||
| (CCGGCA)2 | 39.4 ± 3.7 | 109.7 ± 12.0 | 5.4 ± 0.1 | 34.8 | 38.4 ± 2.4 | 106.5 ± 7.9 | 5.3 ± 0.1 | 34.3 |
| (CCGGCG)2 | 35.1 ± 1.6 | 94.0 ± 5.3 | 6.0 ± 0.1 | 39.9 | 75.2 ± 8.0 | 221.7 ± 24.3 | 6.6 ± 1.0 | 41.0 |
| (CCGGCC)2 | 37.4 ± 4.2 | 101.1 ± 13.5 | 6.1 ± 0.1 | 40.4 | 40.2 ± 0.8 | 110.3 ± 2.5 | 6.0 ± 0.1 | 39.6 |
| (CCGGCU)2 | 37.9 ± 3.9 | 104.4 ± 12.8 | 5.5 ± 0.1 | 35.7 | 43.6 ± 2.8 | 122.8 ± 9.1 | 5.5 ± 0.1 | 35.7 |
| (CCGGCp)2d | 40.9 | 115 | 5.2 | 33.7 | ||||
| (CCGGUA)2 | 41.0 ± 4.1 | 114.6 ± 13.2 | 5.4 ± 0.1 | 35.1 | 46.1 ± 3.9 | 131.1 ± 12.6 | 5.4 ± 0.1 | 35.1 |
| (CCGGUG)2 | 42.1 ± 5.4 | 117.0 ± 17.4 | 5.8 ± 0.1 | 37.6 | 43.9 ± 2.6 | 122.8 ± 8.6 | 5.8 ± 0.1 | 37.8 |
| (CCGGUC)2 | 39.0 ± 4.1 | 107.4 ± 13.3 | 5.7 ± 0.1 | 37.2 | 43.6 ± 1.8 | 122.5 ± 6.0 | 5.6 ± 0.1 | 36.7 |
| (CCGGUU)2 | 41.3 ± 4.2 | 114.9 ± 13.3 | 5.6 ± 0.1 | 36.7 | 37.5 ± 2.0 | 102.8 ± 6.7 | 5.6 ± 0.1 | 36.3 |
| (CCGGUp)2c | 42.6 | 118.5 | 5.8 | 38.3 | ||||
| (AGCGCUAA)2 | 64.1 ± 3.3 | 172.9 ± 9.1 | 10.5 ± 0.2 | 62.2 | 64.5 ± 5.3 | 174.2 ± 16.8 | 10.5 ± 0.2 | 62.1 |
| (AGCGCUAG)2 | 62.5 ± 3.4 | 168.2 ± 10.3 | 10.4 ± 0.3 | 61.8 | 66.8 ± 6.0 | 181.3 ± 9.2 | 10.6 ± 0.2 | 61.7 |
| (AGCGCUAC)2 | 65.3 ± 3.6 | 175.7 ± 10.7 | 10.8 ± 0.3 | 63.5 | 69.4 ± 1.3 | 187.9 ± 3.9 | 11.2 ± 0.1 | 63.6 |
| (AGCGCUAU)2 | 62.6 ± 2.1 | 168.2 ± 6.3 | 10.4 ± 0.2 | 62.5 | 68.4 ± 2.5 | 185.7 ± 7.8 | 10.8 ± 0.2 | 62.3 |
| (AGCGCUA)2f | 58.0 ± 3.0 | 157.0 ± 8.9 | 9.3 ± 0.3 | 57.9 | 63.2 ± 2.6 | 172.7 ± 8.0 | 9.6 ± 0.2 | 57.8 |
| (AGCGCUGA)2 | 57.5 ± 1.8 | 154.8 ± 5.6 | 9.5 ± 0.2 | 59.3 | 66.9 ± 2.1 | 183.4 ± 6.5 | 10.0 ± 0.1 | 58.7 |
| (AGCGCUGG)2 | 57.1 ± 1.4 | 153.3 ± 4.0 | 9.6 ± 0.2 | 59.7 | 65.4 ± 1.3 | 178.7 ± 4.2 | 10.0 ± 0.1 | 59.0 |
| (AGCGCUGC)2 | 63.2 ± 2.2 | 170.5 ± 6.7 | 10.3 ± 0.2 | 61.3 | 67.6 ± 1.4 | 184.1 ± 4.2 | 10.5 ± 0.1 | 61.1 |
| (AGCGCUGU)2 | 62.8 ± 1.7 | 169.3 ± 5.6 | 10.3 ± 0.1 | 61.7 | 65.5 ± 2.1 | 177.4 ± 6.3 | 10.5 ± 0.1 | 61.5 |
| (AGCGCUG)2f | 59.7 ± 2.8 | 162.2 ± 8.5 | 9.4 ± 0.2 | 57.5 | 67.3 ± 2.5 | 185.3 ± 7.6 | 9.8 ± 0.1 | 57.2 |
| (AGCGCUCA)2 | 57.1 ± 2.5 | 152.4 ± 6.4 | 9.1 ± 0.3 | 56.9 | 63.6 ± 1.3 | 174.9 ± 4.1 | 9.4 ± 0.1 | 56.3 |
| (AGCGCUCG)2 | 56.9 ± 6.5 | 153.5 ± 15.4 | 9.3 ± 0.3 | 58.1 | 47.6 ± 1.5 | 125.1 ± 4.6 | 8.8 ± 0.1 | 59.2 |
| (AGCGCUCC)2 | 56.0 ± 5.4 | 150.6 ± 16.3 | 9.1 ± 0.2 | 58.3 | 58.1 ± 3.9 | 157.3 ± 11.9 | 9.3 ± 0.2 | 57.9 |
| (AGCGCUCU)2 | 53.6 ± 2.9 | 144.1 ± 8.9 | 8.9 ± 0.2 | 56.6 | 59.2 ± 2.6 | 161.3 ± 8.0 | 9.2 ± 0.1 | 56.3 |
| (AGCGCUC)2f | 52.8 ± 4.1 | 142.8 ± 12.6 | 8.6 ± 0.3 | 54.8 | 59.8 ± 3.3 | 164.4 ± 10.1 | 8.8 ± 0.2 | 54.4 |
| (AGCGCUUA)2 | 49.5 ± 3.6 | 133.3 ± 11.8 | 8.2 ± 0.2 | 53.5 | 53.7 ± 3.9 | 146.6 ± 12.3 | 8.3 ± 0.2 | 52.8 |
| (AGCGCUUG)2 | 51.9 ± 4.5 | 139.3 ± 14.0 | 8.7 ± 0.2 | 56.5 | 57.3 ± 1.5 | 155.9 ± 4.5 | 9.0 ± 0.1 | 55.8 |
| (AGCGCUUC)2 | 56.4 ± 5.0 | 152.7 ± 15.2 | 9.1 ± 0.3 | 56.9 | 55.6 ± 1.1 | 150.2 ± 3.5 | 9.0 ± 0.1 | 56.6 |
| (AGCGCUUU)2 | 53.5 ± 4.3 | 144.0 ± 13.3 | 8.8 ± 0.2 | 56.5 | 60.5 ± 2.1 | 165.6 ± 6.7 | 9.2 ± 0.2 | 56.0 |
| (AGCGCUU)2f | 54.6 ± 4.5 | 148.1 ± 13.9 | 8.7 ± 0.3 | 55.0 | 58.4 ± 2.6 | 160.1 ± 8.1 | 8.8 ± 0.1 | 54.5 |
| (UGCGCAAA)2 | 58.4 ± 2.5 | 156.3 ± 7.8 | 9.9 ± 0.2 | 61.2 | 62.9 ± 1.5 | 170.0 ± 4.5 | 10.1 ± 0.1 | 60.9 |
| (UGCGCAAG)2 | 59.7 ± 1.7 | 160.0 ± 5.3 | 10.0 ± 0.1 | 61.5 | 61.4 ± 1.4 | 165.2 ± 4.3 | 10.1 ± 0.1 | 61.4 |
| (UGCGCAAC)2 | 60.9 ± 1.7 | 163.0 ± 5.3 | 10.3 ± 0.2 | 62.7 | 62.5 ± 1.7 | 168.0 ± 5.2 | 10.4 ± 0.1 | 62.6 |
| (UGCGCAAU)2 | 60.8 ± 4.7 | 162.7 ± 14.4 | 10.3 ± 0.3 | 62.7 | 63.5 ± 2.5 | 171.0 ± 7.5 | 10.5 ± 0.2 | 62.5 |
| (UGCGCAAp)2e | 59.5 | 161.0 | 9.6 | 58.8 | ||||
| (UGCGCAGA)2 | 57.5 ± 2.5 | 153.6 ± 7.7 | 9.9 ± 0.3 | 61.4 | 56.4 ± 1.1 | 150.3 ± 3.4 | 9.8 ± 0.1 | 61.6 |
| (UGCGCAGG)2 | 59.4 ± 2.0 | 159.3 ± 6.0 | 10.0 ± 0.2 | 61.3 | 59.9 ± 1.6 | 160.9 ± 5.1 | 10.0 ± 0.1 | 61.3 |
| (UGCGCAGC)2 | 63.1 ± 1.9 | 169.6 ± 5.5 | 10.5 ± 0.2 | 62.6 | 62.3 ± 1.9 | 167.3 ± 5.8 | 10.4 ± 0.1 | 62.6 |
| (UGCGCAGU)2 | 60.2 ± 1.6 | 161.0 ± 4.8 | 10.2 ± 0.1 | 62.5 | 61.3 ± 2.0 | 164.5 ± 6.1 | 10.3 ± 0.1 | 62.4 |
| (UGCGCAGp)2e | 60.6 | 163.8 | 9.8 | 59.3 | ||||
| (UGCGCACA)2 | 52.7 ± 3.0 | 139.5 ± 9.4 | 9.5 ± 0.2 | 61.0 | 53.1 ± 1.9 | 140.7 ± 6.0 | 9.4 ± 0.1 | 60.7 |
| (UGCGCACG)2 | 57.9 ± 7.5 | 154.6 ± 22.5 | 9.9 ± 0.4 | 61.4 | 59.5 ± 2.3 | 156.3 ± 6.9 | 10.0 ± 0.1 | 61.9 |
| (UGCGCACC)2 | 53.5 ± 3.1 | 141.8 ± 9.5 | 9.6 ± 0.2 | 61.3 | 54.6 ± 1.7 | 145.1 ± 5.1 | 9.6 ± 0.1 | 60.9 |
| (UGCGCACU)2 | 55.2 ± 2.9 | 147.4 ± 9.2 | 9.5 ± 0.2 | 60.2 | 57.2 ± 2.3 | 153.4 ± 7.0 | 9.6 ± 0.1 | 59.9 |
| (UGCGCACp)2e | 53.4 | 142.6 | 9.2 | 59.0 | ||||
| (UGCGCAUA)2 | 53.6 ± 2.7 | 142.9 ± 8.2 | 9.3 ± 0.2 | 59.3 | 55.4 ± 1.8 | 148.4 ± 5.5 | 9.4 ± 0.1 | 59.0 |
| (UGCGCAUG)2 | 55.7 ± 3.1 | 149.8 ± 9.7 | 9.2 ± 0.2 | 58.2 | 57.9 ± 1.9 | 156.7 ± 5.9 | 9.3 ± 0.1 | 57.9 |
| (UGCGCAUC)2 | 56.3 ± 1.8 | 150.8 ± 5.5 | 9.6 ± 0.1 | 60.0 | 58.4 ± 1.4 | 157.0 ± 4.5 | 9.7 ± 0.1 | 59.8 |
| (UGCGCAUU)2 | 55.3 ± 3.6 | 147.9 ± 10.6 | 9.4 ± 0.3 | 59.6 | 52.3 ± 1.0 | 138.9 ± 2.9 | 9.2 ± 0.1 | 59.6 |
| (UGCGCAUp)2e | 55.7 | 149.6 | 9.3 | 58.7 | ||||
aSolutions are 1.0 M NaCl, 10 mM sodium cacodylate, 0.5 mM EDTA pH 7.
bCalculated at 10−4 M oligomer concentration.
cPetersheim and Turner (2).
dFreier et al. (1).
eSugimoto et al. (3).
fThis study.
We have shown previously that 3′ double-nucleotide dangling ends increase the stability of RNA duplexes relative to the duplexes with only a single 3′ dangling nucleotide when the first-nucleotide overhang is a purine residue. For the oligomers in this study, (Table 1), the second 3′ dangling nucleotide changes the TM of the helix by −3.2 to −9.2°C (average increase 2.4°C) for a 10−4 M solution. Additional stabilization of the duplex by the second dangling nucleotide is sequence dependent. The average increase in TM for helices where the first dangling nucleotide is a purine is 3.6°C, while helices where the first dangling nucleotide is a pyrimidine, the average increase is 1.3°C (2.6°C for cytodine and 0.0°C for uridine). These increases are within the range seen previously for the addition of a second dangling nucleotide on the 3′ end of a duplex with a CG closing base pair (5).
Free energy parameters for 3′ terminal dangling ends on U/A base pair
Thermodynamic values have been previously determined for all the duplexes in this study containing all possible single 3′ dangling nucleotides (1,3), except for those with the (AGCGCU)2 core. We chose this core because the previously measured single 3′ dangling nucleotides on UA closing base pairs (3) had been measured on duplexes with a core of (AUGCAU)2. However, the terminal 2 bp, AU base pair neighboring a terminal UA base pair, are likely to fray affecting the interaction of the terminal base pair with the dangling ends. The core sequence used in this study (AGCGCU)2, with a CG base pair neighboring a single terminal UA base pair is less likely to fray. In order to determine the stability contributed to the UA oligomer by the 3′ dangling nucleotide, differences in the thermodynamic values between the sequences studied and the corresponding core sequence were also determined for all of the oligomers tested using equations similar to Equation 1. Thermodynamic values for duplex formation of the core sequence are −50.3 kcal/mol, −136.2 eu and −8.0 kcal/mol for ΔHo, ΔSo and , respectively (22).
| 1 |
These results are summarized in Table 2.
Table 2.
Thermodynamic parameters for unpaired 3′ dangling ends on a UA terminal base pair in 1 M NaCl
| −ΔΔH°a (kcal/mol) | −ΔΔS°a (eu) | a (kcal/mol) | |
|---|---|---|---|
| UA | 5.2 ± 2.8 | 14.3 ± 8.4 | 0.7 ± 0.2 |
| A | 5.7 ± 1.0 | 16.4 ± 3.2 | 0.7 ± 0.1 |
| UC | 3.0 ± 2.6 | 8.7 ± 8.0 | 0.4 ± 0.2 |
| A | 0.7 ± 1.0 | 1.8 ± 3.0 | 0.1 ± 0.1 |
| UG | 6.6 ± 3.7 | 18.8 ± 11.4 | 0.8 ± 0.2 |
| A | 5.8 ± 0.8 | 16.4 ± 2.5 | 0.7 ± 0.1 |
| UU | 3.1 ± 3.6 | 8.9 ± 11.0 | 0.4 ± 0.2 |
| A | 2.2 ± 1.6 | 6.8 ± 5.3 | 0.1 ± 0.1 |
aValues calculated as described in text. Top row, this study; bottom row, Sugimoto et al. (3). Error values represent average standard deviation of measured values, this study; half the the difference in values between values from ‘log Ct parameters’ and ‘temperature-independent parameters’, Sugimoto et al. (3).
The effect of terminal fraying is demonstrated by a decreased thermodynamic stabilization with the 3′ terminal overhang on the (AUGCAU)2 core compared to stabilization attributed to 3′ overhangs on other terminal base pairs [(3) and Table 2]. This is particularly evident for the 3′ terminal pyrimidine overhangs where the additional stabilization on the (AUGCAU)2 core was found to be only 0.1 kcal/mol (3). Our measured values for the additional duplex stabilization afforded by the 3′ terminal pyrimidine nucleotides on the (AGCGCU)2 core is 0.4 kcal/mol (Table 2). While our values and the previously measured values are within experimental error of each other, our values are in better accordance with the stabilization found for 3′ terminal pyrimidine nucleotides with other terminal base pairs (23).
Nearest-neighbor analysis and free energy parameters for 3′ dangling double-nucleotide overhangs
In order to determine the stability contributed to the oligomer by the second 3′ dangling nucleotide, differences in the thermodynamic values between the sequences studied and the corresponding sequence containing only the first overhanging nucleotide were also determined for all of the oligomers tested using equations similar to Equation 2.
| 2 |
These results are presented in Table 3.
Table 3.
Stabilization by addition of second 3′ dangling nucleotide in 1 M NaCl
| (kcal/mol) values for addition of second 3′ overhanga | ||||
| CCGGXZ | Z | |||
| X | A | G | C | U |
| A | −0.0 | −0.2 | −0.4 | −0.4 |
| G | −0.3 | −0.1 | −0.8 | −0.5 |
| C | −0.1 | — | −0.4 | −0.2 |
| U | +0.2 | −0.0 | +0.1 | +0.1 |
| AGCGCUXZ | ||||
| A | −0.6 | −0.6 | −0.8 | −0.6 |
| G | −0.3 | −0.3 | −0.4 | −0.4 |
| C | −0.3 | −0.2 | −0.2 | −0.2 |
| U | +0.3 | −0.0 | −0.2 | −0.1 |
| UGCGCAXZ | ||||
| A | −0.2 | −0.2 | −0.4 | −0.4 |
| G | −0.0 | −0.1 | −0.3 | −0.4 |
| C | −0.1 | −0.4 | −0.2 | −0.2 |
| U | +0.0 | −0.0 | −0.1 | −0.1 |
| ΔΔH° (kcal/mol) values for addition of second 3′ overhanga | ||||
| CCGGXZ | Z | |||
| X | A | G | C | U |
| A | −1.9 | −0.7 | −1.8 | −1.4 |
| G | −1.2 | −2.1 | −3.0 | −0.0 |
| C | +1.0 | — | +1.0 | +0.1 |
| U | −0.5 | −0.2 | +0.6 | +1.6 |
| AGCGCUXZ | ||||
| A | −1.9 | −2.0 | −3.4 | −2.4 |
| G | +0.6 | +1.5 | −1.0 | −0.3 |
| C | −2.0 | +2.0 | −0.4 | −0.1 |
| U | +2.4 | +1.0 | +0.2 | −0.2 |
| UGCGCAXZ | ||||
| A | −0.6 | −0.5 | −1.1 | −1.3 |
| G | +1.8 | +0.5 | −1.0 | −0.1 |
| C | +0.2 | −2.6 | −0.3 | −1.4 |
| U | +0.6 | −0.6 | −0.8 | +1.0 |
| ΔΔS° (eu) values for addition of second 3′ overhanga | ||||
| CCGGXZ | Z | |||
| X | A | G | C | U |
| A | −6.1 | −1.7 | −4.1 | −3.5 |
| G | −2.9 | −6.5 | −7.1 | +1.7 |
| C | +3.4 | — | +4.6 | +0.7 |
| U | −2.2 | −0.7 | +1.8 | +4.8 |
| AGCGCUXZ | ||||
| A | −4.4 | −5.0 | −8.5 | −6.1 |
| G | +2.4 | +3.7 | −1.8 | +0.2 |
| C | −5.0 | +7.2 | −0.2 | +0.4 |
| U | +7.1 | +3.2 | +1.3 | −0.4 |
| UGCGCAXZ | ||||
| A | −1.1 | −0.8 | −2.2 | −2.9 |
| G | +5.9 | +1.8 | −2.3 | +0.5 |
| C | +1.2 | −6.4 | −0.4 | −3.9 |
| U | +2.0 | −1.8 | −2.1 | +3.1 |
aValues calculated as described in text.
Previously, the influence of the second 3′ dangling nucleotide on the stability of the duplex, (GGCC)2 was grouped into three categories based upon the identity of 2 nt in the overhang. If the first 3′ nucleotide was a pyrimidine, no additional stabilization is observed upon the addition of the second 3′ dangling nucleotide. When the first nucleotide was a purine, the stacking of the second nucleotide made a significant contribution to the stability of the duplex. Sequences that contained a 3′ double-nucleotide dangling end with a 3′-pur-pyr-3′ had greater stability (0.5 kcal/mol on average) than those sequences containing an overhang with 3′-pur-pur-3′ (0.3 kcal/mol on average).
Analysis of all 64 of the 3′ double dangling nucleotides shows that the influence of the second 3′ dangling nucleotide does depend upon the identity of the terminal base pair (Table 3). This is most evident in considering the 3′ double-purine overhangs; duplexes where the purine of the closing base pair neighbors the double-nucleotide overhang [e.g. (CCGGXY)2] no additional stabilization by the second nucleotide is observed, however a significant contribution to the stability of the duplex by the second dangling nucleotide is observed when the 3′ overhang neighbors the pyrimidine of the closing base pair [e.g. (AGCGCUXY)2]. For pur–pyr double-nucleotide overhangs, the orientation of the terminal base pair does not affect the additional stabilization caused by the second 3′ dangling nucleotide. The additional stabilization of the second nucleotide was not significantly different for the pur–pyr double-nucleotide overhang adjacent to a pyrimidine than it was when adjacent to a purine therefore, the thermodynamic values for these sequences were averaged together to arrive at the stabilization of a duplex attributed to the second 3′ dangling nucleotide. As observed previously (5), if the first nucleotide of the 3′ dangling end is a pyrimidine, no additional stabilization is observed, irrespective of the terminal base pair. These results lead to an improvement in the model to predict RNA secondary structure stability and are presented in Table 4.
Table 4.
Average stabilizationa by addition of second 3′ dangling end in 1 M NaCl
| Sequence of 3′ double dangling ends | (kcal/mol) | ΔΔHo (kcal/mol) | ΔΔSo (eu) |
|---|---|---|---|
| 5′-pur-pyr-X | 0.0 | 0.0 | 0.0 |
| pyr-5′ | |||
| 5′-pur-pur-pur | 0.0 | 0.0 | 0.0 |
| pyr-5′ | |||
| 5′-pur-pur-pyr | −0.5 | −2.5 | −6.7 |
| pyr-5′ | |||
| 5′-pyr-pyr-X | 0.0 | 0.0 | 0.0 |
| pur-5′ | |||
| 5′-pyr-pur-X | −0.5 | −2.5 | −6.7 |
| pur-5′ |
aAverage values including data from O'Toole et al. (5) and this study.
To test the generality of conclusions from this work, thermodynamic parameters were also measured for five test sequences with 3′ double-nucleotide overhangs on different core sequences than those used to develop the model. The measured and predicted (both with and without the influence of the second 3′ dangling end) thermodynamic values for the five sequences are presented in Table 5. Four of the test sequences have only one 3′ double-nucleotide overhang that increases the stability, therefore, the predicted stability () for inclusion of the 3′ double-nucleotide dangling end increase by 0.5 kcal/mol. For the fifth test sequence, both 3′ double-nucleotide dangling ends contribute to the duplex stability and therefore, the predicted stability increases by 1.0 kcal/mol. The inclusion of the stabilization caused by the 3′ double-nucleotide overhangs improves the prediction of the thermodynamic stability for the duplexes in Table 5. For example, the average difference between the measured free energy and the predicted value is 0.6 kcal/mol when the contribution of the second 3′ two nucleotide dangling end is not included in the prediction, and improves to 0.3 kcal/mol when the contribution is included. In a similar fashion, the average difference in the prediction of the melting temperature also improves from 3.3 to 1.6°C with the contribution of the second 3′ double-nucleotide overhang taken into account. This is most strikingly seen for the last duplex in Table 5 where both 3′ double-nucleotide overhangs contribute to the duplex stability. The inclusion of the contribution of the 3′ double-nucleotide overhangs at both ends, improves the prediction of the thermal stability by 1.0 kcal/mol and the melting temperature by >6°C.
Table 5.
Measured and predicted thermodynamic parameters for test sequence duplex formationa
| Average of curve fits | versus log Ct plots | |||||||
|---|---|---|---|---|---|---|---|---|
| Sequences | −ΔH° (kcal/mol) | −ΔS° (eu) | (kcal/mol) | TMb (°C) | −ΔH° (kcal/mol) | −ΔS° (eu) | (kcal/mol) | TMb (°C) |
| CGAGCGG | 56.1 | 152.1 | 9.0 | 51.0 | 56.6 | 153.6 | 8.9 | 50.8 |
| UUGCUCG | (57.0) | (156.6) | (8.4) | (47.7) | ||||
| (59.5) | (163.3) | (8.9) | (49.7) | |||||
| CGAGCGG | 53.6 | 145.8 | 8.4 | 48.4 | 48.9 | 130.9 | 8.3 | 48.7 |
| GUGCUCG | (57.0) | (156.6) | (8.4) | (47.7) | ||||
| (59.5) | (163.3) | (8.9) | (49.7) | |||||
| GCUCGGA | 59.5 | 162.2 | 9.2 | 51.4 | 60.9 | 166.5 | 9.2 | 51.5 |
| CGCGAGC | (59.8) | (163.5) | (9.1) | (50.9) | ||||
| (62.3) | (170.2) | (9.6) | (52.7) | |||||
| CUGUGAA | 61.0 | 173.2 | 7.3 | 40.8 | 56.0 | 157.1 | 7.2 | 40.9 |
| CAGACAC | (54.0) | (151.9) | (6.6) | (38.7) | ||||
| (56.4) | (158.6) | (7.2) | (41.2) | |||||
| UGCUAAC | 71.7 | 208.0 | 7.2 | 39.9 | 64.0 | 183.0 | 7.2 | 40.5 |
| UGACGAU | (42.8) | (120.1) | (5.6) | (30.2) | ||||
| (47.8) | (133.5) | (6.6) | (36.3) | |||||
Values in parenthesis are predicted: top row is the predicted values for single 3′ terminal overhang duplexes and bottom row is the predicted values as described in the text for duplexes with 3′ double overhangs.
aSolutions are 1.0 M NaCl, 10 mM sodium cacodylate, 0.5 mM EDTA pH 7.
bCalculated at 10−4 M oligomer concentration.
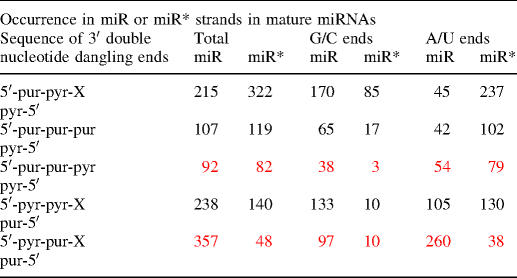
Phylogenetic analysis of experimentally determined miRNA sequences from miRNA database revealed a total of 1290 strand sequences; 1009 of these sequences were terminated with a Watson–Crick closing base pair and had 3′ terminal double-nucleotide overhang consisting of two of the four Watson–Crick bases. The miR strand of all the 1009 miRNAs that we used in this study were experimentally cloned and sequenced, however since the miR* of miRNA duplexes is degraded after the duplex is unwound and the miR strand is incorporated into RISC, the miR* sequences in the database have been predicted based upon the cloned and sequenced miR strand. Frequency of appearance of each of the 64 possible combinations of Watson–Crick closing base pairs neighboring 2 nt 3′ overhangs were determined for both miR and predicted miR* strands. The sequences were divided into categories and analyzed based upon their stability contribution of the 3′ double-nucleotide overhang on duplex stability; results of this search are presented in Table 6. Interestingly, the distribution of sequences into the miR or miR* strand was found to be related to the stability contribution of the 3′ terminal double-nucleotide overhang. For the double-nucleotide overhangs that do not contribute additional stability to the duplex, there is nearly an equal number of sequences found in both the miR and miR* strands (49 and 51%, respectively). For the double-nucleotide overhangs that do contribute to the stability of the duplex (red in the table), >75% were observed on the miR strand while only 25% were on the miR* strand. The identity of the closing base pair does influence the utilization of stabilizing double-nucleotide overhangs. For strands with either a CG closing base pair or a GC closing base pair (G/C base pair) on the end, the 3′ double-nucleotide overhang is almost exclusively (>90%) stabilizing; while for duplexes which end with either an AU closing base pair or a UA closing base pair (A/U), there is nearly a 3:1 ratio of non-stabilizing 3′ double-nucleotide overhangs. It is interesting that the stability of the more stable terminus, G/C, is augmented by the additional stabilization of the 3′ double-nucleotide overhang, while the opposite is seen for the less stable terminus, A/U. The 3′ double-nucleotide overhangs may therefore have been selected to enhance the distinction between the two ends of the miRNA duplex and aid in guide and passenger strand selection and loading into the RNAi effector complex, RISC; a critical step in the RNAi pathway.
Table 6.
Phylogenetic analysis of 3′ double nucleotide overhangs in naturally occurring miRNAsa
aNumber of sequences in the database closed by a Watson–Crick base pair with the corresponding 3′ double overhang (total number = 1009). Values in red represent 3′ double overhangs which contribute to the stability of duplex formation.
Acknowledgments
The authors thank Herve Seitz for providing the 3′ double-nucleotide overhang miRNA data. This work is supported by the Camille and Henry Dreyfus Foundation, National Science Foundation Grant No. MCB-0340958 and National Institutes of Health RGM-068426. Funding to pay the Open Access publication charges for this article was provided by NSF.
Conflict of interest statement. None declared.
REFERENCES
- 1.Freier S.M., Burger B.J., Alkema D., Neilson T., Turner D.H. Effects of 3′ dangling end stacking on the stability of GGCC and CCGG double helicies. Biochemistry. 1983;22:6198–6206. [Google Scholar]
- 2.Petersheim M., Turner D.H. Base-stacking and base-pairing contributions to helix stability: thermodynamics of double-helix formation with CCGG, CCGGp, CCGGAp, ACCGGp, CCGGUp, and ACCGGUp. Biochemistry. 1983;18:256–263. doi: 10.1021/bi00271a004. [DOI] [PubMed] [Google Scholar]
- 3.Sugimoto N., Kierzek R., Turner D.H. Sequence dependence for the energetics of dangling ends and terminal base pairs in ribonucleic acid. Biochemistry. 1987;26:4554–4558. doi: 10.1021/bi00388a058. [DOI] [PubMed] [Google Scholar]
- 4.Ohmichi T., Nakano S., Miyoshi D., Sugimoto N. Long RNA dangling end has large energetic contribution to duplex stability. J. Am. Chem. Soc. 2002;124:10367–10372. doi: 10.1021/ja0255406. [DOI] [PubMed] [Google Scholar]
- 5.O'Toole A.S., Miller S., Serra M.J. Stability of 3′ double nucleotide overhangs which model the 3′ ends of siRNA. RNA. 2005;11:512–516. doi: 10.1261/rna.7254905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 7.Hamilton A.J., Baulcombe D.C. A species of small anti-sense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 8.Zamore P.D., Tuschl T., Sharp P.A., Bartel D.P. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 9.Elbashir S.M., Lendeckel W., Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammond S.M., Caudy A.A., Hannon G.J. Post-transcriptional gene silencing by double-stranded RNA. Nat. Rev. Genet. 2001;2:110–119. doi: 10.1038/35052556. [DOI] [PubMed] [Google Scholar]
- 11.Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:295–296. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 12.Hutvágner G., McLachlan J., Pasquinelli A.E., Balint É., Tuschl T., Zamore P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 13.Knight S.W., Bass B.L. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nykänen A., Haley B., Zamore P.D. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 15.Provost P., Dishart D., Doucet J., Frendewey D., Samuelsson B., Radmark O. Ribonuclease activity and RNA binding of recombinant human dicer. EMBO J. 2002;21:5864–5874. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doench J.G., Petersen C.P., Sharp P.A. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwarz D.S., Hutvágner G., Du T., Xu Z., Aronin N., Zamore P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 18.Khvorova A., Reynolds A., Jeyasena S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 19.Serra M.J., Axenson T.J., Turner D.H. A model for the stabilities of RNA hairpins based on a study of the sequence dependence of stability for hairpins with six nucleotides. Biochemistry. 1994;33:14289–14296. doi: 10.1021/bi00251a042. [DOI] [PubMed] [Google Scholar]
- 20.McDowell J.A., Turner D.H. Investigation of the structural basis for thermodynamic stabilities of tandem GU mismatches: solution structure of (rGAGGUCUC)2 by two-dimensional NMR and simulated annealing. Biochemistry. 1996;35:14077–14089. doi: 10.1021/bi9615710. [DOI] [PubMed] [Google Scholar]
- 21.Freier S.M., Petersheim M., Hickey D.R., Turner D.H. Thermodynamic studies of RNA stability. J. Biomol. Struct. Dyn. 1984;1:1229–1242. doi: 10.1080/07391102.1984.10507514. [DOI] [PubMed] [Google Scholar]
- 22.Freier S.M., Sugimoto N., Sinclair A., Alkema D., Neilson T., Kierzek R., Caruthers M.H., Turner D.H. Stability of XGCGCp, GCGCYp, and XGCGCYp helixes: an empirical estimate of the energetics of hydrogen bonds in nucleic acids. Biochemistry. 1986;25:3214–3219. doi: 10.1021/bi00359a020. [DOI] [PubMed] [Google Scholar]
- 23.Turner D.H., Sugimoto N., Freier S.M. RNA structure prediction. Annu Rev. Biochys. Biophys. Chem. 1988;17:167–192. doi: 10.1146/annurev.bb.17.060188.001123. [DOI] [PubMed] [Google Scholar]