Abstract
Intrinsic flexibility of DNA has hampered the development of efficient protein−DNA docking methods. In this study we extend HADDOCK (High Ambiguity Driven DOCKing) [C. Dominguez, R. Boelens and A. M. J. J. Bonvin (2003) J. Am. Chem. Soc. 125, 1731–1737] to explicitly deal with DNA flexibility. HADDOCK uses non-structural experimental data to drive the docking during a rigid-body energy minimization, and semi-flexible and water refinement stages. The latter allow for flexibility of all DNA nucleotides and the residues of the protein at the predicted interface. We evaluated our approach on the monomeric repressor−DNA complexes formed by bacteriophage 434 Cro, the Escherichia coli Lac headpiece and bacteriophage P22 Arc. Starting from unbound proteins and canonical B-DNA we correctly predict the correct spatial disposition of the complexes and the specific conformation of the DNA in the published complexes. This information is subsequently used to generate a library of pre-bent and twisted DNA structures that served as input for a second docking round. The resulting top ranking solutions exhibit high similarity to the published complexes in terms of root mean square deviations, intermolecular contacts and DNA conformation. Our two-stage docking method is thus able to successfully predict protein−DNA complexes from unbound constituents using non-structural experimental data to drive the docking.
INTRODUCTION
Computational docking has proven to be a valuable tool in the study of biomolecular complexes (1,2). In particular, the field of ‘ab initio’ protein−protein docking has made considerable progress as illustrated by recent results from the community-wide CAPRI experiment [critical assessment of predicted interactions (3,4)]. However, where this field has in many ways matured, the development of docking methods to model protein−DNA interactions has lagged behind. These play an important role in recognition and gene expression (5). Powerful protein−DNA docking methods would thus be of great benefit for their study. However, two particular problems have hampered the development of efficient docking methods: the sparsity of the information to define the DNA-binding interface and the inherent flexibility of DNA. For protein−protein docking there is often enough information available (e.g. from sequence, conservation or biological knowledge) to identify the interaction surfaces of the docking partners. This information can be used to drive the docking (6) and limit the conformational space to be searched. Identification of the interaction surface on DNA is less straightforward than on proteins. There is still no general recognition code and the global conformation of the DNA can play an important role in modulating the eventual interaction surface (7). DNA indeed often exhibits large conformational changes upon binding to a protein, which can greatly alter the shape of the interaction surface. Owing to this, the total conformational space that needs to be searched in order to find favourable conformations becomes even larger. Flexibility in DNA can be separated into global and local components (8). Global flexibility is constrained to two primary motions: bending and twisting. It results from a combination of conformational changes in the flexible base pairs and sugar-phosphate backbone. Allowing for global and local flexibility in DNA during docking while maintaining the relevant conformation is a major challenge in protein−DNA docking.
In the last few years several methods have been developed to solve one or both of these problems, each with varying degrees of success. The program FTDOCK (9) has been used to perform a large search through conformational space by rotating and translating the protein along the DNA while evaluating shape and electrostatic complementarity; an approximation of flexibility was achieved by allowing some degree of overlap between protein and DNA in the scoring. In another approach, a library of pre-bent DNA structures was used to minimize the search through DNA conformational space (10); a selection was made based on structures that could be electrostatically preorientated in the potential of the protein and these were rotated and translated with respect to the protein. To account for some degree of local flexibility protein side chains and DNA base pairs were allowed to move in two separate refinement stages. Knegtel et al. (11) developed MONTY which uses a Monte Carlo search allowing for flexibility in both protein and DNA and experimentally determined contacts to drive the docking. The initial position of the protein in the predicted complex should, however, not deviate too much from that of the actual complex; small deviations in the position of the protein with respect to the interaction interface of the DNA resulted in DNA curling around the protein. Tzou and Hwang (12) modelled the CAP-DNA and Rep-DNA systems from the repressors in their bound conformation and canonical B-DNA in a series of molecular mechanics and dynamics simulations using distance restraints derived from a statistical analysis of homologous protein–DNA complexes. This method successfully introduced DNA bending and local opening of the major groove. All of these docking procedures were able to make predictions that were representative of the published complexes in terms of spatial disposition. Only a few methods allowed for flexibility of the DNA and protein side chains during the docking. They, however, required extensive knowledge to position the two components relative to each other (12) and problems were encountered in the absence of such information (11).
Here we demonstrate that both global and local DNA flexibility can successfully be accounted for in protein−DNA modelling using HADDOCK (High Ambiguity Driven DOCKing) (13), a computational docking approach developed in our group. HADDOCK makes use of available experimental and bioinformatics data to drive the docking process (14). Its successful use in NMR-based structure calculations of protein–DNA and protein–RNA complexes has been shown previously (15–18). Global and local DNA flexibility is introduced in the docking by allowing the DNA sugar-phosphate backbone and DNA base pairs to sample conformations during a semi-flexible refinement stage and by starting the docking from a library of pre-generated DNA structures representing various degrees of conformational flexibility. The latter allows for the sampling of a larger conformational space. Flexibility in the protein is introduced as described previously (13), first along the side chains at the interface and then for both backbone and side chains. We demonstrate here the feasibility of this approach for three repressor complexes in their monomeric form: Cro from bacteriophage 434 (19), the Lac headpiece of Escherichia coli (20) and Arc from bacteriophage P22 (21). The first two recognize the DNA major groove via an α-helix/turn/α-helix motif and the last one via a two-stranded antiparallel β-sheet motif. To drive the docking we make use of mutation data, sequence/structure conservation, DNA footprinting and ethylation interference data. We show that our approach is successful in predicting protein–DNA complexes from unbound constituents by accounting for both global and local DNA flexibility during the docking.
MATERIALS AND METHODS
Initial structures for protein and DNA
The coordinate files of all proteins and protein–DNA complexes were obtained from the RCSB Protein Data Bank (PDB) (22). The PDB entry codes of the respective complexes and their unbound components are as follows: 3CRO, crystal structures of the Cro/O1R complex (19); 1ZUG, NMR ensemble of the unbound Cro monomer (23); 1LCC, NMR ensemble of the Lac/O1 complex (24); 1LQC, NMR ensemble of the unbound monomer of the Lac headpiece (25); 1BDT, crystal structure of the Arc/DNA complex (26) and 1ARQ, NMR ensemble of the unbound Arc monomer (27). The monomeric reference structures for Cro and Arc (right halfside) were extracted from the dimeric PDB structures.
Models of canonical B-DNA were constructed with the nucleic acid analysis and rebuilding program 3DNA (28), using the fiber models provided by Chandrasekaran and Arnott (29). All hydrogens were added according to the standard assigning scheme of CNS followed by a short energy minimization step during the initiation stage in HADDOCK. Base pair and base pair step parameters of the resulting type BII B-DNA starting structures are shown in Table 1. The DNA backbone torsion angles are α = 309°, β = 159°, γ = 37°, δ = 146°, ɛ = 218°, ζ = 191°, χ = 260° and the sugar pseudo-rotation phase angle (P) = 155°, the sugar pucker was thus in the C2′-endo conformation.
Table 1.
Average DNA base pair and base pair step parameters
| Parameters | Cro | Lac | Arc | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ref. | Docking from | Ref. | Docking from | Ref. | Docking from | |||||
| 3CRO | B-DNA | DNA lib. | 1LCC | B-DNA | DNA lib. | 1BDT | B-DNA | DNA lib. | ||
| Twist () | 34.45.3 | 36.41.0 | 34.93.5 | 34.25.4 | 36.80.7 | 36.53.6 | 32.54.1 | 34.61.2 | 35.53.3 | |
| Roll ( ) | 2.53.2 | −0.22.0 | 1.08.1 | 2.611.2 | 0.31.7 | 0.210.4 | 3.35.5 | 4.21.9 | 1.07.7 | |
| Tilt ( ) | 0.53.7 | 0.02.0 | 0.45.4 | −2.77.9 | 0.21.6 | 0.24.9 | −0.33.3 | 0.91.5 | 0.45.9 | |
| Rise (3.40.0 Å) | 3.40.3 | 3.30.2 | 3.40.4 | 3.20.2 | 3.30.2 | 3.30.3 | 3.30.2 | 3.30.1 | 3.30.4 | |
| Slide (0.30.2 Å) | −0.40.4 | 0.00.1 | −0.60.6 | −0.40.7 | 0.20.2 | 0.10.7 | −0.40.7 | 0.01.7 | −0.30.5 | |
| Shift (0.00.1 Å) | 0.00.5 | 0.10.1 | 0.00.6 | −0.10.7 | 0.10.3 | 0.00.5 | −0.10.9 | 0.10.3 | 0.10.8 | |
| Opening (−3.32.5 Å) | −4.54.8 | −4.62.2 | −3.34.0 | −6.77.9 | −2.02.8 | −2.03.8 | 0.44.3 | −0.82.0 | −0.84.7 | |
| Propeller () | −145 | −7.54.4 | −0.912.7 | −14.64.4 | −8.55.0 | −9.310.1 | −4.38.3 | −4.73.8 | −1.114.1 | |
| Buckle ( | 1.08.1 | −1.45.1 | −0.610.8 | −6.913.2 | 4.63.5 | −0.211.8 | −2.76.5 | 5.24.9 | −2.513.5 | |
| Stagger (0.10.0 Å) | −0.10.5 | −0.10.2 | −0.30.6 | 0.10.8 | −0.10.2 | 0.10.5 | 0.00.3 | −0.20.3 | −0.20.5 | |
| Stretch (−0.10.0 Å) | −0.30.2 | −0.10.1 | −0.20.1 | −0.10.2 | −0.20.1 | −0.10.1 | −0.20.1 | −0.10.1 | −0.10.1 | |
| Shear (0.00.1 Å) | 0.20.5 | 0.10.0 | 0.00.3 | −0.30.5 | 0.10.1 | −0.10.2 | −0.10.3 | 0.00.4 | −0.10.2 | |
| Correlations | ||||||||||
| Roll-twist (0.26) | −0.47 | −0.55 | −0.44 | −0.65 | −0.61 | −0.76 | −0.85 | −0.16 | −0.23 | |
| Roll-slide (0.30) | −0.40 | −0.43 | −0.37 | −0.65 | −0.48 | −0.61 | −0.44 | 0.00 | −0.43 | |
Average parameters with standard deviations in subscript are shown for the published complexes (Ref.) and the top five ranking solutions from unbound flexible docking starting from canonical B-DNA (B-DNA) and from a library of pre-bent and twisted DNA structures (DNA lib.). For comparison, the average values for the canonical B-DNA input structure are shown in the left column between brackets next to each parameter.
Custom DNA libraries for the three operator sequences were generated by manipulation of the base pair step parameters of their respective B-DNA structures using 3DNA. The introduction of curvature was accomplished by changing the value of roll using the following equation (30):
where Rn is the roll value for each base pair step in one helical turn, n is the number of base pair steps in one helical turn, k is the average curvature for each base pair step in one helical turn and T is the value for twist. The direction of the curvature in Cartesian space can be controlled by changing the phase (θ) of the cosine function. The positive linear relationship between the value of the slide parameter and the width of the major groove was used to adjust the major groove width.
Restraints used in the docking
Ambiguous Interaction Restraints (AIR) (Table 2): All active residues have a relative solvent accessibility >50% as calculated with NACCESS (31). Residues located in the predicted interaction interface or in a continuous stretch of residues near the predicted interaction interface for which no information is available were defined as passive. AIRs for the protein were defined based on sequence conservation [HSSP (32)] and mutation data. For the DNA only active residues were defined. The recognition sequences of the operators have been determined using DNA-footprinting methods before the experimental structures of the actual complexes became available. This information was used in our docking procedure to define interaction restraints. For those bases shown to be involved in specific interactions with the repressor, only atoms able to interact by hydrogen-bond or non-bonded interactions were defined. Based on ethylation interference experiments, only the oxygen atoms of phosphate groups shown to interact with the repressor were defined as active.
Table 2.
Definition of the AIRs for the three repressor/operator systems
| Protein | DNA | Reference | |
|---|---|---|---|
| Cro – O1R | |||
| Active | K27a,Q29a,S30a,L33a,b | T3c,A4a,c,C5a,A6a,G30cT31a,b,c,T32a,b,c,T33a,G34a,T35a | (50–53) |
| Passive | R10,K40,R41,P42 | — | |
| Lac – O1 | |||
| Active | T5a,b,S16a,Y17b,Q18b,R22b,V30b | T4c,G5a,c,T6a,G7a,A8a,C14c, T15a,c,C16a,A17a,C18a | (54–57) |
| Passive | H29,S31 | — | |
| Arc – operon | |||
| Active | F10a,R13a,S32a | T1c,A2c,T3c,G5c,T6a,A7a,G8a,A9a,A14c,C15c,T16c,C17a,T18a,A19a | (58) |
| passive | Q9,N11,R16,D20,R23 | — | |
The Arc monomer is composed of two symmetric subunits and only the restraints for one subunit are shown.
aConserved residues derived from the database of homology-derived secondary structure of proteins (HSSP).
bMutagenesis data.
cEthylation interference.
DNA restraints: In order to preserve the helical conformation of DNA the following restraints were defined: planarity restraints for the purine and pyrimidine rings were introduced, and the sugar pucker was restrained to the C2′-endo conformation. Watson−Crick base pairs were defined and hydrogen bond lengths of the input structure (either the initial starting DNA conformation or the conformation obtained after semi-flexible refinement prior to water refinement) were measured and restricted to ±0.05 Å. In a similar way the dihedral angles of the sugar-phosphate backbone of the input structure (inp) were measured and used as restraints. (Restricted to α = αinp ± 10°, β = βinp ± 40°, γ = γinp ± 20°, δ = δinp ± 50°, ɛ = ɛinp ± 10° and ζ = ζinp ± 50°).
Docking protocol
Our docking protocol consists of (i) rigid-body docking, (ii) semi-flexible refinement stage and (iii) final refinement in explicit solvent.
Rigid-body docking. A total of 100 structures were generated for each protein−DNA combination from the ensembles of starting structures. Each docking attempt was performed 10 times and the solution with the lowest HADDOCK score was kept. For each protein we used an ensemble of 10 NMR structures; thus 1000 rigid-body docking solutions were generated for each of the three canonical B-DNA docking runs and 5000 structures were generated for each of the DNA library docking runs (5 different pre-bent and twisted DNA structures and 10 protein structures resulting in 50 different combinations). For the docking of the protein and DNA in their bound conformation a total of 1000 structures were generated. Systematic sampling of 180° rotated solutions was used in the rigid-body docking stage to minimize the occurrence of false positives (principles described in Results). This basically doubled the number of docking trials bringing the total to 20 000 and 100 000 evaluations for docking from canonical B-DNA and DNA libraries, respectively.
Semi-flexible refinement. Of all structures generated in the rigid-body docking stage the best 20% based on the HADDOCK score were further refined in the semi-flexible refinement stage consisting of three parts: rigid-body torsion angle dynamics (500 MD steps at 2000 K and 500 MD cooling steps to 500 K with a 8 fs time step), semi-flexible simulated annealing stage (1000 MD steps from 1000 to 50 K with 4 fs time steps) with the side chains of the protein residues at the interface and the complete DNA (excluding terminal base pairs) allowed to move and a final semi-flexible simulated annealing stage (1000 MD steps from 300 to 50 K with 2 fs time steps) with both side chains and backbone of the protein residues at the interface and the complete DNA (excluding terminal base pairs) allowed to move.
Water refinement. This final stage consists of a gentle refinement (100 MD heating steps at 100, 200 and 300 K followed by 750 sampling steps at 300 K and 500 MD cooling steps at 300, 200 and 100 K all with 2 fs time steps) in an 8 Å shell of TIP3P water molecules (33).
Semi-flexible segments for the proteins were defined as residues 7–20, 24–37 for Cro, residues 6–30, 50–56 for Lac and residues 1–17, 54–70 for Arc. In all cases the complete DNA, excluding the terminal base pairs, were defined as semi-flexible.
Scoring
A HADDOCK score is defined to rank the structures after each docking stage. It is a weighted sum of intermolecular electrostatic (Elec), van der Waals (vdW), desolvation (Dsolv) and AIR energies, and a buried surface area (BSA) term: rigid-body score = 1.0 * Elec + 1.0 * vdW − 0.05 * BSA + 1.0 * Dsolv + 1.0 * AIR, final score = 1.0 * Elec + 1.0 * vdW + 1.0 * Dsolv + 1.0 * AIR. A cluster analysis was performed on the final docking solutions using a minimum cluster size of 4. The cut-off for clustering was manually determined for each docking run. The root mean square deviation (r.m.s.d.) matrix was calculated over the backbone atoms of the interface residues of the DNA after fitting on the interface residues of the protein. Final structures within a cluster were selected according to their summed base pair and base pair step deformation energies and the conformation of the helix (classified as B-DNA). Deformation energies were calculated with an extension script of 3DNA (provided by Marc Parisien, University of Montreal, Canada) using the statistical population preferences as determined by Olson et al. (34) and Lankas et al. (35).
Default HADDOCK (version 2.0_devel) parameters were used except for the dielectric constant (epsilon) that was set to 78 for the vacuum part of the protocol. To speed up calculations, non-polar hydrogens were omitted. Inter- and intramolecular energies were evaluated using full electrostatic and van der Waals energy terms with an 8.5 Å distance cut-off. OPLSX non-bonded parameters from the parallhdg5.3.pro parameter file (36) were used for the protein. Topology and linkage parameter files for the DNA were taken from the CNS (37) distribution (dna-rna-allatom.top and dna-rna-allatom.param respectively). The HADDOCK package is freely available to academic users (http://www.nmr.chem.uu.nl/haddock).
Analysis
The r.m.s.d. values of the complexes were calculated using ProFit (A. C. R. Martin, www.bioinf.org.uk/software/profit) All heavy atoms were used to calculate the r.m.s.d. of the total complex, of the DNA and of the interface. The interface was composed of residues 15–44/3–7, 31–37 of Cro/O1R; 6–32/4–10, 13–19 of Lac/O1; and 8–36, 61–89/1–9, 13–21 of Arc/repressor. The backbone r.m.s.d. was calculated using all P and Cα atoms of the complex. Residues in the flexible termini of the protein (having either high B-factors in the X-ray structures or poorly defined in the NMR reference structures) were left out of the calculation. Intermolecular contacts were evaluated using LIGPLOT (38) using a 5 Å cut-off. The fraction of native contacts (Fnat) is defined as the number of native intermolecular contacts on a nucleotide-residue basis (hydrogen-bonded and non-bonded) identified in a docking solution divided by the total number of contacts in the reference structure. Values for base pair and base pair step parameters as well as torsion angles for the sugar-phosphate backbone and the sugar pucker were obtained using the program 3DNA (28). The overall bend-angle of the DNA was calculated using CURVES (39).
Hardware
HADDOCK docking runs were performed on a Transtec (Transtec AG, Tubingen, Germany) computer cluster operating with 32, 2.0 GHz, 64 bit Opteron processors. As a measure of CPU requirements, one complete run starting with 1000 structures in the rigid-body docking stage could be performed in ∼2 h on 32 processors.
RESULTS
Bound rigid-body docking
The use of readily available biochemical and/or biophysical information can alleviate the lack of a general recognition code for protein–DNA interactions. HADDOCK uses this information encoded as AIRs (13) to drive the docking; this reduces the necessary search through interaction space and increases the fraction of unique solutions. In the definition of AIRs we distinguish between active and passive residues. Active residues are defined as those important for the interaction based on conservation [HSSP (32)], mutation or ethylation interference data or any other appropriate experimental data. Passive residues are defined as the solvent-accessible neighbours of active residues (Table 2).
We first evaluated the use of AIRs in protein−DNA docking for the three selected complexes by bound docking (i.e. the reconstruction of the complexes from their separate components). Since the molecules are already in their bound conformation no flexible segments were defined and only rigid-body docking was performed. The best docking solutions for each of the Lac, Arc and Cro repressor in complex with their operators exhibit high similarity with the published complexes based on r.m.s.d. values and intermolecular contacts (Table 3); all base-specific intermolecular contacts are recovered.
Table 3.
The r.m.s.d. values from the target and fraction of native contacts for the top five ranking docking solutions of the best cluster
| r.m.s.d. (Å) | Fnate | ||||
|---|---|---|---|---|---|
| Totala | Interfaceb | Backbonec | DNAd | ||
| Cro–O1R | |||||
| Bound | 0.270.00 | 0.240.00 | 0.280.00 | 0.000.00 | 0.880.00 |
| Unbound rigid | 2.620.01 | 2.370.06 | 1.920.02 | 2.310.00 | 0.530.12 |
| Unbound flex. | 2.300.07 | 2.070.12 | 1.800.09 | 1.970.15 | 0.800.07 |
| DNA lib. | 1.990.05 | 1.690.06 | 1.510.09 | 1.460.07 | 0.940.00 |
| Lac–O1 | |||||
| Bound | 0.340.00 | 0.310.00 | 0.360.00 | 0.000.00 | 0.890.00 |
| Unbound rigid | 2.840.00 | 2.880.00 | 2.560.00 | 1.710.00 | 0.330.00 |
| Unbound flex. | 2.640.10 | 2.560.12 | 2.410.12 | 1.900.18 | 0.510.03 |
| DNA lib. | 2.330.06 | 2.290.08 | 2.060.08 | 1.570.09 | 0.540.01 |
| Arc–operator | |||||
| Bound | 0.220.00 | 0.230.00 | 0.190.00 | 0.000.00 | 0.950.00 |
| Unbound rigid | 2.580.01 | 2.580.01 | 1.970.02 | 2.520.00 | 0.430.00 |
| Unbound flex. | 2.240.08 | 2.130.10 | 1.640.10 | 1.880.15 | 0.500.04 |
| DNA lib. | 2.200.15 | 2.190.19 | 1.730.15 | 1.990.11 | 0.510.08 |
Average r.m.s.d. values (Å, standard deviation in subscript) calculated over the entire complex (a), the interface (b), the backbone (c) and the DNA (d) for the five top ranking solutions. The r.m.s.d. values are reported for the bound rigid-body docking (bound), unbound docking before (unbound rigid) and after semi-flexible refinement (unbound flex.) starting from canonical B-DNA, and unbound semi-flexible docking using a library of pre-bent and twisted DNA as input structures (DNA lib.). eFnat is the fraction of native contacts.
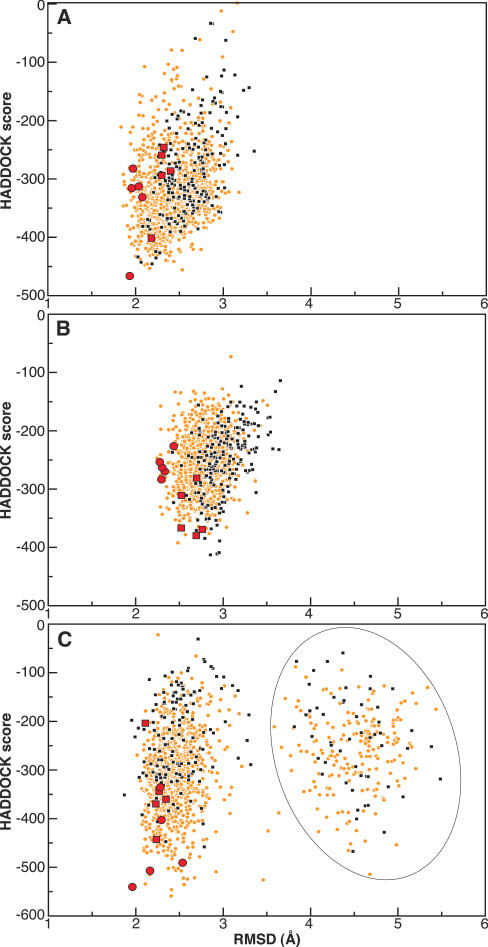
In the biologically relevant complexes the repressors are bound as dimers that are symmetrically oriented on the two recognition sites of the operator. In this study we use the repressors in their monomeric form (in this form the Arc repressor is a symmetrical dimer). Symmetry in the AIR set and in the shape of the protein–DNA interaction surface can result in false positives: these are structures with a favourable HADDOCK score (weighted sum of several energy terms, see Materials and Methods) but with one of the two components rotated 180° with respect to the published complex. To minimize the occurrence of false positives 180° rotated solutions were systematically sampled during the rigid-body docking stage. For this, a 180° rotation around a vector defined by the centres of masses of the interfaces of the protein and DNA was applied and the resulting conformation again minimized. The solution with the lowest HADDOCK score was kept. Using this approach the amount of false positives after the rigid-body docking stage was reduced from ∼70 to ∼40%. In subsequent unbound docking runs including flexibility we selected the best 20% of all solutions from the rigid-body docking stage based on their HADDOCK score. Owing to the sampling of 180° rotations this subset contained no false positives for the Cro and Lac repressor/operator complexes (Figure 1). Because of the intrinsic symmetry of the Arc repressor, 180° rotated symmetrical solutions are similar and can thus not be distinguished. Therefore the problem of rotational false positives does not apply to the Arc repressor. In unbound docking false positives were obtained that correspond to shifted false positives. These are solutions in which the repressor is shifted 1 or 2 bp upstream or downstream of the true interaction surface on the DNA (Figure 1).
Figure 1.
HADDOCK score versus r.m.s.d. from the target (all heavy atoms of the complex) for the Cro (A), Lac (B) and Arc (C) repressors in complex with their operator. Solutions of the unbound flexible docking with canonical B-DNA are shown as small black squares with the five top ranking solutions identified by red squares. Solutions from the docking using a library of pre-bent and twisted DNA structures are shown as small orange circles with the top five ranking solutions identified by red circles. False positives for Arc are shown within a solid ellipse: These correspond to solutions in which the repressor is shifted by 1 or 2 bp along the DNA.
Unbound semi-flexible docking to B-form DNA
We used the AIR sets to dock an ensemble of NMR structures of the unbound repressors to canonical B-DNA (chosen for it's biological relevance). In contrast to the previous bound docking runs in which only rigid-body docking was performed, we now included flexibility in a semi-flexible refinement stage: side chains and backbone of the protein at the predicted interface and the entire DNA were allowed to sample additional conformations. A set of restraints was imposed on the DNA that allowed for local flexibility but preserved the overall helical conformation (Materials and Methods). The final refined structures were clustered based on their pairwise r.m.s.d. matrix. The best cluster was selected based on the HADDOCK score.
The solutions in the selected clusters appeared to be very similar with respect to the protein and the spatial disposition of the complex but less similar on the level of the DNA conformation. An analysis of the base pair and base pair step parameters of the DNA in the selected clusters revealed a higher variation in buckle, propeller, roll and tilt than in other parameters (Table 1). Previous studies have also observed a larger variation for these parameters in both free and bound DNA when it is bending and twisting (5,8,34,40,41). This is not surprising as buckle, propeller, roll and tilt parameters are less restricted by Watson−Crick hydrogen bonds and the conformation of the sugar-phosphate backbone, than is the case with the other parameters. However, their large variation occasionally resulted in an overall loss of B-DNA conformation in the docking solutions as assessed by 3DNA (28). These solutions, however, did not have worse HADDOCK scores than solutions with a smaller variation in the noted parameters. They could, however, in most cases be distinguished by their higher DNA deformation energy. For this we calculated the combined base pair and base pair step deformation energy for every solution in the selected cluster and ranked them according to this energy term (Materials and Methods). The ranked solutions were checked on having a general B-DNA conformation and the best five were selected. This procedure proved successful in selecting solutions that are in better agreement to the published complexes in terms of r.m.s.d. values (Figure 1).
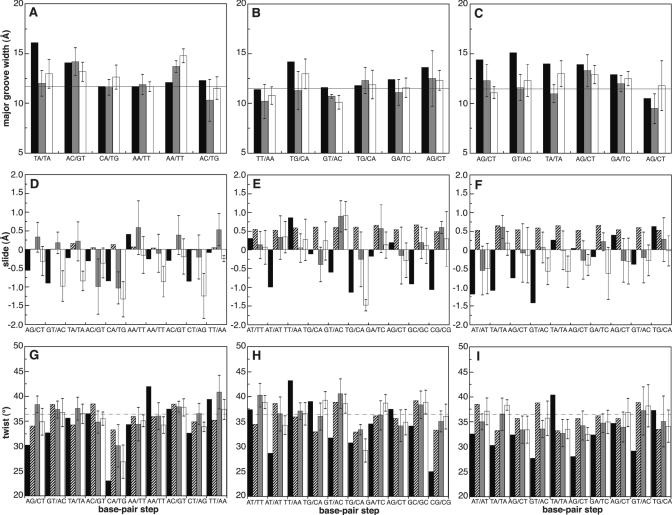
To assess the effect of flexibility on the docking we compared the top ranking solutions after the semi-flexible refinement stage with their initial conformation after rigid-body docking: the results show a clear improvement in r.m.s.d. from the published structure of the complex and fraction of native contacts (Table 3). Analysis of the DNA revealed that the backbone torsion angles were all located in the most populated regions as derived from a statistical analysis of non-complexed DNA structures (42,43) (data not shown). Base pair buckle, propeller, tilt and roll parameters, which are at the origin of overall DNA bending and twisting, showed larger differences between the published complexes and the rigid-body docking solutions than after introduction of flexibility (Table 1). Base pair opening, stagger, stretch and shear parameters and base pair step twist, slide and shift parameters showed little differences. In all three complexes the DNA is slightly bent towards the protein. In this respect tilt rotation is reported to be both statistically and energetically less favourable than roll rotation (44–47). This relationship is observed in the published complexes and the top ranking docking solutions as they show smaller variations in tilt than in roll. Statistical analysis of crystal structures has revealed that a positive change in roll is often accompanied with unwinding and negative slide (46,48,49). In our best solutions we also witness that roll is negatively correlated with both twist and slide (Table 1); more precisely, twist values <36° are often accompanied with negative sliding in bent DNA. This relation is observed at the interface of the top ranking docking solutions (the central 4 bp steps in Figure 2D–I). On a global level the distribution of major groove widths over the different base pair steps followed a trend similar to the published complexes (Figure 2A–C).
Figure 2.
Major groove width, slide and twist parameters of the five top ranking solutions of the Cro (A, D and G), Lac (B, E and H) and Arc (C, F and I) repressor/operator complexes. Average values plus standard deviations for the solutions of the unbound flexible docking with canonical B-DNA are shown as grey bars and those using a library of pre-bent and twisted DNA structures are shown as white bars. The values as measured in the published complexes are presented as black bars and those of the canonical B-DNA input structures as striped bars for slide (D,E,F) and twist (G,H,I) and as a horizontal solid line for the major groove width. All values for the major groove width are corrected by 5.8 Å to account for van der Waals radii of the phosphate groups. A value of 36° twist is presented as a dashed line for clarification of the twist-slide relationship.
Unbound docking from custom-build DNA libraries
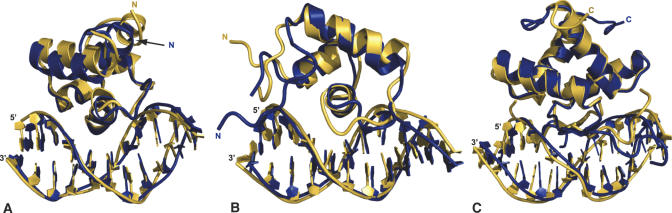
The results above show that the introduction of flexibility results in the prediction of a more native-like complex in comparison with rigid-body docking. To account for even larger DNA conformational changes we explored the possibility of using a library of pre-bent and twisted DNA structures as input structures for the docking procedure. Although the DNA in the best clusters of the flexible docking runs starting from canonical B-DNA showed variation on a local level (e.g. buckle, propeller, roll and tilt parameters) the global conformation of all solutions was quite similar. Analysis of the resulting DNA conformations provided information in the form of bend angles and the width of the major groove, which was used to construct custom DNA libraries. For the Cro/O1R complex the major groove width increased from 11.6 Å (canonical B-DNA) to 12.5 ± 0.5 Å and the DNA adopted a curve towards the protein of 9.4 ± 3.6°. For the Lac and Arc repressors in complex with their operator similar events occur, resulting in major groove widths of 11.3 ± 0.4 and 12.2 ± 0.8 Å and curves towards the protein of 11.3 ± 3.8 and 12.9 ± 5.2°, respectively. Based on this information we constructed for each operator five DNA structures that sample values around the averaged major grooves widths and bend angles from the previous docking runs. Docking from these libraries using the flexible protocol described above resulted in solutions with twist and slide parameters as well as major groove widths in better agreement with those of the published complexes (Figure 2). The overall results (Table 3) demonstrate that the use of a custom library of pre-bent and twisted structures improves the prediction structures of the complexes as assessed by r.m.s.d. values, intermolecular contacts and DNA conformation. Only for the Arc repressor/operator complex did the use of a custom DNA library not result in a significant improvement compared to canonical B-DNA docking. The best docking solutions superimposed onto their reference structure are presented in Figure 3.
Figure 3.
Best solutions of the unbound flexible docking using a library of pre-bent and twisted DNA structures (blue) superimposed on the reference structure (yellow): Cro-O1R (A), Lac-O1 (B) and Arc-operator (C). The structures were superimposed on all heavy atoms of the interface residues (interface r.m.s.d. values: Cro, 1.62 Å; Lac, 2.02 Å; Arc, 1.90 Å). The figures were generated using Pymol (DeLano Scientific, www.pymol.org).
DISCUSSION
Our modelling of protein−DNA complexes is based on AIRs to drive the docking process. These are essential in successfully positioning the protein at the interface of the DNA and, together with flexibility, influence DNA bending in the semi-flexible refinement stage. We used a limited number of easily obtainable experimental data to define the restraints. These were nevertheless sufficient to accurately predict the conformation of the DNA in the complex when starting from canonical B-DNA. This information subsequently allowed us to refine our models by performing docking from a custom-built DNA library instead of canonical B-DNA. This two-stage docking approach significantly improves the conformation of the DNA in the resulting complexes; the protein, however, is less affected and its conformation remains close to the conformation of the respective starting unbound structures.
In this study we did not investigate the effects of a variable number or type of restraints on the docking results. From analogous protein−protein docking studies it is known that the amount of or the ambiguity in the data can influence the reproducibility of the docking. HADDOCK allows the random deletion of a fraction of the restraints for each docking trial to account for errors in their definition, an approach that has proved successful in the past (14). This option was not used in this study. The AIRs were defined with an upper distance limit of 2.0 Å that can affect the packing of the docking solutions. For the Lac/O1 and Arc/operator complexes the BSA was comparable to that of the reference (1496 ± 103 Å versus 1560 Å and 1990 ± 155 Å versus 2072 Å, respectively). For the Cro/O1R complex the BSA of the top ranking solutions was larger than that of the reference (1694 ± 52 Å versus 1453 Å). The tighter packing might contribute to the significant increase in the fraction of native contacts (Table 3) for the Cro/O1R complex, with respect to the other two test systems.
We have demonstrated that the use of readily available non-structural experimental data and the incorporation of DNA flexibility during the docking significantly improve repressor−DNA complex prediction in comparison to rigid-body docking. The method successfully predicted global conformational changes taking place in the DNA upon complexation. The information extracted from these results is sufficient to refine the models by starting a second docking round from custom-built DNA libraries of pre-bent and twisted structures.
The flexible protein−DNA docking approach described in this paper has biological implications since it can benefit studies of protein–DNA interactions at several levels. It can be used to generate models of protein–DNA complexes when the structure of the unbound protein is known and suitable experimental data are available. It is also applicable to study the effects of mutations or different operator sequences on complex formation. In addition, it can assist in experimental structural studies: it can, for example, speed up structure determination of protein–DNA complexes by NMR by providing initial models to guide the tedious NMR analysis and assignment process. In summary, by allowing the inclusion of a large variety of experimental and/or bioinformatics data, together with a flexible description of the DNA, the proposed docking approach should be a useful tool in structural studies of protein–DNA and even protein–RNA interactions provided suitable RNA models are available for the latter.
Acknowledgments
We thank Marc Parisien from the University of Montreal, Canada for his work on the scripts for the calculation of DNA base pair and base pair step deformation energies. Financial support for this research was provided by the European Community, FP6 STREP project ‘ExtendNMR’ (Contract no. LSHG-CT-2005-018988) and by The Netherlands Organisation for Scientific research (NWO/CW grant B81-752 and NWO-TOP grant 700.52.303). Funding to pay the Open Access publication charges for this article was provided by ‘ExtendNMR’ (Contract no. LSHG-CT-2005-018988).
Conflict of interest statement. None declared.
REFERENCES
- 1.Halperin I., Ma B., Wolfson H.J., Nussinov R. Principles of docking: an overview of search algorithms and a guide to scoring functions. Proteins. 2002;47:409–443. doi: 10.1002/prot.10115. [DOI] [PubMed] [Google Scholar]
- 2.Schneidman-Duhovny D., Nussinov R., Wolfson H.J. Predicting molecular interactions in silico: II. Protein−protein and protein−drug docking. Curr. Med. Chem. 2004;11:91–107. doi: 10.2174/0929867043456223. [DOI] [PubMed] [Google Scholar]
- 3.Janin J., Hendrick K., Moult J., Ten Eyck L., Sternberg M.J.E., Vajda S., Vakser I., Wodak S.J. CAPRI: critical assessment of predicted interactions. Proteins. 2003;52:2–9. doi: 10.1002/prot.10381. [DOI] [PubMed] [Google Scholar]
- 4.Méndez R., Leplae R., De Maria L., Wodak S.J. Assessment of blind predictions of protein−protein interactions: current status of docking methods. Proteins. 2003;52:51–67. doi: 10.1002/prot.10393. [DOI] [PubMed] [Google Scholar]
- 5.Rhodes D., Schwabe J.W.R., Chapman L., Fairall L. Towards and understanding of protein–DNA recognition. Phil. Trans. R Soc. Lond. B Biol. Sci. 1996;351:501–509. doi: 10.1098/rstb.1996.0048. [DOI] [PubMed] [Google Scholar]
- 6.van Dijk A.D., Boelens R., Bonvin A.M.J.J. Data-driven docking for the study of biomolecular complexes. FEBS. 2005;272:293–312. doi: 10.1111/j.1742-4658.2004.04473.x. [DOI] [PubMed] [Google Scholar]
- 7.Pabo C.O., Nekludova L. Geometric analysis and comparison of protein−DNA interfaces: why is there no simple code for recognition? J. Mol. Biol. 2000;303:597–624. doi: 10.1006/jmbi.2000.3918. [DOI] [PubMed] [Google Scholar]
- 8.Travers A.A. The structural basis of DNA flexibility. Phil. Trans. R Soc. Lond. A. 2004;362:1423–1438. doi: 10.1098/rsta.2004.1390. [DOI] [PubMed] [Google Scholar]
- 9.Aloy P., Moont G., Gab H.A., Querol E., Aviles F.X., Sternberg M.J.E. Modelling repressor proteins docking to DNA. Proteins. 1998;33:535–549. [PubMed] [Google Scholar]
- 10.Sandmann C., Cordes F., Saenger W. Structure model of a complex between the factor for inversion stimulation (FIS) and DNA: modeling protein–DNA complexes with dyad symmetry and known structures. Proteins. 1996;25:486–500. doi: 10.1002/prot.8. [DOI] [PubMed] [Google Scholar]
- 11.Knegtel R.M., Boelens R., Kaptein R. Monte Carlo docking of protein–DNA complexes: incorperation of DNA flexibility and experimental data. Protein Eng. 1994;7:761–767. doi: 10.1093/protein/7.6.761. [DOI] [PubMed] [Google Scholar]
- 12.Tzou W.H., Hwang M.J. Modeling Helix−Turn−Helix protein-induced DNA bending with knowledge-based distance restraints. Biophys. J. 1999;77:1191–1205. doi: 10.1016/S0006-3495(99)76971-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dominguez C., Boelens R., Bonvin A.M.J.J. HADDOCK: a protein–protein docking approach based on biochemical or biophysical information. J. Am. Chem. Sec. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 14.van Dijk A.D., de Vries S.J., Dominguez C., Chen H., Zhou H.-X., Bonvin A.M.J.J. Data-driven docking: HADDOCK's adventures in CAPRI. Proteins. 2005;60:232–238. doi: 10.1002/prot.20563. [DOI] [PubMed] [Google Scholar]
- 15.Kalodimos C.G., Biris N., Bovin A.M.J.J., Levandoski M.M., Guennuegues M., Boelens R., Kaptein R. Structure and flexibility adaption in nonspecific and specific protein–DNA complexes. Science. 2004;305:386–389. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- 16.Kamphuis M.B., Bonvin A.M.J.J., Monti M.C., Lemonnier M., Munoz-Gomez A., van der Heuvel R.H., Diaz-Orejas R., Boelens R. Model of RNA binding and the catalytic site of the RNase kid of the bacterial parD toxin-antitoxin system. J. Mol. Biol. 2006;357:115–126. doi: 10.1016/j.jmb.2005.12.033. [DOI] [PubMed] [Google Scholar]
- 17.Kopke Salinas R., Folkers G.E., Bonvin A.M.J.J., Das D., Boelens R., Kaptein R. Altered specificity in DNA binding by the lac repressor: a mutant lac headpiece that mimics the gal repressor. ChemBioChem. 2005;6:1628–1637. doi: 10.1002/cbic.200500049. [DOI] [PubMed] [Google Scholar]
- 18.Volpon L., D'Orso I., Young C.R., Frasch A., Gehring K. NMR structural study of TcUBP1, a single RRM domain protein from Trypanosoma cruzi: contribution of a beta-hairpin to RNA binding. Biochemistry. 2005;44:3708–3717. doi: 10.1021/bi047450e. [DOI] [PubMed] [Google Scholar]
- 19.Mondragon A., Harrison S.C. The phage 434 Cro/OR1 complex at 2.5 Å resolution. J. Mol. Biol. 1991;219:321–334. doi: 10.1016/0022-2836(91)90568-q. [DOI] [PubMed] [Google Scholar]
- 20.Spronk C.A.E.M., Bonvin A.M.J.J., Radha P.K., Melacini G., Boelens R., Kaptein R. Structure of Lac repressor headpiece 62 complexed to a symmetrical Lac operator. Structure. 1999;7:1483–1492. doi: 10.1016/s0969-2126(00)88339-2. [DOI] [PubMed] [Google Scholar]
- 21.Raumann B.E., Rould M.A., Pabo C.O., Sauer R.T. DNA recognition by beta-sheets in the Arc repressor-operator crystal structure. Nature. 1994;367:754–757. doi: 10.1038/367754a0. [DOI] [PubMed] [Google Scholar]
- 22.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Padmanabhan S., Jimenez M.A., Gonzalez C., Sanz J.M., Gimenez-Gallego G., Rico M. Three-dimensional solution structure and stability of phage 434 Cro protein. Biochemistry. 1997;36:6424–6436. doi: 10.1021/bi970085p. [DOI] [PubMed] [Google Scholar]
- 24.Chuprina V.P., Rullmann J.A., Lamerichs R.M., van Boom J.H., Boelens R., Kaptein R. Structure of the complex of lac repressor headpiece and an 11 base-pair half-operator determined by nuclear magnetic resonance spectroscopy and restrained molecular dynamics. J. Mol. Biol. 1993;234:446–462. doi: 10.1006/jmbi.1993.1598. [DOI] [PubMed] [Google Scholar]
- 25.Slijper M., Bonvin A.MJ.J., Boelens R., Kaptein R. Refined structure of Lac repressor headpiece (1-56) determined by relaxation matrix calculations from 2D and 3D NOE data: change of tertiary structure upon binding to the Lac operator. J. Mol. Biol. 1996;259:761–773. doi: 10.1006/jmbi.1996.0356. [DOI] [PubMed] [Google Scholar]
- 26.Schildbach J.F., Karzai A.W., Raumann B.E., Sauer R.T. Origins of DNA-binding specificity: role of protein contacts with the DNA backbone. Proc. Natl Acad. Sci. USA. 1999;96:811–817. doi: 10.1073/pnas.96.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bonvin A.M.J.J., Vis H., Burgering M.J.M., Breg J.N., Boelens R., Kaptein R. Nuclear magnetic resonance solution structure of the Arc repressor using relaxation matrix calculations. J. Mol. Biol. 1994;236:328–341. doi: 10.1006/jmbi.1994.1138. [DOI] [PubMed] [Google Scholar]
- 28.Lu X.J., Olson W.K. 3DNA: a software package for the analysis rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chandrasekaran R.A., Arnott S. The structures of DNA and RNA helices in oriented fibres. In: Saenger W., editor. Landolt−Börnstein Numerical Data and functional Relationships in Science and Technology. Vol. VII/1b. Springer-Verlag: 1989. pp. 31–170. [Google Scholar]
- 30.Calladine C.R.D., Drew H.R. The principles of sequence-dependent flexure of DNA. J. Mol. Biol. 1986;192:907–918. doi: 10.1016/0022-2836(86)90036-7. [DOI] [PubMed] [Google Scholar]
- 31.Hubbard S.J., Thornton J.M. ‘NACCESS’, Computer Program, Department of Biochemistry and Molecular Biology. University College London; 1993. [Google Scholar]
- 32.Sander C., Schneider R. Database of homology derived protein structures and the structural meaning of sequence alignment. Proteins. 1991;9:56–68. doi: 10.1002/prot.340090107. [DOI] [PubMed] [Google Scholar]
- 33.Jorgensen W.L., Chandrasekhar J., Madura J.D., Imprey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 34.Olson W.K., Gorin A.A., Lu X.J., Hock L.M., Zhurkin V.B. DNA sequence-dependent deformability deduced from protein–DNA crystal complexes. Proc. Natl Acad. Sci. USA. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lankas F., Sponer J., Langowski J., Cheatham T.E. DNA deformability at the base pair level. J. Am. Chem. Soc. 2004;126:4124–4125. doi: 10.1021/ja0390449. [DOI] [PubMed] [Google Scholar]
- 36.Linge J.P., Williams M.A., Spronk C.A., Bonvin A.M.J.J., Nilges M. Refinement of protein structures in explicit solvent. Proteins. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- 37.Brunger A.T., Adems P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., et al. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 38.Wallace A.C., Laskowski R.A., Thornton J.M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 39.Lavery R.S., Sklenar H. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki M., Yagi N., Gerstein M. DNA recognition and superstructure formation by helix−turn−helix proteins. Protein Eng. 1995;8:329–338. doi: 10.1093/protein/8.4.329. [DOI] [PubMed] [Google Scholar]
- 41.Dickerson R.E., Chiu T.K. Helix bending as a factor in protein/DNA recognition. Biopolymers. 1997;44:361–403. doi: 10.1002/(SICI)1097-0282(1997)44:4<361::AID-BIP4>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 42.Schneider B., Neidle S., Berman H.M. Conformations of the sugar-phosphate backbone in helical DNA crystal structures. Biopolymers. 1997;42:113–124. doi: 10.1002/(sici)1097-0282(199707)42:1<113::aid-bip10>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 43.Berman H.M. Crystal studies of B-DNA: the answers and the questions. Biopolymers. 1997;44:23–44. doi: 10.1002/(SICI)1097-0282(1997)44:1<23::AID-BIP3>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 44.Zhurkin V.B., Ulyanov N.B., Gorin A.A., Jernigan R.L. Static and statistical bending of DNA evaluated by Monte Carlo simulations. Proc. Natl Acad. Sci. USA. 1991;88:7046–7050. doi: 10.1073/pnas.88.16.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grzeskowiak K., Goodsell D.S., Kaczor-Grezeskowiak M., Cascio D., Dickerson R.E. Crystallographic analysis of C-C-A-A-G-C-T-T-G-G and its implications for bending in B-DNA. Biochemistry. 1993;32:8923–8931. doi: 10.1021/bi00085a025. [DOI] [PubMed] [Google Scholar]
- 46.El Hassan M.A., Calladine C.R. Conformational characteristics of DNA: empirical classifications and a hypothesis for the conformational behaviour of dinucleotide steps. Phil. Trans. R Soc. Lond. A. 1997;355:43–100. [Google Scholar]
- 47.Dickerson R.E. DNA bending: the prevalence of kinkness and the virtues of normality. Nucleic Acids Res. 1998;26:1906–1926. doi: 10.1093/nar/26.8.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suzuki M., Amano N., Kukinua J., Tateno M. Use of a 3D structure data base for understanding sequence-dependent conformational aspects of DNA. J. Mol. Biol. 1997;274:421–435. doi: 10.1006/jmbi.1997.1406. [DOI] [PubMed] [Google Scholar]
- 49.Gorin A.A., Zhurkin B., Olson W.K. B-DNA twisting correlates with base-pair morphology. J. Mol. Biol. 1995;247:34–48. doi: 10.1006/jmbi.1994.0120. [DOI] [PubMed] [Google Scholar]
- 50.Wharton R.P., Brown E.L., Ptashne M. Substituting an alpha-helix switches the sequence-specific DNA interactions of a repressor. Cell. 1984;38:361–369. doi: 10.1016/0092-8674(84)90491-4. [DOI] [PubMed] [Google Scholar]
- 51.Koudelka G.B.L., Lam C.Y. Differential recognition of Or1 and Or3 by bacteriophage 434 repressor and Cro. J. Biol. Chem. 1993;268:23812–23817. [PubMed] [Google Scholar]
- 52.Harrison S.C., Anderson J.E., Koudelka G.B., Mondragon A., Subbiah S., Wharton R.P., Wolberger C., Ptashne M. Recognition of DNA sequences by the repressor of bacteriophage 434. Biophys. Chem. 1988;29:31–37. doi: 10.1016/0301-4622(88)87022-4. [DOI] [PubMed] [Google Scholar]
- 53.Bushman F.D., Anderson J.E., Harrison S.C., Ptashne M. Ethylation interference and X-ray crystallography identify similar interactions between 434 repressor and operator. Nature. 1985;316:651–653. doi: 10.1038/316651a0. [DOI] [PubMed] [Google Scholar]
- 54.Markiewicz P., Kleina L.G., Cruz C., Ehret S., Miller J.H. Genetic studies of the Lac repressor. XIV. Analysis of 4000 Altered Escherichia coli repressors reveals essential and non-essential residues as well as ‘spacers’ which do not require a specific sequence. J. Mol. Biol. 1994;240:421–433. doi: 10.1006/jmbi.1994.1458. [DOI] [PubMed] [Google Scholar]
- 55.Kisters-Woike B., Lehming N., Sartorius J., Wilcken-Bergmann B., Müller-Hill B. A model of the Lac repressor−operator complex based on physical and genetic data. Eur. J. Biochem. 1991;198:411–419. doi: 10.1111/j.1432-1033.1991.tb16030.x. [DOI] [PubMed] [Google Scholar]
- 56.Falcon C.M.M., Matthews K.S. Operator DNA sequence variation enhances high affinity binding by hinge helix mutants of lactose repressor protein. Biochemistry. 2000;39:11074–11083. doi: 10.1021/bi000924z. [DOI] [PubMed] [Google Scholar]
- 57.Barkley M.D., Bourgeois S. In: The operon. Miller J.H., Reznikoff W.S., editors. Cold Spring Harbor Laboratory, Cold Spring Harbor; 1978. pp. 177–220. Chapter 7. [Google Scholar]
- 58.Vershon A.K., Kelley R.D., Sauer R.T. Sequence-specific binding of arc repressor to DNA. Effects of operator mutations and modifications. J. Biol. Chem. 1989;264:3267–3273. [PubMed] [Google Scholar]