Abstract
The corepressor complex that includes Ebi and SMRTER is a target of epidermal growth factor (EGF) and Notch signaling pathways and regulates Delta (Dl)-mediated induction of support cells adjacent to photoreceptor neurons of the Drosophila eye. We describe a mechanism by which the Ebi/SMRTER corepressor complex maintains Dl expression. We identified a gene, charlatan (chn), which encodes a C2H2-type zinc-finger protein resembling human neuronal restricted silencing factor/repressor element RE-1 silencing transcription factor (NRSF/REST). The Ebi/SMRTER corepressor complex represses chn transcription by competing with the activation complex that includes the Notch intracellular domain (NICD). Chn represses Dl expression and is critical for the initiation of eye development. Thus, under EGF signaling, double negative regulation mediated by the Ebi/SMRTER corepressor complex and an NRSF/REST-like factor, Chn, maintains inductive activity in developing photoreceptor cells by promoting Dl expression.
Keywords: Drosophila , Ebi, EGF, Notch, REST
Introduction
Highly ordered arrays of neuronal cells in sensory organs and in the central nervous system are produced by sequential inductive events involving intercellular signaling and cell fate determination (Stern, 2001). Intercellular signaling coordinates temporal and spatial expression of genes required for neuronal differentiation (Lee and Pfaff, 2001). The Notch signaling pathway plays important roles in the regulation of cell differentiation, proliferation and apoptosis in many developmental systems (reviewed in Lai, 2004). Binding of a ligand to the Notch receptor induces proteolytic cleavage of the transmembrane domain, with the subsequent release of the Notch intracellular domain (NICD) (Struhl and Adachi, 1998). NICD translocates to the nucleus where it forms a complex with the transcription factor, Suppressor of Hairless (Su(H)), which then activates gene transcription (Struhl and Adachi, 1998). In the absence of activated Notch, Su(H) acts as a transcriptional repressor of various genes involved in the development of mechanosensory cells and photoreceptor neurons in Drosophila (Barolo et al, 2000; Li and Baker, 2001). This Su(H)-mediated repression system is evolutionarily conserved. A human homolog of Su(H), RBPJκ, acts as either a transcriptional activator or repressor for many types of genes, including IL-6, Hes1, p52/NFκB2 and glial fibrillary acidic protein (GFAP) (Kannabiran et al, 1997; Kao et al, 1998; Oswald et al, 1998; Hermanson et al, 2002).
RBPJκ-mediated repression requires the nuclear receptor corepressor (N-CoR) (Horlein et al, 1995) and/or the closely related silencing mediator for retinoic acid and thyroid hormone receptor (SMRT) (Chen and Evans, 1995) to recruit histone deacetylases to target promoters (Kao et al, 1998; Hermanson et al, 2002; reviewed in Jepsen and Rosenfeld, 2002). N-CoR/SMRT corepressor function requires its association with transducin β-like 1 (TBL1; Guenther et al, 2000; Yoon et al, 2003; Perissi et al, 2004). TBL1 is an evolutionarily conserved F-box/WD40-repeat-containing protein originally identified as a candidate gene responsible for X-linked ocular albinism with late-onset sensorineural deafness (Bassi et al, 1999; Dong et al, 1999). It was recently shown that TBL1 function is not limited to transcriptional repression; it also converts transcriptional cofactors between repressive and stimulative states (Perissi et al, 2004).
The Drosophila homolog of TBL1, Ebi, was first identified as a downstream component of epidermal growth factor receptor (EGFR) signaling during eye development (Dong et al, 1999). Ebi activity in photoreceptor cells promotes expression of a Notch ligand, Delta (Dl), which is subsequently used to convert adjacent non-neural cells to cone cell fate (Flores et al, 2000; Tsuda et al, 2002). EGFR positively regulates Dl in photoreceptor cells, whereas Notch downregulates Dl (Tsuda et al, 2002). Previous studies have shown that a protein complex that includes Ebi, SMRTER (N-CoR/SMRT-related molecule in Drosophila; Tsai et al, 1999), Su(H) and strawberry notch (Sno; Majumdar et al, 1997) mediates EGFR signaling and positively regulates Dl expression (Tsuda et al, 2002). It is not clear how the protein complex containing Ebi and SMRTER, which primarily acts as a repressor, stimulates Dl because direct targets of this complex have not yet been identified.
In this study, we report the identification of a gene, charlatan (chn), that is a direct target of the Ebi/SMRTER corepressor complex. Chn encodes a C2H2-type zinc-finger protein and was originally identified as a gene required for normal development of the peripheral nervous system (PNS; Kania et al, 1995). We found that Chn shares several characteristics with human neuronal restricted silencing factor/repressor element RE-1 silencing transcription factor (NRSF/REST), a repressor of various neuron-restricted genes (reviewed in Jones and Meech, 1999). Dl was identified as a direct target of Chn repressor. Taken together, these results show that Ebi/SMRTER/Su(H) corepressor complex mediates the crosstalk between EGFR and Notch by downregulating the NRSF/REST-like repressor, Chn, during photoreceptor cell development.
Results
Ebi modulates the dual role of Su(H) as a repressor and an activator of chn expression during eye development
The corepressor complex that includes Ebi, SMRTER and Su(H) is required for expression of Dl in Drosophila photoreceptor cells (Tsuda et al, 2002). To identify genetic loci that are transcriptionally repressed by the Ebi corepressor, we set up a screen using an ectopic gene expression system (Gene Search System; Toba et al, 1999). Insertion of a Gene Search (GS) vector, a modified P-element carrying the Gal4 upstream activating sequence (UASG) near its 3′ end, causes overexpression of a nearby gene under the control of the Gal4-UASG system (Toba et al, 1999). We identified GS insertions into the chn locus (Kania et al, 1995), whose overexpression phenotype in the eye using an eye-specific Gal4 driver (GMR-Gal4) was modified by reducing ebi activity (Figure 1D and E). We thus studied the regulation of chn by Ebi-dependent transcriptional repression.
Figure 1.
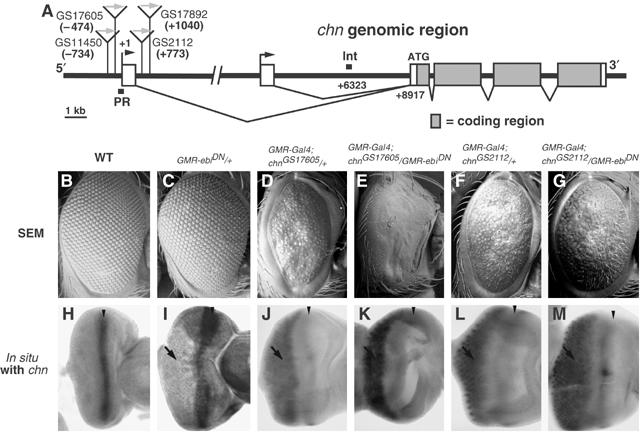
Mapping of the target site of Ebi-dependent repression in the chn promoter. (A) Genomic region of chn. Boxes indicate exons, and filled portions represent the coding region. The locations of the four GS insertion sites are indicated with gray arrows. PR and Int represent the regions used for ChIP analysis. (B–G) Scanning electron micrographs of adult eyes. (H–M) chn mRNA expression patterns in eye imaginal discs. Samples shown in panels H and I were processed with a four-fold longer reaction time for RNA detection. One copy of GMR-ebiDN caused a slight increase in chn expression (I) and a mild rough eye phenotype (C). Both chnGS17605 and chnGS2112, driven by GMR-Gal4, caused abnormal eye morphologies (D, F) as well as ectopic expression of chn posterior to the MF (J, L, arrows). GMR-ebiDN markedly enhanced both the abnormal eye phenotype (E) and expression of chnGS17605 (K). The phenotype caused by chnGS2112 was not significantly altered by GMR-ebiDN (G, M). Anterior is at right. Arrowheads in panels H–M indicate the MF, and arrows indicate the region of ectopic chn expression.
In third instar larval-stage eye discs, the chn transcript was highly expressed in the morphogenetic furrow (MF), where photoreceptor differentiation initiates, but was downregulated in cells in the later stage photoreceptor development (Figures 2A, A′ and 7D). In ebi mutant eye discs, however, chn expression became detectable in differentiating photoreceptor cells, and its expression in the MF was increased (Figure 2B and B′), suggesting that Su(H) in association with Ebi and SMRTER represses chn transcription in the eye disc.
Figure 2.
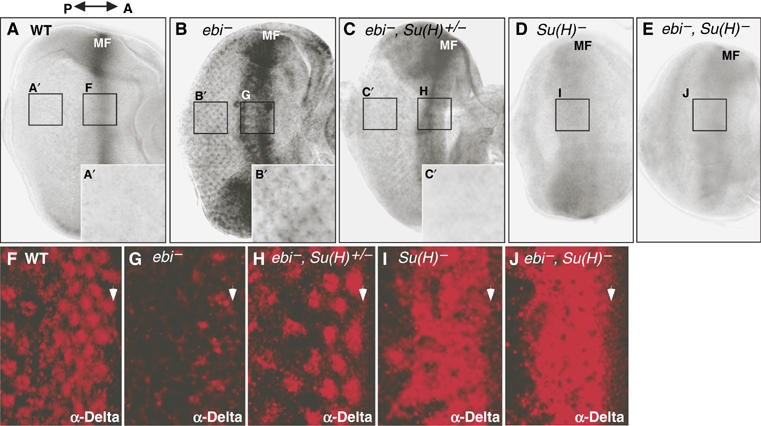
Regulation of chn and Dl expression by ebi and Su(H). (A–E) In situ hybridization patterns of chn mRNA expression in late third instar eye imaginal discs. The double arrow indicates anterior (A) and posterior (P). (A) In wild-type discs, chn is highly expressed around the MF. chn expression was reduced in differentiating ommatidia (A′). (B) ebiP7/ebiE4. In ebi mutants, chn expression was increased around the MF, and ectopic chn expression became detectable in differentiating photoreceptor cells (B′). (C) ebiP7Su(H)2/ebiE4,+. (D) Su(H)1/Su(H)2. (E) ebiP7Su(H)2/ebiE4Su(H)1. (F–I) Dl expression was monitored by staining with anti-Dl. (F) Dl was detected in the MF (arrowhead) and in differentiating photoreceptor cells in wild-type eye discs. (G) In the ebi mutant, Dl levels were reduced in the MF and in photoreceptor cells. (H) ebiP7Su(H)2/ebiE4+. (I) Su(H)1/Su(H)2. (J) ebiP7Su(H)2/ebiE4Su(H)1.
To reveal the role of Su(H) as an activator, we examined chn expression when the level of Su(H) expression was reduced. It was shown previously that removing one copy of Su(H) suppresses the loss-of-Dl expression phenotype in ebi mutants (Figure 2F–H; Tsuda et al, 2002). We found that reducing one copy of Su(H) suppresses ectopic chn expression in ebi mutants (Figure 2C and C′), suggesting that ectopic expression of chn in ebi mutants (Figure 2B) is Su(H)-dependent. RT–PCR analysis of chn expression in ebi− eye discs differing in the dosage of Su(H) gene also supported these results (Supplementary Figure 1). Strong reduction of Su(H) expression alone reduced expression of chn in the MF, which became weaker and was slightly broader (Figure 2D). The phenotype of ebi, Su(H) double mutants was almost the same as Su(H) single mutants (Figure 2E, compare with B and D), suggesting that Su(H) acts as an activator in the absence of Ebi. This might be due to dual functions of Su(H) as an activator or repressor (Barolo et al, 2000; Li and Baker, 2001). Hence, reducing the amount of Ebi in the corepressor complex involving Su(H) might convert Su(H) to an activator by permitting the replacement of the corepressor complex with NICD. The following molecular data support this hypothesis.
Mapping of the target site of Ebi-mediated repression in the chn promoter region
To reveal the molecular nature of transcriptional regulation of chn by Su(H), we searched for Su(H) target sites in the genomic region of chn (Figure 1A). As Su(H) binds slightly degenerate sequences (Flores et al, 2000), it was not easy to identify the functional Su(H) binding region from a simple genomic search. We thus took an alternative approach to map the chn genomic region, which is regulated by Su(H) in the normal chromosomal context. Ebi-mediated repression involves SMRTER, a corepressor that recruits histone deacetylases and induces the formation of inactive chromatin (Tsai et al, 1999), which spreads from the site where Su(H) recruits the corepressor complex. Promoters near the Su(H)-binding site are thus expected to be downregulated in an Ebi-dependent manner. We identified four insertion lines of the GS vector in the chn promoter region (Figure 1A). All these GS lines caused ectopic expression of chn with consequent abnormal eye morphology when they were crossed with GMR-Gal4 (Figure 1D, F, J and L; data not shown). If the effect of the Ebi/SMRTER corepressor complex reaches the UASG in those insertions, reduction of Ebi activity will derepress UASG and further enhance activation by GMR-Gal4. One copy of a dominant-negative construct of ebi (GMR-ebiDN; Dong et al, 1999) caused only a mild defect in eye morphology and weak, if any, ectopic expression of chn (Figure 1C and I). GMR-ebiDN strongly enhanced the overexpression phenotype of chnGS17605 and chnGS11450, which contained GS vector insertions (−474 and −734, respectively) upstream of the transcriptional start site (Figure 1A, and compare E and K to D and J; data not shown). However, GMR-ebiDN failed to enhance the overexpression phenotype of other GS lines (chnGS2112 and chnGS17892) that were inserted downstream (+773 and +1040, respectively) of the first exon (compare Figure 1G and M to F and L; data not shown). As a control, we determined whether GMR-ebiDN affects the overexpression phenotype of UASG-chn cDNA inserted outside of the chn locus. No strong genetic interaction was observed (data not shown). From these results, we concluded that Ebi-dependent transcriptional repression is targeted to the proximity of the transcriptional initiation site of the chn promoter.
Ebi and SMRTER associate with the chn promoter in vivo
We searched for potential Su(H)-binding sites around the transcriptional initiation site of chn and found two sequences, S1 and S2, with similarity to the consensus sequences for Su(H) binding (5′-RTGRGAR-3′; Nellesen et al, 1999) within the fragment PR(chn) (Figures 1A, 3A and B). The S1 and S2 sites were indeed bound by His-Su(H) in an electrophoretic mobility shift assay (EMSA; Figure 3C). The binding was competed out by unlabeled oligonucleotides containing S1, S2 or a consensus Su(H) sequence (E(spl)mγ), but not by mutant forms of S1, S2 or E(spl)mγ sequences (Figure 3B and C), suggesting that Su(H) binds specifically to S1 and S2 sites.
Figure 3.
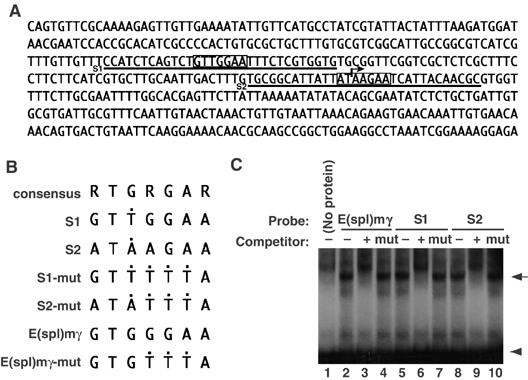
Su(H) binds to the promoter region of chn. (A) The sequence of PR(chn) in the promoter region of chn (Figure 2A). Degenerate Su(H)-binding consensus sites are boxed (S1 and S2, sequences used for EMSA, are underlined). The arrow represents a putative transcriptional initiation site. (B) Core sequences of oligonucleotides used for EMSA. Black dots indicate positions that differ from the Su(H)-binding consensus (5′-RTGRGAR-3′). (C) EMSA using a His fusion of recombinant Su(H). E(spl)mγ is a positive control previously known to bind to Su(H) (lane 1, arrow). Ten-fold excess of unlabeled probe inhibited complex formation (lane 2), but the mutant form of the probe failed to compete (lane 3). S1 and S2 sequences also formed a complex with Su(H) (lanes 4 and 7). Ten-fold excess of unlabeled probes inhibited complex formation (lanes 5 and 8), but competitor containing consensus sequence mutations abrogated the competition (lanes 6 and 9). Arrowhead represents a position of free probe.
We used chromatin immunoprecipitation (ChIP) to test whether PR(chn) is a target of Ebi-mediated repression in vivo. Eye disc chromatin from third instar larvae was fixed and immunoprecipitated by anti-Ebi. This procedure efficiently recovered fragment PR(chn) but not the intron fragment of chn genomic region Int(chn), which was used as a negative control (Figure 4A). Using the same procedure, we detected the association of SMRTER with PR(chn), as expected because Ebi and SMRTER associate with Su(H) (Tsuda et al, 2002). Association of Ebi and SMRTER was also detected using the promoter region of hsp27(hsp27), to which SMRTER is recruited by the ecdysone receptor (Figure 4A; Tsai et al, 1999; Sawatsubashi et al, 2004).
Figure 4.
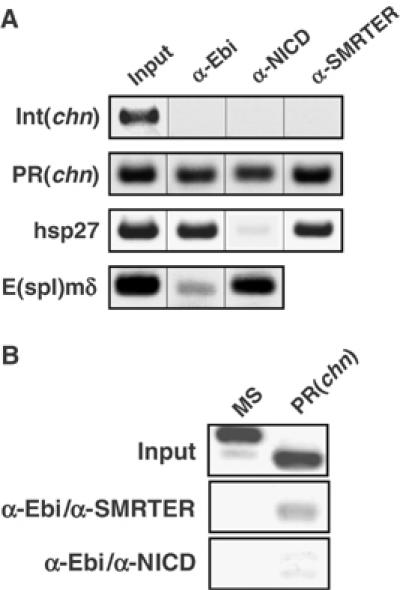
Occupancy of the chn promoter by Ebi, SMRTER and NICD in vivo. (A) ChIP assay of third instar eye imaginal discs using anti-Ebi, anti-SMRTER or anti-NICD. PR(chn) and Int(chn) are derived from the chn genomic region (Figures 1A and 3A), and hsp27 is a fragment of the hsp27 promoter region (Tsai et al, 1999; Sawatsubashi et al, 2004). E(spl)mδ is a fragment of the E(spl)mδ promoter region (Cooper and Bray, 1999). (B) Sequential ChIP assay. Crosslinked chromatin was immunoprecipitated with anti-Ebi followed by a second immunoprecipitation with anti-SMRTER or anti-NICD. MS is the sequence from microsatellite DNA (Sawatsubashi et al, 2004). Although anti-SMRTER precipitated PR(chn) from Ebi-containing chromatin, anti-NICD did not.
Mutually exclusive binding of NICD and Ebi/SMRTER to the Su(H)-binding site of the chn promoter
We showed that chn is positively regulated by Su(H) at the MF (Figure 2). We therefore analyzed whether NICD, a positive regulator of Su(H) under control of Notch signaling, is also recruited to PR(chn) in the eye disc. Using the antibody against NICD, we showed that NICD is indeed recruited to the promoter region of E(spl)mδ,which is shown to be a target site of NICD/Su(H) in the eye imaginal disc (Cooper and Bray, 1999; Figure 4A). We also detected that NICD is recruited to PR(chn) and but not to Int(chn) or hsp27 (Figure 4A). To determine the relationship between NICD and the Ebi/SMRTER complex bound to PR(chn), we conducted a sequential ChIP analysis (Figure 4B). PR(chn) was specifically recovered after two rounds of immunoprecipitation—with anti-Ebi followed by anti-SMRTER—although the microsatellite sequence, which was used for negative control, was not, suggesting that Ebi and SMRTER form a complex bound to PR(chn) in the eye disc. On the other hand, sequential immunoprecipitation with anti-Ebi and anti-NICD failed to recover PR(chn) (Figure 4B). These results suggest that an activator, NICD, and a corepressor, Ebi/SMRTER, are targeted to a common Su(H)-binding site, PR(chn), in the chn promoter in a mutually exclusive manner.
chn encodes a C2H2-type zinc-finger protein with similarity to human NRSF/REST
Chn is a 1108-amino-acid protein with multiple C2H2-type zinc-finger motifs (Figure 5A; Escudero et al, 2005; Reeves and Posakony, 2005). Although we could not detect any highly homologous gene within the mammalian genome using BLAST, we found a small sequence of similarity between the N-terminal zinc-finger motif of Chn and the fifth zinc-finger of human NRSF/REST (Figure 5B). We found that Chn has several structural and functional similarities to human NRSF/REST, as follows. First, Chn and NRSF/REST each contain an N-terminal region with multiple zinc-finger motifs (five motifs in 264 residues in Chn and eight motifs in 251 residues in NRSF/REST), followed by a cluster of S/T-P motifs (serine or threonine followed by a proline) and a single zinc-finger motif at the C terminus (Figure 5A). Second, the C-terminal region of NRSF/REST binds a corepressor, CoREST, which serves as an adaptor molecule to recruit a complex that imposes silencing activities (Andres et al, 1999; Lunyak et al, 2002). We found that the Drosophila homolog of CoREST (dCoREST) (Andres et al, 1999; Dallman et al, 2004) can associate with the C-terminal half of Chn in cultured S2 cells (Figure 5C). Finally, NRSF/REST binds to NRSE/RE1, a 21-bp sequence located in the promoter region of many types of neuron-restricted genes, via the N-terminal zinc-finger motifs (Chong et al, 1995; Schoenherr and Anderson, 1995). We found that a recombinant protein containing the N-terminal zinc-fingers of Chn bound specifically to the NRSE/RE1 sequence in vitro (Figure 5D, left panel). Thus, the structural similarity to NRSF/REST, binding to dCoREST and the DNA-binding specificity of Chn suggest that it is a candidate for a functional Drosophila homolog of NRSF/REST.
Figure 5.
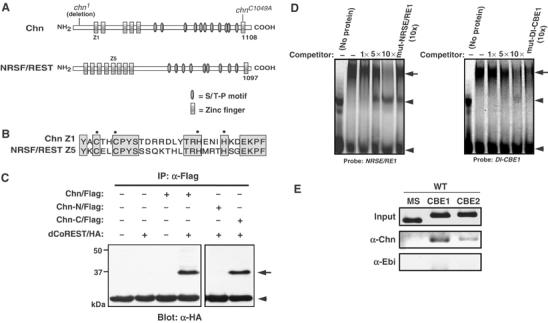
Common properties of Chn and NRSF/REST. (A) Schematic diagram of Chn and human NRSF/REST. Mutation sites of EMS-induced (chn1) and mutagenized (chnC1049A) chn are shown above the Chn diagram. (B) Sequence comparison between the first zinc-finger motif in Chn (Chn Z1) and the fifth zinc-finger motif in human NRSF/REST (NRSF/REST Z5). (C) Immunoprecipitation assay. Chn and dCoREST associate in cultured S2 cells. Chn-N/Flag and Chn-C/Flag contain residues 1–654 and 655–1108, respectively, and are tagged with a single Flag epitope at the C-terminus. dCoREST/HA contains the smallest splice variant of dCoREST, which has EML2 and SANT domains (Dallman et al, 2004) and a single HA epitope at the N-terminus. The arrow indicates the position of dCoREST/HA. The arrowhead indicates the position of IgG. (D) Chn binds to the NRSE/RE1 and Dl-CBE1 sequences. EMSA was performed using NRSE/RE1, a 21-bp recognition sequence of NRSF/REST (Table I), or Dl-CBE1 (Table I) as a probe. A GST fusion of N-terminal zinc-finger domains of Chn formed a specific complex with the probe (arrow). Arrowheads denote a nonspecific shifted band (upper) and free probe (lower). Up to 10-fold molar excess of unlabeled probe was used as competitor. The mutant forms of NRSE/RE1 or Dl-CBE1 failed to compete. (E) ChIP assay of third instar eye imaginal discs. Anti-Chn preferentially precipitated the genomic regions containing CBE1 (Dl CBE1) or CBE2 (Dl CBE2) (Table I), but anti-Ebi did not (WT).
Identification of putative Chn target genes
If Chn acts as a regulator of neural-related functions, as suggested for NRSF/REST, then Chn would be expected to bind to a regulatory region common to many types of neural-related genes in Drosophila. We identified numerous sequences similar to NRSE/RE1 in the Drosophila genome, and their binding to Chn was assessed by EMSA (data not shown). Using these sequences, we derived a consensus binding sequence for Chn (Chn-binding element (CBE), 5′-BBHASMVMMVCNGACVKNNCC-3′). We searched for and identified 26 CBEs within 10 kb of annotated genes from the Drosophila genome (version 3.2) (Table I). Binding to Chn was confirmed for 18 CBEs using EMSA competition assay (Table I). Genes containing the CBE include dopamine receptor 2 (DopR2) and the potassium channel, ether-a-go-go, for which the mammalian homologs are target genes of NRSF/REST (Schoenherr et al, 1996; Chen et al, 1998; Lunyak et al, 2002; Bruce et al, 2004). These results suggest that the CBE is a good indicator of Chn binding sites and that Chn regulates many types of neural-related genes, as is implicated for NRSF/REST (Jones and Meech, 1999; Bruce et al, 2004). However, we found that divergent forms of CBE adjacent to hairy and extramacrochaetae were bound specifically by Chn (Table I, asterisks). Likewise, some of the CBE sites failed to bind to Chn. Thus, a further refinement will be necessary to predict a definitive set of Chn binding sites in the Drosophila genome.
Table 1.
Putative Chn target genes containing CBE and/or its variant sequences adjacent to the coding sequence, based on informatics and binding activity
| Genes (target sites) | Sequence comparison | Binding | Location/direction |
|---|---|---|---|
| (NRSE/RE1) | TTCAGCACCACGGACAGCGCC | + | |
| (CBE) | BBHASMVMMVCNGACVKNNCC | ||
| Neural functions/expressions | |||
| Mnt | GTTACCGACGCCGACGGTACC | + | 1st intron/R |
| HMG protein Z | CCCACCCCCCCTGACCGACCC | + | 1st intron/R |
| allatostatin C receptor2 | TTTAGCCAAGCCGACAGATCC | − | 1st intron/R |
| Arrestin 2 | TGAAGACCCACGGACAGGGCC | + | 0 kb upstream/F |
| Olfactory-specific E | TCCACCCCCACCGACGTTCCC | + | 3 kb upstream/R |
| Dystroglycan | CTTAGAACAGCCGACGTCCCC | + | 1st intron/R |
| nAcRβ-64B | CGCACCCAAACTGACGGCTCC | − | 5′ UTR/F |
| Lar | CGAAGAACAACTGACGTAGCC | + | 4 kb upstream/F |
| ether-a-go-go | GTCACAGACACAGACCTAGCC | + | 1st intron/R |
| Dopamine receptor2 | CCAAGACAAACAGACGGCCCC | + | 3 kb downstream/F |
| kekkon-2 | GCCACCCACACCGACATTTCC | + | 5′ coding/F |
| shaking B | TTAACAGCACCTGACCGACCC | + | 2nd intron/F |
| unc-115 | TCTACACACCCCGACATGGCC | + | 0 kb upstream/R |
| brinker | GCAACAGCAGCAGACATTTCC | − | 3′ coding/F |
| bifid | GGCAGCGACGCTGACGTCGCC | + | 2nd intron/R |
| Deformed | TGCACCACCGCCGACGGCACC | + | 5′ coding/R |
| Socs 16D | TCCAGCGAACCGGACGGCTCC | − | 5th exon/F |
| Saliva | TTTAGCAAACCTGACATTGCC | − | 3rd intron/R |
| hairy (CBE1) | CGCAGCAACACAGACCGCCCC | + | 9 kb upstream/F |
| *hairy (CBE2) | GCCACCAAAAAAGACGTAAGT | + | 2nd exon/F |
| emc (CBE1) | TCCACCAAAGCAGACCTCTCC | + | 5′ UTR/F |
| *emc (CBE2) | CGCACCACCCATGACAGAGCC | + | 3 kb downstream/R |
| *Dl (CBE1) | TTCAGCACCACCGCCATTGGT | + | 2nd intron/F |
| *Dl (CBE2) | CCCAGCCCCACGGCCATATCC | + | 10 kb downstream/F |
| Oogenesis/cytoskeleton | |||
| spire | TGAAGAAAAACCGACATAACC | − | 1st intron/F |
| orb | CCCACAGCAGCCGACCGACCC | + | 1st intron/R |
| lkb1 | TCAACAGAAACGGACAGATCC | − | 0.1 kb upstream/R |
| Death caspase-1 | CGCACCCACACGGACGTACCC | + | 1 kb upstream/R |
| D/V axis/defence | |||
| Tehao | GCAACAAAAACTGACGTCTCC | − | 10 kb upstream/F |
| Cardiac cell development | |||
| tincar | CCAACCCAACCTGACCTGACC | + | 7 kb upstream/R |
| (+) Positive binding to Chn zinc-finger domain, based on EMSA. (−) Very weak binding, if detectable, to Chn zinc-finger domain. Asterisks represent variant CBE sequences. Black dots indicate positions that differ from the CBE consensus. F and R represent the forward and reverse directions, respectively, against the coding sequences of the candidate target genes. nAcRβ-64B, nicotinic acetylcholine receptor beta 64B; Socs16D, suppressor of cytokine signaling at 16D; emc, extra macrochaetae; orb, oo18 RNA-binding protein. Single letter abbreviations: B=C, G or T; H=A, C or T; S=G or C; M=A or C; V=A, C or G; N=A, G, C or T; K=G or T. | |||
Chn represses Dl in photoreceptor neurons
We found that the genomic region of Dl has two divergent CBEs (Table I), suggesting that Dl is a target of Chn (Figure 5D, right panel; data not shown). Dl was normally expressed in photoreceptor cells, where chn is repressed (Figure 2A and F). Upon loss of ebi function, chn was ectopically expressed and Dl levels were reduced (Figure 2B and G, and Supplementary Figure 1). Dl expression was elevated in a double mutant of ebi with Su(H) or with mutant Su(H), in which chn expression in the MF was reduced (Figure 2D, E, I and J). The apparent inverse relationship between chn and Dl expression suggests that Chn might repress Dl. To confirm this hypothesis, we ectopically expressed chn in photoreceptor cells after commitment to neuronal differentiation and monitored the resultant Dl levels. Ectopic expression of chn reduced Dl levels (Figure 6D, compare to A). Furthermore, Cut expression in cone cells, which requires Dl as an inductive signal from photoreceptor cells (Flores et al, 2000; Tsuda et al, 2002), was strongly reduced (Figure 6F, compare to C), and cone cells failed to develop as evidenced by defective lens secretion owing to overexpression of chn (Figure 1D and F). In contrast, Cut expression in glia, which are located beneath the photoreceptor cells and recruited from the brain region (Hummel et al, 2002), was not affected (Figure 6I and J). Despite the strong reduction in the Dl level, chn overexpression did not affect the expression of the neuronal marker Elav (Figure 6E). These results indicate that chn specifically blocks cone cell induction by photoreceptor cells by repressing Dl while leaving neuronal differentiation intact. Using ChIP assays, we detected preferential association of Chn with the divergent CBEs in the Dl genomic region (Dl CBE1 and CBE2; Table I and Figure 5E), whereas no significant association with Ebi was detected, which was used as a negative control (Figure 5E). These results suggest that Chn can interact with CBEs on the genomic region of Dl and regulate the expression level of Dl.
Figure 6.
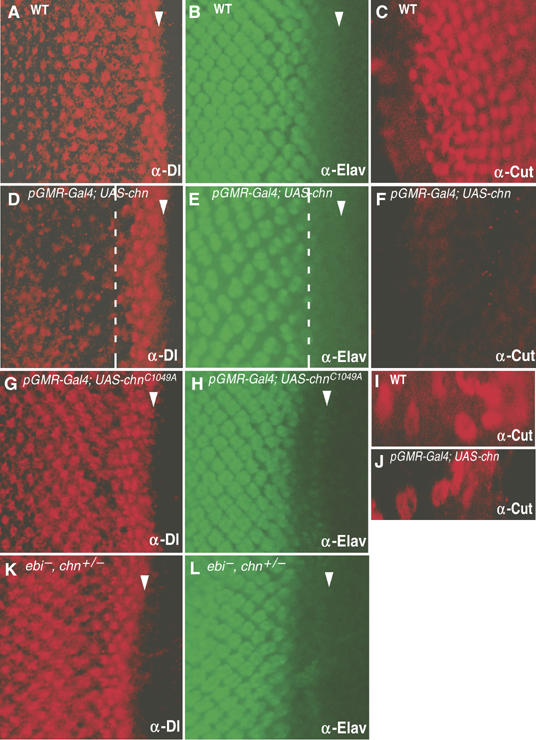
chn represses Dl expression in photoreceptor cells. Eye imaginal discs in third instar larvae stained for Dl (A, D, G, K), Elav (a neuronal marker; B, E, H, L) or Cut (C, F, I, J). Panels A–C and I, wild type; panels D–F and J, GMR-Gal4; UAS-chn/+; panels G and H, GMR-Gal4; UAS- chnC1049A/+. The Dl level was severely reduced by chn overexpression (D), whereas the Elav level was unchanged (E). The level of Cut in cone cells was reduced by chn overexpression (F) but was not affected in glia cells (I, J). A mutated form of chn (chnC1049A) could not repress Dl expression (G), whereas the Elav level was unchanged (H). (K, L) ebiE4, chn1/ebil11, +. Reducing a copy of chn suppressed the defect of Dl expression in the ebi mutant. Arrowheads indicate the MF. The broken white line represents the point at which GMR-Gal4 expression began. Anterior is at right.
The C-terminal zinc-finger motif of NRSF/REST is essential for its repression activities (Tapia-Ramirez et al, 1997). We found that a point mutation introduced into the C-terminal zinc-finger motif of Chn (Figure 5A, chnC1049A) abrogated its ability to repress Dl expression (Figure 6G, compare to D), demonstrating another correlation between Chn and NRSF/REST.
Double-negative regulation of Dl by Ebi and Chn
To obtain in vivo evidence for the role of Ebi-mediated chn repression in eye development, we isolated new chn mutations (see Materials and methods). chn1 contained a small deletion that caused a frame shift after amino acid 94, likely yielding a null allele (Figure 5A). Interference of Ebi function in photoreceptor cells by the strong hypomorphic allele of ebi abrogated Dl expression (Figure 2F and G); however, reducing one copy of chn completely suppressed this phenotype (Figure 6K and L). This strong genetic interaction between ebi and chn suggests that the downregulation of chn activity in photoreceptor cells seems to be responsible for the proper development of photoreceptor neurons. Combined with the strong repression of Dl by Chn, these results suggest that an important function of Ebi in photoreceptor cells is to repress chn to permit expression of Dl; this, in turn, induces cone cell differentiation.
To determine if Chn is required at the stage when photoreceptor cells express Dl to induce cone cell development, we studied phenotypes of small clones of chn1 cells. The chn1 clones present within or behind the MF were normal, as assessed by Elav expression, ommatidium assembly and progression of the MF (data not shown), suggesting that eye development can proceed without chn function once the MF has initiated. Although chn is highly expressed in the MF (Figure 2A), the apparent lack of a requirement for chn in MF progression might be owing to genes with overlapping functions or to a possible long half-life of Chn.
Chn-mediated repression of Dl is essential for the initiation of eye development
In the eye disc at the early third instar, Chn was expressed at the edge of the eye disc, the site of MF initiation (Figure 7A). To address the role of chn in the early stage of the eye development, large clones of cells homozygous for chn1 were induced in the eye by the FLP/FRT technique. Mutant animals had substantially smaller eyes (Figure 7B), similar to the mutant phenotype of thick vein (tkv; Figure 7C). Tkv is a receptor for Decapentaplegic and is required for the initiation of the MF (Borod and Heberlein, 1998), suggesting that chn is also required at the early stage of photoreceptor cell development. In eye discs bearing chn1 mutant clones, the number of photoreceptor cells was greatly reduced and the remaining photoreceptor cells were always associated with Chn-expressing cells that escaped FLP recombination (Figure 7E, arrowheads and arrows). Although chn1 is embryonic lethal, chn1/chn9 heterozygote yielded a few escapers that survived until the late third instar larval stage. Eye discs of those larvae were small, and the MF was absent (Figure 7G, compare to F). Only a few patches of Elav-positive neurons were observed, which apparently formed independently of the MF (Figure 7G, arrow).
Figure 7.
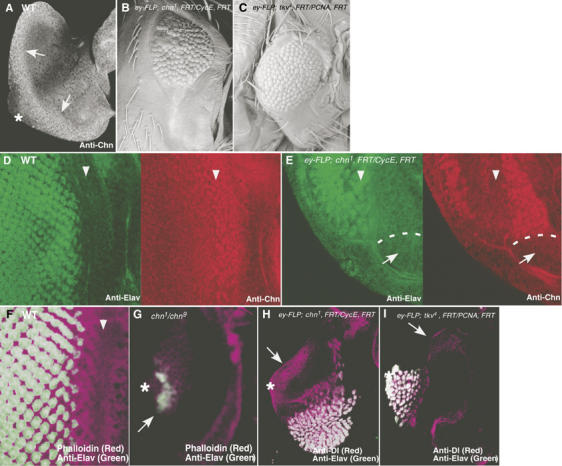
chn is required for eye morphogenesis. (A) In wild-type eye discs from early third instar larvae, Chn expression was detected at the posterior edge of the disc using anti-Chn antibody (arrows). (B, C) Adult eye morphology. (B) ey-FLP; chn1, FRT42D/PCNA775, FRT42D. Large clones of chn1 were formed and caused the small eye phenotype. (C) ey-FLP; tkv4, FRT40A/CycEAR95, FRT40A. A large clone of tkv4 showed a similar eye defect. (D) In wild-type eye disc, Elav (green) was expressed behind the MF (left panel), marked with Chn (red; right panel). (E) In eye discs with large clones of chn1, Elav-positive cells were greatly reduced in number (green; left panel, arrowheads) and were always associated with residual chn expression (red; right panel). White dots represent the border of chn1 clone (arrows). (F) Wild-type eye disc stained with phalloidin (purple) and anti-Elav (green). (G) chn1/chn9. Strong chn mutations eliminated the MF, whereas a few Elav-positive cells can be observed (arrow). (H) In eye discs with large clones of chn1, ectopic expression of Dl was detected at the posterior edge of the disc (arrow). (I) Large clone of tkv4 did not induce ectopic Dl expression (arrow). Asterisks represent the position of the optic stalk where the MF propagation initiates, and arrowheads represent the position of the MF.
These phenotypes suggest that eye development cannot initiate without chn. Furthermore, we found that Dl was substantially increased at the posterior edge of eye discs having large chn1 mutant clones (Figure 7H, arrow). This ectopic expression of Dl likely accounts for the failure to initiate eye development, as a high level of Dl in cells autonomously inhibits Notch (Jacobsen et al, 1998), which is required to initiate the MF (Kumar and Moses, 2001). On the other hand, Dl remained repressed in discs containing the tkv mutant clone (Figure 7I, arrow).
Discussion
Functional similarity of Chn to human NRSF/REST
Although it has been established that mammalian NRSF/REST is a key regulator of neuron-specific genes (Jones and Meech, 1999), attempts to isolate invertebrate homolog of NRSF/REST have so far failed to identify a true homologous factor in invertebrates (Dallman et al, 2004). The properties of Chn, including the similarity in DNA-binding specificity, association with CoREST and transcriptional repressor activity, suggest that Chn is a strong candidate for a functional Drosophila homolog of NRSF/REST. chn was originally identified by its requirement in the development of the PNS (Kania et al, 1995; Escudero et al, 2005; Reeves and Posakony, 2005). We identified a number of candidate target genes of Chn, a large fraction of which is implicated in neural function and/or gene expression (Table I). We expect that further analysis of these candidates will provide valuable information about chn function in vivo, which may be extended to the understanding of NRSF/REST.
Regulation of Dl and the initiation of eye development by chn
The Chn mutation blocked eye development by preventing the initiation of MF, a process requiring Notch signaling (Kumar and Moses, 2001; Figure 7). This phenotype is likely owing to a loss of Notch function, because elevated Dl expression is known to block Notch signaling. The function of Chn during the early stage of eye development might be to regulate Notch signaling at an appropriate level by downregulating Dl (Figure 8B). It is possible that Chn-mediated modulation places a variety of Notch functions in eye (Baonza and Freeman, 2001; Kumar and Moses, 2001; Li and Baker, 2001; Tsuda et al, 2002; Kenyon et al, 2003; Baonza and Freeman, 2005) under the influence of EGFR signaling and provides flexibility in its regulation.
Figure 8.
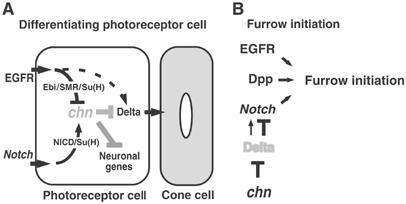
Models for chn function during photoreceptor cell development. (A) chn is not expressed in photoreceptor cells because of repression by Ebi/SMRTER(SMR)/Su(H), which antagonizes activation by NICD/Su(H). In the absence of Chn, Dl is derepressed and induces cone cell differentiation. Elevation of EGFR signaling causes ectopic expression of Dl (Tsuda et al, 2002), suggesting that EGFR signaling activates Dl expression independently of Ebi/SMR/Su(H) activity (dotted line). (B) At the initiation of the MF, Dl expression seems to be kept at a low level by chn function. Elevation of Dl in the chn mutant might downregulate Notch, which is required for the initiation of the MF (Kumar and Moses, 2001).
Although chn is expressed in the MF (Figure 2), our genetic analyses show that small clones of chn mutant cells permit progression of the MF and photoreceptor differentiation (data not shown). We speculate that the repressive effect of Chn is overcome by other signals in the MF, such as hedgehog signaling, which strongly induces Dl (Greenwood and Struhl, 1999).
chn and the crosstalk between EGFR and Notch signaling
Developing photoreceptor cells are exposed to the EGFR ligand, Spitz, and the Notch ligand, Dl, and each cell must assess the level of the two signals and respond appropriately to perform each task of photoreceptor cell specification and induction of non-neural cone cells. We investigated this question by studying the expression of Dl in photoreceptor cells. We identified chn as a direct target of Ebi/SMRTER-dependent transcriptional repression and as a repressor of Dl expression. The abrogated expression of Dl in ebi mutants was recovered by reducing one copy of chn (Figure 6K), suggesting that the negative regulation of chn by ebi is indeed prerequisite for photoreceptor cell development.
Our genetic data suggest that Su(H) may activate or repress chn expression (Figure 2). This idea is supported by data showing that Ebi/SMRTER and NICD are recruited to the promoter region of chn (Figure 4A). The Ebi/SMRTER complex formed in this region did not contain any detectable level of NICD, suggesting that the binding of Ebi/SMRTER and NICD to this region may be mutually exclusive (Figure 4B), and therefore we expect that a regulatory system controls the balance between the active and repressive states of Su(H) (Figure 8A). Taken together, these results suggest that chn is a key factor in the crosstalk between two major signal transduction pathways: the EGFR-dependent pathway and the Notch/Delta-dependent pathway (Figure 8A).
In the mammalian system, competition between SMRT and NICD for interaction with RBPJκ determines the state of RBPJκ-dependent transcriptional activity (Kao et al, 1998). Extracellular signaling may modulate this competition, as diverse signaling pathways modulate the functions of N-CoR/SMRT (Lavinsky et al, 1998). Our findings would prompt investigations of potential interaction of two repression systems of NRSF/REST and N-CoR/SMRT, and their regulation by Notch and EGF signaling in mammalian neuronal differentiation.
Materials and methods
Drosophila stocks
The following stocks were used in this study: Oregon-R, GMR-ebiDN, ebiE4, ebip7 and ebil11 (Dong et al, 1999; Tsuda et al, 2002); Su(H)2 and (Su(H)1 (Schweisguth and Posakony, 1992); tkv4 (Burke and Basler, 1996); ey-FLP (Newsome et al, 2000); da-Gal4 (Nakao and Campos-Ortega, 1996). GMR-Gal4 (Hay et al, 1994) was kindly provided by Dr M Yamaguchi (Kyoto Institute of Technology). cycEAR95FRT40A/CyO and PCNA775FRT42D/CyO were kindly provided by Dr CH Lee (NIH).
Genetics
Genetic modifiers of ebiDN-induced rough eye phenotype were identified by mating males of GS insertion lines with GMR-Gal4; GMR-ebiDN/CyO females. From the progeny of ∼6500 crosses, we identified 17 GS lines in which overexpression-based rough eye phenotype was altered by ebiDN.
Ethylmethanesulfonate (EMS)-induced chn mutations were isolated by reversion of lethality due to chn overexpression. chnGS2112 males treated with EMS were crossed to da-Gal4 females, and four revertant lines were established. The mutated site in chn1 was determined by RT–PCR and sequencing, which identified a small deletion between 280 and 296 bp that caused a frame shift mutation. chn9 is a genetically strong hypomorphic allele.
A large clone of chn or tkv was induced as previously described (Clandinin et al, 2001). Eye-specific mosaic flies were generated by FLP/FRT-induced sister chromatid recombination using ey-FLP and a recessive cell-lethal mutation (cycEAR95 or PCNA775) to eliminate the twin-spot (Stowers and Schwarz, 1999; Newsome et al, 2000).
Molecular biology and biochemistry
A 3.3-kb chn coding region was isolated using PCR from a 0–4 h embryonic cDNA library (Brown and Kafatos, 1988) and cloned into pUAST. The Chn zinc-finger domain (residues 297–610) was also amplified by PCR and cloned into pGEX4T (Amersham Biosciences), and the GST fusion protein was isolated using the standard protocol. The Su(H) DNA-binding domain (residues 126–528) was also cloned into pET28(a+) (Novagen), and the His-Su(H) fusion protein was isolated and used for EMSA. Full length (chn; 1–3327 bp), N-terminal (chn-N; 1–1962 bp) and C-terminal (chn-C; 1963–3327 bp) regions of Chn with flag epitope were amplified by PCR and cloned into pUAST for transfection into S2 cells. Immunoprecipitation was performed essentially as described (Lim et al, 2000). S2 cell transfection experiments were performed as the manufacturer's protocol (Qiagen).
Oligonucleotides
For the Chn binding assay, we chemically synthesized partially double-stranded 31-bp oligonucleotides containing a 21-bp NRSE (or NRSE-like) region and 5 bp each of terminal sequences. This double-stranded oligonucleotide was labeled with Cy5-dCTP and Klenow fragment of DNA polymerase I. For the Su(H) binding assay, we used 32P-labeled double-stranded 31-bp oligonucleotides containing 7-bp Su(H) recognition sequences (or alternative forms) and a 24-bp linker sequence. Oligonucleotide sequences are presented in Supplementary data.
Antibodies
The following antibodies were used: rabbit anti-Ebi (Dong et al, 1999), rabbit anti-SMRTER (a gift from Dr R Evans, Salk Institute), mouse anti-NICD (Developmental Studies Hybridoma Bank (DSHB), University of Iowa), mouse anti-Dl (Parks et al, 1995), rat anti-Elav (O'Neill et al, 1994) and mouse anti-Cut (DSHB). Secondary antibodies were conjugated with Alexa488 or Cy3 (Jackson Labs). Polyclonal anti-Chn was raised (MBL Ltd) by injecting rabbits with purified Chn zinc-finger domain (residues 297–610) fused with GST.
In situ hybridization and immunocytochemistry
In situ hybridization on whole-mount Drosophila eye discs was performed according to Tautz and Pfeifle (1989). Single-stranded antisense digoxigenin-containing RNA probes were prepared using the Genius kit (Boehringer Mannheim). Immunocytochemistry on imaginal discs was performed essentially as described (Majumdar et al, 1997).
EMSA
In vitro DNA and protein interactions were detected by EMSA. Up to 1 μg purified GST-fusion protein was incubated on ice for 2 min in 20 μl of buffer (10 mM HEPES pH 7.6, 1 mM EDTA, 1 mM DDT, 32 mM KCl, 10% (v/v) glycerol containing 5% Ficoll 400 (Sigma), 500 ng/ml poly(dI-dC) and 1 μg of sonicated calf thymus DNA (average size, 0.2 kb). When necessary, unlabeled oligonucleotide competitors were added at this step. Then, double-stranded Cy5-labeled synthetic oligonucleotide (50 pg) was added, and the mixture was incubated for 10 min on ice. DNA–protein complexes were resolved on a 5% polyacrylamide gel in TAE at 4°C. Signals were detected by a fluorescent gel scanner (Typhoon, Amersham Biosciences). In the case of Su(H), we used 32P-labeled oligonucleotide (50 pg) probe and electrophoresis in a 1% agarose gel in TAE at 4°C. Signals were detected by autoradiography. CBEs listed in Table I were tested for binding to Chn by performing EMSA using Cy5-labeled probes under a condition without nonspecific competitors. Those probes that showed association to Chn were further verified by EMSA competition assays in the standard condition.
ChIP analysis
Third instar larval eye discs (200) of wild type (Oregon R) or pGMR-Gal4; UAS-chn were dissected in PBS from the eye–antenna complex, fixed with 1% formaldehyde for 15 min at room temperature and treated as described (Hecht and Grunstein, 1999). Crosslinked adducts were resuspended and sonicated, resulting in DNA fragments of 500–1000 bp. Immunoprecipitation was performed using the antibodies described above. Protein-bound, immunoprecipitated DNA was dissolved in TE buffer and incubated at 65°C for 6 h. Digestion buffer (10 mM Tris–HCl, 100 mM NaCl, 25 mM EDTA, pH 8.0) was added to the sample and incubated for 1 h at 45°C with 0.1 mg/ml proteinase K (Sigma). DNA was purified using a Qiagen column (Qiagen Ltd) and used as template for 40 cycles of PCR.
Sequential immunoprecipitation was performed by eluting the antibody–chromatin–DNA complex from the protein-A column using DTT treatment before the addition of the second antibody.
mRNA quantification
Poly A+ mRNA, isolated from 20 eye-antennal discs of third instar larvae using an mRNA purification kit (Amersham Biosciences), was reverse transcribed with SuperScript II (Invitrogen) reverse transcriptase using oligo-dT as a primer. mRNA was quantified using a Smert Cycler (TAKARA) with SYBR Green PCR Master mix (TOYOBO). Data obtained from duplicate mRNA preparations were standardized using elav mRNA as a control.
Supplementary Material
Supplementary Figure 1
Supplementary Materials
Acknowledgments
We thank Masamitsu Yamaguchi, Chihon Lee and the Bloomington Stock Center for fly stocks, and Housei Wada for germline transformation. We also thank Hitoshi Ueda, Fumiko Hirose and Kenichi Takeyama for technical advice. For genetic screening support, we acknowledge Masayo Shindo, Tadashi Sakata, Ai Akimoto, Yoshihiko Umesono, Yukiko Sado and Misako Taniguchi. We acknowledge all the members of our laboratory for helpful discussions. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas (B) from the Ministry of Education, Culture, Sports, Science and Technology of Japan to LT and SH.
References
- Andres ME, Burger C, Peral-Rubio MJ, Battaglioli E, Anderson ME, Grimes J, Dallman J, Ballas N, Mandel G (1999) CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc Natl Acad Sci USA 96: 9873–9878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baonza A, Freeman M (2001) Notch signaling and the initiation of neural development in the Drosophila eye. Development 128: 3889–3898 [DOI] [PubMed] [Google Scholar]
- Baonza A, Freeman M (2005) Control of cell proliferation in the Drosophila eye by Notch Signaling. Dev Cell 8: 529–539 [DOI] [PubMed] [Google Scholar]
- Barolo S, Walker RG, Polyanovsky AD, Freschi G, Keil T, Posakony JW (2000) A notch-independent activity of suppressor of hairless is required for normal mechanoreceptor physiology. Cell 103: 957–969 [DOI] [PubMed] [Google Scholar]
- Bassi MT, Ramesar RS, Caciotti B, Winship IM, De Grandi A, Riboni M, Townes PL, Beighton P, Ballabio A, Borsani G (1999) X-linked late-onset sensorineural deafness caused by a deletion involving OA1 and a novel gene containing WD-40 repeats. Am J Hum Genet 64: 1604–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borod ER, Heberlein U (1998) Mutual regulation of decapentaplegic and hedgehog during the Initiation of differentiation in the Drosophila retina. Dev Biol 197: 187–197 [DOI] [PubMed] [Google Scholar]
- Brown NH, Kafatos FC (1988) Functional cDNA libraries from Drosophila embryos. J Mol Biol 203: 425–437 [DOI] [PubMed] [Google Scholar]
- Bruce AW, Donaldson IJ, Wood IC, Yerbury SA, Sadowski MI, Chapman M, Gottgens B, Buckley NJ (2004) Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc Natl Acad Sci USA 101: 10458–10463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke R, Basler K (1996) Hedgehog-dependent patterning in the Drosophila eye can occur in the absence of Dpp signaling. Dev Biol 179: 360–368 [DOI] [PubMed] [Google Scholar]
- Chen JD, Evans RM (1995) A transcriptional corepressor that interacts with nuclear hormone receptors. Nature 377: 454–457 [DOI] [PubMed] [Google Scholar]
- Chen ZF, Paquette AJ, Anderson DJ (1998) NRSF/REST is required for repression of multiple neuronal target genes during embryogenesis. Nat Genet 20: 136–142 [DOI] [PubMed] [Google Scholar]
- Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G (1995) REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell 80: 949–957 [DOI] [PubMed] [Google Scholar]
- Clandinin TR, Lee CH, Herman T, Lee RC, Yang AY, Ovasapyan S, Zipursky SL (2001) Drosophila LAR regulates R1–R6 and R7 target specificity in the visual system. Neuron 32: 237–248 [DOI] [PubMed] [Google Scholar]
- Cooper MY, Bray SJ (1999) Frizzled regulation of Notch signalling polarizes cell fate in the Drosophila eye. Nature 397: 526–530 [DOI] [PubMed] [Google Scholar]
- Dallman JE, Allopenna J, Bassett A, Travers A, Mandel G (2004) A conserved role but different partners for the transcriptional corepressor CoREST in fly and mammalian nervous system formation. J Neurosci 24: 7186–7193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Tsuda L, Zavitz KH, Lin M, Li S, Carthew RW, Zipursky SL (1999) ebi regulates epidermal growth factor receptor signaling pathways in Drosophila. Genes Dev 13: 954–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero LM, Caminero E, Schulze KL, Bellen H, Modolell J (2005) Charlatan, a Zn-finger transcription factor, establishes a novel of regulation of the proneural achaete/scute genes of Drosophila. Development 132: 1211–1222 [DOI] [PubMed] [Google Scholar]
- Flores GV, Duan H, Yan H, Nagaraj R, Fu W, Zou Y, Noll M, Banerjee U (2000) Combinatorial signaling in the specification of unique cell fates. Cell 103: 75–85 [DOI] [PubMed] [Google Scholar]
- Greenwood S, Struhl G (1999) Progression of the morphogenetic furrow in the Drosophila eye: the roles of Hedgehog, Decapentaplegic and the Raf pathway. Development 126: 5795–5808 [DOI] [PubMed] [Google Scholar]
- Guenther MG, Lane WS, Fischle W, Verdin E, Lazar MA, Shiekhattar R (2000) A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev 14: 1048–1057 [PMC free article] [PubMed] [Google Scholar]
- Hay BA, Wolff T, Rubin GM (1994) Expression of baculovirus P35 prevents cell death in Drosophila. Development 120: 2121–2129 [DOI] [PubMed] [Google Scholar]
- Hecht A, Grunstein M (1999) Mapping DNA interaction sites of chromosomal proteins using immunoprecipitation and polymerase chain reaction. Methods Enzymol 304: 399–414 [DOI] [PubMed] [Google Scholar]
- Hermanson O, Jepsen K, Rosenfeld MG (2002) N-CoR controls differentiation of neural stem cells into astrocytes. Nature 419: 934–939 [DOI] [PubMed] [Google Scholar]
- Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK, Rosenfeld MG (1995) Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor corepressor. Nature 377: 397–404 [DOI] [PubMed] [Google Scholar]
- Hummel T, Attix S, Gunning D, Zipursky SL (2002) Temporal control of glial migration in the Drosophila eye requires gilgamesh, hedgehog, and eye specification genes. Neuron 33: 193–203 [DOI] [PubMed] [Google Scholar]
- Jacobsen TL, Brennan K, Arias AM, Muskavitch MA (1998) Cis-interactions between Delta and Notch modulate neurogenic signalling in Drosophila. Development 125: 4531–4540 [DOI] [PubMed] [Google Scholar]
- Jepsen K, Rosenfeld MG (2002) Biological roles and mechanistic actions of corepressor complexes. J Cell Sci 115: 689–698 [DOI] [PubMed] [Google Scholar]
- Jones FS, Meech R (1999) Knockout of REST/NRSF shows that the protein is a potent repressor of neuronally expressed genes in non-neural tissues. BioEssays 21: 372–376 [DOI] [PubMed] [Google Scholar]
- Kania A, Salzberg A, Bhat M, D'Evelyn D, He Y, Kiss I, Bellen HJ (1995) P-element mutations affecting embryonic peripheral nervous system development in Drosophila melanogaster. Genetics 139: 1663–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannabiran C, Zeng X, Vales LD (1997) The mammalian transcriptional repressor RBP (CBF1) regulates interleukin-6 gene expression. Mol Cell Biol 17: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao HY, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner CR, Evans RM, Kadesch T (1998) A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev 12: 2269–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon KL, Ranade SS, Curtiss J, Mlodzik M, Pignoni F (2003) Coordinating proliferation and tissue specification to promote regional identity in the Drosophila head. Dev Cell 5: 403–414 [DOI] [PubMed] [Google Scholar]
- Kumar JP, Moses K (2001) The EGF receptor and notch signaling pathways control the initiation of the morphogenetic furrow during Drosophila eye development. Development 128: 2689–2697 [DOI] [PubMed] [Google Scholar]
- Lai EC (2004) Notch signaling: control of cell communication and cell fate. Development 131: 965–973 [DOI] [PubMed] [Google Scholar]
- Lavinsky RM, Jepsen K, Heinzel T, Torchia J, Mulien TM, Schiff R, Del-Rio AL, Ricote M, Ngo S, Gemsch J, Hilsenbeck SG, Osborne CK, Glass CK, Rosenfeld MG, Rose DW (1998) Diverse signaling pathways modulate nuclear receptor recruitment of N-CoR and SMRT complexes. Proc Natl Acad Sci USA 95: 2920–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Pfaff SL (2001) Transcriptional networks regulating neuronal identity in the developing spinal cord. Nat Neurosci 4: 1183–1191 [DOI] [PubMed] [Google Scholar]
- Li Y, Baker NE (2001) Proneural enhancement by Notch overcomes Suppressor-of-Hairless repressor function in the developing Drosophila eye. Curr Biol 11: 330–338 [DOI] [PubMed] [Google Scholar]
- Lim YM, Wong S, Lau G, Witte ON, Colicelli J (2000) BCR/ABL inhibition by an escort/phosphatase fusion protein. Proc Natl Acad Sci USA 97: 12233–12238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze SH, Chenoweth J, Schwartz P, Pevzner PA, Glass C, Mandel G et al. (2002) Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science 298: 1747–1752 [DOI] [PubMed] [Google Scholar]
- Majumdar A, Nagaraj R, Banerjee U (1997) strawberry notch encodes a conserved nuclear protein that functions downstream of Notch and regulates gene expression along the developing wing margin of Drosophila. Genes Dev 11: 1341–1353 [DOI] [PubMed] [Google Scholar]
- Nakao K, Campos-Ortega JA (1996) Persistent expression of genes of the enhancer of split complex suppresses neural development in Drosophila. Neuron 16: 275–286 [DOI] [PubMed] [Google Scholar]
- Nellesen DT, Lai EC, Posakony JW (1999) Discrete enhancer elements mediate selective responsiveness of Enhancer of split Complex genes to common transcriptional activators. Dev Biol 213: 33–53 [DOI] [PubMed] [Google Scholar]
- Newsome TP, Asling B, Dickson BJ (2000) Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development 127: 851–860 [DOI] [PubMed] [Google Scholar]
- O'Neill EM, Rebay I, Tjian R, Rubin GM (1994) The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell 78: 137–147 [DOI] [PubMed] [Google Scholar]
- Oswald F, Liptay S, Adler G, Schmid RM (1998) NF-kappaB2 is a putative target gene of activated Notch-1 via RBP-Jkappa. Mol Cell Biol 18: 2077–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks AL, Turner FR, Muskavitch MA (1995) Relationships between complex Delta expression and the specification of retinal cell fates during Drosophila eye development. Mech Dev 50: 201–216 [DOI] [PubMed] [Google Scholar]
- Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG (2004) A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell 116: 511–525 [DOI] [PubMed] [Google Scholar]
- Reeves N, Posakony JW (2005) Genetic programs activated by proneural proteins in the developing Drosophila PNS. Dev Cell 8: 413–425 [DOI] [PubMed] [Google Scholar]
- Sawatsubashi S, Maki A, Ito S, Shirode Y, Suzuki E, Zhao Y, Yamagata K, Kouzmenko A, Takeyama K, Kato S (2004) Ecdysone receptor-dependent gene regulation mediates histone poly(ADP-ribosyl)ation. Biochem Biopys Res Commun 320: 268–272 [DOI] [PubMed] [Google Scholar]
- Schoenherr CJ, Anderson DJ (1995) The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science 267: 1360–1363 [DOI] [PubMed] [Google Scholar]
- Schoenherr CJ, Paquette AJ, Anderson DJ (1996) Identification of potential target genes for the neuron-restrictive silencer factor. Proc Natl Acad Sci USA 93: 9881–9886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweisguth F, Posakony JW (1992) Suppressor of Hairless, the Drosophila homologue of the mouse recombination signal-binding protein gene, controls sensory organ cell fates. Cell 69: 1199–1212 [DOI] [PubMed] [Google Scholar]
- Stern CD (2001) Initial patterning of the central nervous system: how many organizers? Nat Rev Neurosci 2: 92–98 [DOI] [PubMed] [Google Scholar]
- Stowers RS, Schwarz TL (1999) A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics 152: 1631–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G, Adachi A (1998) Nuclear access and action of Notch in vivo. Cell 93: 649–660 [DOI] [PubMed] [Google Scholar]
- Tapia-Ramirez J, Eggen BJL, Peral-Rubio MJ, Toledo-Aral JJ, Mandel G (1997) A single zinc finger motif in the silencing factor REST represses the neural-specific type II sodium channel promoter. Proc Natl Acad Sci USA 94: 1177–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C (1989) A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98: 81–85 [DOI] [PubMed] [Google Scholar]
- Toba G, Ohsako T, Miyata N, Ohtsuka T, Seong KH, Aigaki T (1999) The gene search system. A method for efficient detection and rapid molecular identification of genes in Drosophila melanogaster. Genetics 151: 725–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CC, Kao HY, Yao TP, McKeown M, Evans RM (1999) SMRTER, a Drosophila nuclear receptor coregulator, reveals that EcR-mediated repression is critical for development. Mol Cell 4: 175–186 [DOI] [PubMed] [Google Scholar]
- Tsuda L, Nagaraj R, Zipursky SL, Banerjee U (2002) An EGFR/Ebi/Sno pathway promotes Delta expression by inactivating Su(H)/SMRTER repression during inductive Notch signaling. Cell 110: 625–637 [DOI] [PubMed] [Google Scholar]
- Yoon HG, Chan DW, Huang ZQ, Li J, Fondell JD, Qin J, Wong J (2003) Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J 22: 1336–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Materials


