Abstract
The usability of the DNA microarray format for the specific detection of bacteria based on their 16S rRNA genes was systematically evaluated with a model system composed of six environmental strains and 20 oligonucleotide probes. Parameters such as secondary structures of the target molecules and steric hindrance were investigated to better understand the mechanisms underlying a microarray hybridization reaction, with focus on their influence on the specificity of hybridization. With adequate hybridization conditions, false-positive signals could be almost completely prevented, resulting in clear data interpretation. Among 199 potential nonspecific hybridization events, only 1 false-positive signal was observed, whereas false-negative results were more common (17 of 41). Subsequent parameter analysis revealed that this was mainly an effect of reduced accessibility of probe binding sites caused by the secondary structures of the target molecules. False-negative results could be prevented and the overall signal intensities could be adjusted by introducing a new optimization strategy called directed application of capture oligonucleotides. The small number of false-positive signals in our data set is discussed, and a general optimization approach is suggested. Our results show that, compared to standard hybridization formats such as fluorescence in situ hybridization, a large number of oligonucleotide probes with different characteristics can be applied in parallel in a highly specific way without extensive experimental effort.
During the last decade, molecular methods based on comparative analysis of 16S rRNA sequences have yielded new and unexpected insights into the diversity of microbial communities (13, 19, 48). Although diversity can be readily studied by PCR-based 16S rRNA gene libraries, with this approach it is not possible to deduce the composition of the analyzed communities quantitatively (2). With the introduction of fluorescence in situ hybridization (FISH), it first became possible to identify single bacterial cells with labeled oligonucleotide probes targeting the rRNA of selected phylogenetic groups. This allows the quantitative analysis of the spatiotemporal composition of microbial communities (3).
A drawback of the FISH method is the limited number of probes which can be applied in one hybridization experiment. This limitation becomes a distinct bottleneck when this technique is used for community analysis on a high level of phylogenetic resolution. Additionally, there is a need for multiple probes to check for false-positive and false-negative results caused by individual probes used for identification of selected target organisms (2). A highly parallel application of multiple probe sets is facilitated by DNA chips or DNA microarrays. In this reverse hybridization format, matrix-immobilized oligo- or polynucleotides (probes) are used for the specific capture of labeled target molecules. This, in principle, allows the simultaneous application of a nearly unlimited number of probes in a single hybridization experiment (23, 26).
The concept of DNA microarray hybridization was introduced more than a decade ago (5, 14, 29, 31, 45), and the technique has been applied for many years in the pharmaceutical industry (12), clinical diagnostics (25), and many fields of research, e.g., functional genomics (28) and genetic analysis (22). In contrast, DNA microarrays are still not common in microbial ecology. Only a limited number of reports have been published, which mainly showed the “proof of principle” of the method in this field of research (24, 41, 43). What are the reasons? In contrast to “standard” applications in the medical field, in microbial ecology defined nucleic acids have to be identified against an often large and partly unknown genetic background. For this kind of analysis, the discrimination of single mismatches is crucial (27), but the specific hybridization of target molecules to immobilized capture oligonucleotides is one of the major challenges of the DNA microarray approach because large sets of probes with different characteristics are applied under identical hybridization conditions.
Several approaches exist to overcome this problem. An example is the addition of tetramethylammonium chloride (32) or betaine (39) to the hybridization buffer. Both compounds equalize the melting points of different oligonucleotides by stabilization of AT base pairs. Another strategy is based on the acquisition of complete melting curves for every single probe spotted on the chip surface (27), which is a promising but methodically and technically demanding approach. Further methodological investigations and optimizations are needed before DNA microarrays can be used for the fast, simple, and efficient screening of microbial communities. Currently, the usefulness of standard microarray formats is often limited by hard to interpret signal patterns caused by an accumulated number of false-positive signals.
In 1994, Williams et al. (49) pointed out the complexity of a microarray hybridization event and the shortcoming of the existing rules to describe it. In recent years, several systematic studies were conducted on different aspects, such as the secondary structures of target molecules and steric hindrance mediated by the solid support (8, 22, 33, 40, 42, 49), but the complex interaction of all these mechanisms and their influence on hybridization specificity are still mostly unknown. In this study, we systematically evaluated the suitability of the microarray format for 16S rRNA-based detection of bacteria by analyzing a model system including single-mismatch controls. Our objective was to get new insights into the mechanisms underlying a microarray hybridization reaction for the deduction of novel optimization strategies leading to signal patterns with a reduced number of false-positive and false-negative results.
MATERIALS AND METHODS
Bacterial strains.
For the analysis of 16S rRNA gene (16S rDNA) amplicons by DNA microarray hybridization, six bacterial strains (KT0202a, KT1117, JP7.1, JP13.1, KT11ds2, and KT71) were randomly selected from a pool of environmental strains isolated from the surface water of the German Bight by Eilers et al. (15, 16). The accession numbers of the corresponding sequences are AF173971, AF235111, AF305498, AY007676, AY007679, and AY007680.
Preparation of fluorescently labeled target single-stranded DNA.
Labeled target molecules were prepared by PCR amplification of the nearly complete 16S rRNA genes of the reference strains with the general bacterial primers 8F and 1492R (11). Living cells were picked from culture plates, resuspended in 65 μl of PCR-water (Sigma, Deisenhofen, Germany) and treated three times by freeze-thawing in liquid nitrogen for cell lysis. For PCR amplification, 10× PCR buffer (Eppendorf, Hamburg, Germany), 30 μg of bovine serum albumin, deoxyribonucleoside triphosphates (final concentration, 250 μM each), primers (final concentration, 150 nM each), and 2.5 U of MasterTaq polymerase (Eppendorf, Hamburg, Germany) were added to a final volume of 100 μl.
For subsequent preparation of fluorescently labeled, single-stranded DNA targets, 5′-indocarbocyanine-labeled forward primer 8f and 5′-biotin-labeled reverse primer 1492R were applied, both purchased from Thermo Hybaid (Interactiva Division, Ulm, Germany). Amplification was done according to the protocol of Buchholz-Cleven et al. (11) except that a temperature of 48°C was used for annealing. Successful amplification was confirmed by analyzing PCR products on 1.5% agarose gels. PCR products were purified with the QIAquick PCR purification kit (Qiagen, Hilden, Germany). Subsequently, the biotin-labeled DNA strands were extracted with streptavidin-coated paramagnetic beads (Boehringer, Mannheim, Germany) as described by Niemeyer et al. (36). The remaining, fluorescently labeled single-stranded DNAs are equivalent to the 16S rRNAs of the corresponding strains. Finally, the concentration of the amplicons was determined by UV spectrometry. The labeled single-stranded DNA was stored at −18°C.
Oligonucleotide probe set.
Twenty oligonucleotide probes of 15 to 20 nucleotides in length, targeting the bacterial 16S rRNA and originally designed for FISH, were selected from the literature for reliable differentiation of the six model strains based on the multiple-probe approach (4). The probes and their characteristics are listed in Table 1. One of the probes (KT13) was designed within this study with the software package ARB (http://www.arb-home.de). Current specificity of all probes was evaluated by ARB with the rRNA database of the Technical University of Munich (http://www.arb-home.de, release 08/01). For all 20 probes, a control containing one single central mismatch based on a transversion was included.
TABLE 1.
Oligonucleotide probes used in this study and their specificitiesa
| Probe | Specificity | Sequence (5′-3′) | 16S rRNA binding siteb and length (nt) | GC content (%) | No. of mismatches to targetc
|
Reference | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| KT0202a | KT1117 | JP7.1 | JP13.1 | KT11ds2 | KT71 | ||||||
| UNIV 1392 | Universal | ACGGGCGGTGTGTAC | 1,392 (15) | 67 | PM | PM | PM | PM | PM | PM | 37 |
| EUB338 | Bacteria | GCTGCCTCCCGTAGGAGT | 338 (18) | 67 | PM | PM | PM | PM | PM | PM | 3 |
| CF319a | Cytophaga-Flavobacterium group of Bacteroidetes | TGGTCCGTGTCTCAGTAC | 319 (18) | 56 | 2 (4,17) | 2 (4,17) | 2 (4,17) | 2 (4,17) | PM | 3 | 30 |
| CYT1448 | KT11ds2 | CTAGGCCGCTCCTTACGG | 1,448 (18) | 67 | >4 | >4 | >4 | >4 | PM | >4 | 16 |
| NOR5-730 | NOR5 cluster of gamma-proteobacteria | TCGAGCCAGGAGGCCGCC | 730 (18) | 78 | 3 | 3 | 2 (10,12) | >4 | >4 | PM | 16 |
| NOR5-130 | KT71 | CCCCACTACTGGATAGAT | 130 (18) | 50 | >4 | >4 | >4 | >4 | >4 | PM | 16 |
| ALF968 | Alpha-proteobacteria | GGTAAGGTTCTGCGCGTT | 968 (18) | 56 | PM | PM | PM | PM | 3 | 1 (12) | 34 |
| MALF-1 | Marine alpha cluster | GCCGGGGTTTCTTTACCA | 488 (18) | 56 | PM | PM | PM | >4 | >4 | >4 | 21 |
| ROS537 | Marine alpha cluster | CAACGCTAACCCCCTCC | 537 (17) | 65 | PM | PM | PM | 2 (9,11) | 4 | 4 | 16 |
| GRb | Marine alpha cluster | GTCAGTATCGAGCCAGTGAG | 735 (20) | 55 | PM | PM | 1 (18) | >4 | >4 | 4 | 20 |
| RSB67 | Subgroup of marine alpha cluster | CGCTCCACCCGAAGGTAG | 67 (18) | 67 | PM | >4 | >4 | >4 | >4 | >4 | 50 |
| RC1031 | KT0202a | ACCTGTCACTATGTCCCG | 1,031 (18) | 56 | PM | >4 | 4 | 2 (12,18) | >4 | 4 | 16 |
| RC1239 | KT0202a | TAACTCACTGTAGTTGCCAT | 1,239 (20) | 40 | PM | 1 (14) | 1 (14) | 4 | >4 | >4 | 16 |
| KT13 | Subgroup of marine alpha cluster | TAACTCACTGTAGATGCCAT | 1,239 (20) | 40 | 1 (14) | PM | PM | 3 | >4 | >4 | This study |
| KT13-231 | KT1117 | ATCTAATCAAACGCGGGCC | 231 (19) | 53 | 1 (9) | PM | 1 (9) | 1 (9) | >4 | 4 | 16 |
| Ros7-1029 | JP7.1 | CTGTCACTTGGTCTCTTG | 1,029 (18) | 50 | >4 | >4 | PM | >4 | >4 | >4 | 16 |
| RRP1088 | Subgroup of alpha-proteobacteria | CGTTGCCGGACTTAACC | 1,088 (17) | 59 | PM | PM | PM | 1 (7) | 3 | 2 (5,7) | 34 |
| PAR1457 | Subgroup of alpha-proteobacteria | CTACCGTGGTCCGCTGCC | 1,457 (18) | 72 | PM | 3 | 1 (2) | 3 | >4 | >4 | 34 |
| ALF4-1322 | Subgroup of alpha-proteobacteria | TCCGCCTTCATGCTCTCG | 1,322 (18) | 61 | 4 | 4 | 4 | PM | >4 | >4 | 34 |
| EP129 | Subgroup of alpha-proteobacteria | CGAACCTAAAGGCAGGTT | 129 (18) | 50 | >4 | >4 | >4 | PM | >4 | >4 | 34 |
For every probe, a control containing one central mismatch was spotted additionally, i.e., the whole probe set includes a total of 40 different oligonucleotides.
E. coli numbering according to Brosius et al. (10); position of 3′ nucleotide of probe is stated. nt, nucleotide.
For up to two mismatches, the position of the mismatches is also stated (numbers in parentheses counted from the 3′ end). PM, perfect match.
Preparation of glass slides and spotting.
The DNA microarray format used in this study is based on standard microscopic glass slides (Menzel, Braunschweig, Germany). Slides were activated by 1,4-phenylenediisothiocyanate treatment for covalent binding of either 5′- or 3′-amino-modified capture oligonucleotides (Thermo Hybaid, Ulm, Germany) according to Benters et al. (7).
Probes were spotted onto the activated slide surface with the piezo-driven spotting device Robodrop (BIAS, Bremen, Germany). The concentration of the amino-modified oligonucleotides in PCR-water was 10 μM with 1% glycerol. Volumes of deposited probe solutions were about 250 pl, resulting in spots with a diameter of approximately 200 μm.
To complete covalent binding, after being spotted with probe solutions, slides were incubated overnight at room temperature in a wet chamber to restrict evaporation of the spots. Blocking of the slides was performed in 6-amino-1-hexanol (50 mM) and diisopropylethylamine (150 mM) in dimethylformamide as described by Beier et al. (6). Finally, the slides were washed with deionized, particle-free water, air dried, and stored under nitrogen at 4°C.
DNA microarray hybridization.
For hybridization and washing of the microarrays, a standard FISH protocol according to Pernthaler et al. (38) was used. Hybridization time was extended from 1.5 to 3 h. Incubation was performed either at room temperature, at 46°C without formamide, or at 46°C with 20% formamide. In all cases, 3.5 pmol of labeled target single-stranded DNA in a total volume of 200 μl of hybridization buffer was applied to the microarrays. To guarantee a uniform moistening of the slide surface, the sample was covered with a coverslip. Unless otherwise specified, probes were applied for hybridization experiments in the standard way, i.e., all probes immobilized via their 5′ end, applied without spacer, and at the same concentration (10 μM).
To investigate the impact of secondary structures, unlabeled “helper” oligonucleotides (18), purchased from Thermo Hybaid (Ulm, Germany), were applied to the hybridization solution at a final concentration of 5 nM. For all probe binding sites analyzed by helper oligonucleotides, a pair of two perfectly matching 20-mer oligonucleotides was applied, one binding adjacent to the 5′ end of the corresponding probe and one to the 3′ end.
For reducing steric hindrance, capture probes carrying polyadenosine triphosphate spacers of 6, 12, 18, or 24 nucleotides, located at either the 5′ end or the 3′ end of the capture oligonucleotide, depending on selected probe orientation, were used.
Signal detection and data analysis.
Air-dried slides were imaged at a resolution of 10 μm with a GenePix4000 microarray scanner (Axon, Union City, Calif.) at the same laser power and sensitivity level of the photomultiplier for each slide. Therefore, absolute signal intensities (arbitrary units) presented for independent experiments should be directly comparable.
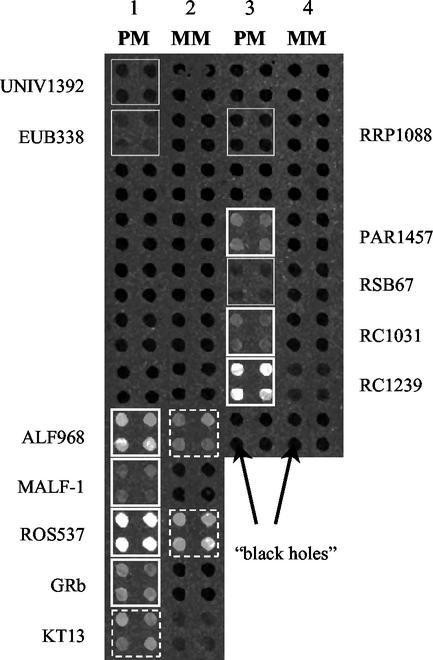
For automatic spot detection and signal quantification, the image analysis software MetaMorph Offline version 4.6 (Universal Imaging Corporation, West Chester, Pa.) was used. Filters for the automatic removal of spots with poor circularity or low uniformity of pixel intensities were established. Signals were considered positive if values were above zero after local background correction. This strategy for assessing the reliability of positive spots was possible because nonspecific binding of labeled target molecules to the slide surface was not observed for regions where oligonucleotides were deposited. This leads to “black holes” for spots where no detectable hybridization occurred, as shown in Fig. 6, and ultimately results in negative measurements for these spots, which were considered zero. We foresee no biases on individual spots by this method because in general we had homogeneous background noise. Otherwise, experiments were not considered.
FIG. 6.
Image of a scanned microarray taken from the experiment shown in Fig. 3A. Only four of the eight replicates spotted for each probe are shown (arranged in blocks). Columns 1 and 3 represent the 20 perfect-match probes, columns 2 and 4 show the corresponding mismatch controls. Highlighted spots are true-positive (solid line), false-positive (dashed line), or false-negative (dotted line) results.
All probes were deposited in eight replicate spots arranged in two blocks of four parallels. Each data point shown represents the arithmetic mean of the replicates analyzed for one probe. Error bars indicate the standard deviation for the analyzed replicates.
RESULTS
Specific detection of environmental bacterial strains with a redundant and hierarchically structured set of probes.
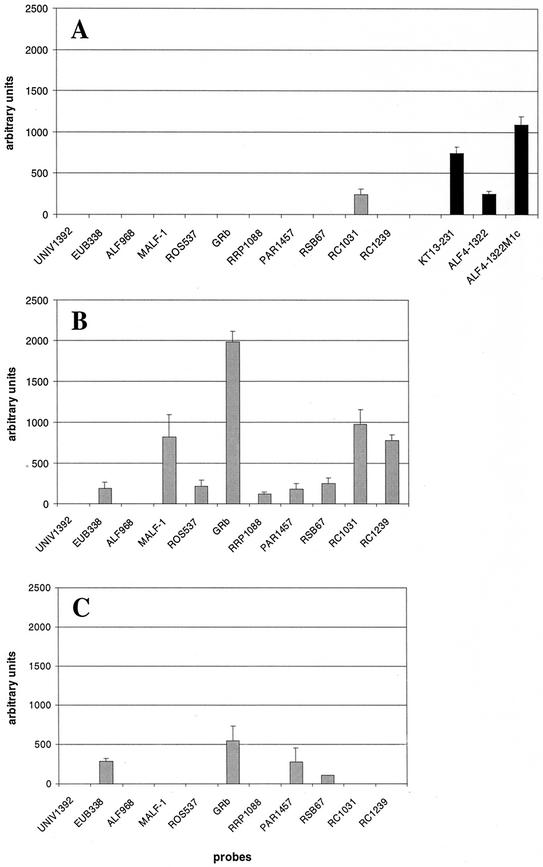
For optimization of global hybridization conditions, different combinations of temperature and formamide concentration were tested with strain KT0202a: hybridization and washing at room temperature without formamide (Fig. 1A), at 46°C without formamide (Fig. 1B), and at 46°C with 20% formamide (Fig. 1C). Hybridization at room temperature led only to one weak true-positive signal for probe RC1031. Additionally, nonspecific (false-positive) signals for probes KT13-231 (one mismatch), ALF4-1322 (four mismatches), and ALF4-1322 M1c (three mismatches) could be observed. After hybridization at 46°C without formamide, 9 of 11 probes targeting strain KT0202a showed a true-positive signal, and two spots were false-negative. Signal intensities ranged from 117 to 1,982 arbitrary units (a.u.) for probes RRP1088 and GRb, respectively. Hybridization at 46°C and 20% formamide led to four true-positive signals, ranging from 107 a.u. for probe RSb67 to 674 a.u. for probe GRb, and seven false-negative results. All further experiments were done at 46°C without formamide.
FIG. 1.
Signal patterns after microarray hybridization of amplified 16S rDNA of strain KT0202a to the complete set of probes shown in Table 1 at room temperature without formamide (A), at 46°C without formamide (B), and at 46°C with 20% formamide (C). Only probes targeting strain KT0202a are shown. Black bars indicate false-positive signals (controls are only shown if signals were detected).
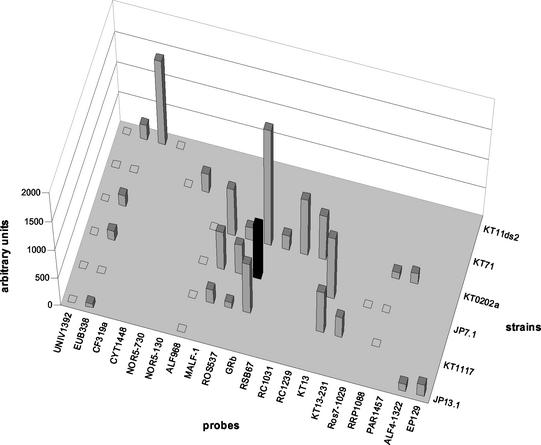
A comprehensive overview of the independent analysis of all six target strains is shown in Fig. 2. The isolates could be clearly differentiated by the signal patterns obtained. For all probes, false-positive signals could be almost completely prevented, even within the controls containing only one central mismatch. Among 199 potential nonspecific hybridization events (20 probes plus 20 controls times 6 targets minus 41 perfect-match situations), only one false-positive signal (0.5%) was observed for probe GRb in combination with target KT1117. However, 17 of 41 expected signals were missing (41.5%). For example, the universal probe UNIV1392 was false-negative with all six targets. Four of six signals for probe EUB338 were just slightly above the detection limit, and the other two were false-negative.
FIG. 2.
Comprehensive overview of signal patterns after microarray hybridization of all six strains analyzed in this study under standard conditions (at 46°C without formamide, all probes 5′ immobilized and applied without spacer). False-positive signals are indicated by black bars; false-negative signals are indicated by open rectangles. For clarity, the single-mismatch controls spotted for all 20 probes are not shown (here, no signals could be observed).
Furthermore, we observed profound differences in the signal intensities of different probes hybridized to the same target. For example, with probe GRb, strain KT0202a had a signal that was 17 times higher than that with probe RRP1088.
Analysis of parameters influencing a microarray hybridization reaction.
Strain KT0202a was used for all further systematic evaluations because it was targeted by the most probes of the probe set applied (compare Table 1).
(i) Secondary structures.
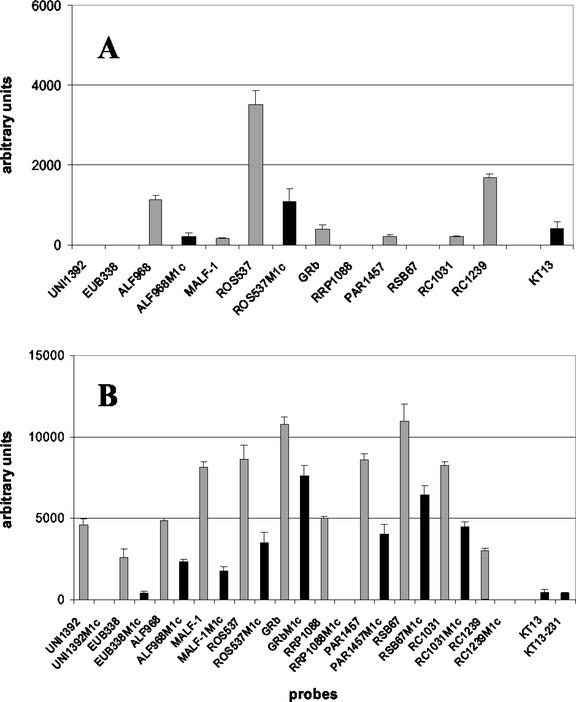
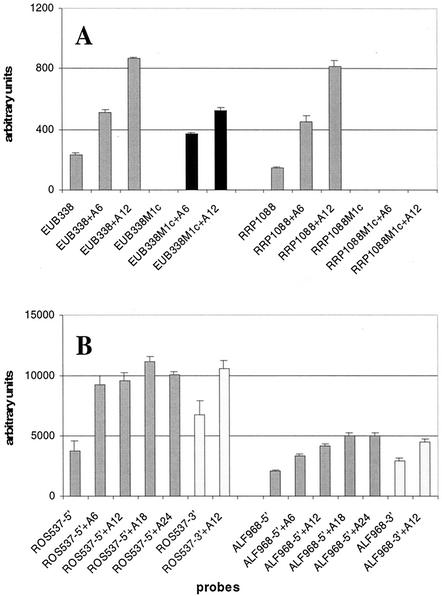
With pairs of helper oligonucleotides, the signals for probes ALF968, ROS537, and RC1239 could be specifically increased (Fig. 3A; see also Fig 6). Compared to the corresponding data generated without helper oligonucleotides (Fig. 1B), the probe ALF968 signal was raised from 0 to 1,126 a.u. The signal for probe ROS537 was increased 16-fold, from 219 to 3,511 a.u., whereas the signal of probe RC1239 only doubled from 775 to 1,670 a.u. Interestingly, application of helper pairs also resulted in signals within the mismatch controls ALF968M1c and ROS537M1c. Also probe KT13, which has the same binding site as probe RC1239 and shows only one central mismatch to strain KT0202a, could no longer be discriminated under these conditions. Signals of probes MALF-1, GRb, and RC1031 decreased from 819 to 160 a.u., from 1,982 to 394 a.u., and from 973 to 197 a.u., respectively, compared to the corresponding experiments without helpers (Fig. 1B).
FIG. 3.
Impact of secondary structures of target molecules on hybridization efficiency. Hybridizations were performed with amplified 16S rDNA of strain KT0202a under standard conditions. (A) For probes ALF968, ROS537, and RC1239, helper oligonucleotides were added. Controls are only shown if signals were detected. (B) For all 11 probes targeting strain KT0202a, helpers were added. All corresponding controls plus two additional probes also featuring false-positive signals are shown.
Subsequently, for all 11 probes targeting KT0202a, helper pairs were applied in parallel (Fig. 3B). For all the probes, absolute signal intensities could be increased, ranging from a factor of 3.8 (from 775 to 2,968 a.u.) for probe RC1239 to a factor of 46.5 (from 184 to 8,565 a.u.) for probe PAR1457. With three exceptions (UNIV1392, RRP1088, and RC1239), all probes showed nonspecific signals within the corresponding mismatch controls. Additionally, probes KT13 and KT13-231 featured weak nonspecific signals (one mismatch for both).
(ii) General steric hindrance.
Poly(A) spacers of different lengths (none, 6-mer, and 12-mer) were tested on probes EUB338 and RRP1088. Both probes showed a linear increase of signal intensities with increasing spacer length (Fig. 4A). For probe EUB338, we observed signals within the corresponding mismatch controls when applying a 6- or 12-mer spacer, whereas for probe RRP1088, the hybridization efficiency could be improved by spacer molecules without getting nonspecific signals.
FIG. 4.
Impact of steric hindrance. (A) Hybridization of the amplified 16S rDNA of strain KT0202a at 46°C without formamide to probes EUB338 and RRP1088, both provided with poly(A) spacers of multiple lengths (none, 6-mer, and 12-mer spacers). The corresponding controls are also shown. (B) Hybridization of strain KT0202a at 46°C without formamide to probes ROS537 and ALF968 in the presence of helper pairs for both probes. Probes were provided with spacers of multiple lengths (none, 6-mer, 12-mer, 18-mer, and 24-mer spacers). Probe variants without a spacer and with a 12-mer spacer were also provided by immobilization via the 3′ end of the probe instead of the 5′ end.
Figure 4B shows the hybridization of strain KT0202a to probes ROS537 and ALF968 provided with no, 6-, 12-, 18-, and 24-mer spacers and under the presence of helper pairs for both probes. Here, for probe ALF968, a saturation was reached with the 18-mer spacer and for probe ROS537 with the 6-mer spacer.
(iii) Orientation of immobilized probe.
Probe orientation experiments with strain KT0202a (Fig. 4B) showed an increase in signal intensities after inversion (3′ instead of 5′ immobilized) from 3,716 to 6,763 a.u. (1.8-fold) and from 2,105 to 2,877 a.u. (1.4-fold) for probes ROS537 and ALF968, respectively. For the 12-mer spacer variants of the same two probes, increases of only about 1.1-fold were observed.
Optimization of signal patterns by directed application of capture oligonucleotides.
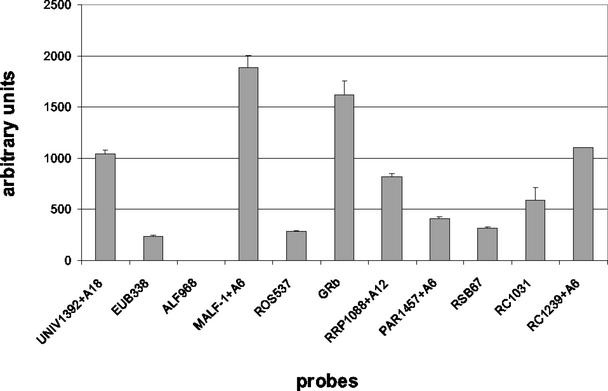
By a directed modification of spacer length based on the signal intensities shown in Fig. 1B for strain KT0202a, probe-specific differences in signal intensity and number of false-negative results should be minimized. For example, the two probes featuring former false-negative signals, UNIV1392 and ALF968, were both tested with 12- and 18-mer spacers, whereas probe GRb was not modified. The results for the hybridization of the 16S rDNA of strain KT0202a with this approach are given in Fig. 5. By elongation of probe UNIV1392 with an 18-mer spacer, the earlier observed false-negative signal could be restored without getting a signal for the corresponding mismatch control. In addition, signal intensities could be increased for probes MALF-1 (with a 6-mer spacer from 41 to 116%, normalized to the GRb signal), RRP1088 (12-mer, from 6 to 50%), PAR1457 (6-mer, from 9 to 25%), and RC1239 (6-mer, from 13 to 68%) without losing the specificity of the hybridization reaction. The sensitivity of probes EUB338, ALF968, ROS537, RSB67, and RC1031 could not be improved without the simultaneous appearance of signals within the corresponding controls.
FIG. 5.
Signal pattern optimization by directed application of capture oligonucleotides. Hybridization of the amplified 16S rDNA of strain KT0202a at 46°C without formamide to the 11 probes of the complete set targeting this strain. Probes were provided with spacers of different lengths, depending on former observed signal intensities (Fig. 1B, reference experiment). Only probe variants where no signals could be observed for the corresponding controls are shown.
DISCUSSION
In our initial experiments, we determined the optimal hybridization conditions for our microarray format in terms of a reduced number of false-positive and false-negative hybridization events. Best results were achieved for reference strain KT0202a at 46°C without formamide (Fig. 1B). Only under these conditions could a sufficient number of true-positive signals in combination with no false-positive signals be detected. This situation allows clear data interpretation. Concurrently, we demonstrated the general transferability of a standard FISH protocol to the DNA microarray format, which enables the easy validation of microarray data by parallel FISH analysis. This is crucial for the further application of microarrays for environmental studies. The fact that the best hybridization results were achieved under moderate stringency is in good agreement with data obtained from FISH experiments (17). Presumably, the lack of hybridization signals at room temperature (Fig. 1A) is due to a more stable secondary structure of the target molecule at lower temperatures, resulting in reduced accessibility of the probe binding sites. For hybridization at 46°C with 20% formamide (Fig. 1C), which equals a hybridization temperature of 56°C (47), we observed signals for only 4 of 11 perfect-match probes, presumably based on melting effects.
The results shown in Fig. 2 indicate that highly specific hybridization of a large number of oligonucleotide probes is feasible in a standard microarray format without any effort to adjust the melting points of the probes. Only one false-positive signal was observed among 199 potential nonspecific hybridization events (0.5%). This particular mismatch was one of the two weakest analyzed in this experimental series, a G:T mismatch three nucleotides away from the 3′ end of the probe (see Table 1). Discrimination of a mismatch near the terminus of a short duplex is hard to achieve (46) even under optimized conditions. The fact that we did not find a single nonspecific hybridization event within the central mismatch controls spotted for all 20 probes seems to be in complete contradiction with experiences from other hybridization formats such as FISH, where every probe has to be applied at appropriate stringency to ensure a specific hybridization reaction (4). Considering the results of the systematic investigations, the underlying mechanisms become more evident.
Parameter analysis.
The addition of helper oligonucleotides had a clear impact on the signal intensities of particular probes (Fig. 3). Helpers are unlabeled oligonucleotides designed to bind adjacent to a probe binding site, resulting in an increase in the accessibility of the corresponding target site (18, 35). As shown for probes ALF968, ROS537, and RC1239, they can resolve the secondary structures of the target rDNAs in a directed and selective way for particular probe binding sites (Fig. 3A and 6). Interestingly, the former strong signals of probes MALF-1, GRb, and RC1031 decreased dramatically in this experiment, suggesting that the opening of selected probe binding sites leads to a reorganization of secondary structures in other target regions. The influence of secondary structures on hybridization efficiency becomes much clearer when we applied helper pairs for all 11 probes targeting strain KT0202a (Fig. 3B): former false-negative signals could be restored, overall signal intensities were dramatically increased, and most of the single mismatch controls now showed strong signals. We do not assume that this is based on a change of Td values by base stacking of probe and helper oligonucleotides because no evidences for such an effect was found by Fuchs et al. (18).
Another outstanding feature of the data set shown in Fig. 2 is the distinct signal heterogeneity of different probes targeting the same preparation of 16S rDNA during parallel hybridization. This is a known phenomenon (44) and may even lead to the situation that signals of mismatch controls for particular probes were significantly higher than perfect match signals of other probes, as shown in Fig. 1A, 2, and 3. Considering the principles of DNA duplex stability prediction (9), measured signal intensities should be primarily connected to the nucleotide sequence of a probe. When applying helper oligonucleotides for all probes targeting an analyzed 16S rDNA, the distribution of measured signal intensities changes significantly (compare Fig. 1B and 3B). This observation is in agreement with the common view that secondary structures of target molecules affect the duplex yield of particular probes selectively (44). In conclusion, the measured signal intensity for a particular probe does not directly correlate with the amount of the corresponding target molecule in solution.
Another parameter that has to be considered in the context of signal limitation is steric hindrance. When hybridizing on a solid support, the binding efficiency of target molecules may be reduced by unfavorable steric interactions mediated by the solid matrix (42). The size of this effect should be correlated with the size of the target nucleic acid and the distance between the support and the capture probe. To investigate the influence of steric hindrance in our system, we applied poly(A) spacers of different lengths (6-, 12-, 18-, and 24-mers) between the slide surface and the probe sequence. The disadvantage of this kind of spacer is of course the potential Watson-Crick interaction between the spacer and the target molecule, but in our system, we did not observe any hybridization events indicating such an effect. Without the addition of helper oligonucleotides, in most cases we found a linear correlation between spacer length and measured signal intensity (Fig. 4A; supplementary data are not shown). When adding helper oligonucleotides, signal saturation was reached even with the shorter spacer variants (Fig. 4B, probe ROS537 [6-mer] and probe ALF968 [18-mer]), indicating a complex interaction of the signal-determining parameters. Interestingly, signal enhancement by reduction of steric hindrance may also lead to signals within the corresponding controls (Fig. 4A, probe EUB338).
Furthermore, it must be assumed that the position of the probe binding site also has an effect on the extent of steric hindrance because every nucleic acid binds in a defined orientation to its complement. To address the significance of this parameter, we tested two variants of probes immobilized via the 5′ and 3′ end. Probe ROS537, whose binding site is located for target KT0202a at nucleotide position 474 of 1428, starting from the 5′ end, shows a higher signal for the 3′-immobilized probe. This is in good agreement with our initial assumption that a 5′-immobilized probe binding to the 5′ end of the target molecule is leading to an unfavorable steric situation because the longer tail of the target nucleic acid is oriented towards the slide surface and vice versa. Nevertheless, probe ALF968 also shows a slightly increased signal for the 3′-immobilized probe, although its binding site for target KT0202a is located at nucleotide position 907 of 1428. Apparently, the secondary and tertiary structures of the target molecules interfere with simple spatial extrapolations. As for application of spacer molecules, we found little influence on signal intensities for reverse probe immobilization compared to the application of helper oligonucleotides. In conclusion, the secondary structure of a target molecule is the main limiting factor for duplex yield considering the parameters investigated in this study.
Mechanisms enabling observed hybridization specificity.
As shown in Fig. 3 and 4A, signals for the single-mismatch controls can be “generated” under identical hybridization conditions if the influence of signal-limiting parameters such as secondary structures and steric hindrance is reduced. Therefore, we conclude that for the data set shown in Fig. 2, mismatch discrimination was mainly achieved by global signal suppression. This is possible because perfect match and mismatch signals react different on changes of signal limiting parameters as shown for secondary structures: without helpers (Fig. 1B), one mismatch signals were selectively shifted below the detection limit compared to the perfect-match signals.
A full-length 16S rDNA molecule and the resulting secondary structure appear to represent a well-suited “calibrator” for global signal suppression under optimized hybridization conditions. When shorter fragments were hybridized under identical hybridization conditions (data not shown), we also observed an overall increase of signal intensities in combination with an accumulated number of false-positive signals. Here, the reduced influence of two factors becomes evident: (i) shorter fragments lead to reduced steric hindrance and (ii) secondary structures of shorter targets are known to be less stable than those of longer fragments (44). Therefore, our results suggest that fragmentation of target molecules can be disadvantageous in terms of preventing false-positive signals.
In summary, we report on a new concept for improving the specificity of microarray hybridizations. It is based on a converse approach compared to current signal enhancement attempts because we “utilize” signal-limiting parameters to selectively shift nonspecific signals below the detection limit. We would like to point out that the overall sensitivity of the detection system is not affected by this strategy because the detection limit is not shifted but hybridization signals are specifically suppressed.
Prevention of false-negative results by a new optimization approach.
As shown in Fig. 2, we found a large number of false-negative results in our data set. Subsequent application of helper pairs for all 11 probes targeting strain KT0202a under identical hybridization conditions led to signals for all 11 probes and nearly all corresponding controls (Fig. 3B). This indicates that false-negative results and discriminated single-mismatch controls were not based on melting effects. We suppose that nonoccurring signals are mainly caused by signal-suppressing parameters such as secondary structure of the target molecules and steric hindrance, selectively acting for different probe binding sites. Considering the fact that the number of 16S rRNA sequence positions is limited, a false-negative results for a particular probe can be a profound restriction for phylogenetic affiliation because additional probes featuring a redundant target specificity are often not available. For example, strain JP7.1 could be clearly assigned to the marine alpha group by the phylogenetically redundant probes MALF-1 and ROS537 (Fig. 2), but the single strain-specific probe available (Ros7-1029) featured a false-negative signal and therefore inhibited a more exact phylogenetic affiliation of the analyzed 16S rDNA.
Based on our systematic investigations we have deduced an approach for further signal pattern optimization called directed application of capture oligonucleotides. Our study shows that (i) single-mismatch discrimination can also be achieved by “proper calibration” of signal intensities for particular probes and (ii) different parameters are available for a probe-specific signal adjustment. As shown in Fig. 3A and 4A, signals can be selectively enhanced by reducing the influence of signal suppressing factors such as secondary structures and steric hindrance. Our intention was to restore former false-negative signals and to adjust overall signal intensities with these options without losing the status of complete discrimination for the single mismatch controls, as shown in Fig. 4A for probe RRP1088. The usability of this approach, based on a directed variation of spacer length, was successfully tested for strain KT0202a, as shown in Fig. 5. This parameter was chosen because it allows a graduated signal adjustment. Another potential mediator would be the amount of immobilized probe, because a direct correlation to duplex yield was also observed for this parameter (data not shown), in agreement with the results of Guo et al. (22). In the context of directed application, the utilization of helper oligonucleotides is not practical for two reasons: (i) the strong signal increase caused by helpers often also results in signals for the corresponding mismatch controls (Fig. 3) and (ii) design of helpers showing the same specificity as the corresponding probe is often not possible (18).
Data normalization.
An important issue when analyzing microarray experiments is data normalization. In the field of microbial ecology published data were either not normalized (24, 43) or domain specific probes were used for normalization (27). For this study we decided to work with absolute signal intensities (arbitrary units) when comparing data generated under different hybridization conditions or analyzing different target molecules. We assume that by using domain-specific probes as internal standards, an additional error will be introduced, because probe binding site accessibility should not be completely comparable for distantly related targets.
Why use a model system based on 16S rDNA targets?
This study was performed with 16S rDNA targets generated by PCR amplification. We are well aware of the possible biases introduced by PCR during analysis of complex communities (3). For the analysis of complex samples, direct extraction and labeling of the rRNA are essential to circumvent possible artifacts. Furthermore, for environmental studies, aspects such as sensitivity and quantification have to be considered more comprehensively. However, we decided to conduct this initial systematic investigation with exactly defined 16S rDNA targets instead of extracted RNA because only single targets were analyzed in our model system. Our results should help to consider new kinds of optimization strategies for the specific detection of 16S rRNA/rDNA molecules by microarray hybridization and for environmental studies. We foresee PCR-based microarray systems in the (i) fast and low-cost screening of isolates for their phylogenetic affiliation and (ii) prescreening of PCR products for diversity or the presence of particular subpopulations (e.g., for generating gene libraries).
Acknowledgments
We thank D. Blohm for providing the spotting device, D. Drutschmann for help in handling the device, and D. Wöhrle and B. Meyer-Schlosser for providing us with activated glass slides. The generous support of J. Pernthaler by setting up the image and data analysis pipeline is gratefully acknowledged.
This work was done in the framework of the Centre of Applied Gensensorik (CAG) at the University of Bremen and supported by the German Ministry of Education and Research (BMBF, contract 0311833A) and by the Max Planck Society.
REFERENCES
- 1.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I. 1995. Fluorescently labelled, rRNA-targeted oligonucleotide probes in the study of microbial ecology. Mol. Ecol. 4:543-554. [Google Scholar]
- 3.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann, R. I., and K.-H. Schleifer. 2001. Nucleic acid probes and their application in environmental microbiology, p. 67-82. In G. Garrity, D. R. Boone, and R. W. Castenholz (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer-Verlag, New York, N.Y.
- 5.Bains, W., and G. C. Smith. 1988. A novel method for nucleic acid sequence determination. J. Theor. Biol. 135:303-308. [DOI] [PubMed] [Google Scholar]
- 6.Beier, M., and D. Hoheisel Jörg. 1999. Versatile derivatisation of solid support media for covalent bonding on DNA-microchips. Nucleic Acids Res. 27:1970-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benters, R., C. M. Niemeyer, D. Drutschmann, D. Blohm, and D. Wöhrle. 2002. DNA microarrays with PAMAM dendritic linker systems. Nucleic Acids Res. 30:e10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blake, R. D., and S. G. Delcourt. 1996. Thermodynamic effects of formamide on DNA stability. Nucleic Acids Res. 24:2095-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breslauer, K. J., R. Frank, H. Blocker, and L. A. Marky. 1986. Predicting DNA duplex stability from the base sequence. Proc. Natl. Acad. Sci. USA 83:3746-3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brosius, J., T. J. Dull, D. D. Sleeter, and H. F. Noller. 1981. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 148:107-127. [DOI] [PubMed] [Google Scholar]
- 11.Buchholz-Cleven, B., B. Rattunde, and K. Straub. 1997. Screening for genetic diversity of isolates of anaerobic Fe(II)-oxidizing bacteria with DGGE and whole-cell hybridization. Syst. Appl. Microbiol. 20:301-309. [Google Scholar]
- 12.Debouck, C., and P. N. Goodfellow. 1999. DNA microarrays in drug discovery and development. Nat. Genet. 21:48-50. [DOI] [PubMed] [Google Scholar]
- 13.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drmanac, R., I. Labat, I. Brukner, and R. Crkvenjakov. 1989. Sequencing of megabase plus DNA by hybridization theory of the method. Genomics 4:114-128. [DOI] [PubMed] [Google Scholar]
- 15.Eilers, H., J. Pernthaler, F. O. Glöckner, and R. Amann. 2000. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol. 66:3044-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eilers, H., J. Pernthaler, J. Peplies, F. O. Glöckner, G. Gerdts, and R. Amann. 2001. Isolation of novel pelagic bacteria from the German bight and their seasonal contributions to surface picoplankton. Appl. Environ. Microbiol. 67:5134-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuchs, B. M., G. Wallner, W. Beisker, I. Schwippl, W. Ludwig, and R. Amann. 1998. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 64:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuchs, B. M., F. O. Glöckner, J. Wulf, and R. Amann. 2000. Unlabeled helper oligonucleotides increase the in situ accessibility of 16S rRNA for fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 66:3603-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60-63. [DOI] [PubMed] [Google Scholar]
- 20.Giuliano, L., M. De Domenico, E. De Domenico, M. G. Höfle, and M. M. Yakimov. 1999. Identification of culturable oligotrophic bacteria within naturally occurring bacterioplankton communities of the Ligurian Sea by 16S rRNA sequencing and probing. Microb. Ecol. 37:77-85. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez, J. M., W. B. Whitman, R. E. Hodson, and M. A. Moran. 1996. Identifying numerically abundant culturable bacteria from complex communities: an example from a lignin enrichment culture. Appl. Environ. Microbiol. 62:4433-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo, Z., R. A. Guilfoyle, A. J. Thiel, R. Wang, and L. M. Smith. 1994. Direct fluorescence analysis of genetic polymorphisms by hybridization with oligonucleotide arrays on glass supports. Nucleic Acids Res. 22:5456-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta, P. K., J. K. Roy, and M. Prasad. 1999. DNA chips, microarrays and genomics. Curr. Sci. 77:875-884. [Google Scholar]
- 24.Guschin, D. Y., B. K. Mobarry, D. Proudnikov, D. A. Stahl, B. E. Rittmann, and A. D. Mirzabekov. 1997. Oligonucleotide microchips as genosensors for determinative and environmental studies in microbiology. Appl. Environ. Microbiol. 63:2397-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hacia, J. G., L. C. Brody, M. S. Chee, S. P. A. Fodor, and F. S. Collins. 1996. Detection of heterozygous mutations in BRCA1 with high density oligonucleotide arrays and two-colour fluorescence analysis. Nat. Genet. 14:441-447. [DOI] [PubMed] [Google Scholar]
- 26.Hoheisel, J. D. 1997. Oligomer-chip technology. Trends Biotechnol. 15:465-469. [Google Scholar]
- 27.Liu, W.-T., A. D. Mirzabekov, and D. A. Stahl. 2001. Optimization of an oligonucleotide microchip for microbial identification studies: a non-equilibrium dissociation approach. Environ. Microbiol. 3:619-629. [DOI] [PubMed] [Google Scholar]
- 28.Lockhart, D. J., H. Dong, M. C. Byrne, M. T. Follettie, M. V. Gallo, M. S. Chee, M. Mittmann, C. Wang, M. Kobayashi, H. Horton, and E. L. Brown. 1996. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14:1675-1680. [DOI] [PubMed] [Google Scholar]
- 29.Lysov, Y. P., V. L. Florentev, A. A. Khorlin, K. R. Khrapko, V. V. Shik, and A. D. Mirzabekov. 1988. Determining the nucleotide sequence of DNA by hybridization with oligonucleotides a new method. Dokl. Akad. Nauk. SSSR 303:1508-1511. (In Russian.) [PubMed]
- 30.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K.-H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 31.Maskos, U., and E. M. Southern. 1992. Parallel analysis of oligodeoxyribonucleotide-oligonucleotide interactions. I. Analysis of factors influencing oligonucleotide duplex formation. Nucleic Acids Res. 20:1675-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maskos, U., and E. M. Southern. 1993. A study of oligonucleotide reassociation with large arrays of oligonucleotides synthesised on a glass support. Nucleic Acids Res. 21:4663-4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mir, K. U., and E. M. Southern. 1999. Determining the influence of structure on hybridization with oligonucleotide arrays. Nat. Biotechnol. 17:788-792. [DOI] [PubMed] [Google Scholar]
- 34.Neef, A. 1997. Ph.D. thesis. Technical University, Munich, Germany.
- 35.Niemeyer, C. M., W. Bürger, and J. Peplies. 1998. Covalent DNA-streptavidin conjugates as building blocks for novel biometallic nanostructures. Angew. Chem. Int. Ed. 37:2265-2268. [DOI] [PubMed] [Google Scholar]
- 36.Niemeyer, C. M., L. Boldt, B. Ceyhan, and D. Blohm. 1999. Evaluation of single-stranded nucleic acids as carriers in the DNA-directed assembly of macromolecules. J. Biomol. Struct. Dyn. 17:527-538. [DOI] [PubMed] [Google Scholar]
- 37.Pace, N. R., D. A. Stahl, D. J. Lane, and G. J. Olsen. 1986. The use of rRNA sequences to characterize natural microbial populations. Adv. Microb. Ecol. 9:1-55. [Google Scholar]
- 38.Pernthaler, J., F. O. Glöckner, W. Schönhuber, and R. Amann. 2001. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. Methods Microbiol. 30:207-226. [Google Scholar]
- 39.Rees, W. A., T. D. Yager, J. Korte, and P. H. von Hippel. 1993. Betaine can eliminate the base pair composition dependence of DNA melting. Biochemistry 32:137-144. [DOI] [PubMed] [Google Scholar]
- 40.Riccelli, P. V., F. Merante, K. T. Leung, S. Bortolin, R. L. Zastawny, R. Janeczko, and A. S. Benight. 2001. Hybridization of single-stranded DNA targets to immobilized complementary DNA probes: comparison of hairpin versus linear capture probes. Nucleic Acids Res. 29:996-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rudi, K., O. M. Skulberg, R. Skulberg, and K. S. Jakobsen. 2000. Application of sequence-specific labeled 16S rRNA gene oligonucleotide probes for genetic profiling of cyanobacterial abundance and diversity by array hybridization. Appl. Environ. Microbiol. 66:4004-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shchepinov, M. S., S. C. Case-Green, and E. M. Southern. 1997. Steric factors influencing hybridisation of nucleic acids to oligonucleotide arrays. Nucleic Acids Res. 25:1155-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Small, J., D. R. Call, F. J. Brockman, T. M. Straub, and D. P. Chandler. 2001. Direct detection of 16S rRNA in soil extracts by with oligonucleotide microarrays. Appl. Environ. Microbiol. 67:4708-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Southern, E., K. Mir, and M. Shchepinov. 1999. Molecular interactions on microarrays. Nat. Genet. 21:5-9. [DOI] [PubMed] [Google Scholar]
- 45.Southern, E. M., U. Maskos, and J. K. Elder. 1992. Analyzing and comparing nucleic acid sequences by hybridization to arrays of oligonucleotides evaluation with experimental model. Genomics 13:1008-1017. [DOI] [PubMed] [Google Scholar]
- 46.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 47.Wahl, G. M., S. L. Berger, and A. R. Kimmel. 1987. Molecular hybridization of immobilized nucleic acids: theoretical concepts and practical considerations. Methods Enzymol. 152:399-407. [DOI] [PubMed] [Google Scholar]
- 48.Ward, D. M., R. Weller, and M. M. Bateson. 1990. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 345:63-65. [DOI] [PubMed] [Google Scholar]
- 49.Williams, J. C., S. C. Case-Green, K. U. Mir, and E. M. Southern. 1994. Studies of oligonucleotide interactions by hybridisation to arrays: the influence of dangling ends on duplex yield. Nucleic Acids Res. 22:1365-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zubkov, M. V., B. M. Fuchs, S. D. Archer, R. P. Kiene, R. Amann, and P. H. Burkill. 2001. Linking the composition of bacterioplankton to rapid turnover of dissolved dimethylsulphoniopropionate in an algal bloom in the North Sea. Environ. Microbiol. 3:304-311. [DOI] [PubMed] [Google Scholar]