Abstract
Cellular CD1 proteins bind lipids that differ in length (C12−80), including antigens that exceed the capacity of the CD1 groove. This could be accomplished by trimming lipids to a uniform length before loading or by inserting each lipid so that it penetrates the groove to a varying extent. New assays to detect antigen fragments generated within human dendritic cells showed that bacterial antigens remained intact, even after delivery to lysosomes, where control lipids were cleaved. Further, recombinant CD1b proteins could bind and present C80 lipid antigens using a mechanism that did not involve cellular enzymes or lipid cleavage, but was regulated by pH in the physiologic range. We conclude that endosomal acidification acts directly, rather than through enzymatic trimming, to insert lipids into CD1b. Lipids are loaded in an intact form, so that they likely protrude through a portal near the bottom of the groove, which represents an escape hatch for long lipids from mycobacterial pathogens.
Keywords: antigen processing, CD1, T cells, tuberculosis
Introduction
Cellular CD1 proteins mediate T-cell activation in response to a variety of lipid, glycolipid and lipopeptide antigens, leading to the emerging view that the CD1 antigen presentation system allows T cells to broadly survey the lipid content of target cells. The mechanism for display of lipid antigens to T-cell receptors (TCR) is illustrated by crystal structures of CD1-β2-microglobulin in complex with phosphatidylinositol, ganglioside GM2, sulfatide, glucose monomycolate (GMM), lipopeptide, phosphatidylcholine and α-galactosyl ceramide (Gadola et al, 2002; Zajonc et al, 2003, 2005a, 2005b; Batuwangala et al, 2004; Giabbai et al, 2005; Koch et al, 2005). The phosphate, sulfate, carbohydrate or peptide moieties lie on the outer, α-helical surface of CD1, where they can directly contact TCRs. The lipid moieties traverse a small portal positioned between the α1- and α2-helices and are inserted deeply within the α1–α2 superdomain of CD1 proteins, where they form extensive hydrophobic interactions with the non-polar residues that line the CD1 groove (Zeng et al, 1997; Gadola et al, 2002). These crystal structures clarify general mechanism of seating lipids within the groove, but there is little understanding of basic questions relating to how CD1 proteins capture antigens with structurally diverse lipid anchors or prioritize among the many types of lipid antigens available within cells.
The lipid anchors in CD1-presented antigens can be composed of mycolic acids (MA) (Beckman et al, 1994; Moody et al, 1997), diacylglycerols (Sieling et al, 1995; Joyce et al, 1998; Rauch et al, 2003; Agea et al, 2005), ceramides (Kawano et al, 1997; Shamshiev et al, 1999, 2002; Kinjo et al, 2005; Mattner et al, 2005), polyisoprenols (Moody et al, 2000b), polyketides (Matsunaga et al, 2004), phthioceranoates (Gilleron et al, 2004) or fatty acyl chains (Moody et al, 2004). These lipids can have either one or two alkyl chains that vary greatly in their overall length. In fact, exogenous lipid antigens presented by CD1-expressing APCs have lipid moieties that are much larger or smaller than the known volume of the antigen binding grooves found in CD1a (1300 Å3), CD1b (2200 Å3) or CD1d proteins (1650 Å3) (Zeng et al, 1997; Gadola et al, 2002; Zajonc et al, 2003). The apparent mismatch of the CD1 groove volume with lipid length is greatest for CD1b proteins, which present diacylglycerols, sphingolipids, mycolates and polyacylated carbohydrates, ranging in length from C12 to C80 (Moody et al, 1997; Shamshiev et al, 1999; Gilleron et al, 2004). The CD1b groove is composed of four contiguous pockets named A′, C′, F′ and T′, which are accessible by two portals located above the F′ pocket and at the bottom of the C′ pocket (Gadola et al, 2002). The crystal structure of CD1b in complex with a GMM antigen shows that a C56 lipid occupies nearly the entire groove (Batuwangala et al, 2004); so the interior of the CD1b groove could optimally accommodate antigens with an overall lipid length of C60–64. However, in vivo studies of naturally occurring mycobacterial lipids show that cellular CD1b proteins mediate T-cell recognition of antigens exceeding this volume, such as C80 MA and C80 GMM (Ulrichs et al, 2003). In addition, acylated sulfotrehaloses may also exceed this limit depending on the number of acyl chains inserted into the groove (Gilleron et al, 2004).
CD1 proteins could present lipids of diverse length by one of two general strategies. Antigens might be trimmed by cells before loading into the CD1 groove. Alternatively, each type of antigen might bind so that the hydrophilic cap sits at the main entrance to the groove, but the lipid moiety present in each type of antigen penetrates the groove to a varying extent. Both hypotheses have some indirect support from studies of lipid antigen presentation in intact cells. In support of the trimming hypothesis, it is known that many CD1-presented antigens must be taken up into late endosomes or lysosomes before their recognition by T cells, which offers an opportunity for exposure to hydrolytically active enzymes before loading antigens into the groove (Porcelli et al, 1992; Sieling et al, 1995; Chiu et al, 1999; Moody et al, 2002; Gilleron et al, 2004). However, the actual molecular events that render these antigens into a recognizable form are not known and might be accounted for by any of several explanations.
Low pH directly promotes the association of recombinant CD1b proteins with lipid antigens in vitro (Ernst et al, 1998), suggesting that low pH generated by vesicular ATPases might relax the α-helical structures that protect the binding groove and thereby directly promote antigen insertion. Secondly, loading of antigens onto CD1b and CD1d is increased by saposin and apolipoprotein lipid transfer proteins, which normally localize to late endosomes and lysosomes (Kang and Cresswell, 2004; Winau et al, 2004; Zhou et al, 2004a; van den Elzen et al, 2005). Last, lysosomal hydrolases, such as α-galactosidase A and β-hexosaminidase, chemically modify the carbohydrate moieties of antigens in ways that create epitopes for TCRs (Prigozy et al, 2001; Zhou et al, 2004b). Although there have been no prior studies of potential modifications of the lipid moieties of antigens, these deglycosylation reactions raise the possibility that other lysosomal enzymes might chemically modify the lipid moieties so that they more closely match the volume of CD1 grooves.
As an alternative to trimming, each lipid antigen may be inserted in an intact form and occupy the CD1 groove to a varying extent, depending on the length and number of its alkyl chains. Antigens with lipid moieties that are significantly smaller than the groove volume, such as gangliosides, sulfatides and shorter forms of GMM (C12 GMM, C32 GMM), might bind together with smaller space-holding lipids, which occupy the remainder of the groove (Gadola et al, 2002). Longer chain antigens might occupy the groove more completely. In the absence of trimming, the largest C80 lipids would fully occupy its volume and then exit from the hydrophobic interior to the outer, solvent-exposed face of CD1b, possibly through a small portal located at the bottom of the C′ pocket (Gadola et al, 2002).
To distinguish among these possibilities, we developed assays to determine whether dendritic cells (DCs) alter the length of long-chain bacterial lipids. Parallel analyses of antigen uptake, generation of antigenic complexes and detection of antigen breakdown products showed that lipid antigen cleavage products could not be detected, even at time periods that greatly exceed those necessary to load long-chain antigens onto CD1b. Further, it was possible to bypass all endosomal factors and form antigenic complexes with long-chain lipids in cell-free conditions, as long as the loading reactions were carried out under a narrow pH range that corresponds to that of late endosomes. We conclude that cellular presentation of long-chain lipids involves acid-mediated loading of intact antigens using a mechanism in which large lipids can enter the α1–α2 superdomain, traverse the antigen binding groove and then likely re-emerge with their termini positioned below the groove on the outer surface of CD1.
Results
Endosomal and non-endosomal antigen loading occur over distinct time frames
Before designing assays for measuring cell-mediated alterations in antigen structure, we first sought to determine the time frame of functional antigen processing by monocyte-derived DCs. We treated DCs with MA and GMM antigens with short (C32), intermediate (C54) or long (C80) lipids and measured the elapsed time before these antigens activated CD1b-restricted T-cell lines. DCs were found to present C32 GMM within 2 min of exposure. In contrast, all of the longer chain GMM and MA antigens were first recognized after more than 30 min of coincubation with DCs (Figure 1B and data not shown). In separate studies of proliferation carried out under conditions of steady-state antigen processing for 72 h, C80 GMM (0.012 μM) and C54 GMM (0.56 μM) were found to half-maximally activate T cells at low concentrations compared to those for C32 GMM (3.6 μM). Therefore, the delayed recognition of long-chain GMM antigens was not accounted for by an intrinsically lower potency.
Figure 1.
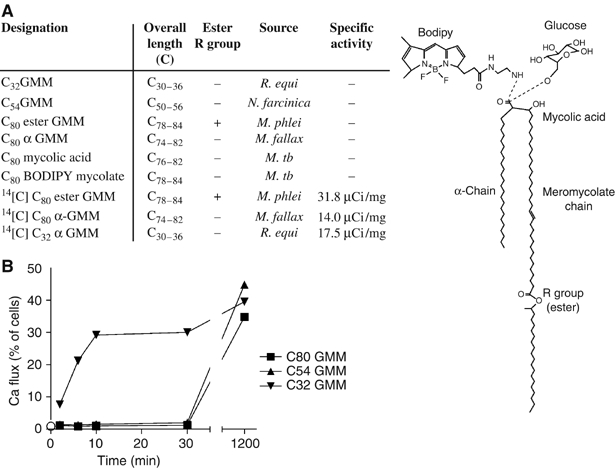
Delayed T-cell recognition of endosomally presented antigens. (A) Free MA and GMM antigens are described as Cx, where x is the combined alkyl chain length of the meromycolate and α-branches of the most abundant species detected by MS. BODIPY or glucose is linked to the carboxyl group as indicated by the dashed line. (B) Human monocyte-derived DCs were preincubated for the indicated time with 20 μM GMM antigens before adding CD1b-restricted LDN5 T cells and assaying for calcium-induced changes in fluorescence as assessed by flow cytometry.
Instead, this paradoxical delay in recognition of the most potent antigens suggested that the T-cell recognition of longer chain antigens was dependent upon processing within endosomes. This conclusion is supported by prior studies showing that C80 and C54 GMM required endosomal processing by CD1b-transfected lymphoblastoid cells, whereas C32 GMM did not (Moody et al, 2002). Thus, the delayed presentation by DCs was seen only for those antigens that were known to require endosomal processing as defined by other criteria. Also, 2 min is not generally sufficient to allow antigens and surface proteins to recycle between the surface and late endosomes. Instead, the 30–60 min delay before T-cell recognition correlates with the time necessary for CD1 and other proteins to recycle to endosomes and back to the surface (Jayawardena-Wolf et al, 2001; Briken et al, 2002; Moody et al, 2002). Thus, kinetic studies of CD1b-mediated antigen presentation clarify the frames necessary for non-endosomal (0–5 min) and late endosomal (>30 min) antigen processing events in DCs.
Mycolyl lipids are transported to CD1b+, MHC II+, LAMP-1+ endosomes
To more directly measure the uptake and subcellular localization of mycolate antigens into DCs, we prepared labeled antigens by coupling a small fluorophore, BODIPY, to free MA (BODIPY-MA) in a process that alters mycolate structure by adding a small label that is comparable in molecular weight to a hexose sugar. Also, we prepared biosynthetically radiolabeled antigens of native structure from mycobacteria grown in the presence of 14[C]acetate (Figure 1A). To measure the uptake of GMM antigens, DCs were cultured with radiolabeled C32 GMM for up to 24 h, washed to remove extracellular antigens and treated with organic solvents to extract lipids. Consistent with prior studies, uptake of GMM by immature DCs was typically in the range of 50 ng/106 cells/24 h (Moody et al, 2002). To more precisely determine the intracellular localization of mycolyl lipids, BODIPY-MA and endosomal markers were visualized after a time period that slightly exceeds the minimal time for processing of long-chain antigens (90 min) using confocal microscopy. BODIPY-MA stained discrete compartments and extensively colocalized with MHC class II, which is expressed broadly within the endosomal network (Figure 2). BODIPY-MA also colocalized with compartments that stained with mAb against CD1b or anti-sera against lysosome-associated membrane protein 1 (LAMP-1 CD107a), markers that more selectively label late endosomes and lysosomes. Thus, within a time frame that is relevant to endosomal processing, mycolyl lipids are internalized into the endosomal network of DCs, including CD1b+ late endosomes.
Figure 2.
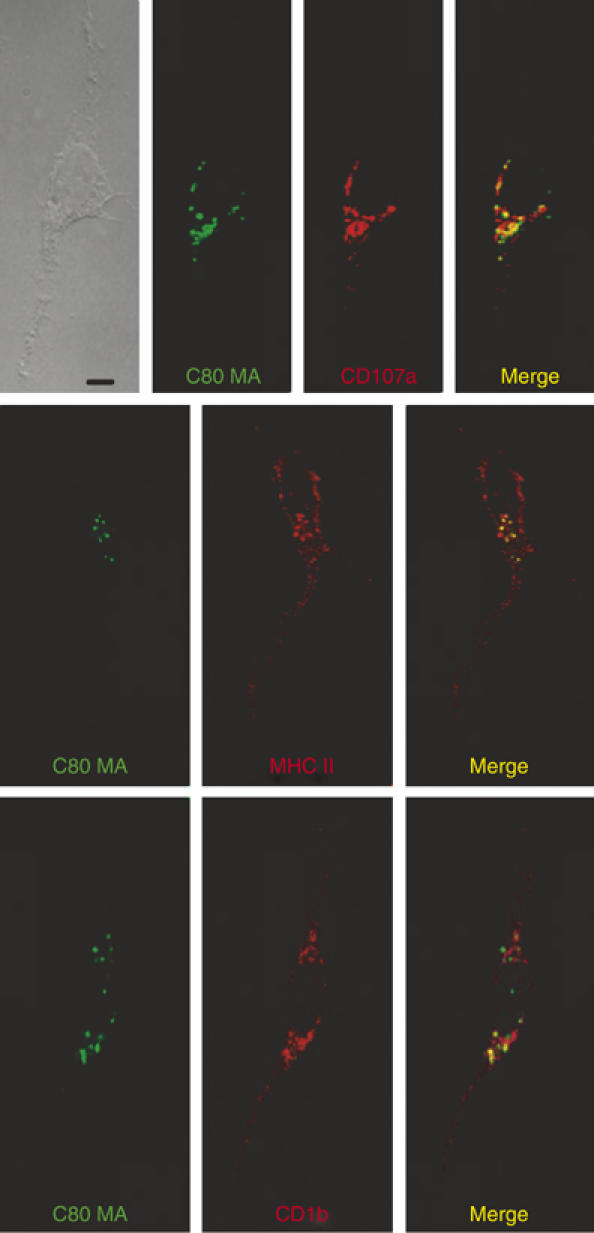
Mycolyl lipids are transported to lysosomes. DCs were incubated with BODIPY-mycolate (green) for 90 min followed by fixation, permeabilization and staining with antibodies against CD1b, MHC II or LAMP-1 (CD107a) and Texas red-conjugated Fab′2 against mouse Ig (red). The phase-contrast light micrograph (upper left) and immunofluorescent confocal micrographs (merge) are shown with a scale bar of 20 μm.
Mycolate antigens persist intact within DCs
To determine whether the lipid moieties of antigens undergo chemical alteration after uptake into the endosomal compartments of DCs, we developed thin-layer chromatographic (TLC) autoradiography assays to detect antigen cleavage products. Preliminary studies showed that the 14[C] label was incorporated into carbohydrate and lipid portions of GMM antigens and that antigens were labeled to a high specific radioactivity (Figure 1A), such that 500 pg of GMM could be detected using a 72 h autoradiograph (Figure 3A and data not shown). Therefore, this assay detected 1 part per 4000 of lipid added to the culture or approximately 1 part per 200 of lipid taken up by DCs in 72 h. This method was used to study short- and long-chain mycolate and GMM antigens, including two forms of long-chain GMM that differed in the chemical structures of their lipid moieties (Figure 3A). One form was designated C80 α-GMM because it contained α-MA that lacks oxygen-containing R groups in their mycolate lipid moieties. The other form, C80 ester GMM, is found only in uncommon or non-pathogenic species of mycobacteria such as Mycobacterium phlei, but was of interest because it contained an internal ester that might be readily subject to chemical cleavage by acid (Figure 3A).
Figure 3.
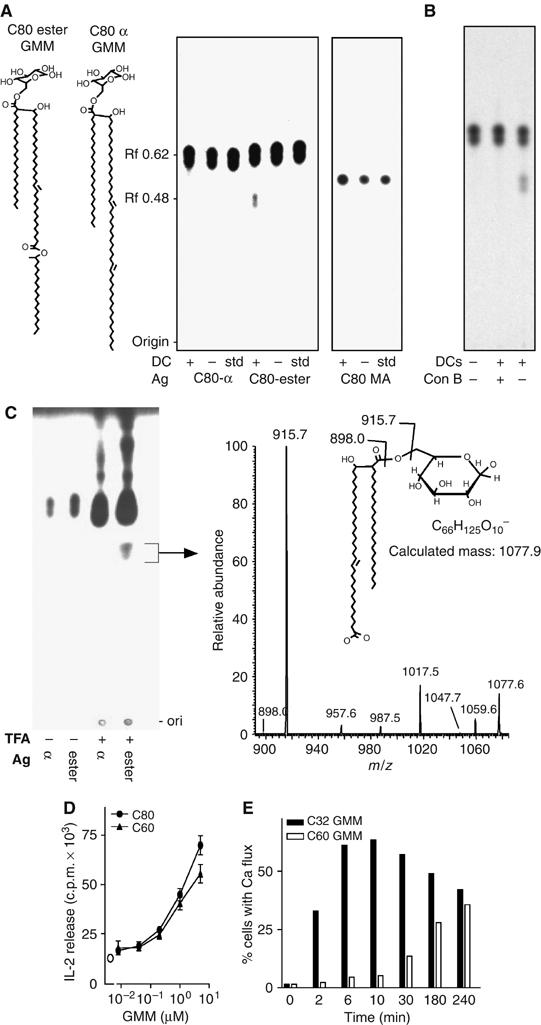
Generation of cleavage products from GMM by cellular processing in DCs. (A) [14C]C80 α-GMM, [14C]C80 ester GMM or [14C]C80 MA were cocultured with DCs mock-treated by culture at 37°C. After 72 h, DCs were washed to remove extracellular lipids and subjected to lipid extraction. After normalizing for equivalent amounts of antigen-derived radiolabel, lipids were developed with silica TLC. Total lipids were detected by charring (not shown) and labeled lipids were detected by autoradiography in comparison with mock-treated lipids (−) and untreated standards. (B) DCs were treated with concanamycin B for 1 h before adding C80 ester GMM and analyzed as in panel A. (C) C80 α-GMM (α) or C80 ester GMM (ester) was treated with TFA and the material with an Rf of 0.48 was isolated by preparative silica TLC. Mass spectrometry (MS) showed a predominant ion at m/z 1077.7, which corresponded to the predicted mass of an anion produced by hydrolysis at the internal ester of the meromycolate chain to generate a dicarboxylate GMM (C60 GMM). CID-MS of the major [M-H]− ion at m/z 1077.6 showed a dominant loss of 162 units consistent with the presence of an intact hexosyl residue on C60 GMM. Additional ions at m/z 957.6, 987.5 and 1017.5 correspond to the incremental loss of CH2O units from the intact molecule, which arise via through-ring cleavage of glucose and give additional evidence that the carbohydrate portion of the molecule survived the side-chain ester hydrolysis without modification. (D) After isolation of the C60 GMM product by preparative TLC, it was compared to the intact precursor, C80 GMM, for IL-2 release by CD1b-restricted T cells. (E) Kinetic analysis of GMM presentation using CD1-transfected cells (C1R.CD1b) was performed as in Figure 1B.
After 72 h of culture with antigen, DCs were washed to remove non-cell-associated antigens and then subjected to a modified Folch extraction and silica TLC with detection of lipids by charring and autoradiography. The C80 ester GMM, which migrates with a retardation factor (Rf) of 0.62, gave rise to a product that migrated at 0.48 (Figure 3A). This fragment was generated in a time-dependent fashion only when DCs were added to the cultures, demonstrating that it was generated by the DCs (Supplementary Figure 1). Because this cleavage reaction occurred after uptake into cells, we hypothesized that the reaction occurred in acidic endosomal or lysosomal compartments. To more directly test this, we preincubated DCs with concanamycin B, a drug that neutralizes the pH of endosomes by inhibition of proton transport by vesicular ATPases. Concanamycin-treated DCs took up [14C]C80 ester GMM, but blocked production of the Rf 0.48 lipid product (Figure 3B).
The Rf 0.48 product was not generated from C80 α-GMM. Because the only difference in chemically reactive groups present in ester GMM and α-GMM is the internal ester, we concluded that the DC-mediated alteration involved cleavage at this site. This conclusion was independently supported by experiments carried out in cell-free conditions in which treatment with trifluoroacetic acid (TFA) under harsh conditions (pH 1.0, 120°C, 120 min) generated a product of Rf 0.48 from C80 ester GMM but not from C80 α-GMM (Figure 3C). This product was isolated and mass spectrometry (MS), yielding a major ion of m/z 1077.6. This ion corresponded to the mass of a product resulting from a cleavage at the internal ester of the meromycolate chain, GMM with a dicarboxyl C60 mycolate (C60 GMM), as predicted. Thus, the concanamycin B-inhibitable cleavage product generated within DCs corresponded to C60 GMM derived from C80 ester GMM. These results indicate that human DCs are capable of altering the lipid length of a model antigen using a selective hydrolysis of an internal ester linkage.
Although this reaction shortened the lipid moiety so that it more closely approximated the CD1b groove volume, this did not account for the ability of long-chain antigens to be loaded onto CD1b proteins in endosomal compartments. Analysis of IL-2 release and calcium flux by T cells in response to the C60 GMM fragment showed that the shortened lipid did not enhance its potency compared to full-length C80 ester GMM, nor did it allow rapid (<5 min) presentation to T cells, as would be expected if this lipid truncation were fully sufficient to allow loading at the cell surface (Figure 3D–E). Most importantly, other naturally occurring MA and GMM antigens with C80 mycolate moieties that lacked the internal ester showed no evidence for antigen cleavage in TLC-autoradiography assays (Figure 3A), even after time periods that greatly exceed those necessary for their presentation to T cells (Figure 1B). After failing to find evidence for lipid trimming, we considered other mechanisms by which endosomal cofactors contribute to processing of long-chain antigens.
pH is a key regulator of antigen complex formation in cells
Other than trimming, a second factor in the lysosomal micoenvironment that contributes to the generation of antigen complexes is acidic pH, which is normally maintained by vesicular ATPases at 4.5–5.5 in DCs (Trombetta et al, 2003). Low pH might act indirectly by activating pH-dependent hydrolases or saposins, or might act directly to promote CD1–lipid association, as suggested by in vitro binding studies (Ernst et al, 1998). Our initial studies of the effects of low pH on GMM and mycolate processing were carried out in intact cells. This approach was guided by the reasoning that if pH could carry out certain effects directly on the CD1b antigen interaction rather than through lysosomally located proteins, then briefly acidifying the extracellular space might be sufficient to bypass the usual endosomal processing requirements.
In contrast to results obtained at pH 7.4, C80 MA, C80 α-GMM and C80 ester GMM antigens were recognized after 5 min of exposure at pH 4.0 with APCs (Figure 4A), such that their kinetic profiles of recognition were comparable to a non-endosomally presented C32 GMM (Figure 4B). Titration of the pH showed that the transition from slow to rapid presentation occurred in the pH range 5.5–5.0, which is in the range normally present in late endosomes (Figure 4C). Treating either antigens alone or APCs alone with acid media had no effect on presentation, suggesting that acidic pH did not somehow activate or cleave CD1b proteins or GMMs to generate a form that could be recognized after subsequent interactions at neutral pH. Instead, two-stage antigen processing assays showed that antigen and APC had to be in physical contact during the acidification to see fast kinetics of recognition (Figure 4D).
Figure 4.
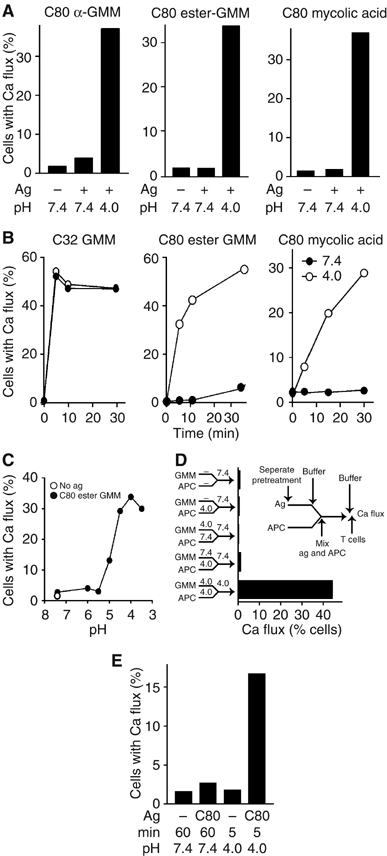
Acidic pH is sufficient to allow rapid presentation of mycolyl glycolipids to T cells. (A–C) B lymphoblastoid cells transfected with CD1b (C1R.CD1b) were preincubated with equipotent doses of C80 α-GMM (0.2 μM), C80 ester GMM (0.2 μM) or C80 MA (40 μM) at either pH 7.4 or 4.0 for 30 min (A), or indicated pH for 15 min (C) or the indicated time (B). The APCs were then neutralized with basic buffers to give pH 7.4 before mixing with T cells and immediately measured by calcium flux. (D) The experimental scheme (inset) shows a two-stage protocol by which cells and antigens were separately incubated at pH 4.0 or 7.4 for 30 min (stage 1), buffered to the indicated pH, mixed, incubated for 5 min (stage 2) and buffered to neutrality before testing T-cell activation by Ca flux. (E) After fixing C1R.CD1b cells under conditions that were previously known to inhibit endosomal uptake (0.02% glutaraldehyde), C80 ester GMM was added for the indicated time before measuring CD1b-mediated Ca flux in T cells.
Because C1R lymphoblastoid cells have particularly inefficient mechanisms for antigen internalization compared to DCs (Moody et al, 2002) and the time periods were short compared to those necessary lysosomal recycling times (Figure 1B), these results were most consistent with pH-induced loading on CD1b proteins at or near the cell surface. However, even in 5 min, antigens or acidic media could be internalized into cells to some degree; so we carried out similar experiments in which cellular membranes were fixed with glutaraldehyde. Although fixation conditions strongly inhibit internalization and recognition of C80 GMM at neutral pH, they did not prevent the acid-mediated presentation of C80 GMM (Figure 4E). These studies suggested that low pH was a key endosomal factor in antigen loading, such that it was sufficient to bypass the need for trafficking of CD1b and antigens to late endosomes and lysosomes, which are normally required for recognition.
Acidic pH promotes CD1b–antigen complex formation in cell-free conditions
To more formally assess whether low pH was sufficient to promote loading in the absence of all cellular factors, we prepared purified, hexahistidine-tagged CD1b proteins and treated them with GMM, recovered them on nickel beads and used GMM-treated CD1b proteins to activate T cells. As seen previously in assays of T-cell activation evaluated with IL-2 release, cytolysis and proliferation (Moody et al, 1997, 2002 and data not shown), control experiments showed that measurement of T-cell activation by calcium flux was absolutely dependent on the presence of antigen, CD1b proteins and a brief centrifugation to initiate contact between T cells and beads coated with CD1 and antigen (Figure 5A). CD1b-coated beads incubated with C32 GMM activated T cells to maximum levels when incubated at neutral pH or mildly acidic pH, but no activation was seen with CD1b proteins treated with C80 α-GMM. At pH 5.0, C80 GMM-treated CD1b proteins were able to stimulate a strong T-cell response that was comparable to levels seen with C32 GMM (Figure 5A).
Figure 5.
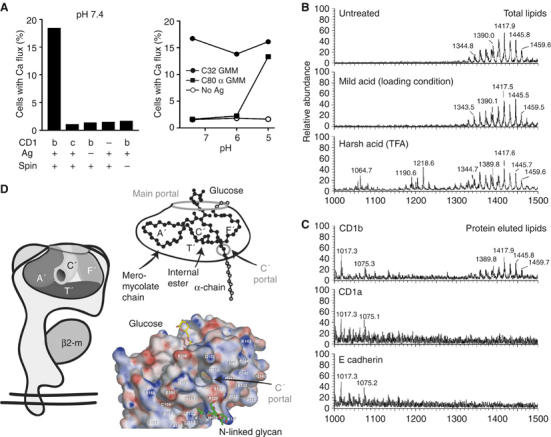
Formation of antigenic CD1b–GMM complexes under cell-free conditions. (A) Recombinant hexahistidine-tagged CD1b or CD1c proteins were coincubated with C80 α-GMM or C32 GMM or no antigen for 20 h at the indicated pH. The mixture was then neutralized to pH 7.4 before adding nickel beads, washing, adding LDN5 T cells and subjecting the mixture to centrifugation (spin) to initiate bead contact with T cells and measuring calcium flux. This result is representative of four experiments and similar results were obtained for C80 ester GMM (not shown). (B, C) His-tagged CD1a, CD1b or E cadherin proteins were incubated with excess C80 ester GMM and α-GMM at pH 4.0 for 5 h, treated with magnetic nickel beads and recovered with a magnet. Total lipids in the supernatant of this mild acid treatment were compared to untreated lipids and lipids treated with harsh acid conditions as in Figure 3C (TFA, pH 1.0, 120°C, 120 min), revealing negative ions corresponding to C60 dicarboxylate GMM (m/z 1064.7 and others), free mycolate (m/z 1190.6 and others) and acetate adducts of intact α-GMM (m/z 1389.8 and others) and ester GMM (m/z 1445.7 and others). Lipids bound to protein–bead complexes showed ions corresponding to intact GMMs (m/z 1417.9 and others). (D) The CD1b groove can be accessed via the main portal or the smaller C′ portal located at the inferior margin of the C′ pocket (Gadola et al, 2002). Based on the crystal structure of CD1b bound to C56 GMM (Batuwangala et al, 2004), this schematic shows how the C56 mycolate moiety (black) lies in the groove with α-branch into the C′ pocket, and the longer meromycolate chain lies in the A′, T′ and F′ pockets. The C77−86 mycolates found in mycobacterial pathogens exceed the interior capacity of the groove, and are predicted to protrude through the portals (gray atoms). An electrostatic surface representation of the CD1b–GMM complex was generated in APBS and illustrates sites of electropositive (blue) and electronegative (red) charge in the range of −15 to +15 kT/e. This view shows the adjacent glycan at N128 and two shallow grooves that lead to relatively non-polar (white) areas on the outer surface of the CD1b protein.
These experiments indicated that CD1b can present GMMs in the absence of all cellular enzymes, so that any possible cleavage reactions would have been chemically mediated. It was unlikely that chemically mediated mycolate cleavage occurred, based on prior experiments in which we had determined that chemical conditions necessary to detectably cleave GMM were significantly more harsh than those used here (Figure 3C). Confirming this, TLC autoradiography of labeled GMMs treated with acid media under conditions that allowed rapid presentation by cells did not detect alterations in GMM mobility (Supplementary Figure 2). Further, we mass spectrometrically analyzed the structures of GMM antigens subjected to the mild conditions that were sufficient for loading in this cell-free assay (pH 5.0, 20°C, 5 h) and the much harsher conditions (pH 1, 120°C, 2 h) necessary to visualize GMM cleavage products in TLC assays (Figure 3C). The chemically susceptible break points in GMM antigens are the internal ester in the meromycolate chain, the ester linkage to glucose and possibly others in glucose moiety or the unsaturation in the meromycolate chain. Unsubstituted alkyl chains are extremely resistant to cleavage. In agreement with these predictions, MS analysis of TFA-treated GMM showed negative ions corresponding to an internal ester-cleaved C60 GMM (m/z 1064.7), free MA (m/z 1190.6) and ions in an intermediate range (m/z∼1300), which may correspond to partial loss of glucose. However, antigens treated with mild acid conditions used for loading gave strong mass spectral signals showing that ions corresponding to intact α-GMMs (m/z 1386 and others) and ester GMMs were unaltered compared to starting material (Figure 5B).
After scale up of the in vitro loading reaction, we used nano-electro-spray MS to specifically analyze lipids eluted from CD1b, CD1a and E cadherin proteins treated with GMM under mild conditions that are necessary for loading (Figure 5C). MS analysis of highly concentrated lipid eluents showed that CD1b yielded ions corresponding to intact α-GMM and ester GMM (m/z 1389.8–1459.7). Although several ions that did not match the mass of GMMs (m/z 1017.3, 1075.3), they did not correspond to known products of GMM cleavage, and they were also present in CD1a and E cadherin eluents, suggesting that they are contaminants introduced during the elution and drying process. Thus, we found no evidence for GMM antigens with reduced lipid length, either in the supernatants or bound to CD1b, generated under the mild acid conditions necessary to form antigenic GMM complexes.
Discussion
Both proteins and lipids undergo antigen processing in the endosomes of APCs before their recognition as antigen complexes by T cells. For peptides, an understanding of the differences in structures of unprocessed proteins and processed peptides was achieved by mass spectrometric detection of peptide fragments eluted from cellular MHC–peptide complexes (Rotzschke et al, 1990; Van Bleek and Nathenson, 1990; Rudensky et al, 1991). For lipids, it has been unclear as to whether endosomal processing reactions normally involve the alteration of the lipid moieties of antigens. Although lipids can be eluted from cellular CD1 proteins (Joyce et al, 1998), this approach has not been widely applied to CD1–lipid complexes formed after antigen processing in intact cells because detergents used to extract the transmembrane domains of CD1 proteins from cellular membranes can disrupt CD1–lipid complexes. Therefore, we sought to produce antigenic complexes in vitro with antigens whose lipid moieties exceed the volume of CD1b grooves and analyze the structure of bound lipids. In parallel, we measured lipid cleavage reactions in intact DCs using radiolabled antigens. The latter approach was considered feasible because mammalian cells cannot trim lipids from the alkyl terminus using ω-oxidation. Therefore, any modification of lipid structure would likely occur by β-oxidation at the carboxylate or chemical attack at discernable chemical substitutions on alkyl chains, resulting in relatively simple and interpretable patterns of antigen-derived breakdown products.
Our results show that human DCs rapidly internalize labeled mycolyl lipids and deliver them to lysosomes that coexpress LAMP-1, CD1b or MHC class II. One of the antigens studied, C80 ester GMM, was cleaved by hydrolysis of an internal ester in its meromycolate chain to yield a product with a C60 lipid moiety and an intact TCR epitope. Thus, DCs can take up lipids into endosomes and modify their length by selectively cleaving them in a way that does not otherwise degrade the antigen. The cleavage reaction does not occur at pH 4.0 outside of cells (Figure 5C; Supplementary Figure 2), suggesting that it is not generated by low pH alone, but instead is carried out by an as yet unknown lysosomal enzyme. This ester hydrolysis reaction is equivalent of that which is necessary for deacylation of diacylglycerols or sphingolipids; so this kind of reaction may be of importance in altering the lipid moieties of other antigens. However, among various types of mycolates, wax-ester forms are known to be present in M. phlei and other saprophytes, but not Mycobacterium tuberculosis and Mycobacterium leprae, two pathogens that generate CD1-mediated responses in vivo (Moody et al, 2000a). Therefore, the cleavage of C80 ester GMM provides a serendipitous positive control, indicating that all tested GMM and free mycolic antigens entered into cells and were exposed to the hydrolytic environment within late endosomal and lysosomal compartments. However, the more general conclusion is that MA and GMM antigens are not detectably altered in chemical structure when they are taken up into DCs for time periods that greatly exceed the 5–30 min necessary to generate antigenic complexes (Figures 1B and 3A) or under conditions needed to form antigenic complexes in cell-free conditions. Even a sensitive cellular assay might have failed to detect all fragments generated by cellular metabolism, but the generation of antigenic CD1b–GMM complexes under cell-free conditions rules out enzymatic cleavage reactions to alter lipid length before loading (Figure 5).
Instead of lipid trimming, low pH is necessary and sufficient to promote the formation of antigen complexes of CD1b with each of three long-chain bacterial lipid antigens. Low pH might act through cellular factors such as lipid transfer proteins (Kang and Cresswell, 2004; Zhou et al, 2004a), especially saposin C and apolipoprotein E, which are normally located in endosomes and have been shown to enhance mycolyl lipid presentation by CD1b (Winau et al, 2004; van den Elzen et al, 2005). Whereas some of the effects of these two lipid transfer proteins occur at neutral pH, the lipid-transferring properties of saposins are enhanced at low pH (Zhou et al, 2004a). Our results make clear that low pH, acting as a single variable, mediates all or nothing differences in antigen recognition under cell-free conditions. Thus, low pH carries out key effects directly on the CD1b–antigen interaction that are separate from any pH-mediated effects on lipid transfer proteins.
Because C80 mycobacterial mycolates exceed the volume of the CD1b groove, a basic question arises as to what happens to any excess lipid that cannot fit within the groove. These results and the crystal structure of the CD1b–GMM complex point to an explicit and plausible mechanism (Figure 5D). In the crystal structure, the carbohydrate moiety of GMM rests near the entrance of the groove and the C56 mycolate lipid is positioned so that the shorter α-branch occupies the small C′ pocket and the longer meromycolate branch lies in the A′T′F′ superchannel (Batuwangala et al, 2004). If cellular CD1b proteins also bind antigens in this way, then loading of intact C80 mycolate moieties indicates that the termini of the meromycolate chain and the α-chain protrude through the main portal and C′ portal, respectively (Figure 5D, gray atoms). This model predicts that certain CD1b-presented lipids are bound entirely within the confines of the groove, but the C′ portal functions as an escape hatch that allows CD1b proteins to bind particularly long bacterial lipids such that their termini protrude to the outer surface of the protein.
This more complex binding mechanism may allow CD1b to mediate T-cell recognition of highly diverse classes of lipids by re-routing excess lipid away from the TCR interaction site. The position of the protruding lipid is not yet known from crystal structures, but two small concavities on the outer surface of CD1b near the C′ portal could guide the lipid toward isoleucine 149 or tyrosine 163 (Figure 5D). Such excess lipid may have no substantial effect on recognition, akin to the mechanism by which the ragged ends of long peptides extend beyond the MHC II platform. Alternatively, the protruding lipid may interact with the outer surface of CD1 proteins in ways that affect lipid retention within the groove, as suggested by studies in which long-chain lipid antigens show particularly high potency and long half-lives compared to short-chain antigens (Beckman et al, 1994; Shamshiev et al, 1999, 2002; Moody et al, 2002; Gilleron et al, 2004). These considerations point to a testable model whereby the CD1b protein is a pH-regulated lipid clamp. CD1b likely binds low-affinity, shorter chain self-lipids during its transit through the secretory pathway, but then pH-induced relaxation of the α1 and α2 domain structure allows more extensive access to the groove by intact long-chain lipids during trafficking through acidic endosomes, which are then retained more effectively in post-endosomal compartments. Because long chain length distinguishes mycolyl and phthioceranoate lipids present in the cell wall of mycobacteria from smaller mammalian lipids, the ability of endosomally localized CD1b proteins to clamp down on long-chain mycobacterial lipids could serve to enhance self–nonself lipid antigen discrimination.
Materials and methods
Lipid antigens
M. phlei, Nocardia farcinica, Rhodococcus equi and M. fallax were cultured in 7H9 medium supplemented with 0.5 g/l Tween 80 and 10 g/l D-glucose. 14C-labeled lipids were produced by adding 1 mCi/l of [14C]acetate in early log phase for 24 h. GMMs were purified as described previously (Moody et al, 1997) or by one-dimensional preparative silica TLC (Scientific Adsorbents Inc., Atlanta, GA) resolved with chloroform/methanol/water (60/16/2). R. equi, N. farcinica, M. phlei and M. fallax produced C32 GMM, C54 GMM, C80 ester GMM and C80 α-GMM, respectively. M. tuberculosis MA (C80 MA) was purchased from Sigma or purified from saponificates of M. tuberculosis.
T-cell cultures and cellular assays
LDN5 and DN1 T cell lines (Porcelli et al, 1992; Beckman et al, 1994; Moody et al, 1997) were activated by C1R B lymphoblastoid cells transfected with full-length human CD1b (C1R.CD1b) or human monocyte-derived DCs prepared from the peripheral blood by centrifugation over Ficoll–Hypaque, adherence to tissue-culture flasks, culture of adherent cells with 300 U/ml GM-CSF and 200 U/ml IL-4 for 72–96 h, followed by irradiation (5000 rad). IL-2 release from LDN5 T cells or J.RT-3 cells transfected with the αβ-chains of LDN5 T cells (10 ng/ml PMA) was measured by culturing T cells with DCs or C1R (1:1 ratio) cells plus antigen in 200 μl/well. After 24 h, 50 μl of supernatant was transferred to wells containing 125 μl of media and 104 HT-2 cells, which were cultured for 24 h before adding 1 μCi [3H]thymidine for an additional 6–24 h of culture, followed by harvesting and counting β emissions.
To determine the kinetic profiles for T-cell activation, T cells were loaded with fluorophores, Fura-red (4 μM) and Fluo-4 (2 μM), in Hank's balanced salt solution (HBSS) and subjected to flow cytometric (BD FacSort) analysis of calcium-induced changes in their fluorescence emission spectra. Antigen loading onto APCs at neutral pH was accomplished by incubating 106 cells/ml for 1 min to 24 h, followed by mixing 1:1 with antigen-loaded C1R cells or DCs, centrifugation (∼1000 g) for 60 s, vortexing and FACS analysis for 20 s or 2000 events (Moody et al, 2002). For fixation experiments, C1R.CD1b was fixed with 0.02% glutaraldehyde for 30 s and then the reaction was quenched with excess lysine (0.2 M) as described (Moody et al, 2002).
Antigen loading was measured after either one- or two-stage protocols for adjusting the pH. For one-stage loading, APCs (2 × 106/ml) and antigens were mixed together at a 1:1 ratio in HBSS with 1% BSA, 0.35 g sodium bicarbonate and 20 ml Hepes that had been adjusted to pH 3.5, 4.0, 4.5, 5.0, 5.5, 6.0 or 7.4 using HCl or NaOH and confirmed by a electronic pH meter. Before adding T cells, the medium was neutralized by adding an equal volume of basic medium to adjust pH to 7.4. Two-stage experiments were carried out by first incubating the APCs alone and antigen alone in separate tubes under defined pH conditions for 30 min before mixing the APCs and antigen (stage 1). The mixture of APCs and antigens were adjusted to pH to 7.4 or 4.0 followed by an additional incubation for 5 min (stage 2), pH neutralization, adding T cells and immediately measuring T-cell activation by calcium flux.
Recombinant protein–antigen complexes
The extracellular domains of CD1a, CD1b and CD1c heavy chains linked to β2M were amplified from their corresponding cDNA by PCR and separately cloned into the expression vector pMT/BiP/V5-His B (Invitrogen), which carries a C-terminal hexahistidine tag sequence. A stop codon was introduced at the 3′ end of the β2M sequence via the PCR primer and the resulting plasmids were cotransfected at equimolar ratios together with a 1:40 molar ratio of a blasticidin resistance gene into Drosophila melanogaster S2 cells using Effectene (Sigma). CD1-β2M was purified from by Ion Metal Affinity Chromatography on a Ni-NTA resin (Qiagen), anion exchange chromatography on MonoQ HR5/5 in 10 mM Tris–HCl pH 8.0 and size-exclusion chromatography on Superdex S200 16/60 in 50 mM Hepes pH 7.5 and 150 mM NaCl using a fast-performance liquid chromatography system (GE Healthcare). Recombinant His-tagged proteins comprised of the first two extracellular domains of E cadherin were the gift of Emilio Parisini and Jonathon Higgins.
For cell-free presentation of antigens to T cells, His-tagged CD1 or E cadherin was incubated for 5–20 h with 40- to 75-fold molar excess of GMM in 100 μl HBS at pH values ranging from 5 to 7.4. Acidic buffers were neutralized and incubated with 60 μl of Ni-NTA agarose bead for 20–45 min, and pilot experiments using flow cytometry showed that the level of CD1b adherent to beads was not affected by pH treatments. The resulting complexes were added to T cells for measuring calcium flux (6 μg, 1.3 μM) or used in larger amounts (100 μg) for elution for MS. CD1b-bound lipids were separated from total lipids by centrifugation and magnetic capture of nickel beads, followed by three washes, extraction in chloroform/methanol (2:1), drying under nitrogen and re-dissolving in 1/20th of the starting volume of 100 mM NH4C2H3O2 of C/M (2:1) solution for nanospray MS. Total lipids subjected to mild acid but not bound to CD1b were analyzed in the same way except that supernatants were subjected to Folch extraction.
Mass spectrometry
Nanospray ESI-MS (ThermoFinnigan LCQ Advantage) was performed using borosilicate glass pipettes pulled to a 2-μm orifice. Preparative TLC eluates were analyzed by CID-MS/MS using 8:2 (V/V) isopropyl alcohol:120 mM NH4C2H3O2 aqueous solution.
Lipid cleavage assays
Immature DCs (600 000) were incubated with 1 μM of [14C]C32 GMM, [14C]C80 α-GMM, [14C]C80 ester GMM or [14C]C80 MA for 4–72 h, washed with PBS (5% FBS), detached with 0.05% trypsin and 0.53 mM EDTA in PBS in a total volume of 300 μl and then transferred to a 15 ml glass tube. CHCl3/MeOH (2:1; 700 μl) was added to the tube followed by vortexing and centrifugation (2000 g for 10 min). The upper aqueous phases lack detectable scintillation. The lower organic phase was collected and washed twice with an excess volume of water and developed on silica TLC with CHCl3/MeOH/H2O 60/16/2 (V/V/V) and exposed to film for 24–72 h. [14C]MA-treated cells were analyzed in the same way except that silica plates were resolved with petroleum ether/ether/acetic acid 65/35/1 (V/V/V). Plates were charred with cupric acetate solution (3% cupric acetate in 8% phosphoric acid) at 160°C for 15 min. In selected experiments, concanamycin B (10 nM) was added 1 h before adding antigens. For cell-free antigen cleavage under harsh conditions, 400 μg of C80 ester GMM or C80 α-GMM was dried under a stream of nitrogen in a 15 ml glass tube. A 1 ml volume of 2 M TFA (Sigma-Aldrich) was added and heated at 120°C for 2 h and cooled. Then, 1 ml of chloroform was added, extensively vortexed and subjected to centrifugation (2500 r.p.m., 10 min) to give two layers. The lower layer was dried on the silica TLC plate and resolved with chloroform:methanol:water (60:16:2).
Confocal microscopy
DCs were cultured in complete medium with 4 μM BODIPY-GMM at 37°C for 30 min, adhered to fibronectin (20 μg/ml)-coated coverslips for 1 h before fixing with 2% paraformaldehyde in Hank's media for 2 min at 20°C. Cells were permeabilized with 0.2% saponin and stained with P3, BCD1B.3 (anti-CD1b), L243 (anti-MHC II) or antisera against LAMP-1 followed by Texas red-conjugated donkey F(ab′)2 anti-rabbit IgG or Texas red conjugated donkey F(ab′)2 anti-mouse IgG and analyzed with a Nikon Eclipse TE 2000-U confocal microscope.
Computer modeling of CD1b
Surfaces and electrostatic potentials of the CD1b–GMM complex were generated with the programs PyMol (Delano Scientific, San Corlos, CA, 2002) by Delano and APBS (Baker et al, 2001) using CD1b coordinates from 1UQS (Batuwangala et al, 2004).
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure Legends
Acknowledgments
We gratefully acknowledge William Biddison for advice, John Belisle and Patrick Brennan for M. tuberculosis lipids and Masahiko Sugita for providing concanamycin B. Charles Anthony Debono analyzed BODIPY-mycolates and Carme Roura-mir helped with microscopic analysis of lipids. This work was supported by the Pew Foundation, the American College of Rheumatology Research and Education Foundation, the Mizutani Foundation, the Cancer Research Institute, the Lister Institute, the Personal Research Chair from James Burdick, the Medical Research Council, the Wellcome Trust (GSB) and the NIH AI49313 (DBM), AR48632 (DBM), GM62116 (IAW) and CA 58896 (IAW).
References
- Agea E, Russano A, Bistoni O, Mannucci R, Nicoletti I, Corazzi L, Postle AD, De Libero G, Porcelli SA, Spinozzi F (2005) Human CD1-restricted T cell recognition of lipids from pollens. J Exp Med 202: 295–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA 98: 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batuwangala T, Shepard D, Gadola SD, Gibson KJC, Zaccai NR, Besra GS, Cerundolo V, Jones EY (2004) The crystal structure of human CD1b with a bound bacterial glycolipid. J Immunol 172: 2382–2388 [DOI] [PubMed] [Google Scholar]
- Beckman EM, Porcelli SA, Morita CT, Behar SM, Furlong ST, Brenner MB (1994) Recognition of a lipid antigen by CD1-restricted αβ T cells. Nature 372: 691–694 [DOI] [PubMed] [Google Scholar]
- Briken V, Jackman RM, Dasgupta S, Hoening S, Porcelli SA (2002) Intracellular trafficking pathway of newly synthesized CD1b molecules. EMBO J 21: 825–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YH, Jayawardena J, Weiss A, Lee D, Park SH, Dautry-Varsat A, Bendelac A (1999) Distinct subsets of CD1d-restricted T cells recognize self-antigens loaded in different cellular compartments. J Exp Med 189: 103–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst WA, Maher J, Cho S, Niazi KR, Chatterjee D, Moody DB, Besra GS, Watanabe Y, Jensen PE, Porcelli SA, Kronenberg M, Modlin RL (1998) Molecular interaction of CD1b with lipoglycan antigens. Immunity 8: 331–340 [DOI] [PubMed] [Google Scholar]
- Gadola SD, Zaccai NR, Harlos K, Shepherd D, Castro-Palomino JC, Ritter G, Schmidt RR, Jones EY, Cerundolo V (2002) Structure of human CD1b with bound ligands at 2.3 Å, a maze for alkyl chains. Nat Immunol 3: 721–726 [DOI] [PubMed] [Google Scholar]
- Giabbai B, Sidobre S, Crispin MD, Sanchez-Ruiz Y, Bachi A, Kronenberg M, Wilson IA, Degano M (2005) Crystal structure of mouse CD1d bound to the self ligand phosphatidylcholine: a molecular basis for NKT cell activation. J Immunol 175: 977–984 [DOI] [PubMed] [Google Scholar]
- Gilleron M, Stenger S, Mazorra Z, Wittke F, Mariotti S, Bohmer G, Prandi J, Mori L, Puzo G, De Libero G (2004) Diacylated sulfoglycolipids are novel mycobacterial antigens stimulating CD1-restricted T cells during infection with Mycobacterium tuberculosis. J Exp Med 199: 649–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayawardena-Wolf J, Benlagha K, Chiu YH, Mehr R, Bendelac A (2001) CD1d endosomal trafficking is independently regulated by an intrinsic CD1d-encoded tyrosine motif and by the invariant chain. Immunity 15: 897–908 [DOI] [PubMed] [Google Scholar]
- Joyce S, Woods AS, Yewdell JW, Bennink JR, De SA, Boesteanu A, Balk SP, Cotter RJ, Brutkiewicz RR (1998) Natural ligand of mouse CD1d1: cellular glycosylphosphatidylinositol. Science 279: 1541–1544 [DOI] [PubMed] [Google Scholar]
- Kang SJ, Cresswell P (2004) Saposins facilitate CD1d-restricted presentation of an exogenous lipid antigen to T cells. Nat Immunol 5: 175–181 [DOI] [PubMed] [Google Scholar]
- Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, Ueno H, Nakagawa R, Sato H, Kondo E, Koseki H, Taniguchi M (1997) CD1d-restricted and TCR-mediated activation of Vα14 NKT cells by glycosylceramides. Science 278: 1626–1629 [DOI] [PubMed] [Google Scholar]
- Kinjo Y, Wu D, Kim G, Xing GW, Poles MA, Ho DD, Tsuji M, Kawahara K, Wong CH, Kronenberg M (2005) Recognition of bacterial glycosphingolipids by natural killer T cells. Nature 434: 520–525 [DOI] [PubMed] [Google Scholar]
- Koch M, Stronge VS, Shepherd D, Gadola SD, Mathew B, Ritter G, Fersht AR, Besra GS, Schmidt RR, Jones EY, Cerundolo V (2005) The crystal structure of human CD1d with and without alpha-galactosylceramide. Nat Immunol 6: 819–826 [DOI] [PubMed] [Google Scholar]
- Matsunaga I, Bhatt A, Young DC, Cheng TY, Eyles SJ, Besra GS, Briken V, Porcelli SA, Costello CE, Jacobs WR Jr, Moody DB (2004) Mycobacterium tuberculosis pks12 produces a novel polyketide presented by CD1c to T cells. J Exp Med 200: 1559–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattner J, Debord KL, Ismail N, Goff RD, Cantu C III, Zhou D, Saint-Mezard P, Wang V, Gao Y, Yin N, Hoebe K, Schneewind O, Walker D, Beutler B, Teyton L, Savage PB, Bendelac A (2005) Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature 434: 525–529 [DOI] [PubMed] [Google Scholar]
- Moody DB, Briken V, Cheng TY, Roura-Mir C, Guy MR, Geho DH, Tykocinski ML, Besra GS, Porcelli SA (2002) Lipid length controls antigen entry into endosomal and nonendosomal pathways for CD1b presentation. Nat Immunol 3: 435–442 [DOI] [PubMed] [Google Scholar]
- Moody DB, Guy MR, Grant E, Cheng TY, Brenner MB, Besra GS, Porcelli SA (2000a) CD1b-mediated T cell recognition of a glycolipid antigen generated from mycobacterial lipid and host carbohydrate during infection. J Exp Med 192: 965–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody DB, Reinhold BB, Guy MR, Beckman EM, Frederique DE, Furlong ST, Ye S, Reinhold VN, Sieling PA, Modlin RL, Besra GS, Porcelli SA (1997) Structural requirements for glycolipid antigen recognition by CD1b-restricted T cells. Science 278: 283–286 [DOI] [PubMed] [Google Scholar]
- Moody DB, Ulrichs T, Muhlecker W, Young DC, Gurcha SS, Grant E, Rosat JP, Brenner MB, Costello CE, Besra GS, Porcelli SA (2000b) CD1c-mediated T-cell recognition of isoprenoid glycolipids in Mycobacterium tuberculosis infection. Nature 404: 884–888 [DOI] [PubMed] [Google Scholar]
- Moody DB, Young DC, Cheng TY, Rosat JP, Roura-Mir C, O'Connor PB, Zajonc DM, Walz A, Miller MJ, Levery SB, Wilson IA, Costello CE, Brenner MB (2004) T cell activation by lipopeptide antigens. Science 303: 527–531 [DOI] [PubMed] [Google Scholar]
- Porcelli S, Morita CT, Brenner MB (1992) CD1b restricts the response of human CD4−8− T lymphocytes to a microbial antigen. Nature 360: 593–597 [DOI] [PubMed] [Google Scholar]
- Prigozy TI, Naidenko O, Qasba P, Elewaut D, Brossay L, Khurana A, Natori T, Koezuka Y, Kulkarni A, Kronenberg M (2001) Glycolipid antigen processing for presentation by CD1d molecules. Science 291: 664–667 [DOI] [PubMed] [Google Scholar]
- Rauch J, Gumperz J, Robinson C, Skold M, Roy C, Young DC, Lafleur M, Moody DB, Brenner MB, Costello CE, Behar SM (2003) Structural features of the acyl chain determine self-phospholipid antigen recognition by a CD1d-restricted invariant NKT (iNKT) cell. J Biol Chem 278: 47508–47515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotzschke O, Falk K, Deres K, Schild H, Norda M, Metzger J, Jung G, Rammensee HG (1990) Isolation and analysis of naturally processed viral peptides as recognized by cytotoxic T cells. Nature 348: 252–254 [DOI] [PubMed] [Google Scholar]
- Rudensky AY, Preston-Hurlburt P, Hong SC, Barlow A, Janeway CA Jr (1991) Sequence analysis of peptides bound to MHC class II molecules. Nature 353: 622–627 [DOI] [PubMed] [Google Scholar]
- Shamshiev A, Donda A, Carena I, Mori L, Kappos L, De Libero G (1999) Self glycolipids as T-cell autoantigens. Eur J Immunol 29: 1667–1675 [DOI] [PubMed] [Google Scholar]
- Shamshiev A, Gober HJ, Donda A, Mazorra Z, Mori L, De Libero G (2002) Presentation of the same glycolipid by different CD1 molecules. J Exp Med 195: 1013–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieling PA, Chatterjee D, Porcelli SA, Prigozy TI, Mazzaccaro RJ, Soriano T, Bloom BR, Brenner MB, Kronenberg M, Brennan PJ (1995) CD1-restricted T cell recognition of microbial lipoglycan antigens. Science 269: 227–230 [DOI] [PubMed] [Google Scholar]
- Trombetta ES, Ebersold M, Garrett W, Pypaert M, Mellman I (2003) Activation of lysosomal function during dendritic cell maturation. Science 299: 1400–1403 [DOI] [PubMed] [Google Scholar]
- Ulrichs T, Moody DB, Grant E, Kaufmann SH, Porcelli SA (2003) T-cell responses to CD1-presented lipid antigens in humans with Mycobacterium tuberculosis infection. Infect Immun 71: 3076–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bleek GM, Nathenson SG (1990) Isolation of an endogenously processed immunodominant viral peptide from the class I H-2Kb molecule. Nature 348: 213–216 [DOI] [PubMed] [Google Scholar]
- van den Elzen P, Garg S, Leon L, Brigl M, Leadbetter EA, Gumperz JE, Dascher CC, Cheng TY, Sacks FM, Illarionov PA, Besra GS, Kent SC, Moody DB, Brenner MB (2005) Apolipoprotein-mediated pathways of lipid antigen presentation. Nature 437: 906–910 [DOI] [PubMed] [Google Scholar]
- Winau F, Schwierzeck V, Hurwitz R, Remmel N, Sieling PA, Modlin RL, Porcelli SA, Brinkmann V, Sugita M, Sandhoff K, Kaufmann SH, Schaible UE (2004) Saposin C is required for lipid presentation by human CD1b. Nat Immunol 5: 169–174 [DOI] [PubMed] [Google Scholar]
- Zajonc DM, Cantu C, Mattner J, Zhou D, Savage PB, Bendelac A, Wilson IA, Teyton L (2005a) Structure and function of a potent agonist for the semi-invariant natural killer T cell receptor. Nat Immunol 6: 810–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajonc DM, Crispin MD, Bowden TA, Young DC, Cheng TY, Hu J, Costello CE, Rudd PM, Dwek RA, Miller MJ, Brenner MB, Moody DB, Wilson IA (2005b) Molecular mechanism of lipopeptide presentation by CD1a. Immunity 22: 209–219 [DOI] [PubMed] [Google Scholar]
- Zajonc DM, Elsliger MA, Teyton L, Wilson IA (2003) Crystal structure of CD1a in complex with a sulfatide self antigen at a resolution of 2.15 Å. Nat Immunol 4: 808–815 [DOI] [PubMed] [Google Scholar]
- Zeng Z, Castaño AR, Segelke BW, Stura EA, Peterson PA, Wilson IA (1997) Crystal structure of mouse CD1: an MHC-like fold with a large hydrophobic binding groove. Science 277: 339–345 [DOI] [PubMed] [Google Scholar]
- Zhou D, Cantu C III, Sagiv Y, Schrantz N, Kulkarni AB, Qi X, Mahuran DJ, Morales CR, Grabowski GA, Benlagha K, Savage P, Bendelac A, Teyton L (2004a) Editing of CD1d-bound lipid antigens by endosomal lipid transfer proteins. Science 303: 523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Mattner J, Cantu C III, Schrantz N, Yin N, Gao Y, Sagiv Y, Hudspeth K, Wu YP, Yamashita T, Teneberg S, Wang D, Proia RL, Levery SB, Savage PB, Teyton L, Bendelac A (2004b) Lysosomal glycosphingolipid recognition by NKT cells. Science 306: 1786–1789 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure Legends


