Abstract
The protein kinase Chk2 (checkpoint kinase 2) is a major effector of the replication checkpoint. Chk2 activation is initiated by phosphorylation of Thr68, in the serine–glutamine/threonine–glutamine cluster domain (SCD), by ATM. The phosphorylated SCD-segment binds to the FHA domain of a second Chk2 molecule, promoting dimerisation of the protein and triggering phosphorylation of the activation segment/T-loop in the kinase domain. We have now determined the structure of the kinase domain of human Chk2 in complexes with ADP and a small-molecule inhibitor debromohymenialdisine. The structure reveals a remarkable dimeric arrangement in which T-loops are exchanged between protomers, to form an active kinase conformation in trans. Biochemical data suggest that this dimer is the biologically active state promoted by ATM-phosphorylation, and also suggests a mechanism for dimerisation-driven activation of Chk2 by trans-phosphorylation.
Keywords: cancer, CHEK2, CHK2, inhibitor, kinase
Introduction
DNA double-strand breaks, generated by ionising radiation, genotoxic chemicals or collapsed replication forks, are the most serious type of DNA damage with which the eukaryotic cell must contend. Survival depends on a coordinated response whereby the DNA lesion is repaired, and the progress of the cell cycle is halted to allow that repair to occur. Key to this response is the activation of a signalling network, dependent on the phosphatidylinositol kinase-like kinases (PIKKs), ATM and ATR, which are primary sensors of DNA damage (McGowan and Russell, 2004; Lavin et al, 2005) (Figure 1A). ATM/ATR driven signals modulate transcription through stabilisation of p53, and directly regulate cell cycle progression by promoting the degradation of the Cdc25 phosphatases that activate Cdk2/CyclinE complexes (Falck et al, 2001). An immediate downstream target of the ATM branch of the network is checkpoint kinase 2 (Chk2) (Cds1), a Ser/Thr kinase consisting of an N-terminal serine–glutamine/threonine–glutamine cluster domain (SCD), a middle phosphothreonine-binding FHA domain (Li et al, 2002) and a C-terminal catalytic domain (Figure 1B). Activated Chk2 in turn phosphorylates a range of downstream targets including Cdc25A and Cdc25C, BRCA1 and p53 (Bartek and Lukas, 2003). Chk2 activation is a multistep process, initiated by ATM, which phosphorylates Chk2 on Thr68 in a segment of the protein upstream of the FHA domain (Ahn et al, 2000; Matsuoka et al, 2000; Melchionna et al, 2000). Once phosphorylated, the pThr68 segment of Chk2 can bind in trans to the FHA domain of another molecule promoting dimerisation and trans-activating phosphorylation of Thr383 and Thr387 in the activation segment or ‘T-loop' of the catalytic domain (Ahn et al, 2002; Xu et al, 2002). However, the mechanism by which dimerisation promotes T-loop phosphorylation is not understood.
Figure 1.
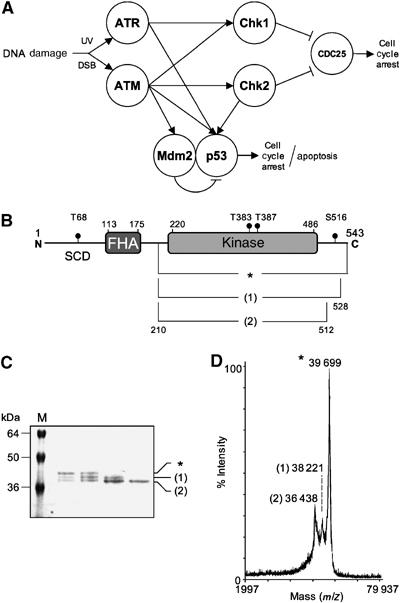
Chk2 in the DNA damage signalling network. (A) Simplified diagram of the ATM/ATR modulated signalling pathway. The PIKKs ATM and ATR are activated by DNA damage; ATM principally by DSB in DNA and ATR by UV damage and its associated lesions. Activated ATM phosphorylates the ‘transducer' kinases Chk1 and Chk2, which in turn phosphorylate the CDC25 phosphatases promoting their degradation. Subsequent downstream events lead to cell cycle arrest, facilitating DNA repair processes. ATR similarly promotes CDC25 degradation, but only via Chk1 activity. The ‘effector' protein p53, in undamaged cells, is rapidly degraded due to Mdm2-mediated ubiquitylation targeting the protein to the proteasome. Upon DNA damage p53 is phosphorylated (ATM, ATR, Chk2) inhibiting its interaction with Mdm2, hence stabilising the protein and allowing it to function as a transcription factor. Phosphorylation of Mdm2 by ATM also prevents the protein from shuttling p53 from the nucleus to the cytoplasm for degradation. (B) Schematic representation of the functional domains of Chk2. SCD=serine–glutamine/threonine–glutamine cluster domain, FHA=Forkhead-associated domain and KINASE=kinase domain. Predicted amino-acid ranges for each domain are indicated (from UniProt entry CHK2_HUMAN), along with phosphorylation sites (T69, T383, T387 and S516). The approximate amino-acid ranges for each protein/proteolysis product are also shown. (C) SDS–PAGE of selected gel filtration fractions. M=molecular size marker, with sizes indicated in kilodaltons (kDa). (D) MALDI-TOF analysis of proteolysis products. Molecular masses determined for each peak are shown. In (B–D), the full-length protein (amino acids 210–531) is labelled with an asterisk (*) and the proteolysis products are labelled (1) and (2) respectively.
The role played by Chk2 in mediating the cellular response to DNA damage gives it an important function as a tumour suppressor (McGowan, 2002). Truncations and missense mutations of Chk2 have been identified in a wide range of tumours, particularly breast (reviewed in Bartek and Lukas, 2003). Although Chk2 is formally a tumour suppressor, there is a growing interest in its inhibition in the treatment of cancer, both to radio-sensitise tumour cells already defective in other damage checkpoints (e.g. p53−/−), and to inhibit radiation induced apoptosis in sensitive collateral tissues (Bartek and Lukas, 2003; Collins and Garrett, 2005; Pommier et al, 2005).
Towards understanding the molecular mechanism for Chk2 activation and to facilitate development of specific inhibitors, we have now determined the crystal structure of a catalytic domain construct of human Chk2 in complex with Mg-ADP, and with the ATP-competitive kinase inhibitor debromohymenialdisine (DBQ) (Meijer et al, 2000; Sharma and Tepe, 2004). The structure of the Chk2 kinase domain reveals an unusual dimeric arrangement involving exchange of T-loops, and suggests a mechanism for dimerisation-driven activation of Chk2 by trans-phosphorylation.
Results
Chk2 kinase domain constructs
As attempts to crystallise full-length Chk2 were unsuccessful, we sought smaller constructs that would encapsulate the catalytic domain. Initial trials, based on sequence threading predictions, used an N-terminally His6-tagged construct running from Ser210 to the natural C-terminus at Leu543, but these were also unsuccessful. During purification of this protein, two proteolysis products were observed in SDS–PAGE. Western blotting with an anti-His6 antibody confirmed the retention of the N-terminal tag indicating that these products were C-terminally truncated (Figure 1C). The masses of the truncated proteins were determined using MALDI-TOF mass spectrometry (Figure 1D). The mass for the larger of the two bands equated to a protein containing amino acids 210–528, while the smaller equated to amino acids 210–512. A new recombinant construct based on the larger of these (210–531) was constructed as a protease-cleavable N-terminal GST fusion (see Materials and methods), which expressed at high levels, could be fully released from the fusion protein, and was readily purified to homogeneity.
Crystal structure of the binary Chk2 kinase domain/ADP complex
The purified Chk2(210-531) protein (CHK2-KD) crystallised as a binary complex with Mg-ADP. The structure was solved by molecular replacement using the structure of PKA as a search model (see Materials and methods), then rebuilt and refined at 2.25 Å. Statistics for the structure determination and refinement are given in Table I. Clear interpretable electron density is seen for residues 210–504 of the molecule, except for the loop comprised of residues 255–268 (connecting strand β3 to helix αC), which appears to be poorly ordered in these crystals. Similarly, at the C-terminus residues 505–531 have no discernable electron density and are also considered to be disordered. As expected, CHK2-KD has the two-lobe architecture of the canonical kinase fold, with the nucleotide-binding site residing between the two lobes (Figure 2). The N-lobe is folded around a five-stranded antiparallel β-sheet and contains the αC helix and the glycine-rich loop (residues 227–232 in Chk2), which is fully ordered in the Mg–ADP complex. The C-lobe is predominantly helical and contains the highly conserved DFG and APE motifs, and the intervening T-loop (residues 368–394); the hinge region comprised of residues 303–321 links the two lobes.
Table 1.
Data collection and refinement statistics
| CHK2-KD/MgADP | CHK2-KD/DBQ | |
|---|---|---|
| Data collection | ||
| Space group | P3221 | P3221 |
| Cell dimensions | ||
| a, b, c (Å) | 90.87, 90.87, 92.91 | 88.65, 88.65, 88.90 |
| α, β, γ (deg) | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å) | 40.83–2.25 (2.31–2.25)a | 39.65–2.70 (2.85–2.70) |
| Rmerge | 0.076 (0.474) | 0.067 (0.533) |
| Mn I/σI | 16.5 (3.2) | 16.0 (2.8) |
| Completeness (%) | 100 (100) | 98.7 (100) |
| Redundancy | 5.5 (5.6) | 4.3 (4.3) |
| Refinement | ||
| Resolution (Å) | 2.25 | 2.70 |
| No. of reflections | 20 374 | 10 709 |
| Rwork/Rfree | 0.20/0.24 | 0.21/0.27 |
| No. of atoms | ||
| Protein | 2271 | 2153 |
| Ligand/ion | 1(Cl−), 2(Mg2+), 4(NO3−), 27(ADP) | 1(Mg2+), 4(NO3−), 18 (DBQ) |
| Water | 156 | 44 |
| B-factors | ||
| Protein | 46.6 | 55.0 |
| Ligand/ion | 43.5(Cl−), 48.1(Mg2+), 40.0(NO3−), 42.3(ADP) | 65.8(Mg2+), 74.0(NO3−), 48.6(DBQ) |
| Water | 47.6 | 63.2 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.02 | 0.02 |
| Bond angles (deg) | 1.73 | 2.23 |
| aHighest resolution shell is shown in parenthesis. | ||
Figure 2.
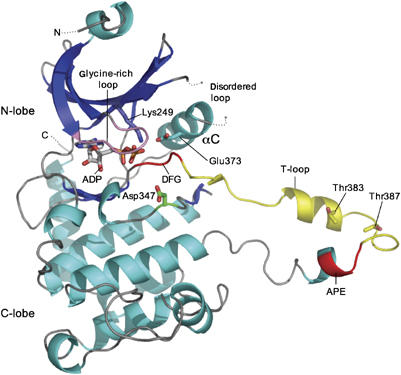
Structure of Chk2 kinase domain. Secondary structure cartoon of the binary complex of CHK2-KD with ADP. The glycine-rich loop is coloured pink, the DFG and APE motifs red, and the T-loop yellow. Secondary structure elements corresponding to β-strands are coloured blue and α-helices cyan. ADP is shown as a ‘stick' model. The catalytic residue Asp347, conserved residues Lys249 and Glu373, and autophosphorylation sites Thr383 and Thr387 are also highlighted. Molecular images in this and subsequent figures were generated using PyMOL (www.pymol.org).
T-loop exchange
The most immediately striking feature of the CHK2-KD crystal structure is the presence of an ordered T-loop or activation segment, projecting from the body of the kinase to interact with a second Chk2 molecule, forming a two-fold symmetric dimer interaction (Figure 3A and B). As well as its unusual location, the structure of the T-loop itself departs from the norm significantly, consisting of an antiparallel pair of close-packed amphipathic α-helices (377–386, 392–402) connected by an extended but well-ordered loop. The residues at the tip of the loop (Pro388, Thr389 and Tyr390) and the beginning of the second helix (Ala392, Pro393, Leu396 and Val397) pack into a hydrophobic recess on the dimer-related Chk2 lined by residues from the α-helices 405–423 and 435–442, and the loop from 466 to 476 (Figure 3C). Additionally, Glu394 (from the conserved APE motif) on the second helix of the projecting T-loop makes a bidentate ion-pair/hydrogen bond interaction with Arg474 (Figure 3D). The first helix of the projecting T-loop, however, makes no interactions with the dimer-related Chk2 molecule and has significantly higher temperature factors than the second helix.
Figure 3.
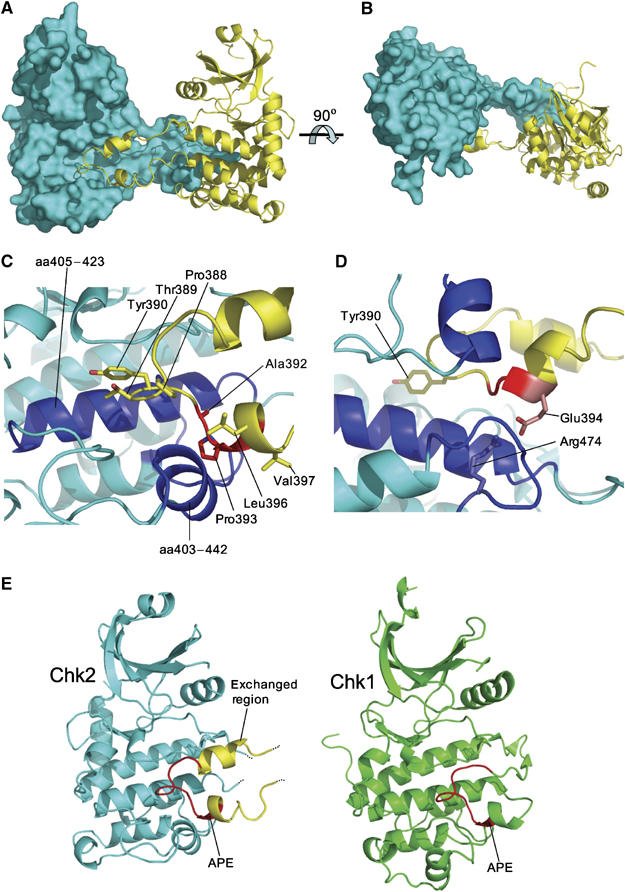
Chk2 T-loop conformation and exchange. (A) T-loop exchanged dimer. One molecule of the dimer is shown as a molecular surface representation (coloured cyan), and the second as a secondary structure cartoon (yellow). (B) As in (A) but rotated 90° around the indicated axis. (C) Molecular details of T-loop exchange. Residues Pro388, Thr389 and Tyr390 comprising the tip of the exchanged loop (coloured yellow), along with residues Ala392, Pro393, Leu396 and Val397 forming the start of a helix (red) pack into a hydrophobic recess in a dimer-related CHK2-KD molecule (cyan). This recess is lined by two helices (residues 405–423 and 435–442) and the loop comprised of residues 466–476 (blue). (D) APE motif: Glu394 of the APE motif (coloured red) is hydrogen-bonded to the canonically conserved arginine (Arg474) despite coming from an exchanged T-loop (yellow). Helices and loops comprising the hydrophobic recess are coloured blue, as in (C). Tyr390 is also shown, as a guide for orientating the figure. (E) Chk2/Chk1 comparison. Despite the exchange of T-loops between Chk2 molecules, the orientation, position and conformation of the C-terminal part of this loop (coloured red) is very similar to that of Chk1 (PDB: 1ia8).
CHK2-KD is in an ‘active' state
Comparison of the CHK2-KD structure with other known protein structures using DALI (Holm and Sander, 1993) (www.ebi.ac.uk/dali), highlighted the Ser/Thr protein kinases PKA and Chk1 as the most similar, with root-mean-squared deviations of 1.64 and 1.75 Å between 205 and 204 Cα positions, respectively (Krissinel and Henrick, 2004) (PDBs: 1cdk and 1ia8).
The CHK2-KD structure is judged to be in an ‘active' conformation, both by comparison to the PKA and Chk1 structures (also in active conformations (Bossemeyer et al, 1993; Chen et al, 2000)) and through several features conserved throughout protein kinases (Johnson et al, 1996): the essential catalytic residues Lys249, Asp368 and Asp347 (equivalent to Lys72, Asp166 and Asp184 in PKA, respectively) are conserved both in identity and spatial arrangement, the conserved lysine–glutamate salt-bridge (Lys249—Glu273) is maintained, the nucleotide binding site is open and accessible, and the relative orientation of the two lobes with respect to each other is consistent with an active state. Most significantly, the T-loop is fully ordered, with the APE motif and loop tip properly engaged in interactions with the body of the C-lobe, albeit in the dimer-related Chk2 molecule (Figure 3E). The orientation, position and conformation of this C-terminal part of the T-loop in Chk2 is extremely similar to that of PKA and Chk1. However, the helical conformation of the N-terminal side of the Chk2 T-loop is radically different to that of PKA/Chk1, and indeed to all other kinases we are aware of, for which ‘active' structures have been described.
Nucleotide and inhibitor binding
Clear electron density is visible for the ADP molecule located in the nucleotide-binding site of CHK2-KD and for the glycine-rich loop, which is fully ordered in the complex and contacts the phosphate groups of the bound ADP (Figure 4A and C). The phosphates additionally receive hydrogen bonds from the side chains of Lys249, Asp368 and Asn352, and are coordinated by one of the two magnesium ions located in the catalytic site. The phenylalanine residue (Phe369) of the adjacent DFG motif is in a ‘flipped out' conformation. The adenine ring of ADP makes hydrogen bonds to the backbone groups of hinge residues Glu302 and Met304, while the oxygen of the ribose moiety make hydrogen bonds to the side chain of Glu308, and the backbone of Glu351.
Figure 4.
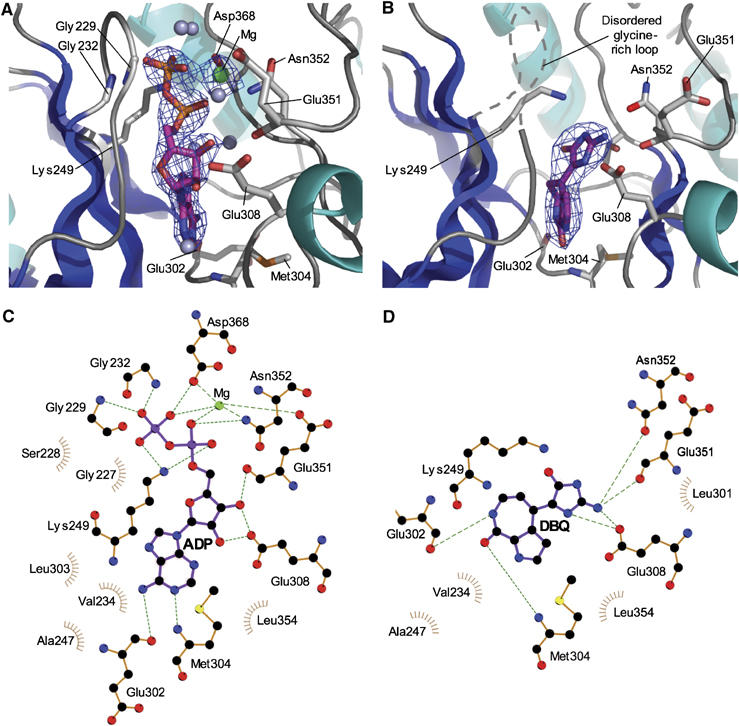
Nucleotide and drug binding to Chk2 kinase domain. (A) Electron density for ADP bound to CHK2-KD, contoured at a level of 4.5σ from an Fo−Fc omit map. (B) Electron density for the inhibitor DBQ bound to CHK2-KD, contoured at a level of 3σ from an Fo−Fc omit map. (C) LIGPLOT diagram (Wallace et al, 1995) detailing the interactions made between ADP and CHK2-KD. (D) LIGPLOT diagram detailing the interactions made between DBQ and CHK2-KD. In (A) and (B), residues involved in hydrogen bonds to the bound ligand are shown in ‘stick' representation. Water molecules are shown as grey spheres, and magnesium ions as green spheres. Secondary structure elements corresponding to β-sheets are coloured blue, and α-helices cyan. In (C) and (D), green dotted lines represent hydrogen-bonds (<3.5 Å) whereas brown ‘sunbursts' represent hydrophobic-type interactions between the protein and ligand.
While there is growing interest in Chk2 as a therapeutic target in cancer (Bartek and Lukas, 2003; Collins and Garrett, 2005; Pommier et al, 2005), few highly potent and specific inhibitors have been described. Hymenialdisine is a natural product found in marine sponges, which is an ATP-competitive inhibitor of several protein kinases (Meijer et al, 2000) and derivatives of this are showing promise as Chk2 inhibitors (Sharma and Tepe, 2004). Therefore, as a possible starting point for developing specific Chk2 inhibitors, we have determined the structure of a binary complex between CHK2-KD and DBQ at 2.7 Å resolution; a compound with an IC50 of 3.5 μM (Curman et al, 2001).
As expected from biochemical studies, the compound occupies the nucleotide-binding pocket, making hydrogen bonds to several key residues (Figure 4B and D). Principally, two hydrogen bonds are made to the backbone of hinge residues Glu302 and Met304, mimicking the interactions made by the adenine group of bound ADP, a binding-mode equivalent to that reported for other kinase/DBQ complexes (Meijer et al, 2000; Lougheed et al, 2004). Hydrogen bonds are also made between the amino group of the dihydroimidazolone moiety and the side-chains of Glu308 and Asn352, and with the backbone oxygen of Asn352, similar to the contacts made by the ribose and phosphate groups of bound ADP. An additional hydrogen bond is made between the N4H position of DBQ and the side-chain of Glu308. In contrast to other kinase/DBQ complexes, however, no polar interactions, direct or water-mediated, are made to either Asp368 of the DFG motif, or its conserved salt-bridge partner Lys249. Furthermore, no interactions are made between DBQ and the glycine-rich loop, which is disordered in the drug complex. One effect of the extensive hydrogen-bonding network involving Glu308, Asn352 and DBQ is to change the orientation of the dihydroimidazolone moiety with respect to the rest of the DBQ molecule, compared to the conformation in other kinases, a difference that might be exploited in drug development.
Chk2 dimerisation and activation
The current model for Chk2 activation in response to DNA damage involves two distinct phases of phosphorylation. Initially, Chk2 is primed for activation by phosphorylation of Thr68 within its SCD, principally by the ATM kinase (Matsuoka et al, 2000; Melchionna et al, 2000). Thr68-phosphorylation of Chk2 converts it to a state where it is competent to autophosphorylate two residues in its T-loop, Thr383 and Thr387 (Lee and Chung, 2001). Phosphorylation of Thr68 had been proposed to activate Chk2 by promoting allosteric changes in the kinase domain that facilitate autophosphorylation (Bartek et al, 2001). However, a body of subsequent data have shown that the main effect of Thr68 phosphorylation is to promote homo-dimerisation of Chk2 via interaction of the phospho-Thr68 segment of one molecule with the FHA domain of the other (Ahn et al, 2002; Xu et al, 2002). Once dimerised, one Chk2 molecule can phosphorylate the T-loop in the other Chk2 molecule, to give a fully active species which may then be able to cis-phosphorylate at Thr68 and Ser516 (Schwarz et al, 2003), although the significance of this latter phosphorylation is unknown.
The activated conformation and dimeric interactions of CHK2-KD in the crystal structure suggests that it is (or closely resembles) the trans-phosphorylation-competent state engendered by Thr68 phosphorylation. In support of this idea we first sought to determine that the Chk2 kinase domain does indeed have an inherent propensity to dimerise and that the observed dimer is not merely the result of crystallisation. We therefore incubated CHK2-KD in dilute solution (0.1 mg/ml, compared to 20 mg/ml used for crystallisation) in the presence of increasing concentrations of a lysine-specific crosslinking reagent, and analysed the result on SDS–PAGE as previously described (Dajani et al, 2001). At all concentrations of the crosslinker, we observed a consistent crosslinked dimer band, but with no significant higher bands that would indicate nonspecific aggregation of the protein at this concentration, suggesting that CHK2-KD does exist in a monomer–dimer equilibrium in solution (Figure 5A). Although this does not prove that the T-loop exchanged dimer in the crystals is that which forms in solution, the surface area buried by formation of this interface (1344 Å2 per monomer, 2688 Å2 in total) is in the range typically found for functional interactions (Ponstingl et al, 2000) and significantly larger than any other intermolecular contact observed in the crystals. The equilibrium for the isolated kinase domain clearly favours the monomer over the dimer at this low protein concentration (8 μM). However in the intact protein, tethering of two Chk2 molecules via pThr68–FHA domain interactions would massively increase the effective concentration of the associated kinase domains, shifting the equilibrium significantly towards the dimerised state.
Figure 5.
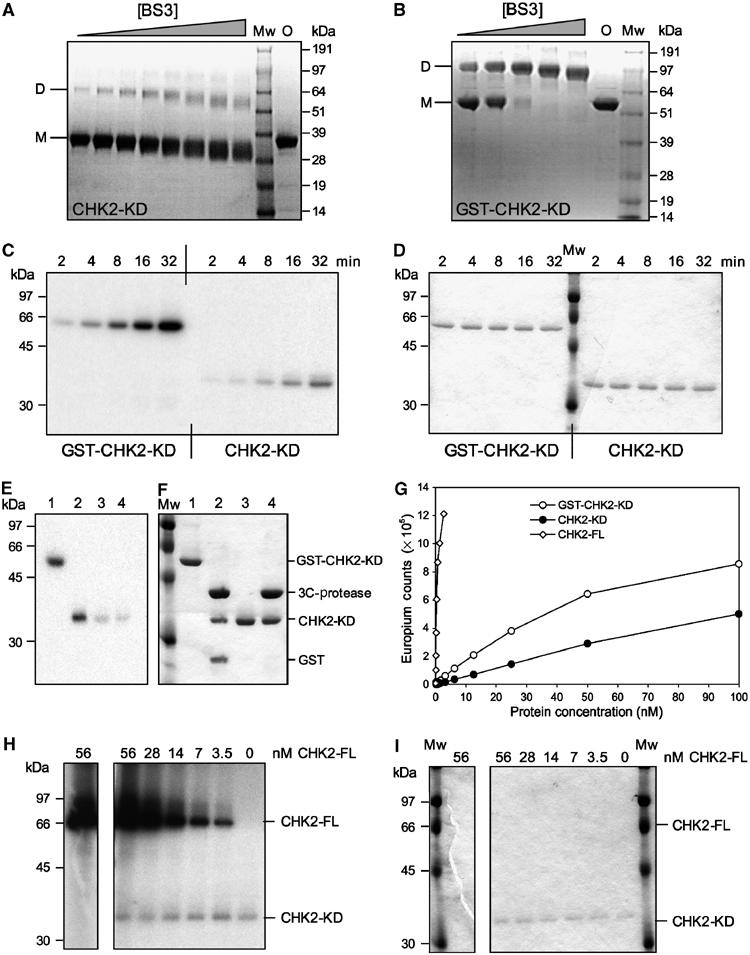
Dimerisation, Auto- and substrate-phosphorylation of CHK2-KD. (A) SDS–PAGE analysis of CHK2-KD crosslinking. (B) SDS–PAGE analysis of GST-CHK2-KD crosslinking. In each case, a fixed concentration of protein was incubated with increasing concentrations of the homo-bifunctional crosslinker BS3. Mw=molecular mass marker, O=protein not exposed to crosslinking agent, D=dimeric species, M=monomeric species. (C) Autophosphorylation assay. Fixed concentrations of GST-CHK2-KD or CHK2-KD were incubated with γ-32P-labelled ATP for increasing periods of time (as indicated). Samples were analysed by SDS–PAGE, visualised by phosphorimager and autoradiography. (D) Coomassie-stained SDS–PAGE gel of (C). (E) Autophosphorylation specifically occurs within the kinase domain of Chk2. Autophosphorylated samples of GST-CHK2-KD (Lane 1) and CHK2-KD (3) were incubated with rhinovirus 3C-protease (Lanes 2 and 4, respectively) then analysed by SDS–PAGE. Incorporation of γ-32P-labelled ATP was visualised by autoradiography. (F) Coomassie-stained SDS–PAGE gel of (E). (G) DELFIA assay. Samples of CHK2-FL, GST-CHK2-KD and CHK2-KD at varying concentrations were incubated, in the presence of ATP, with a substrate peptide (corresponding to the sequence flanking the Ser216 phosphorylation site of CDC25C). Phosphorylation was detected by incubation with a polyclonal anti-CDC25C(Ser216) antibody, followed by a Europium-labelled secondary antibody. A fluorescent plate reader was then used to quantitate the level of peptide phosphorylation (as Europium counts). (H) Trans-phosphorylation assay. Increasing amounts of CHK2-FL were incubated with a fixed concentration of CHK2-KD (as indicated) in the presence of γ-32P-labelled ATP. Samples were analysed by SDS–PAGE, then visualised by phosphorimager and autoradiography. (I) Coomassie-stained SDS–PAGE gel of (H).
We next asked whether the dimeric arrangement of the kinase domains was architecturally consistent with the proposed activation mechanism in which the phosphorylated Thr68 segments of each Chk2 protomer could bind to the FHA domain of the other and promote kinase domain association. The visible C-terminus of the crystal structure of the Chk2 FHA domain (Asp207) is only separated by a couple of residues from the visible N-terminus of the CHK2-KD structure (Ser210), so that the juxtaposition of the two domains is closely constrained, and the FHA domain and N-terminal lobe of the kinase, are likely to be in contact. With this constraint, we constructed a structure-based model for a Chk2 FHA-KD segment dimer. Taking the pThr residue bound to the FHA domain structure to represent pThr68, 20 residues separate the C-terminal end of the phosphopeptide (equivalent to Chk2 residue 72) bound to the FHA domain on one chain, and the visible N-terminus of the FHA domain (Pro92) on the other (Figure 6). The distance between residues 72 and 92 (∼50–70 Å) could be spanned comfortably by this amount of polypeptide chain, so that the model is geometrically reasonable. While this is of course a hypothetical model, that it fits comfortably within the architectural constraints of Thr68-driven dimerisation is reassuring, and further supports the idea that the kinase-domain dimer in the crystals is biologically authentic.
Figure 6.
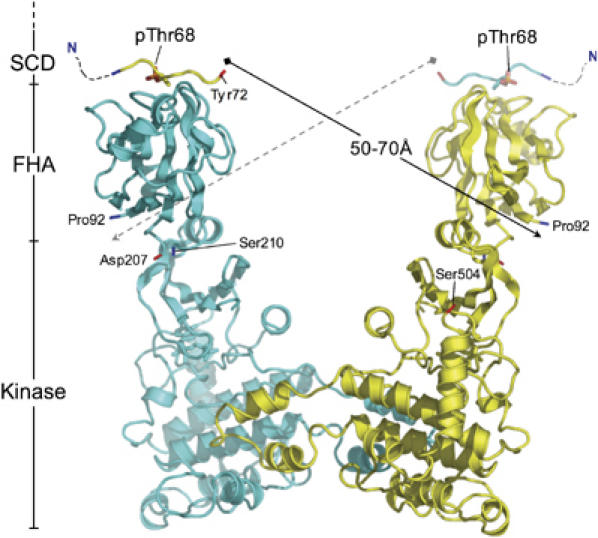
Model of the trans-activating Chk2 dimer. Schematic model of a full-length Chk2 dimer. The X-ray structure of the Chk2 FHA domain and its associated phospho-peptide (PDB: 1GXC), along with the CHK2 kinase domain reported here, were used to build a theoretical model for the full-length Chk2 molecule. Predictive modelling of the FHA/Kinase domain junction is facilitated due to the short (2 amino acid) distance between the reported visible termini in each structure (Asp207 and Ser210 respectively), which suggests a spatially constrained arrangement. The position of the FHA-bound phosphothreonine is taken to represent Thr68. The visible C-terminus of the bound peptide therefore equates to Tyr72. The distance between this residue and the visible N-terminus of a dimer-related FHA molecule (Pro92) is estimated to be between 50 and 70 Å. A connecting polypeptide chain of 30 amino acids would be therefore be sufficient to connect the two termini. See Figure 1 for SCD, FHA and KINASE abbreviations. N and C termini for each polypeptide chain are coloured blue and red, respectively.
We then sought to determine whether dimerisation per se had any effect on the ability of Chk2 to autophosphorylate. To do this, we compared the autophosphorylation activity of CHK2-KD and a GST-CHK2-KD construct. GST is itself a dimer and we verified that the GST-CHK2-KD fusion protein is also strongly dimeric (Figure 5B). Examination of the crystal structure of S. japonicum GST shows that the two C-termini in the GST dimer present at the same face of the structure, so that although they would not be optimally positioned, the attached kinase domains would not be prevented from exchanging T-loops in the fusion protein. We reasoned that if our model was correct, the surrogate dimerisation provided by GST would enhance the ability of CHK2-KD to autophosphorylate to some degree, compared with the unconstrained isolated CHK2-KD. Both constructs showed some ability to autophosphorylate on incubation with γ-32P-labelled ATP (Figure 5C and D); however, consistent with our model, the GST-fused kinase was at least four-fold more active in autophosphorylation than the isolated kinase domain. To determine whether this enhanced activity was specific, we cleaved the GST-CHK2-KD fusion protein postincubation at a rhinovirus 3C-protease site between the C-terminus of the GST and the N-terminus of the Chk2 kinase domain, and analysed the products by SDS–PAGE and autoradiography (Figure 5E and F). All the incorporated 32P resided in the Chk2 component and not the GST, indicating that dimeric tethering of the kinase had promoted a specific autophosphorylation reaction. We then compared the ability of the bacterially expressed and unphosphorylated CHK2-KD, GST-CHK2-KD proteins and the full-length baculovirus expressed Chk2 (CHK2-FL) to phosphorylate a substrate peptide derived from Cdc25C. The baculovirus expressed protein, which contains high-levels of pThr68 (as measured by phospho-specific antibody) and is therefore likely to be substantially phosphorylated on T-loop residues Thr383 and Thr387, was found to be very significantly (100-fold compared to GST-CHK2-KD, 400-fold to CHK2-KD) more active than either of the kinase domain constructs over the course of the assay period (Figure 5G). However, with the unphosphorylated proteins, the hetero-phosphorylation of the Cdc25C peptide is dependent on progressive activation of the protein by autophosphorylation during the assay. The higher rate of Cdc25C phosphorylation by the GST-dimerised protein is then fully consistent with enforced dimerisation by GST enhancing specific T-loop trans-phosphorylation. Finally, we analysed the ability of this activated full-length Chk2 to phosphorylate a separate CHK2-KD. When incubated with ATP, CHK2-FL did autophosphorylate to some degree, suggesting that not all possible sites were fully phosphorylated. However, despite its high activity against Cdc25C substrate, CHK2-FL showed no ability to phosphorylate a separate Chk2 kinase domain presented as an untethered substrate (Figure 5H and I).
Discussion
Phosphorylation of residues within the T-loop/activation segment is a key determinant of activation of many protein kinases (Johnson et al, 1996), in particular those of the ‘RD' group (Krupa et al, 2004) of which Chk2 is a member. Typically, the unphosphorylated T-loop is disordered, or is locked into an inactive alternative conformation. Phosphorylation on one or more residues fixes the T-loop into an active conformation in which it forms the substrate-binding site. The conserved arginine of the RD motif, immediately preceding the catalytic aspartate, interacts with the phosphorylated residue(s) in the T-loop, orientating both the DFG motif and other catalytic/substrate-binding residues to facilitate the phospho-transfer reaction.
In many phospho-regulated kinases, such as those in MAP-kinase cascades, phosphorylation of the T-loop is performed by a different upstream kinase. However, there are a significant number of enzymes, which like Chk2, have been shown unambiguously to be responsible for their own T-loop phosphorylation; in some cases by intramolecular reactions coupled to protein folding (Lochhead et al, 2005), but most commonly by intermolecular reactions. Although earlier models had suggested a mechanism of intramolecular autophosphorylation allosterically triggered by phosphorylation of Thr68 by ATM (Bartek et al, 2001), subsequent work suggests that T-loop phosphorylation of Chk2 is an intermolecular event dependent on pThr68-induced dimerisation (Ahn et al, 2002; Xu et al, 2002). This latter model is strongly supported by our observation that fusion of a heterologous dimerisation domain (GST) to CHK2-KD gives at least a four-fold increase in the rate of autophosphorylation.
Intermolecular autophosphorylation by a kinase that requires phosphorylation for activity presents an interesting logical paradox. This is conventionally resolved by assuming that the T-loop in the unphosphorylated kinase can transiently adopt the activated conformation rarely, but sufficiently often, for one molecule to bind and phosphorylate the T-loop of a second molecule, initiating a chain-reaction. In these circumstances, the T-loop of the substrate kinase molecule binds in the same way as a heterologous substrate and its amino-acid sequence typically conforms to the same consensus as other target sites (Johnson et al, 1996). However, the sequences surrounding both phosphorylation sites (Thr383 and Thr387) in the T-loop sequence of Chk2 present very poor matches to the consensus sequences of known Chk2 phosphorylation sites in targets such as E2F1, Cdc25A/C, PML and BRCA1, and to consensus substrate peptides derived there from, lacking in particular the preferred Arg at −3 and Leu at −5 (Seo et al, 2003) (Figure 7A). Consistent with this we find that the Chk2 kinase domain is not significantly phosphorylated when presented as a conventional substrate to an activated Chk2 molecule, compared to a Cdc25C sequence that conforms to the specificity. This argues strongly that the process of autophosphorylation proceeds via a mechanism that is distinct from the process of hetero-phosphorylation of a substrate.
Figure 7.
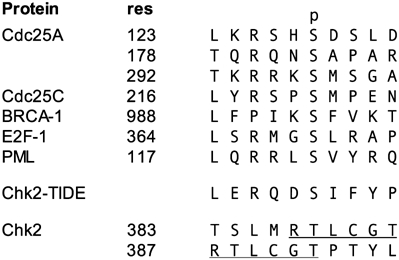
Chk2 T-loop autophosphorylation sites do not resemble endogenous substrates. Alignment of endogenous Chk2 substrate sequences (CDC25A, CDC25C, BRCA-1, E2F-1, PML) with an optimised substrate sequence (Chk2-TIDE) and T-loop autophosphorylation sites. For each sequence, the residue number of the first amino acid is listed (res). The autophosphorylation sites of Chk2 overlap, as indicated by the underlined regions. The ‘p' column header indicates the phosphorylated amino acid.
The crystal structure of the Chk2 kinase domain and the biochemical data described here suggest a mechanism for dimerisation-driven trans-phosphorylation that simultaneously circumvents the problem of specificity mismatch by the Chk2 T-loop, and the starting paradox that bedevils all autophosphorylation mechanisms, as follows (summarised in Figure 7B). Initially, phosphorylation of Chk2 Thr68 by ATM promotes interaction with the FHA domain of a second Chk2 molecule, generating a symmetrical Thr68-FHA interaction when both Chk2 molecules are phosphorylated on Thr68. The proximity of the kinase domains generated by this interaction would favour formation of a kinase-domain dimer as we observe in the crystal structure; the inherent ability of the kinase domains to dimerise is clearly seen from the crosslinking data. Formation of the dimer stabilises a conformation for the T-loop from each Chk2 molecule whereby they project out from the body of one kinase to interact with the other. The exchanged T-loop segments have an ordered structure despite their lack of phosphorylation, and make all the key interactions with the body of the other kinase that would typically be made by a phosphorylated T-loop segment. The body of the kinase domain with which the exchanged T-loop interacts is also in a conformation fully indicative of an activated state, so that the T-loop exchanged dimer appears to be catalytically competent.
When the CHK2-KD structure is compared to structures of activated kinases with bound substrate peptides, it is immediately clear that the unusual helical conformation of the N-terminal side of the T-loop is inimical to the conventional mode of substrate peptide-binding by protein kinases, which typically involves β-strand main-chain hydrogen bonding between the activation segment and the +1 and +3 residues of the substrate (Figure 7C). In any event, even if the activated dimer were able to bind a Chk2 substrate in a conventional manner, this could only achieve phosphorylation of a third Chk2 molecule, not the trans-phosphorylation of its mate in the Thr68-FHA linked dimer. However, within the dimer, Thr383 and Thr387 in the exchanged T-loop are each between 5.6 and 7.3 Å from the optimal position for a phosphorylatable Ser or Thr residue on a conventionally bound substrate. Thus, given the active conformation of the rest of the protein, a relatively small movement of the T-loop and partial unwinding of the helix on its N-terminal side would allow phosphoryl-transfer to either of these residues from the ATP bound to the other Chk2 molecule, thereby achieving trans-phosphorylation. Once phosphorylated, especially at Thr383, the helical conformation for the N-terminal side of the T-loop would be greatly destabilised and the T-loop could adopt a conventional extended conformation in which the phosphate of pThr383 interacts with the side chains of Arg346 and Lys373 in an essentially identical manner to the interaction of pThr197 with Arg165 and Lys189 in PKA (Knighton et al, 1991) (Figure 7D).
Given retention of Thr68 phosphorylation and consequent maintenance of N-terminal dimerisation, there is no clear structural reason why dimerisation of the T-loop exchanged kinase domains should not be maintained after trans-phosphorylation. With the T-loop stabilised in a conventional active conformation consistent with productive binding of hetero-substrates, and the rest of the protein in an active conformation, each Chk2 molecule in the T-loop exchanged dimer would in principle be a fully competent protein kinase so long as the presence of the other Chk2 molecule did not obstruct binding of a substrate protein (Figure 8). Alternatively, the activated kinase domains might operate independently despite N-terminal dimerisation, with the phosphorylated T-loop segment folding back onto the body of its own kinase domain to give a conventional protein kinase structure. Further work will be required to distinguish these possibilities.
Figure 8.
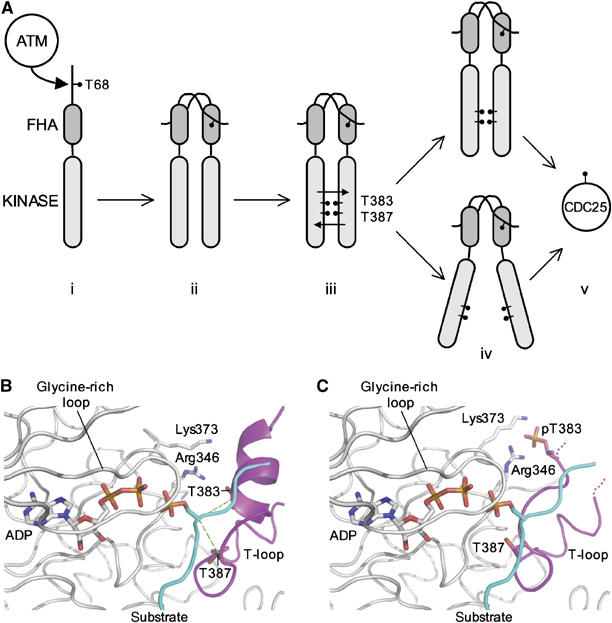
Proposed mechanism for Chk2 activation. (A) (i) Activated ATM phosphorylates Thr68 of Chk2. (ii) Dimerisation of Chk2 driven by FHA-pThr68 interactions. (iii) Auto-phosphorylation of Thr383 and Thr387 by T-loop exchange. (iv) Either auto-phosphorylated Chk2 remains as an active T-loop exchanged dimer, or the T-loops fold back to give two conventional kinase domain structures. The endogenous CDC25 phosphatase substrates can now be phosphorylated. (B) In the T-loop exchanged dimer, the Ca positions of Thr387 and Thr383 are at a distance of 5.85 and 7.29 Å, respectively, from the Ca position of a modelled phosphorylated substrate. A relatively small movement of the T-loop and partial helix unwinding would facilitate phosphoryl-transfer to either residue. However, in this conformation, the helix containing the autophosphorylation sites would occlude binding of a conventional peptide substrate. (C) Upon autophosphorylation of Thr387, the T-loop could be remodelled into a standard extended β-strand, forming canonical interactions between the phosphate group and the side chains of Lys373 and Arg346. The substrate binding channel would now be fully accessible. The position of the substrate peptide was modelled by superimposition of the crystal structure of a PKA-substrate complex (PDB: 1JLU) onto CHK2-KD.
During review of this manuscript, additional evidence supporting our proposed model of Chk2 activation by T-loop exchange has been published by Xu et al (2006) on Cds1, the fission yeast homolog of Chk2.
Materials and methods
Mass spectroscopy
MALDI-TOF experiments were carried out using standard protocols on a Voyager-DE STR Biospectrometry Workstation (Applied Biosystems, Foster City, CA).
Cloning, expression and purification
The IMAGE clone AU20-2, was used as a template for PCR, as it contained a full-length human CHK2 cDNA. Primers were designed to amplify the region encompassing the kinase domain (residues 210–531), with additional flanking restriction enzymes sites (NdeI and EcoRI) to facilitate cloning into the expression vector pTHREE-E (a GST-fusion vector modified to contain the multiple cloning site of pET-17b, following a encoded rhinovirus 3C-protease site). The vector pTHREE-Chk2 (210–531) was transformed into the Escherichia coli expression host Rosetta2 (DE3) pLysS (EMD Bioscience, Inc., San Diego, USA). A 250 ml flask containing 50 ml of L-broth (1% w/v tryptone, 0.5% w/v NaCl, 0.5% w/v yeast extract) supplemented with 50 μg/ml of carbenicillin and 34 μg/ml of chloramphenicol was inoculated with a single transformed bacterial colony. This was grown in an orbital incubator set at 37°C and 225 r.p.m., until the A600 of the culture reached 0.6. This ‘starter' culture was then stored overnight at 4°C. From the starter culture, 10 ml was used to inoculate a 2 l flask containing 1 l of L-broth supplemented as before with antibiotics. The culture was grown in an orbital incubator set at 37°C and 200 r.p.m. Once the A600 of the cell culture had reached 0.6, expression of the Chk2 kinase domain was induced by the addition of IPTG to a final concentration of 0.2 mM. Cultures were grown for a further 19 h at a reduced temperature of 20°C, after which the cells were harvested by centrifugation (5000 g, 10 min, 10°C). Cell pellets were stored at −80°C until required.
The cell pellet arising from 3 l of culture was resuspended, on ice, in 100 ml of buffer A (50 mM HEPES NaOH pH 7.5, 250 mM NaCl, 1 mM EDTA, 10 mM DTT, supplemented with protease inhibitors (Roche, Lewes, East Sussex, UK)). Cells were then lysed by sonication (15 × 5 s bursts, on ice, at 80% amplitude, with a large parallel probe; Jencons Ultrasonic Processor), after which cell debris and insoluble materials were removed by centrifugation (40 000 g, 60 min at 10°C). The supernatant arising from this step was filtered through a 4-μm filter (Whatman plc, Brentford, Middlesex, UK), then applied to a batch/gravity column containing 25 ml of Glutathione Sepharose 4 resin (Amerham Biosciences, Little Chalfont, Buckinghamshire, UK) equilibrated in Buffer A. The column containing the cell extract and resin was rotated/rolled at 4°C, for a period of 1 h to facilitate protein binding. The resin was allowed to pack under gravity, and then washed with successive applications of buffer A (approximately 250 ml in total). It was then resuspended in buffer A to give an approximate 50% resin/buffer slurry, transferred to a 50 ml Falcon tube (BD Biosciences, Le Pont De Claix, France), and 150 ml of rhinovirus 3C-protease (PreScission protease, Amersham Biosciences) added. This was incubated overnight with constant rotation/rolling at 4°C. The resin was again allowed to pack under gravity, and the flow-through collected. Fractions containing the Chk2 kinase domain (CHK2-KD) were identified by SDS–PAGE, then pooled and concentrated to a final volume of 10 ml (Vivaspin 20, 10 kD MWCO; Vivascience AG, Hannover, Germany). The concentrated protein was applied to a Superdex 75 (Amersham Biosciences) size exclusion column equilibrated in buffer B (10 mM HEPES NaOH pH 7.5, 250 mM NaCl, 1 mM EDTA, 10 mM DTT). Again, fractions containing CHK2-KD were identified by SDS–PAGE, then pooled and concentrated to a final concentration of 20 mg/ml (as determined by UV spectroscopy). The purified protein was flash-frozen on dry-ice and stored at −80°C until required.
Crystallisation
Initial crystallisation trials were carried out at a concentration of 20 mg/ml in microbatch experiments, under oil, at 4°C. Small crystals were observed, after a period of several days, in condition 64 of the ‘The PEGs Suite' from Nextal Biotechnologies (Montreal, Quebec, Canada). This condition was optimised in hanging drop experiments at 4°C, to mixing 1 μl of protein (20 mg/ml in 10 mM HEPES NaOH pH 7.5, 250 mM NaCl, 10 mM DTT, 1 mM EDTA) supplemented with either 2.5 mM ADP or 2.5 mM DBQ, with 1 μl of precipitant containing 0.1 M HEPES NaOH pH 7.6, 0.2 M Mg(NO3)2, 22% w/v PEG 3350, 16% v/v ethylene glycol. The mother liquor from these experiments was sufficient to cryo-protect the crystals during data collection.
Data collection and processing
Data to 2.25 Å were collected for the CHK2-KD/ADP binary complex on station ID29 at the ESRF (European Synchrotron Radiation Facility, Grenoble, France) from a single crystal at 100 K, recorded on an ADSC CCD detector. This complex crystallised in spacegroup P3221, with a single molecule comprising the asymmetric unit, and unit cell dimensions a, b=90.87 Å, c=92.91 Å. Data to 2.7 Å were collected for the CHK2-KD complex with DBQ on station 14.2 at the SRS (Synchrotron Radiation Source, Daresbury, UK) from a single crystal at 100 K, recorded on an ADSC CCD detector. This complex also crystallised in the spacegroup P3221, with a single molecule in asymmetric unit, but with a smaller unit cell of a, b=88.65 Å, c=88.90 Å. Images for both data sets were integrated using MOSFLM (Leslie, 1995), and reduced/scaled using programs from the CCP4 suite (Collaborative Computational Project, 1994).
The structures of the Chk2 complexes were solved by molecular replacement using PHASER (McCoy et al, 2005), with the X-ray structure of PKA as a search model (PDB: 1ATP). Difference maps were used to extend and rebuild the initial models using Coot (Emsley and Cowtan, 2004). Iterative cycles of refinement (REFMAC5; Collaborative Computational Project, 1994) and manual intervention produced the current models for each complex, CHK2-KD/ADP: which contains 2271 protein atoms, 1 nitrate ion, 2 magnesium ions, 1 ADP molecule, and 156 solvent atoms, and CHK2-KD/debromohymenisaldisine: which contains 2153 protein atoms, 1 nitrate ion, 1 magnesium ion, 1 DBQ molecule and 44 solvent atoms. Crystallographic statistics are given in Table I. Coordinates and structure factors for both complexes have been deposited in the RSCB Protein Data Bank, with codes 2CN5 and 2CN8 for the CHK2-KD/ADP and CHK2-KD/DBQ complexes respectively.
BS3 crosslinking
Experiments were carried out at a protein concentration of 8 μM in 20 mM HEPES NaOH pH 7.5, 250 mM NaCl. A 50 mM stock solution of BS3 (Pierce Biotechnology Inc., IL, USA) was serially diluted, then added to each of several protein samples. Crosslinking was allowed to proceed for 30 min at room temperature, before the samples were analysed by SDS–PAGE. Final BS3 concentrations ranged between 5 mM and 8 μM. BS3 (Bis[sulfosuccinimidyl] suberate) is a homobifunctional primary-amine reactive crosslinking agent.
Phosphorylation assays
Equimolar amounts (60 nM) of either purified GST-CHK2-KD or CHK2-KD were incubated at 25°C in a final reaction volume of 24 μl of assay buffer (40 mM HEPES NaOH pH 7.4, 40 mM KCl, 10 mM DTT, 2 mM MgCl2, 0.02% (v/v) Tween-20) containing 10 μM ATP and 0.1 μCi γ-32P-labelled ATP. Reactions were stopped by the addition of SDS loading buffer and proteins resolved by SDS–PAGE before autoradiography or analysis by phosphorimager.
For the rhinovirus 3C-protease digestions, phosphorylation reactions were incubated as before, for 20 min, then stopped by the addition of EDTA (50 mM final concentration) and the phosphatase inhibitor NaF (10 mM final concentration). Reactions were split and incubated with or without protease for 30 min on ice prior to addition of SDS loading buffer.
For the CHK2-FL trans-phosphorylation assay, increasing amounts of purified CHK2-FL were incubated with a fixed (120 nM) concentration of CHK2-KD, at 25°C, in assay buffer containing 10 μM ATP and 0.1 μCi γ-32P-ATP, for 20 min.
DELFIA assays
Kinase activity was measured in a DELFIA® assay that monitors phosphorylation of a CDC25C peptide using a specific phospho-antibody. The enzyme reaction was carried out in polypropylene plates (Greiner Bio-One Ltd, Gloucestershire, UK). The reaction mix (25 μl) contained enzyme (10 μl), peptide (Biotin-KKKVSRSGLYRSPSMPENLNRPR) (1 μM, 5 μl), ATP (30 μM, 5 μl) in assay buffer (see phosphorylation assays). The reaction was incubated for 30 min at room temperature and stopped by the addition of buffer (125 μl) containing 40 mM EDTA, 0.05% v/v Tween-20, 0.1% w/v BSA in TBS. An aliquot (100 μl) of the reaction mix was transferred to a black neutravidin-coated plate (Perbio Science UK Ltd, Northumberland, UK) and incubated for 1 h on a shaker at room temperature. The plates were washed four times with wash buffer (25 mM Tris pH 8, 150 mM NaCl and 0.1% v/v Tween-20) and incubated for 1 h as before with antibody mix (100 μl) consisting of anti-phospho CDC25C (1.25 nM, #9528, Cell Signalling Technology Inc., MA, USA) and europium-labelled anti-rabbit IgG (0.3 μg/ml, AD0105, PerkinElmer Life Sciences) diluted in DELFIA assay buffer (PerkinElmer Life Sciences, MA, USA). The plates were washed a further four times with wash buffer before the addition of enhancement solution (100 μl/well, PerkinElmer Life Sciences). The plate was read on a Victor2 1420 multilabel counter (PerkinElmer Life Sciences) using a time-resolved measurement mode reading fluorescence at 615 nm.
Acknowledgments
We are very grateful to our colleagues: Mark Roe for advice and assistance with data processing and model refinement. David Komander, Markus Hassler, Andy Doré, Mairi Kilkenny and Wyatt Yue for assistance with synchrotron data collection, and Matthew Tall for assistance with the DELFIA assays. We acknowledge both the European Synchrotron Radiation Facility and the Daresbury Synchrotron Radiation Source for provision of synchrotron radiation facilities. This work was supported by a Programme Grant from Cancer Research UK (LHP).
References
- Ahn JY, Li X, Davis HL, Canman CE (2002) Phosphorylation of threonine 68 promotes oligomerization and autophosphorylation of the Chk2 protein kinase via the forkhead-associated domain. J Biol Chem 277: 19389–19395 [DOI] [PubMed] [Google Scholar]
- Ahn JY, Schwarz JK, Piwnica-Worms H, Canman CE (2000) Threonine 68 phosphorylation by ataxia telangiectasia mutated is required for efficient activation of Chk2 in response to ionizing radiation. Cancer Res 60: 5934–5936 [PubMed] [Google Scholar]
- Bartek J, Falck J, Lukas J (2001) CHK2 kinase—a busy messenger. Nat Rev Mol Cell Biol 2: 877–886 [DOI] [PubMed] [Google Scholar]
- Bartek J, Lukas J (2003) Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 3: 421–429 [DOI] [PubMed] [Google Scholar]
- Bossemeyer D, Engh RA, Kinzel V, Ponstingl H, Huber R (1993) Phosphotransferase and substrate binding mechanism of the cAMP-dependent protein kinase catalytic subunit from porcine heart as deduced from the 2.0 A structure of the complex with Mn2+ adenylyl imidodiphosphate and inhibitor peptide PKI(5–24). EMBO J 12: 849–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Luo C, Deng Y, Ryan K, Register J, Margosiak S, Tempczyk-Russell A, Nguyen B, Myers P, Lundgren K, Kan CC, O'Connor PM (2000) The 1.7 A crystal structure of human cell cycle checkpoint kinase Chk1: implications for Chk1 regulation. Cell 100: 681–692 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, N (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallographica, D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Collins I, Garrett MD (2005) Targeting the cell division cycle in cancer: CDK and cell cycle checkpoint kinase inhibitors. Curr Opin Pharmacol 5: 366–373 [DOI] [PubMed] [Google Scholar]
- Curman D, Cinel B, Williams DE, Rundle N, Block WD, Goodarzi AA, Hutchins JR, Clarke PR, Zhou B-B, Lees-Miller SP, Anderson RJ, Roberge M (2001) Inhibition of the G2 DNA damage checkpoint and of protein kinases Chk1 and Chk2 by the marine sponge alkaloid debromohymenialdisine. J Biol Chem 276: 17914–17919 [DOI] [PubMed] [Google Scholar]
- Dajani R, Fraser E, Roe SM, Young N, Good V, Dale TC, Pearl LH (2001) Crystal structure of glycogen synthase kinase 3 beta: structural basis for phosphate-primed substrate specificity and autoinhibition. Cell 105: 721–732 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallograph D 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Falck J, Mailand N, Syljuasen RG, Bartek J, Lukas J (2001) The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 410: 842–847 [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C (1993) Protein structure comparison by alignment of distance matrices. J Mol Biol 233: 123–138 [DOI] [PubMed] [Google Scholar]
- Johnson LN, Noble ME, Owen DJ (1996) Active and inactive protein kinases: structural basis for regulation. Cell 85: 149–158 [DOI] [PubMed] [Google Scholar]
- Knighton DR, Zheng J, Ten Eyck LF, Ashford VA, Xuong N-H, Taylor SS, Sowadski JM (1991) Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253: 407–413 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallograph D 60: 2256–2268 [DOI] [PubMed] [Google Scholar]
- Krupa A, Preethi G, Srinivasan N (2004) Structural modes of stabilization of permissive phosphorylation sites in protein kinases: distinct strategies in Ser/Thr and Tyr kinases. J Mol Biol 339: 1025–1039 [DOI] [PubMed] [Google Scholar]
- Lavin MF, Birrell G, Chen P, Kozlov S, Scott S, Gueven N (2005) ATM signaling and genomic stability in response to DNA damage. Mutat Res 569: 123–132 [DOI] [PubMed] [Google Scholar]
- Lee CH, Chung JH (2001) The hCds1 (Chk2)-FHA domain is essential for a chain of phosphorylation events on hCds1 that is induced by ionizing radiation. J Biol Chem 276: 30537–30541 [DOI] [PubMed] [Google Scholar]
- Leslie AGW (1995) MOSFLM Users Guide. Cambridge, UK: MRC Laboratory of Molecular Biology [Google Scholar]
- Li J, Williams BL, Haire LF, Goldberg M, Wilker E, Durocher D, Yaffe MB, Jackson SP, Smerdon SJ (2002) Structural and functional versatility of the FHA domain in DNA-damage signaling by the tumor suppressor kinase Chk2. Mol Cell 9: 1045–1054 [DOI] [PubMed] [Google Scholar]
- Lochhead PA, Sibbet G, Morrice N, Cleghon V (2005) Activation-loop autophosphorylation is mediated by a novel transitional intermediate form of DYRKs. Cell 121: 925–936 [DOI] [PubMed] [Google Scholar]
- Lougheed JC, Chen RH, Mak P, Stout TJ (2004) Crystal structures of the phosphorylated and unphosphorylated kinase domains of the Cdc42-associated tyrosine kinase ACK1. J Biol Chem 279: 44039–44045 [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Rotman G, Ogawa A, Shiloh Y, Tamai K, Elledge SJ (2000) Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc Natl Acad Sci USA 97: 10389–10394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ (2005) Likelihood-enhanced fast translation functions. Acta Crystallographica, D 61: 458–464 [DOI] [PubMed] [Google Scholar]
- McGowan CH (2002) Checking in on Cds1 (Chk2): a checkpoint kinase and tumor suppressor. Bioessays 24: 502–511 [DOI] [PubMed] [Google Scholar]
- McGowan CH, Russell P (2004) The DNA damage response: sensing and signaling. Curr Opin Cell Biol 16: 629–633 [DOI] [PubMed] [Google Scholar]
- Meijer L, Thunnissen AM, White AW, Garnier M, Nikolic M, Tsai LH, Walter J, Cleverley KE, Salinas PC, Wu YZ, Biernat J, Mandelkow EM, Kim SH, Pettit GR (2000) Inhibition of cyclin-dependent kinases, GSK-3beta and CK1 by hymenialdisine, a marine sponge constituent. Chem Biol 7: 51–63 [DOI] [PubMed] [Google Scholar]
- Melchionna R, Chen XB, Blasina A, McGowan CH (2000) Threonine 68 is required for radiation-induced phosphorylation and activation of Cds1. Nat Cell Biol 2: 762–765 [DOI] [PubMed] [Google Scholar]
- Pommier Y, Sordet O, Rao VA, Zhang H, Kohn KW (2005) Targeting chk2 kinase: molecular interaction maps and therapeutic rationale. Curr Pharm Des 11: 2855–2872 [DOI] [PubMed] [Google Scholar]
- Ponstingl H, Henrick K, Thornton JM (2000) Discriminating between homodimeric and monomeric proteins in the crystalline state. Proteins 41: 47–57 [DOI] [PubMed] [Google Scholar]
- Schwarz JK, Lovly CM, Piwnica-Worms H (2003) Regulation of the Chk2 protein kinase by oligomerization-mediated cis- and trans-phosphorylation. Mol Cancer Res 1: 598–609 [PubMed] [Google Scholar]
- Seo GJ, Kim SE, Lee YM, Lee JW, Lee JR, Hahn MJ, Kim ST (2003) Determination of substrate specificity and putative substrates of Chk2 kinase. Biochem Biophys Res Commun 304: 339–343 [DOI] [PubMed] [Google Scholar]
- Sharma V, Tepe JJ (2004) Potent inhibition of checkpoint kinase activity by a hymenialdisine-derived indoloazepine. Bioorg Med Chem Lett 14: 4319–4321 [DOI] [PubMed] [Google Scholar]
- Wallace AC, Laskowski RA, Thornton JM (1995) LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng 8: 127–134 [DOI] [PubMed] [Google Scholar]
- Xu X, Tsvetkov LM, Stern DF (2002) Chk2 activation and phosphorylation-dependent oligomerization. Mol Cell Biol 22: 4419–4432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YJ, Davenport M, Kelly TJ (2006) Two stage-mechanism for activation of the DNA replication checkpoint kinase Cds1 in fission yeast. Genes Dev 20: 990–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]


