Abstract
Many Fusarium species produce one or more agriculturally important trichothecene mycotoxins, and the relative level of toxicity of these compounds is determined by the pattern of oxygenations and acetylations or esterifications on the core trichothecene structure. Previous studies with UV-induced Fusarium sporotrichioides NRRL 3299 trichothecene mutants defined the Tri1 gene and demonstrated that it was required for addition of the oxygen at the C-8 position during trichothecene biosynthesis. We have cloned and characterized the Tri1 gene from NRRL 3299 and found that it encodes a cytochrome P450 monooxygenase. The disruption of Tri1 blocks production of C-8-oxygenated trichothecenes and leads to the accumulation of 4,15-diacetoxyscirpenol, the same phenotype observed in the tri1 UV-induced mutants MB1716 and MB1370. The Tri1 disruptants and the tri1 UV-induced mutants do not complement one another when coinoculated, and the Tri1 gene sequence restores T-2 toxin production in both MB1716 and MB1370. The DNA sequence flanking Tri1 contains another new Tri gene. Thus, Tri1 encodes a C-8 hydroxylase and is located either in a new distal portion of the trichothecene gene cluster or in a second separate trichothecene gene cluster.
Trichothecene mycotoxins are sesquiterpenoid secondary metabolites synthesized by several genera of filamentous fungi, including Fusarium, Stachybotrys, Myrothecium, Trichothecium, and Trichoderma (11, 23). Interest in these compounds stems from their occurrence as contaminants in major crops and their acute toxicity in humans and domesticated animals upon consumption (19, 23, 41). Many of the cytotoxic characteristics of the trichothecenes are attributed to their ability to inhibit protein synthesis (29) and to induce apoptosis in eukaryotic cells (30). They also act as immunosuppressors (34) and neurotoxins (24). These mycotoxins significantly enhance the virulence of trichothecene-producing fungal pathogens for some plant hosts (12, 14, 32). Accordingly, considerable efforts to understand the biosynthesis and genetic control of trichothecene production are in progress.
Biosynthetically, trichothecenes are derived from the primary metabolite farnesyl pyrophosphate (FPP). Following the cyclization of FPP to trichodiene, 14 additional steps culminate in the formation of T-2 toxin, the primary trichothecene produced by Fusarium sporotrichioides NRRL 3299. Although most of the T-2 toxin biosynthetic pathway is known, several essential reactions lack an identified gene, and not all of the identified trichothecene (Tri) genes have been cloned (3, 7, 8, 9, 10, 18, 28).
Tri1 (formerly Tox1) is the only known gene for a specific step in the T-2 toxin biosynthetic pathway that has not been cloned. Tri1 was identified following a UV mutagenesis of F. sporotrichioides NRRL 3299 that produced mutants blocked in T-2 toxin production (3). The reference tri1 mutant strain MB1716 accumulates 4,15-diacetoxyscirpenol (4,15-DAS) and lacks only the ability to add a hydroxyl group at C-8, demonstrating that Tri1 is required for C-8 hydroxylation (3, 7). This step utilizes molecular oxygen (13), so Tri1 probably encodes a cytochrome P450 monooxygenase that catalyzes this reaction. However, other possibilities exist, and cloning Tri1 will help define its precise role in T-2 toxin production.
We recently used a new approach to clone Tri1 and additional genes involved in trichothecene biosynthesis (A. W. Peplow, A. G. Tag, G. F. Garifullina, and M. N. Beremand, submitted for publication). Tri10 controls T-2 toxin production by controlling gene transcript levels. Disruption of Tri10 severely reduces the accumulation of all the tested Tri genes, while the overexpression of Tri10 has the opposite effect (38). We therefore used cDNA samples prepared from the Tri10 deletion (ΔTri10) and overproducing (↑Tri10) strains to differentially screen a cDNA library constructed from RNA from the ↑Tri10 strain to obtain new genes controlled by Tri10. One of the new cDNA sequences we identified encodes a cytochrome P450 monooxygenase (Peplow et al., submitted). Our objectives in this study were to determine if the corresponding P450 gene is required for trichothecene biosynthesis and to investigate its gene neighborhood. As a result, we cloned Tri1 and found that it both encodes a C-8 hydroxylase and resides immediately upstream of another new Tri gene.
MATERIALS AND METHODS
Strains, culture conditions, and cloning vectors.
The F. sporotrichioides wild-type strain NRRL 3299 was originally obtained from the USDA Agricultural Research Service Culture Collection at the National Center for Agricultural Utilization Research (Peoria, Ill.) (3). The T-2 toxin-negative strains MB1370 (tri1-2), MB1716 (tri1-1), and MB5493 (tri4) were previously isolated following UV mutagenesis of NRRL 3299 (3, 5). The NRRL 3299-derived T-2 toxin-hyperproducing transformant FsTri10-1-20 (↑Tri10) and the two T-2 toxin-negative transformants FsTri10-1-12 (ΔTri10) and NN4 (ΔTri6) have been previously described (33, 38). Transformants were inoculated onto V-8 juice agar (37) containing 300 μg of hygromycin B (Calbiochem, San Diego, Calif.)/ml and grown in continuous darkness with alternating temperature (25°C for 12 h and 20°C for 12 h). The remaining strains were inoculated onto V-8 juice agar and grown under alternating temperature and light conditions (25°C, light for 12 h and 20°C, dark for 12 h) (3). Strains were grown in liquid shake cultures in YEPD-2G medium (0.3% yeast extract, 1% peptone, 2% glucose) for DNA isolation (34) and in YEPD-5G medium (0.1% yeast extract, 0.1% peptone, 5% glucose) for trichothecene analysis and RNA isolation (39). All strains were maintained at −80°C as frozen conidial stocks in 15:85 glycerol-water. The vector pT7Blue-2 was purchased from Novagen (Madison, Wis.), and the vector pCR-Script was purchased from Stratagene (La Jolla, Calif.).
Fungal nucleic acid extraction and analysis.
Genomic DNA extractions, RNA isolations, Southern blotting, and Northern blotting, including subsequent hybridizations and autoradiography, were conducted by using standard protocols (36, 39). RNA was prepared from mycelia collected 23 h postinoculation from F. sporotrichioides strains. RNA blots were hybridized with five α-32P-labeled probes: Tri1, Tri3, Tri5, Tri6, and Tri8. Probes were amplified by PCR from F. sporotrichioides NRRL 3299 genomic DNA with gene-specific primers. All PCR products were purified with a Qiaquick gel extraction kit (Qiagen Inc., Valencia, Calif.) and subsequently analyzed and quantitated on a 1% agarose (Roche Molecular Biochemicals, Mannheim, Germany) gel.
Genomic clone construction.
A promoterless 2.0-kb genomic fragment of Tri1, representing the potential amino acid coding region plus introns, was amplified by PCR using primers A-74 (5′-CCAGTGGCATCTGCAGCTTTG-3′) and A-75 (5′-CAAGCGCTATTCAACATCAGATAATC-3′) and then ligated to pCR-Script to produce plasmid pAWP84. Sequences flanking this Tri1 fragment were cloned by chromosome crawling via inverse PCR (36). Plasmid pIKE1 contains an upstream fragment, while plasmids pAWP72 and pIKE5 contain slightly overlapping downstream sequences (Table 1). All PCR amplifications utilized DNA isolated from F. sporotrichioides NRRL 3299 and consisted of 30 cycles of 0.5 min at 95°C, 1 min at 50 to 55°C, and 1.5 min at 72°C, followed by 1 cycle of 10 min at 72°C. Prior to ligation to the cloning vector, each PCR product was separated on a 1% agarose (Roche Molecular Biochemicals) gel and purified with the Qiaquick gel extraction kit (Qiagen). Restriction enzymes were purchased from New England Biolabs (Beverly, Mass.). T4 DNA ligase was purchased from Promega (Madison, Wis.).
TABLE 1.
Construction information for plasmids carrying Tri1 flanking regions
| Plasmid | Cloning vector | Restriction enzymea | Size of PCR insert (kb) | Primers (sequences [5′-3′]) |
|---|---|---|---|---|
| pIKE1 | pCR-Script | KpnI | 2.1 | A-78 (GGTAGTGATCAGGGTTCGTC), A-65 (CATATCGAACCGGAAGTCCTCC) |
| pIKE5 | pCR-Script | XbaI | 2.5 | A-79 (GGTAGTGATGCCGAATAGAGG), A-81 (CTATGGTAGTGTGACCCTCTTG) |
| pAWP72 | pT7Blue-2 | PstI | 1.5 | A-65 (CATATCGAACCGGAAGTCCTCC), A-66 (GGCGATGTTCAAATCTGCCTGG) |
Restriction enzyme used to prepare genomic DNA samples for chromosome crawling via inverse PCR.
Genomic and cDNA sequencing analyses.
The above vector inserts were prepared for sequencing by using an exonuclease-driven Erase-a-Base system (Promega) to produce a nested series of directionally digested subclones. Sequencing reactions were performed by using vector-based universal primers and the BigDye terminator cycle sequencing core kit (Perkin-Elmer Corporation, Gaithersburg, Md.). Reactions were analyzed on a model 373A or a model 377 DNA sequencer (Applied Biosystems Inc., Foster City, Calif.) at the Gene Technologies Laboratory at Texas A&M University. The Tri1 cDNA sequence was provided by the Advanced Center for Genome Technology at the University of Oklahoma.
Plasmid construction and fungal transformation.
The Tri1 disruption plasmid, pIKE4, was constructed from pAWP84, which contains the promoterless 2.0-kb genomic fragment of Tri1, by inserting a fragment containing Cochliobolus heterostrophus promoter 1 fused to the hygromycin B phosphotransferase coding region (40) into a unique MscI site located 918 bp from the beginning of the largest possible open reading frame (ORF) in Tri1. The Tri1 complementation plasmid, pIKE9, was constructed by inserting a PCR product that contained an intact genomic copy of the Tri1 coding region plus approximately 900 bp upstream and 300 bp downstream of the putative ORF into pUCH3. Specifically, the PCR product was amplified from F. sporotrichioides NRRL 3299 genomic DNA with Platinum Pfx DNA polymerase (Invitrogen, Carlsbad, Calif.) and primers A-81 (5′-CTATGGTAGTGTGACCCTCCTG-3′) and A-83 (5′-ATCAGAATTCAGGACACTGC-3′); it was then purified with the Qiaquick gel extraction kit (Qiagen), digested with SphI, and ligated to the fungal cloning vector pUCH3, which was digested with SmaI and SphI. The pUCH3 vector contains the P1:hygB cassette (27).
Previously published procedures for protoplast production (39) and fungal transformation (35) were followed. PCR analyses of the ΔTri1 transformants were conducted using primers A-81, A-82 (5′-GTAGGCCTAAGTCAAGTTAC-3′), and S-26 (5′-GTTAGCAATTTAACTGTGATAAACTACCGC-3′).
Chemical analysis of toxin production.
Trichothecene identity and production in liquid cultures were determined by gas chromatography-mass spectrometry. Samples were prepared by vortexing (90 s) 1 volume of ethyl acetate with 1 volume of whole culture or culture filtrate. The ethyl acetate layers were collected, and a 2-μl sample was analyzed. Two methods of analysis with minor modifications (39) were used. Method 1 was as follows. Trichothecene profiles for the Tri1-disrupted strains were obtained with a Hewlett-Packard 5890 gas chromatograph equipped with an HP 5972 MS engine. An HP5-MS bonded stationary-phase capillary column (30 m by 2.5 mm [inside diameter]; film thickness, 0.25 μm) was used. The oven temperature program was as follows: initial oven temperature of 90°C for 2 min, followed by an increase in temperature at a rate of 23°C/min to 275°C for 2 min and then by an increase in temperature at a rate of 30°C/min to 290°C for 4.7 min. Helium was used as the carrier gas at a constant flow rate of 0.75 ml/min. Mass-selective detector parameters were as follows: full-scan mode, ionization energy set at 70 eV, and ion source temperature set at 180°C. Method 2 was as follows. Trichothecene data were obtained for the Tri1-complemented strains as described above with the following differences: (i) the Hewlett-Packard 5890 gas chromatograph was equipped with an HP 5970A MS engine, (ii) the injection port temperature was 300°C, and (iii) the oven temperature program included an initial oven temperature of 60°C and an initial temperature ramp of 7°C/min to 310°C for 8 min. Compound identifications were based on retention times and comparison to a mass-spectral library. T-2 toxin, 4,15-DAS, and neosolaniol (NEO) reference standards were purchased from Sigma Chemical Co. (St. Louis, Mo.) and used without further purification. T-2 toxin concentrations were determined by using appropriate standard curves.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the Tri1 genomic and cDNA sequences are AY040587 and AY032743, respectively.
RESULTS
Characterization of the coding and promoter regions.
Comparative analysis of the genomic and cDNA sequences for the P450 gene revealed the presence of four small introns (50, 66, 58, and 56 bp). All four contain the conserved GTNNGT::NAGsplice site (18). The largest potential ORF is 1,626 bp and could encode a 542-amino-acid protein. However, there are five potential AUG start codons in the first 100 bp of the largest possible ORF. The fourth of these five AUG codons has the most characteristic translation initiation context. This start site results in a 1,542-bp ORF and a protein containing 514 amino acids.
Regardless of the AUG codon that serves as the start site, the predicted protein contains the characteristic cytochrome P450 monooxygenase heme-binding motif sequence (FGYGIHVCPG), including the universally conserved cysteine residue. The amino acids in boldface represent the highly conserved motif residues. Based on the original analysis of the cDNA sequence, the predicted protein was placed in the cytochrome P450 subfamily CYP68C1 (Peplow et al., submitted). By BLAST analysis (2), this protein has the highest identity (45%) to a putative lovastatin (encoded by lovA) homolog from Neurospora crassa and had homology to the lovA-encoded homologs from Fusarium sambucinum and Aspergillus terreus.
Four putative TRI6 binding sites with the consensus sequence YNAGGCC (17) are located in the upstream flanking sequence within 350 bp of the first potential translation start site. Tri6 encodes a transcriptional activator (17), and the presence of the putative TRI6 binding sites is consistent with the finding that the expression of this P450 gene is regulated by the expression of Tri6 (Peplow et al., submitted).
Gene disruption results in 4,15-DAS accumulation.
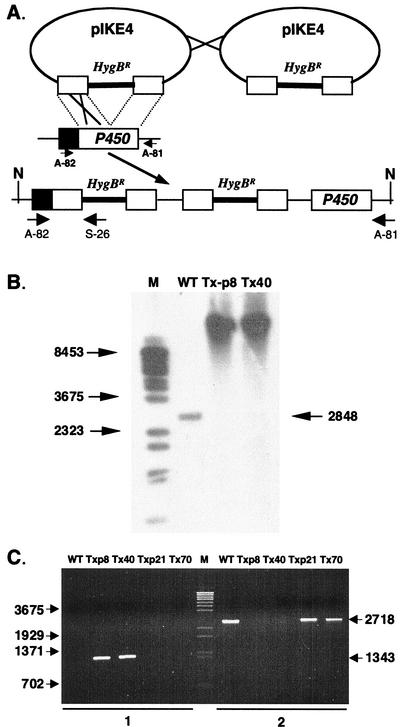
The P450 gene was disrupted by transforming F. sporotrichioides NRRL 3299 with pIKE4, a vector that contains a promoterless fragment of the P450 gene coding region interrupted by the P1:hygB cassette (Fig. 1A). Twelve of the 81 recovered transformants lacked the wild-type gene fragment, but in no case was the vector integrated via a homologous double crossover, which would have replaced the wild-type gene with a disrupted copy. However, as determined by gel blot and PCR analyses, two of the transformants resulted from an upstream homologous tandem integration of pIKE4. This type of integration should yield only nonfunctional copies of the P450 gene: an upstream disrupted copy, a downstream promoterless copy, and promoterless, disrupted copies in the tandem vector sequences (Fig. 1A). Southern blots for transformants Tx-p8 and Tx40 contained a high-molecular-weight band (instead of the wild type DNA fragment) that hybridized with the P450 gene probe, indicating that the transforming vector integrated as multiple, tandem copies at only a single site (Fig. 1B). Only transformants resulting from an upstream homologous crossover event could produce a PCR product with a primer (A-82) from the P450 gene promoter region and a primer (S-26) from within the P1:hygB fragment (Fig. 1A). Transformants Tx-p8 and Tx40 had the expected 1.3-kb PCR product (Fig. 1C). Conversely, primers that amplified a native P450 gene PCR product (2.7 kb) from the wild-type parent strain, as well as from transformants in which the vector integrated ectopically, produced no product from the Tx-p8 and Tx40 transformants as expected. Accordingly, the two disruptants presumably carry only nonfunctional copies of the P450 gene. Both Tx-p8 and Tx40 accumulated only 4,15-DAS in contrast to the wild-type parent strain, NRRL 3299, which produced primarily T-2 toxin but which also synthesized low levels of 4,15-DAS, NEO, 8-propylneosolaniol (P-NEO), and 8-butylneosolaniol (B-NEO) (Fig. 2A). Since the synthesis of NEO, P-NEO, and B-NEO and T-2 toxin, but not 4,15-DAS, requires C-8 hydroxylation, these results indicated that this cytochrome P450 monooxygenase catalyzes the C-8 hydroxylation reaction during trichothecene synthesis in F. sporotrichioides and suggested that this P450 gene might be Tri1.
FIG. 1.
Disruption of the P450 gene in F. sporotrichioides NRRL 3299. (A) Diagram of the production of inactive copies of the P450 gene as a result of the tandem integration of the disruption vector pIKE4 via a homologous upstream crossover event. Solid boxes, P450 gene promoter region; open boxes, region of the P450 gene present in pIKE4. N, NruI restriction sites. (B) Southern blot analysis of the wild-type (WT) parent NRRL 3299 and P450 gene disruption transformants. Genomic DNA was digested with NruI and probed with a P450 gene fragment prepared by using primers A-74 and A-75. The molecular weight ladder (M) is lambda DNA cut with BstEII. Arrows, band sizes. (C) PCR analysis of F. sporotrichioides NRRL 3299 and transformants. PCRs were conducted with primers A-82 and S-26 (lanes 1) and primers A-82 and A-81 (lanes 2). The primer positions are shown in panel A. PCR products were resolved on an agarose gel and stained with ethidium bromide. The molecular marker (M) is lambda DNA cut with BstEII. Arrows, band sizes.
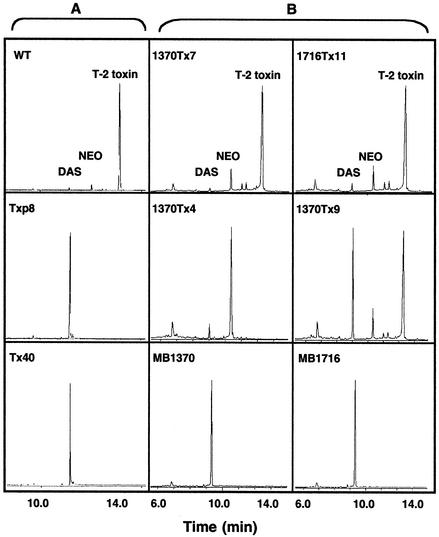
FIG. 2.
Chromatograms of ethyl acetate extracts of F. sporotrichioides cultures. (A) Wild-type (WT) NRRL 3299 and P450 disruption transformants. (B) tri1 UV-induced mutant MB1370 and MB1716 strains and transformants carrying the P450 (Tri1) gene complementation vector pIKE9.
Complementation analysis and rescue of the tri1 UV-induced mutants.
In previous studies, the two tri1 4,15-DAS-accumulating UV-induced mutants, MB1370 and MB1716, behaved like allelic strains during complementation tests (3, 7). We repeated the coinoculation tests but included the two new 4,15-DAS-accumulating disruptant strains, Tx-p8 and Tx40. All four 4,15-DAS-accumulating strains were able to complement with the isogenic trichodiene-accumulating tri4 mutant strain MB5493 to produce T-2 toxin. When these same four 4,15-DAS-accumulating strains were coinoculated with one another, however, none of the six pairs was able to produce T-2 toxin. Thus the P450 gene disruptants and tri1 mutants could be allelic.
We also transformed the tri1 UV-induced mutants MB1370 and MB1716 with pIKE9, which contains an intact copy of the P450 gene. Approximately 64% of the 39 recovered transformants (28 for MB1370 and 11 for MB1716) in each strain produced wild-type trichothecene profiles (Fig. 2B). The C-8 hydroxylation reaction was also restored in two additional MB1370 transformants, but they had somewhat different toxin profiles. One transformant, 1370Tx9, produced nearly equal amounts of 4,15-DAS and T-2 toxin, while another, 1370Tx4, produced primarily NEO but no T-2 toxin (Fig. 2B). The restoration of C-8 hydroxylation and T-2 toxin production in both tri1 UV-induced mutants provides critical evidence that this P450 gene is Tri1.
Northern characterization of Tri1 UV mutants and transformants.
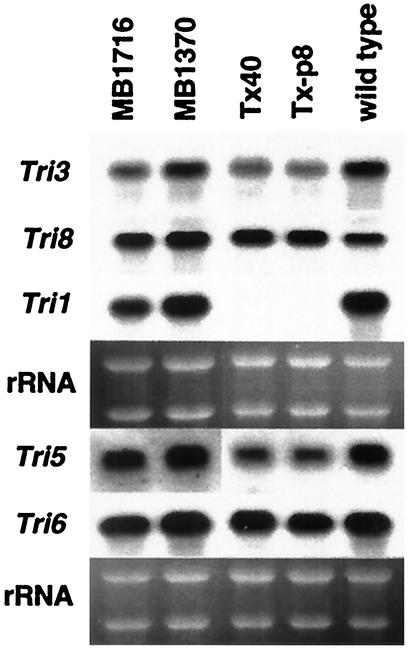
We prepared RNA from mycelia collected 23 h postinoculation for strains MB1370, MB1716, Tx-p8, Tx40, and NRRL 3299. This time point was selected because it corresponds to a high level of trichothecene gene expression (38). Northern blots were hybridized with probes for five genes: Tri1, Tri3, Tri5, Tri6, and Tri8 (Fig. 3). The Tri1 transcript was absent in both Tx-p8 and Tx40, confirming that they are Tri1-disrupted (ΔTri1) transformants. However, the presence of a Tri1 transcript in both tri1 UV-induced mutants indicates that the accompanying mutation in each of these strains does not affect transcription, but instead alters the production and/or function of the translated product. Transcript levels for Tri3, Tri5, Tri6, and Tri8 in the wild-type parent and all four tri1 mutant strains appeared similar. These data are consistent with both toxin production levels and previous intermediate feeding studies, which indicated that MB1716 retains all of the regulatory and enzymatic steps required for trichothecene synthesis except for the ability to hydroxylate the C-8 position (3, 7).
FIG. 3.
Northern blot analysis of gene transcript profiles for the tri1 UV mutants and ΔTri1 transformants. RNA blots, prepared with total RNA isolated for each strain from mycelia collected from 23-h YEPD-5G liquid shake cultures, were hybridized with α-32P-labeled gene-specific probes as indicated. Blots were stripped before rehybridization with another probe. rRNA bands in the corresponding ethidium bromide-stained gel prior to blotting are shown under each hybridization panel.
Evidence for a new gene, adjacent to Tri1, coregulated by Tri10 and Tri6.
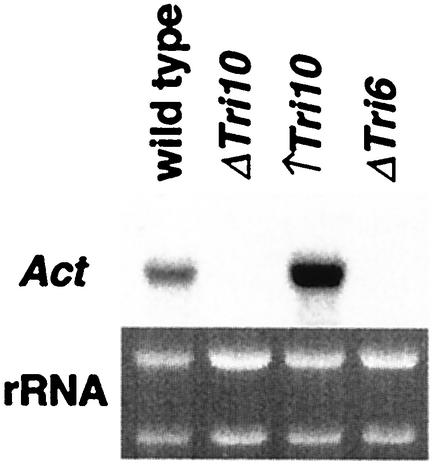
Sequence analysis of the DNA regions flanking Tri1 identified a potential gene for a Gal4-like transcription factor 1.0 kb upstream from Tri1 and another potential gene for a putative acetyltransferase 2.5 kb downstream from Tri1. Northern blots, which contained RNA from the wild-type parent strain, a Tri10-negative strain, a Tri10-overexpressing strain, and a Tri6-negative strain, were probed with a DNA sequence specific for each putative gene. Expression of the putative Gal4-like transcription factor was not observed under these conditions (data not shown). The putative acetyltransferase was not expressed in either the Tri10-negative strain or Tri6-negative strain and was overexpressed in the Tri10-overexpression strain (Fig. 4). This pattern of gene expression is characteristic for Tri genes (Peplow et al., submitted).
FIG. 4.
Northern hybridization analysis of the new acetyltransferase gene transcript in the wild-type NRRL 3299 strain, the Tri10-disrupted transformant (ΔTri10), the Tri10-overproducing transformant (↑Tri10), and the Tri6-disrupted transformant (ΔTri6). RNA blots, prepared with total RNA isolated for each strain from mycelia collected from 23-h YEPD-5G liquid shake cultures, were hybridized with an α-32P-labeled gene-specific probe. rRNA bands in the corresponding ethidium bromide-stained gel prior to blotting are shown.
DISCUSSION
Tri1 (formerly Tox1), the first Tri gene identified (3), has now been cloned as determined both by complementation of the tri1 UV-induced mutants with an intact cloned P450 gene sequence and by the molecular generation of 4,15-DAS-accumulating disruptants. The presented data demonstrate that Tri1 encodes a cytochrome P450 monooxygenase that catalyzes hydroxylation of the C-8 position during trichothecene biosynthesis.
Tri1 is the fourth cytochrome P450 gene involved in trichothecene biosynthesis to be cloned (1, 10, 15, 22). The predicted TRI1 amino acid sequence has 45% identity with a putative lovA gene-encoded lovastatin from Neurospora crassa but has less than 20% identity with the product of either Tri4, Tri11, or Tri13, suggesting that Tri1 did not arise by duplication of any of the other known trichothecene-specific P450 genes.
If Tri1 is used as a probe in Southern hybridizations, there is a positive signal with DNA from F. sambucinum (Gibberella pulicaris) R-6380, but not with DNA from Fusarium graminearum (Gibberella zeae) GZ3639 (Peplow et al., submitted), suggesting that F. sambucinum carries a homolog to Tri1 that is either missing in F. graminearum or insufficiently conserved at the nucleotide level to be detected. F. sporotrichioides and F. sambucinum both produce A-type trichothecenes and therefore would be expected to carry a similar gene for C-8 hydroxylation. F. graminearum produces B-type trichothecenes that have a keto group at the C-8 position and may therefore carry a divergent or different gene for addition of the oxygen at C-8.
Crosses between natural variants of F. sambucinum produce C-8+:C-8− progeny in a ratio of either 1:1 or 1:3, with the latter ratio indicating that two unlinked loci are required for C-8 hydroxylation (4, 6). The C-8 hydroxylation locus identified by the 1:1 segregation ratio in a cross between strains R-6380 and R-5455 was designated Tox1, but it is not known if this gene and Tri1 are homologs. This question can now be addressed by cloning the Tri1 homolog from F. sambucinum.
A gene in F. graminearum has also been designated Tox1 (20). However, this locus controls trichothecene toxin levels, not C-8 hydroxylation, and therefore is not a homolog of Tri1 or the designated Tox1 locus in F. sambucinum.
We sequenced the DNA flanking Tri1 because the determination of Tri1's position in the genome has been elusive. While 12 Tri genes reside in a 26-kb gene cluster (10, 16), cosmids covering a 60-kb region that evenly spans this cluster failed to complement the tri1 UV-induced mutant MB1716 (18). Consequently, Tri1 is at least 15 kb away from this trichothecene gene cluster. The noncluster gene Tri101 is positioned between an ammonia ligase gene and a phosphate permease gene (21). Thus, none of the 13 previously cloned Tri genes are adjacent to Tri1.
Tri1 is flanked by two new potential genes. A putative gene for a Gal4-like transcription activator is upstream of Tri1. It is not yet known if this gene is involved in trichothecene biosynthesis. The acetyltransferase gene downstream of Tri1 is regulated by both Tri10 and Tri6 and could be involved in trichothecene biosynthesis. In fact, we have determined that this gene is required for C-8 esterification (A. W. Peplow, I. B. Meek, M. Wiles, T. D. Phillips, and M. N. Beremand, unpublished data). Thus Tri1 is adjacent to another Tri gene. Altogether, the above data indicate that Tri1 is either in a new, distal region of the core trichothecene gene cluster or, more likely, resides in a separate, newly defined, second trichothecene gene cluster.
The above gene organization could also account for the recovery of the NEO-accumulating transformant 1370Tx4 following the transformation of MB1370 with the Tri1 complementation vector (Fig. 2B). In addition to restoring Tri1 gene expression, this vector may have undergone a homologous integration event that inactivated the adjacent C-8 acetyltransferase gene. However, regardless of its origin, the NEO-accumulating transformant provides the first evidence that the trichothecene biosynthetic pathway can be blocked at this point in F. sporotrichioides and is consistent with the possibility that the same enzyme catalyzes all three C-8 esterifications for the production of P-NEO, B-NEO, and T-2 toxin (8).
The isolation of the Tri1 gene raises additional interesting points regarding the trichothecene biosynthetic pathway. The hypothesized substrate for TRI1 is 3,14,15-triacetoxyscirpenol (Fig. 5). However, the tri11, tri13, and tri7 mutants readily produce C-8-oxygenated trichothecenes as demonstrated by the respective production of 8-hydroxyisotrichodermol in addition to isotrichodermin (1), 4-deoxy-T-2 toxin (10), and HT-2 toxin (9). Thus, TRI1 can apparently efficiently utilize calonectrin or 3,15-DAS and, to a lesser extent, isotrichodermin as substrates. This utilization pattern suggests that the TRI1 active site has a relaxed specificity that allows the binding and hydroxylation of these compounds, depending on their relative availabilities. At the same time, the tri3 mutants accumulate 15-calonectrin and 3,15-didecalonectrin (5, 27, 31), the TRI13 enzyme does not appear to efficiently utilize isotrichodermin (1), and TRI8 apparently deacetylates all substrates acelytated at C-4 (25) while TRI101 presumably competitively reverses these reactions (21, 26). Accordingly, we propose the metabolic grid shown in Fig. 5 for the biosynthesis of A-type trichothecenes. The key factor in this grid is the apparent broad substrate specificity displayed by TRI1.
FIG. 5.
Proposed metabolic grid for biosynthesis of the type A trichothecenes. The preferred T-2 toxin biosynthetic pathway (boldface arrows) has been previously established in F. sporotrichioides NRRL 3299 (7, 8, 9, 10, 28). Thin arrows, major alternative biosynthetic routes; dashed arrows, minor routes. Any one of the alternative routes can become the predominant route in response to limitations imposed by a variety of mechanisms, including mutations or other perturbations that alter substrate availability or inhibit the activity of one or more of the TRI enzymes. For example, the routes indicated by the thin arrows define the preferred alternative routes in response to single gene mutations that inactivate either Tri11 (1), Tri13 (10), or Tri7 (9). MAS, monoacetoxyscirpenol.
It is possible that the TRI1-catalyzed C-8 hydroxylation reaction may partially or fully precede the TRI13- or TRI7-catalyzed reaction or both during T-2 toxin biosynthesis. The coaccumulation of 4,15-DAS and T-2 toxin in the Tri1-complemented MB1370 transformant 1370Tx9 suggests that TRI1 activity levels are partially reduced in this isolate (Fig. 2B). This coproduction phenotype could result from a reduction in the ability of TRI1 to compete for a common substrate with TRI13, TRI7, or TRI8 or all three, depending on the order of these reactions relative to the TRI1-catalyzed step. These possibilities can be tested by independently overexpressing Tri7, Tri8, and Tri13 in the wild-type and 1370Tx9 strains.
Both the continued analysis of Tri1 in other species and the further characterization of the new genes flanking Tri1 will provide additional information about the regulation and evolution of the genes and gene clusters for the synthesis of these interesting secondary metabolites.
Acknowledgments
We thank Qun Ren and Bruce A. Roe at the University of Oklahoma for providing the Tri1 cDNA sequence data and Andrew Tag for helpful discussions.
This study was supported in part by USDA/CREES NRICGP grant no. 9503702. A.W.P. received additional support from a National Science Foundation grant to the Program for the Biology of Filamentous Fungi at Texas A&M University.
REFERENCES
- 1.Alexander, N. J., T. M. Hohn, and S. P. McCormick. 1998. The Tri11 gene of Fusarium sporotrichioides encodes a cytochrome P450 monooxygenase required for C-15 hydroxylation in trichothecene biosynthesis. Appl. Environ. Microbiol. 64:221-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Beremand, M. N. 1987. Isolation and characterization of mutants blocked in T-2 toxin biosynthesis. Appl. Environ. Microbiol. 53:1855-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beremand, M. N. 1989. Genetic and mutational tools for investigating the genetics and molecular biology of trichothecene production in Gibberella pulicaris (Fusarium sambucinum). Mycopathologia 107:67-74. [DOI] [PubMed] [Google Scholar]
- 5.Beremand, M. N., P. J. Black, and R. D. Plattner. 1988. Isolation and characterization of two new mutants blocked in T-2 toxin biosynthesis. J. Cell. Biochem. Suppl. 12C:261.
- 6.Beremand, M. N., and A. E. Desjardins. 1988. Trichothecene biosynthesis in Gibberella pulicaris: inheritance of C-8 hydroxylation. J. Ind. Microbiol. 3:167-174. [Google Scholar]
- 7.Beremand, M. N., and S. P. McCormick. 1992. Biosynthesis and regulation of trichothecene production by Fusarium species, p. 359-384. In D. Bhatnagar, E. B. Lillehoj, and D. K. Arora (ed.), Handbook of applied microbiology, vol. 5. Marcel Dekker, Inc., New York, N.Y.
- 8.Beremand, M. N., F. Van Middlesworth, S. Taylor, R. D. Plattner, and D. Weisleder. 1988. Leucine auxotrophy specifically alters the pattern of trichothecene production in a T-2 toxin-producing strain of Fusarium sporotrichioides. Appl. Environ. Microbiol. 54:2759-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, D. W., S. P. McCormick, N. J. Alexander, R. H. Proctor, and A. E. Desjardins. 2001. A genetic and biochemical approach to study trichothecene diversity in Fusarium sporotrichioides and Fusarium graminearum. Fungal Genet. Biol. 32:121-133. [DOI] [PubMed] [Google Scholar]
- 10.Brown, D. W., S. P. McCormick, N. J. Alexander, R. H. Proctor, and A. E. Desjardins. 2002. Inactivation of a cytochrome P-450 is a determinant of trichothecene diversity in Fusarium species. Fungal Genet. Biol. 36:224-233. [DOI] [PubMed] [Google Scholar]
- 11.Bu'Lock, J. D. 1980. Mycotoxins as secondary metabolites, p. 1-16. In P. S. Steyn (ed.), The biosynthesis of mycotoxins: a study in secondary metabolism. Academic Press, Inc., New York, N.Y.
- 12.Desjardins, A. E., G. F. Spencer, R. D. Plattner, and M. N. Beremand. 1989. Furanocoumarin phytoalexins, trichothecene toxins, and infection of Pastinaca sativa by Fusarium sporotrichioides. Phytopathology 79:170-175. [Google Scholar]
- 13.Desjardins, A. E., R. D. Plattner, and F. Van Middlesworth. 1986. Trichothecene biosynthesis in Fusarium sporotrichioides: origin of the oxygen atoms of T-2 toxin. Appl. Environ. Microbiol. 51:493-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris, L. J., A. E. Desjardins, R. D. Plattner, P. G. Nicholson, G. Butler, J. C. Young, G. Weston, R. H. Proctor, and T. M. Hohn. 1999. Possible role of trichothecene mycotoxins in virulence of Fusarium graminearum on maize. Plant Dis. 83:954-960. [DOI] [PubMed] [Google Scholar]
- 15.Hohn, T. M., A. E. Desjardins, and S. P. McCormick. 1995. The Tri4 gene of Fusarium sporotrichioides encodes a cytochrome P450 monooxygenase involved in trichothecene biosynthesis. Mol. Gen. Genet. 248:95-102. [DOI] [PubMed] [Google Scholar]
- 16.Hohn, T. M., A. E. Desjardins, S. P. McCormick, and R. H. Proctor. 1995. Biosynthesis of trichothecenes, genetic and molecular aspects, p. 239-248. In M. Eklund, J. L. Richard, and K. Mise (ed.), Molecular approaches to food safety: issues involving toxic microorganisms. Alaken, Inc., Ft. Collins, Colo.
- 17.Hohn, T. M., R. Krishna, and R. H. Proctor. 1999. Characterization of a transcriptional activator controlling trichothecene toxin biosynthesis. Fungal Genet. Biol. 26:224-235. [DOI] [PubMed] [Google Scholar]
- 18.Hohn, T. M., S. P. McCormick, and A. E. Desjardins. 1993. Evidence for a gene cluster involving trichothecene-pathway biosynthetic genes in Fusarium sporotrichioides. Curr. Genet. 24:291-295. [DOI] [PubMed] [Google Scholar]
- 19.Hsu, I. C., E. B. Smalley, F. M. Strong, and W. E. Ribelin. 1972. Identification of T-2 toxin in moldy corn associated with a lethal toxicosis in dairy cattle. Appl. Microbiol. 24:684-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jurgenson, J. E., R. L. Bowden, K. A. Zeller, J. F. Leslie, N. J. Alexander, and R. D. Plattner. 2002. A genetic map of Gibberella zeae (Fusarium graminearum). Genetics 160:1451-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura, M., G. Matsumoto, Y. Shingu, K. Yoneyama, and I. Yamaguchi. 1998. The mystery of the trichothecene 3-O-acetyltransferase gene. Analysis of the region around Tri101 and characterization of its homologue from Fusarium sporotrichioides. FEBS Lett. 435:163-168. [DOI] [PubMed] [Google Scholar]
- 22.Lee, T., Y.-K. Han, K.-H. Kim, S.-H. Yun, and Y.-W. Lee. 2002. Tri13 and Tri7 determine deoxynivalenol- and nivalenol-producing chemotypes in Gibberella zeae. Appl. Environ. Microbiol. 68:2148-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marasas, W. F. O., P. E. Nelson, and T. A. Toussoun. 1984. Toxigenic Fusarium species: identify and mycotoxicology. Pennsylvania State University Press, University Park.
- 24.Martin, L. J., J. A. Doebler, and A. Anthony. 1986. Scanning cytophotometric analysis of brain neuronal nuclear chromatin changes in acute T-2 toxin-treated rats. Toxicol. Appl. Pharmacol. 85:207-214. [DOI] [PubMed] [Google Scholar]
- 25.McCormick, S. P., and N. J. Alexander. 2002. Fusarium Tri8 encodes a trichothecene C-3 esterase. Appl. Environ. Microbiol. 68:2959-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCormick, S. P., N. J. Alexander, S. E. Trapp, and T. M. Hohn. 1999. Disruption of Tri101, the gene encoding trichothecene 3-O-acetyltransferase, from Fusarium sporotrichioides. Appl. Environ. Microbiol. 65:5252-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCormick, S. P., T. M. Hohn, and A. E. Desjardins. 1996. Isolation and characterization of Tri3, a gene encoding 15-O-acetyltransferase from Fusarium sporotrichioides. Appl. Environ. Microbiol. 62:353-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCormick, S. P., S. L. Taylor, R. D. Plattner, and M. N. Beremand. 1990. Bioconversion of possible T-2 toxin precursors by a mutant strain of Fusarium sporotrichioides NRRL 3299. Appl. Environ. Microbiol. 56:702-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLaughlin, C. S., M. H. Vaughn, J. M. Campbell, C. M. Wei, and M. E. Stafford. 1977. Inhibition of protein synthesis by trichothecenes, p. 263-273. In J. V. Rodricks, C. W. Hesseltine, and M. A. Mehlman (ed.), Mycotoxins in human and animal health. Pathotoxin Publishers, Park Forest, Ill.
- 30.Okumwai, H., N. Yoshino, Y. Suglura, M. Sugamata, E. L. Hintikka, B. Jarvis, and Y. Ueno. 1999. Trichothecenes as potent inducers of apoptosis, p. 221-231. In E. Johanning (ed.), Bioaerosols, fungi and mycotoxins: health effects, assessment, prevention, and control. Boyd Printing Co., Inc., Albany, N.Y.
- 31.Plattner, R. D., L. W. Tjarks, and M. N. Beremand. 1989. Trichothecenes accumulated in liquid culture of a mutant of Fusarium sporotrichioides NRRL 3299. Appl. Environ. Microbiol. 55:2190-2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Proctor, R. H., T. M. Hohn, and S. P. McCormick. 1995. Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol. Plant-Microbe Interact. 8:593-601. [DOI] [PubMed] [Google Scholar]
- 33.Proctor, R. H., T. M. Hohn, S. P. McCormick, and A. E. Desjardins. 1995. Tri6 encodes an unusual zinc finger protein involved in regulation of trichothecene biosynthesis in Fusarium sporotrichioides. Appl. Environ. Microbiol. 61:1923-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rotter, B. A., D. B. Prelusky, and J. J. Pestka. 1996. Toxicology of deoxynivalenol (vomitoxin). J. Toxicol. Environ. Health 48:1-34. [DOI] [PubMed] [Google Scholar]
- 35.Salch, Y. P., and M. N. Beremand. 1993. Gibberella pulicaris transformants: state of transforming DNA during asexual and sexual growth. Curr. Genet. 23:343-350. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 37.Stevens, R. R. 1974. Mycology guidebook, p. 703. University of Washington Press, Seattle.
- 38.Tag, A. G., G. F. Garifullina, A. W. Peplow, C. Ake, Jr., T. D. Phillips, T. M. Hohn, and M. N. Beremand. 2001. A novel regulatory gene, Tri10, controls trichothecene toxin production and gene expression. Appl. Environ. Microbiol. 67:5294-5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tag, A. G., J. Hicks, G. Garifullina, C. Ake, Jr., T. D. Phillips, M. N. Beremand, and N. Keller. 2000. G-protein signaling mediates differential production of toxic secondary metabolites. Mol. Microbiol. 38:658-665. [DOI] [PubMed] [Google Scholar]
- 40.Turgeon, B. G., R. C. Garber, and O. C. Yoder. 1987. Development of a fungal transformation system based on selection of sequences with promoter activity. Mol. Cell. Biol. 7:3297-3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ueno, Y. 1977. Trichothecenes: overview address, p. 189-207. In J. V. Rodricks, C. W. Hesseltine, and M. A. Mehlman (ed.), Mycotoxins in human and animal health. Pathotox Publishers, Inc., Park Forest South, Ill.