Abstract
The surface of Aspergillus fumigatus conidia, the first structure recognized by the host immune system, is covered by rodlets. We report that this outer cell wall layer contains two hydrophobins, RodAp and RodBp, which are found as highly insoluble complexes. The RODA gene was previously characterized, and ΔrodA conidia do not display a rodlet layer (N. Thau, M. Monod, B. Crestani, C. Rolland, G. Tronchin, J. P. Latgé, and S. Paris, Infect. Immun. 62:4380-4388, 1994). The RODB gene was cloned and disrupted. RodBp was highly homologous to RodAp and different from DewAp of A. nidulans. ΔrodB conidia had a rodlet layer similar to that of the wild-type conidia. Therefore, unlike RodAp, RodBp is not required for rodlet formation. The surface of ΔrodA conidia is granular; in contrast, an amorphous layer is present at the surface of the conidia of the ΔrodA ΔrodB double mutant. These data show that RodBp plays a role in the structure of the conidial cell wall. Moreover, rodletless mutants are more sensitive to killing by alveolar macrophages, suggesting that RodAp or the rodlet structure is involved in the resistance to host cells.
The surface of many fungal conidia is covered by a thin layer of regularly arranged rodlets. This structure, which favors air buoyancy and dispersion of the conidia by air currents (2), is mainly proteinaceous (3, 8-10, 16). The proteins present in the cell wall of aerial structures of fungi responsible for this rodlet configuration are the hydrophobins, a family of small, moderately hydrophobic proteins characterized by the conserved spacing of eight cysteine residues (42, 44). For the human opportunistic pathogen Aspergillus fumigatus, the presence of a rodlet layer has been visualized and the RODA gene has been previously shown to be involved in the formation of the rodlets of its conidia (41). In plants, hydrophobins have been associated with the virulence of phytopathogenic fungi (38). Although it has been repeatedly shown that cell wall and associated structures help human fungal pathogens to resist host defense reactions (22), to date no studies have analyzed the role of the rodlet layer in the resistance of the conidia to phagocytosis. Even though the rodlet layer of the conidia of Neurospora crassa, Beauveria bassiana, and Magnaporthe grisea contained a single hydrophobin (5, 39, 40), A. nidulans, a species phylogenetically close to A. fumigatus, has two conidial hydrophobins, RodAp and DewAp (35, 36). These data have prompted us to reexamine the surface layer of the conidia of A. fumigatus with a view to (i) analyzing exhaustively hydrophobins present on the surface of the conidia and (ii) studying their role in resistance to phagocytosis. A. fumigatus is a good model for the later study, since conidia, which are a main component of the airborne thermophilic fungal florae (1), are all engulfed and killed by lung alveolar macrophages (AM) following their inhalation (11; B. Philippe, O. Ibrahim-Granet, M. C. Prévost, M. A. Gougerot-Pocidalo, J. Roes, M. Sanchez-Perez, A. Van der Meeren, and J. P. Latgé, submitted for publication).
We report here that the conidia of A. fumigatus contain two hydrophobins: RodAp, a 16-kDa protein encoded by the previously described RODA gene, and RodBp, a 14-kDa protein encoded by the RODB gene that is different from DewAp of A. nidulans. Disruption of the RODB gene showed that this second hydrophobin, RodBp, although homologous to RodAp, is not involved in rodlet formation. Using single and double mutants, we show that RodAp but not RodBp protects conidia against conidial killing by AM.
MATERIALS AND METHODS
Strains and culture conditions.
The A. fumigatus strains used for this study were previously characterized: G10, a nitrate reductase mutant of strain CBS 144-89 (27), and ΔrodA-47 mutant (41). Conidia were harvested from 1-week-old culture grown at 25°C on 2% malt extract agar. Mycelia for DNA preparation were grown at 37°C in Sabouraud liquid medium (2% glucose, 1% Mycopeptone [Biokar, Beauvais, France]). The wettability of strains was tested essentially as described by Stringer and Timberlake (36).
All plasmid subcloning experiments were performed using Escherichia coli DH5α.
Preparation of the rodlet extract.
The rodlet layer was dislodged from the spore surface by sonicating the conidial suspension at 140 W (3-mm-diameter microtip, 50% duty cycle) for 2 × 10 min in a Sonifier cell disrupter B-30 (Branson Ultrasons, Rungis, France). Conidia were removed by low-speed centrifugation, and the supernatant was ultracentrifuged for 1 h at 50,000 × g. The pellet was boiled in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (2% SDS, 5% β-mercaptoethanol, 10% glycerol in 62 mM Tris-HCl, pH 6.8) and washed twice with sample buffer and three times with distilled water. The resulting pellet was freeze dried. The freeze-dried material was subsequently treated for 10 min at room temperature with 100% trifluoroacetic acid (TFA) (13). The acid was removed under a stream of nitrogen, and dried extracts were stored at room temperature under dry air.
Electrophoresis and immunoblotting.
After boiling the sample for 15 min in sample buffer, proteins were subjected to SDS-PAGE (15% polyacrylamide) according to the method of Laemmli (21) and visualized by silver nitrate following standard protocols. After using peroxidase-conjugated concanavalin A (ConA) (14) for electrotransfer of proteins to nitrocellulose membranes with Cat1 protein as a positive control (7), putative glycosylation of hydrophobins was assayed.
Purification and amino acid sequence analysis of RodBp.
The rodlet extract of ΔrodA-47 strain was fractionated using reverse-phase high-performance liquid chromatography essentially as described by Templeton et al. (40). The dried pellet was resuspended in 40% (vol/vol) acetonitrile containing 0.1% (vol/vol) TFA and was applied to a C18 column. The sample was eluted in a 20 to 80% (vol/vol) acetonitrile gradient containing 5% (vol/vol) methanol at a flow rate of 0.5 ml/min. The fractions corresponding to the major peak were pooled, dried, and subjected to SDS-PAGE. For internal peptides, tryptic fragments were generated by in-gel digestion of the 14-kDa band essentially as described by Kawasaki et al. (20). Amino terminal sequencing was carried out on a cutout band from the gel blot (ProBlott membrane; Applied Biosystem, Courtaboeuf, France) (25). Sequencing was performed by J. D'Alayer and M. Davi (Plateau Technique d'Analyze et de Microséquençage des protéines, Institut Pasteur).
Standard DNA procedures.
Agarose gel electrophoresis, Southern blotting, and subcloning of genomic DNA fragments into pBluescript SK plasmid (Stratagene, La Jolla, Calif.) were performed according to standard protocols (33). Using the procedure of Girardin et al. (15), genomic DNA was extracted from fungal mycelium. DNA hybridization probes were labeled, using the Ready Prime DNA labeling kit (Amersham, les Ullis, France), by the random primer method. Nucleotide sequences were determined on both strands by E.S.G.S. (Cybergene, Evry, France), and sequence analysis was performed using University of Wisconsin Genetics Computer Group programs (12).
Cloning and disruption of RODB.
We used a previously constructed pCosAX cosmid library of A. fumigatus as kindly provided by P. Borgia (6). Recombinant clones of the genomic library were immobilized on nylon membranes (Zeta-probe; Bio-Rad) and probed with 32P-labeled oligonucleotides (Amersham) as described by Jaton-Ogay et al. (19). Using the codon bias derived from published sequences of structural genes of A. fumigatus, the N-terminal sequence and an internal peptide were chosen for use in designing the following two degenerate oligonucleotides: n term (5′ GGY GTI GTI CAC CCT ACC TTC GCY TCY GCY GAY AAG TAC AC [41 bp]) and intern 1 (5′ GTC GTC RAC RCC GAT RAG RGC GGT [24 bp]).
The pAN8-1 plasmid (26) was digested with XbaI, the XbaI site was filled with the Klenow enzyme, and the linearized plasmid was cut with XhoI, giving a 2.3-kb fragment containing the phleomycin resistance marker with one blunt end and one XhoI site at the other end. The deletion construct pΔrodB was obtained by replacing the 0.2-kb SalI-EcoRV fragment of the RODB coding region with the 2.3-kb fragment. Employing lysing enzymes from Trichoderma harzianum (Sigma, St-Quentin Fallavier, France), the 7-kb KpnI-XbaI fragment of pΔrodB was used to transform A. fumigatus protoplasts as previously described (27). Gene replacement was verified by Southern blot analysis.
Production of recombinant RodAp.
Recombinant RodAp was produced as a hexahistidine-tagged protein in E. coli M15 by following a strategy used for other A. fumigatus proteins (28). An intron-free coding sequence was assembled from the published genomic sequence (GenBank accession no. U06121) by PCR. In a first step, the region spanning amino acids 1 to 109 was amplified using the 5′ primer GA AGA TCT ATG AAG TTC TCT TTG AGC GCT GC and the 3′ primer TGG CTC GAC GTT GGA GCA CTG GTT GAA, introducing BglII and SalI restriction sites. The region spanning amino acids 110 to 159 was amplified with the 5′ primer CCA GTC GAG CTC CAG ATC CCC GTC ATT GGT ATT CCA ATC C and the 3′ primer CCC AAG CTT TTA CAG GAT AGA ACC AAG GGC AAT GCA AGG AAG ACC CAG TCC AAT GAG GGA ACC GCT GGC ATC GGA AGG AGA GTT CTG GCT GC, introducing SalI and HindIII restriction sites. Both PCR products were purified from agarose gels, restricted with SalI, and ligated in a 1:1 ratio. The ligation product, corresponding to the expected size of 888 bp, was isolated from the agarose gel, restricted with BglII and HindIII, and subcloned into a pDS 56 high-level-expression vector (37) to produce RodAp protein.
Murine-specific antibodies.
RodAp purified by Ni2+-chelate affinity chromatography and electroeluted RodBp was used to immunize Swiss mice intracutaneously. Several booster injections were performed every 2 weeks after the first injection, and the immunization was followed by immunoblot analysis with peroxidase-conjugated anti-mouse immunoglobulin. Mice were sacrificed after blots gave positive results at 1:1,000 (RodAp) and 1:100 (RodBp) serum dilutions.
Field emission scanning electron microscopy.
Conidia of the different mutants and of the parental wild-type strain were analyzed with a JEOL JSM-6700F apparatus, which is an ultra-high-resolution field emission scanning electron microscope (FESEM) equipped with a cold-field-emission gun and a strongly excited conical lens. The secondary-electron-image resolution was 1 nm at 15 kV and 2.2 nm at 1 kV. Conidia or pieces of cultures were frozen using a Gatan Alto 2500 cryo-stage and cryo-preparation chamber dedicated for use in FESEM. The preparation chamber was cooled with liquid nitrogen, and a gas-cooled SEM stage module contained an anticontaminator system and employed temperature control between −185°C and +100°C. Samples were frozen in slush nitrogen and cryotransferred under vacuum to the preparation chamber for ice sublimation and gold palladium sputter coating (2 nm). Material was then transferred (under secondary vacuum) from the Gatan preparation chamber to the SEM stage for sample observation. The working temperature with the SEM stage module was −110°C. SEM working conditions were as follows: accelerating voltage, 1 or 2 kV; probe current, 30 pA; semi-in-lens secondary electron image; working distance, 1.5 nm.
Transmission electron microscopy.
For ultrastructural observation of the cell wall, conidia were fixed in a 0.1 M cacodylate buffer (pH 7.4) containing 3% glutaraldehyde and 0.075% ruthenium red. Washings and postfixation in 1% OsO4 were done in the presence of 0.075% ruthenium red (17). Embedding was performed in Epon. Ultrathin sections (20 nm) were stained with uranyl acetate and lead citrate before observation with a Jeol 1200 Ex transmission electron microscope operating at 80 kV.
AM killing assay.
Male outbred Swiss OF1 mice (Iffa Credo, Saint Germain sur l'Arbresle, France) 6 to 8 weeks of age (32 to 34 g of body weight) were intranasally infected with 4 × 106 fluorescein isothiocyanate-labeled conidia of each strain (Philippe et al., submitted). At 24 h after infection, AM were recovered from the bronchoalveolar lavages by a 5-min centrifugation (400 × g). The cell pellet was resuspended in 0.2 ml of water to lyse the AM and was then incubated for 6 to 8 h at 37°C after the addition of 200 μl of 2× Sabouraud medium containing 0.1% chloramphenicol to induce germination of the living conidia. The percentage of killing was defined as the number of nongerminated spores per 100 counted fluorescein isothiocyanate conidia as estimated under a light-fluorescent microscope. Using SuperANOVA software (Abacus Concepts, Berkeley, Calif.), data were evaluated by variance analysis.
Nucleotide sequence accession number.
The nucleotide sequence of the RODB gene has been submitted to the GenBank database under accession number AY057385.
RESULTS
Hydrophobins of the outer cell wall layer of A. fumigatus.
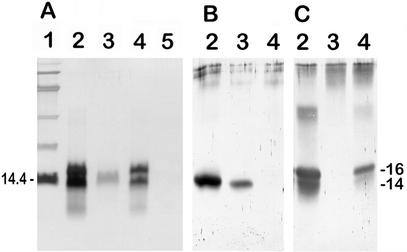
Ultrasonic treatment of conidia removed SDS-insoluble material without affecting the morphology of the conidia. The SDS-insoluble material was solubilized with TFA and analyzed by SDS-PAGE. Two proteins with apparent molecular masses of 16 and 14 kDa were identified in the wild-type G10 strain (Fig. 1A). The absence of the 16-kDa protein from the conidial extract of the ΔrodA-47 strain indicated that this polypeptide was the RodA protein. The 30-kDa band was a dimer of RodAp that reacted positively with the anti-RodAp antiserum (Fig. 1C). Dimerization of hydrophobins in an SDS-PAGE gel due to the difficulty of dissolving hydrophobins into monomers and the presence of strong noncovalent interactions responsible for the formation of insoluble complexes of hydrophobins (13) was also observed by other authors (39, 42). The 14-kDa RodBp polypeptide was purified by reverse-phase liquid chromatography from the rodlet extract of ΔrodA-47 strain. The major peak eluted at 65% acetonitrile was subjected to SDS-PAGE and used for amino acid sequencing. Two peptide sequences were obtained: LPNAGVVHPTFASADKYTLQ for the N terminus and ISVTALIGVDDLLNK for an internal tryptic fragment.
FIG. 1.
Analysis of SDS-insoluble TFA-soluble material from ultrasonicate extract of A. fumigatus conidia. SDS-PAGE (15% polyacrylamide) gels were stained with silver nitrate (A) or transferred to nitrocellulose and probed with monospecific anti-RodBp antibodies (B) or anti-RodAp antibodies (C). Lane 1, size markers are protein standards (in kilodaltons); lanes 2, G10 strain (RODA RODB); lanes 3, transformant ΔrodA-47 (rodA RODB), lanes 4, transformants ΔrodB-02 (RODA rodB), lane 5, transformant ΔrodA ΔrodB-26 (rodA rodB).
Cloning and characterization of the RODB gene.
The amino acid sequences GVVHPTFASADKYT of the N terminus and TALIGVDD of the internal peptide of the 14-kDa band were used to design degenerate primers. The screening of the cosmid library with the degenerated oligonucleotides revealed a 0.9-kb PstI-EcoRI fragment that contained an open reading frame coding for a predicted protein of 140 amino acids. The presence of two putative introns in the nucleotide sequence was confirmed by amino acid sequencing of two internal peptides of the 14-kDa protein. These two peptides, ISVT/ALIGVDDLLNK and SVA/TGG, overlap the introns (positions shown by slashes). The N-terminal amino acid sequence LPNAGVV of the 14-kDa protein was preceded by a hydrophobic peptide of 16 amino acid residues, showing the existence of a signal sequence for secretion. The predicted molecular mass of the mature protein RodBp is 12.7 kDa, which is consistent with the observed 14-kDa molecular mass on the SDS gel. Although there is a putative N-glycosylation site (NKS) at position 117 (C terminus), RodBp did not react with the ConA-peroxidase conjugate, showing that RodBp did not have an N-glycan moiety (data not shown).
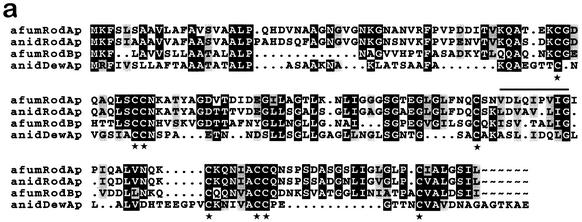
RodB protein shared all the hallmarks of class I hydrophobins: (i) it is a small protein (100 ± 25 amino acids), (ii) it is secreted (signal sequence), and (iii) it features conserved cysteine spacing (C-X5-8-C-C X17-39-C-X8-23-C-X5-6-CC-X6-18-C-X2-13) (44). An amino acid sequence comparison (Fig. 2a) revealed that RodBp was homologous to RodAp of A. nidulans (47% similarity) and to RodAp of A. fumigatus (44% similarity) but showed only 26% similarity to the 14-kDa DewAp hydrophobin of A. nidulans. Interestingly, the signal sequence of RodBp shared 86% similarity with that of DewAp but only 44 and 50% similarity with those of RodAp of A. fumigatus and A. nidulans, respectively.
FIG. 2.
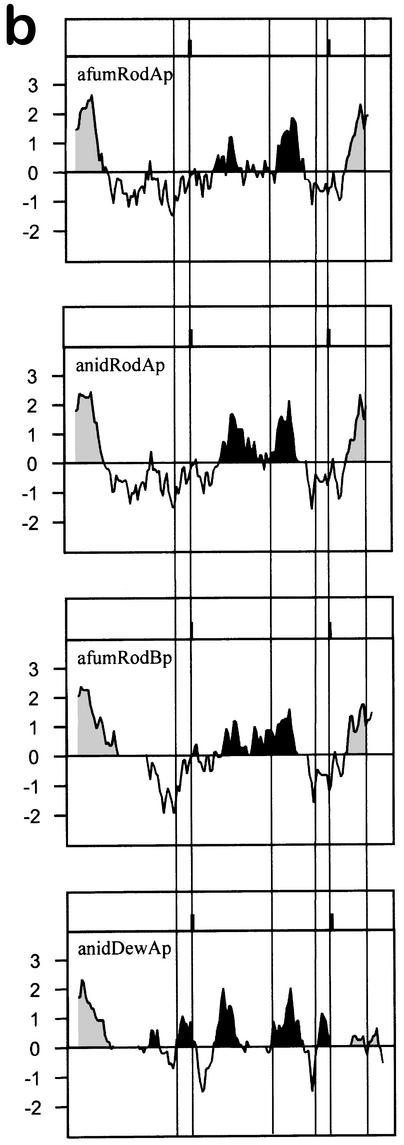
(a) Alignment of RodBp with the other conidial hydrophobins of Aspergillus species. The protein sequences are those of A. fumigatus RodAp (afumRodAp; GenBank accession no. U06121 [41]) and RodBp (afumRodBp), A. nidulans RodAp (anidRodAp; GenBank accession no. M61113 [35]), and A. nidulans DewAp (anidDewAp; GenBank accession no. U07935 [36]). Peptide sequences were aligned with GCG Pileup and Boxshade software customized to force alignment of the cysteine residues. Identical residues are shown on a black background; similar residues are shaded. Gaps introduced to optimize alignment are indicated by dots. The conserved cysteine residues are indicated by stars below the sequences. The sequence of the internal peptide of the 14-kDa band of the ΔrodB-02 mutant is indicated by a horizontal line (see Results). (b) Hydrophobicity plots of conidial hydrophobins from A. fumigatus (RodAp and RodBp) and A. nidulans (RodAp and DewAp), derived using the Kyte and Doolittle algorithm of the DNA Strider program (24). The sequences were aligned at the cysteine residues (vertical lines), with gaps introduced as described for panel a. Hydrophobic N- and C-terminal domains are shaded, while central hydrophobic domains are in black.
The hydropathy plot of RodBp (Fig. 2b) was similar to those of RodAp of A. fumigatus and A. nidulans but showed some differences from that of DewAp of A. nidulans. After the hydrophobic signal peptide, RodAp and RodBp had a neutral-to-hydrophilic domain, a hydrophobic central core followed by a small hydrophilic region (around the fifth to seventh cysteine residues), and a highly hydrophobic C terminus. In contrast, DewAp was characterized by hydrophobic and hydrophilic domains dispersed along its sequence without hydrophobicity at the C terminus.
Disruption and analysis of ΔrodB transformants.
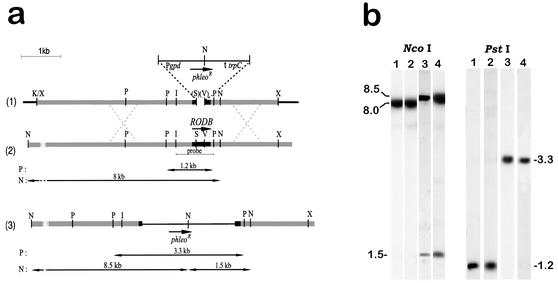
To investigate the function of the RodB protein, we disrupted the RODB gene by substituting the phleomycin cassette for the 216-bp SalI-EcoRV fragment of the RODB open reading frame (Fig. 3a). The 7-kb KpnI-XbaI fragment was used to transform recipient strains. G10 was transformed to obtain a rodB mutant, and ΔrodA-47 was transformed to create a double rodA rodB mutant. Phleomycin-resistant transformants were analyzed by Southern blotting to look for the deletion replacement event (Fig. 3b). Southern blot analysis of A. fumigatus transformants showed the replacement of the wild-type RODB gene with the disrupted gene in two ΔrodB transformants (ΔrodB-02 and ΔrodB-42) and in one ΔrodA ΔrodB transformant (ΔrodA ΔrodB-26). By using the deleted 0.2-kbp SalI-EcoRV fragment as a probe, the deletion event was confirmed by Southern blotting. No positive band was observed in any single or double ΔrodB mutant (data not shown).
FIG. 3.
Disruption of RODB. (a) Construction of plasmid pΔrodB. RODB was cloned on a 6-kb genomic XbaI fragment (see Materials and Methods), and a 216-bp SalI-EcoRV fragment was removed from the coding region and replaced with a 2.3-kb blunt XhoI fragment containing the phleomycin resistance gene (26), thereby removing the restriction enzymes (indicated in parentheses). A linear 7-kb XbaI-KpnI fragment from the resulting construct was used to transform G10 and ΔrodA-47 strains. The thin line at the top of the panel indicates a phleomycin cassette; the black dotted lines indicate an A. fumigatus DNA flanking the coding region of the RODB gene (black box). I, EcoRI; V, EcoRV; K, KpnI; N, NcoI; P, PstI; S, SalI; X, XbaI. (b) Southern hybridization of NcoI- and PstI-digested genomic DNAs of the wild-type G10 strain (lanes 1) and of transformants ΔrodA-47, ΔrodB-02, and ΔrodA ΔrodB-26 (lanes 2, 3, and 4, respectively), all probed with the 1-kb EcoRI-PstI fragment of the RODB gene. Sizes are given in kilobases.
Sporulating colonies of the wild-type G10 and ΔrodB-02 strains were morphologically similar: they were characterized by a light green color and did not wet when water or a dilute detergent solution (0.2% SDS, 50 mM EDTA) was dropped on the colony surface. The ΔrodA ΔrodB-26 transformant displayed a dark color and conidia easily wetted in distilled water, as with the ΔrodA-47 recipient strain (data not shown).
Analysis by SDS-PAGE of material extracted from conidia of the single and double ΔrodB mutants confirmed the deletion of the RODB gene. No SDS-insoluble TFA-soluble protein was recovered from the double mutant ΔrodA ΔrodB-26, as shown by the absence of both RodAp and RodBp polypeptides on SDS-PAGE (Fig. 1A). Unexpectedly, two polypeptides with apparent molecular masses of 16 and 14 kDa, similar to those present in wild-type extracts, were detected in conidial extracts of the simple mutant ΔrodB-02. The hypothesis of the presence of both a glycosylated (16 kDa) and an unglycosylated (14 kDa) RodAp in the ΔrodB-02 mutant was ruled out, because there are no potential N-glycosylation sites in RodAp and ConA did not bind to RodAp at either the 16- or the 14-kDa band (data not shown). Peptide sequencing of the 14-kDa band of the ΔrodB-02 mutant revealed a sequence of RodAp VDLQIPVIG (Fig. 2a), suggesting that the 14-kDa band in the ΔrodB-02 mutant is a modified form of RodAp. This hypothesis was confirmed by Western blot analysis. The anti-RodAp and anti-RodBp antisera obtained were monospecific and did not cross-react, as the ΔrodA-47 and ΔrodB-02 extracts did not give a positive band with the anti-RodAp and anti-RodBp antibodies, respectively (Fig. 1B and C). The anti-RodBp antibody bound to the 14-kDa band of G10 extract, but it did not bind to the protein of similar molecular mass in the ΔrodB-02 extract (Fig. 1B). Moreover, the anti-RodAp antiserum bound to the 16- and 14-kDa bands of G10 and ΔrodB-02 extracts (Fig. 1C). This absence of binding of anti-RodBp antiserum in ΔrodB transformants and the positive binding with anti-RodAp serum confirmed that the 14-kDa band of the ΔrodB transformants represents a form of RodAp.
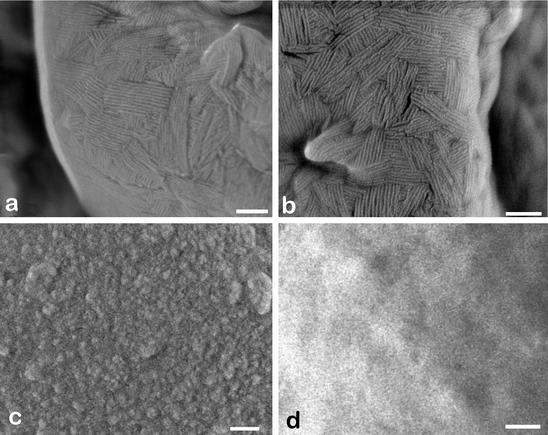
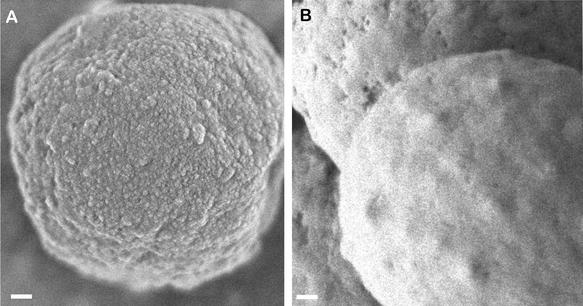
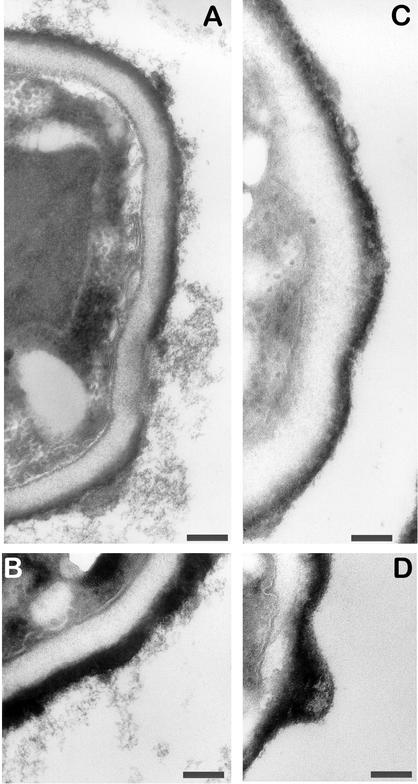
High-resolution SEM showed that RodBp was not responsible for the formation of the rodlet layer in A. fumigatus, as ΔrodB-02 conidia possessed a rodlet layer (Fig. 4b) with an arrangement of bundles of rod similar to that of the wild-type G10 recipient strain (Fig. 4a). However, the absence of RodBp in the double mutant was associated with modification of the conidial surface morphology. The ΔrodA ΔrodB-26 conidia showed a smooth surface (Fig. 4d and 5B), whereas the recipient ΔrodA-47 mutant displayed a granular aspect (Fig. 4c and 5A). ΔrodA-47 conidia stained with ruthenium red and observed by transmission electron microscopy were covered with cell wall material (mostly fibrillar), whereas ΔrodA ΔrodB-26 conidia had no fibrils on their surface (Fig. 6).
FIG. 4.
Scanning electron micrographs of A. fumigatus conidia of the wild-type G10 strain (a) and of transformants ΔrodB-02 (b), ΔrodA-47 (c), and ΔrodA ΔrodB-26 (d). Size bar, 100 nm.
FIG. 5.
Scanning electron micrographs of A. fumigatus conidia of transformants ΔrodA-47 (A) and ΔrodA ΔrodB-26 (B). Size bar, 100 nm.
FIG. 6.
Transmission electron micrographs of A. fumigatus conidia of transformants ΔrodA-47 (A and B) and ΔrodA ΔrodB-26 (C and D) after ruthenium red staining. Size bar, 200 nm.
AM killing of mutant conidia.
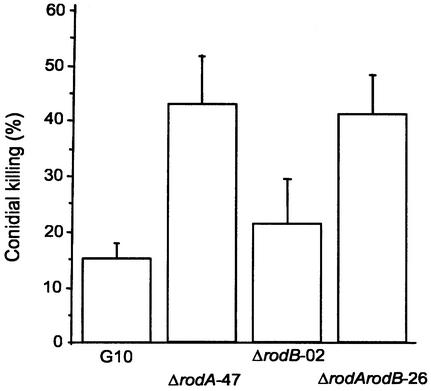
The sensitivity of the mutant and parental conidia to killing by AM was evaluated: 43% of ΔrodA and 41% of ΔrodA ΔrodB conidia were killed by AM, while only 15% of G10 and 21% of ΔrodB conidia were killed (Fig. 7). Rodletless conidia (ΔrodA and ΔrodA ΔrodB) were significantly more sensitive to killing by macrophages than ΔrodB and G10 conidia (P = 0.01), while there was no difference between G10 and ΔrodB conidia or ΔrodA and ΔrodA ΔrodB conidia in sensitivity to killing (P > 0.5).
FIG. 7.
In vivo conidial killing estimated in murine AM recovered from mice infected intranasally with 105 conidia of the wild-type G10 strain and of transformants ΔrodA-47, ΔrodB-02, and ΔrodA ΔrodB-26. Means of three experiments ± standard errors are shown.
DISCUSSION
Hydrophobins are proteins presumably present as highly insoluble complexes in the outermost layer of the fungal wall (42, 44). While numerous genes encoding putative hydrophobins expressed during conidiation have been cloned (4, 23, 31, 35, 36, 39, 41), the hydrophobin proteins of the conidial rodlet layer have been isolated in only three fungal species: N. crassa, B. bassiana, and M. grisea (5, 39, 40). In these studies only one SDS-insoluble TFA-soluble protein of low molecular mass (7, 10, and 15 kDa for Neurospora crassa, Beauveria bassiana, and Magnaporthe grisea, respectively) was found in rodlets of conidia. In our present study, two SDS-insoluble TFA-soluble proteins, RodAp and RodBp, with 16- and 14-kDa molecular masses, respectively, were isolated from the conidial outer wall of A. fumigatus (Fig. 1). In A. nidulans and A. niger, two proteins of 16 and 14 kDa were also present as insoluble complexes in the conidial cell wall (unpublished data). In contrast to the conidial rodlet layers of N. crassa and B. bassiana, the rodlet layer of Aspergillus conidia contains two hydrophobins. The two hydrophobins RodAp and RodBp are present only in conidia and were not found in the mycelium (unpublished data), suggesting that these two hydrophobins are developmentally regulated in a manner similar to that by which RodAp and DewAp of A. nidulans are developmentally regulated.
RodAp was previously shown to be an essential component of the rodlet layer of A. nidulans and A. fumigatus, because it is required for rodlet formation (35, 41). The SEM study of ΔrodB conidia revealed the presence of rodlets at their surfaces (Fig. 4) and showed that disruption of RODB was not associated with a modification of the rodlet structure. Therefore, in contrast to RodAp but like DewAp in A. nidulans (36), RodBp is not essential for rodlet formation. This result was unexpected, since an amino acid sequence comparison showed a high similarity between RodBp and RodAp of A. nidulans and A. fumigatus (47% and 44%, respectively). Parta et al. (31) have shown that the RODA gene of A. fumigatus under the control of its own promoter was able to complement the homologous rodA mutation in A. nidulans. Our attempts to complement the rodA mutation of A. nidulans with the A. fumigatus RODB gene under its native promoter failed: the RODB mRNA was expressed during conidiogenesis of A. nidulans transformants, but no RodB protein, either as an SDS-soluble or SDS-insoluble TFA-soluble form, was found in the rodlet fraction or in the cell wall of conidia or phialides (unpublished data). The poor homology between the signal sequences of A. nidulans RodAp and A. fumigatus RodBp suggests that the A. fumigatus RodBp secretion sequence does not operate properly in the A. nidulans rodA mutant and that RodBp is degraded.
Even though A. nidulans and A. fumigatus have two hydrophobic proteins per species, only one protein was responsible for rodlet formation. The proteins DewAp of A. nidulans and RodBp of A. fumigatus have the same molecular mass (14 kDa; unpublished data) and share similar signal sequences. However, they show differences: (i) they share only 26% similarity of their amino acid sequences (Fig. 2a); (ii) their hydropathy profiles are different (Fig. 2b); (iii) anti-RodBp antibodies do not bind to DewAp (unpublished data); and (iv) the A. fumigatus RODB gene did not complement the A. nidulans dewA mutation. Since RodBp and DewAp were isolated with RodAp in the rodlet extract, we expected to find RodBp of A. fumigatus with RodAp of A. nidulans in the rodlet extract of the A. nidulans dewA mutant transformed with the A. fumigatus RODB gene. However, even though RODB mRNA and RodB protein were produced in the A. nidulans dewA transformant, RodBp had only an intracellular localization, as shown using an anti-RodBp antiserum (unpublished data). These results suggested that even though the secretion sequence of RodBp is similar to that of A. nidulans DewAp, RodBp was not targeted to the outer cell wall of the conidia of A. nidulans as an SDS-insoluble complex.
The presence of a low-molecular-mass protein band in transformants ΔrodB-02 and ΔrodB-42 with an apparent molecular mass similar to that of RodBp was unexpected. Our data showed that the 16- and 14-kDa bands in ΔrodB mutants are RODA gene products and that the 14-kDa band is a consequence of the partial degradation of RodAp resulting from the harsh treatments required for hydrophobin extraction (ultrasonication, TFA solubilization, etc.). Similar observations with other hydrophobins have been previously reported: two bands on SDS-PAGE having the same N-terminal sequence were found in Trichoderma reesei, Pleurotus ostreatus, and Schizophyllum commune hydrophobin extracts (30, 32, 43).
The high sensitivity of the rodletless ΔrodA and ΔrodA ΔrodB conidia to killing by AM shows that the rodlet layer or RodAp protects the conidia against the defense mechanisms of the AM. The simplest explanation for this sensitivity is that hydrophilic killing molecules such as reactive oxidants, produced by the macrophage following activation, do not easily cross the hydrophobic layer. However, a direct trapping of the deleterious host molecules by RodAp might also occur in a manner similar to that suggested for melanin (18). These data are in agreement with the results of Shibuya et al. (34), who showed that pulmonary lesions induced by the rodletless mutant ΔrodA-47 were limited compared to those induced by the G10 strain. In contrast, RodBp is not directly involved in the resistance of conidia to killing. Neither RodAp nor RodBp played a role in protecting conidia from desiccation, as was suggested by Stringer et al. and Parta et al. (31, 35): 6-month-old ΔrodA ΔrodB conidia germinated as well as wild-type conidia (unpublished data). Moreover, mutant conidia are as resistant to acid pH (3.0 and 4.5) as wild-type conidia (unpublished data). No specific function has been associated to date to the RodB protein. Other examples of dispensable cell wall-specific proteins with unknown function and no obvious phenotype following gene disruption have been previously recorded for A. fumigatus (29). Analysis of the slimy material present on the surface of the ΔrodB conidia might give some clue to the cellular role of RodBp.
The results presented here show that RodAp and RodBp are two hydrophobins present as SDS-insoluble complexes in the outer conidial wall. RodBp does not form rodlets at the surface of A. fumigatus conidia but is involved in the building of the conidial outer cell wall. Although the contribution of RodBp alone in the structure of the outer layer of conidia cell wall seems minor, RodBp interacts in some way with the cell wall components and alters the surface properties of ΔrodA mutants. While RodBp is specific to the species A. fumigatus, it does not protect conidia against killing by AM and its biological significance for the fungus remains an open question.
Acknowledgments
Plasmid pAN8-1 was kindly provided by P. J. Punt (TNO Medical Biological Laboratory, Rijswijk, The Netherlands). We thank P. T. Borgia (Department of Medical Microbiology and Immunology, Southern Illinois University School of Medicine, Springfield) for providing the A. fumigatus genomic cosmid library and Catherine Dotigny and Marc Tournaire for their technical assistance. We are grateful to Rich Calderone for his critical reading of the manuscript and to Marinette Cormier for typing the manuscript.
REFERENCES
- 1.Al-Doory, Y. 1984. Airborne fungi, p. 27-40. In Y. Al-Doory and J. F. Domson (ed.), Mould allergy. Lea & Febiger, Philadelphia, Pa.
- 2.Beever, R. E., and G. P. Dempsey. 1978. Function of rodlets on the surface of fungal spores. Nature (London) 272:608-610. [DOI] [PubMed] [Google Scholar]
- 3.Beever, R. E., R. J. Redgwell, and G. P. Dempsey. 1979. Purification and chemical characterization of the rodlet layer of Neurospora crassa conidia. J. Bacteriol. 140:1063-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell-Pedersen, D., J. C. Dunlap, and J. J. Loros. 1992. The Neurospora circadian clock-controlled gene, ccg-2, is allelic to eas and encodes a fungal hydrophobin required for formation of the conidial rodlet layer. Genes Dev. 6:2382-2394. [DOI] [PubMed] [Google Scholar]
- 5.Bidochka, M. J., R. J. St. Leger, L. Joshi, and D. W. Roberts. 1995. The rodlet layer from aerial and submerged conidia of the entomopathogenic fungus Beauveria bassiana contains hydrophobin. Mycol. Res. 99:403-406. [Google Scholar]
- 6.Borgia, P. T., C. L. Dodge, L. E. Eagleton, and T. H. Adams. 1994. Bidirectional gene transfer between Aspergillus fumigatus and Aspergillus nidulans. FEMS Microbiol. Lett. 122:227-232. [DOI] [PubMed] [Google Scholar]
- 7.Calera, J. A., S. Paris, M. Monod, A. J. Hamilton, J. P. Debeaupuis, M. Diaquin, R. Lopez-Medrano, F. Leal, and J. P. Latgé. 1997. Cloning and disruption of the antigenic catalase gene of Aspergillus fumigatus. Infect. Immun. 65:4718-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claverie-Martin, F., M. R. Diaz-Torres, and M. J. Geoghegan. 1986. Chemical composition and electron microscopy of the rodlet layer of Aspergillus nidulans conidia. Curr. Microbiol. 14:221-225. [Google Scholar]
- 9.Cole, G. T., and L. M. Pope. 1981. Surface wall components of Aspergillus niger conidia, p. 195-215. In G. Turian and H. R. Hohl (ed.), The fungal spore: morphogenetic controls. Academic Press, London, United Kingdom.
- 10.Cole, G. T., T. Sekiya, R. Kasai, T. Yokoyama, and Y. Nozawa. 1979. Surface ultrastructure and chemical composition of the cell walls of conidial fungi. Exp. Mycol. 3:132-156. [Google Scholar]
- 11.Denning, D. W. 1998. Invasive aspergillosis. Clin. Infect. Dis. 26:781-804. [DOI] [PubMed] [Google Scholar]
- 12.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Vries, O. M. H., M. P. Fekkes, H. A. B. Wösten, and J. G. H. Wessels. 1993. Insoluble hydrophobin complexes in the walls of Schizophyllum commune and other filamentous fungi. Arch. Microbiol. 159:330-335. [Google Scholar]
- 14.Fontaine, T., R. P. Hartland, A. Beauvais, M. Diaquin, and J. P. Latgé. 1997. Purification and characterization of an endo-β-1,3-glucanase from Aspergillus fumigatus. Eur. J. Biochem. 243:315-321. [DOI] [PubMed] [Google Scholar]
- 15.Girardin, H., J. P. Latgé, T. Srikantha, B. Morrow, and D. R. Soll. 1993. Development of DNA probes for fingerprinting Aspergillus fumigatus. J. Clin. Microbiol. 31:1547-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hashimoto, T., C. D. Wu-Yuan, and H. J. Blumenthal. 1976. Isolation and characterization of the rodlet layer of Trichophyton mentagrophytes microconidial wall. J. Bacteriol. 127:1543-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacques, M., and L. Graham. 1989. Improved preservation of bacterial capsule for electron microscopy. J. Electron Microsc. Tech. 11:167-169. [DOI] [PubMed] [Google Scholar]
- 18.Jahn, B., A. Koch, A. Schmidt, G. Wanner, H. Gehringer, S. Bhakdi, and A. A. Brakhage. 1997. Isolation and characterization of a pigmentless-conidium mutant of Aspergillus fumigatus with altered conidial surface and reduced virulence. Infect. Immun. 65:5110-5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaton-Ogay, K., M. Suter, R. Crameri, R. Falchetto, A. Fatih, and M. Monod. 1992. Nucleotide sequence of a genomic and a cDNA clone encoding an extracellular alkaline protease of Aspergillus fumigatus. FEMS Microbiol. Lett. 92:163-168. [DOI] [PubMed] [Google Scholar]
- 20.Kawasaki, H., Y. Emori, and K. Suzuki. 1990. Production and separation of peptides from proteins stained with Coomassie brilliant blue R-250 after separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Anal. Biochem. 191:332-336. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Latgé, J.-P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lauter, F.-R., V. E. A. Russo, and C. Yanovsky. 1992. Developmental and light regulation of eas, the structural gene for the rodlet protein of Neurospora. Genes Dev. 6:2373-2381. [DOI] [PubMed] [Google Scholar]
- 24.Marck, C. 1988. ′DNA Strider': a ′C' program for the fast analysis of DNA and protein sequences for the Apple Macintosh family of computers. Nucleic Acids Res. 16:1829-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsudaira, P. (ed.) 1993. A practical guide to protein and peptide purification for microsequencing. Academic Press, Orlando, Fla.
- 26.Mattern, I. E., P. J. Punt, and C. A. M. J. J. Van den Hondel. 1988. A vector of Aspergillus transformation conferring phleomycin resistance. Fungal Genet. Newsl. 35:25. [Google Scholar]
- 27.Monod, M., S. Paris, J. Sarfati, K. Jaton-Ogay, P. Ave, and J. P. Latgé. 1993. Virulence of alkaline protease-deficient mutants of Aspergillus fumigatus. FEMS Microbiol. Lett. 106:39-46. [DOI] [PubMed] [Google Scholar]
- 28.Moser, M., R. Crameri, G. Menz, T. Schneider, T. Dudler, C. Virchow, M. Gmachl, K. Blaser, and M. Suter. 1992. Cloning and expression of recombinant Aspergillus fumigatus allergen I/a (rAspfI/a) with IgE binding and type I skin test activity. J. Immunol. 149:454-460. [PubMed] [Google Scholar]
- 29.Mouyna, I., R. P. Hartland, T. Fontaine, M. Diaquin, and J. P. Latgé. 1998. A β(1-3) glucanosyltransferase isolated from the cell wall of Aspergillus fumigatus is an homolog of the yeast Bgl2p. Microbiology 144:3171-3180.9846753 [Google Scholar]
- 30.Nakari-Setälä, T., N. Aro, M. Ilmen, G. Munoz, N. Kalkkinen, and M. Penttilä. 1997. Differential expression of the vegetative and spore-bound hydrophobins of Trichoderma reesei: cloning and characterization of the hfb2 gene. Eur. J. Biochem. 248:415-423. [DOI] [PubMed] [Google Scholar]
- 31.Parta, M., Y. Chang, S. Rulong, P. Pinto-DaSilva, and K. J. Kwon-Chung. 1994. HYP1, a hydrophobin gene from Aspergillus fumigatus, complements the rodletless phenotype in Aspergillus nidulans. Infect. Immun. 62:4389-4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peñas, M. M., S. A. Ásgeirsdóttir, I. Lasa, F. A. Culiañez-Macià, A. G. Pisabarro, J. G. H. Wessels, and L. Ramírez. 1998. Identification, characterization, and in situ detection of a fruit-body-specific hydrophobin of Pleurotus ostreatus. Appl. Environ. Microbiol. 64:4028-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Shibuya, K., M. Takaoka, K. Uchida, M. Wakayama, H. Yamaguchi, K. Takahashi, S. Paris, J. P. Latge, and S. Naoe. 1999. Histopathology of experimental invasive pulmonary aspergillosis in rats: pathological comparison of pulmonary lesions induced by specific virulent factor deficient mutants. Microb. Pathog. 27:123-131. [DOI] [PubMed] [Google Scholar]
- 35.Stringer, M. A., R. A. Dean, T. C. Sewall, and W. E. Timberlake. 1991. Rodletless, a new Aspergillus developmental mutant induced by directed gene inactivation. Genes Dev. 5:1161-1171. [DOI] [PubMed] [Google Scholar]
- 36.Stringer, M. A., and W. E. Timberlake. 1995. dewA encodes a fungal hydrophobin component of the Aspergillus spore wall. Mol. Microbiol. 16:33-44. [DOI] [PubMed] [Google Scholar]
- 37.Stüber, D., H. Matile, and G. Garotta. 1990. System for high level production in Escherichia coli and rapid purification of recombinant proteins: application to epitope mapping, preparation of antibodies and structure-function analysis, p. 121-152. In I. Lefkovits and B. Pernis (ed.), Immunological methods, 4th ed., vol. 4. Academic Press, San Diego, Calif.
- 38.Talbot, N. J., D. J. Ebbole, and J. E. Hamer. 1993. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 5:1575-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Talbot, N. J., M. J. Kershaw, G. E. Wakley, O. M. H. De Vries, J. G. H. Wessels, and J. E. Hamer. 1996. MPG1 encodes a fungal hydrophobin involved in surface interactions during infection-related development of Magnaporthe grisea. Plant Cell 8:985-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Templeton, M. D., D. R. Greenwood, and R. E. Beever. 1995. Solubilization of Neurospora crassa rodlet proteins and identification of the predominant protein as the proteolytically processed eas (ccg-2) gene product. Exp. Mycol. 19:166-169. [DOI] [PubMed] [Google Scholar]
- 41.Thau, N., M. Monod, B. Crestani, C. Rolland, G. Tronchin, J. P. Latgé, and S. Paris. 1994. rodletless mutants of Aspergillus fumigatus. Infect. Immun. 62:4380-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wessels, J. G. H. 1997. Hydrophobins: proteins that change the nature of the fungal surface, p. 1-45. In R. K. Poole (ed.), Advances in microbial physiology, vol. 38. Academic Press, London, United Kingdom. [DOI] [PubMed]
- 43.Wessels, J. G. H., O. M. H. de Vries, S. A. Àsgeirsdòttir, and J. Springer. 1991. The thn mutation of Schizophyllum commune, which suppresses formation of aerial hyphae, affects expression of the hydrophobin gene. J. Gen. Microbiol. 137:2439-2445. [DOI] [PubMed] [Google Scholar]
- 44.Wösten, H. A. B., and M. L. de Vocht. 2000. Hydrophobins, the fungal coat unravelled. Biochim. Biophys. Acta 1469:79-86. [DOI] [PubMed] [Google Scholar]