Abstract
A unique metabolite with a molecular mass of 119 Da (C2H5N3O3) accumulated during biotransformation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) by Rhodococcus sp. strain DN22 (D. Fournier, A. Halasz, J. C. Spain, P. Fiurasek, and J. Hawari, Appl. Environ. Microbiol. 68:166-172, 2002). The structure of the molecule and the reactions that led to its synthesis were not known. In the present study, we produced and purified the unknown metabolite by biotransformation of RDX with Rhodococcus sp. strain DN22 and identified the molecule as 4-nitro-2,4-diazabutanal using nuclear magnetic resonance and elemental analyses. Furthermore, we tested the hypothesis that a cytochrome P450 enzyme was responsible for RDX biotransformation by strain DN22. A cytochrome P450 2B4 from rabbit liver catalyzed a very similar biotransformation of RDX to 4-nitro-2,4-diazabutanal. Both the cytochrome P450 2B4 and intact cells of Rhodococcus sp. strain DN22 catalyzed the release of two nitrite ions from each reacted RDX molecule. A comparative study of cytochrome P450 2B4 and Rhodococcus sp. strain DN22 revealed substantial similarities in the product distribution and inhibition by cytochrome P450 inhibitors. The experimental evidence led us to propose that cytochrome P450 2B4 can catalyze two single electron transfers to RDX, thereby causing double denitration, which leads to spontaneous hydrolytic ring cleavage and decomposition to produce 4-nitro-2,4-diazabutanal. Our results provide strong evidence that a cytochrome P450 enzyme is the key enzyme responsible for RDX biotransformation by Rhodococcus sp. strain DN22.
Hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) is a cyclic nitramine explosive commonly used for military and commercial purposes worldwide. The extensive manufacturing, use, and disposal of RDX have resulted in severe environmental contamination (7, 14). RDX is toxic, mutagenic, and carcinogenic for humans and other biological systems (9, 19); hence, there is an urgent need for safe removal of this compound from the environment. In several reports workers have described biodegradation of cyclic nitramine explosives, such as RDX, under both aerobic (2, 3, 5, 18) and anaerobic (8, 10, 12, 13, 15, 20) conditions, but little information is available on the initial enzymatic processes involved in RDX degradation. Such information is necessary for understanding the pathway by which RDX can serve as a nitrogen source for aerobic bacteria.
Unlike 2,4,6-trinitrotoluene, the cyclic nitramine compounds lack the electronic stability of aromatic compounds. Therefore, a successful microbial or chemical attack on one of the —NO2 or —CH2— groups of RDX is sufficient for ring cleavage and subsequent spontaneous decomposition. Binks et al. (2) provided strong evidence that RDX ring cleavage occurs during aerobic degradation of RDX by Stenotrophomonas maltophilia PB1. Previously, we proved that initial denitration of RDX by Klebsiella sp. strain SCZ-1, an isolate from anaerobic sludge (21), and Rhodococcus sp. strain DN22, a soil isolate (5), can lead to ring cleavage and spontaneous decomposition to HCHO, CO2, N2O, and NH3. However, we did not determine the enzymes responsible for initiating the biotransformation of RDX. During RDX degradation with Rhodococcus sp. strain DN22, we detected a dead end product with a molecular mass of 119 Da corresponding to the empirical formula C2H5N3O3. This product was tentatively identified as either 4-nitro-2,4-diazabutanal or 2-nitro-3-amino-2-azapropanal (5).
In the present study, we evaluated the role of a cytochrome P450 enzyme in RDX biotransformation by using cytochrome P450 2B4 (EC 1.14.14.1) from rabbit liver. We carried out this study with a model system in order to identify the metabolites produced and to understand the initial reaction mechanism(s) involved in the degradation process. We also purified and characterized the dead end metabolite C2H5N3O3 produced during RDX degradation by Rhodococcus sp. stain DN22. We compared cytochrome P450 2B4-catalyzed RDX biotransformation with RDX biotransformation by Rhodococcus sp. strain DN22 to obtain insight into the RDX biotransformation pathways of the two systems.
MATERIALS AND METHODS
Chemicals.
Commercial grade RDX (purity, >99%) and [15N]RDX (purity, >98%) were provided by Defense Research and Development Canada, Valcartier, Quebec, Canada. Hexahydro-1-nitroso-3,5-dinitro-1,3,5-triazine (MNX) was obtained from SRI International, Menlo Park, Calif. Pentoxyresorufin, cytochrome c, NADPH, 1-aminobenzotriazole, and ellipticine were purchased from Sigma Chemicals, Canada. Methylenedinitramine, 2-methyl-1,2-di-3-pyridyl-1-propanone (metyrapone), and phenylhydrazine were purchased from Aldrich, Canada. 18O-labeled water (enrichment, 95 atom%) and 18O-labeled molecular oxygen (minimum, 99 atom%) were purchased from Isotec Inc., Miamisburg, Ohio. Nitrous oxide was purchased from Scott Specialty Gases, Sarnia, Ontario, Canada. Carbon monoxide was purchased from Aldrich Chemical Company, Milwaukee, Wis. All other chemicals were of the highest purity available.
Production and purification of the RDX metabolite C2H5N3O3 by using strain DN22.
The dead end metabolite with a molecular mass of 119 Da was produced by biotransformation of RDX by Rhodococcus sp. strain DN22 in a 3-liter bioreactor (Applicon Inc., Foster City, Calif.) containing 2 liters of growth medium. Rhodococcus sp. strain DN22 was grown in 2 liters of medium containing the following ingredients (initial amounts): 2.44 g of K2HPO4, 1.22 g of KH2PO4, 66 mg of CaCl2 · 2H2O, 0.4 g of MgCl2, 6 ml of a trace element solution (17), 10.8 g of sodium succinate · 6H2O as the sole carbon source, and 80 mg of RDX as the sole nitrogen source. The bioreactor was constantly aerated and stirred at 800 rpm at 30°C. Sodium succinate · 6H2O (10.8 and 4.7 g) and a trace element solution (6.0 and 2.6 ml) were added twice at 28 and 190.5 h, respectively, to the bioreactor. RDX was added to the bioreactor as a stock solution in acetone (40 mg/ml) at an initial concentration of 180 μM, and thereafter it was added periodically (average amount, 122 mg/day) to the bioreactor as soon as the concentration was less than 180 μM. The total amount of RDX added (including the initial amount) was 1.1 g. The bioreactor was operated for 9 days (the optical density at 530 nm increased from 0.07 to 5.11), until there was no further increase in the concentration of the dead end metabolite.
Cells were filtered out of the broth (Pellicon cassette system; Millipore, Bedford, Mass.), and the filtrate was passed through C18 (Sep-Pak; 10 g; Waters, Milford, Mass.) and SAX (Mega Bond Elut; 10 g; Varian, Harbor City, Calif.) columns. The aqueous phase that was collected was concentrated under reduced pressure by using a rotary evaporator (Buchi, Falwil, Switzerland). The concentrated residue was washed eight times with 10 to 20 ml of acetonitrile each time in order to extract the organic metabolite from the inorganic salts. The fractions containing the metabolite were pooled, evaporated to dryness, and stored at 4°C before analysis.
NMR and elemental analysis.
1H and 13C nuclear magnetic resonance (NMR) spectra of the RDX ring cleavage metabolite (molecular mass, 119 Da) were obtained in dimethyl sulfoxide by using Bruker AV-400 and DMX-600 spectrometers at the University of Montreal, Montreal, Quebec, Canada. Several different NMR techniques were used, including 1H and 13C NMR, distortionless enhancement by polarization transfer (DEPT) 135, homonuclear J-correlated spectroscopy (COSY), and two-dimensional exchange-nuclear Overhauser effect spectroscopy (NOESY) to assign proton and carbon chemical shifts (δ). Elemental analysis of the metabolite was performed at the University of Montreal by using a Carlo Erba EA1108 analyzer.
Enzymes.
Cytochrome P450 2B4 (EC 1.14.14.1) and cytochrome P450 reductase (EC 1.6.2.4) from rabbit liver were obtained from Sigma Chemicals, Canada. The protein concentration was measured with a bicinchoninic acid kit (Sigma Chemicals, Canada) by using bovine serum albumin as the standard. The native enzyme activities were estimated by using the manufacturer's guidelines.
RDX biotransformation assays.
RDX biotransformation assays with cytochrome P450 2B4 and cytochrome P450 reductase were performed under both aerobic and anaerobic conditions in 6-ml glass vials. Anaerobic conditions were created by purging all the solutions with argon three times (15 min each time) and replacing the headspace air with argon in sealed vials. Each vial was charged with 1 ml of an assay mixture containing RDX (100 μM), NADPH (150 μM), and cytochrome P450 2B4 (100 μg) or cytochrome P450 reductase (10 μg) in potassium phosphate buffer (50 mM; pH 7.2). Reactions were performed at 37°C. Three different controls were prepared by omitting the enzyme or NADPH or both from the assay mixture. Samples were withdrawn from the liquid and gas phases in the vials periodically to analyze RDX and the biotransformation products as described below. NADPH contents were determined as described previously (1). The RDX biotransformation activity of an enzyme was expressed in nanomoles of RDX biotransformed per minute per milligram of protein unless otherwise stated.
Incorporation of labeled 18O into RDX metabolite.
H218O was mixed with potassium phosphate buffer (100 mM; pH 7.2) at a ratio of 7:3. All other assay ingredients and conditions were the same as those described above for the RDX biotransformation assays. When the reaction was carried out under anaerobic conditions, reactants were added to the vial through a rubber septum with a syringe and needle.
For the labeling experiments with 18O2, the headspace and the aqueous phase were flushed with argon three times (15 min each time) at 10-min intervals, and the headspace was overlaid with 18O2 (approximately 21% [vol/vol]). After this, the reaction was performed as described above.
Analytical procedures.
RDX was analyzed with a high-performance liquid chromatograph connected to photodiode array detector (λ, 254 nm). Samples (50 μl) were injected into a Supelcosil LC-CN column (inside diameter, 4.6 mm; length, 25 cm; Supelco, Oakville, Ontario, Canada), and the analytes were eluted by using a methanol-water gradient at a flow rate of 1.5 ml/min (8).
Liquid chromatography-mass spectrometry was performed with a Micromass bench top single quadrupole mass detector attached to a Hewlett-Packard 1100 series high-performance liquid chromatograph system equipped with a photodiode array detector (5). Ionization was carried out in the negative electrospray ionization mode (ES−), which produced mainly the deprotonated molecular mass ions [M-H]. The RDX metabolite was detected at 118 Da.
Other RDX metabolites, including methylenedinitramine, MNX, formamide, and ammonium (NH4+), were analyzed as described previously (1). Nitrite (NO2−), formaldehyde (HCHO), and nitrous oxide (N2O) were analyzed by using previously described methods (5, 8).
RESULTS AND DISCUSSION
Molecular structure of RDX metabolite from Rhodococcus sp. strain DN22.
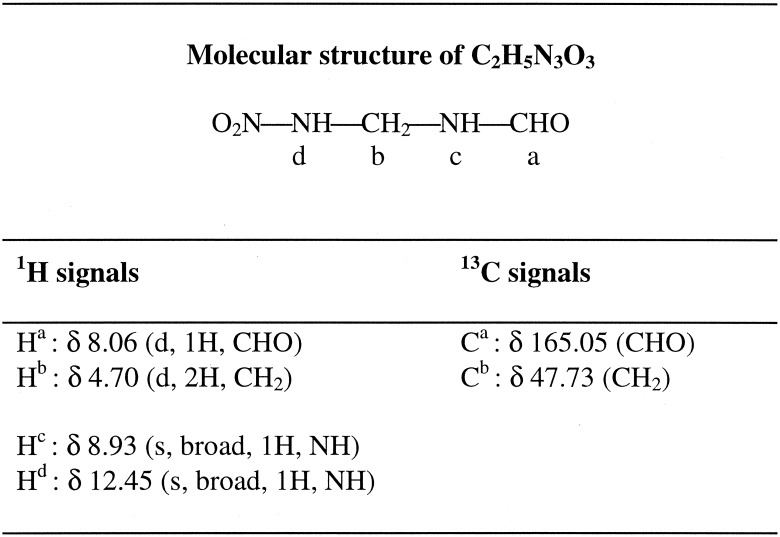
Elemental analysis data (Table 1) and liquid chromatography-mass spectrometry (ES−) data for the dead end metabolite produced during RDX biodegradation by strain DN22 were consistent with an empirical formula of C2H5N3O3. In a previous study, the metabolite was tentatively identified as either 4-nitro-2,4-diazabutanal (O2NNHCH2NHCHO) or 2-nitro-3-amino-2-azapropanal (NH2CH2NNO2CHO) (5). In the present study, NMR spectroscopy was used to elucidate the structure of the metabolite. NMR spectra were collected in d6-dimethyl sulfoxide. The 1H spectrum had two broad signals at 12.45 ppm (1H) and 8.93 ppm (1H) assignable to exchangeable NH protons and two duplet signals at 8.06 ppm (1H) and 4.70 ppm (2H) assignable to CHO and CH2 groups, respectively (Table 2). A COSY experiment demonstrated that the splitting of the two latter signals was due to coupling with the proton at 8.92 ppm (Table 2). A two-dimensional exchange-NOESY experiment indicated that there was fast exchange between the proton at 12.45 ppm and the signal of residual water, thus indicating that this proton was not coupled with other protons. Carbon spectra had two signals at 47.73 and 165.05 ppm, which were assigned to CH2 and CHO groups, respectively, as confirmed by a DEPT 135 experiment. All of the results described above and especially the two distinct NH signals are consistent with the structure of 4-nitro-2,4-diazabutanal (Table 2) rather than the structure of 2-nitro-3-amino-2-azapropanal.
TABLE 1.
Elemental analysis of 4-nitro-2,4-diazabutanala
| Value | % Nitrogen (SD) | % Carbon (SD) | % Hydrogen (SD) |
|---|---|---|---|
| Measured | 34.25 (0.028) | 20.27 (0.021) | 4.19 (0.005) |
| Calculated | 35.29 | 20.17 | 4.23 |
The suggested molecular formula is C2H5N3O3 (molecular weight, 119.08).
TABLE 2.
Molecular structure of 4-nitro-2,4-diazabutanal as elucidated by NMR analysis in d6-dimethyl sulfoxide
RDX biotransformation by rabbit liver cytochrome P450.
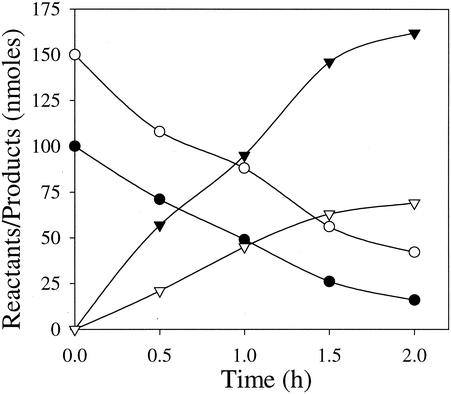
A time course study of the biotransformation of RDX catalyzed by cytochrome P450 2B4 showed that there was simultaneous release of nitrite and HCHO from RDX at the expense of the electron donor NADPH (Fig. 1). RDX was biotransformed at rates of 9.0 and 2.8 nmol min−1 mg of protein−1 at pH 7.2 and 37°C under anaerobic and aerobic conditions, respectively. In comparison, the reaction rate of cytochrome P450 2B4 with the standard substrate, pentoxyresorufin, was 0.88 nmol min−1 mg of protein−1 under aerobic conditions, which was about threefold less than the RDX biotransformation rate. The metabolites produced during RDX biotransformation by cytochrome P450 2B4 were 4-nitro-2,4-diazabutanal, nitrite, formaldehyde, and ammonium (Table 3). In addition, traces of MNX and methylenedinitramine were also detected as transient metabolites, probably due to the presence of cytochrome P450 reductase activity in cytochrome P450 2B4 (as indicated in the product description obtained from the manufacturer). For instance, when RDX was incubated with cytochrome P450 reductase and NADPH as an electron donor under anaerobic conditions at 37°C, the compound was biotransformed at a rate of 0.42 μmol min−1 mg of protein−1, and MNX and methylenedinitramine were produced as transient intermediates and N2O, HCHO, and NH4+ were produced as end products (data not shown). In comparison, the reaction rate of cytochrome P450 reductase with cytochrome c was 32.50 μmol min−1 mg of protein−1, which was about 77-fold higher than the RDX biotransformation rate under similar reaction conditions.
FIG. 1.
Time course study of biotransformation of RDX catalyzed by rabbit liver cytochrome P450 2B4. The data are means of duplicate experiments, and the standard deviations were within 6% of the mean absolute values (n = 2). Symbols: •, RDX; ○, NADPH; ▾, NO2−; ▿, HCHO.
TABLE 3.
Carbon and nitrogen mass balance and stoichiometry of reactants consumed and metabolites produced during RDX biotransformation catalyzed by rabbit liver cytochrome P450 2B4 (100 μg/ml) at pH 7.2 and 37°C for 2 h under anaerobic conditions
| Reactants or metabolite | Amt (nmol) | % Carbon recoverya | % Nitrogen recoverya |
|---|---|---|---|
| Reactants consumed | |||
| RDX | 84 | 100 | 100 |
| NADPH | 108 | NAb | NA |
| Metabolites produced | |||
| Nitrite (NO2−) | 162 | NA | 32.0 |
| 4-Nitro-2,4-diazabutanal | 68 | 54 | 40 |
| Formaldehyde (HCHO) | 69 | 27 | NA |
| Ammonium (NH4+) | 65 | NA | 13 |
Recovery values were calculated from the total carbon and nitrogen masses in the biotransformed RDX (84 nmol). The initial amounts of RDX and NADPH were 100 and 150 nmol, respectively. The data are means for duplicate experiments, and the standard deviations were within 6% of the mean absolute values (n = 2). The levels of total mass recovery were 81% for carbon and 85% for nitrogen.
NA, not applicable.
As mentioned above, the rate of RDX biotransformation by cytochrome P450 2B4 was about threefold higher under anaerobic conditions than under aerobic conditions. Our experimental finding that anaerobic conditions favored RDX biotransformation by cytochrome P450 2B4 can be explained on the basis of a single-electron transfer mechanism catalyzed by a cytochrome P450 enzyme (6). According to this mechanism, the substrate either binds directly at the Fe2+ site of the prosthetic heme group or binds at a nearby site (e.g., heme ligand position 6) and undergoes single-electron reduction by oxidizing the Fe2+. In addition to the substrate, the Fe2+ of the prosthetic heme is also a binding site for O2. Therefore, O2 competes with the substrate for binding at the same site, and this provides a probable explanation for the inhibition of RDX biotransformation by cytochrome P450 2B4 under aerobic conditions. In the case of Rhodococcus sp. strain DN22, although aerobic conditions are needed for bacterial growth, RDX transformation does not involve the incorporation of molecular oxygen. When RDX was incubated with strain DN22 in the presence of 18O2, we detected the metabolite, 4-nitro-2,4-diazabutanal, without 18O (5). In the presence of H218O we detected 4-nitro-2,4-diazabutanal (molecular weight, 121; [M + 2]) with one 18O atom. Similarly, in the case of cytochrome P450 2B4, we observed that 18O was incorporated into 4-nitro-2,4-diazabutanal from H218O but not from 18O2.
Inhibition study.
A comparative inhibition study was carried out with cytochrome P450 2B4 and Rhodococcus sp. strain DN22 by using various cytochrome P450 inhibitors. At a concentration of 200 μM, all of the inhibitors tested inhibited the RDX biotransformation activity of cytochrome P450 2B4, as well as the activity of Rhodococcus sp. strain DN22 (Table 4). These inhibitors act on the exposed prosthetic heme group of cytochrome P450 (16). The inhibition study provided additional support for the hypothesis that the enzyme responsible for the RDX biotransformation activity of Rhodococcus sp. strain DN22 is cytochrome P450. Coleman et al. (4) previously reported that metyrapone and menadione strongly inhibited the RDX biotransformation activity of Rhodococcus sp. strain DN22, which indicated that the cytochrome P450 enzyme was probably involved in RDX biotransformation by Rhodococcus sp. strain DN22. Seth-Smith et al. (18) also described metyrapone-mediated inhibition of RDX biotransformation by Rhodococcus rhodochrous strain 11Y expressing a cytochrome P450 gene.
TABLE 4.
Effects of cytochrome P450 inhibitors on RDX biotransformation activities of rabbit liver cytochrome P450 2B4 and Rhodococcus sp. strain DN22a
| Inhibitor (200 μM) | % Inhibition of activity |
|
|---|---|---|
| Cytochrome P450 2B4 (100 μg/ml) | Rhodococcus sp. strain DN22 (5 mg [wet wt]/ml)b | |
| Control (no inhibitor) | 0 | 0 |
| Ellipticine | 75 ± 4d | 76 ± 7 |
| Metyrapone | 68 ± 8 | 60 ± 6 |
| Phenylhydrazine | 70 ± 7 | 77 ± 5 |
| 1-Aminobenzotriazole | 55 ± 7 | 43 ± 7 |
| Carbon monoxidec | 82 ± 6 | 48 ± 5 |
The RDX transformation activity without inhibitor was considered 0% inhibition; the 100% activities for cytochrome P450 2B4 and Rhodococcus sp. strain DN22 were 9.0 nmol min−1 mg of protein−1 and 0.12 nmol min−1 mg of biomass−1, respectively.
A washed and centrifuged cell pellet was used as wet biomass.
Carbon monoxide was bubbled through the aqueous phase and headspace for 60 s in sealed vials.
The values are means ± standard deviations (n = 3).
Stoichiometry and mass balance of RDX biotransformation by cytochrome P450 2B4.
We found that 84 nmol of RDX was transformed at the expense of 108 nmol of NADPH, suggesting that the stoichiometry is 1:1. The remaining 24 nmol of NADPH was presumably consumed by cytochrome P450 reductase present in the cytochrome P450 2B4 preparation (see above), and/or a small fraction of NADPH could also have reacted directly with RDX or its metabolites. The total recovered carbon mass balance was 81% and was distributed as 4-nitro-2,4-diazabutanal (54%) and HCHO (27%) (Table 3), and the total nitrogen mass recovery was 85% and was distributed as nitrite (32%), 4-nitro-2,4-diazabutanal (40%), and ammonium (13%) (Table 3).
Based on the product distribution and mass balance of RDX transformation by cytochrome P450 2B4, we concluded that of the six nitrogen atoms and three carbon atoms present in one RDX molecule, two nitrogen atoms were recovered as nitrite ions, whereas three nitrogen atoms and two carbon atoms were recovered in a dead end metabolite, 4-nitro-2,4-diazabutanal. The remaining one nitrogen atom and one carbon atom were present in NH4+ and HCHO (Table 3).
Taken together, the results described above provided several lines of evidence which support the hypothesis that a cytochrome P450 type of enzyme is responsible for RDX transformation by Rhodococcus sp. strain DN22. A comparison of the biotransformation of RDX catalyzed by rabbit liver cytochrome P450 2B4 and the biotransformation of RDX by Rhodococcus sp. strain DN22 (5) revealed that the product distributions and stoichiometries were strikingly similar. In a previous study, cytochrome P450s from R. rhodochrous participated in the degradation of 2-ethoxyphenol and 4-methoxybenzoate (11). Our enzyme inhibition studies are consistent with those of Coleman et al. (4) regarding involvement of cytochrome P450 in RDX degradation by Rhodococcus sp. strain DN22. Finally, Seth-Smith et al. (18) recently provided strong molecular evidence that the constitutively expressed cytochrome P450-like gene xplA from R. rhodochrous strain 11Y is responsible for RDX degradation.
Proposed mechanism.
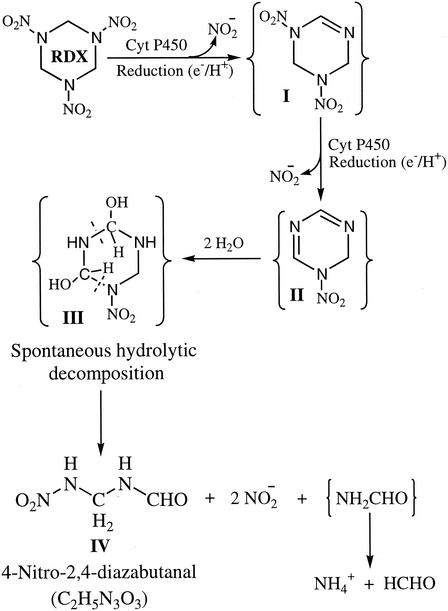
Based on stoichiometry and mass balance studies and the key observation that 18O was incorporated into 4-nitro-2,4-diazabutanal from H218O but not from 18O2, we suggest a plausible mechanism for the initial biotransformation of RDX by cytochrome P450 2B4. According to this mechanism, cytochrome P450 2B4 catalyzes sequential transfer of two single electrons to RDX; the first electron causes denitration to form compound I (Fig. 2), and the second electron causes a second denitration to produce compound II. The latter product is unstable in water and should be hydrolyzed by incorporation of two 18OH groups from two H218O molecules to give hypothetical compound III. The spontaneous decomposition of compound III produces 4-nitro-2,4-diazabutanal (Fig. 2).
FIG. 2.
Proposed pathway for RDX biotransformation catalyzed by rabbit liver cytochrome P450 2B4 (Cyt P450). Products in brackets were not detected.
In conclusion, we provide direct biochemical evidence that a rabbit liver cytochrome P450 catalyzed the biotransformation of RDX. We propose a plausible mechanism for the initial enzymatic attack on RDX by cytochrome P450, which eventually produced the same products as the products produced by Rhodococcus sp. strain DN22. This mechanism is consistent with our observation that two electrons (approximately one NADPH molecule) were consumed and two nitrite ions were produced per reacted RDX molecule. Several lines of evidence in the present study and the results of previous studies (4, 5, 18) support the conclusion that an enzyme(s) belonging to the cytochrome P450 family is responsible for RDX biotransformation by Rhodococcus sp. strain DN22.
Acknowledgments
We thank the Natural Sciences and Engineering Research Council (NSERC) and the National Research Council (NRC) of Canada for awarding a visiting fellowship to B. Bhushan and the U.S. Strategic Environmental Research and Development Program (SERDP) for funding this project (grant CU 1213). We also thank the Department of National Defense, Canada, for its support.
We especially thank S. Bilodeau, P. V. M. Tan, H. Dinel, and F. Bélanger-Gariépy of the University of Montreal and Fanny Monteil of BRI, Montreal, Quebec, Canada, for conducting NMR studies and elemental analysis of 4-nitro-2,4-diazabutanal. We sincerely acknowledge the analytical and technical support of C. Beaulieu, C. Groom, A. Corriveau, and S. Deschamps. Special thanks also go to S. Nishino for her technical assistance. We thank Neil Bruce, University of York, York, United Kingdom, for sharing unpublished results concerning RDX biodegradation by R. rhodochrous. Finally, we thank N. Coleman for providing Rhodococcus sp. strain DN22.
REFERENCES
- 1.Bhushan, B., A. Halasz, J. Spain, S. Thiboutot, G. Ampleman, and J. Hawari. 2002. Biotransformation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) catalyzed by a NAD(P)H:nitrate oxidoreductase from Aspergillus niger. Environ. Sci. Technol. 36:3104-3108. [DOI] [PubMed] [Google Scholar]
- 2.Binks, P. R., S. Nicklin, and N. C. Bruce. 1995. Degradation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) by Stenotrophomonas maltophilia PB1. Appl. Environ. Microbiol. 61:1318-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coleman, N. V., D. R. Nelson, and T. Duxbury. 1998. Aerobic biodegradation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) as a nitrogen source by a Rhodococcus sp., strain DN22. Soil Biol. Biochem. 30:1159-1167. [Google Scholar]
- 4.Coleman, N. V., J. C. Spain, and T. Duxbury. 2002. Evidence that RDX biodegradation by Rhodococcus strain DN22 is plasmid-borne and involves a cytochrome p-450. J. Appl. Microbiol. 93:463-472. [DOI] [PubMed] [Google Scholar]
- 5.Fournier, D., A. Halasz, J. C. Spain, P. Fiurasek, and J. Hawari. 2002. Determination of key metabolites during biodegradation of hexahydro-1,3,5-trinitro-1,3,5-triazine with Rhodococcus sp. strain DN22. Appl. Environ. Microbiol. 68:166-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guengerich, P. F. 2001. Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem. Res. Toxicol. 14:611-650. [DOI] [PubMed] [Google Scholar]
- 7.Haas, R., E. V. Löw Schreiber, and G. Stork. 1990. Conception for the investigation of contaminated munitions plants. 2. Investigation of former RDX-plants and filling stations. Fresenius J. Anal. Chem. 338:41-45. [Google Scholar]
- 8.Halasz, A., J. Spain, L. Paquet, C. Beaulieu, and J. Hawari. 2002. Insights into the formation and degradation of methylenedinitramine during the incubation of RDX with anaerobic sludge. Environ. Sci. Technol. 36: 633-638. [DOI] [PubMed] [Google Scholar]
- 9.Hawari, J. 2000. Biodegradation of RDX and HMX: from basic research to field application, p. 277-310. In J. C. Spain, J. B. Hughes, and H.-J. Knackmuss (ed.), Biodegradation of nitroaromatic compounds and explosives. CRC Press, Boca Raton, Fla.
- 10.Hawari, J., A. Halasz, T. Sheremata, S. Beaudet, C. Groom, L. Paquet, C. Rhofir, G. Ampleman, and S. Thiboutot. 2000. Characterization of metabolites during biodegradation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) with municipal anaerobic sludge. Appl. Environ. Microbiol. 66:2652-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karlson, U., D. F. Dwyer, S. W. Hooper, E. R. B. Moore, K. N. Timmis, and L. D. Eltis. 1993. Two independently regulated cytochromes P-450 in a Rhodococcus rhodochrous strain that degrades 2-ethoxyphenol and 4-methoxybenzoate. J. Bacteriol. 175:1467-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitts, C. L., C. E. Green, R. A. Otley, M. A. Alvarez, and P. J. Unkefer. 2000. Type I nitroreductase in soil enterobacteria reduce TNT (2,4,6-trinitrotoluene) and RDX (hexahydro-1,3,5-trinitro-1,3,5-triazine). Can. J. Microbiol. 46:278-282. [DOI] [PubMed] [Google Scholar]
- 13.McCormick, N. G., J. H. Cornell, and A. M. Kaplan. 1981. Biodegradation of hexahydro-1,3,5-trinitro-1,3,5-triazine. Appl. Environ. Microbiol. 42:817-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Myler, C. A., and W. Sisk. 1991. Bioremediation of explosives contaminated soils (scientific questions/ engineering realities), p. 137-146. In G. S. Sayler, R. Fox, and J. W. Blackburn (ed.), Environmental bio/technology for waste treatment. Plenum Press, New York, N.Y.
- 15.Oh, B.-T., C. L. Just, and P. J. J. Alvarez. 2001. Hexahydro-1,3,5-trinitro-1,3,5-triazine mineralization by zero-valent iron and mixed anaerobic cultures. Environ. Sci. Technol. 35:4341-4346. [DOI] [PubMed] [Google Scholar]
- 16.Ortiz de Montellano, P. R., and M. A. Correia. 1995. Inhibition of cytochrome P450 enzymes, p. 305-366. In P. R. Ortiz de Montellano (ed.), Cytochrome P450: structure, mechanism and biochemistry, 2nd ed. Plenum Press, New York, N.Y.
- 17.Owens, J. D., and R. M. Keddie. 1969. The nitrogen nutrition of soil and herbage coryneform bacteria. J. Appl. Bacteriol. 32:338-347. [DOI] [PubMed] [Google Scholar]
- 18.Seth-Smith, H. M. B., S. J. Rosser, A. Basran, E. R. Travis, E. R. Dabbs, S. Nicklin, and N. C. Bruce. 2002. Cloning, sequencing and characterization of the hexahydro-1,3,5-trinitro-1,3,5-triazine degradation gene cluster from Rhodococcus rhodochrous. Appl. Environ. Microbiol. 68:4764-4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Talmage, S. S., D. M. Opresko, C. J. Maxwel, C. J. E. Welsh, F. M. Cretella, P. H. Reno, and F. B. Daniel. 1999. Nitroaromatic munition compounds: environment effects and screening values. Rev. Environ. Contam. Toxicol. 161:1-156. [DOI] [PubMed] [Google Scholar]
- 20.Young, D. M., C. L. Kitts, P. J. Unkefer, and K. L. Ogden. 1997. Biotransformation of hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) by a prospective consortium and its most effective isolate, Serratia marcescens. Biotechnol. Bioeng. 53:515-522. [DOI] [PubMed] [Google Scholar]
- 21.Zhao, J. S., A. Halasz, L. Paquet, C. Beaulieu, and J. Hawari. 2002. Biodegradation of hexahydro-1,3,5-trinitro-1,3,5-triazine and its mononitroso derivative hexahydro-1-nitroso-3,5-dinitro-1,3,5-triazine by Klebsiella pneumoniae strain SCZ-1 isolated from an anaerobic sludge. Appl. Environ. Microbiol. 68:5336-5341. [DOI] [PMC free article] [PubMed] [Google Scholar]