Abstract
In its natural environment, Escherichia coli is exposed to short-chain fatty acids, such as acetic acid or propionic acid, which can be utilized as carbon sources but which inhibit growth at higher concentrations. DNA microarray experiments revealed expression changes during exponential growth on complex medium due to the presence of sodium acetate or sodium propionate at a neutral external pH. The adaptive responses to acetate and propionate were similar and involved genes in three categories. First, the RNA levels for chemotaxis and flagellum genes increased. Accordingly, the expression of chromosomal fliC′-′lacZ and flhDC′-′lacZ fusions and swimming motility increased after adaptation to acetate or propionate. Second, the expression of many genes that are involved in the uptake and utilization of carbon sources decreased, indicating some kind of catabolite repression by acetate and propionate. Third, the expression of some genes of the general stress response increased, but the increases were more pronounced after short-term exposure for this response than for the adaptive response. Adaptation to propionate but not to acetate involved increased expression of threonine and isoleucine biosynthetic genes. The gene expression changes after adaptation to acetate or propionate were not caused solely by uncoupling or osmotic effects but represented specific characteristics of the long-term response of E. coli to either compound.
Enteric bacteria colonizing their mammalian hosts are exposed to acid stress in the stomach and to high concentrations of short-chain fatty acids in the intestine. The responses of Escherichia coli to low pHs and to short-chain fatty acids are thought to largely overlap as, for example, survival at an extremely low pH is increased after exposure either to an acidic pH or to acetate at a neutral pH (4, 61). The toxicity of short-chain fatty acids has been attributed to their uncoupling effect on the transmembrane pH gradient and thus to interference with efficient energy metabolism (3, 8). In their neutral form, these uncoupling agents diffuse across the plasma membrane; once they reach the cytoplasm, the anionic form dominates, releasing a proton and decreasing the transmembrane proton gradient (5). The inhibitory effects of acetate on growth rates and maximum cell densities vary with the carbon source (39). A low pH or treatment with growth-inhibiting concentrations of acetate, salicylate, or benzoate affects the expression of many genes, in particular, those of the transcription and translation machineries and of the general stress response (4, 11, 35, 38, 52). RpoS was shown to be essential for acid tolerance induced by either entry into the stationary growth phase or growth at a low pH or with acetate (4, 35, 62). The growth-inhibiting effects of acetate can be relieved by the addition of methionine, indicating homocysteine toxicity and impaired methionine biosynthesis as their bases (55).
Acetate and propionate serve as sole carbon sources for the growth of E. coli. Acetate is transported into the cell and activated to acetyl coenzyme A (acetyl-CoA) by acetyl-CoA synthetase (encoded by acs). The expression of acs is increased in the presence of acetate and is regulated by cyclic AMP (cAMP)-cAMP receptor protein (CRP) and FNR as well as by IclR and its activator, FadR (36). Acetyl-CoA is further metabolized through the glyoxylate shunt and the tricarboxylic acid cycle. The expression of the aceBAK operon, which codes for the key enzymes of the glyoxylate cycle, isocitrate lyase and malate synthase, and for isocitrate dehydrogenase kinase/phosphatase, is activated by FruR and IHF and is repressed by IclR, which in turn is activated by FadR (16). Compared to glucose-grown cells, acetate-grown E. coli possess increased RNA levels for aceBAK, acs, the malic enzyme gene maeB, several other gluconeogenic genes, genes involved in glycol formation, genes of the d-glycerate pathway, and many glucose-repressed genes (50). Propionate, after uptake into the cell, is activated to propionyl-CoA by prpE-encoded propionyl-CoA synthetase in enteric bacteria (27). Propionyl-CoA is converted to pyruvate in the methylcitrate cycle (68). The expression of the prpBCDE operon of enteric bacteria, which codes for 2-methylcitrate synthase, 2-methylcitrate dehydratase, and 2-methylcitrate lyase, which are key enzymes of the methylcitrate cycle, and propionyl-CoA synthetase, depends on PrpR, IHF, and NtrA (10, 27, 51, 68).
Acetate is also a major by-product of E. coli metabolism. Fermentation by E. coli leads to the production of acetate, formate, d-lactate, and succinate (12, 32, 56). At low pHs, mainly lactate (14) and H2 and CO2 (56) are produced by E. coli. At neutral and basic pHs, acetate, ethanol, and formate are the major fermentation products (32). Whereas the production of acetate is maximal anaerobically (32), aerobically growing cultures also excrete acetate (13, 15) and formate (1, 66). At present, acetate excretion during growth on glucose in the presence of sufficient availability of oxygen is not fully understood. Insufficient carbon flux through the citric acid cycle due to repression of this cycle in rapidly growing cells has been proposed to prompt acetate production (40, 49).
As acetate toxicity is believed to limit biotechnological production, for example, of recombinant proteins by E. coli, strategies to reduce acetate formation through medium design and feeding control were developed (reviewed in reference 42). Metabolic engineering to reduce acetate formation mainly resulted in the redirection of metabolism toward other by-products (3, 8, 15, 20, 22, 75). mlc, originally identified in a genetic screen for reduced acetate production (28), encodes a negative regulator of the glucose-specific ptsG gene, of the general phosphotransferase system (PTS) gene ptsIH, of malT and thus the mal regulon, and of the mannose degradation operon manXYZ. Mlc activity is regulated by its sequestration to the EIIBCGlc component of the glucose PTS (41, 48, 67), and its overexpression leads to reduced sugar uptake and subsequently to less acetate formation (28).
As acetate affects the physiology of E. coli in several ways, namely, as a carbon source for growth, as a metabolic product, and as a toxic compound inhibiting growth, in this study we performed DNA microarray experiments to assess gene expression patterns due exclusively to the presence of acetate. We studied E. coli cells growing exponentially for many generations on Luria-Bertani (LB) complex medium in the presence or absence of sodium acetate at a neutral pH. Under these conditions, acetate neither was a significant carbon source nor was produced as a metabolic by-product; in addition, at the low acetate concentration used, growth was not substantially impaired. Similar experiments were performed with sodium propionate. The differential expression patterns were distinguished from short-term adaptation to either sodium acetate or sodium propionate and from adaptation to osmotic effects due to increased medium osmolality. The effects of transmembrane pH decoupling were analyzed by comparing the effects of acetate or propionate on global gene expression to those of the uncoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP).
MATERIALS AND METHODS
Strains, media, and culture conditions.
E. coli wild-type strain MG1655 (CGSC 6300) (http://cgsc.biology.yale.edu/), its lrhA disruption strain IMW331 [Δ(argF-lac)169 zah-Tn10 (Tetr) lrhA::Spcr) (43), its fliC′-′lacZ-carrying derivative IMW356 {Δ(argF-lac)169 zah-Tn10 (Tetr) λ [φ(flic′-′lacZ)(Hyb)]} (43), and its flhDC′-′lacZ-carrying derivative CP992 [Δlac λSS10 (flhDC′-′lacZ) Ampr] (60) were used. All cultures were grown at 37°C on LB complex medium in 500-ml baffled shake flasks with agitation at 130 rpm (57). Cell growth was monitored as the optical density at 600 nm (OD600) (UV-1202 spectral photometer; Shimadzu). When needed, cultures contained 20 mM sodium acetate (pH 7.4), 20 mM sodium propionate (pH 7.4), or 40 μM CCCP (dissolved in ethanol). The reference culture was given an equal amount of water or ethanol. During cultivation, the pH remained constant at between 6.6 and 6.9. Precultures were inoculated into main cultures and contained the same additives as the main cultures. For adaptation experiments, cultures were grown exponentially for ∼20 generations by repeated dilution to ensure full adaptation. For pulse experiments, acetate, propionate, or CCCP was added 5 min before cell harvest for RNA preparation.
Assays of β-galactosidase and glutamic acid decarboxylase activities.
Aliquots from replicate cultures in the exponential growth phase were withdrawn and specific β-galactosidase activities (Miller units) were measured and calculated as described previously (47). For measurement of glutamic acid decarboxylase activities, cells were collected by centrifugation and resuspended in 1 mM pyridoxal 5′-phosphate-1 mM dithiothreitol in water. Cells were disrupted by sonication and enzyme activity in crude extracts was measured as described previously (17).
Determination of acetate, propionate, pyruvate, and d-glucose levels.
The cell-free culture medium obtained by centrifugation of cell culture aliquots (3 min, 4,500 × g, 4°C) was assayed for acetate, propionate, pyruvate, and d-glucose levels. Acetate and d-glucose levels were determined enzymatically by using kits according to the manufacturer's instructions (R-Biopharm, Darmstadt, Germany). Pyruvate levels were determined by converting pyruvate to lactate with pig lactate dehydrogenase (Boehringer Mannheim) in triethanolamine buffer (0.5 M triethanolamine [pH 7.6], 5 mM EDTA) containing NADH, and NADH consumption was measured at 340 nm. For the determination of propionate levels, reverse-phase high-pressure liquid chromatography with a Hitachi D-7000 HPLC system (Merck) equipped with an Aminex HPX-87H column (150 by 7.8 mm; Bio-Rad) was used. Isocratic elution was performed with 6 mM H2SO4 at a flow rate of 0.6 ml/min, and absorption at 215 nm was used for detection.
Swimming motility assay.
The medium used for the swimming motility assay was LB medium containing 0.3% (wt/vol) agar (Difco) and, if necessary, 20 mM sodium acetate or sodium propionate at a neutral pH. Plates were inoculated by placing a 5-μl volume of bacteria from adapted cultures (OD600, 0.4) in the center of the plates. The plates were wrapped to prevent dehydration and incubated at 37°C. After 18 h, the colony diameters on each plate were determined.
Generation of E. coli DNA microarrays.
To characterize changes in the patterns of gene expression due to the presence of each additive in complex medium, mRNA levels for nearly every gene (94%) of the E. coli MG1655 genome were compared by using DNA microarrays. These arrays were made by robotically spotting PCR products that were generated with an ORFmer primer set (Genosys Biotechnologies, Cambridgeshire, England) onto glass microscope slides as described previously (76). These primers amplify each entire open reading frame. As a random test, a microarray of each generated batch was hybridized to ascertain whether sufficient amounts of PCR products were immobilized on the glass surface. Fluorescence-labeled DNA derived from random priming of 2 μg of E. coli genomic DNA with the large fragment of DNA polymerase I (Gibco BRL, Life Technologies) was hybridized. Hybridization signals exceeding noise by a factor of 3 were observed for 4,035 (98.2%) of 4,108 amplified and arrayed E. coli genes, indicating that the immobilized PCR products (spots) allowed for sufficient hybridization (data not shown).
Preparation of total RNA and cDNA synthesis.
Portions (∼35 ml) of exponentially growing E. coli cultures (OD600, 0.3 to 0.5) were added to 15 g of ice (−20°C), and cells were harvested immediately by centrifugation (3 min, 4,500 × g, 4°C) (72). Cells were suspended in 350 μl of RLT buffer from an RNeasy system (Qiagen, Hilden, Germany). For mechanical disruption, cells were added to 0.5 g of 0.1-mm Zirconia/Silica beads (Roth, Karlsruhe, Germany), followed by bead beating five times for 30 s each time with a Silamat S5 apparatus (Vivadent, Ellwangen, Germany). After centrifugation (1 min, 13,000 × g, room temperature), the supernatant was processed with the RNeasy system as recommended by the manufacturer. The isolated RNA was treated with 20 U of DNase I (RNase free; Roche Diagnostics GmbH, Mannheim, Germany) in DNase I buffer (1 M sodium acetate, 50 mM MgSO4 [pH 5.0]) for 20 min at 37°C, incubated for 10 min at 70°C to inactivate DNase I, and purified by phenol-chloroform-isoamyl alcohol (25:24:1) and chloroform-isoamyl alcohol (24:1) extractions followed by ethanol precipitation (57). RNA concentrations were determined photometrically, and RNA quality was checked on formamide gels as described previously (57).
Equal amounts of total RNA (15 to 25 μg) were used for random hexamer-primed synthesis of fluorescence-labeled cDNA with the fluorescent nucleotide analogues Cy3-dUTP (green) and Cy5-dUTP (red) (Amersham Pharmacia) as described previously (72).
DNA microarray hybridization and washing.
Mixtures of Cy3- and Cy5-labeled cDNA probes containing 1 μg of poly(A) (Sigma, Taufkirchen, Germany)/μl as a competitor, 3× SSC (1× SSC is 0.15 M sodium chloride plus 0.015 M sodium citrate; nuclease free), and 25 mM HEPES were hybridized to the microarrays for 5 to 10 h at 65°C. After hybridization, the arrays were washed in a solution containing 1× SSC and 0.03% sodium dodecyl sulfate and then in 0.05× SSC. After washing, the slides were dried by centrifugation (5 min, 50 × g). Detailed procedures for microarray hybridization and stringent washing are described in the MGuide (http://cmgm.stanford.edu/pbrown/protocols/index.html).
DNA microarray scanning, data normalization, and gene expression analyses.
Immediately after drying, the fluorescence at 532 nm (Cy3-dUTP) and 635 nm (Cy5-dUTP) was determined at a 10-μm resolution by using an Axon Inc. GenePix 4000 laser scanner. Acquired raw fluorescence data were stored as image files in TIFF format by using GenePix Pro 3.0 software (Axon). TIFF images were analyzed quantitatively by using GenePix Pro 3.0 software. TIFF images contained measured Cy3 and Cy5 fluorescence signals for each image pixel. Typically, at a 10-μm scan resolution, each spot area comprised ∼100 image pixels, and ∼600 image pixels around the spot area were considered background. The median Cy3 and the median Cy5 fluorescence signals were used to calculate the background subtracted Cy5/Cy3 ratio for the median for each spot (GenePix Pro 3.0 software). The ratios were log transformed and normalized by multiplying by a constant normalization factor to normalize the Cy3 fluorescence to the Cy5 fluorescence. The factor was calculated individually for each microarray experiment by using the log-transformed ratios of genomic DNA spots of E. coli MG1655 whose averages of log-transformed and normalized ratios were zero (33, 43, 72, 76). In each microarray experiment, the ratio variance for genomic DNA spots was 10 to 20%. The log-transformed and normalized Cy5/Cy3 ratio for the median was taken to reflect relative RNA level changes. Typically, 2,500 to 4,000 genes could be detected (by the Axon Genepix software) on the basis of spot fluorescence signals exceeding spot background noise. With a more rigorous criterion, RNA levels for 2,000 to 3,600 genes could be determined under the conditions tested on the basis of hybridization signals exceeding background noise by at least a factor of 3. When Cy3 and Cy5 fluorescence signals were less than threefold above the background, signals were considered too weak to be analyzed quantitatively. Replicate experiments typically correlated well, as determined from scatter plots (correlation coefficient, ∼0.9).
For statistical analysis of global gene expression (2, 26), P values were calculated by use of Student's t test with log-transformed RNA levels determined in independent replicate experiments or log-transformed ratios of genomic DNA spots normalized to zero (43).
For hierarchical cluster analysis of gene expression data by using the average linkage clustering method (18), normalized and log-transformed mRNA levels for 4,108 amplified and arrayed genes of E. coli were selected by using the following criteria. (i) Reliable detection was based on signal-to-noise ratios exceeding a factor of 3. (ii) In a paired Student's t test, relative RNA levels were significantly different from the levels of the genomic DNA controls (P < 0.05) (43). (iii) Relative mRNA levels were greater than 2 or less than 0.5 in at least one DNA microarray experiment. (iv) In addition to these genes, we included genes that belonged to the same operon but that did not fulfill each of the above criteria. (v) Genes were excluded when reliable signals were detected in less than two-thirds of all experiments. The color image (Fig. 1) shows the log-transformed numerical relative mRNA levels encoded by color according to the method introduced by Eisen et al. (18), with grey indicating that no reliable signal was measured.
FIG.1.
Hierarchical cluster analysis of gene expression changes in response to sodium acetate, sodium propionate, sodium chloride, and CCCP. Gene expression data from 27 microarray experiments (columns) and 179 genes (lines) are represented. Microarray experiments involved comparing gene expression by E. coli MG1655 on LB medium (reference) to gene expression 5 min (Ac_pulse_#I and II) and 50 min (Ac_50 min_#I to VI) after the addition of acetate, after adaptation to acetate (Ac_adapt_#I to IV), 5 min after the addition of propionate (Prop_pulse), after adaptation to propionate (Prop_adapt_#I to III), after adaptation to increased sodium chloride concentrations (NaCl_#I to III), and 5 min (CCCP_pulse_#I and II) and 3 h (CCCP_adapt_#I and II) after the addition of CCCP. Gene expression after the adaptation of MG1655 to acetate was compared directly to that after adaptation to propionate (Ac_vs_Prop_adapt_#I to III). Gene expression by the lrhA disruption strain IMW331 on LB medium (reference) was compared to gene expression after adaptation to acetate (Ac_adapt_delta_LrhA). Green indicates decreased RNA levels and red indicates increased RNA levels in the test culture compared to the reference culture. (A) Graph depicting the whole cluster (see the text for details). Subclusters 1 and 2 are highlighted in red, and subcluster 3 is highlighted in green. The scale bar indicates color coding of the RNA levels. (B to D) Subclusters 1 to 3, respectively, from panel A with corresponding gene names.
RESULTS
Analyses of genome-wide expression in response to acetate or propionate.
In order to determine global gene expression changes in E. coli MG1655 exponentially growing on LB complex medium after adaptation to sodium acetate and sodium propionate, we performed four and three independent DNA microarray experiments, respectively. E. coli MG1655 was cultivated exponentially by repeated dilution on LB complex medium for about 20 generations in the absence or in the presence of neutralized 20 mM sodium acetate or 20 mM sodium propionate. Under these conditions, the medium pH remained approximately constant at 6.6 to 6.9 during cultivation. The presence of sodium acetate or sodium propionate led to only slight decreases in growth, as doubling times were 26 min on LB medium, 34 min on LB medium plus acetate, and 34 min on LB medium plus propionate. When cells were harvested at an OD600 of 0.4 to 0.5 for RNA preparation, 20 ± 1 mM acetate or 20 ± 1 mM propionate (mean and standard deviation) was detected in the medium, indicating that neither compound was utilized to a large extent. Moreover, on LB medium plus acetate, 6 mM pyruvate had accumulated, compared to 1 mM on LB medium and 3.5 mM on LB medium plus propionate. The addition of sodium acetate and sodium propionate resulted in changed RNA levels for about 3% of the E. coli genes (Table 1 and data not shown).
TABLE 1.
Relative RNA levels for genes in DNA microarray comparisons of global gene expression in E. coli MG1655 grown on various mediaa
| Gene | Operon organization | Function | Avg relative mRNA levels obtained on LB medium with or without:
|
|||
|---|---|---|---|---|---|---|
| Designation | Name | Acetate | Propionate | NaCl | ||
| b0002 | thrA | ↓ | Aspartokinase I, homoserine dehydrogenase I | 1.18 | 2.04* | 1.06 |
| b0003 | thrB | ↓ | Homoserine kinase | 1.01 | 2.43 | 1.16 |
| b0004 | thrC | ↓ | Threonine synthase | 1.07 | 2.03 | 0.99 |
| b0030 | yaaF | ↓ | ORF, hypothetical protein | 0.35* | 0.42* | 0.93 |
| b0064 | araC | ↓ | Transcriptional regulator for ara operon | 0.36* | 0.49* | 0.83* |
| b0124 | gcd | ↑ | Glucose dehydrogenase | 3.11* | 3.36* | 1.14 |
| b0162 | yaeG | ↓ | ORF, hypothetical protein | 0.42* | 0.72 | 1.00 |
| b0259 | yi52_1 | ↑ | IS5 transposase | 0.61* | 0.50* | 1.04 |
| b0268 | yagE | ↓ | Putative lyase/synthase | 0.25* | 0.33* | 0.85 |
| b0269 | yagF | ↓ | Putative dehydratase | |||
| b0306 | ykgE | ↓ | Putative dehydrogenase subunit | 0.33 | 0.75 | 0.60 |
| b0307 | ykgF | ↓ | ORF, hypothetical protein | 0.38* | 0.76 | 0.25 |
| b0308 | ykgG | ↓ | Putative transporter | 0.62* | 0.78 | 0.80 |
| b0311 | betA | ↑ | Choline dehydrogenase, a flavoprotein | 1.77* | 2.10* | 0.78 |
| b0312 | betB | ↑ | NAD+-dependent betaine aldehyde | 1.84* | 2.14* | 0.94 |
| b0313 | betI | ↑ | Probably transcriptional repressor of bet genes | 1.63* | 2.07* | 0.87 |
| b0342 | lacA | ↑ | Thiogalactoside acetyltransferase | 0.88 | 0.93 | 0.91 |
| b0343 | lacY | ↑ | Galactoside permease (M protein) | 0.58* | 0.48 | 0.62* |
| b0344 | lacZ | ↑ | β-d-Galactosidase | 0.66 | 0.48* | 0.75 |
| b0522 | purK | ↑ | Phosphoribosylaminoimidazole carboxylase, CO2-fixing subunit | 2.11* | 1.24 | 0.98 |
| b0523 | purE | ↑ | Phosphoribosylaminoimidazole carboxylase, catalytic subunit | 1.90* | 1.29 | 0.99 |
| b0598 | cstA | ↓ | Carbon starvation protein | 0.34* | 0.53* | 0.75 |
| b0679 | nagE | ↓ | PTS, N-acetylglucosamine specific | 0.44* | 0.48* | 0.95* |
| b0681 | b0681 | ↓ | ORF, hypothetical protein | 0.39* | 0.53* | 0.91 |
| b0682 | b0682 | ↓ | ORF, hypothetical protein | 1.82 | ||
| b0756 | galM | ↑ | Galactose-1-epimerase (mutarotase) | 0.79 | 0.91 | 1.02 |
| b0757 | galK | ↑ | Galactokinase | 0.61* | 0.83 | 0.83 |
| b0758 | galT | ↑ | Galactose-1-phosphate uridylytransferase | 0.49* | 0.69 | 0.67 |
| b0759 | galE | ↑ | UDP-galactose-4-epimerase | 0.54* | 0.77 | 0.78 |
| b0797 | rhlE | ↓ | Putative ATP-dependent RNA helicase | 2.05* | 2.27* | 1.00 |
| b0812 | dps | ↑ | Global regulator, starvation conditions | 2.14* | 2.03 | 1.49* |
| b0869 | b0869 | ↑ | Putative dTDP-glucose enzyme | 0.89 | 1.03 | 1.06* |
| b0870 | b0870 | ↑ | Putative arylsulfatase | 1.07 | 1.01 | 1.32* |
| b0871 | poxB | ↑ | Pyruvate oxidase | 2.06* | 2.99 | 1.42 |
| b1015 | putP | ↓ | Major sodium/proline symporter | 0.33* | 0.50* | 0.77 |
| b1070 | flgN | ↑ | Protein of flagellar biosynthesis Fli A | |||
| b1071 | flgM | ↑ | Anti-FliA (anti-sigma) factor; also known as RflB | 2.33* | 2.70* | 0.95 |
| b1072 | flgA | ↑ | Flagellar biosynthesis, assembly of basal body | 2.94* | 3.19* | 0.95 |
| b1073 | flgB | ↓ | Flagellar biosynthesis, cell-proximal portion | 3.97* | 5.12* | 1.02 |
| b1074 | flgC | ↓ | Flagellar biosynthesis, cell-proximal portion | 3.88* | 5.61* | 0.98 |
| b1075 | flgD | ↓ | Flagellar biosynthesis, initiation of hook | 3.95* | 5.67* | 1.09 |
| b1076 | flgE | ↓ | Flagellar biosynthesis, hook protein | 3.70* | 6.10* | 1.05 |
| b1077 | flgF | ↓ | Flagellar biosynthesis, cell-proximal portion | 2.57* | 3.73* | 0.88 |
| b1078 | flgG | ↓ | Flagellar biosynthesis, cell-distal portion | |||
| b1079 | flgH | ↓ | Flagellar biosynthesis, basal body outer portion | 2.67* | 3.79* | 0.67 |
| b1080 | flgI | ↓ | Homolog of Salmonella P-ring of flagellum basal body | 2.10 | 3.04* | |
| b1081 | flgJ | ↓ | Flagellar biosynthesis | 2.56* | 3.63* | 0.83 |
| b1082 | flgK | ↓ | Flagellar biosynthesis, hook-filament junction | 3.35* | 4.53* | 0.92 |
| b1083 | flgL | ↓ | Flagellar biosynthesis, hook-filament junction | 2.80* | 3.45* | 1.12 |
| b1156 | ycfA | ↑ | ORF, hypothetical protein | |||
| b1157 | b1157 | ↑ | Putative fail fiber protein | 0.60* | 0.49* | 0.90* |
| b1198 | ycgC | ↑ | Putative PTS enzyme I | 0.22* | 0.67 | 1.10 |
| b1199 | b1199 | ↑ | Putative dihydroxyacetone kinase (EC 2.7.1.2) | 0.31* | 0.62 | 1.27 |
| b1200 | b1200 | ↑ | Putative dihydroxyacetone kinase (EC 2.7.1.2) | 0.19* | 0.51 | 0.95 |
| b1243 | oppA | ↓ | Oligopeptide transport, periplasmic binding | 1.47* | 2.19* | 1.15* |
| b1244 | oppB | ↓ | Oligopeptide transport, permease protein | 1.54* | 2.31* | 1.06 |
| b1245 | oppC | ↓ | Oligopeptide transport permease protein | 1.50* | 2.18* | 0.93 |
| b1246 | oppD | ↓ | ATP-binding protein | 1.49* | 2.07 | 0.90 |
| b1329 | b1329 | ↓ | Putative transport periplasmic protein | 0.46* | 0.59* | 0.91 |
| b1370 | yi52_5 | ↑ | IS5 transposase | 0.57* | 0.47* | 0.96 |
| b1380 | IdhA | ↑ | Fermentative d-lactate dehydrogenase, NAD | 3.27* | 1.78 | 1.80* |
| b1480 | rpsV | ↑ | 30S ribosomal subunit protein S22 | 2.57* | 2.06 | 1.50 |
| b1494 | yddC | ↑ | Putative peptidase | 0.46 | 0.26 | |
| b1495 | yddB | ↑ | ORF, hypothetical protein | 0.64 | 0.69 | 0.98 |
| b1496 | yddA | ↑ | Putative ATP-binding component of a transport system | 0.68* | 0.50* | 0.85 |
| b1530 | marR | ↓ | Multiple antibiotic resistance protein, repressor | 1.25* | 2.08* | 0.82 |
| b1531 | marA | ↓ | Multiple antibiotic resistance, transcription | 1.16 | 2.16* | 1.15* |
| b1532 | marB | ↓ | Multiple antibiotic resistance protein | 1.32 | 1.72* | 0.95 |
| b1621 | malX | ↓ | PTS, maltose- and glucose-specific enzyme II | 0.38* | 0.57* | 0.82* |
| b1622 | malY | ↓ | Enzyme that may degrade or block biosynthesis | 0.53* | 0.66* | 0.60 |
| b1722 | b1722 | ↑ | ORF, hypothetical protein | 2.32* | 2.21 | 0.89 |
| b1729 | b1729 | ↓ | Part of a kinase | 0.96 | 2.28* | 0.91 |
| b1878 | b1878 | ↑ | Flagellar protein | 3.33 | ||
| b1879 | flhA | ↑ | Flagellar biosynthesis, possible export of flagellar proteins | 2.35* | 2.52* | 0.99 |
| b1880 | b1880 | ↑ | Putative part of export apparatus for flagellar proteins | 3.06* | 3.56* | 0.91 |
| b1881 | cheZ | ↑ | Chemotactic response, CheY protein | 2.21 | 2.70* | 0.94 |
| b1882 | cheY | ↑ | Chemotaxis regulator, transmits chemoreceptor | 3.54 | ||
| b1883 | cheB | ↑ | Response regulator for chemotaxis (cheA) | 1.80 | ||
| b1884 | cheR | ↑ | Response regulator for chemotaxis, protein | 2.91* | 2.79 | |
| b1885 | tap | ↑ | Methyl-accepting chemotaxis protein IV | 6.82 | 7.81 | |
| b1886 | tar | ↑ | Methyl-accepting chemotaxis protein II | 2.84* | 2.31* | 1.12* |
| b1891 | flhC | ↑ | Regulator of flagellar biosynthesis | 1.61* | 2.04* | 1.08 |
| b1892 | flhD | ↑ | Regulator of flagellar biosynthesis | 1.54* | 1.92* | 0.94 |
| b1896 | otsA | ↑ | Trehalose-6-phosphate synthase | 2.23* | 2.13 | 1.31 |
| b1897 | otsB | ↑ | Trehalose-6-phosphate phophatase | 2.38* | 2.14 | 1.40 |
| b1917 | b1917 | ↑ | Putative ATP-binding component of a transport system | 1.27 | 1.46 | |
| b1918 | yecC | ↑ | Putative transport system permease protein | 1.25 | 0.80 | |
| b1919 | b1919 | ↑ | Putative 1-aminocyclopropane-1-carboxylate | 1.30 | 1.57* | 0.73 |
| b1920 | fliY | ↑ | Putative periplasmic binding protein | 1.92* | 2.10* | 1.09 |
| b1921 | fliZ | ↑ | ORF, hypothetical protein | 3.38* | 3.80* | 1.09* |
| b1922 | fliA | ↑ | Flagellar biosynthesis, alternative sigma factor | 4.82* | 5.86* | 1.26* |
| b1923 | fliC | ↑ | Flagellar biosynthesis, flagellin, filament | 21.46 | 10.39 | 1.03 |
| b1924 | fliD | ↓ | Flagellar biosynthesis, filament capping protein | 5.87* | 5.48* | |
| b1925 | fliS | ↓ | Flagellar biosynthesis, repressor of class 3a | 2.89* | 2.76* | 0.91 |
| b1926 | fliT | ↓ | Flagellar biosynthesis, repressor of class 3a | 1.09 | 1.21 | 0.82 |
| b1936 | b1936 | ↓ | ORF, hypotheticla protein | 2.50* | 2.93* | 1.05 |
| b1937 | fliE | ↑ | Flagellar biosynthesis, basal body component | 4.07* | 4.75* | 1.23 |
| b1938 | fliF | ↓ | Flagellar biosynthesis, basal body | 2.01* | 2.90* | 0.91 |
| b1939 | fliG | ↓ | Flagellar biosynthesis, component of motor | 2.96* | 3.75* | 1.10 |
| b1940 | fliH | ↓ | Flagellar biosynthesis, export of flagellar proteins? | 2.28* | 2.92* | 0.91 |
| b1941 | fliI | ↓ | Flagellum-specific ATP synthase | 3.36* | 3.94* | 0.76 |
| b1942 | fliJ | ↓ | Flagellar fliJ protein | 3.00* | 3.74* | 0.71 |
| b1943 | fliK | ↓ | Flagellar hook-length control protein | 3.12* | 4.76* | 0.78 |
| b1944 | fliL | ↓ | Flagellar biosynthesis | 2.27* | 3.25* | 0.85 |
| b1945 | fliM | ↓ | Flagellar biosynthesis, component of motor | 1.96* | 2.61* | 0.78 |
| b1946 | fliN | ↓ | Flagellar biosynthesis, component of motor | |||
| b1947 | fliO | ↓ | Flagellar biosynthesis | 2.25* | 3.05* | 0.77 |
| b1948 | fliP | ↓ | Flagellar biosynthesis | 2.26* | 2.79* | 0.66 |
| b1949 | fliQ | ↓ | Flagellar biosynthesis | 2.61* | 3.22* | |
| b1950 | fliR | ↓ | Flagellar biosynthesis | 3.73 | 4.05* | |
| b1976 | b1976 | ↓ | ORF, hypothetical protein | 0.49* | 0.56 | 1.25 |
| b1994 | yi52_6 | ↑ | IS5 transposase | 0.69* | 0.50* | 1.09 |
| b2146 | b2146 | ↓ | Putative oxidoreductase | 0.27 | 0.69* | |
| b2147 | yeiA | ↓ | Putative oxidoreductase | 0.47* | 0.87 | 0.67 |
| b2148 | mgIC | ↑ | Methyl-galactoside transport and galactose taxis | 0.29* | 0.48 | 0.60* |
| b2149 | mgIA | ↑ | ATP-binding component of methyl-galactoside | 0.28* | 0.48 | 0.62* |
| b2150 | mgIB | ↑ | Galactose-binding transport protein, receptor for galactose taxis | 0.22* | 0.43 | 0.70* |
| b2151 | galS | ↑ | mgl repressor, galactose operon inducer | 0.30* | 0.45* | 0.83 |
| b2192 | yi52_8 | ↑ | IS5 transposase | 0.64* | 0.50* | 1.01 |
| b2210 | yojH | ↑ | ORF, hypothetical protein | 2.11* | 2.93* | 1.02 |
| b2239 | glpQ | ↑ | Glycerophosphodiester phosphodiesterase | 0.59* | 0.75 | 1.39 |
| b2240 | glpT | ↑ | sn-Glycerol-3-phosphate permease | 0.49* | 0.68 | 1.39 |
| b2241 | glpA | ↓ | sn-Glycerol-3-phosphate dehydrogenase | 0.32* | 0.51 | 0.66* |
| b2242 | glpB | ↓ | sn-Glycerol-3-phosphate dehydrogenase | 0.37* | 0.56 | 0.78 |
| b2243 | glpC | ↓ | sn-Glycerol-3-phosphate dehydrogenase | 0.39* | 0.65 | 0.67* |
| b2341 | b2341 | ↑ | Putative enzyme | 0.45* | 0.60* | 0.85* |
| b2342 | b2342 | ↑ | Putative acyltransferase | 0.38* | 0.63* | 0.76* |
| b2364 | dsdC | ↑ | d-Serine dehydratase (deaminase) | 0.40* | 0.60* | 1.01 |
| b2365 | dsdX | ↓ | Transport system permease (serine?) | 1.00 | ||
| b2366 | dsdA | ↓ | d-Serine dehydratase (deaminase) | 0.21* | 0.85 | 0.90 |
| b2388 | glk | ↑ | Glucokinase | 0.42* | 0.48* | 1.48 |
| b2428 | b2428 | ↓ | Putative regulator | 0.59* | 0.66 | 0.74 |
| b2429 | b2429 | ↓ | Putative PTS enzyme II | 0.35* | 0.40* | 0.50 |
| b2430 | b2430 | ↓ | Putative β-lactamase | 0.48* | 0.54* | 0.70* |
| b2535 | csiE | ↓ | ORF, hypothetical protein | 0.38* | 0.55 | 0.69 |
| b2537 | hcaR | ↑ | Transcriptional activator of hca cluster | 0.38* | 0.53* | 0.82 |
| b2595 | b2595 | ↓ | ORF, hypothetical protein | 1.38 | 1.27 | 1.55* |
| b2596 | b2596 | ↓ | ORF, hypothetical protein | 0.43* | 0.67 | 1.02 |
| b2597 | yfiA | ↓ | Putative yhbH sigma 54 modulator | 0.69 | 0.69 | 1.78* |
| b2677 | proV | ↓ | ATP-binding component of transport system | 1.56* | 2.25 | 1.85* |
| b2678 | proW | ↓ | High-affinity transport system for glycine | 1.55* | 2.24* | 1.74* |
| b2679 | proX | ↓ | High-affinity transport system for glycine | 1.85* | 2.78* | 1.85* |
| b2680 | b2680 | ↓ | ORF, hypothetical protein | 1.65* | 2.33* | 1.20 |
| b2681 | b2681 | ↓ | Putative transport protein | 0.97 | 1.54* | 0.87 |
| b2702 | srlA1 | ↓ | PTS, glucitol/sorbitol-specific IIC component | 0.14* | 0.38* | 0.93 |
| b2703 | srlA2 | ↓ | PTS, glucitol/sorbitol-specific IIB component | 0.15* | 0.42* | 0.91 |
| b2704 | srlB | ↓ | PTS, glucitol/sorbitol-specific enzyme | 0.18* | 0.50* | 0.95 |
| b2705 | srlD | ↓ | Glucitol (sorbitol)-6-phosphate dehydrogenase | 0.30* | 0.72 | 1.14* |
| b2706 | gutM | ↓ | Glucitol operon activator | 0.25* | 0.54 | 0.68* |
| b2707 | srlR | ↓ | Regulator for gut (srl), glucitol operon | 0.43* | 0.62* | 0.84* |
| b2708 | gutQ | ↓ | ORF, hypothetical protein | 0.77 | 0.77* | 0.82 |
| b2797 | sdaB | ↓ | l-Serine dehydratase (deaminase) | 0.43 | 0.27* | 0.71* |
| b2798 | exo | ↓ | 5′-3′ Exonuclease | 0.77 | 0.66* | 0.94 |
| b2801 | fucP | ↓ | Fucose permease | 0.51* | 0.65 | 0.82 |
| b2802 | fucI | ↓ | l-Fucose isomerase | 0.66 | 0.67 | 0.83 |
| b2803 | fucK | ↓ | l-Fuculokinase | 0.30 | ||
| b2804 | fucU | ↓ | Protein of fucose operon | 0.81 | 1.01 | 1.14* |
| b2805 | fucR | ↓ | Positive regulator of the fuc operon | 0.42* | 0.48* | 1.18* |
| b2844 | b2844 | ↑ | Putative acyltransferase | 0.41* | 0.55* | 0.83 |
| b2869 | b2869 | ↑ | Putative transcriptional regulator | 0.41 | 0.49* | 1.25 |
| b2924 | yggB | ↑ | Component of MscS | 2.15* | 1.56* | 1.59* |
| b2973 | b2973 | ↑ | ORF, hypothetical protein | 0.32* | 0.25* | 0.86 |
| b2974 | b2974 | ↑ | Putative endoglucanase | 0.27* | 0.23 | 0.92 |
| b3072 | air | ↑ | Aerotaxis sensor receptor, flavoprotein | 1.65* | 2.08* | 0.99 |
| b3076 | ebgA | ↓ | Evolved β-d-galactosidase, alpha subunit | 0.39* | 0.55 | 0.52 |
| b3077 | ebgC | ↓ | Evolved β-d-galactosidase, beta subunit | |||
| b3093 | exuT | ↓ | Transport of hexuronates | 0.32* | 0.48* | 0.58 |
| b3112 | tdcG | ↑ | l-Serine deaminase 3 | 0.80 | 0.86* | |
| b3113 | tdcF | ↑ | ORF, hypothetical protein | 0.71* | 1.01 | 0.86 |
| b3114 | tdcE | ↑ | 2-Ketobutyrate formate lyase | 0.74* | 0.77 | 0.83* |
| b3115 | tdcD | ↑ | Propionate kinase/acetate kinase C | |||
| b3116 | tdcC | ↑ | Anaerobically inducible l-threonine, l-serine | 0.26* | 0.47* | 0.80 |
| b3117 | tdcB | ↑ | Threonine dehydratase, catabolic | 0.31 | 0.07* | 0.38 |
| b3118 | tdcA | ↑ | Transcriptional activator of tdc operon | 0.49* | 0.37 | 1.02 |
| b3218 | yi52_10 | ↑ | IS5 transposase | 0.61* | 0.50* | 1.04 |
| b3221 | yhcH | ↑ | ORF, hypothetical protein | 0.27* | 0.47 | 0.86 |
| b3222 | nanK | ↑ | ManNAc kinase | 0.22* | 0.29 | 0.78* |
| b3223 | nanE | ↑ | ManNAc epimerase | 0.21* | 0.32 | 0.81* |
| b3224 | nanT | ↑ | Sialic acid transporter | 0.15* | 0.26 | 0.67* |
| b3225 | nanA | ↑ | N-Acetylneuraminate lyase (aldolase) | 0.13* | 0.29 | 0.76* |
| b3418 | malT | ↓ | Positive regulator of mal regulon | 0.42* | 0.62* | 1.11* |
| b3509 | hdeB | ↑ | ORF, hypothetical protein | 4.26* | 2.30* | 1.21* |
| b3510 | hdeA | ↑ | ORF, hypothetical protein | 6.16* | 3.82* | 1.35 |
| b3525 | yhjH | ↑ | ORF, hypothetical protein | 2.17* | 2.42* | 0.92 |
| b3528 | dctA | ↑ | Uptake of C4-dicarboxylic acids | 0.38* | 0.63 | 0.65* |
| b3603 | IldP | ↓ | l-Lactate permease | 0.34* | 0.46 | 0.51* |
| b3686 | hslS | ↑ | Heat shock protein | 1.23 | 2.06* | 1.12* |
| b3708 | tnaA | ↓ | Tryptophanase | 0.16* | 0.31 | 1.41 |
| b3709 | tnaB | ↓ | Low-affinity tryptophan permease | 0.10* | 0.26 | 0.89 |
| b3748 | rbsD | ↓ | d-Ribose high-affinity transport system | 0.49* | 0.73* | 0.97 |
| b3749 | rbsA | ↓ | ATP-binding component of d-ribose high-affinity transport system | 0.41 | 0.62 | 0.75 |
| b3750 | rbsC | ↓ | d-Ribose high-affinity transport system | 0.51* | 0.80 | 0.65* |
| b3751 | rbsB | ↓ | Periplasmic d-ribose binding protein | 0.62* | 0.98 | 1.26* |
| b3752 | rbsK | ↓ | Ribokinase | 0.61* | 0.89 | 0.98 |
| b3845 | fadA | ↑ | Thiolase I, 3-ketoacyl-CoA thiolase, acetyl-CoA | 0.67* | 1.10 | 0.83 |
| b3846 | fadB | ↑ | Four-enzyme protein, 3-hydroxyacyl-CoA | 0.39* | 0.71 | 0.81* |
| b3872 | b3872 | ↓ | Putative transcriptional regulator | 0.54* | 0.67* | 0.89* |
| b3873 | yihM | ↓ | ORF, hypothetical protein | 0.38* | 0.48* | 1.25* |
| b3907 | rhaT | ↑ | Rhamnose transport | 0.14* | 0.46 | 0.84 |
| b3926 | glpK | ↑ | Glycerol kinase | 0.49* | 0.67 | 0.80 |
| b3927 | glpF | ↑ | Facilitated diffusion of glycerol | 0.41* | 0.57 | 0.72 |
| b4014 | aceB | ↓ | Malate synthase A | 1.59 | 3.31* | 1.35* |
| b4015 | aceA | ↓ | Isocitrate lyase | 1.57* | 2.67* | 1.46* |
| b4016 | aceK | ↓ | Isocitrate dehydrogenase kinase/phosphatase | 1.50* | 2.39 | 1.45 |
| b4032 | malG | ↑ | Part of maltose permease, inner membrane | 0.03 | 0.09 | 1.32 |
| b4033 | malF | ↑ | Part of maltose permease, periplasmic | 0.18 | 0.21 | 0.79 |
| b4034 | malE | ↑ | Periplasmic maltose-binding protein, substrate | 0.46* | 0.58* | 1.18 |
| b4055 | yjbP | ↓ | Diadenosine tetraphosphatase | 0.40* | 0.50* | 1.09 |
| b4118 | melR | ↑ | Regulator of melibiose operon | 0.47* | 0.73* | 0.69 |
| b4139 | aspA | ↑ | Aspartate ammonia lyase (aspartase) | 0.37* | 0.45* | 1.38 |
| b4188 | yjfN | ↑ | ORF, hypothetical protein | 0.39* | 0.50 | 0.67 |
| b4189 | yjfO | ↑ | ORF, hypothetical protein | 0.28* | 0.40* | 0.88 |
| b4239 | treC | ↑ | Trehalase-6-phosphate hydrolase | 0.31* | 0.38 | 0.77* |
| b4240 | treB | ↑ | PTS enzyme II, trehalose specific | 0.23 | 0.25* | 0.98 |
| b4312 | fimB | ↓ | Recombinase involved in phase variation | 1.32 | 2.42 | 1.38* |
| b4313 | fimE | ↓ | Recombinase involved in phase variation | |||
| b4314 | fimA | ↓ | Major type 1 subunit fimbrin (pilin) | 2.00 | 2.06* | 1.02 |
| b4315 | fimI | ↓ | Fimbrial protein | 4.04* | 5.23* | 0.94 |
| b4316 | fimC | ↓ | Periplasmic chaperone, required for type 1 fimbriae | 3.91* | 4.32* | 0.71 |
| b4317 | fimD | ↓ | Outer membrane protein, export and assembly | 0.86 | 1.30 | 0.82 |
| b4318 | fimF | ↓ | Fimbrial morphology | 1.88* | 2.32* | 0.76 |
| b4319 | fimG | ↓ | Fimbrial morphology | 3.04* | 3.01* | 0.75 |
| b4320 | fimH | ↓ | Minor fimbrial subunit, d-mannose specific | 2.19* | 2.35* | 0.78 |
| b4322 | uxuA | ↓ | Mannonate hydrolase | 0.29* | 0.43* | 0.86 |
| b4323 | uxuB | ↓ | d-Mannonate oxidoreductase | 0.74 | 0.75 | 0.99 |
| b4353 | yjiX | ↑ | ORF, hypothetical protein | 0.45* | 0.53* | 0.89 |
| b4354 | yjiY | ↑ | Putative carbon starvation protein | 0.44* | 0.46 | 0.90 |
| b4357 | yjjM | ↑ | ORF, hypothetical protein | 0.32 | 0.48* | |
| b4372 | holD | ↓ | DNA polymerase III, psi subunit | 1.44* | 1.53 | 0.83 |
| b4373 | rimI | ↓ | Acyltransferase for 30S ribosomal subunit | 1.13 | 1.32 | 0.83 |
| b4374 | yjjG | ↓ | Putative phosphatase | 1.26* | 1.41 | 0.97 |
| b4375 | prfC | ↓ | Peptide chain release factor RF-3 | 1.22 | 1.49 | 0.88 |
| b4376 | osmY | ↓ | Hyperosmotically inducible periplasmic protein | 3.52* | 3.02* | 1.69* |
Media tested were LB, LB-20 mM sodium acetate, LB-20 mM sodium propionate, and LB-20 mM NaCl. Asterisks indicate significant RNA level differences (P value, <0.05; Student's t test) that exceeded 2-fold or were below 0.5-fold. For operons, RNA levels for all genes are given. At least three independent cultivations and DNA microarray experiments were performed. ORF, open reading frame.
To address gene expression changes elicited by increased sodium concentrations or increased medium osmolarity, we performed three DNA microarray experiments comparing relative RNA levels from cultures grown for 20 generations with or without sodium chloride at 20 mM. However, the addition of sodium chloride caused only very minor expression changes (Table 1).
Gene expression data from these 10 DNA microarray experiments (including replicates) carried out to study adaptation to sodium acetate, sodium propionate, or sodium chloride were analyzed statistically (for details, see Materials and Methods). Spot fluorescence signals exceeded spot background fluorescence by at least a factor of 3, and changes in relative mRNA levels were significant (P value, <0.05, as determined by a paired Student's t test) (43) and were greater than 2-fold or less than 0.5-fold in at least one experiment. In this set of experiments, 153 genes fulfilled these criteria; if they belong to an operon, they are listed together with all genes of that operon (233 genes in total) in Table 1.
To determine whether these expression changes were specific for long-term adaptation to acetate or propionate, several control experiments were performed and included in the hierarchical cluster analysis. In order to assess the immediate response of E. coli MG1655 to either compound, gene expression changes 5 and 50 min after the addition of 20 mM sodium acetate (two and six microarray experiments, respectively) and 5 min after the addition of sodium propionate (one microarray experiment) were determined. As acetic acid and propionic acid are weak acids known to inhibit growth presumably by uncoupling of the transmembrane pH gradient (5), we determined gene expression changes 5 min and 3 h after the addition of the uncoupler CCCP (two microarray experiments each). To minimize effects due to growth inhibition, CCCP was added to a final concentration of 40 μM, which led to a 41-min doubling time, compared to 26 min on LB medium. The hierarchical cluster calculations were performed by using the expression data obtained for the 233 genes listed in Table 1 from a total of 27 DNA microarray experiments. Of these, 179 genes were measured reliably in two-thirds or more of all experiments and were included in the clustering analysis by the algorithm described by Eisen et al. (18) (Fig. 1).
Clustering of experimental conditions revealed that gene expression changes after adaptation to acetate or to propionate were similar to each other but were distinct from gene expression changes after short exposure to either acetate or propionate and distinct from the response to CCCP (Fig. 1). Clustering of genes showed similar RNA level differences for genes in three subclusters (Fig. 1). Most notably, a group of 33 genes which included 27 chemotaxis and flagellum genes showed 1.5- to 21-fold increases in mRNA levels after adaptation to acetate and to propionate but not under any other conditions (Fig. 1, subcluster 2). A large group of genes exhibited ∼0.67- to 0.1-fold decreases in mRNA abundances after adaptation to either acetate or propionate. Most of these 70 genes are involved in the uptake and catabolism of carbon and energy sources other than glucose (Fig. 1, subcluster 3). Furthermore, a group of 12 genes, most of which are RpoS-dependent general stress response genes, exhibited increases in RNA levels after adaptation to either acetate or propionate. However, the extent of expression increases for these genes was greater shortly after the addition of acetate, propionate, or CCCP (Fig. 1, subcluster 1).
Increased expression of the chemotaxis and flagellum genes after long-term adaptation to acetate and propionate.
In the hierarchical cluster analysis, 27 chemotaxis and flagellum genes were part of subcluster 2, with increased RNA levels only after adaptation to acetate or propionate (Fig. 1, subcluster 2). In addition, this subcluster contained four genes of the type I fimbria operon and genes b1936 and b2680, encoding hypothetical proteins. To verify the DNA microarray expression data for the chemotaxis and flagellum genes, we used translational fusions of lacZ with the promoter regions of flhDC and fliC (43). The flhDC operon (class I) codes for the master regulator of flagellar operons, whereas fliC (class III gene of the flhDC regulon) codes for the major flagellar subunit flagellin (46). The expression of the chromosomal flhDC′-′lacZ fusion increased 3- and 8-fold in cells adapted to acetate and propionate, respectively (Table 2), whereas 1.5- to 2-fold increases in RNA abundances were determined in DNA microarray experiments (Table 1). After adaptation to acetate and propionate, the expression of the chromosomal fliC′-′lacZ fusion increased 5- and 6-fold, respectively (Table 2), whereas 21- and 10-fold increases in RNA levels were determined in DNA microarray experiments (Table 1). Thus, DNA microarray determination of increased RNA levels for a class I gene and a class III gene of the flagellar regulon could be verified independently by analysis of the expression of lacZ translational fusions.
TABLE 2.
Expression of chromosomal fliC′-′lacZ and flhDC′-′lacZ translational fusions in E. coli IMW356 and CP992, respectively, and mRNA levels in E. coli MG1655 grown for ∼20 generations on various mediaa
| Gene | β-Galactosidase activity (MU)b on the following medium:
|
β-Galactosidase activity ratio for:
|
RNA levelc for:
|
||||
|---|---|---|---|---|---|---|---|
| LB | LB + acetate | LB + propionate | LB + acetate/B | LB + propionate/LB | LB + acetate/LB | LB + propionate/LB | |
| fliC | 574 | 2,853 | 3,390 | 5.0 | 5.9 | 2.1 | 10 |
| flhDC | 6 | 16 | 50 | 2.7 | 8.3 | 1.5 | 1.9 |
Media tested were LB, LB-20 mM sodium acetate, and LB-20 mM sodium propionate.
β-Galactosidase activities were obtained from three or more independent cultivations of strains IMW356 and CP992 and varied less than 20% Mu, Miller units.
RNA levels were obtained from three or more independent cultivations of strain MG1655 and are taken from Table 1.
To determine whether the increased expression of chemotaxis and flagellum genes after adaptation to acetate or propionate has an effect on swimming behavior, we tested the swimming of E. coli MG1655 on soft tryptone agar with or without 20 mM sodium acetate (pH 7) or 20 mM sodium propionate (pH 7). E. coli MG1655 showed increased colony sizes on soft tryptone agar with 20 mM acetate or propionate compared to the colony sizes seen on plates without either compound (Fig. 2). Thus, adaptation to sodium acetate or sodium propionate included a positive effect on the motility of E. coli MG1655.
FIG. 2.
Swimming motility assays on LB semisolid agar plates. When needed, plates contained neutralized 20 mM sodium acetate or 20 mM sodium propionate. Plates were sealed to avoid evaporation and were incubated at 37°C. Diameters of swim colonies were determined 18 h after inoculation. Means and standard deviations of 10 replicates of three independent determinations are shown.
Long-term adaptation to acetate and propionate included decreased expression of genes for uptake and utilization of sugars, amino sugars, and amino acids.
A number of genes showed decreased relative RNA abundances after long-term adaptation to acetate as well as to propionate (Table 2). Most of these genes belong to operons coding for proteins involved in the uptake and initial degradation of carbon sources other than glucose, e.g., for the sugars maltose (malXY, malT, malEFG, and glk), trehalose (treBC operon), galactose (galETKM, galS, and mglBAC, with the adjacent, divergently oriented putative b2146-b2147 operon), melibiose (melR), rhamnose (rhaT), fucose (fucPIKUR), ribose (rbsDACBK), arabinose (araC), the sugar alcohols glycerol (glpACB, glpTQ, and glpKF) and sorbitol (srlA1A2BD-gutM-srlR-gutQ), the hexuronates (exuT and uxuA), and the amino sugars N-acetylglucosamine (nagE) and N-acetylneuraminate (nanATEK-yhcH) (Table 1). Moreover, genes coding for the uptake and/or catabolism of the amino acids serine (dsdC, dsdA, sdaB), threonine (tdcABCDEFG), aspartate (aspA), proline (putP), and tryptophan (tnaAB) showed decreased RNA levels (Table 1). The expression of genes coding for the uptake of lactate (lldP) and dicarboxylic acids (dctA) as well as the fatty acid utilization operon fadAB was decreased in the presence of acetate or propionate. A similar reduction in RNA levels was found for genes involved in carbon starvation (cstA, b4353-b4354, and csiE, with neighboring hcaR).
Short-term exposure and long-term adaptation to acetate and propionate resulted in increased expression of genes of the general stress response.
As revealed by the hierarchical cluster analysis, a number of genes involved in the RpoS-dependent general stress response (osmY, otsAB, poxB, dps, and hdeAB) showed increased expression after challenge with acetate or propionate (Fig. 1, subcluster 1). In general, the increase in RNA levels was most pronounced in the experiment comparing gene expression before and gene expression 50 min after the addition of acetate or propionate and was not as high in cultures fully adapted to acetate or propionate. This pattern of expression was seen for osmY, coding for a hyperosmotically inducible periplasmic protein; the otsAB operon, including trehalose biosynthesis genes; the purEK operon, coding for phosphoribosylaminoimidazole carboxylase; rpsV, coding for small ribosomal subunit protein S22; σ32-dependent heat shock gene hslS (ibpB); dps, coding for a protein involved in DNA protection during starvation; and the pyruvate-oxidase gene poxB. RNA levels for the hdeAB operon and for ldhA, encoding d-lactate dehydrogenase, were increased 50 min after the addition of acetate and increased further in fully adapted cells.
The σS-controlled gadA and gadB genes, encoding two isoforms of glutamic acid decarboxylase involved in extreme acid resistance, exhibited slightly increased relative mRNA levels (1.3- to 2.4-fold; statistically not significant; P value, >0.1) after adaptation to acetate or propionate. These findings were corroborated by slightly increased enzymatic glutamic acid decarboxylase activities measured in crude extracts (22 mU/mg on LB medium, 39 mU/mg on LB medium plus 20 mM acetate, and 82 mU/mg on LB medium plus 20 mM propionate).
The expression of isoleucine and threonine biosynthesis genes differed between acetate- and propionate-adapted E. coli cells.
The gene expression changes after adaptation to sodium acetate and to sodium propionate largely overlapped in DNA microarray experiments comparing cells grown on LB medium in the absence or presence of either compound (Table 1). We therefore compared RNAs prepared from cells grown for 20 generations in the presence of either 20 mM sodium acetate or 20 mM sodium propionate directly (Table 3). Propionate-adapted E. coli cells exhibited increased RNA levels for the threonine biosynthetic operon thrABC, the isoleucine biosynthetic gene ilvC, and the multiple antibiotic resistance operon marRAB (Tables 1 and 3 and data not shown). Changes in the expression of the sorbitol operon srlA1A2BD-gutM-srlR-gutQ and of hdeAB occurred in both acetate- and propionate-adapted cells compared to cells grown on LB medium but were more extreme after adaptation to acetate (Tables 1 and 3).
TABLE 3.
Relative RNA levels of genes in DNA microarray comparisons of global gene expression in E. coli MG1655 cells grown for ∼20 generations on various mediaa
| Gene | Operon organization | Function | Avg relative mRNA levels for acetate adaptation/propionate adaptation | |
|---|---|---|---|---|
| Designation | Name | |||
| b0001 | thrL | ↓ | thr operon leader peptide | 0.93 |
| b0002 | thrA | ↓ | Aspartokinase I, homoserine dehydrogenase I | 0.53* |
| b0003 | thrB | ↓ | Homoserine kinase | 0.51* |
| b0004 | thrC | ↓ | Threonine synthase | 0.60* |
| b0929 | ompF | ↑ | Outer membrane protein 1a (la;b;F) | 2.18* |
| b2702 | srlA1 | ↓ | PTS, glucitol/sorbitol-specific IIC component | 0.32* |
| b2703 | srlA2 | ↓ | PTS, glucitol/sorbitol-specific IIB component | 0.37* |
| b2704 | srlB | ↓ | PTS, glucitol/sorbitol-specific enzyme IIA component | 0.38* |
| b2705 | srlD | ↓ | Glucitol (sorbitol)-6-phosphate dehydrogenase | 0.41* |
| b2706 | gutM | ↓ | Glucitol operon activator | 0.55 |
| b2707 | srlR | ↓ | Regulator for gut (srl), glucitol operon | 0.65* |
| b2708 | gutQ | ↓ | ORF, hypothetical protein | 0.93 |
| b3509 | hdeB | ↑ | ORF, hypothetical protein | 1.64* |
| b3510 | hdeA | ↑ | ORF, hypothetical protein | 2.10* |
| b3774 | ilvC | ↓ | Ketol-acid reductoisomerase | 0.19* |
Media tested were LB-20 mM sodium acetate and LB-20 mM sodium propionate. Asterisks indicate significant RNA level differences (P value, <0.05; Student's t test) that exceeded 2-fold or were below 0.5-fold. For operons, RNA levels for all genes are given. Three independent cultivations and DNA microarray experiments were performed.
DISCUSSION
We have identified global gene expression changes due to long-term adaptation to acetate or propionate at a neutral external pH. To avoid effects due to growth inhibition, low concentrations of acetate or propionate were used. Previously, such effects, which included reduced expression of many ribosomal protein genes, were determined for enterohemorrhagic E. coli O157:H7 30 min after challenge with a growth-inhibiting concentration of acetate (100 mM) (4) and by comparison of glucose-grown and acetate-grown E. coli MC4100 (50). The short-term response to the addition of high concentrations of acetate was characterized and revealed increased expression of some genes of the general stress response (4, 35, 50). Our study confirmed these results (Table 1) and revealed a transient character of these expression changes, as fully adapted E. coli cells exhibited expression increases to a lesser extent than did cells 50 min after the addition of acetate (Fig. 1, Table 1, and data not shown). The global gene expression analyses in this work identified increased expression of the chemotaxis and flagellum genes as well as decreased expression of a large group of catabolic genes and operons as novel characteristics of the adaptive response of E. coli MG1655 to acetate or propionate.
Changes in the expression of chemotaxis and flagellum genes.
Our experiments revealed that increased expression of the chemotaxis and flagellum genes was specific to adaptation to acetate and propionate, as the short-term responses of E. coli to low pHs (11), to high acetate concentrations (4), or to low acetate concentrations (Fig. 1) did not reveal such expression changes. Acetate and propionate belong to one class of repellents and are active in the negative chemotaxis of E. coli above threshold concentrations of 0.3 and 0.2 mM, respectively (34, 69). E. coli monitors the chemical composition of its surroundings through different transmembrane chemoreceptors: the aspartate receptor Tar, the serine receptor Tsr, the receptor Trg (which mediates the response to the attractants ribose and galactose as well as to the repellent phenol), the peptide receptor Tap, and the aerotaxis receptor Aer (21, 44, 74). The chemoreceptors function in complexes with a histidine protein kinase, CheA, and an adapter protein, CheW. Repellents enhance the autophosphorylation of CheA and the subsequent phosphorylation of CheY, whereas attractants favor the opposite reactions. Phosphorylated CheY binds to the flagellar switch component FliM and enhances the tumbling frequency. It is not known which chemoreceptor senses acetate or propionate. However, acetate metabolism influences switching of the flagellar motor, an effect which is attributed to the activation of CheY through adenylation by acetyl-CoA synthetase (6, 7, 53, 73).
The complex regulation of chemotaxis and flagellum gene expression has been well studied (reviewed in reference 46), but the presence of acetate or propionate is not known to increase the expression of these genes. The master regulator FlhD2C2 is at the top level of the hierarchical control of chemotaxis and flagellum gene expression (for reviews, see references 31 and 46). FlhD2C2, encoded by the class I flhDC operon, activates the transcription of class II promoters (45). Class II genes encode proteins for the basal body and hook of the flagellum as well as for the sigma factor FliA. FliA is necessary for the transcription of class III genes, which code for the anti-sigma factor FliM (29) and proteins needed for flagellar assembly, motor activity, and chemotaxis. Full expression of flhCD and thus of the chemotaxis and flagellum genes depends on cAMP-CRP (37, 63); the carbon storage regulator Csr (70); the nucleoid-associated protein H-NS (9, 26, 64); the heat shock proteins DnaK, DnaJ, and GrpE (59); the regulators of phosphatidylethanolamine biosynthesis—Pss and Psd (58); polyphosphate (54); and the quorum-sensing regulator QseBC (65). At a high medium osmolality, acetylphosphate, an intermediate in the interconversion of acetate and acetyl-CoA by acetate kinase and phosphotransacetylase, has a negative influence on the expression of the chemotaxis and flagellum genes, as under these conditions, phosphorylated OmpR inhibits the expression of flhDC (60). A high gene dosage for the quorum-sensing regulator SdiA caused reduced expression of flhDC and class II and class III chemotaxis and flagellum genes (71). Recently, the LysR-type regulator LrhA (43) was shown to repress flhDC transcription and thus motility.
None of the known regulators of flagellum and chemotaxis gene expression has been shown to bind either acetate or propionate. The relative mRNA levels for flhDC are increased 1.5- to 2-fold in acetate- and propionate-adapted E. coli (Table 1), so it is reasonable to assume that regulation occurs via modulation of FlhD2C2 levels. RNA levels for genes regulated by OmpR did not change (ompC) or were decreased (fadL and ompF) rather than showing an increase, like those for the chemotaxis and flagellum genes. CsrA represses gluconeogenesis and glycogen metabolism but activates glycolysis and acetate metabolism and stimulates flhDC expression. Similarly, steady-state RNA levels for acs and glgCAP, which are regulated by CsrA, did not change after adaptation to propionate or acetate in exponentially growing E. coli. RNA levels for the genes for the regulators Pss, Psd, GrpE, CsrA, SdiA, CRP, H-NS, DnaK, DnaJ, and QseBC (b3025-b3026) were not changed significantly after adaptation to acetate or propionate (Table 1 and data not shown). Cultures adapted to acetate or propionate showed 1.8-fold decreases in RNA levels for the LrhA regulator gene (Table 1 and data not shown). However, an lrhA disruption strain also showed increased expression of the chemotaxis and flagellum genes (Fig. 1, Table 1, and data not shown). Thus, increased expression of these genes after adaptation to acetate is not dependent on regulation by LrhA, and it remains to be shown how this regulation is mediated. We expect that increased expression of chemotaxis and flagellum genes after adaptation to propionate similarly is not dependent on regulation by LrhA, although we did not test this notion.
Changes in the expression of central metabolic genes.
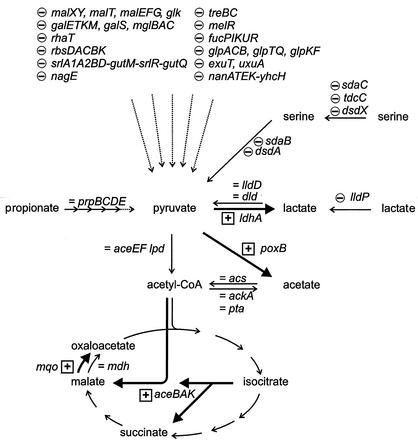
In our experiments, neither acetate nor propionate served as a primary carbon source. Neither the acetyl-CoA synthetase gene acs (16, 36) nor the propionate utilization operon prpBCDE (10, 27, 51, 68) showed increased expression after adaptation to acetate or propionate (Table 1 and Fig. 3). However, the expression of the glyoxylate cycle operon aceBAK which is essential for the utilization of acetate and of propionate as sole carbon sources (16, 68), increased after adaptation to acetate and propionate (1.5- and 2.5-fold, respectively) (Table 1 and Fig. 3). As under the conditions used in this study glucose could not be detected (<0.5 mM), genes subject to glucose repression, e.g., the gluconeogenesis genes pckA, ppsA, maeB (b2463), and fbp, did not show a significant change in expression (Table 1 and data not shown), whereas they obviously did in a comparison of glucose-grown and acetate-grown E. coli cultures (50).
FIG. 3.
Changes in the expression of central metabolic genes in the adaptive responses to acetate and to propionate. Enzyme reactions or pathways are depicted as arrows, and names of the genes for the proteins involved are given next to them. Broken arrows indicate several reactions of uptake and/or initial catabolism, and solid arrows indicate single reactions. Decreased (−), unchanged (=), and increased (+) RNA levels after adaptation to acetate or propionate compared to growth on LB medium are shown.
The global gene expression changes seen after adaptation to external acetate (and to propionate) suggest a metabolic response aimed at avoiding acetate formation by E. coli itself. Many genes involved in the uptake and degradation of potential carbon sources, such as sugars (e.g., malEFG), amino sugars (e.g., nanATEK-yhcH), sugar alcohols (e.g., srlA1A2BD-gutM-srlR-gutQ), amino acids (e.g., tnaAB), lactate (lldP), dicarboxylic acids (dctA), and fatty acids (fadAB), showed significantly decreased gene expression after adaptation to acetate (Table 1 and Fig. 3). Accordingly, the expression of carbon starvation genes also was reduced (e.g., cstA).
Moderate increases in ldhA expression (2- to 3-fold) were seen after adaptation to sodium acetate, sodium propionate, and sodium chloride (Table 1 and Fig. 3). The expression of the NAD-dependent d-lactate dehydrogenase gene ldhA is maximal with pyruvate, at a low pH, and with a low level of oxygen (30). A role in rerouting fermentative metabolism from acetate and formate toward lactate, hydrogen, and carbon dioxide has been described (14, 56), and this mechanism may be related to increased ldhA expression after adaptation to external acetate. Besides ldhA, the genes coding for the pyruvate-converting enzyme pyruvate oxidase (poxB) were expressed at higher levels, whereas lactate uptake permease gene (lldP) expression was decreased (Table 1). The expression of aceEF-lpd, encoding pyruvate dehydrogenase, was not changed significantly (Fig. 3, Table 1, and data not shown). Increased extracellular pyruvate concentrations (6 mM on LB medium plus acetate and 3.5 mM on LB medium plus propionate compared to 1 mM on LB medium) correlated with these expression changes and may indicate an important role for intracellular pyruvate in the regulation of carbon metabolism.
Many of the genes showing decreased RNA levels after adaptation to acetate or propionate are known to be regulated by cAMP-CRP. Recently, glycerol was shown to induce catabolite repression in E. coli growing on tryptone broth complex medium (19), but temporary dephosphorylation of the PTS component EIIAGlc phosphate was not the determining factor. Glycerol-3-phosphate, which is derived from glycerol, inhibits the stimulation of adenylate cyclase activity by EIIAGlc phosphate, a mechanism which may be the basis for catabolite repression by other non-PTS carbon sources (19). Thus, it is conceivable that the addition of acetate or propionate resulted in a similar kind of catabolite repression.
Changes in the expression of stress genes.
The inhibitory effects of acetic acid and propionic acid or of their deprotonated forms on E. coli are well known and are potentiated at low pHs. In the experiments presented here, all cultures were kept in the exponential growth phase by repeated dilution, and the pH during cultivation remained constant at between 6.6 and 6.9. Therefore, we did not primarily interrogate the response of E. coli MG1655 to stress from a basic or an acidic external pH. The response of E. coli to low external pHs involves amino acid decarboxylation and export of basic amines (61). The expression of cadAB, encoding lysine decarboxylase, along with the cadaverine exporter gene lysU and the ornithine and arginine decarboxylase genes speF and adi is increased anaerobically at low pHs but remained unchanged under the conditions used in this study. Similarly, the expression of yfiD, ahpC, manX, ptsH, and gatY was not increased after adaptation to acetate or propionate, whereas increased levels of the encoded proteins were found at low pHs (11). Fermentation genes, like the formate hydrogen lyase system genes, show anaerobic acid regulation. As expected, the expression of the formate hydrogen lyase system genes remained unaffected in the aerobic acetate and propionate adaptation experiments described here (Table 1).
Proteome as well as transcriptome analyses have been performed to assess the effect of acetate on E. coli at a neutral pH (4, 35). For pathogenic E. coli O157:H7, increased RNA levels for 26 genes and decreased RNA levels for 60 genes were determined 1 h after challenge with a growth-inhibiting concentration of acetate (100 mM) by expression profiling with membrane macroarrays (4). Most of the genes showing reduced expression encode components of the transcription-translation machinery, reflecting the growth-inhibiting effect (4), and the expression of these genes was not affected by the addition of only 20 mM acetate (Table 1). The addition of acetate led to increased expression of members of the rpoS regulon (4, 11) (dps, hdeAB, osmY, poxB, and otsAB in Table 1). RpoS is necessary but not sufficient for the induction of acid tolerance at entry into the stationary growth phase, during growth at low pHs, or during growth with acetate (4, 61). RpoS is also important for the virulence of pathogenic E. coli strains and the expression of specific virulence genes (24). The RpoS regulon comprises about 70 genes (24), but only a subset of them showed statistically significant expression changes by adaptation to acetate or propionate (Table 1) (4, 35). These results might be explained by the fact that the regulation of most rpoS-controlled genes is complex and involves, e.g., additional transcription factors (23).
Gene expression changes as a consequence of an increase in the sodium chloride concentration in LB medium to 20 mM and thus in medium osmolality were very small, as no gene exhibited twofold increases or decreases in RNA levels (Table 1). However, the adaptive response to acetate or propionate included expression changes for genes involved in osmotic homeostasis. Besides the expression of RpoS-regulated osmY, the expression of the betIBA operon for the synthesis of the compatible solute glycine betaine from choline, of the proVWX operon for the uptake of proline and glycine betaine, and of the otsAB operon for the synthesis of the compatible solute trehalose was increased after adaptation to acetate or propionate. The expression of the treBC operon for trehalose degradation was decreased (Table 1). Thus, sodium acetate and sodium propionate elicit different or stronger expression changes for genes for osmoadaptation compared to an equal increase in the sodium chloride concentration.
Differences between adaptation to acetate and adaptation to propionate.
Although gene expression changes after adaptation to acetate and to propionate were very similar and included gene expression changes for chemotaxis and flagellum genes, for RpoS-regulated genes, and for catabolic genes, only adaptation to propionate led to higher levels of expression of threonine and isoleucine biosynthetic genes (thrABC and ilvC) as well as to increased RNA levels for the multiple antibiotic resistance operon marRAB. Propionate is a degradation product of threonine and is formed by enzymes encoded by the tdcABCDEFG operon via 2-ketobutyrate and propionyl-CoA (25). The expression of the tdcABCDEFG operon, which is strongly catabolite repressed (25), was decreased in the presence of propionate or acetate (Table 1). Conversely, the threonine biosynthetic thrABC operon showed higher RNA levels after adaptation to propionate. Isoleucine limitation is more effective in derepression of the thrABC operon than is threonine limitation. Propionate adaptation also led to higher ilvC RNA levels. iC codes for the second enzyme (acetohydroxyacid isomeroreductase) in the common pathway for the biosynthesis of isoleucine and valine from pyruvate and threonine. IlvY activates the transcription of ilvC when α-acetolactate or α-aceto-α-hydroxybutyrate, each a substrate for acetohydroxyacid isomeroreductase, is present as a coinducer. It is conceivable that α-aceto-α-hydoxybutyrate levels increase as a consequence of higher levels of thrABC expression and thus lead to the activation of ilvC transcription by IlvY.
In summary, our study of the long-term adaptation of E. coli MG1655 to acetate and to propionate revealed increased expression of the chemotaxis and flagellum genes as a novel aspect of the response to these compounds. As a consequence, E. coli gains motility, a result which adds to acetate and propionate both being known repellents. Moreover, besides eliciting at least parts of the general stress response, gene expression changes suggest regulation of the carbon metabolism of E. coli to avoid the further formation of acetate or propionate.
Acknowledgments
This work was supported in part by Deutsche Bundesstiftung Umwelt, Verbundprojekt Biokatalyse, AZ 13040/16.
We thank Daniela Lehnen and Gottfried Unden for providing strains carrying fliC′-′lacZ and flhDC′-′lacZ fusions and an lrhA disruption strain prior to publication. We thank Michael Bott and Steffen Schaffer for critical reading of the manuscript.
REFERENCES
- 1.Alexeeva, S., B. de Kort, G. Sawers, K. J. Hellingwerf, and M. J. de Mattos. 2000. Effects of limited aeration and of the ArcAB system on intermediary pyruvate catabolism in Escherichia coli. J. Bacteriol. 182:4934-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arfin, S. M., A. D. Long, E. T. Ito, L. Tolleri, M. M. Riehle, E. S. Paegle, and G. W. Hatfield. 2000. Global gene expression profiling in Escherichia coli K12. The effects of integration host factor. J. Biol. Chem. 275:29672-29684. [DOI] [PubMed] [Google Scholar]
- 3.Aristidou, A. A., K. Y. San, and G. N. Bennett. 1995. Metabolic engineering of Escherichia coli to enhance recombinant protein production through acetate reduction. Biotechnol. Prog. 11:475-478. [DOI] [PubMed] [Google Scholar]
- 4.Arnold, C. N., J. McElhanon, A. Lee, R. Leonhart, and D. A. Siegele. 2001. Global analysis of Escherichia coli gene expression during the acetate-induced acid tolerance response. J. Bacteriol. 183:2178-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Axe, D. D., and J. E. Bailey. 1995. Transport of lactate and acetate through the energized cytoplasmic membrane of Escherichia coli. Biotechnol. Bioeng. 47:8-19. [DOI] [PubMed] [Google Scholar]
- 6.Barak, R., and M. Eisenbach. 2001. Acetylation of the response regulator, CheY, is involved in bacterial chemotaxis. Mol. Microbiol. 40:731-743. [DOI] [PubMed] [Google Scholar]
- 7.Barak, R., M. Welch, A. Yanovsky, K. Oosawa, and M. Eisenbach. 1992. Acetyladenylate or its derivative acetylates the chemotaxis protein CheY in vitro and increases its activity at the flagellar switch. Biochemistry 31:10099-10107. [DOI] [PubMed] [Google Scholar]
- 8.Bermejo, L. L., N. E. Welker, and E. T. Papoutsakis. 1998. Expression of Clostridium acetobutylicum ATCC 824 genes in Escherichia coli for acetone production and acetate detoxification. Appl. Environ. Microbiol. 64:1079-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertin, P., E. Terao, E. H. Lee, P. Lejeune, C. Colson, A. Danchin, and E. Collatz. 1994. The H-NS protein is involved in the biogenesis of flagella in Escherichia coli. J. Bacteriol. 176:5537-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blank, L., J. Green, and J. R. Guest. 2002. AcnC of Escherichia coli is a 2-methylcitrate dehydratase (PrpD) that can use citrate and isocitrate as substrates. Microbiology 148:133-146. [DOI] [PubMed] [Google Scholar]
- 11.Blankenhorn, D., J. Phillips, and J. L. Slonczewski. 1999. Acid- and base-induced proteins during aerobic and anaerobic growth of Escherichia coli revealed by two-dimensional gel electrophoresis. J. Bacteriol. 181:2209-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Böck, A., and G. Sawers. 1996. Fermentation, p. 262-282. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 13.Brown, T. D. K., M. C. Jones-Mortimer, and H. L. Kornberg. 1977. The enzymic interconversion of acetate and acetyl-coenzyme A in Escherichia coli. J. Gen. Microbiol. 102:327-336. [DOI] [PubMed] [Google Scholar]
- 14.Bunch, P. K., F. Mat-Jan, N. Lee, and D. P. Clark. 1997. The ldhA gene encoding the fermentative lactate dehydrogenase of Escherichia coli. Microbiology 143:187-195. [DOI] [PubMed] [Google Scholar]
- 15.Chang, D. E., S. Shin, J. S. Rhee, and J. G. Pan. 1999. Acetate metabolism in a pta mutant of Escherichia coli W3110: importance of maintaining acetyl coenzyme A flux for growth and survival. J. Bacteriol. 181:6656-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark, D. P., and J. E. Cronan. 1996. Two-carbon compounds and fatty acids as carbon sources, p. 343-357. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 17.De Biase, D., A. Tramonti, R. A. John, and F. Bossa. 1996. Isolation, overexpression, and biochemical characterization of the two isoforms of glutamic acid decarboxylase from Escherichia coli. Protein Expr. Purif. 8:430-438. [DOI] [PubMed] [Google Scholar]
- 18.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eppler, T., P. Postma, A. Schutz, U. Volker, and W. Boos. 2002. Glycerol-3-phosphate-induced catabolite repression in Escherichia coli. J. Bacteriol. 184:3044-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farmer, W. R., and J. C. Liao. 1997. Reduction of aerobic acetate production by Escherichia coli. Appl. Environ. Microbiol. 63:3205-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gestwicki, J. E., and L. L. Kiessling. 2002. Inter-receptor communication through arrays of bacterial chemoreceptors. Nature 415:81-84. [DOI] [PubMed] [Google Scholar]
- 22.Gokarn, R. R., M. A. Eiteman, and E. Altman. 2000. Metabolic analysis of Escherichia coli in the presence and absence of the carboxylating enzymes phosphoenolpyruvate carboxylase and pyruvate carboxylase. Appl. Environ. Microbiol. 66:1844-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hengge-Aronis, R. 2002. Recent insights into the general stress response regulatory network in Escherichia coli. J. Mol. Microbiol. Biotechnol. 4:341-346. [PubMed] [Google Scholar]
- 24.Hengge-Aronis, R. 2000. A role for the sigma S subunit of RNA polymerase in the regulation of bacterial virulence. Adv. Exp. Med. Biol. 485:85-93. [DOI] [PubMed] [Google Scholar]
- 25.Hesslinger, C., S. A. Fairhurst, and G. Sawers. 1998. Novel keto acid formate lyase and propionate kinase enzymes are components of an anaerobic pathway in Escherichia coli that degrades l-threonine to propionate. Mol. Microbiol. 27:477-492. [DOI] [PubMed] [Google Scholar]
- 26.Hommais, F., E. Krin, C. Laurent-Winter, O. Soutourina, A. Malpertuy, J. P. Le Caer, A. Danchin, and P. Bertin. 2001. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 40:20-36. [DOI] [PubMed] [Google Scholar]
- 27.Horswill, A. R., and J. C. Escalante-Semerena. 1999. The prpE gene of Salmonella typhimurium LT2 encodes propionyl-CoA synthetase. Microbiology 145:1381-1388. [DOI] [PubMed] [Google Scholar]
- 28.Hosono, K., H. Kakuda, and S. Ichihara. 1995. Decreasing accumulation of acetate in a rich medium by Escherichia coli on introduction of genes on a multicopy plasmid. Biosci. Biotechnol. Biochem. 59:256-261. [DOI] [PubMed] [Google Scholar]
- 29.Hughes, K. T., K. L. Gillen, M. J. Semon, and J. E. Karlinsey. 1993. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science 262:1277-1280. [DOI] [PubMed] [Google Scholar]
- 30.Jiang, G. R., S. Nikolova, and D. P. Clark. 2001. Regulation of the ldhA gene, encoding the fermentative lactate dehydrogenase of Escherichia coli. Microbiology 147:2437-2446. [DOI] [PubMed] [Google Scholar]
- 31.Kalir, S., J. McClure, K. Pabbaraju, C. Southward, M. Ronen, S. Leibler, M. G. Surette, and U. Alon. 2001. Ordering genes in a flagella pathway by analysis of expression kinetics from living bacteria. Science 292:2080-2083. [DOI] [PubMed] [Google Scholar]
- 32.Kessler, D., and J. Knappe. 1996. Anaerobic dissimilation of pyruvate, p. 199-204. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 33.Khodursky, A. B., B. J. Peter, N. R. Cozzarelli, D. Botstein, P. O. Brown, and C. Yanofsky. 2000. DNA microarray analysis of gene expression in response to physiological and genetic changes that affect tryptophan metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:12170-12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kihara, M., and R. M. Macnab. 1981. Cytoplasmic pH mediates pH taxis and weak-acid repellent taxis of bacteria. J. Bacteriol. 145:1209-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirkpatrick, C., L. M. Maurer, N. E. Oyelakin, Y. N. Yoncheva, R. Maurer, and J. L. Slonczewski. 2001. Acetate and formate stress: opposite responses in the proteome of Escherichia coli. J. Bacteriol. 183:6466-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumari, S., C. M. Beatty, D. F. Browning, S. J. Busby, E. J. Simel, G. Hovel-Miner, and A. J. Wolfe. 2000. Regulation of acetyl coenzyme A synthetase in Escherichia coli. J. Bacteriol. 182:4173-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kutsukake, K. 1997. Autogenous and global control of the flagellar master operon, flhD, in Salmonella typhimurium. Mol. Gen. Genet. 254:440-448. [DOI] [PubMed] [Google Scholar]
- 38.Lambert, L. A., K. Abshire, D. Blankenhorn, and J. L. Slonczewski. 1997. Proteins induced in Escherichia coli by benzoic acid. J. Bacteriol. 179:7595-7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lasko, D. R., N. Zamboni, and U. Sauer. 2000. Bacterial response to acetate challenge: a comparison of tolerance among species. Appl. Microbiol. Biotechnol. 54:243-247. [DOI] [PubMed] [Google Scholar]
- 40.Lee, J., A. Goel, M. M. Ataai, and M. M. Domach. 1994. Flux adaptations of citrate synthase-deficient Escherichia coli. Ann. N. Y. Acad. Sci. 745:35-50. [DOI] [PubMed] [Google Scholar]
- 41.Lee, S. J., W. Boos, J. P. Bouche, and J. Plumbridge. 2000. Signal transduction between a membrane-bound transporter, PtsG, and a soluble transcription factor, Mlc, of Escherichia coli. EMBO J. 19:5353-5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee, S. Y. 1996. High cell-density culture of Escherichia coli. Trends Biotechnol. 14:98-105. [DOI] [PubMed] [Google Scholar]
- 43.Lehnen, D., C. Blumer, T. Polen, B. Wackwitz, V. F. Wendisch, and G. Unden. 2002. LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli. Mol. Microbiol. 45:521-532. [DOI] [PubMed] [Google Scholar]
- 44.Levit, M. N., Y. Liu, and J. B. Stock. 1998. Stimulus response coupling in bacterial chemotaxis: receptor dimers in signalling arrays. Mol. Microbiol. 30:459-466. [DOI] [PubMed] [Google Scholar]
- 45.Liu, X., and P. Matsumura. 1994. The FlhD-FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J. Bacteriol. 176:7345-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macnab, R. M. 1996. Flagella and motility, p. 123-145. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 47.Miller, J. H. 1992. A short course in bacterial genetics, p. 72-74. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Nam, T. W., S. H. Cho, D. Shin, J. H. Kim, J. Y. Jeong, J. H. Lee, J. H. Roe, A. Peterkofsky, S. O. Kang, S. Ryu, and Y. J. Seok. 2001. The Escherichia coli glucose transporter enzyme IICB(Glc) recruits the global repressor Mlc. EMBO J. 20:491-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noronha, S. B., H. J. Yeh, T. F. Spande, and J. Shiloach. 2000. Investigation of the TCA cycle and the glyoxylate shunt in Escherichia coli BL21 and JM109 using (13)C-NMR/MS. Biotechnol. Bioeng. 68:316-327. [PubMed] [Google Scholar]
- 50.Oh, M. K., L. Rohlin, K. C. Kao, and J. C. Liao. 2002. Global expression profiling of acetate-grown Escherichia coli. J. Biol. Chem. 277:13175-13183. [DOI] [PubMed] [Google Scholar]
- 51.Palacios, S., and J. C. Escalante-Semerena. 2000. prpR, ntrA, and ihf functions are required for expression of the prpBCDE operon, encoding enzymes that catabolize propionate in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 182:905-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pomposiello, P. J., M. H. Bennik, and B. Demple. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183:3890-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramakrishnan, R., M. Schuster, and R. B. Bourret. 1998. Acetylation at Lys-92 enhances signaling by the chemotaxis response regulator protein CheY. Proc. Natl. Acad. Sci. USA 95:4918-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rashid, M. H., and A. Kornberg. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:4885-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roe, A. J., C. O'Byrne, D. McLaggan, and I. R. Booth. 2002. Inhibition of Escherichia coli growth by acetic acid: a problem with methionine biosynthesis and homocysteine toxicity. Microbiology 148:2215-2222. [DOI] [PubMed] [Google Scholar]
- 56.Rossmann, R., G. Sawers, and A. Bock. 1991. Mechanism of regulation of the formate-hydrogenlyase pathway by oxygen, nitrate, and pH: definition of the formate regulon. Mol. Microbiol. 5:2807-2814. [DOI] [PubMed] [Google Scholar]
- 57.Sambrock, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 58.Shi, W., M. Bogdanov, W. Dowhan, and D. R. Zusman. 1993. The pss and psd genes are required for motility and chemotaxis in Escherichia coli. J. Bacteriol. 175:7711-7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shi, W., Y. Zhou, J. Wild, J. Adler, and C. A. Gross. 1992. DnaK, DnaJ, and GrpE are required for flagellum synthesis in Escherichia coli. J. Bacteriol. 174:6256-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shin, S., and C. Park. 1995. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J. Bacteriol. 177:4696-4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Slonczewski, J. L., and J. W. Foster. 1996. pH-regulated genes and survival at extreme pH, p. 1539-1549. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 62.Small, P., D. Blankenhorn, D. Welty, E. Zinser, and J. L. Slonczewski. 1994. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J. Bacteriol. 176:1729-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Soutourina, O., A. Kolb, E. Krin, C. Laurent-Winter, S. Rimsky, A. Danchin, and P. Bertin. 1999. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J. Bacteriol. 181:7500-7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Soutourina, O. A., E. Krin, C. Laurent-Winter, F. Hommais, A. Danchin, and P. N. Bertin. 2002. Regulation of bacterial motility in response to low pH in Escherichia coli: the role of H-NS protein. Microbiology 148:1543-1551. [DOI] [PubMed] [Google Scholar]
- 65.Sperandio, V., A. G. Torres, and J. B. Kaper. 2002. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol. 43:809-821. [DOI] [PubMed] [Google Scholar]
- 66.Suppmann, B., and G. Sawers. 1994. Isolation and characterization of hypophosphite-resistant mutants of Escherichia coli: identification of the FocA protein, encoded by the pfl operon, as a putative formate transporter. Mol. Microbiol. 11:965-982. [DOI] [PubMed] [Google Scholar]
- 67.Tanaka, Y., K. Kimata, and H. Aiba. 2000. A novel regulatory role of glucose transporter of Escherichia coli: membrane sequestration of a global repressor Mlc. EMBO J. 19:5344-5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Textor, S., V. F. Wendisch, A. A. De Graaf, U. Muller, M. I. Linder, D. Linder, and W. Buckel. 1997. Propionate oxidation in Escherichia coli: evidence for operation of a methylcitrate cycle in bacteria. Arch. Microbiol. 168:428-436. [DOI] [PubMed] [Google Scholar]
- 69.Tso, W. W., and J. Adler. 1974. Negative chemotaxis in Escherichia coli. J. Bacteriol. 118:560-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wei, B. L., A. M. Brun-Zinkernagel, J. W. Simecka, B. M. Pruss, P. Babitzke, and T. Romeo. 2001. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol. Microbiol. 40:245-256. [DOI] [PubMed] [Google Scholar]
- 71.Wei, Y., J. M. Lee, D. R. Smulski, and R. A. LaRossa. 2001. Global impact of sdiA amplification revealed by comprehensive gene expression profiling of Escherichia coli. J. Bacteriol. 183:2265-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wendisch, V. F., D. P. Zimmer, A. Khodursky, B. Peter, N. Cozzarelli, and S. Kustu. 2001. Isolation of Escherichia coli mRNA and comparison of expression using mRNA and total RNA on DNA microarrays. Anal. Biochem. 290:205-213. [DOI] [PubMed] [Google Scholar]
- 73.Wolfe, S. A., and J. M. Smith. 1988. Nucleotide sequence and analysis of the purA gene encoding adenylosuccinate synthetase of Escherichia coli K12. J. Biol. Chem. 263:19147-19153. [PubMed] [Google Scholar]
- 74.Yamamoto, K., R. M. Macnab, and Y. Imae. 1990. Repellent response functions of the Trg and Tap chemoreceptors of Escherichia coli. J. Bacteriol. 172:383-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang, Y. T., A. A. Aristidou, K. Y. San, and G. N. Bennett. 1999. Metabolic flux analysis of Escherichia coli deficient in the acetate production pathway and expressing the Bacillus subtilis acetolactate synthase. Metab. Eng. 1:26-34. [DOI] [PubMed] [Google Scholar]
- 76.Zimmer, D. P., E. Soupene, H. L. Lee, V. F. Wendisch, A. B. Khodursky, B. J. Peter, R. A. Bender, and S. Kustu. 2000. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc. Natl. Acad. Sci. USA 97:14674-14679. [DOI] [PMC free article] [PubMed] [Google Scholar]