Abstract
We describe the first freshwater members of the class Actinobacteria that have been isolated. Nine ultramicro-size (<0.1 μm3) strains were isolated from five freshwater habitats in Europe and Asia. These habitats represent a broad spectrum of ecosystems, ranging from deep oligotrophic lakes to shallow hypertrophic lakes. Even when the isolated strains were grown in very rich media, the cell size was <0.1 μm3 and was indistinguishable from the cell sizes of bacteria belonging to the smaller size classes of natural lake bacterioplankton. Hybridization of the isolates with oligonucleotide probes and phylogenetic analysis of the 16S rRNA gene sequences of the isolated strains revealed that they are affiliated with the class Actinobacteria and the family Microbacteriaceae. The previously described species with the highest levels of sequence similarity are Clavibacter michiganensis and Rathayibacter tritici, two phytopathogens of terrestrial plants. The 16S rRNA gene sequences of the nine isolates examined are more closely related to cloned sequences from uncultured freshwater bacteria than to the sequences of any previously isolated bacteria. The nine ultramicrobacteria isolated form, together with several uncultured bacteria, a diverse phylogenetic cluster (Luna cluster) consisting exclusively of freshwater bacteria. Isolates obtained from lakes that are ecologically different and geographically separated by great distances possess identical 16S rRNA gene sequences but have clearly different ecophysiological and phenotypic traits. Predator-prey experiments demonstrated that at least one of the ultramicro-size isolates is protected against predation by the bacterivorous nanoflagellate Ochromonas sp. strain DS.
The majority of the bacterial cells present in the bacterioplankton of marine and freshwater habitats are small (cell length, <1.5 μm; volume, <0.3 μm3). Even very small bacteria, which are termed ultramicrobacteria (<0.1 μm3), are always present, and they frequently represent the numerically predominant fraction of typical marine and freshwater bacterioplankton. In contrast, in culture the cells of almost all bacteria isolated from bacterioplankton are much larger (cell length, >1.5 μm; volume, >0.3 μm3) than most planktonic bacteria. In the case of several normally large-cell-size bacterial species, it has been shown that under strong starvation conditions the cell size decreases to dimensions typical of the majority of planktonic bacteria (23, 26, 27, 31). There are only a few bacterial strains that have been isolated from marine and soil habitats whose cells are in the ultramicro size range (volume, <0.1 μm3) during growth under lab conditions (4, 6, 19, 20, 29, 32, 33, 38). The most intensively investigated ultramicrobacterium is Sphingopyxis alaskensis (formerly Sphingomonas alsakensis [10, 37]), a marine bacterium belonging to the class Alphaproteobacteria. Strains of this species have been isolated from Resurrection Bay, Alaska, from the North Sea, and from coastal waters near Japan (6, 32, 33). Recently, Rappé et al. (29) isolated ultramicrobacterial strains from marine bacterioplankton which belong to the ubiquitous SAR11 cluster (Alphaproteobacteria). Iizuka et al. (19) isolated several ultramicrobacterial strains from urban soil. These strains are affiliated with the Betaproteobacteria, the Cytophaga-Flavobacterium-Bacteroides group, and the class Actinobacteria. Ultramicrobacteria belonging to the order Verrucomicrobiales have been isolated from anoxic rice paddy soil (20). To our knowledge, however, there have been no previous reports of isolation of ultramicrobacteria from freshwater sites.
Zwart and coworkers recently identified 34 putative phylogenetic clusters of bacteria which seem to contain typical freshwater inhabitants (41); 5 of these 34 clusters are affiliated with the class Actinobacteria (high-G+C-content gram-positive bacteria). Several studies demonstrated by cultivation-independent methods that these freshwater actinobacteria are present in a wide spectrum of ecologically different and globally distributed freshwater habitats (5, 9, 18, 24, 41, 42). In situ hybridization experiments with group-specific oligonucleotide probes revealed that actinobacteria may account for a large fraction (up to 60%) of the freshwater bacterioplankton (9). Furthermore, in this study the workers demonstrated that the cells of most of the hybridized freshwater actinobacteria are very small in situ.
In an intensive study Glöckner and coworkers (9) attempted to isolate typical freshwater actinobacteria but obtained no isolates. Pernthaler et al. (28) successfully enriched a phylotype of one of the freshwater clusters of actinobacteria in a continuous culture system but could not isolate the strain. Thus, despite the high numbers of cells of actinobacteria observed in freshwater samples and the regular occurrence of these organisms in clone libraries, no representatives of the freshwater actinobacterial lineages have been isolated so far (41).
We observed that up to 20% of the bacterioplankton cells in Lake Mondsee belong to a morphotype which is characterized by a selenoid morphology (vibrio) and an ultramicrobacterial cell size. We physically separated cells of this morphotype from the majority of the other cells by filtration through 0.2-μm-pore-size filters. Bacteria in the filtrate were slowly and stepwise acclimatized to artificial culture conditions. Large numbers of cultures were set up, and microscopic screening was used for identification of cultures containing higher numbers of selenoid morphotypes. The cell sizes of all nine strains of actinobacteria that were isolated were typical of the cell sizes of ultramicrobacteria (volume, <0.1 μm3).
It is well known that predation by bacterivorous protists is one of the factors responsible for structuring bacterioplankton communities (15, 35, 36). Therefore, we investigated the interaction of one of the ultramicrobacteria isolated with the bacterivorous predator Ochromonas sp. strain DS. Surprisingly, the predator was completely unable to graze on the bacterial strain offered. This is the first time that we have observed complete protection of a bacterial strain against grazing by the bacterivorous flagellate Ochromonas sp. strain DS.
MATERIALS AND METHODS
Media used for isolation and maintenance of bacteria.
Nutrient broth soyotone yeast extract (NSY) medium in several versions was used for isolation and maintenance of bacterial strains. NSY medium consists of an inorganic basal medium [75 mg of MgSO4 · 7H2O liter−1, 43 mg of Ca(NO3)2 · 4H2O liter−1, 16 mg of NaHCO3 liter−1, 5 mg of KCl liter−1, 3.7 mg of K2HPO4 · 3H2O liter−1, 4.4 mg of Na2EDTA liter−1, 3.2 mg of FeCl3 · 6H2O liter−1, 1.0 mg of H3BO3 liter−1, 0.2 mg of MnCl2 · 4H2O liter−1, 0.02 mg of ZnSO4 · 7H2O liter−1, 0.01 mg of CuSO4 · 6H2O liter−1, 0.01 mg of CoCl2 · 6H2O liter−1, 0.006 mg of Na2MoO4 · 2H2O liter−1, 0.1 mg of NiCl2 · 6H2O liter−1; pH 7.2] supplemented with equal amounts of nutrient broth, soyotone, peptone, and yeast extract (all obtained from Difco). The modifications of NSY medium were restricted to the concentrations of the organic supplements; the composition of the inorganic basal medium was kept constant.
For initial dilution in the isolation experiments (see below) and for maintenance of axenic Ochromonas sp. strain DS cultures, the inorganic basal medium (see above) was used. For the subsequent isolation procedure NSY media containing increasing concentrations of the organic supplements were used, and for maintenance of the isolated strains solid (15% agar) or liquid 3-g liter−1 NSY medium (NSY medium containing 1 g of each complex medium component liter−1) was used. For some purposes, 9-g liter−1 NSY medium (containing 3 g of each complex medium component liter−1) was used.
Sampling sites and isolation of actinobacteria.
We obtained samples from four ecologically contrasting lakes and one small pond (Table 1). Lake Mondsee and Lake Wolfgangsee are deep alpine lakes located in the Salzkammergut area near Salzburg, Austria. Pond 1 is a shallow artificial pond situated only 50 m from the shore of Lake Mondsee. Lake Constance (surrounded by Germany, Austria, and Switzerland) is the second largest prealpine lake. Lake Taihu, located in the Shanghai area, is very shallow, but it is third largest lake in the People's Republic of China.
TABLE 1.
Characteristics of aquatic habitats from which actinobacterial strains were isolated
| Habitat | Country(ies) | Trophic state | Mixing type | Surface area (km2) | Maximum depth (m) | Vol (106 × m3) | Residence time (yr) | Distance from Lake Mondsee (km) |
|---|---|---|---|---|---|---|---|---|
| Lake Mondsee | Austria | Oligomesotrophic | Dimictic | 14 | 68 | 510 | 1.8 | |
| Pond 1 | Austria | Eutrophic | Homogeneous | 0.0002 | 1.3 | 2 × 10−5 | NKa | 0.05 |
| Lake Wolfgangsee | Austria | Oligotrophic | Dimictic | 13 | 114 | 619 | 3.9 | 3.5 |
| Lake Constance | Austria, Germany, Switzerland | Oligomesotrophic | Monomictic | 571 | 252 | 48,530 | 4.5 | 300 |
| Lake Taihu | People's Republic of China | Hypertrophic | Homogeneous | 2,428 | 2.6 | 5,140 | 0.8 | 10,000 |
NK, not known.
Five- to twenty-milliliter water samples were filtered through sterile 0.2-μm-pore-size filters (Minisart syringe filter; Sartorius, Goettingen, Germany) for isolation of bacteria. Subsamples (0.01, 0.1, 0.5, and 1.0 ml) of the filtrate were transferred to wells of sterile 24-well cell culture plates. The volume in each well was adjusted to 1 ml by adding the appropriate volume of temperature-adapted inorganic basal medium. One to five 24-well plates were prepared for each of the four inoculum volumes. The diluted subsamples were acclimatized over periods of a few days in 2°C steps from the in situ temperature to 15°C. After temperature acclimatization was completed, the plates were incubated at 15°C. Addition of organic substrates (NSY medium) was begun during the temperature acclimatization procedure. Each well received increasing doses of 5, 10, 20, 50, 100, 300, 600, and 1,000 mg of the NSY medium organic substrates liter−1 (assumed concentrations in the wells after substrate addition) by addition of appropriate volumes of 3-g liter−1 NSY medium. Substrate addition was carried out stepwise at 2- to 4-day intervals. Microscopic screening of cultures for growth of ultramicro-size selenoid bacteria was started a few days after substrate addition was finished. Subsamples from single wells were stained with 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma), filtered onto 0.2-μm-pore-size Nuclepore filters (Millipore), and inspected by epifluorescence microscopy (Zeiss Axioplan) at a magnification of ×1,250 (16). Subsamples of positive cultures were diluted with 3 g-liter−1 NSY medium and plated on 3-g liter−1 NSY agar plates. Samples of colonies on agar plates were suspended in sterile liquid medium and microscopically checked for the presence of small selenoid morphotypes. Positive colonies were recultured in liquid NSY medium and plated again on NSY agar. The colonies were subcultured until a pure culture was obtained. Established pure cultures were stored at −70°C in 3-g liter−1 NSY medium containing 10% glycerol. All work done after sampling was carried out under sterile conditions.
Electron microscopy.
Mid-log-phase cells of four isolates grown in 9- or 3-g liter−1 NSY medium were conventionally fixed and embedded in epoxy resin, as described by Yakimov et al. (40). For Pt-C shadow casting, prefixed cells were adsorbed to Formvar-coated 300-mesh copper grids for 15 to 90 s, blotted with filter paper (Schleicher and Schuell, Dueren, Germany), and air dried. Samples were unidirectionally shadowed at a 15° angle with an approximately 4-nm Pt-C layer by using a MED 020 evaporation unit (BAL-TEC; Balzers, Liechtenstein). Shadow-cast samples and ultrathin sections were analyzed by using a transmission electron microscope with an integrated energy filter (Zeiss CEM 902) at an acceleration voltage of 80 kV, and zero-loss images (energy selective slit, 30 eV; objective aperture, 30 μm; condenser aperture, 400 μm) were recorded at primary magnifications of ×7,000 to ×30,000.
Sequencing of 16S rRNA genes and phylogenetic analysis.
Amplification and sequencing of the 16S rRNA gene of the isolates were performed as described by Moore et al. (25). To search for related sequences with high similarity values, the program BLAST (1; http://www.ncbi.nlm.nih.gov/BLAST/) was used. A comparative sequence analysis was performed by using the program package ARB (http://www.arb-home.de). The ARB database, which already contained ca. 12,000 aligned 16S rRNA gene sequences, was supplemented with missing high-similarity sequences found by using BLAST. Sequences imported into the ARB database were aligned by using the automatic alignment function of ARB and then were visually checked for correct alignment. Phylogenetic trees were reconstructed on the basis of homologous 1,439-nucleotide sequence stretches. Alignment positions at which less than 50% of the sequences in the entire data set had the same residues were excluded from the calculations. Evolutionary distances were corrected for multiple substitutions by using the algorithm of Jukes and Cantor (22). Partial sequences (<1,439 nucleotides) which were found by detailed phylogenetic analyses to be closely related to the sequences of the isolates analyzed were added to the existing neighbor-joining tree by a parsimony procedure.
Determination of microbial numbers and sizes.
Bacterial and flagellate numbers were determined as described previously (13). For determination of bacterial cell volumes, images of >100 cells per sample (pure cultures of isolates) or >500 cells per sample (environmental samples) were collected with a monochrome charge-coupled device camera (Hitachi Denshi), and sizes were determined with the LUCIA D image analysis systems (Lucia 4.51; resolution, 750 by 520 pixels; 256 grey levels; Laboratory Imaging, Prague, Czech Republic). Bacterial cell volumes were calculated by using the formula of Andersson et al. (2).
Flagellate predation of strain MWH-Mo1. (i) Microorganisms.
In two experiments we examined the influence of grazing by the flagellate Ochromonas sp. strain DS (13) on isolate MWH-Mo1 (Table 2). In controls or as an alternative food for the flagellate, we used Pseudomonas sp. and Brevundimonas diminuta strains.
TABLE 2.
Characterization of the ultramicrobacterial strains investigated
| Strain | Origin | Pigmentation | Cell vol (μm3)a | Growth rate (h−1)e
|
16S rRNA genotype | Accession no. | |
|---|---|---|---|---|---|---|---|
| Mean | SD | ||||||
| MWH-Mo1 | Lake Mondsee | Yellow | 0.052 | 0.152 | 0.009 | 1 | AJ507460 |
| 0.056 ± 0.002b | |||||||
| 0.056 ± 0.001c | |||||||
| MWH-Mo2 | Lake Mondsee | Yellow | 0.057 | NDf | 3 | AJ507461 | |
| MWH-Mo3 | Lake Mondsee | Yellow | 0.056d | 0.11 | 0.003 | 4 | AJ507462 |
| MWH-Po1 | Pond 1 | Yellow | 0.058 | ND | 5 | AJ507463 | |
| MWH-Wo1 | Lake Wolfgangsee | Yellow | 0.054 | 0.164 | 0.006 | 1 | AJ507464 |
| MWH-Bo1 | Lake Constance | Yellow | 0.061 | ND | 5 | AJ507465 | |
| MWH-Ta1 | Lake Taihu | Yellow | 0.051 | 0.177 | 0.001 | 1 | AJ507466 |
| MWH-Ta2 | Lake Taihu | Yellow | 0.056 | 0.183 | 0.008 | 2 | AJ507467 |
| MWH-Ta3 | Lake Taihu | Red | 0.083 | 0.151 | 0.001 | 6 | AJ507468 |
Exponential phase of growth in 9-g liter−1 NSY medium at room temperature (ca. 22°C), unless indicated otherwise.
Exponential phase of growth in 3-g liter−1 NSY medium at room temperature (ca. 20°C). Average ± standard deviation for three replicates.
Exponential phase of growth in 9-mg liter−1 NSY medium at room temperature (ca. 20°C). Average ± standard deviation for three replicates.
Exponential phase of growth in 3-g liter−1 NSY medium at room temperature (ca. 22°C). The strain cannot grow in 9-g liter−1 NSY medium.
Growth rate in 3-g liter−1 NSY medium at 20°C. Triplicate preparations were used in this analysis.
ND, not determined.
(ii) Precultures.
The bacterivorous flagellate Ochromonas sp. strain DS was cultured axenically in inorganic basal medium enriched with heat-killed Pseudomonas sp. cells. B. diminuta, Pseudomonas sp., and strain MWH-Mo1 were precultured in liquid 9-g liter−1 NSY medium until they reached the stationary phase; then they were harvested by centrifugation, washed, and resuspended in inorganic basal medium. For better comparability the size of the usually large (length, >2 μm) Pseudomonas cells was decreased by starvation to a mean cell length of 1.15 μm ± 0.30 μm. The small Pseudomonas cells were harvested by centrifugation, washed, resuspended in inorganic basal medium, killed with heat (70°C, 2 h), and stored at −20°C until they were used. Stationary-phase cells of B. diminuta were relatively small (cell length, <1.5 μm); therefore, the cell size was not decreased by starvation.
(iii) Experimental design.
Two similar batch experiments were performed (Table 3). In experiment 1 the batches were incubated with illumination, and in experiment 2 the batches were incubated without illumination. Each experiment consisted of triplicate batches containing the flagellate Ochromonas sp. strain DS and isolate MWH-Mo1, as well as controls which tested the grazing activity of the flagellate. In experiment 1 the control (control 1) contained the flagellate and heat-killed Pseudomonas sp. cells. In experiment 2 four different control experiments were performed. The control 2a− preparation contained Ochromonas sp. strain DS and heat-killed Pseudomonas sp. Control 2b− contained the flagellate, and B. diminuta was added as prey instead of Pseudomonas sp. The other two controls (controls 2a+ and 2b+) contained in addition to either Pseudomonas sp. or B. diminuta (prey 1) low numbers of isolate MWH-Mo1 (prey 2). In both of these controls the initial number of MWH-Mo1 cells was approximately 1% of the number of prey 1 cells. The two bacterial populations present in these treatments were clearly distinguishable microscopically due to pronounced differences in cell morphology (rods versus selenoid cells) and cell size. Details concerning the initial and final numbers of prey organisms tested are shown in Table 3.
TABLE 3.
Batch experiments to examine predation by and growth of the flagellate Ochromonas sp. strain DS with strain MWH-Mo1a
| Prepn | No. of replicates | Incubation conditions | Prey 1
|
Prey 2
|
||||
|---|---|---|---|---|---|---|---|---|
| Strain | Initial no. (106 cells ml−1) (SD) | Final no. (106 cells ml−1) (SD) | Strain | Initial no. (106 cells ml−1) | Final no. (106 cells ml−1) | |||
| Expt 1 | ||||||||
| Expt 1 | 3 | Illumination | MWH-Mo1 | 14.0 (0.6) | 10.1 (1.2)b | None | ||
| Control 1 | 3 | Illumination | Pseudomonas sp. | 13.8 (1.4) | 0.0c | None | ||
| Expt 2 | ||||||||
| Expt 2 | 3 | Darkness | MWH-Mo1 | 18.2 (1.7) | 12.7 (0.4)d | None | ||
| Control 2a− | None | Darkness | Pseudomonas sp. | 7.4 | 0.0e | None | ||
| Control 2a+ | None | Darkness | Pseudomonas sp. | 8.1 | 0.0e | MWH-Mo1 | 0.08 | 10.4f |
| Control 2b− | None | Darkness | B. diminuta | 7.1 | 0.0f | None | ||
| Control 2b+ | None | Darkness | B. diminuta | 7.0 | 0.0f | MWH-Mo1 | 0.07 | 3.2f |
All preparations were initially inoculated with low numbers of Ochromonas sp. strain DS cells (experiment 1, 1.3 × 103 cells ml−1; experiment 2, 2.7 × 103 cells ml−1). Two of the control preparations received in addition to the prey bacteria low numbers of strain MWH-Mo1 cells. All preparations were incubated at 15°C without shaking. Sampling of the control preparations was stopped after 20 days, while samples were obtained from the experimental preparations for 38 days (experiment 1) and 35 days (experiment 2).
Data obtained on day 38.
Data obtained on day 4.
Data obtained on day 20 (before addition of heat-killed Pseudomonas sp. [Fig. 4]).
Data obtained on day 5.
Data obtained on day 20.
(iv) Performance of experiments.
Erlenmeyer flasks filled with 300 ml of inorganic basal medium were inoculated with high numbers of bacterial cells and low numbers of Ochromonas sp. strain DS cells (Table 3). After the initial inoculation with bacteria only the three experiment 2 preparations (Table 3) received additional inocula 20 days after the start of the experiment. This treatment consisted of addition of high numbers of heat-killed Pseudomonas sp. cells (see Fig. 4). No organic substrates which could support the growth of bacteria were added. The preparations were incubated at 15°C without permanent shaking. The development of the bacterial and flagellate populations was monitored for 5 weeks. Samples were removed, fixed with formaldehyde, and analyzed microscopically.
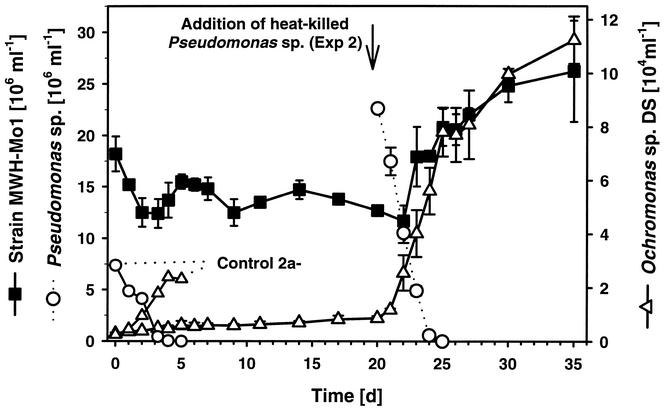
FIG. 4.
Predation of strain MWH-Mo1 by Ochromonas sp. in experiment 2. See Table 3. The development of the flagellate and the MWH-Mo1 population in experiment 2 (based on triplicate preparations) and the development of Ochromonas sp. strain DS and heat-killed Pseudomonas sp. in control 2a− are shown. Data from the other three controls (controls 2a+, 2b−, and 2b+) are not shown. In experiment 2 the three preparations were initially inoculated with isolate MWH-Mo1, and 20 days after the start of the experiment they were inoculated with heat-killed Pseudomonas sp. cells. For experiment 2 the average cell numbers and standard deviations (error bars) for the three parallel preparations are shown.
Nucleotide sequence accession numbers.
The nucleotide sequence accession numbers for the 16S rRNA gene sequences of the strains investigated are shown in Table 2.
RESULTS
Isolation.
Water samples from oligomesotrophic Lake Mondsee (Table 1) contained up to 3 × 105 small (volume, <0.1 μm3) selenoid bacterial cells per ml. These cells accounted for up to ca. 20% of the bacterioplankton cells. Filtration of samples from Lake Mondsee through 0.2-μm-pore-size filters decreased the number of the selenoid bacteria to 10 to 20% of the number before filtration. However, the more important result of the filtration analysis was the almost complete physical separation of the small selenoid bacteria from the other planktonic bacteria. In filtrates microscopically checked immediately after filtration, only the small selenoid bacteria could be detected. Enumeration of the bacteria in the 0.2-μm-pore-size filtrates by using 0.02-μm-pore-size Anodisc filters yielded slightly higher but not significantly different numbers of bacteria compared to the numbers obtained by using 0.2-μm-pore-size Nuclepore filters. The small selenoid bacteria in the filtrates were very frequently overgrown by initially undetectable large filamentous bacteria. Therefore, we split the filtrates into smaller inocula and diluted these inocula with sterile medium (see Materials and Methods). This treatment reduced the percentage of cultures which were overgrown from nearly 100% to 0 to 70% (depending on the dilution).
For successful enrichment and isolation of the target bacteria, growing a large number of liquid cultures was necessary. For more than 95% of the cultures that were established and screened enrichment or isolation of the target bacteria was not successful. Furthermore, we could not predict which dilution would result in the highest percentage of cultures dominated by the small selenoid bacteria. In the case of liquid cultures which contained, after the acclimatization process was complete, high numbers of selenoid cells and no or only a few other morphotypes, the probability of obtaining colonies from the selenoid bacteria was high. All of the selenoid ultramicrobacteria that were finally isolated grew slowly on agar plates (at 20 to 25°C), and most of the strains formed only small or very small colonies. For some strains handling single colonies was very difficult. Usually, successful isolation of the selenoid bacteria took several weeks (i.e., from initial filtration until establishment of a pure culture growing on agar plates).
In total, we isolated more than 30 strains with ultramicro cell sizes and selenoid morphology. Most of these strains are yellow pigmented (which is not clearly visible in the case of small colonies), and only a few are red pigmented. The red-pigmented strains were isolated only from Lake Taihu, while yellow-pigmented strains were obtained from all five ecosystems investigated. Fluorescence in situ hybridization with the oligonucleotide probe HGC69a, which is group specific for the class Actinobacteria (30), revealed that all of the pigmented selenoid strains isolated belong to this class (data not shown). In order to avoid studying identical clones, we restricted our investigation to nine isolates which were either isolated from different study sites or were isolated from different samples taken from the same site. These nine isolates include eight yellow-pigmented strains and one red-pigmented strain.
Electron microscopy.
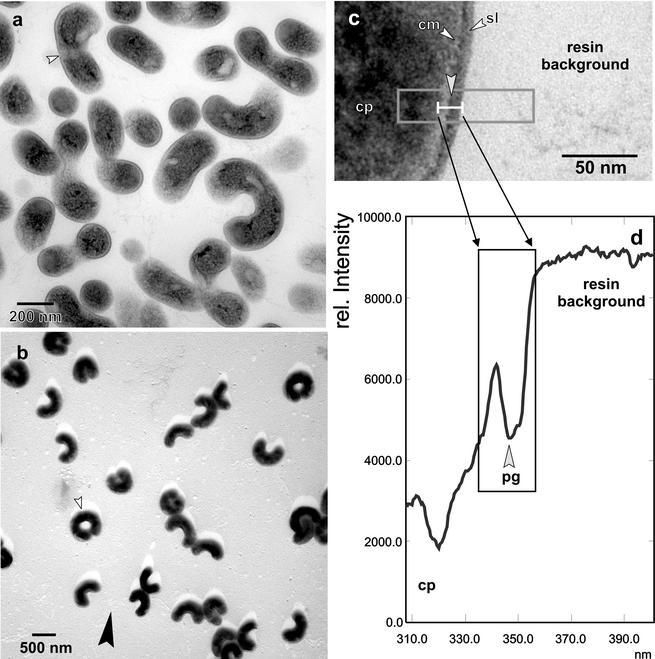
Mid-log-phase cells of isolates MWH-Mo1, MWH-Mo3, and MWH-Ta1 and the red-pigmented strain MWH-Ta3, which represent three different 16S rRNA genotypes (see below), were investigated by transmission electron microscopy. This investigation revealed almost no significant differences in cell shape and cellular ultrastructure for these four isolates. The only clear difference found was the larger cell size of strain MWH-Ta3. The latter finding is in accordance with the results of the epifluorescence microscopic size analysis (Table 2). Due to the overall similarities the following ultrastructural description is restricted to isolate MWH-Ta1 (Fig. 1).
FIG. 1.
(a and b) Survey views of ultrathin-sectioned (a) and shadow-cast (b) cells of strain MWH-Ta1. The small arrowheads indicate sites of binary fission. The large arrowhead in panel b indicates the direction of shadow casting. (c) Detailed view of a cross section of the cell wall. The cytoplasm (cp), the cytoplasmic membrane (cm), and the outer surface-like layer (sl) are indicated. The area enclosed by a box represents the measured area, and an averaged line scan density profile of this area is shown in panel d. The density features of the cell wall are shown in the area enclosed by a box in panel d. pg, peptidoglycan.
The cell width of strain MWH-Ta1 generally ranged from 154.2 to 244.8 nm, with a mean (n = 79) of 190.9 ± 15.6 nm. The overall shape was selenoid, and occasionally cells formed rings, when dissociation did not occur after septum formation was completed (Fig. 1b). When ultrathin sections were examined, the cytoplasm showed the general characteristics and did not contain electron-dense or translucent deposits (Fig. 1a). The cytoplasm was outlined by the cytoplasmic membrane (Fig. 1c), which did not exhibit the characteristic double track upon conventional osmium fixation but appeared as a bright single-layer rim. The cytoplasmic membrane was covered by an electron-dense layer, which was in lieu of a murein meshwork and was resolved as an individual peak in the averaged density profile (Fig. 1d). The outermost layer often exhibited a particulate substructure and did not have a characteristic membranous architecture, which ruled out the existence of an outer membrane. Therefore, this constituent of the cell wall has to be regarded as an S-layer-like surface (Fig. 1c). Thus, strain MWH-Ta1 has a characteristic gram-positive cell wall architecture, and the overall thickness of the cell wall, including the cytoplasmic membrane, ranges from 11.1 to 27.8 nm, with an average of 16.5 ± 3.3 nm (n = 72).
Size characteristics of the isolates and natural bacterioplankton cells.
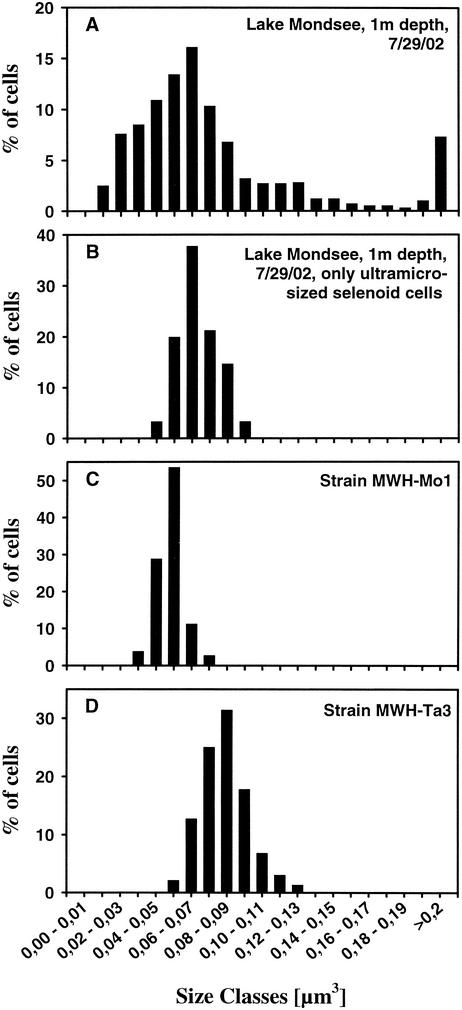
While cell lengths of isolates measured on electron microscopic and epifluorescence microscopic images were similar, cells widths determined by epifluorescence microscopy were approximately 50% greater than the cell widths determined by electron microscopy for the same strains. Calculation of cell volumes was based on cell lengths and widths determined by epifluorescence microscopy. Thus, the cell volumes of ultramicro-size selenoid cells shown in Table 2 and Fig. 2 may overestimate the real cell volumes of the cells. The cells of all nine isolates characterized, however, clearly are ultramicro size (volume, <0.1 μm3), even when they are grown in rich organic medium (Table 2). The cell sizes determined for all eight yellow-pigmented isolates were very similar, while the cells of red-pigmented isolate MWH-Ta3 were markedly larger. No impact of substrate concentration on the sizes of the cells of the isolated strains was observed. When the organism was grown in medium with a low substrate concentration (9 mg liter−1), the size of MWH-Mo1 cells was not different from the size of cells grown in the same medium with a 333-fold-higher substrate concentration (Table 2).
FIG. 2.
Bacterial size distribution of bacterioplankton in Lake Mondsee (including selenoid cells) (A), of ultramicro-size selenoid bacteria in Lake Mondsee (B), of isolate MWH-Mo1 grown in 9-g liter−1 NSY medium (C), and of isolate MWH-Ta3 grown in 9-g liter−1 NSY medium (D).
The selenoid portion of the Lake Mondsee bacterioplankton could be divided into two subgroups, a ultramicro-size subgroup (Fig. 2B) and a subgroup containing organisms whose cell volumes were more than 0.1 μm3. The morphology and cell sizes of the selenoid bacteria belonging to the ultramicro-size class were identical to the morphology and cell sizes of the strains isolated (Fig. 2).
Comparative analysis of 16S rRNA gene sequences.
The almost complete 16S rRNA gene sequences obtained for the nine isolates investigated clearly showed that all these strains are affiliated with the class Actinobacteria and thus support the results obtained by fluorescence in situ hybridization. The previously described species which are the most closely related to the isolates belong to the family Microbacteriaceae (Fig. 3).
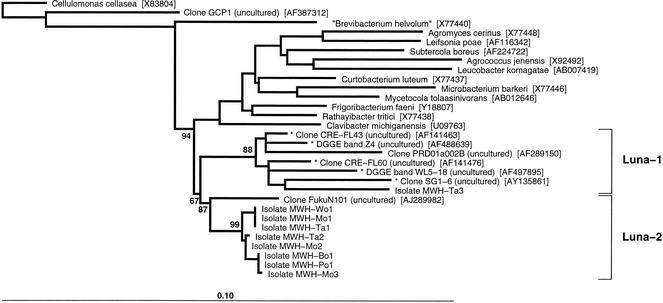
FIG. 3.
Neighbor-joining tree based on homologous 1,439-nucleotide sequence stretches of the 16S rRNA gene. Shorter partial sequences which were added by parsimony to the existing neighbor-joining tree are indicated by asterisks. Only the bootstrap values (percentage, 1,000 replicates) relevant for the Luna cluster are shown. The tree was rooted by using the sequence of Cellulomonas cellasea (Cellulomonadaceae). Except for the C. cellasea, clone GCP1, and “Brevibacterium helvolum” sequences, all the reference sequences are sequences of members of the family Microbacteriaceae. The positions of the Luna cluster and its two subclusters (Luna-1 and Luna-2) are indicated by brackets. The Luna cluster was named after the ecosystem from which the first isolate was obtained (Lake Mondsee [German Mond = English moon = Latin luna]). Bar = 1 nucleotide substitution per 10 nucleotides. DGGE, denaturing gradient gel electrophoresis.
The eight yellow-pigmented ultramicrobacteria are closely related to each other and have levels of 16S rRNA gene similarity of 99.4 to 100%. The sequence variability among these isolates was exclusively in a 337-nucleotide fragment (positions 75 to 412, Escherichia coli numbering) of the gene and was restricted to only nine nucleotide positions. Based on the 16S rRNA gene sequences, five genotypes could be distinguished (Table 2). One genotype was represented by three isolates with identical 16S rRNA gene sequences, and another genotype was represented by two isolates with identical 16S rRNA gene sequences. The different isolates with identical sequences were obtained from different ecosystems; e.g., isolate MWH-Mo1 was obtained from the deep oligomesotrophic Lake Mondsee in Austria, and isolate MWH-Ta1 was obtained from the very shallow hypertrophic Lake Taihu in the People's Republic of China. We sequenced a fragment of the 16S rRNA gene which included the fragment containing all nine variable sequence positions of the eight strains three times (strain MWH-Mo1) and two times (strain MWH-Ta1) without finding any differences in the sequences. Furthermore, the yellow-pigmented isolates showed sequence similarities of 97.0 to 97.5% with the sequence of the uncultured bacterium FukuN101 obtained from Lake Fuchskuhle in northern Germany (9). The previously described species with the highest levels of similarity to the yellow-pigmented isolates were two phytophathogens of terrestrial plants, Clavibacter michiganensis (95.7 to 96.0%) and Rathayibacter tritici (95.7 to 95.9%).
The red-pigmented isolate MWH-Ta3 exhibited levels of 16S rRNA similarity of 94.5 to 94.6% to the yellow-pigmented isolates and of 94.1% to the uncultured freshwater bacterium PRD01a002B from Parker River in Massachusetts (41). Five smaller partial sequences (466 to 588 bp), which also clustered with MWH-Ta3 (Fig. 3), exhibited levels of sequence similarity of 94.1 to 97.5%. All five of these sequences have been obtained from freshwater habitats (reference 5 and three unpublished studies). Two other partial sequences exhibited levels of sequence similarity of ca. 93% but did not consistently cluster with MWH-Ta3 in all of the analyses performed. These two partial sequences were obtained by sequencing denaturing gradient gel electrophoresis bands from bacteria which inhabited a high-salinity solar saltern pond (3).
The 16S rRNA gene sequences of all nine ultramicrobacteria isolated form, together with the sequences of the uncultured freshwater actinobacteria mentioned above, a phylogenetic cluster (Luna cluster) (Fig. 3). Depending on the sequence data set used for analysis, the Luna cluster is supported by bootstrap values (1,000 replicates) ranging from 67 to 94%. The two subclusters (Luna-1 and Luna-2) (Fig. 3) tended in most of the analyses to be supported by higher bootstrap values than the whole Luna cluster.
In contrast to the Luna cluster, which currently consists exclusively of isolates or sequences obtained from freshwater sites, all other known organisms or sequences which are related to the Luna cluster have been obtained from terrestrial sites.
Predation of strain MWH-Mo1.
In both predation experiments the level of MWH-Mo1 decreased slightly during the first few days but never dropped below 10 × 106 cells ml−1 (Fig. 4 and Table 3). In contrast, in the control treatments containing only Pseudomonas sp. or B. diminuta (controls 1, 2a−, and 2b−) the flagellate decreased the number of bacteria to less than 0.1 × 106 cells ml−1 within 4 days (heat-killed Pseudomonas sp.) and 9 days (live B. diminuta) (Table 3). In the control treatments containing in addition to the main prey species small numbers of strain MWH-Mo1 (controls 2a+ and 2b+), the decreases in the numbers of Pseudomonas sp. and B. diminuta were indistinguishable from the decreases in the numbers of these organisms in the controls which did not contain MWH-Mo1. In these control treatments, however, strong increases in the numbers of strain MWH-Mo1 cells to final concentrations of 3.2 × 106 cells ml−1 (control 2b+) and 10.4 × 106 cells ml−1 (control 2a+) were observed. Due to the growth of strain MWH-Mo1 the total bacterial concentrations in both controls with two prey strains were never less than 2 × 106 cells ml−1.
An increase in the number of Ochromonas sp. strain DS cells was observed for all treatments. The increase was quick and large for all control treatments but slow and small for the treatments which contained only strain MWH-Mo1 as prey. In experiment 2 (incubation in the dark) the numbers of Ochromonas sp. cells increased during the first 20 days only inconsiderably from the initial concentration of 0.3 × 104 cells ml−1 to 0.8 × 104 cells ml−1 (Fig. 4). After addition of an alternative prey, heat-killed Pseudomonas sp., the number of flagellate cells quickly increased to >8 × 104 cells ml−1 (Fig. 4). Simultaneous with this increase the number of Pseudomonas sp. cells dropped below the detection limit and the number of MWH-Mo1 cells doubled.
DISCUSSION
Isolation.
It is well known that bacteria with selenoid morphologies (vibrios) frequently comprise high percentages of the freshwater bacterioplankton cells (8). We focused in this study on isolation of ultramicro-size selenoid bacteria. In our isolation strategy we combined physical separation of target bacteria with stepwise acclimatization of bacteria to artificial culture conditions. Filtration through 0.2-μm-pore-size filters separated the target bacteria from the majority of bacterioplankton cells; however, separation of the target bacteria remained incomplete. Therefore, further dilution steps were necessary for better separation of the target cells from bacteria able to overgrow the target bacteria.
We found 25-year-old reports on the occurrence of small selenoid bacteria in filtrates, which passed through 0.1- to 0.45-μm-pore-size filters (21), but we found no reports of successful isolation of bacteria with such morphology. Vybiral et al. (39) isolated a bacterial strain from marine water samples filtered through 0.2-μm-pore-size filters. In contrast to our isolates, the cells of this strain were large when the organism was grown in culture.
Sixth cluster of freshwater members of the class Actinobacteria.
Zwart et al. (41) suggested five putative clusters of freshwater actinobacteria. In general, these authors defined a putative cluster of typical freshwater bacteria as a monophyletic group of 16S rRNA gene sequences containing at least two sequences that are at least 95% identical and originating from the pelagic zones of at least two different freshwater sites. The Luna cluster fulfills all these criteria; thus, this cluster can be suggested to be a sixth putative cluster of freshwater actinobacteria. The monophyletic nature of the Luna cluster is indicated by bootstrap values that range from 67 to 94% (depending on data set used for analysis). The lower bootstrap values observed in some analyses and the observed minimum sequence similarity of 93.8% within the whole Luna cluster may indicate that the cluster will probably fall apart with the appearance of new sequences.
Data currently available for members of the Luna cluster clearly demonstrate that this cluster contains bacteria distributed in diverse freshwater ecosystems at least in Europe, Asia, and North America. Despite this wide distribution no data on abundance of members of the Luna cluster in freshwater ecosystems are currently available. Specific in situ studies are needed to reveal the contributions of these bacteria to the bacterioplankton of different freshwater ecosystems.
Isolation of strains with identical 16S rRNA gene sequences from ecologically contrasting lakes.
The occurrence of bacteria with identical or very similar 16S rRNA gene sequences in diverse freshwater ecosystems separated by great distances was previously observed by Zwart et al. (42). We isolated from five different ecosystems five strains which shared an identical 16S rRNA gene sequence with at least one other strain (Table 2). Two strains with identical sequences (MWH-Mo1 and MWH-Ta1) were isolated from the most dissimilar ecosystems included in our study (Table 1). These two strains had clear differences in phenotypic and ecophysiological traits; they had clearly different colony morphologies, significantly (P < 0.01, as determined by a t test) different growth rates at 20°C (Table 2), and clearly different temperature optima (data not shown). Thus, we assume that these two isolates do not represent the same genetic clone.
Grazing protection of strain MWH-Mo1.
We observed no vulnerability of strain MWH-Mo1 to grazing by Ochromonas sp. strain DS. On the contrary, in the presence of the flagellate strain MWH-Mo1 responded to grazing on alternative prey with a strong increase in the number of cells (Fig. 4 and Table 3). This was observed in experiment 2, as well as in two independent control treatments. The increase in the number of MWH-Mo1 cells was more pronounced in the presence of heat-killed alternative prey. Obviously, strain MWH-Mo1 benefited from the release of organic matter during the grazing of the flagellate. The presence of the potential competitor B. diminuta may have reduced this benefit.
The observed small and slow increase in the number of cells of the mixotrophic flagellate Ochromonas sp. strain DS without a simultaneous decrease in the number of strain MWH-Mo1 cells is thought to result from phototrophy (experiment 1, with illumination) and starvation-induced reductive cell division (experiment 2, without illumination). Reductive cell division by starved Ochromonas sp. strain DS cells has been observed previously (17), and this phenomenon has also been observed with other flagellate species (7).
The observed slight decrease in the number of strain MWH-Mo1 cells during the first few days in both experiments might have been a result of the transfer from a rich organic medium to the inorganic basal medium. Furthermore, centrifugation, washing, and resuspension may have had a negative influence on these ultramicrobacteria. However, we observed no correlation between the initial decrease in the number of MWH-Mo1 cells and the increase in the number of flagellate cells (Fig. 4). Thus, it is unlikely that the initial decrease in the number of bacteria was a result of flagellate grazing. The persistence of ≥1 × 107 MWH-Mo1 cells ml−1 despite the presence of 0.1 × 104 to 11.0 × 104 cells of actively grazing Ochromonas sp. strain DS ml−1 clearly demonstrates that strain MWH-Mo1 was protected against grazing by Ochromonas sp. strain DS. It seems that this grazing protection cannot be explained solely by the small cell size of strain MWH-Mo1. The theory of size selectivity in interception feeding of protists suggests that the efficiency of grazing decreases as the prey size decreases (11), but no lower prey size limit principally restricts uptake of small prey particles. Actually, uptake of virus particles (12) and high-molecular-weight polysaccharides (34) has been demonstrated. We previously observed ingestion of 0.5-μm-diameter latex beads, whose volume was similar to that of MWH-Mo1 cells, by Ochromonas sp. strain DS (14). Even particles having diameters less then 0.4 μm can be ingested by Ochromonas sp. strain DS (Boenigk, unpublished data). We also observed in an experiment performed with a complex bacterial community that Ochromonas sp. strain DS was able to decrease the absolute numbers of cells in all bacterial size classes present in the experiment (14). This included four ultramicro size classes which include the cell sizes determined for strain MWH-Mo1. Thus, we concluded that the protection of strain MWH-Mo1 against predation by Ochromonas sp. strain DS is at least partially due to a size-independent trait.
Despite the fact that the flagellate Ochromonas sp. strain DS has been used for grazing experiments with a phylogenetically wide range of bacterial strains (13, 14, 15, 16, 17; unpublished data), grazing protection like that observed for strain MWH-Mo1 has not been found before either for grazing by Ochromonas sp. strain DS or for grazing by another bacterivorous protist species (15). Pernthaler et al. (28) observed in experiments with a bacterial community that predation by Ochromonas sp. strain DS increased the absolute abundance of a phylotype of a member (Ac1) of one of the other clusters of freshwater actinobacteria (ACK-M1) (41). The increase was very similar to the increases which we observed in our experiments with strain MWH-Mo1. It is not known, however, whether the increase in the abundance of Ac1 was the result of grazing protection or whether Ac1 simply overcompensated for grazing losses by fast growth. In contrast to the experiment of Pernthaler et al., the actinobacterial population investigated in our experiments received no substrates. Thus, we eliminated the possibility that the observed grazing protection of strain MWH-Mo1 was the result of fast growth.
Further experiments are needed to reveal whether the protection of strain MWH-Mo1 is restricted to grazing by Ochromonas sp. strain DS and whether other strains belonging to the Luna cluster or other freshwater actinobacteria also have grazing protection.
Access to members of the globally distributed Luna cluster in culture should provide an unusual opportunity for further studies of the ecophysiology of both freshwater ultramicrobacteria and freshwater actinobacteria. The relatively close phylogenetic relationship of the freshwater Luna cluster strains to several typical terrestrial actinobacteria may provide an opportunity to gain more insight into the evolutionary processes which enabled soil actinobacteria to successfully invade freshwater ecosystems. The observed relatively thin cell walls of the strains isolated is probably one evolutionary adaptation which contributed to the success of this invasion. We assume, however, that many more evolutionary adaptations are necessary for invasion of habitats which are ecologically very different from the ecosystems inhabited by the organisms in the past.
Acknowledgments
We thank Elke Barth (GBF) for skillful preparation of electron microscopic specimens, Julia Bötel (GBF) for skillful sequencing of most of the isolates, and Nicholas D. Crosbie for correcting the English.
This work was supported and M.S. was partially supported by the Austrian Science Fund (project P15655 awarded to M.W.H.). Q.W. was supported by a scholarship from the North-South Dialogue Program, Ministry of Foreign Affairs, Austria.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, A., U. Larsson, and Å. Hagström. 1986. Size-selective grazing by a microflagellate on pelagic bacteria. Mar. Ecol. Prog. Ser. 33:51-57. [Google Scholar]
- 3.Benlloch, S., A. Lopez-Lopez, E. O. Casamayor, L. Ovreas, V. Goddard, F. L. Daae, G. Smerdon, R. Massana, I. Jiont, F. Thingstad, C. Pedros-Alio, and F. Rodriguez-Valera. 2002. Prokaryotic genetic diversity throughout the salinity gradient of a coastal solar saltern. Environ. Microbiol. 4:349-360. [DOI] [PubMed] [Google Scholar]
- 4.Connon, S., and S. J. Giovannoni. 2002. High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl. Environ. Microbiol. 68:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crump, B. C., E. V. Armbrust, and J. A. Baross. 1999. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl. Environ. Microbiol. 65:3192-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eguchi, M., M. Ostrowski, F. Fegatella, J. Bowman, D. Nichols, T. Nishino, and R. Cavicchioli. 2001. Sphingomonas alaskensis strain AFO1, an abundant oligotrophic ultramicrobacterium from the North Pacific. Appl. Environ. Microbiol. 67:4945-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fenchel, T. 1982. Ecology of heterotrophic microflagellates. III. Adaptation to heterogeneous environments. Mar. Ecol. Prog. Ser. 8:25-33. [Google Scholar]
- 8.Fischer, U. R., and B. Velimirov. 2000. Comparative study of the abundance of various bacterial morphotypes in a eutrophic freshwater environment determined by AODC and TEM. J. Microbiol. Methods 39:213-224. [DOI] [PubMed] [Google Scholar]
- 9.Glöckner, F. O., E. Zaichikov, N. Belkova, L. Denissova, J. Pernthaler, A. Pernthaler, and R. Amann. 2000. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl. Environ. Microbiol. 66:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Godoy, F., M. Vancanneyt, M. Martínez, A. Steinbuchel, J. Swings, and B. H. A. Rehm. Sphingopyxis chilensis sp. nov., a chlorophenol-degrading bacterium which accumulates polyhydroxyalkanoate, and transfer of Sphingomonas alaskensis to Sphingopixis alaskensis comb. nov. Int. J. Syst. Evol. Microbiol., in press. [DOI] [PubMed]
- 11.Gonzáles, J. M. 1996. Efficient size-selective bacterivory by phagotrophic nanoflagellates in aquatic ecosystems. Mar. Biol. 126:785-789. [Google Scholar]
- 12.Gonzáles, J. M., and C. A. Suttle. 1993. Grazing by marine nanoflagellates on viruses and virus-sized particles: ingestion and digestion. Mar. Ecol. Prog. Ser. 94:1-10. [Google Scholar]
- 13.Hahn, M. W., and M. G. Höfle. 1998. Grazing pressure by a bacterivorous flagellate reverses the relative abundance of Comamonas acidovorans PX54 and Vibrio strain CB5 in chemostat cocultures. Appl. Environ. Microbiol. 64:1910-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn, M. W., and M. G. Höfle. 1999. Flagellate predation on a bacterial model community: interplay of size-selective grazing, specific bacterial cell size, and bacterial community composition. Appl. Environ. Microbiol. 65:4863-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hahn, M. W., and M. G. Höfle. 2001. Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microbiol. Ecol. 35:113-121. [DOI] [PubMed] [Google Scholar]
- 16.Hahn, M. W., E. R. B. Moore, and M. G. Höfle. 1999. Bacterial filament formation, a defense mechanism against flagellate grazing, is growth rate controlled in bacteria of different phyla. Appl. Environ. Microbiol. 65:25-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hahn, M. W., E. R. B. Moore, and M. G. Höfle. 2000. Role of microcolony formation in the protistan grazing defense of the aquatic bacterium Pseudomonas sp. MWH1. Microb. Ecol. 39:175-185. [DOI] [PubMed] [Google Scholar]
- 18.Hirons, W. D., B. A. Methé, S. A. Nierzwickibauer, and J. P. Zehr. 1997. Bacterial diversity in Adirondack mountain lakes as revealed by 16S rRNA gene sequences. Appl. Environ. Microbiol. 63:2957-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iizuka, T., S. Yamanaka, T. Nishiyama, and A. Hiraishi. 1998. Isolation and phylogenetic analysis of aerobic copiotrophic ultramicrobacteria from urban soil. J. Gen. Appl. Microbiol. 44:75-84. [DOI] [PubMed] [Google Scholar]
- 20.Janssen, P. H., A. Schuhmann, E. Morschel, and F. A. Rainey. 1997. Novel anaerobic ultramicrobacteria belonging to the Verrucomicrobiales lineage of bacterial descent isolated by dilution culture from anoxic rice paddy soil. Appl. Environ. Microbiol. 63:1382-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, R. C. 1977. The spirochetes. Annu. Rev. Microbiol. 31:89-106. [DOI] [PubMed] [Google Scholar]
- 22.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism, vol. 3. Academic Press, New York, N.Y.
- 23.Kjelleberg, S., N. Albertson, K. Flärdh, L. Holmquist, Å. Jouper-Jaan, R. Marouga, J. Östling, B. Svenblad, and D. Weichart. 1993. How do nondifferentiating bacteria adapt to starvation? Antonie Leeuwenhoek 63:333-341. [DOI] [PubMed] [Google Scholar]
- 24.Lindström, E. S., and E. Leskinen. 2002. Do neighboring lakes share common taxa of bacterioplankton? Comparison of 16S rRNA gene fingerprints and sequences from three geographic regions. Microb. Ecol. 44:1-9. [DOI] [PubMed] [Google Scholar]
- 25.Moore, E. R. B., M. Mau, A. Arnscheidt, E. C. Bittger, R. A. Hutson, M. D. Collins, Y. Van de peer, R. De Wachter, and K. N. Timmis. 1996. The determination and comparison of the 16S rRNA gene sequences of species of the genus Pseudomonas (sensu stricto) and estimation of the natural intrageneric relationships. Syst. Appl. Microbiol. 19:478-492. [Google Scholar]
- 26.Morita, R. Y. 1985. Starvation and miniaturisation of heterotrophs, with special emphasis on maintenance of the starved viable state, p. 111-130. In M. Fletcher and G. D. Floodgate (ed.), Bacteria in their natural environment. Academic Press, London, England.
- 27.Morita, R. Y. 1997. Bacteria in oligotrophic environments. Starvation-survival lifestyle. Chapman & Hall, New York, N.Y.
- 28.Pernthaler, J., T. Posch, K. Simek, J. Vrba, A. Pernthaler, F. O. Glöckner, U. Nübel, R. Psenner, and R. Amann. 2001. Predator-specific enrichment of actinobacteria from a cosmopolitan freshwater clade in mixed continuous culture. Appl. Environ. Microbiol. 67:2145-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rappé, M. S., S. A. Connon, K. L. Vergin, and S. J. Giovannoni. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630-633. [DOI] [PubMed] [Google Scholar]
- 30.Roller, C., M. Wagner, R. Amann, W. Ludwig, and K.-H. Schleifer. 1994. In situ probing of Gram-positive bacteria with high DNA G+C content using 23S rRNA-targeted oligonucleotides. Microbiology 140:2849-2858. [DOI] [PubMed] [Google Scholar]
- 31.Roszak, D. B., and R. R. Colwell. 1987. Survival strategies of bacteria in the natural environment. Microbiol. Rev. 51:365-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schut, F., E. J. Devries, J. C. Gottschal, B. R. Robertson, W. Harder, R. A. Prins, and D. K. Button. 1993. Isolation of typical marine bacteria by dilution culture: growth, maintenance, and characteristics of isolates under laboratory conditions. Appl. Environ. Microbiol. 59:2150-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schut, F., J. C. Gottschal, and R. A. Prins. 1997. Isolation and characterization of the marine ultramicrobacterium Sphingomonas sp. strain RB2256. FEMS Microbiol. Rev. 20:363-369. [Google Scholar]
- 34.Sherr, E. B. 1988. Direct use of high molecular weight polysaccharide by heterotrophic flagellates. Nature 335:348-351. [Google Scholar]
- 35.Simek, K., J. Armengol, M. Comerma, J. C. Garcia, P. Kojecka, J. Nedoma, and J. Hejzlar. 2001. Changes in the epilimnetic bacterial community composition, production, and protist-induced mortality along the longitudinal axis of a highly eutrophic reservoir. Microb. Ecol. 42:359-371. [DOI] [PubMed] [Google Scholar]
- 36.Simek, K., J. Pernthaler, M. G. Weinbauer, K. Hornak, J. R. Dolan, J. Nedoma, M. Masin, and R. Amann. 2001. Changes in bacterial community composition and dynamics and viral mortality rates associated with enhanced flagellate grazing in a mesoeutrophic reservoir. Appl. Environ. Microbiol. 67:2723-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vancanneyt, M., F. Schut, C. Snauwaert, J. Goris, J. Swings, and J. C. Gottschal. 2001. Sphingomonas alaskensis sp. nov., a dominant bacterium from a marine oligotrophic environment. Int. J. Syst. Evol. Microbiol. 51:73-79. [DOI] [PubMed] [Google Scholar]
- 38.Velimirov, B. 2001. Nanobacteria, ultramicrobacteria and starvation forms: a search for the smallest metabolizing bacterium. Microbes Environ. 16:67-77. [Google Scholar]
- 39.Vybiral, D., E. B. M. Denner, C. M. Haller, H. J. Busse, A. Witte, M. G. Höfle, and B. Velimirov. 1999. Polyphasic classification of 0.2 μm filterable bacteria from the western Mediterranean Sea. Syst. Appl. Microbiol. 22:635-646. [DOI] [PubMed] [Google Scholar]
- 40.Yakimov, M. M., P. N. Golyshin, S. Lang, E. R. B. Moore, W.-R. Abraham, H. Lünsdorf, and K. N. Timmis. 1998. Alcanivorax borkumensis gen. nov., sp. nov., a new, hydrocarbon-degrading and surfactant-producing marine bacterium. Int. J. Syst. Bacteriol. 48:339-348. [DOI] [PubMed] [Google Scholar]
- 41.Zwart, G., B. C. Crump, M. P. Kamst-van Agterveld, F. Hagen, and S. K. Han. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28:141-155. [Google Scholar]
- 42.Zwart, G., W. D. Hirons, B. A. Methé, M. P. van Agterveld, R. Huismans, S. C. Nold, J. P. Zehr, and H. J. Laanbroek. 1998. Nearly identical 16S rRNA gene sequences recovered from lakes in North America and Europe indicate the existence of clades of globally distributed freshwater bacteria. Syst. Appl. Microbiol. 21:546-556. [DOI] [PubMed] [Google Scholar]