Abstract
The bacterial community composition of a linuron-degrading enrichment culture and the role of the individual strains in linuron degradation have been determined by a combination of methods, such as denaturing gradient gel electrophoresis of the total 16S rRNA gene pool, isolation and identification of strains, and biodegradation assays. Three strains, Variovorax sp. strain WDL1, Delftia acidovorans WDL34, and Pseudomonas sp. strain WDL5, were isolated directly from the linuron-degrading culture. In addition, subculture of this enrichment culture on potential intermediates in the degradation pathway of linuron (i.e., N,O-dimethylhydroxylamine and 3-chloroaniline) resulted in the isolation of, respectively, Hyphomicrobium sulfonivorans WDL6 and Comamonas testosteroni WDL7. Of these five strains, only Variovorax sp. strain WDL1 was able to use linuron as the sole source of C, N, and energy. WDL1 first converted linuron to 3,4-dichloroaniline (3,4-DCA), which transiently accumulated in the medium but was subsequently degraded. To the best of our knowledge, this is the first report of a strain that degrades linuron further than the aromatic intermediates. Interestingly, the rate of linuron degradation by strain WDL1 was lower than that for the consortium, but was clearly increased when WDL1 was coinoculated with each of the other four strains. D. acidovorans WDL34 and C. testosteroni WDL7 were found to be responsible for degradation of the intermediate 3,4-DCA, and H. sulfonivorans WDL6 was the only strain able to degrade N,O-dimethylhydroxylamine. The role of Pseudomonas sp. strain WDL5 needs to be further elucidated. The degradation of linuron can thus be performed by a single isolate, Variovorax sp. strain WDL1, but is stimulated by a synergistic interaction with the other strains isolated from the same linuron-degrading culture.
When pesticides are used correctly, they can save up to 40% in crop losses, but misuse or overuse of these chemicals can have considerable environmental and public health consequences (37). Phenylurea herbicides are among the most widely used herbicides in non-crop areas, as well as in tree crops (49). Given the slow natural attenuation rate in various soils with respect to mineralization of the phenyl structure (half-life in soil of 38 to 67 days) (3) and the potential carcinogenic risk of these herbicides and their potential intermediates such as the chloroanilines (40, 47, 48), there is a serious need to develop remediation processes to eliminate or minimize contamination of surface and groundwater.
Linuron [3-(3,4-dichlorophenyl)-1-methoxy-1-methylurea] is a substituted phenylurea herbicide used widely to selectively control newly established broadleaf weeds and grasses in fruit and field crops, cereals, and shelter belts (3). Although (photo)chemical and physical processes may be involved in the removal of this compound, biodegradation is reported to be the most significant mechanism for its dissipation from soil (3). Several enrichment cultures have indeed been obtained that use linuron as the sole source of nitrogen and carbon (5, 12, 13, 38, 39). Although these consortia rapidly mineralized linuron and demonstrated a stable degradation activity upon subculture, isolation of the key organisms seemed to be difficult. Roberts et al. (38, 39) and El-Fantroussi et al. (13) isolated a linuron-degrading mixed culture from a linuron-treated soil, but failed to obtain a pure culture of the organism or organisms responsible for the degradation of the herbicide. Only Wallnöfer (53) and Cullington et al. (5, 50), respectively, isolated Bacillus sphaericus ATCC 12123 and Arthrobacter globiformis D47, which can use the alkyl chain of linuron as the sole N and C source. However, these pure strains can only partially degrade linuron, since 3,4-dichloroaniline (3,4-DCA), one of the main potential metabolites in the degradation pathway of linuron (Fig. 1), accumulated in the medium. Since the chloroaniline metabolites are more toxic and persistent than their phenylurea mother compounds (47, 48), complete mineralization of these phenylurea herbicides is required and deserves further investigation. One reason for incomplete microbial degradation is the presence of a variety of substituted groups on the aromatic backbone, each of which requires slightly different catabolic enzymes for total breakdown. The collection of these enzymes would more likely be present in consortia than in single bacteria.
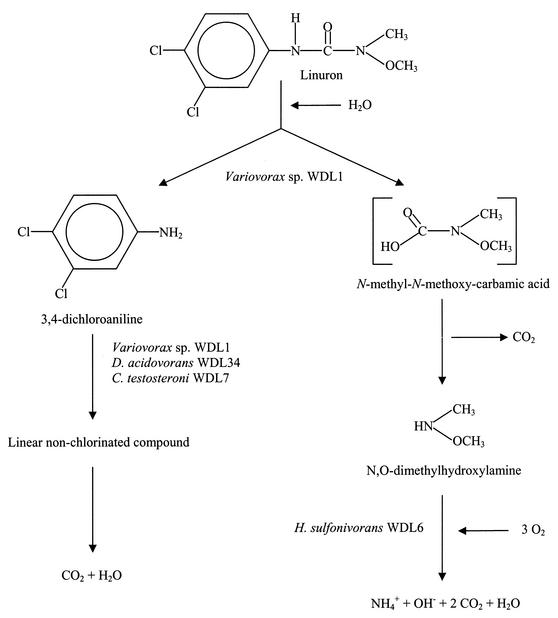
FIG. 1.
Proposed degradation pathway of linuron (14, 53) and flow of the carbon and nitrogen through the different members of the linuron enrichment culture, as suggested by the data presented in our study.
In our laboratory, a linuron-degrading mixed culture was enriched from a soil that had been treated for 10 years with this herbicide (12). Several attempts to isolate the strain(s) responsible for the degradation of this phenylurea herbicide failed, although different isolation conditions were used (13). Therefore, the goal of the present study was to further unravel this mixed culture in order to find the key players responsible for complete degradation of linuron. Several subcultures of the existing linuron-degrading enrichment culture were set up in media with several possible intermediates of the linuron degradation pathway, such as different (chloro)anilines and N,O-dimethylhydroxylamine (N,O-DMHA), used as the sole C and N sources (Fig. 1). From each culture, the most abundant bacteria, as estimated by denaturing gradient gel electrophoresis (DGGE) of 16S rRNA genes, were isolated by plating, and their role in the degradation of linuron was examined.
MATERIALS AND METHODS
Chemicals.
Analytical-grade linuron was purchased from Chem Service (West Chester, Great Britain). Aniline and 3,4-DCA were purchased from Sigma-Aldrich Chemie (Steinheim, Germany). 3-Chloroaniline (3-CA), 4-chloroaniline (4-CA), and hydroxylamine hydrochloride were purchased from Fluka AG Chemische Fabrik (Buchs, Switzerland). Finally, N,O-DMHA was purchased from Janssen Chimica (Geel, Belgium).
Media.
Mineral medium MMN (mineral medium without nitrogen and carbon) is derived from mineral medium MMO by elimination of all nitrogen (2). MMN medium contained 1,419.6 mg of Na2HPO4, 1,360.9 mg of KH2PO4, 98.5 mg of MgSO4, 5.88 mg of CaCl2 · 2H2O, 1.16 mg of H3BO4, 2.78 mg of FeSO4 · 7H2O, 1.15 mg ZnSO4 · 7H2O, 1.69 mg of MnSO4 · H2O, 0.38 mg of CuSO4 · 5H2O, 0.24 mg of CoCl2 · 6H2O, 0.10 mg of MoO3, and 3.2 mg of EDTA in 1 liter of distilled water. Linuron, aniline, 3-CA, 4-CA, 3,4-DCA, and N,O-DMHA were added as the sole N and C sources, while hydroxylamine was used as an N source in the presence of the C source methanol. Succinate was added to the agar medium (2 g/liter) to obtain better colony growth on plates. Nutrient agar (Merck) and Luria Bertani (LB) medium (10 g of Bacto peptone [Difco, Detroit, Mich.], 5 g of Bacto yeast extract [Difco], and 5 g of NaCl in 1 liter of demineralized water) were used as rich media. A 1/10 volume of LB medium contained 1 g of tryptone, 0.5 g of yeast extract, and 5 g of NaCl. When needed, these media were solidified with 15 g of agar per liter: agar-agar hochrein (Merck, Germany) for MMN medium and agar pulvis (Federa-Bufa, Haren, Belgium) for LB medium.
Bacterial enrichment cultures.
An enrichment culture degrading the N′-methoxy-methyl phenylurea herbicides linuron and metobromuron was previously obtained from soil samples (Royal Research Station, Gorsem, Belgium) with a 10-year history of treatment with a mixture of linuron, diuron, and simazine (12). This enrichment culture was maintained in MMN medium with 250 mg of linuron per liter as the sole source of N and C. In addition, different subcultures were set up in MMN medium, in which aniline, 3-CA, 4-CA (gradually increased from 50 to 400 mg/liter over a period of 1 month), 3,4-DCA (increased from 50 to 80 mg/liter), and N,O-DMHA (increased from 50 to 200 mg/liter) were added separately as the sole source of N and C. To initiate these subcultures, a 1% (vol/vol) inoculum from the original linuron-degrading culture was inoculated in these media, and all batch cultures were incubated at 28°C with shaking at 140 rpm. To maintain the cultures (Table 1), a 1% (vol/vol) inoculum was transferred every 2 weeks to fresh MMN medium with the respective compound as the sole N and C source.
TABLE 1.
Bacterial cultures and strains used and isolated in this study
| Culture or straina | Isolated from | Characteristicsb | Source or reference |
|---|---|---|---|
| Cultures | |||
| Linuron-degrading culture | Soil treated with linuron, diuron, and simazine | L+, 3,4-DCA+, 3-CA+, 4-CA+, aniline+, N,O-DMHA+, metobromuron+ | 12 |
| N,O-DMHA-degrading subculture | Linuron-degrading culture | L−, 3,4-DCA−, 3-CA−, 4-CA−, aniline−, N,O-DMHA+ | This study |
| 3,4-DCA-degrading subculture | Linuron-degrading culture | L−, 3,4-DCA+, 3-CA+, 4-CA+, aniline+, N,O-DMHA− | This study |
| 3-CA-degrading subculture | Linuron-degrading culture | L−, 3,4-DCA+, 3-CA+, 4-CA+, aniline+, N,O-DMHA− | This study |
| 4-CA-degrading subculture | Linuron-degrading culture | 4-CA+, other compounds not tested | This study |
| Aniline degrading subculture | Linuron-degrading culture | Aniline+, other compounds not tested | This study |
| Strains | This study | ||
| Variovorax sp. strain WDL1 | Linuron-degrading culture | L+, 3,4-DCA+, 3-CA+, 4-CA+, aniline+, N,O-DMHA? | This study |
| Delftia acidovorans WDL34 | Linuron-degrading culture, | L−, 3,4-DCA+, 3-CA+, 4-CA+, aniline+, N,O-DMHA− | This study |
| 3,4-DCA-culture | |||
| Pseudomonas sp. strain WDL5 | Linuron-degrading culture, 3,4-DCA culture, N,O-DMHA culture | L−, 3,4-DCA−, 3-CA−, 4-CA−, aniline−, N,O-DMHA− | This study |
| H. sulfonivorans WDL6 | N,O-DMHA culture | L−, 3,4-DCA−, 3-CA−, 4-CA−, aniline−, N,O-DMHA+ | This study |
| Comamonas testosteroni WDL7 | 3-CA culture | L−, 3,4-DCA+, 3-CA+, 4-CA+, aniline+, N,O-DMHA− | This study |
Strains were isolated from a linuron-degrading culture that had been transferred over 25 months and from a 3,4-DCA-, 3-CA-, or N,O-DMHA-degrading culture transferred over 4 months.
+ and −, used or not used, respectively, as the sole source of C, N, and energy.
Bacterial strains and cultivation conditions.
The bacterial strains isolated or used in this study are presented in Table 1. All strains were cultured at 28°C. Variovorax sp. strain WDL1 was cultured on MMN agar plates with 2 g of succinate and 100 mg of linuron per liter. Delftia acidovorans WDL34, Pseudomonas sp. strain WDL5, and Comamonas testosteroni WDL7 were grown on MMN agar plates with 2 g of succinate and 75 mg of 3,4-DCA per liter, and Hyphomicrobium sulfonivorans WDL6 was grown on MMN agar plates with 200 mg of N,O-DMHA per liter. Liquid cultures of strains to be used in growth and degradation tests and for isolation of total genomic DNA were always prepared in 1/10 LB medium with 30 mg of linuron, 3,4-DCA, or N,O-DMHA per liter depending on the degradation characteristics of the strains. A fully grown culture of Variovorax sp. strain WDL1 was only obtained after 4 days, while all the other strains were grown overnight. Enrichment cultures were grown for 4 days in MMN medium with the respective compound as the sole N and C source.
Identification of the isolates.
The obtained isolates were identified by sequencing of their nearly complete 16S rRNA gene as described by Coenye et al. (4).
Degradation and growth experiments.
Cultures of all the tested strains, grown in 1/10 LB medium supplemented with linuron, 3,4-DCA, or N,O-DMHA and enrichment cultures grown in MMN medium with the appropriate N and C source were pelleted, washed, and suspended in MMN medium. After the optical density at 590 nm (OD590) had been adjusted to 1.0, a 1% inoculum (1 ml) was brought into 100 ml of MMN medium with a single dosage of 50 mg of linuron or 30 mg of 3,4-DCA per liter. When combinations of strains were used, a 1% (vol/vol) inoculum of each strain was inoculated. Every day, the concentration of linuron and 3,4-DCA was determined by high-performance liquid chromatography (HPLC) analysis (7). To elucidate whether the isolated strains could use N,O-DMHA and hydroxylamine as the sole source of C or N, these compounds were added to 100 ml of MMN medium containing a 1% bacterial inoculum. Every day, the flasks were spiked either with additional doses of 30 mg of N,O-DMHA per liter (as an N and C source) or with 15 or 30 mg of hydroxylamine per liter (as an N source) together with 250 or 500 mg of methanol per liter (as a C source). Growth was determined by measuring the OD590 (Kontron Uvikon spectrophotometer, model 932). All degradation and growth tests were performed in triplicate and repeated at least three times. The results shown and described here are averages of one such experiment performed in triplicate. The results of the repeated experiments were very similar and completely confirmed the results shown here.
Chemical analyses.
Supernatants of bacterial cultures were analyzed by reverse-phase HPLC after the cells were removed by centrifugation (10 min at 5,000 × g). In the Summit HPLC system (Dionex, Wommelgem, Belgium) a Hypersil Green env column (150- by 8-mm inner diameter; 5-μm particle size; Alltech, Deerfield, Ill.), a mobile phase of CH3OH-0.1% H3PO4 (70/30), a flow rate of 0.8 ml/min, and a UV detector set at 210 nm were used (7). The concentrations of chloride ions were determined by ion chromatography after filtration of the samples through a 0.22-μm-pore-diameter filter. The Dionex ion chromatograph analyzer DX-600 (Dionex) consisted of an Ionpac AS9-HC column (250- by 4-mm inner diameter, 9-μm particle size; Dionex) with a Guard AG9-HC pre-column (80- by 4-mm inner diameter; Dionex), and a mobile phase of Na2CO3 (9 mM), with a flow rate of 1 ml/min (7). The detection limits were 1 mg/liter for both HPLC and ion chromatography analysis.
DGGE.
Total genomic DNA was prepared from 100-ml enrichment cultures in MMN medium, from 100 ml of Variovorax sp. strain WDL1 in 1/10 LB medium, and from 5 ml of the other strains in 1/10 LB medium, based on a previously described protocol (1). To obtain the DNA of colony mixtures resuspended from plates, colonies were scraped off with a loop. After washing the plates with 2 ml of water, the total suspension was subjected to DNA extraction. One microliter of total genomic DNA extract was used as a template for 50 μl of PCR master mix with the bacterium-specific 16S rRNA forward primer P63f and the reverse primer P518r (12, 29, 32). The PCR products contain a GC clamp of 40 bases, added to the forward primer, and have a total length of 531 bp (based on the reference strain Escherichia coli K-12). PCR products were subjected to DGGE analysis based on the protocols of Muyzer et al. (31) and El-Fantroussi et al. (12). In brief, PCR samples were run for 16 h at 45 V and 60°C on a 6% (wt/vol) polyacrylamide gel with a denaturing gradient ranging from 45 to 60% (where 100% denaturant contains 7 M urea and 40% formamide). After electrophoresis, the DGGE gels were stained with SYBR GreenI nucleic acid gel stain and photographed.
DNA cloning and sequencing.
The 16S rRNA genes of the different bacteria present in the linuron, 3,4-DCA, and N,O-DMHA cultures were amplified with the primers P63f and P1378r as previously described (18). The amplified products of approximately 1,300 bp were cloned into the PCR-TOPO 2.1 cloning vector (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. The insert of approximately 46 recombinant (white) colonies for each culture was amplified by using the P63f-GC and P518r primers as previously described (12). The DGGE pattern of the clones was compared with those of the respective consortia, and fragments that migrated to the same height in a DGGE gel were supposed to originate from the same species.
Nucleotide sequence accession number.
The nucleotide sequences for the strains WDL1, WDL34, WDL6, WDL5, and WDL7 have been deposited in the GenBank database under accession no. AF538929 to AF538933.
RESULTS
Enrichment of a linuron-degrading mixed culture on possible intermediates of the linuron degradation pathway.
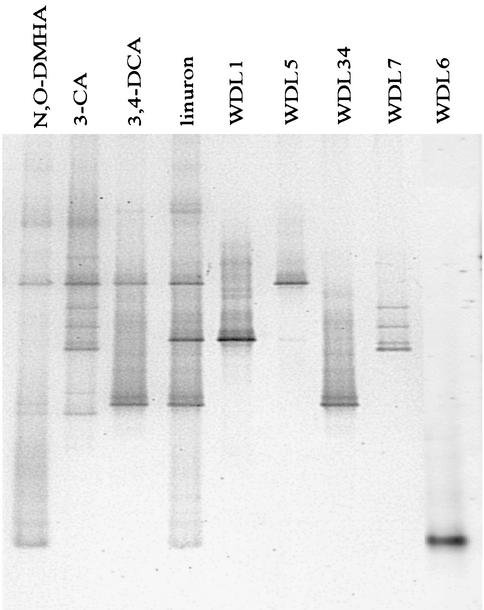
In a previous study, a linuron-degrading mixed culture was enriched from a soil that had been treated with this herbicide for more than 10 years (12), but several attempts to isolate the strain or strains responsible for the degradation of linuron failed (13). In order to unravel the enrichment culture, which still revealed a quite complex DGGE pattern (13), subcultures were made by growing the 15-month-old linuron-degrading culture in medium that contained possible intermediates of the linuron degradation pathway (Fig. 1) as the sole substrate for growth. Growth and degradation tests performed on the different subcultures indicated that the original linuron-degrading culture contained organisms that can use 3,4-DCA (80 mg/liter), 3-CA (400 mg/liter), 4-CA (400 mg/liter), aniline (400 mg/liter), and N,O-DMHA (200 mg/liter) as the sole source of N, C, and energy (Table 1). Interestingly, all of the different subcultures had lost their ability to degrade linuron (25 mg/liter) as quickly as 4 months after being enriched on one of the intermediates (corresponding to 16 transfers; data not shown). In addition, comparison of the DGGE pattern (Fig. 2) of the linuron-degrading culture with those of the subcultures revealed that the latter had microbial community structures different from that of the original linuron-degrading culture. First of all, the very dominant strain WDL1, which was always represented in the DGGE pattern of the linuron enrichment culture, was never noticed in any of the subcultures. Since the same 16S rRNA gene fragment has previously been found to be missing in DGGE patterns of resuspended colonies after plating the linuron-degrading culture on different media or in DGGE patterns of 3,4-DCA-degrading subcultures (13), our data strongly confirmed the previously stated hypothesis (i.e., that the corresponding bacterial strain WDL1 may be the key player in the initial transformation of linuron). Second, certain bacteria of the linuron-degrading culture that were barely visible in the original DGGE pattern, seemed to have been enriched by the addition of N,O-DMHA and 3-CA (see bands WDL6 and WDL7 in Fig. 2). Analysis of a 16S rRNA gene clone library of the 3,4-DCA- and N,O-DMHA-degrading cultures also provided a rough estimate of the numerically dominant members and confirmed the DGGE results (Table 2). Subculture of the linuron-degrading mixed culture on metabolites of the linuron degradation pathway has thus allowed detection of additional bacteria that are potentially involved in degradation of intermediates. We further used the linuron-, 3-CA-, 3,4-DCA-, and N,O-DMHA-degrading cultures to isolate those bacteria and characterize their roles in the linuron-degrading mixed culture.
FIG. 2.
Representation of the DGGE patterns of the different cultures and the isolated strains used in this study. The first four lanes represent the DGGE patterns of the different enrichment cultures, and the last five lanes represent the bacteria isolated from these cultures. The DGGE patterns of the aniline and 4-chloroaniline-degrading cultures were the same as those of the 3,4-DCA-degrading culture (data not shown).
TABLE 2.
Composition of the microbial community structure of the different consortia as determined by 16S rRNA gene clone libraries
| Strain | % of clones with same 16S rRNA gene insert
|
||
|---|---|---|---|
| Linuron-degrading culture (51 mo old) | 3,4-DCA-degrading culture (4 mo old) | N,O-DMHA-degrading culture (4 mo old) | |
| Variovorax sp. strain WDL1 | 24 | ||
| D. acidovorans WDL34 | 38 | 70 | |
| Pseudomonas sp. strain WDL5 | 0a | 30 | |
| H. sulfonivorans WDL6 | 25 | 100 | |
| Other bacteria | 13 | ||
Determined as 0%, although the WDL5 band was clearly visible in the DGGE pattern.
Elucidation of the different enrichment cultures by the isolation of strains through plating.
Since degradation tests and DGGE analysis revealed that the bacterial strain that performed the first step in the degradation of linuron was only present in the linuron-degrading culture, different attempts were made to isolate this strain from the 25-month-old mixed culture by plating on agar media. Only the mixture of bacterial colonies resuspended from the mineral MMN agar medium with linuron (100 mg/liter) and succinate (2 g/liter) could degrade linuron and contained strain WDL1 in its DGGE pattern, while the colony mixtures recovered from LB and nutrient agar plates did not (data not shown). These data confirm that strain WDL1 seems to be responsible for the degradation of linuron and also indicate that this strain only grew sufficiently on mineral medium MMN with linuron and succinate, but not on the rich media. After an incubation period of 2 weeks at 28°C, three dominant colony morphologies were distinguished on the MMN plates. As shown in Fig. 2, the band in the DGGE gel that corresponded to the yellow colony type (called WDL1) migrated to the same position as band WDL1, which was only found in the linuron-degrading culture. The DGGE band of the white glistening circular colony type (called “WDL5” in Fig. 2) is present in all of the different cultures. Finally the DGGE band of the white colony type with irregular edge and elevated center, designated WDL34, corresponded to the lowest bright band, WDL34, present in the pattern of the linuron enrichment culture, in addition to the 3,4-DCA culture (Fig. 2). Plating of a 4-month-old 3,4-DCA-degrading subculture on MMN with 50 mg of 3,4-DCA per liter resulted in the isolation of two of the same colony types, WDL5 and WDL34 (data not shown). These first results suggest that strains WDL1, WDL34, and WDL5 could be key players, respectively, in the initial transformation of linuron and in the degradation of the metabolite 3,4-DCA.
Since the bacteria that seemed very dominant in the 4-month-old N,O-DMHA- and 3-CA-degrading subcultures (corresponding to bands WDL6 and WDL7, respectively, in Fig. 2 and Table 2), were not isolated by plating the linuron-degrading enrichment culture on the media mentioned above, these separate subcultures were plated on MMN medium amended with their respective substrate: i.e., N,O-DMHA and 3-CA. Plating of the 3-CA-degrading culture yielded only one colony type with a DGGE pattern consisting of the four bands that are typical for the bacterium that is unique to the 3-CA subculture (see WDL7, Fig. 2). Plating of the N,O-DMHA-degrading culture yielded two kinds of bacteria. Based on colony morphology and DGGE analysis, one colony type (ca. 5% of the colonies) corresponded to strain WDL5, which had also been found in all other cultures. The numerically dominant colony type (ca. 95% of the colonies) was small and brownish in transmitted light and bright beige or colorless in reflected light and corresponded to the bacterium represented by one band (WDL6) at the bottom of the DGGE pattern of both the N,O-DMHA- and linuron-degrading cultures (Fig. 2). This strain could thus be responsible for N,O-DMHA degradation in the linuron-degrading enrichment culture. Thanks to the isolation of strain WDL6 from the N,O-DMHA subculture, all of the four strains that were represented in the DGGE pattern of the linuron-degrading culture were now isolated as pure cultures.
In summary, plating the 25-month-old linuron-degrading enrichment culture on mineral medium yielded three bacteria, of which one seemed essential for linuron degradation. By using an additional isolation procedure involving subculture of the linuron-degrading culture on possible intermediates in the degradation pathway of linuron, two additional bacteria were enriched and subsequently isolated. Although these bacteria were not directly isolated from the linuron enrichment culture, they may nevertheless play an important role in linuron degradation, such as degradation of the side chain, N,O-DMHA.
Role of the different strains in the degradation of linuron.
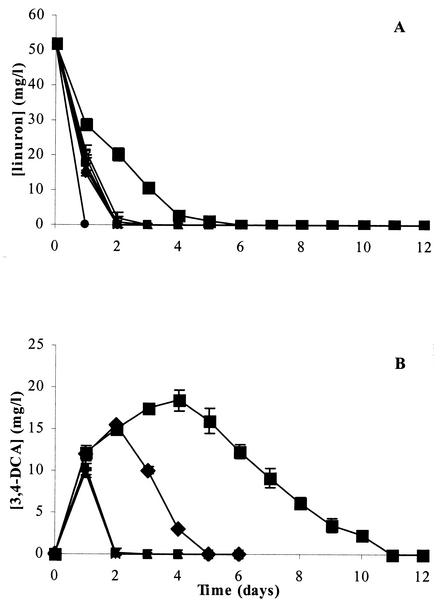
To determine the role of the isolates of the different cultures in the degradation of linuron, growth and degradation tests were performed in liquid MMN medium with 50 mg of linuron per liter as the sole source of N and C. As expected from the previous results, strain WDL1 was the only one of the five isolates that could degrade linuron in pure culture (Fig. 3A) (data not shown). However, strain WDL1 degraded linuron more slowly than the linuron enrichment culture (Fig. 3A), which could be caused in part by a much larger accumulation of 3,4-DCA, as shown in Fig. 3B. During the first 4 days, WDL1 converted linuron (50 mg/liter or 0.2 mM) partially to 3,4-DCA (18 mg/liter [or 0.11 mM] at day 4), known to be one of the main intermediates in the degradation pathway. This intermediate was however subsequently removed to undetectable levels at day 11. After total degradation of 3,4-DCA, no ring structures were detected by HPLC analysis. Moreover, stoichiometric amounts of chloride (ca. 14 mg/liter or 0.4 mM Cl−) were found in the medium (data not shown), corresponding to complete dechlorination of the aromatic ring. Since aerobic dechlorination of chloroanilines has been shown to proceed via a modified ortho-cleavage (55) or meta-cleavage (1) pathway, these data together with the absence of accumulating aromatic intermediates of 3,4-DCA suggest that the ring structure was completely degraded. No abiotic linuron removal was observed (data not shown). As far as we know, strain WDL1, isolated in this study, represents the first pure culture that encodes the total degradation of linuron. Apparently, the degradation of linuron proceeds via 3,4-DCA, which transiently accumulates in the medium, but eventually seems to be completely degraded.
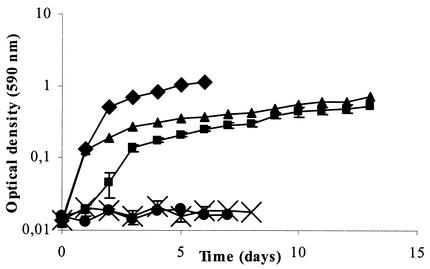
FIG. 3.
Degradation of linuron (A) and accumulation of 3,4-DCA (B) during linuron degradation by Variovorax sp. strain WDL1 and cocultures of WDL1 with the different strains isolated from the linuron-degrading culture or one of its subcultures. The data points and error bars show the means and standard deviations based on data from triplicate measurements. When error bars are not visible, they are hidden behind the symbols. ▪, Variovorax sp. strain WDL1; ▪, Variovorax sp. strain WDL1 plus Pseudomonas sp. strain WDL5; ▴, Variovorax sp. strain WDL1 plus D. acidovorans WDL34; ♦, Variovorax sp. strain WDL1 plus H. sulfonivorans WDL6; −, Variovorax sp. strain WDL1 plus C. testosteroni WDL7; *, Variovorax sp. strain WDL1 plus Pseudomonas sp. strain WDL5 plus D. acidovorans WDL34 plus H. sulfonivorans WDL6 plus C. testosteroni WDL7; •, linuron consortium.
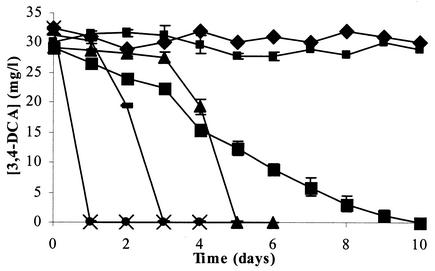
To examine if 3,4-DCA degradation by strain WDL1 could be induced directly by 3,4-DCA as a substrate, WDL1 was incubated with 30 mg of 3,4-DCA per liter (0.19 mM) in MMN medium. This amount was totally degraded in 10 days (Fig. 4) and resulted again in the release of stoichiometric amounts of chloride (ca. 13 mg/liter, 0.37 mM Cl−) (data not shown). Again, no abiotic degradation of this compound was observed (data not shown). Strain WDL1 is thus able to degrade 3,4-DCA in the absence of linuron.
FIG. 4.
Degradation of 3,4-DCA by different strains obtained from the linuron-degrading culture or one of its subcultures. The data points and error bars show the means and standard deviations based on data from triplicate measurements. When error bars are not visible, they are hidden behind the symbols. ▪, Variovorax sp. strain WDL1; ▪, Pseudomonas sp. strain WDL5; ▴, D. acidovorans WDL34; ♦, H. sulfonivorans WDL6; −, C. testosteroni WDL7; •, linuron consortium; ×, 3,4-DCA consortium.
Since the rate of degradation of linuron and 3,4-DCA for strain WDL1 was much lower than that of the linuron-degrading mixed culture, combinations of the different strains with WDL1 were tested for their degradation capacity (Fig. 3A and B). Interestingly, when each of the other strains (WDL34, WDL5, WDL6, and WDL7) was coinoculated separately with strain WDL1 in linuron-containing MMN medium, linuron was always degraded more rapidly than when only strain WDL1 was inoculated (Fig. 3). This indicates that a synergistic interaction between WDL1 and each of the bacteria WDL34, WDL5, WDL6, and WDL7 takes place. As shown in Fig. 3B, the clearly lower concentrations of accumulated 3,4-DCA in these cocultures could be in part responsible for the faster linuron degradation. Although the degradation of linuron by these cocultures was still not as rapid as for the linuron-degrading consortium, even after inoculation of WDL1 with all four strains at once, it was clearly enhanced in comparison to the performance of the pure culture of strain WDL1. The results of repeated experiments were very similar and completely confirmed the results shown here. In addition, this synergistic interaction was also observed when the concentration of linuron was decreased from 50 to 25 mg/liter.
To further determine the role of the strains WDL34, WDL5, WDL6, and WDL7 in the degradation of linuron, their ability to degrade possible intermediates in the degradation pathway of linuron was tested. Strains WDL34 and WDL7, but not strains WDL5 and WDL6, were able to degrade 3,4-DCA. Figure 4 shows that the degradation rates of 3,4-DCA were very similar for WDL34 (0.54 mg/liter · h) and WDL7 (0.63 mg/liter · h), but WDL34 showed a longer lag phase (3 days compared to 1). Both strains degraded 3,4-DCA much more rapidly than the linuron-degrading strain WDL1 (0.12 mg/liter · h) (Fig. 4). These data suggest that strains WDL34 and WDL7 enhance the degradation of linuron in cocultures with strain WDL1 by degrading the accumulating 3,4-DCA more rapidly than strain WDL1 alone. Since only strain WDL34, and not strain WDL7, was clearly present in the linuron-degrading consortium, based on its DGGE pattern (Fig. 2), we postulate that after strain WDL1, strain WDL34 is the second key player in the degradation of linuron by the consortium.
Since the linuron consortium can also grow with N,O-DMHA as the sole C and N source, and WDL6 was isolated from the N,O-DMHA-degrading subculture, this organism is probably responsible for degradation of this side chain. To confirm this assumption, the growth of the five different strains on this compound as the sole C and N source was tested by daily addition of 30 mg of N,O-DMHA per liter to MMN medium and measurements of the culture turbidity. As anticipated, growth was only observed for strain WDL6 (Fig. 5), and the growth curve was very similar to that of the N,O-DMHA-degrading enrichment culture. These results confirm that strain WDL6 seems to be the third key player in the complete degradation of linuron by degrading the side chain, N,O-DMHA (Fig. 1).
FIG. 5.
Growth of H. sulfonivorans WDL6 and the N,O-DMHA consortium on N,O-DMHA (as the sole source of N and C) or hydroxylamine and methanol (as N and C sources, respectively), as measured by the cumulative OD590 in cultures fed daily with 30 mg of N,O-DMHA per liter, 15 mg of hydroxylamine per liter, or 250 mg of methanol per liter as the N and/or C source. The data points and error bars show the means and standard deviations based on data from triplicate measurements. When error bars are not visible, they are hidden behind the symbols. •, MMN plus H. sulfonivorans WDL6; ▪, MMN plus H. sulfonivorans WDL6 plus N,O-DMHA; ▴, MMN plus N,O-DMHA culture plus N,O-DMHA; ×, MMN plus H. sulfonivorans WDL6 plus hydroxylamine plus methanol; ♦, MMN plus H. sulfonivorans WDL6 plus 0.3 mg of (NH4)2SO4 per liter plus methanol.
Identification of strains isolated from the different consortia.
Based on the 16S rRNA gene sequence, the taxonomic situation of isolate WDL1 is not entirely clear. The sequence was 98.1% similar to that of Variovorax paradoxus DSM 66T (accession no. AJ420329), but also 97.4% similar to those of Xylophilus ampelinus DSM 7250T (accession no. AJ420330) and Aquaspirillum delicatum LMG 4328T (accession no. AF078756). We designated strain WDL1 throughout this study as belonging to Variovorax sp. As expected, the 16S rRNA gene sequence of Variovorax sp. strain WDL1 was 99.7% similar to sequence VL3 (accession no. AF180947), a 16S rRNA gene fragment that had previously been amplified and cloned directly from the linuron-degrading consortium (13). The 3,4-DCA-degrading isolates WDL34 and WDL7 showed very high 16S rRNA gene sequence similarities (99.1 and 99.7%, respectively) to Delftia acidovorans ACM 489T (accession no. AF078774) and Comamonas testosteroni ATCC 11996T (accession no. M11224), respectively, and were <95.6% similar to all other named EMBL database entries. Therefore, isolates WDL34 and WDL7 could be identified with high confidence as Delftia acidovorans and Comamonas testosteroni, respectively. The 16S rRNA gene sequence of isolate WDL6 showed extremely high similarity (99.9%) to Hyphomicrobium sulfonivorans S1T (accession no. AF235089). This strain, WDL6, was therefore identified with high confidence as Hyphomicrobium sulfonivorans. Isolate WDL5 showed around 99% 16S rRNA gene sequence similarity to several type strains of the genus Pseudomonas sensu stricto (e.g., Pseudomonas asplenii ATCC 23835T [accession no. AB021397] and Pseudomonas fuscovaginae MAFF 301177T [accession no. AB021381]) and was therefore designated as Pseudomonas sp.
DISCUSSION
By isolating bacteria from a linuron-degrading mixed culture, as well as from subcultures growing on intermediates of the linuron degradation pathway, five potential key players were obtained: Variovorax sp. strain WDL1, D. acidovorans WDL34, Pseudomonas sp. strain WDL5, H. sulfonivorans WDL6, and C. testosteroni WDL7. Coupling of the community structure data of the different enrichment cultures (Fig. 2 and Table 2) to the results obtained from degradation assays for linuron and its main intermediates, 3,4-DCA and N,O-DMHA (Fig. 3, 4, and 5), allowed us to show that four of these five bacteria played a major role in the degradation of linuron (Fig. 1). Variovorax sp. strain WDL1 is the only member of the consortium that is able to metabolize this herbicide. A significant amount of the substrate carbon spills from WDL1 as 3,4-DCA and probably also as N,O-DMHA. 3,4-DCA is very efficiently taken up by D. acidovorans WDL34, which thereby seems to protect WDL1 from the toxicity that would otherwise accrue from a buildup of 3,4-DCA. This metabolite thus presumably reflects a kinetic bottleneck in the degradation pathway of Variovorax sp. strain WDL1. We assume that WDL1 also released an amount of N,O-DMHA, which is very efficiently taken up by H. sulfonivorans WDL6, the only member of the linuron-degrading culture that can grow on this compound. The role of Pseudomonas sp. strain WDL5, which was very dominant in the DGGE patterns of the 3,4-DCA- and linuron-degrading cultures, is currently unknown. Since it does not degrade 3,4-DCA nor N,O-DMHA, further research will have to determine whether other metabolites or compounds released during linuron or 3,4-DCA degradation sustain the presence of this strain in these cultures or why else it was maintained.
The results obtained by DGGE, cloning, and strain isolation together demonstrate that further parallel enrichment of a pollutant-degrading mixed culture on different potential intermediates of the metabolic pathway of that pollutant can successfully enrich specific but originally nondominant consortium members that can degrade these intermediates. These can then subsequently be isolated and used to reconstitute the consortium and understand the metabolic interactions. Kane et al. (23) and Teske et al. (46) used a similar approach to design the specific culture conditions to separate and isolate strains from enrichment cultures capable of sulfate reduction. Other authors have also recognized the advantage of molecular techniques to get a grip on the complex bacterial composition of consortia (26, 35).
To the best of our knowledge, Variovorax sp. strain WDL1 is the first strain reported to be able to degrade the aromatic ring and to remove the chloride ions of linuron. We assume that this strain converts this phenylurea herbicide at least to a linear nonchlorinated compound and probably even to CO2 and H2O. The degradation seems to be initiated by an initial attack at the amide bond of the urea side chain, resulting in the formation of 3,4-DCA and N,O-DMHA (Fig. 1). 3,4-DCA then transiently accumulated in the medium, but in contrast to previously isolated phenylurea herbicide-converting strains (5, 47, 51, 53), it was further degraded by the same strain. The linuron-degrading activity of Variovorax sp. strain WDL1 expands the list of diverse metabolic traits (28, 44) exhibited by this genus, such as chemolithoautotrophic growth (24) and degradation of other pesticides, such as 2,4-D (2,4-dichlorophenoxyacetic acid) (10, 16, 22, 52) and carbofuran (8). Another very interesting characteristic of Variovorax sp. strain WDL1 is its ability to degrade linuron at concentrations as low as 0.04 mg/liter or 160 nM. This was demonstrated by Hulsen et al. (20) with a Lemna minor L.-based bioassay.
Although several groups have been successful in isolating bacterial consortia capable of mineralizing phenylurea herbicides, only a few obtained pure cultures that mostly only cleaved the N′N′-dimethyl or N′-methoxy-methyl urea side chain, but lacked the ability to mineralize the phenyl structure (5, 38, 47). However, only very recently, a Pseudomonas strain (11) and a Sphingomonas sp. strain, SRS2 (43), were isolated that completely degrade diuron and isoproturon (dimethyl urea herbicides), respectively, but not linuron (a methoxymethyl urea herbicide). For these two strains, a transient accumulation of the respective intermediates 3,4-DCA and 3-(4-isopropylphenyl)-1-methylurea was observed, which probably indicates that diuron and isoproturon were, respectively, degraded by an initial attack at the amide bond of the urea side chain or by a successive N demethylation. In the case of Sphingomonas sp. strain SRS2, a clear increase in the efficiency of the degradation of isoproturon (from 10% release of 14CO2 to 48%) was achieved by the addition of particular amino acids to the medium. The most important amino acid seemed to be l-methionine, which was delivered to SRS2 by another bacterial strain of the enrichment culture, which indicates that Sphingomonas sp. strain SRS2 was auxotrophic (42). In contrast to SRS2, Variovorax sp. strain WDL1 did not seem to depend on the presence of compounds provided by the other members, but its ability to degrade linuron was clearly stimulated by them.
The process of isolating of Variovorax sp. strain WDL1 from the linuron-degrading consortium was not very straightforward, and different attempts under different culture conditions were necessary before a pure culture was obtained (13). The choice of isolation medium and presence of linuron were undoubtedly major factors with influence. Apparently the continuous transfer of the consortium in mineral medium with linuron over 25 months clearly enriched strain WDL1 and may have allowed the strain to grow better on agar plates that contained the same medium. This could explain why previous attempts to isolate this strain from a less old (5 months) enrichment culture in the same laboratory were never successful (13). DGGE analysis of this earlier consortium then revealed that the bacteria that corresponded to the most dominant bands in the linuron-degrading culture and were growing on MMN agar plates with 100 mg of linuron per liter corresponded mainly to Pseudomonas species (13). These species, which are known for their fast growth on agar plates (33), probably overgrew strain WDL1 or prevented visible colony formation. Even Pseudomonas sp. strain WDL5, which was shown not to use linuron as an N source, formed colonies on MMN medium with linuron as the sole N source.
Cooperative metabolic activities in bacterial consortia during degradation of organic pollutants may be an interesting phenomenon that is widespread in nature (41) and that involves two mechanisms: (i) complementation of metabolic deficiencies, in which the degrading bacteria are fastidious and depend on secondary strains that provide essential growth factors or nutrients as presented above for the isoproturon-degrading strain Sphingomonas sp. strain SRS2 (42); (ii) associated metabolism, in which cross-feeding with metabolites from the degradation pathway occurs between members of the consortium. For example, De Souza et al. (9) determined that in a four-member atrazine-degrading consortium, Clavibacter michiganese ATZ1 initiates the degradation of this s-triazine herbicide by removing the side chain while Pseudomonas sp. strain CN1 subsequently cleaves the ring. In another study, Pelz et al. (34) examined the degradation route of 4-chlorosalicylate in a four-member community. Their data indicated that the metabolic and physiological deficiencies of the primary degrader were compensated for by the presence of other organisms with the appropriate complementary physiology to build a consortium that was able to extract the maximal metabolic benefit from the available resources. Although DGGE analysis (Fig. 2) and 16S rRNA gene clone libraries (Table 2) revealed in our study that strains WDL1, WDL34, WDL5, and WDL6 were the most dominant species in the linuron-degrading culture, the degradation rate of linuron by an artificial combination of these four strains was lower than that of the original linuron consortium. This could be because the specific relative abundance of each member in the stable degradative consortium was not reconstituted in these artificial combinations (54), or because this four-member mixture did not fully represent the complete degradative consortium. Other 16S rRNA genes than those of WDL1, WDL34, WDL5, and WDL6 were indeed detected in the DGGE pattern (data not shown) and clone library (Table 2) of the linuron consortium that had been enriched for 51 months on linuron. However, each of them only represented between 2 and 4% of the total clones. Although we are aware of the biases of the traditional PCR amplification (45), cloning (17), and sequencing (36) approach, we assume—due to the clear dominance of WDL1, WDL34, WDL5, and WDL6—that these four bacteria represent the most important bacteria of the linuron consortium, and therefore no further attempts to isolate the other bacteria were pursued.
Both D. acidovorans WDL34 and C. testosteroni WDL7 could degrade 3,4-DCA and 3-CA. Although both strains have the same degradation capacities, only WDL34 was isolated from the linuron consortium, while WDL7 was only obtained after subculture of the consortium in 3-CA. Under the conditions of the linuron-degrading mixed culture and the subsequent 3,4-DCA-degrading subculture, strain WDL34 must have been more competitive than strain WDL7, despite its longer lag phase and slightly lower 3,4-DCA degradation rate in pure culture. Apparently the soil from Gorsem (Belgium), from which the linuron-degrading culture was originally enriched, contains several 3-CA-degrading strains. Indeed, after enrichment of the same soil on 3-CA, Boon et al. (2) obtained a different D. acidovorans strain, LME1, and no C. testosteroni strains. Bacteria belonging to the Delftia and Comamonas genera are known to degrade a wide range of xenobiotic compounds (15, 30) and have been often isolated from 3-CA- and 3,4-DCA-degrading enrichment cultures from soil and sludge (2, 7).
H. sulfonivorans WDL6 is the only member of the linuron-degrading culture that can grow on N,O-DMHA. To the best of our knowledge, this is the first report of a bacterial strain that can grow with N,O-DMHA as the sole source of C, N, and energy. Other members of the genus are able to degrade related compounds, such as trimethylamine, dimethylamine, and methylamine (25), as well as halomethanes, methyl sulfates, and methylated phosphates (27). Although it was assumed that N,O-DMHA would be degraded to hydroxylamine, no growth of WDL6 or one of the other strains was observed on hydroxylamine as an N source in the presence of methanol as a C source (Fig. 5). According to other studies, this could not be due to toxicity of hydroxylamine (6, 21) or methanol (19) or lack of induction of degradation enzymes (data not shown). Although we assume that Variovorax sp. strain WDL1 converts linuron to 3,4-DCA and N,O-DMHA, no growth of WDL1 on N,O-DMHA was observed when it was added as the sole substrate. Possible reasons could be (i) the low formation of biomass by WDL1, resulting in an undetectable increase in OD (even in 1/10 LB medium, the OD590 was only 0.3 after 4 days); (ii) the inability of WDL1 to degrade this intermediate; or (iii) lack of induction of the necessary degradation enzymes in the absence of linuron.
In conclusion, DGGE and cloning analysis of the 16S rRNA genes of the linuron-degrading mixed culture, together with degradation assays, indicate that this consortium consists of four major key players: Variovorax sp. strain WDL1, able to degrade linuron by itself, although slowly; D. acidovorans WDL34, which degrades 3,4-DCA and thus accelerates the removal of this metabolite; H. sulfonivorans WDL6, which is the only strain able to degrade N,O-DMHA; and Pseudomonas sp. strain WDL5, whose role is currently unknown. The observation that each of them was able to accelerate the degradation of linuron in coculture with Variovorax sp. strain WDL1 compared to WDL1 alone supports their role in the degradation of this herbicide.
Acknowledgments
This work was supported by a research grant from the Flemish Fund for Scientific Research (FWO-Vlaanderen), the EU-concerted action MECBAD, and Project G.O.A. (1997 to 2002) of the “Ministerie van de Vlaamse Gemeenschap, Bestuur Wetenschappelijk Onderzoek.” E.M.T. was partly supported by the Flemish Fund for Scientific Research (a research associate position until 2000) and partly (since 2002) by a grant from the National Institutes of Health (P20 RR16448-01).
We thank Muhammet Yilmaz for technical assistance and Larissa Hendrickx, Bart Levecke, and Hanne Lievens for useful suggestions on the manuscript.
REFERENCES
- 1.Boon, N., J. Goris, P. De Vos, W. Verstraete, and E. M. Top. 2000. Bioaugmentation of activated sludge by an indigenous 3-chloroaniline-degrading Comamonas testosteroni strain, I2gfp. Appl. Environ. Microbiol. 66:2906-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boon, N., J. Goris, P. De Vos, W. Verstraete, and E. M. Top. 2001. Genetic diversity among 3-chloroaniline- and aniline-degrading strains of the Comamonadaceae. Appl. Environ. Microbiol. 67:1107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caux, P. Y., R. A. Kent, G. T. Fan, and C. Grande. 1998. Canadian water quality guidelines for linuron. Environ. Toxicol. Water Qual. 13:1-41. [Google Scholar]
- 4.Coenye, T., E. Falsen, M. Vancanneyt, B. Hoste, J. R. Govan, K. Kersters, and P. Vandamme. 1999. Classification of Alcaligenes faecalis-like isolates from the environment and human clinical samples as Ralstonia gilardii sp. nov. Int. J. Syst. Bacteriol. 49:405-413. [DOI] [PubMed] [Google Scholar]
- 5.Cullington, J. E., and A. Walker. 1999. Rapid biodegradation of diuron and other phenylurea herbicides by a soil bacterium. Soil Biol. Biochem. 31:677-686. [Google Scholar]
- 6.de Bruijn, P., A. A. van de Graaf, M. S. M. Jetten, L. A. Robertson, and J. G. Kuenen. 1995. Growth of Nitrosomonas europaea on hydroxylamine. FEMS Microbiol. Lett. 125:179-184. [Google Scholar]
- 7.Dejonghe, W., J. Goris, A. Dierickx, V. De Dobbeleer, K. Crul, P. De Vos, W. Verstraete, and E. M. Top. 2002. Diversity of 3-chloroaniline and 3,4-dichloroaniline degrading bacteria isolated from three different soils and involvement of their plasmids in chloroaniline degradation. FEMS Microbiol. Ecol. 42:315-325. [DOI] [PubMed] [Google Scholar]
- 8.Desaint, S., A. Hartmann, N. R. Parekh, and J. C. Fournier. 2000. Genetic diversity of carbofuran-degrading soil bacteria. FEMS Microbiol. Ecol. 34:173-180. [DOI] [PubMed] [Google Scholar]
- 9.De Souza, M. L., D. Newcombe, S. Alvey, D. E. Crowley, A. Hay, M. J. Sadowsky, and L. P. Wackett. 1998. Molecular basis of a bacterial consortium: interspecies catabolism of atrazine. Appl. Environ. Microbiol. 64:178-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunbar, J., I. Zlatkin, and L. Forney. 1995. Variation in genome organization among Variovarax paradoxus clones. J. Cell. Biochem. Suppl. 104. 19A:
- 11.El-Deeb, B. A., S. M. Soltan, A. M. Ali, and K. A. Ali. 2000. Detoxication of the herbicide diuron by Pseudomonas sp. Folia Microbiol. 45:211-216. [DOI] [PubMed] [Google Scholar]
- 12.El-Fantroussi, S., L. Verschuere, W. Verstraete, and E. M. Top. 1999. Effect of phenylurea herbicides on soil microbial communities estimated by analysis of 16S rRNA gene fingerprints and community-level physiological profiles. Appl. Environ. Microbiol. 65:982-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El-Fantroussi, S., W. Verstraete, and E. M. Top. 2000. Enrichment and molecular characterization of a bacterial culture that degrades methoxy-methyl urea herbicides and their aniline derivatives. Appl. Environ. Microbiol. 66:5110-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelhardt, G., P. R. Wallnöfer, and R. Plapp. 1972. Identification of N, O-dimethylhydroxylamine as a microbial degradation product of the herbicide, Linuron. Appl. Microbiol. 23:664-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fedi, S., M. Carnevali, F. Fava, A. Andracchio, S. Zappoli, and D. Zannoni. 2001. Polychlorinated biphenyl degradation activities and hybridization analyses of fifteen aerobic strains isolated from a PCB-contaminated site. Res. Microbiol. 152:583-592. [DOI] [PubMed] [Google Scholar]
- 16.Fisher, P. R., J. Appleton, and J. M. Pemberton. 1978. Isolation and characterization of the pesticide-degrading plasmid pJP1 from Alcaligenes paradoxus. J. Bacteriol. 135:798-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanahan, D. 1983. Studies on transformation of E. coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 18.Heuer, H., M. Krsek, P. Baker, K. Smalla, and E. M. H. Wellington. 1997. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 63:3233-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirsch, P. 1989. Genus Hyphomicrobium Stutzer and Hartleb 1898, 76AL, p. 1897-1904. In J. T. Staley, M. P. Bryant, N. Pfennig, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 3. Williams & Wilkins, Baltimore, Md.
- 20.Hulsen, K., V. Minne, P. Lootens, P. Vandecasteele, and M. Hofte. 2002. A chlorophyll a fluorescence-based Lemna minor bioassay to monitor microbial degradation of nanomolar to micromolar concentrations of linuron. Environ. Microbiol. 4:327-337. [DOI] [PubMed] [Google Scholar]
- 21.Jetten, M. S. M., P. de Bruijn, and J. G. Kuenen. 1997. Hydroxylamine metabolism in Pseudomonas PB16: involvement of a novel hydroxylamine oxidoreductase. Antonie Leeuwenhoek 71:69-74. [DOI] [PubMed] [Google Scholar]
- 22.Kamagata, Y., R. R. Fulthorpe, K. Tamura, H. Takami, L. J. Forney, and J. M. Tiedje. 1997. Pristine environments harbor a new group of oligotrophic 2,4-dichlorophenoxyacetic acid-degrading bacteria. Appl. Environ. Microbiol. 63:2266-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kane, M. D., L. K. Poulson, and D. A. Stahl. 1993. Monitoring the enrichment and isolation of sulfate-reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl. Environ. Microbiol. 59:682-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kersters, K., and J. De Ley. 1984. Genus Alcaligenes Castellani and Chalmers 1919, p. 361-373. In N. R. Krieg, and J. G. Holt (ed.), Bergey's manual of systematic bateriology, vol. 1. Williams & Wilkins, Baltimore, Md.
- 25.Kim, S. G., H. S. Bae, and S. T. Lee. 2001. A novel denitrifying bacterial isolate that degrades trimethylamine both aerobically and anaerobically via two different pathways. Arch. Microbiol. 176:271-277. [DOI] [PubMed] [Google Scholar]
- 26.Koizumi, Y., J. J. Kelly, T. Nakagawa, H. Urakawa, S. El-Fantroussi, S. Al-Muzaini, M. Fukui, Y. Urushigawa, and D. A. Stahl. 2002. Parallel characterization of anaerobic toluene- and ethylbenzene-degrading microbial consortia by PCR-denaturing gradient gel electrophoresis, RNA-DNA membrane hybridization, and DNA microarray technology. Appl. Environ. Microbiol. 68:3215-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Large, P. J., and C. W. Bamforth. 1988. Methylotrophy and biotechnology. Longman Scientific & Technical, New York, N.Y.
- 28.Leadbetter, J. R., and E. P. Greenberg. 2000. Metabolism of acyl-homoserine lactone quorum-sensing signals by Variovorax paradoxus. J. Bacteriol. 182:6921-6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchesi, J. R., T. Sato, A. J. Weightman, T. A. Martin, J. C. Fry, S. J. Hiom, and W. G. Wade. 1998. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 64:795-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muller, R. H., S. Kleinsteuber, and W. Babel. 2001. Physiological and genetic characteristics of two bacterial strains utilizing phenoxypropionate and phenoxyacetate herbicides. Microbiol. Res. 156:121-131. [DOI] [PubMed] [Google Scholar]
- 31.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Øvreas, L., L. Forney, F. L. Daae, and V. Torsvik. 1997. Distribution of bacterioplankton in meromictic Lake Saelevannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl. Environ. Microbiol. 63:3367-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palleroni, N. J. 1984. Genus I. Pseudomonas Migula 1894, 237 AL (Nom. cons. Opin. 5, Jud. Comm. 1952, 237), p. 140-219. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams & Wilkins, Baltimore, Md.
- 34.Pelz, O., M. Tesar, R.-M. Wittich, E. R. B. Moore, K. N. Timmis, and W.-R. Abraham. 1999. Towards elucidation of microbial community metabolic pathways: unravelling the network of carbon sharing in a pollutant-degrading bacterial consortium by immunocapture and isotopic ratio mass spectrometry. Environ. Microbiol. 1:167-174. [DOI] [PubMed] [Google Scholar]
- 35.Phelps, C. D., L. J. Kerkhof, and L. Y. Young. 1998. Molecular characterization of a sulfate-reducing consortium which mineralizes benzene. FEMS Microbiol. Ecol. 27:269-279. [Google Scholar]
- 36.Reysenbach, A.-L., L. Giver, G. S. Wickham, and N. R. Pace. 1992. Differential amplification of rRNA genes by polymerase chain reaction. Appl. Environ. Microbiol. 58:3417-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richardson, M. 1998. Pesticides—friend or foe? Water Sci. Technol. 37:19-25. [Google Scholar]
- 38.Roberts, S. J., A. Walker, L. Cox, and S. J. Welch. 1998. Isolation of isoproturon-degrading bacteria from treated soil via three different routes. J. Appl. Microbiol. 85:309-316. [DOI] [PubMed] [Google Scholar]
- 39.Roberts, S. J., A. Walker, N. R. Parekh, S. J. Welch, and M. J. Waddington. 1993. Studies on a mixed bacterial culture from soil which degrades the herbicide linuron. Pestic. Sci. 39:71-78. [Google Scholar]
- 40.Scassellati-Sforzolini, G., R. Pasquini, M. Moretti, M. Villarini, C. Fatigoni, P. Dolara, S. Monarca, G. Caderni, F. Kuchenmeister, P. Schmezer, and B. L. Pool-Zobel. 1997. In vivo studies on genotoxicity of pure and commercial linuron. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 390:207-221. [DOI] [PubMed] [Google Scholar]
- 41.Slater, J. H., and D. Lovatt. 1984. Biodegradation and the significance of microbial communities, p. 439-485. In D. T. Gibson (ed.), Microbial degradation of organic compounds. Marcel Dekker, New York, N.Y.
- 42.Sørensen, S. R., Z. Ronen, and J. Aamand. 2002. Growth in coculture stimulates metabolism of the phenylurea herbicide isoproturon by Sphingomonas sp. strain SRS2. Appl. Environ. Microbiol. 68:3478-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sørensen, S. R., Z. Ronen, and J. Aamand. 2001. Isolation from agricultural soil and characterization of a Sphingomonas sp. able to mineralize the phenylurea herbicide isoproturon. Appl. Environ. Microbiol. 67:5403-5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suyama, T., H. Hosoya, and Y. Tojkiwa. 1998. Bacterial isolates degrading aliphatic polycarbonates. FEMS Microbiol. Lett. 161:255-261. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teske, A., P. Sigalevich, Y. Cohen, and G. Muyzer. 1996. Molecular identification of bacteria from a coculture by denaturing gradient gel electrophoresis of 16S ribosomal DNA fragments as a tool for isolation in pure cultures. Appl. Environ. Microbiol. 62:4210-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tixier, C., M. Sancelme, S. Ait-Aissa, P. Widehem, F. Bonnemoy, A. Cuer, N. Truffaut, and H. Veschambre. 2002. Biotransformation of phenylurea herbicides by a soil bacterial strain, Arthrobacter sp. N2: structure, ecotoxicity and fate of diuron metabolite with soil fungi. Chemosphere 46:519-526. [DOI] [PubMed] [Google Scholar]
- 48.Tixier, C., M. Sancelme, F. Bonnemoy, A. Cuer, and H. Veschambre. 2001. Degradation products of a phenylurea herbicide, diuron: synthesis, ecotoxicity, and biotransformation. Environ. Toxicol. Chem. 20:1381-1389. [DOI] [PubMed] [Google Scholar]
- 49.Tomlin, C. (ed.) 1994. The pesticide manual, 10th ed. British Crop Protection Council, Farnham, Surrey, United Kingdom.
- 50.Turnbull, G. A., J. E. Cullington, A. Walker, and J. A. W. Morgan. 2001. Identification and characterisation of a diuron-degrading bacterium. Biol. Fertil. Soils 33:472-476. [Google Scholar]
- 51.Turnbull, G. A., M. Ousley, A. Walker, E. Shaw, and J. A. W. Morgan. 2001. Degradation of substituted phenylurea herbicides by Arthrobacter globiformis strain D47 and characterization of a plasmid-associated hydrolase gene, puhA. Appl. Environ. Microbiol. 67:2270-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vallaeys, T., L. Albino, G. Soulas, A. D. Wright, and A. J. Weightman. 1998. Isolation and characterization of a stable 2,4-dichlorophenoxyacetic acid degrading bacterium, Variovorax paradoxus, using chemostat culture. Biotechnol. Lett. 20:1073-1076. [Google Scholar]
- 53.Wällnofer, P. 1969. The decomposition of urea herbicides by Bacillus sphaericus, isolated from soil. Weed Res. 9:333-339. [Google Scholar]
- 54.Wolfaardt, G. M., J. R. Lawrence, R. D. Robarts, and D. E. Caldwell. 1994. The role of interactions, sessile growth, and nutrient amendments on the degradative effieciency of a microbial consortium. Can. J. Microbiol. 40:331-340. [DOI] [PubMed] [Google Scholar]
- 55.Zeyer, J., A. Wasserfallen, and K. N. Timmis. 1985. Microbial mineralization of ring-substituted anilines through an ortho-cleavage pathway. Appl. Environ. Microbiol. 50:447-453. [DOI] [PMC free article] [PubMed] [Google Scholar]