Abstract
Enrichment of microorganisms with special traits and the construction of metagenomic libraries by direct cloning of environmental DNA have great potential for identifying genes and gene products for biotechnological purposes. We have combined these techniques to isolate novel genes conferring oxidation of short-chain (C2 to C4) polyols or reduction of the corresponding carbonyls. In order to favor the growth of microorganisms containing the targeted genes, samples collected from four different environments were incubated in the presence of glycerol and 1,2-propanediol. Subsequently, the DNA was extracted from the four samples and used to construct complex plasmid libraries. Approximately 100,000 Escherichia coli strains of each library per test substrate were screened for the production of carbonyls from polyols on indicator agar. Twenty-four positive E. coli clones were obtained during the initial screen. Sixteen of them contained a plasmid (pAK101 to pAK116) which conferred a stable carbonyl-forming phenotype. Eight of the positive clones exhibited NAD(H)-dependent alcohol oxidoreductase activity with polyols or carbonyls as the substrates in crude extracts. Sequencing revealed that the inserts of pAK101 to pAK116 encoded 36 complete and 17 incomplete presumptive protein-encoding genes. Fifty of these genes showed similarity to sequenced genes from a broad collection of different microorganisms. The genes responsible for the carbonyl formation of E. coli were identified for nine of the plasmids (pAK101, pAK102, pAK105, pAK107 to pAK110, pAK115, and pAK116). Analyses of the amino acid sequences deduced from these genes revealed that three (orf12, orf14, and orf22) encoded novel alcohol dehydrogenases of different types, four (orf5, sucB, fdhD, and yabF) encoded novel putative oxidoreductases belonging to groups distinct from alcohol dehydrogenases, one (glpK) encoded a putative glycerol kinase, and one (orf1) encoded a protein which showed no similarity to any other known gene product.
Interconversion of alcohols, aldehydes, and ketones is involved in an astonishingly wide range of essential metabolic reactions in microorganisms. These redox reactions are catalyzed by oxidoreductases, including dehydrogenases. Most of these enzymes require cofactors such as flavin adenine dinucleotide, pyrroquinoline quinone, and NAD(P)+ (25, 35). The nicotinamide-dependent alcohol dehydrogenases especially are of considerable industrial importance. These enzymes catalyze the reaction alcohol + NAD(P)+ ⇄ aldehyde + NAD(P)H + H+.
Enzymes of this type are useful catalysts, with high enantioselectivity for the synthesis of carbonyl compounds, hydroxy acids, amino acids, and chiral alcohols, which are chemically difficult to prepare (19, 25, 26). Alcohol dehydrogenases are employed for the preparation of deuterium- or tritium-labeled compounds, production of dihydroxyacetone, and as tools for enzymatic analysis of serum lipids (19, 47). For large-scale applications, regeneration of the nicotinamide cofactors by effective regeneration reactions such as the glucose-6-phosphate dehydrogenase or the formate dehydrogenase system is necessary (9). These systems reduce costs and provide an elegant way of driving thermodynamically unfavorable reactions towards the desired product (9, 19, 26).
The isolation of new or improved catalysts from environmental sources can be facilitated by two different approaches. The classical approach to isolate new enzymes by enrichment and screening of a wide variety of microorganisms for the desired activity. The enzymes and the corresponding genes are then recovered from the identified organisms. In this way, a large fraction (>99%) of the microbial diversity in an environment is lost due to difficulties in enriching and isolating microorganisms in pure culture (2). The alternative way is the assessment and exploitation of the entirety of the microbial genomes found in nature (4, 7, 11, 23, 31), which is termed the metagenome by some authors (21, 37). This approach comprises the construction of metagenomic libraries by direct extraction and cloning of DNA from environmental samples. Subsequently, the resulting libraries are screened for the targeted genes (12, 37). The number of positive clones in a screen can be increased by with environmental samples which exhibit an enrichment of microorganisms containing the desired activity for library production (12).
Previous studies have shown that the metagenomic approach offers an almost unlimited pool for encountering novel genes encoding biotechnologically relevant gene products such as lipases (23), cellulases (22), amylases (36, 37), chitinases (11), esterases (23), and enzymes involved in biotin synthesis (17). Nevertheless, the list of enzyme activities encountered in this way is still rather small in comparison to the classical discovery of new enzymes by enrichment and isolation of individual microorganisms.
The main goal of this study was to combine enrichment technology and metagenomics for the identification of a large number of diverse genes which confer the ability to oxidize short-chain polyols or reduce the corresponding carbonyl compounds. The recovered genes can serve as a resource (gene bank) for the rapid identification of suitable genes and gene products during development or improvement of industrial processes. The DNA used for preparation of the libraries was isolated directly from different environmental samples after a short enrichment for polyol-fermenting microorganisms. The resulting DNA libraries were screened for the presence of genes conferring a carbonyl compound-producing phenotype on Escherichia coli ECL707 (41) during incubation in the presence of short-chain (C2 to C4) polyols with adjacent hydroxyl groups. Crude extracts of the positive E. coli clones obtained were analyzed for nicotinamide-dependent dehydrogenase activity. Subsequently, the plasmids encoding the targeted activity were recovered from the positive E. coli strains and sequenced.
MATERIALS AND METHODS
Sample sites, bacterial strains, and plasmids.
For the construction of environmental DNA libraries after enrichment for polyol-consuming microorganisms, soil from a sugar beet field near Göttingen (Germany), sediment from the river Grone (Germany), sediment from Solar Lake (Egypt), and sediment from the Gulf of Eilat (Israel) were collected. E. coli strains DH5α (3) and ECL707 (41) were used as hosts for the cloning experiments and the activity-based screening procedures, respectively. The plasmid pBluescript SK+ (pSK+) (Stratagene, San Diego, Calif.) was employed as the vector for the cloning experiments. The recombinant vectors pRD1 (13, 14) and pFL1 (30) were applied as positive controls during activity-based screening.
Media and growth conditions.
E. coli was routinely grown in Luria-Bertani (LB) medium at 30°C (3). For activity-based screening of environmental libraries, recombinant E. coli strains were grown on carbonyl indicator plates. These plates were prepared according to the method of Conway et al. (10), but ethanol was replaced with 100 mM 1,2-ethanediol, 1,2-propanediol, glycerol, 2,3-butanediol, or combinations of these substrates. The enrichment for polyol-fermenting microorganisms was performed under anaerobic conditions for 24 h in a medium containing (per liter) K2HPO4, 14.0 g; KH2PO4, 6.0 g; (NH4)2SO4, 3.0 g; MgSO4 · 7H2O, 0.2 g; CoCl2 · 6H2O, 0.012 g; yeast extract, 0.2 g; cysteine-HCl, 0.2 g; and trace element solution SL4 (33), 1 ml (pH 7.5). The medium was supplemented with 100 mM glycerol and 1,2-propanediol. The enrichment was initiated by adding 50 g (wet weight) of environmental sample to flasks (1 liter) containing enrichment medium (500 ml). For the preparation of crude extracts, recombinant E. coli strains were grown in LB medium at 37°C. All growth media for E. coli strains harboring plasmids contained 100 μg of ampicillin per ml to maintain the plasmids.
Preparation of cell extracts.
Cells in the stationary growth phase from 500-ml cultures were harvested by centrifugation at 6,000 × g for 20 min, washed once with 100 mM potassium phosphate buffer (pH 8.0), and resuspended in 2 to 3 ml of the same buffer. The cells were disrupted by French pressing (1.38 × 108 Pa), and the extract was cleared by centrifugation at 32,000 × g and 4°C for 30 min.
Enzyme assays.
Alcohol dehydrogenase activity was determined by measuring the NAD(P)H-dependent reduction of carbonyls or by measuring the NAD(P)+-dependent oxidation of alcohols according to Daniel et al. (14) and Ruch et al. (38), respectively. For the oxidation reaction, the substrates were 1,2,4-butanetriol, 2,3-butanediol, 1-butanol, 2-butanol, glycerol, 1,2-propanediol, 1-propanol, 2-propanol, 1,2-ethanediol, ethanol, and methanol at a final concentration of 100 mM; for the reduction reaction, the substrates were diacetyl, methylglyoxal, glyceraldehyde, dihydroxyacetone, hydroxyacetone, acetone, glycolaldehyde, and acetaldehyde at a final concentration of 10 mM. The specific background activities of the E. coli host with the substrates were measured in each case and subtracted from those of the recovered carbonyl-producing E. coli strains.
Protein concentrations were determined by the method of Bradford (6) with bovine serum albumin as the standard.
Molecular procedures.
The isolation and purification of DNA from environmental samples were performed as described previously (12). The purified environmental DNA was partially digested with Bsp143I and, in order to avoid cloning of very small DNA fragments, size fractionated by sucrose density centrifugation (10 to 40% [wt/vol]). Fractions containing DNA fragments of >2 kb were ligated into the BamHI-digested high-copy vector pSK+ (500 to 700 copies per cell). The ligation products were then transformed into E. coli DH5α. To amplify the libraries, the resulting clones were grown on LB agar plates containing ampicillin for 18 h. Subsequently, the colonies were collected, and the plasmids were prepared by employing the Qiagen plasmid maxi kit (Qiagen, Hilden, Germany) as recommended by the manufacturer. To initiate the screening, the recovered plasmids were used to transform E. coli ECL707.
All other manipulations of DNA, PCR, and transformation of plasmids into E. coli were done according to routine procedures (3). The Göttingen Genomics Laboratory (Göttingen, Germany) determined the DNA sequences. Sequence analysis was performed with the Genetics Computer Group program package (16).
Nucleotide sequence accession numbers.
The nucleotide sequences of the inserts of pAK101 to pAK116 have been deposited in the GenBank database. The accession numbers are given in Table 3.
TABLE 3.
Identified genes, GenBank accession numbers, and observed similaritiesa
| Plasmid | Gene identified (accession no.) | Similar protein | Organism (accession no. of similar protein) | % Identity | No. of similar amino acids | Score |
|---|---|---|---|---|---|---|
| pAK101 | orf1 (AF543467) | No similar protein | ||||
| pAK102 | orf2∗ (AF543469) | Hypothetical protein | Yersinia pestis (NP_406777) | 77 | 268 | 424 |
| glpK (AF543469) | Glycerol kinase | Y. pestis (NP_406778) | 46 | 498 | 427 | |
| orf3 (AF543469) | Putative acetyltransferase | Y. pestis (NP_405514) | 44 | 222 | 201 | |
| orf4∗ (AF543469) | Conserved hypothetical protein | Salmonella enterica (NP_455160) | 83 | 356 | 626 | |
| pAK103 | sbmC (AF543472) | SbmC protein, unknown function | Escherichia coli (NP_288514) | 74 | 155 | 254 |
| dacD (AF543472) | Carboxypeptidase | Salmonella enterica serovar Typhimurium (NP_461007) | 90 | 390 | 723 | |
| phsC∗ (AF543472) | Thiosulfate reductase, cytochrome b subunit | S. enterica (NP_456611) | 85 | 71 | 128 | |
| pAK104 | rnb∗ (AF543475) | RNase II | S. enterica serovar Typhimurium (NP_460661) | 88 | 536 | 957 |
| yciR (AF543475) | Hypothetical protein | E. coli (NP_415802) | 66 | 106 | 152 | |
| sbmC (AF543475) | SbmC protein, unknown function | E. coli (NP_288514) | 74 | 155 | 254 | |
| pAK105 | orf5 (AF543478) | Probable monooxygenase/oxidoreductase | Sinorhizobium meliloti (T46818) | 62 | 380 | 499 |
| pAK106 | sodA∗ (AF543480) | Superoxide dismutase | Thermoanaerobacter tengcongensis (NP_622509) | 34 | 184 | 91 |
| greA (AF543480) | Transcription elongation factor | Caulobacter crescentus (NP_421644) | 31 | 151 | 65 | |
| mutY (AF543480) | Hypothetical adenine glycosylase | Ralstonia solanacearum (NP_518522) | 45 | 365 | 286 | |
| orf6 (AF543480) | Hypothetical protein | Pseudomonas aeruginosa (NP_254046) | 51 | 111 | 112 | |
| orf7 (AF543480) | No similar protein | |||||
| orf8 (AF543480) | Putative aminopeptidase | Agrobacterium tumefaciens (NP_355456) | 39 | 241 | 144 | |
| orf9 (AF543480) | Probable response regulator | Rhizobium solanacearum (NP_518671) | 52 | 91 | 100 | |
| orf10 (AF543480) | Hypothetical protein | Mesorhizobium loti (NP_104373) | 33 | 106 | 58 | |
| orf11∗ (AF543480) | No similar protein | |||||
| pAK107 | sucB (AF543482) | 2-Oxoglutarate dehydrogenase, E2 subunit | Vibrio cholerae (NP_231718) | 60 | 225 | 251 |
| sucA∗ (AF543482) | 2-Oxoglutarate dehydrogenase, E1 subunit | Vibrio cholerae (NP_231719) | 74 | 123 | 198 | |
| pAK108 | orf12∗ (AF543483) | Putative zinc-type alcohol dehydrogenase | Mesorhizobium loti (NP_104464) | 31 | 217 | 118 |
| pAK109 | fdhD (AF543481) | Putative FdhD protein | S. enterica (NP_458012) | 88 | 218 | 402 |
| orf13 (AF543481) | Putative membrane protein | Y. pestis (NP_404224) | 24 | 148 | 57 | |
| pAK110 | trrA (AF543479) | Putative transcriptional regulator | Bacillus cereus (CAB69794) | 25 | 415 | 121 |
| yqhC (AF543479) | AraC-type regulatory protein | E. coli (NP_417483) | 89 | 299 | 531 | |
| orf14 (AF543479) | Iron-containing alcohol dehydrogenase | S. enterica (NP_457557) | 90 | 387 | 704 | |
| dkgA∗ (AF543479) | 2,5-Diketo-d-gluconic acid reductase | E. coli (Q56857) | 86 | 140 | 254 | |
| pAK111 | aroD∗ (AF543477) | 3-Dehydroquinate dehydratase | Salmonella enterica serovar Enteritidis (Q9RN77) | 88 | 90 | 157 |
| ydiF (AF543477) | Probable membrane protein | E. coli (NP_416209) | 85 | 103 | 182 | |
| orf15 (AF543479) | Sensor histidine kinase, PleD homologue | Synechocystis sp. (NP_441080) | 38 | 221 | 150 | |
| yedA (AF543477) | IS10, transposase | Plasmid R100 (NP_052934) | 99 | 401 | 818 | |
| orf16∗ (AF543477) | Hypothetical protein | Pseudomonas aeruginosa (NP_251460) | 46 | 228 | 189 | |
| pAK112 | yjhP∗ (AF543476) | Putative methyltransferase | E. coli (NP_418726) | 93 | 114 | 225 |
| orf17 (AF543476) | Hypothetical protein | Y. pestis (NP_405552) | 84 | 277 | 498 | |
| orf18 (AF543476) | Putative transmembrane efflux protein | Streptomyces coelicolor (NP_628101) | 46 | 183 | 160 | |
| orf19 (AF543476) | Probable integral membrane protein | Clostridium perfringens (NP_560977) | 95 | 221 | 403 | |
| pAK113 | hutH∗ (AF543474) | Histidine ammonia-lyase | S. enterica serovar Typhimurium (NP_459769) | 83 | 361 | 606 |
| deoR (AF543474) | Transcriptional regulator | S. enterica serovar Typhimurium (NP_459841) | 88 | 252 | 454 | |
| pAK114 | aroD∗ (AF543473) | 3-Dehydroquinate dehydratase | S. enterica (Q9RN77) | 88 | 90 | 157 |
| ydiF (AF543473) | Probable membrane protein | E. coli (NP_416209) | 85 | 103 | 182 | |
| orf20 (AF543473) | Sensor histidine kinase | Synechocystis sp.(NP_441080) | 38 | 221 | 150 | |
| orf21∗ (AF543473) | Probable oxidoreductase | Xanthomonas axonopodis (NP_641091) | 41 | 244 | 164 | |
| pAK115 | glpX∗ (AF543471) | Glycerol metabolism, unknown function | S. enterica serovar Typhimurium (NP_462966) | 94 | 308 | 561 |
| glpK (AF543471) | Glycerol kinase | E. coli (NP_290555) | 92 | 502 | 952 | |
| glpF (AF543471) | Glycerol uptake facilitator protein | S. enterica (NP_457965) | 86 | 281 | 490 | |
| orf22 (AF543471) | Putative short-chain alcohol dehydrogenase | Agrobacterium tumefaciens (NP_396110) | 55 | 241 | 243 | |
| pAK116 | yaaU∗ (AF543470) | Putative transport protein | E. coli (NP_414587) | 78 | 66 | 109 |
| orf23 (AF543470) | Putative lipoprotein | S. enterica (NP_454690) | 83 | 77 | 129 | |
| yabF (AF543470) | Probable NAD(P)H oxidoreductase | E. coli (NP_414588) | 89 | 176 | 355 | |
| secF (AF543470) | Protein export protein | E. coli (AAB40165) | 88 | 171 | 311 |
The identification of proteins similar to the presumptive gene products encoded by the inserts of pAK101 to pAK116 and the determination of the values for the percent identity, region of similar amino acids, and score were performed by using the Blast programs (1). Incomplete genes are marked by asterisks. The names of genes identified as being responsible for carbonyl formation are shown in bold. The apparent gene organizations are given in Fig. 1.
RESULTS AND DISCUSSION
Enrichment for polyol-fermenting microbial consortia and construction of metagenomic DNA libraries.
The enrichments for polyol-utilizing microorganisms were performed with samples derived from four different environments (soil from a sugar beet field and sediments from the Grone River, Solar Lake, and the Gulf of Eilat). Generally, the use of enrichment steps is associated with the loss of major portions of the microbial populations present in environmental samples (8). In order to maintain a high microbial diversity in the samples during the enrichment for microorganisms harboring the desired genes, we applied mild selective pressure for a short period. In addition, the subsequent DNA isolation was performed by the direct lysis approach. This method is less biased than the fractionation approach, in which the microbial cells are separated from the environmental matrix substances prior to DNA isolation (for a review, see reference 12).
The enrichment medium contained glycerol and 1,2-propanediol as selective agents. These substrates were chosen because several different nicotinamide-dependent dehydrogenases, such as propanol dehydrogenases, propionaldehyde dehydrogenases, 1,2-propanediol dehydrogenases, 1,3-propanediol oxidoreductases, and glycerol dehydrogenases, are known to be involved in the degradation of these substrates (15, 34, 38, 42, 45). In addition, the latter three types of enzymes exhibit a rather broad substrate specificity and are most active with short-chain polyols and the correlating carbonyl compounds (13, 14, 28, 29, 42, 43, 45). After inoculation of the enrichment cultures and growth for 1 day at 30°C, microorganisms and environmental matrix substances were pelleted, used to inoculate fresh medium, and incubated for another day. The enrichment of polyol-utilizing microorganisms was indicated by recording significant substrate consumption in the four different cultures (data not shown). A high degree of diversity within the four enrichment cultures was microscopically verified. Different forms of microbial cells, including cocci, rods, endospore-forming microbes, and chains of short rods, were observed.
Subsequently, the cells and the remaining environmental matrix substances were harvested by centrifugation, and the resulting pellets were used as starting material for the direct isolation of genomic DNA, which was performed as described previously (12, 24). Approximately 1 to 2 mg of DNA was recovered in each case. The DNA obtained was partially digested and size fractionated. DNA fragments of >2 kb were ligated in pSK+ and then transformed into E. coli. The vector pSK+ was selected for library construction because of its high copy number. This allows the detection of even weakly expressed genes in the subsequently performed activity-based screening of the four libraries constructed, because heterologous expression of environmental genes in E. coli is often dependent upon the native promoters (12). The quality of the four different libraries produced was controlled by determination of the average insert size and the percentage of recombinant plasmids containing inserts. The four libraries revealed average insert sizes of 3.0 to 5.6 kb (Table 1). The percentage of plasmids containing inserts was approximately 70 to 80%.
TABLE 1.
Activity-based screening of four environmental librarics for genes conferring the ability to form carbonyl compounds from polyols on E. colia
| Test substrate | Library (avg insert size) | Sample site | No. of carbonyl-forming E. coli clones after initial screen | No. of carbonyl-forming E. coli clones with stable phenotype (plasmid[s]) |
|---|---|---|---|---|
| Glycerol/1,2-propanediol | I (3.3 kb) | River Grone | 5 | 3 (pAK101-pAK103) |
| II (5.4 kb) | Sugar beet field | 6 | 6 (pAK111-pAK116) | |
| III (3.0 kb) | Solar Lake | 1 | 0 | |
| IV (5.6 kb) | Gulf of Eilat | 1 | 1 (pAK108) | |
| 1,2-Ethanediol | I (3.3 kb) | River Grone | 3 | 2 (pAK106-pAK107) |
| II (5.4 kb) | Sugar beet field | 0 | 0 | |
| III (3.0 kb) | Solar Lake | 0 | 0 | |
| IV (5.6 kb) | Gulf of Eilat | 0 | 0 | |
| 2,3-Butanediol | I (3.3 kb) | River Grone | 3 | 2 (pAK104-pAK105) |
| II (5.4 kb) | Sugar beet field | 1 | 0 | |
| III (3.0 kb) | Solar Lake | 4 | 2 (pAK109-pAK110) | |
| IV (5.6 kb) | Gulf of Eilat | 0 | 0 |
Approximately 100,000 E. coli clones from each library per test substrate were screened for carbonyl formation.
Screening for genes conferring alcohol oxidoreductase activity.
In order to access the alcohol oxidoreductase-encoding genes of the four different environmentally derived libraries, the recombinant E. coli strains of each library were screened for production of carbonyl compounds from short-chain (C2 to C4) polyols with adjacent hydroxyl groups. The screening was performed on indicator plates (10) that contained 1,2-ethanediol, 2,3-butanediol, or a mixture of 1,2-propanediol and glycerol as test substrates and a mixture of pararosaniline and bisulfite for the detection of carbonyl compounds formed by the E. coli clones. Upon production of carbonyls from the test substrates by recombinant E. coli strains, an intensely red Schiff base is formed. Thus, E. coli colonies capable of carbonyl formation appeared red on indicator medium and were surrounded by a red zone, whereas colonies failing to produce carbonyl compounds remained uncolored. To avoid a high background coloring of the indicator plates by glycerol dehydrogenase activity of E. coli, the glycerol-minus mutant E. coli ECL707 (41) was used as the host for the libraries during the screening procedure.
Positive colonies were best visualized after overnight incubation at 37°C. Such colonies completely colored the indicator plates after prolonged incubation. The reliability of the entire activity-based screening procedure was controlled by transferring pRD1 (13, 14) or pFL1 (30), harboring the genes encoding NAD+-dependent alcohol dehydrogenases from Citrobacter freundii and Clostridium pasteurianum, respectively, into E. coli ECL707. Both resulting recombinant E. coli strains showed a strong reaction on the indicator plates, in contrast to E. coli ECL707. Subsequently, the screening of the four metagenomic libraries was initiated by transformation of the recombinant plasmids of each constructed library in E. coli ECL707, followed by transfer of the resulting E. coli clones on indicator plates containing the different types of test substrates (see above). Approximately 100,000 E. coli clones of each library per test substrate were screened for formation of carbonyls.
Twenty-four E. coli clones were positive during the initial screen of the four libraries constructed (Table 1). Most of the clones were obtained from libraries I and II and with the glycerol/1,3-propanediol mixture as the test substrate (Table 1). In order to confirm that the carbonyl-forming phenotype of the positive clones was plasmid encoded and stable, the recombinant plasmids were isolated and retransformed into E. coli, and the resulting E. coli strains were screened again on appropriate indicator plates. Sixteen different recombinant plasmids, designated pAK101 to pAK116, conferred a stable carbonyl-forming phenotype on the resulting recombinant E. coli strains. Seven of these plasmids were derived from library I, six from library II, two from library III, and one from library IV (Table 1). The insert sizes of pAK101 to pAK116 were in the range of 651 to 8,249 bp (Fig. 1). The corresponding E. coli strains (E. coli/pAK101 to E. coli/pAK116) were studied further.
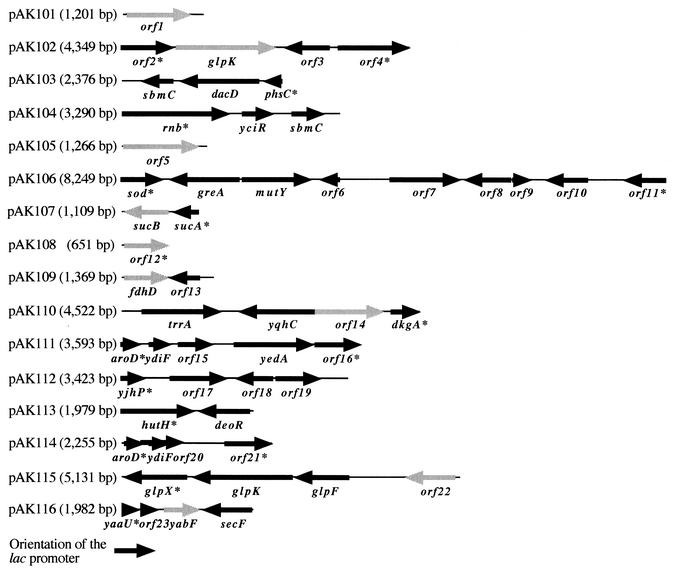
FIG. 1.
Genetic organization and length of the inserts of pAK101 to pAK116. The insert sizes of the plasmids are given in brackets. Arrows and arrowheads represent the lengths, locations, and orientations of the potential genes within the inserts. The arrows of potential genes which were identified as being responsible for the carbonyl-forming phenotype of the E. coli clones are shaded gray. The gene designations are written below the arrows. Incomplete open reading frames are marked with asterisks. Observed similarities of the deduced amino acid sequences to sequences available in the NCBI databases are listed in Table 3.
Biochemical characterization of carbonyl-forming E. coli clones.
Crude extracts of the 16 positive E. coli clones obtained during the screening procedure were prepared and tested for NAD(P)+-dependent oxidation and NAD(P)H-dependent reduction of various short-chain alcohols and carbonyl compounds, respectively. NADP+- or NADPH-dependent reactions with the tested substrates were not detectable in the crude extracts of all E. coli clones. E. coli/pAK103, E. coli/pAK104, E. coli/pAK108, E. coli/pAK109, E. coli/pAK110, E. coli/pAK112, E. coli/pAK113, and E. coli/pAK116 showed NAD+-dependent oxidation or NADH-dependent reduction of one or more substrates (Table 2). Substrate specificity studies revealed that the enzymes produced by these clones were capable of catalyzing a number of oxidation and reduction reactions except the enzyme encoded by pAK116, which was specific for 1,2-ethanediol under the assay conditions employed (Table 2).
TABLE 2.
Substrate specificity of nicotinamide-dependent dehydrogenase reactions in crude extracts of E. coli carrying various plasmidsa
| Reaction and substrate | Relative sp act in crude extracts of corresponding clones (%)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| pAK103 | pAK104 | pAK108 | pAK109 | pAK110 | pAK112 | pAK113 | pAK116 | |
| Oxidation | ||||||||
| 1,2-Ethanediol | — | — | — | — | — | 5 | — | 100 |
| 1,2-Propanediol | 81 | 100 | 100 | 100 | 100 | 100 | 100 | — |
| 2,3-Butanediol | — | 22 | 50 | 31 | 44 | 32 | 43 | — |
| Glycerol | 100 | 35 | 91 | 36 | 40 | 44 | 68 | — |
| 1,2,4-Butanetriol | 46 | 14 | 27 | 26 | 27 | 16 | 36 | — |
| Reduction | ||||||||
| Dihydroxyacetone | 76 | 100 | — | 78 | 100 | 100 | 100 | — |
| Hydroxyacetone | 9 | 39 | — | 41 | 70 | — | — | — |
| Glycolaldehyde | 100 | 19 | — | 100 | 93 | — | 53 | — |
| Methylglyoxal | 33 | — | — | — | — | — | — | — |
The specific alcohol dehydrogenase activities were determined with various short-chain (C2 to C4) alcohols or carbonyls as substrates and NAD+ or NADH, respectively, as the cofactor. The oxidation of the alcohols and the reduction of the carbonyl compounds were assayed as described in Materials and Methods. The specific background activities of the E. coli host containing the cloning vector were measured in each case and subtracted. The specific activities in oxidation reactions were expressed relative to that obtained with the substrate yielding the highest value in each case (pAK103, 0.024 U/mg; pAK104, 0.033 U/mg; pAK108, 0.022 U/mg; pAK109, 0.039 U/mg; pAK110, 0.048; pAK112, 0.057 U/mg; pAK113, 0.028 U/mg; and pAK116, 0.040). —, no detectable activity. The specific activities in reduction reactions were expressed relative to that obtained with the substrate yielding the highest value in each case (pAK103, 0.054 U/mg; pAK104, 0.080 U/mg; pAK109, 0.044 U/mg; pAK110, 0.137 U/mg; pAK112, 0.032 U/mg; and pAK113, 0.068 U/mg).
The enzymes were most active with polyols containing two adjacent alcohol groups. The preferred substrates were glycerol (pAK103), 1,2-propanediol (pAK104, pAK108, pAK109, pAK110, pAK112, and pAK113), and 1,2-ethanediol (pAK116). A significant oxidation of primary or secondary alcohols containing single hydroxyl groups such as ethanol, 1-propanol, 2-propanol, 1-butanol, and 2-butanol was not detected (data not shown). The reduction reaction was more specific (Table 2). The enzymes produced were most active with dihydroxyacetone (pAK104, pAK110, pAK112, and pAK113) or glycolaldehyde (pAK103 and pAK109). No significant reduction of glyceraldehyde, diacetyl, acetaldehyde, or acetone was observed. Although E. coli/pAK108 and E. coli/pAK116 exhibited significant activity in oxidation reactions, NADH-dependent reductions of the tested carbonyl compounds were not detected under these assay conditions.
In contrast to the above-mentioned carbonyl-forming E. coli clones, the other eight positive clones (E. coli/pAK101, E. coli/pAK102, E. coli/pAK105, E. coli/pAK106, E. coli/pAK107, E. coli/pAK111, E. coli/pAK114, and E. coli/pAK115) exhibited nicotinamide-dependent activities with the tested substrates that did not significantly exceed the background activities of the E. coli control harboring the cloning vector (data not shown). This result indicated that these strains did not contain genes encoding an NAD(H)-dependent dehydrogenase activity. The carbonyl-forming phenotype of these E. coli clones on indicator plates was probably caused by the activities of other classes of enzymes acting on alcohols, such as metallooxidases, dehydratases, and flavin adenine dinucleotide-dependent or pyrroloquinoline quinone-dependent dehydrogenases (15, 19, 25, 40). In addition, the assay conditions employed may not be suitable for the activity of the gene products produced by these clones and may also account for the lack of activity of the dehydrogenase activity-exhibiting clones with certain substrates.
Molecular analyses.
The inserts of pAK101 to pAK116 were sequenced and compared to the sequences available in the National Center for Biotechnology Information (NCBI) databases. The identified genes and the observed similarities of the deduced amino acid sequences to known gene products are summarized in Table 3 and Fig. 1. Thirty-six complete and 17 incomplete protein-encoding genes were predicted within the sequences of all inserts, with 50 showing significant similarity to sequenced genes from other organisms. All were most similar to bacterial homologues derived from a wide variety of different genera, i.e., Salmonella, Yersinia, Thermoanaerobacter, Pseudomonas, Sinorhizobium, Agrobacterium, and Ralstonia.
Although the observed similarities of some of the identified genes to known genes, especially to genes from enteric bacteria, were surprisingly high, the sequences of almost all predicted genes showed significant differences from sequenced genes from individual microorganisms. Thus, we have no evidence indicating from which species the inserts of the recovered plasmids were derived. The similarity to genes from enteric bacteria could also be a result of the activity screen, which was based on the ability of E. coli clones to form carbonyl compounds from polyols. This allowed only the detection of cloned genes whose promoters were recognized by the E. coli host strain or of genes expressed via the plasmid-encoded lac promoter, which is induced in the presence of isopropyl-1-thio-β-d-galactopyranoside (IPTG). The activity of the lac promoter was probably very low under the growth conditions employed, since IPTG was not added to the growth medium. In addition, the latter mode of expression was only possible for genes oriented in the same direction as the vector-encoded lac promoter (Fig. 1).
The use of other hosts, such as Bacillus or Streptomyces, and mobilizable expression vectors might have increased the diversity and number of resulting clones. However, in all published reports, E. coli has been used as the host for metagenomic libraries during activity-based screening programs. Sequencing of more than one recombinant plasmid recovered from the positive E. coli clones has been performed in only a few studies (23, 24, 31, 46). The results obtained have shown that activity-based screens of metagenomic libraries are suitable for the detection of genes which are similar to genes derived from microorganisms weakly related to the E. coli host. For example, our screening of metagenomic libraries derived from soil samples led to the identification of genes encoding 4-hydroxybutyrate dehydrogenases which were most similar to the genes from Ralstonia eutropha, Clostridium acetobutylicum, and Synechocystis sp. strain PCC6803 (24).
The known and sequenced alcohol oxidoreductases can be divided into three major categories (35). (i) The NAD(P)+-dependent dehydrogenases are the best characterized of the three groups. These can in turn be separated into type I long-chain zinc-containing enzymes, such as alcohol dehydrogenase I of Saccharomyces cerevisiae and Zymomonas mobilis (44); type II short-chain zinc-independent enzymes, such as ribitol dehydrogenase of Enterobacter aerogenes and glucose dehydrogenase of Bacillus megaterium (32); and type III iron-dependent enzymes, such as alcohol dehydrogenase II of Z. mobilis (10) and glycerol dehydrogenase of Citrobacter freundii (13). Most of the characterized alcohol oxidoreductases, which are involved in the degradation of glycerol and 1,2-propanediol by individual microorganisms such as Citrobacter freundii (13, 14), Clostridium pasteurianum (30), Erwinia aroidea (43), Desulfovibrio sp. strain HDv (34), Cellulomonas sp. (47), Lactobacillus reuteri (45), and Klebsiella pneumoniae (28), belong to the last family of NAD(H)-dependent alcohol dehydrogenases (13, 14, 28, 30). (ii) NAD(P)+-independent enzymes use other cofactors, such as pyrroloquinoline quinone or F420, exemplified by the ethanol dehydrogenase of Acetobacter aceti (27) and the alcohol dehydrogenase of Methanobacterium palustre (5). (iii) Oxidases that catalyze an essentially irreversible oxidation of alcohols, such as the flavin adenine dinucleotide-dependent alcohol oxidase 1 of Candida boidinii (39).
DNA sequence analyses of the inserts of pAK101 (1,201 bp), pAK105 (1,266 bp), and pAK108 (651 bp) revealed in each case one large potential gene of 969, 1,137, and 534 bp, respectively, within the sequence (Fig. 1). The amino acid sequence deduced from orf1 located on pAK101 exhibited no significant similarities to the above-mentioned alcohol oxidoreductases or to sequences and signatures of proteins available in the NCBI and PROSITE databases, whereas the proteins deduced from orf5 (pAK105) and orf12 (pAK108) showed similarities to oxidoreductases from other organisms. The orf5 gene product showed 62% identity to a putative monooxygenase/oxidoreductase of Sinorhizobium meliloti, and the partial orf12 gene product showed 31% identity to a putative NAD+-dependent zinc-containing (type I) alcohol dehydrogenase of Mesorhizobium loti (Table 3). These results corresponded well with those obtained during the determination of the alcohol dehydrogenase activity in crude extracts of the correlating E. coli clones, since solely E. coli/pAK108 harboring orf12 exhibited NAD+-dependent dehydrogenase activity with polyols as the substrates (Table 2).
Sequencing of the other 13 plasmids revealed in each case more than one potential gene within the sequence of the insert (Fig. 1). In order to identify the genes on the plasmids that were responsible for the formation of carbonyl compounds by the corresponding recombinant E. coli strains, the inserts were partially subcloned by restriction digestion with various enzymes or PCR with primers derived from sequencing and subsequent ligation into pSK+. The resulting constructs were transformed into E. coli, and the recombinant E. coli strains were screened again on indicator plates containing 1,2-ethanediol, glycerol/1,2-propanediol, or 2,3-butanediol as the substrate. This method was successful for the inserts of pAK102, pAK107, pAK109, pAK110, pAK115, and pAK116. The corresponding subclones were indistinguishable from the original clones with respect to NAD(H)-dependent alcohol dehydrogenase activities in crude extracts. In all six cases, single genes were responsible for the carbonyl-forming phenotype (Fig. 1). All attempts to subclone a DNA region conferring the carbonyl-forming phenotype of pAK103, pAK104, pAK106, pAK111, pAK112, pAK113, and pAK114 failed. For most of these plasmids, we were unable to recover all of the possible subclones. Since subcloning can lead to changes in expression levels of genes, this may have resulted from toxic effects caused by increased levels of the foreign gene products in E. coli.
All six identified genes conferring the carbonyl-forming phenotype were preceded by a potential ribosome-binding site appropriately spaced from the start codon. The deduced gene products of all genes except the one located on pAK102 revealed significant similarities to known oxidoreductases (Table 3). The protein deduced from the glpK gene of pAK2 (502 amino acids) was 46% identical to the glycerol kinase of Yersinia pestis. This indicated that production of the carbonyl compounds from glycerol on indicator agar by E. coli ECL707/pAK102 was initiated by phosphorylation of glycerol catalyzed by the heterologously produced kinase. Subsequently, the resulting glycerol 3-phosphate is oxidized by a dehydrogenase, i.e., glycerol 3-phosphate dehydrogenase, provided by the E. coli host. This mode of glycerol consumption can also account for the lack of NAD(H)-dependent enzyme activity with the tested substrates in crude extracts of the E. coli clone. We cannot exclude that indirect reactions like this also account for the carbonyl-forming phenotype of some other positive clones, since the introduction of unknown environmental DNA into the E. coli host can yield hybrid products whose synthesis is directed in part by the host genome and in part by the introduced DNA.
Interestingly, one of the proteins deduced from the potential genes located on pAK115 was also highly similar to glycerol kinases of enteric bacteria, but the orf22 gene product (247 amino acids) identified as being responsible for the carbonyl-forming phenotype of E. coli ECL707 was related to a type II short-chain alcohol dehydrogenase of Agrobacterium tumefaciens (55% identity). The short-chain dehydrogenase/reductase family is very large, and most members are proteins of approximately 250 to 300 amino acids. The signature pattern for this protein family (LIVSPADNK-x12-Y-PSTAGNCV-STAGNQCIVM-STAGC-K-PC-SAGFYR-LIVMSTAGD-x2-LIVMFYW-x3-LIVMFYWGAPTHQ-GSACQRHM) includes two perfectly conserved residues, a tyrosine and a lysine. The tyrosine residue participates in the catalytic mechanism (18). This motif was retained in the deduced amino acid sequence of orf22 (amino acids 143 to 171) except that the first amino acid residue (LIVSPADNK) was replaced by a cytosine residue (see GenBank accession).
In addition to this gene product, the one responsible for the carbonyl-forming phenotype encoded by pAK110 (Orf14; 387 amino acids) was also similar to one of the above-mentioned categories of alcohol oxidoreductases. The deduced Orf14 protein sequence showed 90% identity to the putative iron-containing type III alcohol dehydrogenase of Salmonella enterica. For the detection of type III alcohol dehydrogenases, two specific fingerprint patterns are available in the PROSITE database (18); the first one is STALIV-LIVF-x-DE-x6, 7-P-x4-ALIV-x-GST-x2-D-TAIVM-LIVMF-x4-E and the second one is GSW-x-LIVTSACD-GH-x2-GSAE-GSHYQ-x-LIVTP-GAST-GAS-x3-LIVMT-x-HNS-GA-x-GTAC. The orf14 gene product showed both fingerprint patterns (amino acids 173 to 201 and 264 to 284). In addition, the correlating E. coli clone (Table 2) and the purified gene product (data not shown) exhibited NAD+-dependent activity with a variety of polyols, which is typical of some type III alcohol dehydrogenases, such as glycerol dehydrogenases and 1,3-propanediol dehydrogenases (13, 14, 28, 30).
The deduced amino acid sequences of the other three presumptive oxidoreductase-encoding genes sucB, fdhD, and yabF identified during subcloning of pAK107, pAK109, and pAK116, respectively, were similar to those of members of the groups of oxidoreductases, which were different from the above-described alcohol oxidoreductases. The amino acid sequences deduced for the sucB, yabF, and fdhD products (214, 176, and 212 amino acids, respectively) were most similar to a part of the 2-oxoglutarate dehydrogenase enzyme complex of Vibrio cholerae, a probable NAD(P)H oxidoreductase from E. coli, and a putative protein required for formate dehydrogenase activity from Salmonella enterica. The last putative gene product is found in other organisms, such as Xanthomonas campestris (NP_637709) and Mesorhizobium loti (NP_106066) as part of the formate dehydrogenase. The yabF and fdhD gene products conferred NAD(H)-dependent activity with short-chain polyols or carbonyl compounds as substrates on the E. coli clones (Table 2). To our knowledge, the reactions with these substrates have not been reported for the related proteins of other organisms.
The sequence analyses of the seven plasmids for which the identification of a single gene responsible for the carbonyl-forming phenotype failed (see above) revealed for open reading frames (phsC and orf21, respectively) encoding putative enzymes similar to known oxidoreductases/reductases in pAK103, and pAK114, respectively, but both putative genes were incomplete (Table 3). In addition to these two deduced gene products, the inserts of the correlating plasmids and those of the remaining five plasmids (pAK104, pAK106, pAK111, pAK112, and pAK113) encoded putative gene products similar to proteins with unknown functions or to gene products involved in a broad range of different metabolic reactions (Table 3). Although pAK103, pAK104, pAK112, and pAK113 conferred significant NAD(H)-dependent dehydrogenase activities on E. coli, none of the plasmids encoded complete genes similar to those encoding oxidoreductases. These may therefore be entirely new genes encoding products mediating these reactions. Another possibility is that the gene products facilitate the dehydrogenase activity of the recombinant E. coli strains and the carbonyl-forming phenotype in a not yet understood way. The function of these genes will be unraveled by characterization of the purified gene products.
In summary, nine novel genes were identified as being responsible for the formation of carbonyls from short-chain polyols by recombinant E. coli strains during subcloning and sequence analyses. Three (orf12, orf14, and orf22) encoded presumptive alcohol dehydrogenases, four (orf5, sucB, fdhD, and yabF) encoded putative oxidoreductases belonging to groups different from alcohol dehydrogenases, one (glpK) encoded a putative glycerol kinase, and one (orf1) encoded a protein unrelated to known gene products. Thus, the majority of the gene products encountered were similar to groups of enzymes for which the formation of carbonyls from the polyols employed in this study have not been reported. In addition, type III alcohol dehydrogenases are involved in the consumption of glycerol and 1,2-propanediol by the characterized microorganisms (see above), but only one of the identified gene products (Orf14) was related to this enzyme type. These results are typical for activity-based screening programs of libraries derived from complex microbial assemblages.
Most of the genes and the corresponding gene products discovered are entirely novel, similar to genes with unknown functions, and weakly related or unrelated to known genes encoding the targeted functions (20, 23, 24, 37, 46). The novelty of the biocatalysts encountered in this line of work arises from the widely unexplored enormous genetic and metabolic diversity of uncultured microorganisms (12, 20, 37). Nevertheless, the involvement of enrichment steps yields metagenomic libraries that are more biased than those constructed without these steps, but the results presented here and by other authors (17, 22) showed that a diverse set of genes conferring the targeted reaction were still recovered by this approach. Thus, the coupling of classical enrichment and modern metagenomic technologies in combination with simple activity-based screening systems is a good way to isolate a large number of diverse genes conferring redox reactions with polyols and the corresponding carbonyls. The number of genes and gene products performing this reaction can be extended by increasing the number of clones screened. In this way, gene banks consisting of several hundred genes conferring certain types of industrially important reactions can be prepared rapidly. These gene banks or the corresponding clones can serve as starting material for the development of novel processes and products. Future studies targeting other biocatalysts will define the true scope and general usefulness of this approach.
Acknowledgments
We thank Gerhard Gottschalk for generous support and helpful discussions.
This work was supported by the Bundesministerium für Bildung und Forschung within the Göttinger Competence Center “Genomforschung an Bakterien für die Analyze der Biodiversität und die Nutzung zur Entwicklung neuer Produktionsverfahren.”
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 4.Béjà, O., M. T. Suzuki, E. V. Koonin, L. Aravind, A. Hadd, L. P. Nguyen, M. Amjadi, C. Garrigues, S. B. Jovanovich, R. A. Feldman, and E. F. DeLong. 2000. Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol. 2:516-529. [DOI] [PubMed] [Google Scholar]
- 5.Bleicher, K., and J. Winter. 1991. Purification and properties of F420 and NADP+-dependent alcohol dehydrogenases of Methanogenium liminitans and Methanobacterium palustre specific for secondary alcohols. Eur. J. Biochem. 200:43-51. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Brady, S. F., C. J. Chao, J. Handelsman, and J. Clardy. 2001. Cloning and heterologous expression of a natural product biosynthetic gene cluster from eDNA. Org. Lett. 3:1981-1984. [DOI] [PubMed] [Google Scholar]
- 8.Caldwell, D. E., G. M. Wolfaardts, D. R. Kober, and J. R. Lawrence. 1997. Cultivation of microbial consortia and communities, p. 79-90. In J. H. Hurst, G. R. Knudsen, M. J. McInerey, L. D. Stetzenbach, and M. V. Walter (ed.), Manual of environmental microbiology. ASM Press, Washington, D.C.
- 9.Chenault, H. K., and G. M. Whitesides. 1987. Regeneration of nicotinamide cofactors for use in organic synthesis. Appl. Biochem. Biotechnol. 14:147-197. [DOI] [PubMed] [Google Scholar]
- 10.Conway, T., G. W. Sewell, Y. A. Osman, and L. O. Ingram. 1987. Cloning and sequencing of the alcohol dehydrogenase II gene from Zymomonas mobilis. J. Bacteriol. 169:2591-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cottrell, M. T., J. A. Moore, and D. L. Kirchman. 1999. Chitinases from uncultured microorganisms. Appl. Environ. Microbiol. 65:2553-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daniel, R. 2002. Construction of environmental libraries for functional screening of enzyme activity, p. 63-78. In S. Brakmann and K. Johnsson (ed.), Directed molecular evolution of proteins. Wiley-VCH Verlag GmbH, Weinheim, Germany.
- 13.Daniel, R., K. Stuertz, and G. Gottschalk. 1995. Biochemical and molecular characterization of the oxidative branch of glycerol utilization by Citrobacter freundii. J. Bacteriol. 177:4392-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daniel, R., R. Boenigk, and G. Gottschalk. 1995. Purification of the 1,3-propanediol dehydrogenase from Citrobacter freundii and cloning, sequencing and overexpression of the corresponding gene in Escherichia coli. J. Bacteriol. 177:2151-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daniel, R., T. A. Bobik, and G. Gottschalk. 1998. Biochemistry of coenzyme B12-dependent glycerol and diol dehydratases and organization of the encoding genes. FEMS Microbiol. Rev. 22:553-566. [DOI] [PubMed] [Google Scholar]
- 16.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Entcheva, P., W. Liebl, A. Johann, T. Hartsch, and W. Streit. 2001. Direct cloning from enrichment cultures, a reliable strategy for isolation of complete operons and genes from microbial consortia. Appl. Environ. Microbiol. 67:89-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falquet, L., M. Pagni, P. Bucher, N. Hulo, C. J. Sigrist, K. Hofmann, and A. Bairoch. 2002. The PROSITE database: its status in 2002. Nucleic Acids Res. 30:235-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang, J.-M., C.-H. Lin, C. W. Bradshaw, and C.-H. Wong. 1995. Enzymes in organic synthesis: oxidoreductions. J. Chem. Soc. Perkin Trans. 1:967-978. [Google Scholar]
- 20.Gupta, R., Q. K. Berg, and P. Lorenz. 2002. Bacterial alkaline proteases: molecular approaches and industrial applications. Appl. Microbiol. Biotechnol. 59:15-32. [DOI] [PubMed] [Google Scholar]
- 21.Handelsman, J., M. R. Rondon, S. F. Brady, J. Clardy, and R. M. Goodman. 1998. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol. 5:R245-249. [DOI] [PubMed] [Google Scholar]
- 22.Healy, F. G., R. M. Ray, H. C. Aldrich, A. C. Wilkie, L. O. Ingram, and K. T. Shanmugam. 1995. Direct isolation of functional genes encoding cellulases from the microbial consortia in a thermophilic, anaerobic digester maintained on lignocellulose. Appl. Microbiol. Biotechnol. 43:667-674. [DOI] [PubMed] [Google Scholar]
- 23.Henne, A., R. A. Schmitz, M. Bömeke, G. Gottschalk, and R. Daniel. 2000. Screening of environmental DNA libraries for the presence of genes conferring lipolytic activity on Escherichia coli. Appl. Environ. Microbiol. 66:3113-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henne, A., R. Daniel, R. A., Schmitz, and G. Gottschalk. 1999. Construction of environmental DNA libraries in Escherichia coli and screening for the presence of genes conferring utilization of 4-hydroxybutyrate. Appl. Environ. Microbiol. 65:3901-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hummel, W. 1997. New alcohol dehydrogenases for the synthesis of chiral compounds. Adv. Biochem. Eng. Biotechnol. 58:145-184. [DOI] [PubMed] [Google Scholar]
- 26.Hummel, W. 1999. Large-scale applications of NAD(P)-dependent oxidoreductases: recent developments. Trends Biotechnol. 17:487-492. [DOI] [PubMed] [Google Scholar]
- 27.Inoue, T., M. Sunagawa, A. Mori, A. Imai, C. Imai, M. Fukuda, M. Takagi, and K. Yano. 1992. Nucleotide sequencing and characterization of the gene encoding the 45-kilodalton subunit of alcohol dehydrogenase from Acetobacter aceti. J. Ferment. Bioeng. 73:419-424. [Google Scholar]
- 28.Johnson, E. A., and E. C. C. Lin. 1987. Klebsiella pneumoniae 1,3-propanediol:NAD+-oxidoreductase. J. Bacteriol. 169:2050-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelley, J. L., and E. E. Dekker. 1985. Identity of Escherichia coli D-1-amino-2-propanol:NAD+ with E. coli glycerol dehydrogenase but not with Neisseria gonorrhoeae 1,2-propanediol:NAD+ oxidoreductase. J. Bacteriol. 162:170-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luers, F., M. Seyfried, R. Daniel, and G. Gottschalk. 1997. Glycerol conversion to 1,3-propanediol by Clostridium pasteurianum: cloning and expression of the gene encoding 1,3-propanediol dehydrogenase. FEMS Microbiol. Lett. 154:337-345. [DOI] [PubMed] [Google Scholar]
- 31.Majernik, A., G. Gottschalk, and R. Daniel. 2001. Screening of environmental DNA libraries for the presence of genes conferring Na+(Li+)/H+ antiporter activity on Escherichia coli: characterization of the recovered genes and the corresponding gene products. J. Bacteriol. 183:6645-6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Persson, B., M. Krook, and H. Jörnvall. 1991. Characteristics of short-chain alcohol dehydrogenases and related enzymes. Eur. J. Biochem. 200:537-543. [DOI] [PubMed] [Google Scholar]
- 33.Pfennig, N., and K. D. Lippert. 1966. Über das Vitamin B12-Bedürfnis phototropher Schwefelbakterien. Arch. Microbiol. 55:245-256. [Google Scholar]
- 34.Quattara, A. S., N. Cuzin, A. S. Traore, and J.-L. Garcia. 1992. Anaerobic degradation of 1,2-propanediol by a new Desulfovibrio strain and D. alcoholvorans. Arch. Microbiol. 158:218-225. [DOI] [PubMed] [Google Scholar]
- 35.Reid, M. F., and C. A. Fewson. 1994. Molecular characterization of microbial alcohol dehydrogenases. Crit. Rev. Microbiol. 20:13-56. [DOI] [PubMed] [Google Scholar]
- 36.Richardson, T. H., X. Tan, G. Frey, W. Callen, M. Cabell, D. Lam, J. Macomber, J. M. Short, D. E. Robertson, and C. Miller. 2002. A novel, high performance enzyme for starch liquefaction. Discovery and optimization of a low-pH, thermostable alpha-amylase. J. Biol. Chem. 277:26501-26507. [DOI] [PubMed] [Google Scholar]
- 37.Rondon, M. R., P. R. August, A. D. Bettermann, S. F. Brady, T. H. Grossman, M. R. Liles, K. A. Loiacono, B. A. Lynch, L. A. MacNeil, C. Minor, C. L. Tiong, M. Gilman, M. S. Osburne, J. Clardy, J. Handelsman, and R. M. Goodman. 2000. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 66:2541-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruch, F. E., J. Lengeler, and E. C. C. Lin. 1974. Regulation of glycerol catabolism in Klebsiella aerogenes. J. Bacteriol. 119:50-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakai, Y., and Y. Tani. 1992. Cloning and sequencing of the alcohol oxidase-encoding gene (AOD1) from the formaldehyde-producing asporogenous methylotrophic yeast Candida boidinii S2. Gene 114:67-73. [DOI] [PubMed] [Google Scholar]
- 40.Seyfried, M., R. Daniel, and G. Gottschalk. 1996. Cloning, sequencing and overexpression of the genes encoding coenzyme B12-dependent glycerol dehydratase of Citrobacter freundii. J. Bacteriol. 178:5793-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sprenger, G. A., B. A Hammer, E. A. Johnson, and E. C. C. Lin. 1989. Anaerobic growth of Escherichia coli on glycerol by importing genes of the dha regulon from Klebsiella pneumoniae. J. Gen. Microbiol. 135:1255-1262. [DOI] [PubMed] [Google Scholar]
- 42.Sridhara, S., T. T. Wu, T. M. Chused, and E. C. C. Lin. 1969. Ferrous-activated nicotinamide adenine dinucleotide-linked dehydrogenase from a mutant of Escherichia coli capable of growth on 1,2-propanediol. J. Bacteriol. 98:87-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sugiura, M., T. Oikawa, K. Hirano, H. Shimizu, and F. Hirata. 1978. Purification and some properties of glycerol dehydrogenase from Erwinia aroideae. Chem. Pharm. Bull 26:716-721. [DOI] [PubMed] [Google Scholar]
- 44.Sun, H.-W., and B. V. Plapp. 1992. Progressive sequence alignment and molecular evolution of the Zn-containing alcohol dehydrogenase family. J. Mol. Evol. 34:522-535. [DOI] [PubMed] [Google Scholar]
- 45.Talarico, T. L., L. T. Axelsson, J. Novotny, M. Fiuzat, and W. J. Dobrogosz. 1990. Utilization of glycerol as hydrogen acceptor by Lactobacillus reuteri: purification of 1,3-propanediol:NAD+ oxidoreductase. Appl. Environ. Microbiol. 56:943-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilkinson, D. E., T. Jeanicke, and D. A. Cowan. 2002. Efficient molecular cloning of environmental DNA from geothermal sediments. Biotechnol. Lett. 24:155-161. [Google Scholar]
- 47.Yamada, H., A. Nagao, H. Nishise, and Y. Tani. 1982. Formation of glycerol dehydrogenase by microorganisms. Agric. Biol. Chem. 46:2325-2331. [Google Scholar]