Abstract
Sporulation in yeast requires that a modified form of chromosome segregation be coupled to the development of a specialized cell type, a process akin to gametogenesis. Mps1p is a dual-specificity protein kinase essential for spindle pole body (SPB) duplication and required for the spindle assembly checkpoint in mitotically dividing cells. Four conditional mutant alleles of MPS1 disrupt sporulation, producing two distinct phenotypic classes. Class I alleles of mps1 prevent SPB duplication at the restrictive temperature without affecting premeiotic DNA synthesis and recombination. Class II MPS1 alleles progress through both meiotic divisions in 30–50% of the population, but the asci are incapable of forming mature spores. Although mutations in many other genes block spore wall formation, the cells produce viable haploid progeny, whereas mps1 class II spores are unable to germinate. We have used fluorescently marked chromosomes to demonstrate that mps1 mutant cells have a dramatically increased frequency of chromosome missegregation, suggesting that loss of viability is due to a defect in spindle function. Overall, our cytological data suggest that MPS1 is required for meiotic SPB duplication, chromosome segregation, and spore wall formation.
INTRODUCTION
Gamete formation in sexually reproducing organisms consists of meiotic chromosome segregation to generate haploid cells coupled with a developmental pathway to generate specialized cell bodies equipped for fertilization. Meiotic cell division in the budding yeast Saccharomyces cerevisiae is initiated when a diploid cell of a/α mating type is starved for a fermentable carbon source and nitrogen. Starvation of yeast initiates a transcriptional program that controls meiotic DNA synthesis, recombination, and chromosome segregation and coordinates these events with the generation of spore bodies (Malone, 1990; Chu et al., 1998). Successful sporulation in yeast requires the proper coordination of meiotic spindle formation and segregation of chromosomes into the four developing spore bodies.
Two significant distinctions between meiotic and mitotic chromosome segregation include the duplication of the spindle pole body (SPB; the yeast equivalent of the centrosome) and the formation and function of the meiotic spindles. During meiotic chromosome segregation, two rounds of spindle formation occur after a single round of DNA synthesis, resulting in a reduction of the chromosome number by half in the progeny spores. Presumably, the physical duplication of the SPB requires the same fundamental assemblage of components during meiosis and mitosis; however, regulation of the process must differ significantly because of the requirement of two successive duplications of the meiotic SPBs. Furthermore, the SPBs become the site of prospore wall initiation after a modification to the cytoplasmic face of this organelle at the time of the second duplication (Moens and Rappaport, 1971). Not only are the spindle poles required to build spindles for chromosome segregation, as they are in mitosis, but they are also modified for spore formation, a process unique to meiosis. These observations suggest that the SPB may be an organelle of central importance in the coordination of meiosis and spore formation.
A host of meiosis-specific genes are required for spore wall formation and could potentially affect SPB function because of its role in prospore wall initiation. A meiosis-specific gene that has a role in SPB duplication is SPO1 (Moens et al., 1974; Tevzadze et al., 2000). The spo1 mutation causes a largely monopolar phenotype in sporulating yeast, and sequence analysis revealed strong similarity to the phospholipase B–encoding gene PLB1 (Tevzadze et al., 1996). A better characterized set of genes regulate and carry out spore wall formation. A sporulation-specific MAPK, SMK1, was demonstrated to regulate spore formation (Krisak et al., 1994). Two upstream kinases, SPS1 and CAK1, modulate the function of Smk1p, although CAK1 is not a meiosis-specific gene (Wagner et al., 1997). Also, the recently discovered gene SWM1 is a regulator of spore wall formation (Ufano et al., 1999). SMK1, SPS1, CAK1, and SWM1 control spore wall formation through transcriptional regulation of a set of effector genes represented by DIT1, DIT2, SPS100, and others. Double mutant analysis has demonstrated that SMK1 and SPS1 function in a pathway separate from SWM1 (Ufano et al., 1999). Furthermore, Sps1p may have a meiotic function in addition to its proposed role as a Smk1p upstream activating kinase (Friesen et al., 1994), suggesting that a complex signaling network regulates spore wall initiation and assembly.
Many of the genes required during the vegetative cell cycle are also used in the context of meiosis and spore formation (Simchen, 1974). The effects of two mutations that disrupt the process of SPB duplication in mitotic cells have been studied in meiotic cells. In vegetatively growing cells, both cdc31-1 and ndc1-1, which define the early and late steps, respectively, of SPB duplication, prevent mitotic SPB duplication and trigger cell cycle arrest by activating the spindle assembly checkpoint (Winey et al., 1993; Weiss and Winey, 1996). The meiotic effects of the cdc31-1 and ndc1-1 mutations result in viable dyads (two-spored asci) as a result of the failure in the second of the two rounds of SPB duplication (Byers, 1981; Thomas and Botstein, 1986). Predominantly, the chromosomes undergo reductional segregation of paired homologues, indicating a frequent failure of meiosis II. Nonetheless, strains carrying either mutation execute one of two rounds of SPB duplication during meiosis and form viable diploid spores.
An intermediate step of mitotic SPB duplication is revealed by mutation of the MPS1 gene (Winey et al., 1991). MPS1 encodes an essential dual-specificity kinase that performs two known roles during mitosis (Lauzéet al., 1995). Mps1p is a positive-acting regulator of SPB duplication and a component of the spindle assembly checkpoint (Weiss and Winey, 1996). Recently, a set of conditional mutations in the kinase domain of the MPS1 gene was analyzed (Schutz and Winey, 1998). All of the mutant alleles of MPS1 disrupt both mitotic functions at the restrictive temperature, producing a loss of viability associated with monopolar mitosis in the absence of a checkpoint. Here we report our phenotypic analysis of four mps1 mutant alleles that form two phenotypic classes during meiotic chromosome segregation and spore formation.
MATERIALS AND METHODS
Strain Construction
Yeast strains used are listed in Table 1. Original temperature-sensitive strains were constructed by the two-step allele replacement method (Scherer and Davis, 1979). URA3-marked integrative (pRS306-derived) plasmids, containing each mps1 allele, were cut with the MroI restriction enzyme to direct integration at the endogenous MPS1 locus. DNA fragments were transformed with the use of the EZ Transformation kit (Zymo Research, Orange, CA). Ura+ transformants were selected and subsequently streaked to synthetic complete (SC)-ura plates containing 5-FOA (1 g/l) to select for strains having excised part of the integrated DNA by homologous recombination. Temperature-sensitive colonies, having retained the mutation contained in the integrated allele, were selected by failure to grow at 37°C and subsequently tested for complementation of the mps1-1 allele by mating. Noncomplementing strains were used for further study according to the integrated allele of mps1. The majority of experiments presented here were done with SK-1–derived strains, but the sporulation phenotypes as described by light microscopic analysis (phase and fluorescence data) were recapitulated in W303-derived strains.
Table 1.
Yeast strains
| Strain | Genotype | Source |
|---|---|---|
| S490 | MATα, ura3, leu2Δ (asp718-ecoR1), arg4-bgl, ade2-bgl | N. Hollingsworth |
| S483 | MATa, ura3, leu2Δ (asp718-ecoR1), arg4-nsp, ade2-bgl | N. Hollingsworth |
| RKY1145 | MATa, ura3, leu2Δ∷hisG, his4-X | M. Lichten |
| RKY1146 | MATα, ura3, leu2Δ∷hisG, his4-B | M. Lichten |
| MAS119 | MATa, ho∷LYS2, lys2, ura3, leu2∷hisG | M. Shonn |
| MAS140 | MATα, LacOpr∶LEU2, Cyc1pLacI-GFP13∶URA3, ho∷LYS2 | M. Shonn |
| MAS142 | MATa, LacOpr∶LEU2, ho∷LYS2 | M. Shonn |
| YUMY069 | MATa/α, mps1-1/mps1-1, ura3/ura3, HIS4/his4-X, ADE2/ade2-B, ARG4/arg4-B | This study |
| YUMY070 | MATa/α, mps1-3796/mps1-3796, ura3/ura3, leu2Δ/leu2Δ, arg4-N/arg4-B, ade2-B/ade2- | This study |
| YUMY071 | MATa/α, mps1-1237/mps1-1237, ura3/ura3, leu2∷hisG/leu2Δ, HIS4/his4-X, ARG4/arg4-B, ADE2/ade2-B | This study |
| YUMY072 | MATa/α, mps1-412/mps1-412, ura3/ura3, leu2∷hisG/leu2Δ, ARG4/arg4-N, ADE2/ade2-B, HIS4/his4-B | This study |
| YUMY1D1 | MATa, ura3, leu2Δ (asp718-ecoR1), arg4-bgl, ade2-bgl | This study |
| YUMY102 | MATα, mps1-1237, Cyc1pLacI-GFP13∶URA3, LacOpr∶LEU2, arg4-N | This study |
| YUMY104 | MATa, mps1-1237, Cyc1pLacI-GFP13∶URA3, LacOpr∶LEU2, arg4-B | This study |
| YUMY110 | MATa, mps1-1237, ura3, leu2Δ (asp718-ecoR1), arg4-B | This study |
| YUMY119 | MATα, mps1-1237, ura3, leu2Δ (asp718-ecoR1), arg4-nsp | This study |
| YUMY120 | MATa, mps1-1237, ura3, leu2Δ (asp718-ecoR1), arg4-bgl | This study |
| YUMY125 | MATα, ura3, leu2Δ (asp718-ecoR1), arg4-bgl | This study |
| YUMY126 | MATa, ura3, leu2Δ (asp718-ecoR1), arg4-nsp | This study |
| YUMY131 | MATα, ura3, leu2Δ (asp718-ecoR1), arg4-nsp, his4-bgl | This study |
| YUMY1980 | MATa/α, mps1-412/mps1-412, ura3/ura3, leu2D/leu2D, arg4-N/arg4-B, ade2-B/ade2-B | This study |
| YUMY3E3 | MATa/α, MPS1/MPS1, ura3/ura3, ho∷LYS2/ho∷LYS2, leu2D/leu2D, arg4-N/arg4-B, ade2-B/ade2-B | This study |
| YUMY3I1 | MATa/α, mps1-1237/mps1-1237, ura3/ura3, ho∷LYS2/ho∷LYS2, leu2D/leu2D, arg4-N/arg4-B, ade2-B/ade2-B | This study |
| YUMY3I8 | MATa/α, mps1-3796/mps1-3796, ura3/ura3, ho∷LYS2/ho∷LYS2, leu2D/leu2D, arg4-N/arg4-B, ade2-B/ade2-B | This study |
| YUMY4B1 | MATa/α, MPS1/MPS1, GFP-TUB1∶URA3/GFP-TUB1∶URA3, leu2Δ (asp718-ecoR1)/leu2-K, HIS4/his4-X, ARG4/arg4-N | This study |
| YUMY4B5 | MATa/α, mps1-412/mps1-412, GFP-TUB1∶URA3/GFP-TUB1∶URA3, leu2D∷hisG/leu2D(asp718-ecoR1), ADE2/ade2-B, arg4-N/arg4-N, HIS4/his4-X | This study |
| YUMY4C2 | MATa/α, mps1-3796/mps1-3796, GFP-TUB1∶URA3/GFP-TUB1∶URA3, leu2D/leu2D, arg4-N/arg4-B, ade2-B/ade2-B | This study |
| YUMY4C6 | MATa/α, mps1-1237/mps1-1237, GFP-TUB1∶URA3/GFP-TUB1∶URA3, leu2-R/leu2Δ (asp718-ecoR1), arg4-N/arg4-N | This study |
| YUMY4C9 | MATa/α, mps1-1/mps1-1, GFP-TUB1∶URA3/GFP-TUB1∶URA3, leu2Δ (asp718-ecoR1)/leu2-K, HIS4/his4-X, ARG4/arg4-N | This study |
| YUMY4F4 | MATa/α, MPS1/MPS1, ura3/ura3, leu2∷hisG/leu2∷hisG, his4-X/his4-B | This study |
| TM002a | MATa/α, MPS1/MPS1, ura3-1/ura3-1, leu2-3, 112/leu2-3, 112, his3-11/his3-11, ade2-1/ade2-1, trp1-1/trp1-1, canr/CANs, cyhr/CYHs | T. McKenna |
| TM0019a | MATa/α, mps1-3796/mps1-3796, ura3-1/ura3-1, leu2-3, 112/leu2-3, 112, his3-11/his3-11, ade2-1/ade2-1, trp1-1/trp1-1, canr/CANs, cyhr/CYHs | T. McKenna |
| TM027a | MATa/α, mps1-1237/mps1-1237, ura3-1/ura3-1, leu2-3, 112/leu2-3, 112, his3-11/his3-11, ade2-1/ade2-1, trp1-1/trp1-1, canr/CANs, cyhr/CYHs | T. McKenna |
These strains are W303 background; all others are SK-1.
Yeast Cultures and Sporulation
Yeast culture and genetic techniques were as described by Rose et al. (1990) with the following exceptions. Synchronous sporulation in liquid medium was performed as described by Alani et al. (1990) with modifications for use of temperature-sensitive strains. Cultures were grown overnight in 5 ml of YPD to stationary phase. Cells were diluted in YEPA (1% potassium acetate) to an OD600 of 0.30–0.35 and allowed to grow at room temperature for 13.5 h. Cultures of density > OD600 = 1.2 (∼3 × 107 cells/ml) were then shifted to sporulation medium (0.3–1% potassium acetate) at the restrictive temperature, maintaining the same cell density. Medium was supplemented with amino acids to cover auxotrophies at one-fourth the concentration recommended for synthetic medium (Rose et al., 1990). Can/Cyh assays were performed with the use of SC medium without arginine containing 3 μg/ml cycloheximide and 60 μg/ml l-canavanine.
Light Microscopy
Fluorescence imaging consisted of visualization of DNA with the use of 1 μg/ml DAPI, tubulin in a green fluorescent protein GFP–TUB1 fusion (Straight et al., 1996), or centromere-proximal marked chromosomes with the use of a GFP–LacI repressor fusion (Straight et al., 1996). Microscopy was performed on a Leica DMRXA/RF4/V fluorescence microscope (Leica, Solma, Germany) equipped with a Cooke Sensicam charge-coupled device (CCD) camera (Tonawanda, NY) and a motorized stage for three-dimensional imaging. Image processing was performed with the use of the Slidebook program by Intelligent Imaging Innovations (Denver, CO). All images were subjected to deconvolution with the use of the nearest-neighbors algorithm. Sporulated cells were either visualized live as a wet mount in 1% potassium acetate on poly-l-lysine–coated microscope slides (Polysciences, Warrington, PA) or fixed in 70% ethanol for 15 min to 1 h and resuspended in PBS containing 0.1 μg/ml DAPI. Cells used for DAPI exclusion assays were fixed in a 3.7% formaldehyde solution for 15 min, stained with 1 μg/ml DAPI for <5 min, resuspended in PBS, and placed on poly-l-lysine–coated slides.
Dityrosine Fluorescence Assay
Assays for spore wall fluorescence were performed with the use of a method similar to that described by Esposito et al. (1991). Patches of cells were sporulated on nitrocellulose filters (Schleicher & Schuell, Keene, NH) incubated at 23 and 34°C. Filters were subsequently treated with lysis buffer (350 μl of 0.1 M Na-citrate, 0.01 M EDTA, pH 5.8, 70 μl of Glusulase [DuPont NEE-154], 15 μl of β-mercaptoethanol) at 37°C for 4 h and then treated with 300 μl of concentrated ammonium hydroxide. Fluorescence was detected by excitation under UV light (UVP, San Gabriel, CA), and images were captured with the use of a digital camera with a blue filter (Wratten #98, Kodak, Rochester, NY).
Electron Microscopy
Samples for electron microscopy were prepared with the use of a modification of the method used by Winey et al. (1995). At selected times, 5- to 10-ml aliquots were removed from a synchronously sporulating culture, and cells were collected by vacuum filtration to form a yeast paste. The cell paste was cryofixed in a BAL-TEC (Balzers, Liechtenstein) HPM-010 high-pressure freezer. The samples were then processed by freeze-substitution in 2% osmium tetroxide and 0.1% uranyl acetate in acetone at −80°C for 3 d, followed by equilibration to room temperature and embedding in Spurr's resin. Thin sections were stained with uranyl acetate and lead citrate and examined in a Philips (Eindhoven, the Netherlands) CM10 electron microscope. Images were captured with the use of a CCD camera (Gatan, Pleasanton, CA) and processed with Digital Micrograph version 2.5 (Gatan, Pleasanton, CA).
Commitment to Recombination
Assays for commitment to recombination were performed with the use of strains heteroallelic for different point mutations at arg4 (Wu and Lichten, 1995). Wild-type (YUMY3E3), mps1-1237 (YUMY3I1), and mps1-3796 (YUMY3I8) strains were synchronously sporulated at the restrictive temperature, and aliquots were removed at 2-h time points, diluted in 1% potassium acetate, and plated to SC for viability and to SC-arginine for commitment to recombination. Viability was measured as colony-forming units (CFU) at each time point per CFU at time zero. Interhomologue recombination frequency is expressed as the number of Arg+ colonies per total CFU. Each sample was plated in triplicate.
Chromosome Segregation Assays
Chromosome III was marked at the LEU2 locus by integration of pAFS59, containing 256 tandem repeats of the LacO (Lac operator) (Straight et al., 1996). Visualization of the marked chromosome was made possible by integration at the URA3 locus of a GFP–LacI repressor fusion. Diploid strains were made both homozygous and hemizygous for the LacO array by mating marked to marked, and marked to unmarked, a and α haploids, respectively. Homozygous strains were generated by crossing wild-type strains (MAS140 × MAS142) and mps1-1237 strains (YUMY102 × YUMY104). The hemizygous strains were used for microscopy and quantitation of the phenotype in this study. Marked MPS1 (MAS140 × MAS119) and mps1-1237 (YUMY102 × YUMY110) strains were induced to undergo synchronous sporulation as described previously. Samples were removed at 1-h time points from 6–12 h after initiation of sporulation. Cells were visualized either live (GFP/differential interference contrast [DIC] panels) or after fixation in 70% ethanol (GFP/DAPI). Several planes of a whole field of cells were captured with the use of a motorized stage and a CCD camera. Digital images were then used to perform data acquisition.
Northern Analysis
Total RNA was isolated from synchronously sporulating cultures of the mps1-1237/mps1-1237 (YUMY119 × YUMY120) and MPS1/MPS1 (YUMY125 × YUMY126) strain backgrounds according to yeast RNA isolation procedures from Ausubel et al. (1997). Twenty micrograms of RNA was loaded per lane in a 1% agarose–formaldehyde gel. rRNA was stained with ethidium bromide (25 μg/ml) and photographed before transfer of RNA to Hybond-N by a capillary blotting procedure according to the manufacturer's specifications (Amersham International, Buckingham, United Kingdom). RNA was UV cross-linked to the membrane with the use of a UV Stratalinker (Stratagene, La Jolla, CA), and the blots were dried at 80°C for 1 h. DNA probes were labeled with [α-32P]dCTP with the use of the Prime-It II kit (Stratagene). PCR products, used as templates, were generated for SMK1, SWM1, SPS100, and ACT1 by amplification from isolated SK-1 genomic DNA. Blots were stripped between probing by boiling in 10 mM Tris (pH 7.5), 1 mM EDTA, and 0.1% SDS and allowing the solution to cool to room temperature.
RESULTS
Loss of Sporulation in Temperature-sensitive mps1 Strains
To study the sporulation phenotypes caused by mutation in MPS1, several mutant alleles of the gene were introduced into the efficiently sporulating strain SK-1 (Kane and Roth, 1974) and the W303 strain of Saccharomyces cerevisiae by two-step integration (see MATERIALS AND METHODS). To confirm proper integration of the mutant alleles, strains were assayed mitotically for their ability to complement the mps1-1 mutation. With this method, strains of both a and α mating type were constructed that contain the following conditional alleles of mps1: mps1-1, mps1-412, mps1-1237, and mps1-3796. All of the alleles tested have mutations that fall within the kinase domain of MPS1. Molecular characterization of the mitotic phenotypes of these strains has been described (Schutz and Winey, 1998). In vegetatively growing cells, all four alleles of mps1 cause a failure of SPB duplication and are unable to activate the spindle assembly checkpoint at the restrictive temperature. Homozygous diploid (W303 and SK-1) strains for each allele of mps1 were made and assayed for their ability to sporulate at 33°C (Table 1). Also, the minimal restrictive temperature for all four strains during vegetative growth was determined. All four strains are completely inviable at 33°C in mitotic culture, indicating that 33°C is a fully restrictive temperature for the meiotic experiments (our unpublished results).
A role for Mps1p during meiosis was suspected based on a report that the expression of MPS1, constant with respect to vegetative growth, is enhanced specifically during meiosis (Poch et al., 1994). Furthermore, the upstream activating sequence (UAS) of MPS1 contains two meiosis specific element (MSE) elements used for meiosis-specific up-regulation of transcription (Chu and Herskowitz, 1998). Initially, the ability of the mps1 mutant strains to form spores at the restrictive temperature was assessed by phase microscopy. All of the alleles of MPS1 prevented wild-type levels of spore formation at this temperature, reducing the number of asci to between 1 and 30% of the population (our unpublished results). Furthermore, the extent of the defect depends on the allele present in the strain. Strains homozygous for the mps1-412 and mps1-1237 alleles produce a higher percentage of two- to four-spored asci (∼30–50% of the population) than strains of the mps1-1 (∼0–2%) and mps1-3796 (∼5–15%) types. However, all of the spores that could be dissected did not form viable haploid colonies. Germination in the absence of cell wall digestive enzymes was checked by isolation of whole asci on rich medium. Spores bearing mps1 mutations were unable to form colonies on rich medium, indicating that passage through the events of meiosis and spore formation under conditions of compromised Mps1p function is lethal to the cells (mps1-1237: 2 of 30 germinated, 1 of 30 colony forming; mps1-412: 3 of 30 germinated, 0 of 30 colony forming).
Loss of Mps1p Function Is Lethal in the Presence and Absence of Spore Formation
Strains carrying mps1 mutations were sporulated at the restrictive temperature and at times tested during the course of 12 h to assess viability in the population. Whereas wild-type strains maintained high viability during the experiment, cells containing the mutant alleles of MPS1 lost viability dramatically (Figure 1A). Extending the analysis of viability to 24–48 h demonstrated that the loss of viability in mps1 strains was nearly complete (<1% colony forming; our unpublished results). To demonstrate that any viable cells left in mps1 cultures were not from strains that carry out sporulation, viability was tested with the use of the highly sensitive Canr, Cyhr haploidization assay (Figure 1C). Homozygous mps1-3796 strains that are heterozygous for two recessive alleles conferring resistance to canavanine and cycloheximide are unable to produce any Canr, Cyhr progeny, indicating that these strains produce no viable haploid spores (Hollingsworth and Byers, 1989). Similar strains homozygous for mps1-1237 produce very few papillae within a patch, indicating a severe loss of viability in these strains.
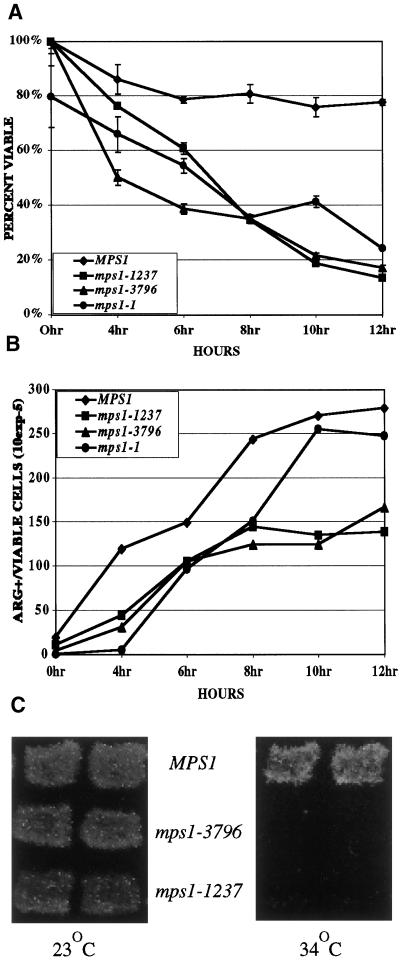
Figure 1.
Return-to-growth assays were done to determine viability (A) and commitment to recombination (B) with the use of wild-type (YUMY3E3), mps1-1 (YUMY131 × YUMY1D1), mps1-1237 (YUMY3I1), and mps1-3796 (YUMY3I8) homozygous diploid mutant strains. Strains were synchronously sporulated, and aliquots of cells were plated to SC or SC-arginine medium at the times indicated. Viability was measured as the number of CFU at each time point as a percentage of CFU at 0 h. Commitment to recombinationwas determined with the use of arg4 heteroallelic diploids and plating to SC-arginine medium. Recombinant frequency is expressed as Arg+ colonies per CFU at each time point according to the method described by Esposito and Esposito (1974). Recombination assays were performed three times with triplicate samples. One representative example is shown. (C) Viability of cells sporulated at the restrictive temperature was assessed with the use of Can/Cyh resistance generated by haploidization of two recessive drug-resistance markers. Wild-type (TM002), mps1-3796 (TM019), and mps1-1237 (TM027) strains were sporulated at room temperature and 34°C. Patches were replica plated to medium containing cycloheximide and l-canavanine (see MATERIALS AND METHODS) and incubated at room temperature for 2–5 d to assay viable haploid cell production caused by haploidization of both can1-100 and cyhr markers.
Mutations in genes required for recombination during meiosis, such as RAD50 and SPO11, are often lethal (Klapholz et al., 1985; Alani et al., 1990). Because mutation in mps1 results in a loss of viability, commitment to recombination was measured in diploid strains homozygous for mps1-1, mps1-1237, and mps1-3796 mutations. Strains mutant for mps1 and heteroallelic at the ARG4 locus were sporulated at the restrictive temperature and plated to SC-arginine or SC to assess production of Arg+ recombinants (Figure 1B). This assay measures the level of commitment to recombination within the sporulating population (Esposito and Esposito, 1974). Cells that have initiated meiotic recombination can complete recombination and revert to vegetative growth as diploids when returned to glucose-containing medium before anaphase I. Using this assay, we can measure the timing and level of entry into the meiotic recombination process in our strains. The mps1 mutant strains are competent to become committed to recombination, because they produced Arg+ colonies with a slight reduction compared with MPS1. The loss of viability and the absence of recombination checkpoint arrest in these strains suggest that the mps1 phenotypes are not attributable to a defect in recombination.
Mutation in MPS1 Produces Two Phenotypic Classes
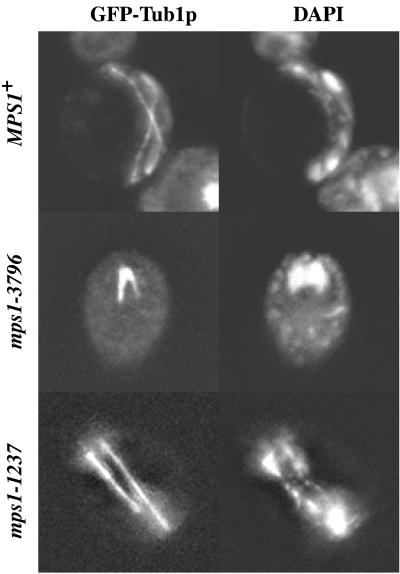
We performed a cytological analysis of the mutant strains to characterize the physiological defect caused by mutation of mps1 in sporulating cells. Strains containing mutant alleles of mps1 were transformed with a GFP-tagged copy of the α-tubulin gene (TUB1) integrated at the URA3 locus for visualization of spindle formation. Cells containing the GFP–TUB1 fusion gene were induced to undergo sporulation synchronously at the restrictive temperature (Alani et al., 1990). Aliquots were removed from the sporulation cultures, fixed, and stained with DAPI to monitor DNA segregation. Flow cytometry demonstrates that in all mps1 mutant strains, premeiotic DNA synthesis is completed with similar timing as in wild-type strains (our unpublished results). After 12 h of sporulation at the restrictive temperature, wild-type strains showed ∼80% completion of meiosis and many mature ascospores, e.g., 161 of 200 three- to four-spored asci. Spindle formation during meiosis I and meiosis II was clearly visible in these strains with the use of the GFP–Tub1p chimera (Figure 2). A distinction could be made between two levels of progression through sporulation in strains carrying different mutant alleles of mps1. Cells harboring the mps1-3796 and mps1-1 alleles failed to form spindles (e.g., ∼1–5% spindle formation, 12 of 200 three- to four-spored asci) or segregate DNA during meiosis, as seen by DAPI staining (Figure 2; our unpublished results). We define these cells as exhibiting the class I meiotic phenotype of mps1. On the other hand, cells containing the mps1-1237 and mps1-412 alleles revealed spindle formation at times between 6 and 12 h in a significant proportion (e.g., 30–50%, 56 of 200 two- to four-spored asci) of cells in the culture (Figure 2; our unpublished results). The timing of spindle formation was similar to that of wild-type cells, with the first meiotic spindles formed roughly 6 h after introduction into sporulation medium. Therefore, the mps1-1237 and mps1-412 alleles result in the class II phenotype. The observation of phenotypic differences between alleles may reveal two distinct roles for the kinase during meiosis and spore formation.
Figure 2.
Cytological analysis of mps1 mutants during meiosis at the restrictive temperature. GFP–Tub1p and DAPI staining of DNA demonstrate allele-dependent phenotypes. Cultures were sporulated at 33°C for 8 h (MPS1, YUMY4B1; mps1-1237, YUMY4C6) or 12 h (mps1-3796, YUMY4C2), fixed with 3.7% formaldehyde for up to 1 h, and stained with 1 μg/ml DAPI for 5 min. Wild-type cells proceed through two rounds of spindle formation and chromosome segregation, as seen by GFP and DAPI, respectively. The mps1-3796 strain fails to form spindles over the course of meiosis and remains mononucleate. However, ∼30% of cells harboring the mps1-1237 allele successfully complete spindle formation and carry out chromosome segregation.
Mps1p Is Required for Meiotic SPB Duplication
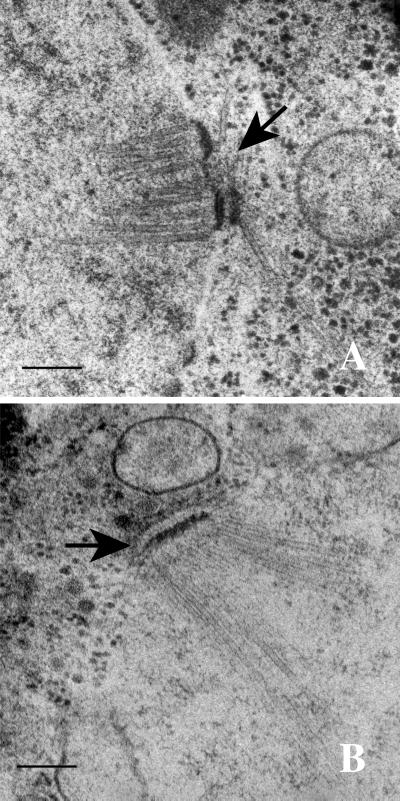
The class I phenotype caused by mps1 mutation in sporulating cells suggests a conserved requirement for Mps1p in SPB duplication during meiosis. Figure 3B shows an electron micrograph of a nucleus in a mps1-3796 cell after 12 h of incubation in sporulation medium at 33°C. Whereas wild-type cells duplicate their SPBs early in meiosis (Figure 3A), near the onset of premeiotic DNA synthesis, the mps1-3796 and mps1-1 strains maintain a single SPB after sufficient time for cells to complete sporulation has passed. To demonstrate that the mononucleate cells in the culture did not represent only cells that failed to enter the sporulation process, several cells from an earlier time (8 h) were examined by electron microscopy. Prophase meiotic nucleoplasm can be distinguished visually from mitotic nucleoplasm by a variety of meiosis-specific characteristics with the use of electron microscopy (Moens and Rappaport, 1971; Zickler and Olson, 1975). Wild-type meiotic prophase I nuclei, in addition to duplicated side-by-side SPBs, typically contain synaptonemal complex and polycomplex. The SPBs in meiotic prophase nuclei of class I phenotype were examined by serial section electron microscopy. The entire nucleus was examined to account for all SPBs. In every nucleus of a mps1-3796 strain (n = 16) and a mps1-1 strain (n = 10), only a single SPB was found within the nuclear envelope. Therefore, we conclude that the major defect in strains exhibiting the class I phenotype appears to be the failure to duplicate the SPBs.
Figure 3.
Thin-section electron microscopy of MPS1 (YUMY4F4) and mps1-3796 (YUMY070) strains during meiosis at the restrictive temperature reveals a SPB duplication defect in the mps1 mutant strain. (A) MPS1 strain after 6 h at 33°C in sporulation medium. Prophase meiotic nuclei have duplicated side-by-side SPBs (arrow) in the nuclear envelope. (B) mps1-3796 strain after 12 h at 33°C. Spindle pole bodies fail to duplicate during meiosis in both the mps1-3796 and mps1-1 (YUMY069) strains, leading to the class I phenotype. Bars, 0.2 μm.
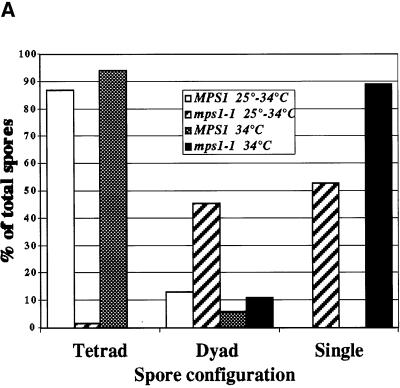
We hypothesized that if both meiosis I and meiosis II SPB duplications require Mps1p, it would be possible to disrupt the second duplication event specifically with the use of a timed shift to the restrictive temperature. Cells of the mps1-1 genotype and wild-type cells were sporulated synchronously at 34 or 25°C for 6 h and then shifted to 34°C for the remainder of the 24-h time course (Figure 4). After 6 h in sporulation medium, the cells are nearing the end of prophase and contain duplicated SPBs (Alani et al., 1990). The mps1-1 cells that were shifted to 34°C after 6 h produced nearly 50% two-spored asci, indicating successful completion of the first SPB duplication and failure in the second SPB duplication. Less than 2% three- to four-spored asci were detected in the mps1-1 strain, revealing a requirement for Mps1p after the shift to higher temperature. Because dyads are formed with a timed temperature shift, we conclude that Mps1p is required for the completion of both meiosis I and meiosis II SPB duplication events.
Figure 4.
Timed temperature shift of an mps1-1 strain produces two-spored asci. Wild-type (YUMY4B1) and mps1-1 (YUMY4C9) strains were sporulated at 34 or 25°C for 6 h and then shifted to 34°C. Spore morphology was counted for 200 cells of each culture after 24 h of sporulation. White bars, wild-type, 25–34°C; striped bars, mps1-1, 25–34°C; gray bars, wild-type, 34°C; black bars, mps1-1, 34°C. The chart shows the percentages of cell morphology within the spore-forming population of the culture; n = 200 for each culture. Sporulation efficiency (two or more spores per ascus) was as follows: wild-type, 25–34°C = 91%; mps1-1, 25–34°C = 29.5%; wild- type, 34°C = 78%; mps1-1, 34°C = 2%.
Mps1p Is Required for Normal Spore Wall Formation
Meiotically dividing cells homozygous for mps1-412 or mps1-1237 mutations display the second class of mutant phenotypes, involving progression through sporulation. DAPI staining of DNA and GFP–Tub1p autofluorescence of microtubules reveal that these cells are capable of chromosome segregation by meiotic spindles (Figure 2). However, DAPI-treated mutant cells could be distinguished from wild-type cells by the ability of the stain to penetrate the interior of the spore (Figure 5). We used wild-type asci, fixed for 15 min with formaldehyde and then stained for 3 min with DAPI, to demonstrate that the mature spore walls are resistant to the permeation of DAPI (Figure 5A). In contrast, the interiors of spores from either mps1-1237– or mps1-412–containing strains are readily penetrated by DAPI, resulting in stained nuclear material (Figure 5, B and C). This difference between spores in wild-type and mps1 strains suggested a defect in spore wall assembly.
Figure 5.
DAPI staining and electron microscopy of wild-type and mps1 mutant strains reveals a spore wall defect. Cells were fixed for 15 min with 3.7% formaldehyde and stained with 1 μg/ml DAPI for <5 min (see MATERIALS AND METHODS). DAPI permeation into the interior of the spore is retarded by mature spore walls in wild-type MPS1 (YUMY4B1) (A). Lack of complete spore wall formation in the mps1 mutant strains allows DAPI to readily stain DNA within individual spores of mps1-1237 (YUMY4C6) (B), mps1-412 (YUMY4B5) (C), mps1-1 (YUMY4C9) (D), and mps1-3796 (YUMY4C2) (E). Note that in D and E, meiotic chromosome segregation fails as expected, and the cells build defective spore walls. Also note the fragmentation of DAPI-staining material within the mutant spores. Thin-section electron microscopy of MPS1 (YUMY4F4) (F), mps1-1237 (YUMY071) (G), and mps1-3796 (YUMY070) (H) strains displays the heterogeneous nature of the spore wall defect caused by mps1 mutation. (F) Wild-type spores have a four-layered structure that is relatively uniform in thickness. Mature spores are tightly associated in a tetrahedral array within a condensed ascus. Darkly staining material defines the outer, dityrosine-containing layer of the spore wall, and electron-dense material is seen adjoining individual spores (arrow). (G) Two to four spores are produced in mps1-1237 strains, and individual spores lack the organization of the spore wall seen in wild-type. Spore wall thickness is more variable than in wild- type, with component layers of spore wall material not organized as in wild-type. The electron-dense outer layer of the mutant spores is often missing, and the asci tend not to condense around tightly adherent spores. Note that the very electron-dense material connecting the MPS1 spores is disrupted in the mps1-1237 ascus (arrow). Up to 15% of cells containing the mps1-3796 allele eventually form an aberrant single spore body (H, arrowhead). The spore produced in these cells reveals similar structural defects as in a mps1-1237 strain. Bars, 0.7 μm
We used electron microscopy to examine the sporulation products of diploid cells homozygous for the mps1-1237 (Figure 5, F–H) or mps1-412 alleles (our unpublished results). A mature wild-type spore wall is composed of four layers: the prospore membrane, the electron-translucent second layer, the chitin/chitosan layer, and the outer darkly staining dityrosine-containing layer (Briza et al., 1986). Whereas wild-type spores contain all four spore wall layers in their proper organization (Figure 5F), mps1-1237 (Figure 5G) and mps1-412 spore walls are defective, lacking the uniform four-layered structure seen in wild-type spores. This phenotype is similar to that of homozygous smk1 and sps1 strains (Friesen et al., 1994; Krisak et al., 1994). Also, similar spore wall defects are detected in mps1-1 and mps1-3796 (class I) strains in a small percentage of the cells (up to 15%) that attempt to form an aberrant single spore (Figure 5, D, E, and H).
In mps1 homozygous mutant strains, the following defects are detected: some of the spores contain fewer than four layers, the ordering of the component layers is frequently abnormal, and the overall thickness of the mutant spore walls is variable. The electron-dense outer layer of the spore walls is often missing, and when present it fails to develop the uniformity of the wild-type outer layer. Furthermore, the electron-dense material adjoining individual spores in a wild-type ascus is disorganized and fragmented in the mps1 mutant strains, and the ascus fails to condense during maturation. However, unlike the spore walls in smk1Δ strains, the defective spores formed in mps1 strains often contain spore wall structures detectable by phase microscopy (our unpublished results), indicating a different type of defect. To determine if a gross structural defect in SPB morphology could be the cause of deficient spore production, we examined several SPBs from homozygous mps1-1237 and mps1-412 strains. No visible defect in SPB morphology was detected in either strain (our unpublished results).
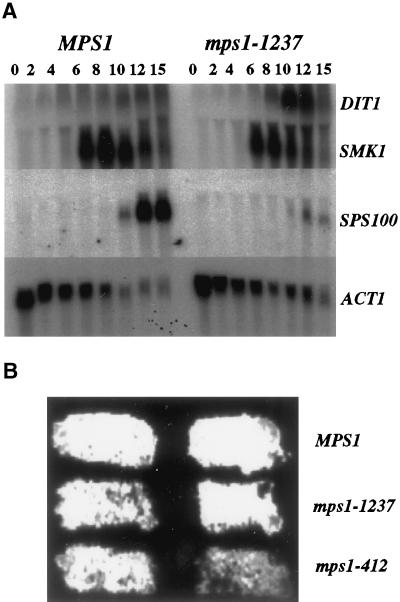
Genes required for spore wall formation are regulated by a sporulation-specific transcriptional program (Chu et al., 1998). To investigate the role of Mps1p in spore wall formation, we performed Northern analysis to determine if mutations in MPS1 affected induction of genes involved in the regulation or structural composition of developing spores. A strain containing the mps1-1237 allele, which produces the class II phenotype, was sporulated at the restrictive temperature. Transcriptional induction of midlate genes such as SMK1, SWM1 (Ufano et al., 1999), and DIT1 was assayed as well as the late spore wall–specific gene SPS100 (Figure 6A). Analysis of the abundance of these transcripts in sporulating cells of the mps1-1237 genotype revealed no dramatic change in the expression of SMK1 or SWM1 (our unpublished results), but the abundance of the SPS100 transcript was reduced dramatically at the late times. The level of DIT1 transcript was consistently higher in the mps1-1237 strain than in wild type, similar to the effect of sps1 mutation (Friesen et al., 1994). Furthermore, these strains were assayed for dityrosine production with the use of a dityrosine fluorescence assay (Esposito et al., 1991) and found to produce a fluorescent signal similar to wild type, indicating that the function of the DIT1 gene is intact (Figure 6B). Therefore, we conclude that the defects in spore wall assembly seen in mps1 mutant strains are independent of Smk1p kinase and Swm1p but may exert their effect through Sps1p or some other mechanism.
Figure 6.
Northern analysis of sporulation-specific gene expression and dityrosine fluorescence in mps1 mutant strains. (A) MPS1 (YUMY125 × YUMY126) and mps1-1237 (YUMY119 × YUMY120) strains were synchronously sporulated, and aliquots were removed for RNA isolation. Filters were probed for expression of three midlate sporulation-specific genes, SMK1, SWM1, and DIT1, the late spore wall–specific gene SPS100, and the ACT1 gene as a loading control. Expression of SMK1 and SWM1 (our unpublished results) is unaffected in the mps1-1237 strain compared with wild type. The expression of the SPS100 gene is severely diminished in the mutant strains, whereas DIT1 expression is enhanced above wild-type levels. (B) Dityrosine fluorescence is detected in both mps1 mutant strains producing two- to four-spored asci. Patches of MPS1 (YUMY3E3), mps1-1237 (YUMY3I1), and mps1-412 (YUMY 1980) were sporulated at the restrictive temperature and then treated to activate dityrosine fluorescence. The presence of fluorescent material in wild-type and mps1 mutant strains indicates that insoluble fluorescent dityrosine is produced efficiently in both strain types.
Finally, to determine if the class II phenotype results from the mps1-1237 or mps1-412 alleles being hypomorphic, we assayed the ability of a centromeric plasmid containing the mps1-1237 allele to rescue the mps1-1237 sporulation phenotype. Two independent experiments were carried out with the use of a mps1-1237 homozygous strain (YUMY119 × YUMY120; Table 1) transformed with either pMPS1-1237 containing the mps1-1237 allele (Schutz and Winey, 1998) or the pRS316 empty vector. The strains were sporulated at the restrictive temperature for 48 h, fixed, and examined for spore wall integrity with the use of the DAPI permeation assay. Neither plasmid was capable of rescuing the phenotype (mps1-1237, 6.3% normal spores, number of DAPI-resistant per total in two experiments: 0 of 52 and 5 of 23; vector, 7.3% normal spores, number of DAPI-resistant per total in two experiments: 1 of 34 and 5 of 42). On the other hand, a plasmid carrying wild-type MPS1 recovered the DAPI resistance of spores (72%, 36 of 50). Extra copies of the mps1-1237 allele did not rescue the mutant phenotype, suggesting that this is not a hypomorphic allele of MPS1.
Chromosome Missegregation Is Increased in MPS1 Mutant Strains
The high viability of spores with mutations in any one of several late-sporulation genes involved specifically in spore wall formation demonstrates that disruption of spore wall production is not an inherently lethal event (Krisak et al., 1994). The defective spores produced in many of these strains, for example smk1, are hypersensitive to environmental assault such as heat shock, Glusulase, or ether (Krisak et al., 1994). Nonetheless, the spores are capable of germinating at near wild-type levels in the absence of external perturbation. In the case of class II mps1 mutant strains, the spores produced at the restrictive temperature show low viability under all circumstances. An early observation with the use of DAPI staining (Figure 5) indicated that chromosomes in the MPS1 mutant strains may be unequally partitioned to individual spores during meiosis.
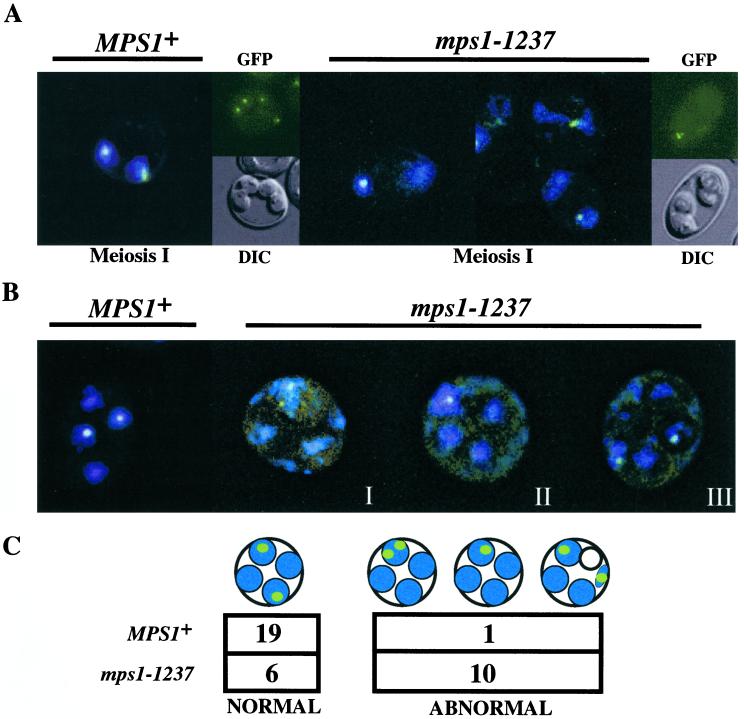
To test directly the hypothesis that mutation in MPS1 causes increased levels of chromosome missegregation in meiotically dividing strains, we used a system of chromosome III marked with a Lac operator array near the centromere and visualized by fluorescence microscopy with the use of a GFP–LacI repressor fusion (Straight et al., 1996). As the homozygous mps1-1237 mutant strains progressed through meiosis, it became evident that in many cases the labeled centromeres failed to segregate properly (Figure 7, A and B). By means of a combination of a single marked centromere of chromosome III and marked copies of both centromeres on homologous pairs of chromosome III, missegregation of chromosomes at either meiosis I (Figure 7A) or meiosis II (Figure 7B) was observed. Cells progressing through the first meiotic division are seen as binucleates by DAPI staining. GFP marking of both chromosome III homologues in binucleate cells demonstrated that meiosis I often segregated homologues to a single pole. Meiosis I and II missegregation, resulting in all four chromatids segregated to a single spore, was detected in mps1-1237 cells, as shown in GFP/DIC panels (Figure 7A). To obtain a quantitative assessment of the level of missegregation in these cells, we used a single marked chromosome to monitor meiosis II specifically (Figure 7, B and C). More than half (62%) of the mps1-1237 mutant cells that carry out both meiosis I and meiosis II and show at least four DAPI masses within the ascus inappropriately segregate sister chromatids during meiosis II (Figure 7B). In many cases, DAPI staining revealed chromosomal material outside the developing spore body. Moreover, in some of these cells, spore formation excluded the marked chromosomes (Figure 7, far right), leaving one or more GFP foci in the ascal or “epiplasmic” space. Therefore, it is probable that missegregation and misincorporation of chromosomes into spore bodies contributes to the loss of viability in cells harboring mutations at the MPS1 locus.
Figure 7.
Loss of chromosome segregation fidelity in the class II mps1 mutant strains. Chromosome segregation was monitored in mps1-1237 and wild-type strains with the use of LacOpr-marked centromere III (LEU2) visualized with a GFP–LacI repressor protein fusion (Straight et al., 1996). (A) Chromosome segregation during meiosis I was monitored with the use of marked homologous chromosomes (four chromatids). Binucleate cells in a sporulating population were examined for segregation of paired homologues to opposite poles. Left panels, wild-type binucleate (GFP/DAPI) and four-spored (GFP/DIC) asci; right panels, examples of meiosis I missegregation in mps1-1237 binucleates (GFP/DAPI), which occurred in up to 80% of cells that entered meiosis I in a given experiment, and a four-spored ascus (GFP/DIC), showing no meiosis I or meiosis II segregation. (B and C) A single copy of chromosome III was marked to follow meiosis II (sister chromatid separation) specifically. Wild-type MPS1 strains show four DAPI-staining spore nuclei, two of which contain copies of the marked chromosome III (left panel). Right panels, mps1-1237 cells display a variety of aberrant segregation patterns. Three examples are shown. (I) Both marked centromeres reside in one of four detectable spore bodies. DAPI staining is seen within and outside of spore bodies. (II) A single GFP signal is associated with one of four DAPI-staining foci, suggesting that sister chromatids remain paired and unresolvable. (III) Many instances of DAPI staining outside of nascent spore bodies can be seen in mps1 cells. In some cases, one of the marked chromatids is found outside any of the four spores. To quantitate missegregation in these cells, we restricted our analysis to the few mutant cells with a single pair of marked chromatids and at least four visible DAPI foci. This restriction allowed us to unambiguously assign normal versus abnormal segregation, but it also resulted in a small number of scoreable cells due to the percentage of segregation in the population and weakened expression of GFP in sporulating (autofluorescent) cells. Nonetheless, the difference between MPS1 and mps1-1237 is statistically significant (C; p < 0.005 by χ2 analysis).
DISCUSSION
The essential Mps1p kinase is a regulator of SPB duplication during mitosis (Winey et al., 1991) and a component of the spindle assembly checkpoint in budding yeast (Hardwick et al., 1996; Weiss and Winey, 1996). We have shown that mutation in the Mps1p kinase prevents wild-type levels of spore formation. Four alleles of mps1, previously described in the context of their mitotic phenotypes (Schutz and Winey, 1998), were studied, and their effects on meiosis revealed multiple phenotypes, indicating more than one function for the kinase during sporulation (Table 2). The multiple requirements for Mps1p during sporulation reflect functions either shared with mitosis (SPB duplication) or unique to sporulation (chromosome segregation and spore formation). The mps1 mutations have no effect on premeiotic DNA synthesis, and the mps1 strains show meiotic commitment to recombination. Nonetheless, Mps1p kinase is essential to the completion of meiosis and spore formation, as demonstrated by the severe loss of viable progeny in temperature-sensitive strains sporulated at the restrictive temperature.
Table 2.
Mutant alleles of MPS1
| Allele | Subdomaina | Mutationb | Restrictive
temperature
|
Phosphorylationbd | Kinase activitybe | Mitoticb
|
Meioticc
|
|||
|---|---|---|---|---|---|---|---|---|---|---|
| S288cb | SK-1c | SPB duplication | Checkpoint | SPB duplication | Spore wallsc | |||||
| 1 | XI | C696Y | 30°C | 30°C | Reduced | Reduced | Fails | Fails | Fails | Defect |
| 6 | IX | C642Y | 30°C | Lethal | Slight reduction | WT | Fails | Fails | N/D | N/D |
| 412 | VI | E546K | 34°C | 33°C | WT | Reduced | Fails | Fails | OK | Defect |
| 737 | III | E486K | 34°C | Lethal | Slight reduction | WT | Failsf | Fails | N/D | N/D |
| 1237 | VIII | P608S | 34°C | 33°C | Slight reduction | WT | Fails | Fails | OK | Defect |
| 3796 | V | E517K | 34°C | 30°C | WT | Reduced | Fails | Fails | Failsg | Defect |
WT, wild type; N/D, not detected.
This study.
Data from Western analysis of Mps1p banding patterns corresponding to phosphorylated forms of GST-Mps1p.
In vitro kinase activity was monitored by autophosphorylation of GST-Mps1p.
A distinct step of SPB duplication; see Schutz and Winey (1998).
Intermediate phenotype.
The role of Mps1p in SPB duplication is retained during sporulation. The class I phenotypes resulting from the mps1-3796 and mps1-1 mutations are caused by a failure in SPB duplication early during meiosis. The end product of sporulation in this case is a mononucleate cell that proceeds through prophase I but fails to segregate chromosomes because of a lack of spindle formation. This phenotype contrasts with that of mutations in other known SPB duplication genes, such as cdc31-1 and ndc1-1 (Byers, 1981; Thomas and Botstein, 1986), that result in the production of viable dyad asci. It is possible that this phenotype is unique to the experimental conditions or the particular alleles of cdc31 and ndc1 used. In fact, cells harboring the mps1-1 allele, when shifted to the restrictive temperature after 6 h of sporulation at the permissive temperature, generate many normal dyad asci, demonstrating a requirement for Mps1p during the second SPB duplication event. In addition to CDC31, NDC1, and MPS1, a meiosis-specific gene that plays a role in SPB duplication is SPO1. Deletion of the SPO1 gene produces a predominantly monopolar phenotype during sporulation (Moens et al., 1974; Tevzadze et al., 2000). Unlike MPS1, SPO1 is not essential for vegetative growth. Therefore, Mps1p is the only protein known to regulate mitotic SPB duplication that is also required during both meiotic SPB duplication events.
An unexpected second class of sporulation phenotypes arises in homozygous mps1-1237 and mps1-412 strains. In these strains, 30–50% of the cells in the population progress through both meioses, and the initiation of the spore wall formation process appears to occur normally. However, electron microscopy analysis showed that in these strains the morphology of the spore walls is abnormal, suggesting a defect in the assembly and maturation of spore wall components. A mid-meiotic function for Mps1p was predicted from the meiosis-specific transcriptional induction of MPS1 (Poch et al., 1994; Chu et al., 1998). Furthermore, the abnormal spores produced are inviable because of extensive missegregation of chromosomes, indicating a role for Mps1p in spindle function during meiosis.
The morphological characteristics of the mps1 class II spore defects are shared with several known regulators of spore wall formation. Northern analysis demonstrated that the mps1 class II phenotype does not involve a delay or reduction in the expression of either of the key regulators, SMK1 or SWM1. However, the abundance of the late meiotic marker transcript SPS100 is dramatically reduced, suggesting that the Mps1p spore wall function is downstream or in a separate pathway from SMK1 or SWM1. In addition to loss of SPS100 transcription, the mid-late meiotic marker transcript DIT1 is consistently more abundant in the mps1-1237 strain than it is in wild-type strains, both phenotypes shared with the sps1 mutation. SPS1 encodes a Ste20p kinase homologue that functions upstream of the Smk1p MAPK, but Sps1p is thought to have a Smk1p-independent function based on the observed loss of DIT1 regulation (Friesen et al., 1994; Krisak et al., 1994). Therefore, the mps1 class II defect reveals that Mps1p may function in regulating the late events of spore wall maturation in a pathway shared with Sps1p.
Disruption of spore wall formation is accompanied by a loss of viability in the class II mps1 strains. Fluorescence microscopy of GFP-marked chromosomes demonstrated that the fidelity of chromosome segregation is compromised as a result of defective Mps1p. Chromosome missegregation at the level of a single pair of sister chromatids was quantitated as both chromatids segregated to a single pole during meiosis II. Also, aberrant patterns of chromosome segregation were detected during meiosis I. Extreme chromosome missegregation, revealed by marked chromosomes outside of spore bodies, indicates either lagging chromosomes during spore wall formation or a loss of coordination between spindle function and spore formation. The consequence of this disruption is a failure to position chromosomes in close proximity to the SPBs as spores are formed. These observations indicate a defect in spindle function that compromises the fidelity of chromosome segregation in class II strains. The genetic interaction of MPS1 with DAM1, a spindle-associated protein that does not show a synthetic interaction with other spindle checkpoint genes, hints at a role for Mps1p in spindle function (developed further by Jones et al., 1999). In the context of meiosis, this putative Mps1p function may be essential, a hypothesis requiring further experimentation.
The defects in chromosome segregation seen in mps1 mutant strains are likely to be the primary cause of lethality in these strains, because spore wall defects do not result in severe loss of viability (Krisak et al., 1994). The second mitotic function of Mps1p, a component of the spindle assembly checkpoint, could become an essential function during meiosis. A MAD2-dependent checkpoint monitors meiosis I specifically during sporulation (Shonn and Murray, 2000). In fact, cells lacking Mad2p undergo increased levels of meiosis I nondisjunction without spindle perturbation, indicative of an enhanced requirement for checkpoint function during meiosis I (Shonn and Murray, 2000). This first demonstration of the Mad2p-dependent checkpoint during meiosis reveals that the checkpoint is important but not absolutely required for all meioses. The meiotic checkpoint function of Mps1p cannot be tested with the existing alleles of mps1 that exhibit multiple defects. However, we can conclude that Mps1p has a role in meiotic chromosome segregation separate from the Mad2p checkpoint based on the severity of chromosome missegregation during both meiosis I and meiosis II in mps1 class II strains.
The two classes of mps1 phenotypes demonstrate that Mps1p is an important regulator of multiple meiosis and spore formation processes: SPB duplication, high-fidelity chromosome segregation, and spore wall assembly. Although all three processes involve the SPB at some level, they are likely to require Mps1p independently. We have demonstrated that the class II mutant phenotype is not the result of a hypomorphic allele of mps1, and Northern data implicate Mps1p in transcriptional regulation of spore wall assembly. Furthermore, the spore wall defect is not likely a consequence of chromosome missegregation, because other mutations that lead to missegregation, such as recombination minus (rec−) mutants (e.g., rad50Δ), produce normal spore walls, as does a mad2Δ strain. However, using the mutant alleles at hand, we cannot determine whether Mps1p carries out its spore wall function early in meiosis at the time of SPB duplication or if Mps1p interacts directly with the regulatory networks that control spore wall maturation late during sporulation. We favor the second model because the MPS1 transcript accumulates during mid-meiosis, and our Northern data reveal a phenotype similar to sps1. Further experimentation examining the interaction of Mps1p with the late sporulation gene products (e.g., Sps1p, Smk1p) and possibly the generation of meiosis-specific separation-of-function alleles of mps1 will likely resolve this issue.
In summary, this study demonstrates that the essential regulatory kinase Mps1p, which is required for mitotic SPB duplication and the activation of the spindle assembly checkpoint, is also essential for meiosis and spore formation. Furthermore, the protein is necessary for completion of multiple processes during sporulation, including SPB duplication, spore formation, and the fidelity of meiotic chromosome segregation. Future study of Mps1p function during meiosis may enhance our understanding of the regulatory networks that ensure that properly segregated chromosomes are incorporated into the specialized cell types of spores in yeast and gametes in other eukaryotes.
ACKNOWLEDGMENTS
We thank Nancy Hollingsworth for providing SK-1 strains. We thank Sean Burgess and Saul Honigberg for advice on the use of SK-1 for meiotic analysis. We thank Aaron Straight, Marion Shonn, and Andrew Murray for GFP reagents, strains, and communication of unpublished results. We thank Taryn McKenna and Garry Morgan for technical assistance. We are very grateful to Dean Dawson and Michele Jones for critical reading of the manuscript. P.D.S. was supported in part by a National Institutes of Health training grant (GM-07135). Deconvolution microscopy was made possible in part by a gift from Virginia and Mel Clark. This work was supported by the March of Dimes Birth Defects Foundation through a grant to M.W. (FY98.409/FY99.617).
Abbreviations used:
- CCD
charge-coupled device
- DIC
differential interference contrast
- GFP
green fluorescent protein
- SC
synthetic complete
- SPB
spindle pole body
REFERENCES
- Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1997. [Google Scholar]
- Briza P, Winkler G, Kalchhauser H, Breitenbach M. Dityrosine is a prominent component of the yeast ascospore wall. J Biol Chem. 1986;261:4288–4294. [PubMed] [Google Scholar]
- Byers B. Multiple roles of the spindle pole bodies in the life cycle of Saccharomyces cerevisiae. In: von Wettstein D, Stenderup A, Kielland-Brandt M, Friis J, editors. Molecular Genetics in Yeast: Alfred Benzon Symposia. Vol. 16. Copenhagen: Munksgaard; 1981. pp. 119–133. [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Chu S, Herskowitz I. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol Cell. 1998;1:685–696. doi: 10.1016/s1097-2765(00)80068-4. [DOI] [PubMed] [Google Scholar]
- Esposito R, Esposito M. Genetic recombination and commitment to meiosis in Saccharomyces. Proc Natl Acad Sci USA. 1974;71:3172–3176. doi: 10.1073/pnas.71.8.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito RE, Dresser M, Breitenbach M. Identifying sporulation genes, visualizing synaptonemal complexes, and large scale spore and spore wall purification. Methods Enzymol. 1991;194:110–131. doi: 10.1016/0076-6879(91)94010-a. [DOI] [PubMed] [Google Scholar]
- Friesen H, Lunz R, Doyle S, Segall J. Mutation of the SPS1-encoded protein kinase of Saccharomyces cerevisiae leads to defects in transcription and morphology during spore formation. Genes Dev. 1994;8:2162–2175. doi: 10.1101/gad.8.18.2162. [DOI] [PubMed] [Google Scholar]
- Hanks S, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- Hardwick KG, Weiss E, Luca F, Winey M, Murray AW. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- Hollingsworth N, Byers B. HOP1: a yeast meiotic pairing gene. Genetics. 1989;121:445–462. doi: 10.1093/genetics/121.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M, Bachant J, Castillo A, Giddings TJ, Winey M. Yeast Dam1p is required to maintain spindle integrity during mitosis and interacts with the Mps1p kinase. Mol Biol Cell. 1999;10:2377–2391. doi: 10.1091/mbc.10.7.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane S, Roth R. Carbohydrate metabolism during ascospore development in yeast. J Bacteriol. 1974;118:8–14. doi: 10.1128/jb.118.1.8-14.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapholz S, Waddell C, Esposito R. The role of the SPO11 gene in meiotic recombination in yeast. Genetics. 1985;110:187–216. doi: 10.1093/genetics/110.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krisak L, Strich R, Winters RS, Hall JP, Mallory MJ, Kreitzer D, Tuan RS, Winter E. SMK1, a developmentally regulated MAP kinase, is required for spore wall assembly in Saccharomyces cerevisiae. Genes Dev. 1994;8:2151–2161. doi: 10.1101/gad.8.18.2151. [DOI] [PubMed] [Google Scholar]
- Lauzé E, Stoelcker B, Luca FC, Weiss E, Schutz AR, Winey M. Yeast spindle pole body duplication gene MPS1 encodes an essential dual specificity protein kinase. EMBO J. 1995;14:1655–1663. doi: 10.1002/j.1460-2075.1995.tb07154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone RE. Dual regulation of meiosis in yeast. Cell. 1990;61:375–378. doi: 10.1016/0092-8674(90)90517-i. [DOI] [PubMed] [Google Scholar]
- Moens PB, Esposito RE, Esposito MS. Aberrant nuclear behavior at meiosis and anucleate spore formation by sporulation-deficient (SPO) mutants of Saccharomyces cerevisiae. Exp Cell Res. 1974;83:166–174. doi: 10.1016/0014-4827(74)90700-9. [DOI] [PubMed] [Google Scholar]
- Moens PB, Rappaport E. Spindles, spindle plaques, and meiosis in the yeast Saccharomyces cerevisiae. J Cell Biol. 1971;50:344–361. doi: 10.1083/jcb.50.2.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poch O, Schwob E, de Fraipont F, Camasses A, Bordonné R, Martin RP. RPK1, an essential yeast protein kinase involved in the regulation of the onset of mitosis, shows homology to mammalian dual-specificity kinases. Mol Gen Genet. 1994;243:641–653. doi: 10.1007/BF00279573. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1990. pp. 177–187. [Google Scholar]
- Scherer S, Davis RW. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci USA. 1979;76:4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutz AR, Winey M. New alleles of the yeast MPS1 gene reveal multiple requirements in spindle pole body duplication. Mol Biol Cell. 1998;9:759–774. doi: 10.1091/mbc.9.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shonn M, McCarroll R, Murray A. Requirement of the spindle checkpoint for proper chromosome segregation in budding yeast meiosis. Science. 2000;289:300–303. doi: 10.1126/science.289.5477.300. [DOI] [PubMed] [Google Scholar]
- Simchen G. Are mitotic functions required in meiosis? Genetics. 1974;76:745–753. doi: 10.1093/genetics/76.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- Tevzadze G, Swift H, Esposito R. Spo1, a phospholipase B homolog, is required for spindle pole body duplication during meiosis in Saccharomyces cerevisiae. Chromosoma. 2000;109:72–85. doi: 10.1007/s004120050414. [DOI] [PubMed] [Google Scholar]
- Tevzadze GG, Mushegian AR, Esposito RE. The SPO1 gene product required for meiosis in yeast has a high similarity to phospholipase B enzymes. Gene. 1996;177:253–255. doi: 10.1016/0378-1119(96)00261-2. [DOI] [PubMed] [Google Scholar]
- Thomas JH, Botstein D. A gene required for the separation of chromosomes on the spindle apparatus in yeast. Cell. 1986;44:65–76. doi: 10.1016/0092-8674(86)90485-x. [DOI] [PubMed] [Google Scholar]
- Ufano S, San-Segundo P, Rey FD, Aldana CRVD. SWM1, a developmentally regulated gene, is required for spore wall assembly in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:2118–2129. doi: 10.1128/mcb.19.3.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M, Pierce M, Winter E. The CDK-activating kinase CAK1 can dosage suppress sporulation defects of smk1 MAP kinase mutants and is required for spore wall morphogenesis in Saccharomyces cerevisiae. EMBO J. 1997;16:1305–1317. doi: 10.1093/emboj/16.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E, Winey M. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Goetsch L, Baum P, Byers B. MPS1 and MPS2: novel yeast genes defining distinct steps of spindle pole body duplication. J Cell Biol. 1991;144:745–754. doi: 10.1083/jcb.114.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Hoyt MA, Chan C, Goetsch L, Botstein D, Byers B. NDC1: a nuclear periphery component required for yeast spindle pole body duplication. J Cell Biol. 1993;122:743–751. doi: 10.1083/jcb.122.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Mamay C, O'Toole E, Mastronarde D, Giddings TJ, McDonald K, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T-C, Lichten M. Factors that affect the location and frequency of meiosis-induced double-strand breaks in Saccharomyces cerevisiae. Genetics. 1995;140:55–66. doi: 10.1093/genetics/140.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D, Olson L. The synaptonemal complex and the spindle plaque during meiosis in yeast. Chromosoma. 1975;50:1–23. doi: 10.1007/BF00284959. [DOI] [PubMed] [Google Scholar]